Abstract
CCAAT enhancer binding protein α (C/EBPα) plays an essential role in cellular differentiation, growth, and energy metabolism. Here, we investigate the correlation between C/EBPα and hepatocellular carcinoma (HCC) patient outcomes and how C/EBPα protects cells against energy starvation. Expression of C/EBPα protein was increased in the majority of HCCs examined (191 pairs) compared with adjacent nontumor liver tissues in HCC tissue microarrays. Its upregulation was correlated significantly with poorer overall patient survival in both Kaplan-Meier survival (P = 0.017) and multivariate Cox regression (P = 0.028) analyses. Stable C/EBPα-silenced cells failed to establish xenograft tumors in nude mice due to extensive necrosis, consistent with increased necrosis in human C/EBPα-deficient HCC nodules. Expression of C/EBPα protected HCC cells in vitro from glucose and glutamine starvation–induced cell death through autophagy-involved lipid catabolism. Firstly, C/EBPα promoted lipid catabolism during starvation, while inhibition of fatty acid beta-oxidation significantly sensitized cell death. Secondly, autophagy was activated in C/EBPα-expressing cells, and the inhibition of autophagy by ATG7 knockdown or chloroquine treatment attenuated lipid catabolism and subsequently sensitized cell death. Finally, we identified TMEM166 as a key player in C/EBPα-mediated autophagy induction and protection against starvation. Conclusion: The C/EBPα gene is important in that it links HCC carcinogenesis to autophagy-mediated lipid metabolism and resistance to energy starvation; its expression in HCC predicts poorer patient prognosis. (Hepatology 2015;61:965–978)
Hepatocellular carcinoma (HCC) is the third leading cause of cancer deaths worldwide. Patients diagnosed with HCC have a very low 5-year survival rate (<15%) because of late diagnosis and impaired liver function.1 It is refractory to most chemotherapeutic treatment regimes, and there is a lack of an effective molecularly targeted therapy. Sorafenib, for example, extends survival unsatisfactorily by approximately 2 months, according to two large multicenter randomized clinical studies.2, 3 Furthermore, there have been no oncogenic addiction loops discovered for HCC so far,1 in contrast to other cancers such as lung (with ALK rearrangement) and breast (BRCA1/2 mutations) cancers, which likely reflects the complexity of gene interactions and diverse pathways involved in HCC. Thus, understanding the mechanisms underlying the development and progression of HCC is a critical first step toward novel effective HCC treatments.
The importance of metabolic reprogramming in HCC and other cancers has been increasingly appreciated in the past decade.4 Several recent metabolomic studies have shown an increase in glycolysis (Warburg effect) and accelerated beta-oxidation in HCC tumors compared with adjacent nontumor tissues.5–7 The idea of targeting cancer metabolism for HCC treatment is further supported by clinical findings that embolization therapy, especially transarterial chemoembolization, which deprives blood supply predominantly to the tumor, improves 2-year survival of patients with unresectable HCC.8
A transcription factor belonging to the CCAAT/enhancer-binding protein family, C/EBPα, is involved in cellular differentiation and energy metabolism.9 Previous studies found that C/EBPα-null mice died postnatally of hypoglycemia,10 while liver-specific C/EBPα-null mice exhibited glucose intolerance, serum cholesterol reduction, and steatotic livers.11 We recently reported that C/EBPα was up-regulated in a subset of human HCCs and promoted cell growth in HCC cell lines.12 We hypothesized that C/EBPα up-regulation may play an important role in cancer cell metabolism during HCC development.
Materials and Methods
The full methods for the sub-G1 assay, western blotting, quantitative reverse-transcription polymerase chain reaction, gene microarray profiling, and small interfering RNA (siRNA) sequences for gene knockdown assays are available in the Supporting Information.
Samples and Immunohistochemical Staining
The study was approved by the National University of Singapore's Institutional Review Board (NUS-IRB 09-244). The HCC tissue microarrays consisted of 191 patients with HCC who were diagnosed between the years 1990 and 2009 at the National University Hospital of Singapore. Expression of C/EBPα protein was determined by immunohistochemistry using 1:50 diluted rabbit polyclonal antibody (#2295; Cell Signaling Technology, Danvers, MA), as described.12 Immunostaining was scored on a four-tiered grading system, with <10% of liver cells staining indicating negative and 10%–40% positively scored as 1, 41%–70% as 2, and 71%–100% as 3.
Statistical Analysis
Experimental differences were analyzed by SPSS software package (version 22.0) using the two-tailed t test or chi-squared analysis. Survival analyses were performed using Kaplan-Meier and multivariate Cox regression models. Patients who died within 2 weeks after surgery were excluded from the survival analysis. Statistical significance was defined as P < 0.05.
Ectopic Xenograft Tumor Development
The protocol was approved by National University of Singapore's Institutional Animal Care and Use Committee (092-04). A total of 16 male Balb/c nude mice at 7 weeks old were purchased from Biological Resource Center (Singapore) and separated randomly into two groups. Five million C/EBPα-expressing cells (Hep3B and stable negative control shNC) and C/EBPα-silenced stable cells (sh4 and sh7) were inoculated subcutaneously into the left or right flank of mice. Xenograft tumor growth was observed for 30 days.
Cell Culture and Treatments
Cell lines were cultured in Dulbecco's modified Eagle's medium (with 4.5 g/L glucose and 4.0 mM l-glutamine) with 10% fetal bovine serum.12 Dulbecco's modified Eagle's medium (glucose- and glutamine-free), Earle's Balanced Salt Solution (with 1 g/L glucose) medium, and chemicals (simvastatin, C75, diethylum-belliferyl phosphate, etomoxir, ranolazine, chloroquine) were purchased from Sigma-Aldrich (St. Louis, MO). Cells were washed with phosphate-buffered saline twice before starvation.
Gene Silencing and Overexpression
Stable Hep3B and PLC/PRF/5 cells silenced for C/EBPα expression were generated as described previously12 and maintained using 2 µg/mL puromycin (Sigma-Aldrich). Stable HCC-M cells with inducible C/EBPα expression were generated using an inducible plasmid downstream of metallothionein promoter, a kind gift from Dr. A.D. Friedman (Johns Hopkins University, Baltimore, MD).13 Cells were maintained in 500 µg/mL Geneticin G418 (Sigma-Aldrich) and induced by treatment of 100 µM zinc chloride. Synthetic Stealth or Silencer siRNA targeting C/EBPα, ATG7, or TMEM166 was purchased from Life Technologies (Carlsbad, CA) and transducted as before.12 The plasmids used for TMEM166 overexpression were kindly provided by Dr. Yingyu Chen (Peking University, Beijing, China).14
Biochemical Determination of Intracellular Metabolites
Five million cells were starved and homogenized. Biochemical assays (Biovision, Milpitas, CA) of adenosine triphosphate, pyruvate, glycogen, triglyceride, and acetyl-coenzyme A were conducted according to prescribed protocols and measured using a Varioskan Flash multimode plate reader (ThermoFisher Scientific, Waltham, MA). Fatty acid beta-oxidation rates were determined as described.15
Metabolic Measurement of Oxygen Consumption Rate
Oxygen consumption rates (OCR) were measured with XF-24 Extracellular Flux Analyzer (Seahorse Bioscience, North Billerica, MA). Twenty thousand cells were assayed in the XF medium (nonbuffered Dulbecco's modified Eagle's medium with or without 25 mM glucose and 2 mM l-glutamine) under basal conditions and in response to 100 µM ranolazine (or chloroquine [CQ]) and mitochondrial poisons (1 µM oligomycin, 1 µM fluorocarbonyl cyanide phenylhydrazone, 1 µM rotenone, and 1 µM antimycin A).
Results
Up-Regulation of C/EBPα Predicted Poorer HCC Prognosis
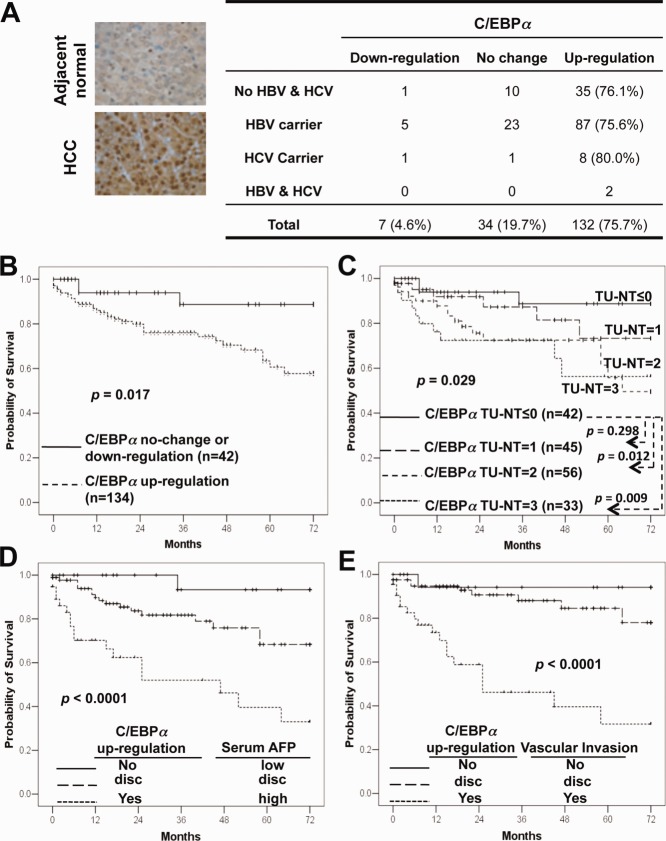
We analyzed C/EBPα protein expression in 191 pairs of human primary HCC and adjacent nontumor liver tissues by immunohistochemistry. The C/EBPα protein was up-regulated in 76.4% of HCC samples (Supporting Table S1), independent of hepatitis B and C viral infection status (Fig. 1A), and other clinicopathological parameters (Supporting Table S2). More importantly, HCC patients with up-regulated C/EBPα had poorer overall prognoses than those without (P = 0.017; Fig. 1B). The higher the C/EBPα expressed in HCCs compared with adjacent nontumor tissues, the poorer the prognosis (P = 0.029; Fig. 1C). The patients with the highest up-regulation of C/EBPα expression (difference between tumor and nontumor tissue = 3) had the worst prognosis (P = 0.009) compared to those without expression (difference between tumor and nontumor tissue ≤ 0). In addition, the level of C/EBPα protein expressed in HCC tissues itself (P = 0.018; Supporting Fig. S1), but not that in adjacent nontumor liver tissues (P = 0.543), was correlated to poorer survival.
Figure 1.
Up-regulation of C/EBPα in human primary HCC tissues predicted poorer patient survival. (A) The C/EBPα protein was determined by immunohistochemical staining in HCC tissue microarrays and is summarized in the right panel. (B) A Kaplan-Meier survival analysis showed that the patients with upregulated C/EBPα had poorer overall survival. (C) The HCC patients were ranked into four different groups according to the difference between C/EBPα expression in tumor (TU) and in nontumor (NT) tissues, with 0 for no difference and 3 for the highest up-regulation. The number of patients analyzed in each group and P values are indicated. (D,E) Kaplan-Meier curves showed the overall survival of HCC patients subgrouped by C/EBPα and serum AFP level (D) or C/EBPα and vascular invasion (E). Disc, discordant risk assessments: high C/EBPα expression and low risk predicted by AFP (<300 ng/mL)/vascular invasion or vice versa. Abbreviations: HBV, hepatitis B virus; HCV, hepatitis C virus.
We performed a multivariate Cox regression analysis to determine whether the correlation of C/EBPα with patient survival was confounded by underlying clinical parameters. Up-regulation of C/EBPα (hazard ratio = 5.116, 95% confidence interval 1.188-22.026; P = 0.028) as well as serum α-fetoprotein (AFP) level and microscopic vascular invasion were independent prognosticators of patient survival (Table1, lower panel). Next, we determined whether C/EBPα could improve prognostic prediction by combining with AFP or vascular invasion. The combination of C/EBPα up-regulation with either high AFP levels or vascular invasion predicted poorer prognosis compared to the absence of C/EBPα up-regulation with either low AFP levels or no vascular invasion and the discordant groups (P < 0.0001; Fig. 1D, E). Specifically, there were trends that C/EBPα up-regulation discriminated poorer survival outcomes among patients with low serum AFP of <300 ng/mL (P = 0.062; Supporting Fig. S1C) or microscopic vascular invasion (P = 0.039; Supporting Fig. S1F). Taken together, C/EBPα up-regulation may improve HCC clinical risk stratification.
Table 1.
C/EBPα Independently Predicted Poorer Overall Survival in Primary HCC Patients
| Molecular and Clinical Variables | Number of Cases | Hazard Ratio(95% Confidence Interval) | P Value |
|---|---|---|---|
| Univariate analysis (Cox: enter) | |||
| C/EBPα (no change vs. up-regulation) | 42/137 | 3.690 (1.131-12.038) | 0.030 |
| Serum AFP (<300 vs. ≥300 ng/mL) | 114/48 | 3.637 (1.866-7.013) | 0.0002 |
| Age (<50 vs. ≥50) | 47/135 | 0.739 (0.371-1.472) | 0.390 |
| Gender (male vs. female) | 150/37 | 0.456 (0.232-0.897) | 0.023 |
| Diabetes (no vs. yes) | 133/47 | 0.504 (0.210-1.213) | 0.126 |
| HBV (no vs. yes) | 60/117 | 1.062 (0.522-2.159) | 0.868 |
| HCV (no vs. yes) | 167/12 | 0.494 (0.068-3.603) | 0.486 |
| Cirrhosis (no vs. yes) | 87/100 | 1.056 (0.553-2.015) | 0.868 |
| Tumor size (<5 vs. ≥5 cm) | 89/95 | 2.581 (1.263-5.275) | 0.009 |
| Number of nodules (single vs. multiple) | 129/58 | 0.619 (0.314-1.222) | 0.167 |
| TNM staging (I vs. II, III, and IV) | 87/91 | 2.952 (1.477-5.897) | 0.002 |
| Microscopic lymphatic invasion (no vs. yes) | 76/14 | 1.410 (0.312-6.373) | 0.655 |
| Microscopic vascular invasion (no vs. yes) | 99/57 | 4.836 (2.252-10.386) | <0.0001 |
| Steatosis (< 5% vs. ≥5%) | 163/24 | 0.560 (0.172-1.825) | 0.336 |
| Fibrosis (no vs. yes) | 151/36 | 0.874 (0.399-1.916) | 0.874 |
| Tumor encapsulation (no vs. yes) | 40/68 | 0.576 (0.209-1.590) | 0.287 |
| Tumor necrosis (no vs. yes) | 93/93 | 1.028 (0.539-1.961) | 0.933 |
| Multivariate analysis (Cox: forward LR) | |||
| C/EBPα (no change vs. up-regulation) | 29/103 | 5.116 (1.188-22.026) | 0.028 |
| Serum AFP (≤300 vs. >300 ng/mL) | 94/38 | 4.238 (1.958-9.170) | 0.0002 |
| Microscopic vascular invasion (no vs. yes) | 82/50 | 5.577 (2.391-13.012) | <0.0001 |
Correlations of HCC patient overall survival with C/EBPα expression level and clinical factors were analyzed by univariate and multivariate Cox analyses. The factors significant in the univariate Cox model were included in the multivariate Cox model, and only three factors including C/EBPα were finally enrolled.
Abbreviations: HBV, hepatitis B virus; HCV, hepatitis C virus.
To exclude the possibility that C/EBPα was mutated in HCC as observed in acute myeloid leukemia (AML),9 we sequenced the full-length coding region of CEBPA genomic DNA from 26 pairs of HCC samples and found no mutations in any of the samples (data not shown). Furthermore, the protein expression of C/EBPα in HCCs was correlated with mRNA levels (Supporting Fig. S2), supporting our conclusion that C/EBPα was up-regulated in HCC.
C/EBPα Protected Cells Against Cell Death In Vivo and In Vitro
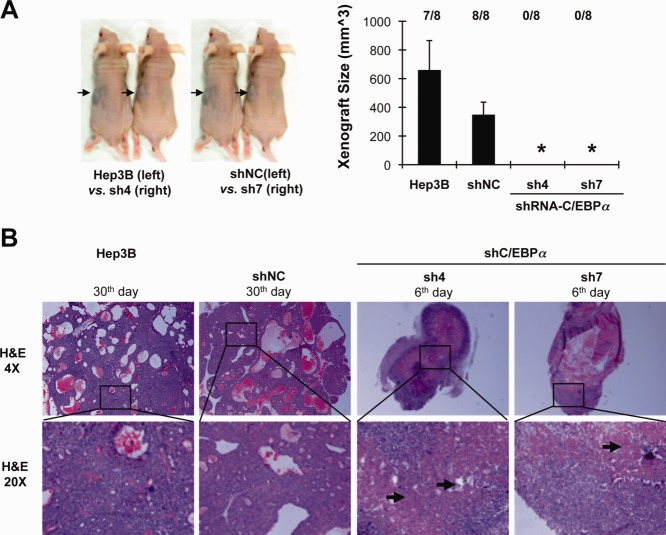
We used an ectopic xenograft model in Balb/c nude mice to investigate the effect of C/EBPα on tumor development in vivo. Cells expressing C/EBPα (Hep3B, shNC—“control” Hep3B cells with stable incorporation of scrambled shRNA) and C/EBPα-silenced cells (sh4 and sh7—Hep3B cells stably silenced for C/EBPα expression) were inoculated subcutaneously into the left or right flank of nude mice, respectively. After 30 days, solid tumor nodules developed only in the side injected with C/EBPα-expressing cells (Hep3B and shNC) but not in the side with C/EBPα-silenced cells (sh4 and sh7; Fig. 2A). Interestingly, the silenced cells were able to develop five transient small tumor nodules from a total of 16 inoculations. However, tumor growth was not sustained and disappeared within 2 weeks after inoculation. Histopathological examination in a follow-up study showed that the transient nodules which developed from C/EBPα-silenced cells had extensive necrosis in the nodular core (Fig. 2B).
Figure 2.
We found that C/EBPα was required for generation of xenograft tumor. (A) Five million C/EBPα-expressing cells (Hep3B and stable negative control shNC) and C/EBPα–silenced stable cells (sh4 and sh7) were inoculated into the left or right flank, respectively, in Balb/c nude mice. After 30 days of observation, solid tumor nodules were removed from dead mice. Tumor size was determined by a caliper and calculated using the formula volume = (width × width × length)/2. *P < 0.01. The numbers of solid nodules developed at day 30 are shown above. (B) Solid nodules were collected from dead mice at different days and subjected to hematoxylin and eosin staining. Representative histopathological hematoxylin and eosin–stained images with 4× and 20× magnification are shown. Arrows indicate regions impaired by necrotic cell death. H&E, hematoxylin and eosin.
Nutrient deprivation may occur during solid tumor development, at a phase prior to neoangiogenesis, or in cells that are farthest away from the blood supply. In this context, it is noteworthy that the absence of C/EBPα protein in human HCCs tended to be correlated with necrosis, especially in small-sized HCC nodules (Table2). Among HCCs without C/EBPα expression, tumor necrosis was found in no less than 50% of all tumor sizes compared with HCCs expressing C/EBPα (14.3% for tumor size <1.5 cm [P = 0.023], 22.7% for 3.0 cm [P = 0.058], and 34.9% for 5.0 cm [P = 0.227]). Although limited by the small sample size, these preliminary findings suggest that C/EBPα may protect HCC cells from cell death in tumor development.
Table 2.
More Necrosis Was Observed in C/EBPα-Absent Primary HCC Tissues
| Tumor Necrosis | C/EBPα in Tumor | |
|---|---|---|
| Absent | Present | |
| Tumor Size ≤1.5 cm (P = 0.023) | ||
| No | 0 | 6 |
| Yes | 2 (100%) | 1 (14.3%) |
| Tumor Size ≤3.0 cm (P = 0.058) | ||
| No | 3 | 34 |
| Yes | 4 (57.1%) | 10 (22.7%) |
| Tumor Size ≤5.0 cm (P = 0.277) | ||
| No | 7 | 56 |
| Yes | 7 (50.0%) | 30 (34.9%) |
We compared the percentage of necrosis found between HCC tumors with and without C/EBPα. The statistical difference was determined by Pearson's chi-squared analysis. The number of cases involved and P values are indicated.
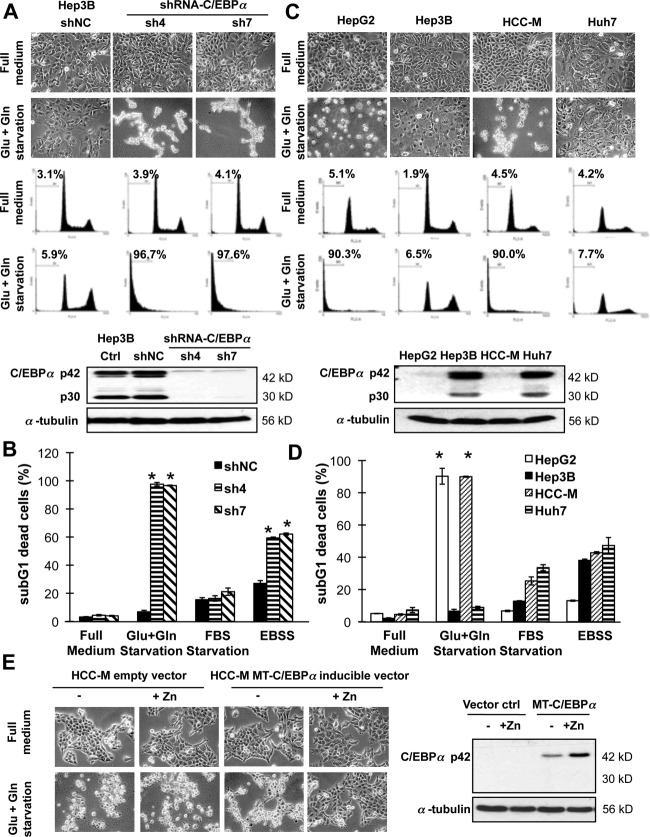
We next observed that C/EBPα-expressing cells were able to survive in nutrient-poor conditions in vitro when the culture medium was not changed or supplemented for weeks, in contrast to C/EBPα-silenced cells, which completely died (Supporting Fig. S3A). We therefore hypothesized that C/EBPα endowed cells with a metabolic advantage, especially in a nutrient-poor environment, during tumor development. We demonstrated that knockdown of C/EBPα in Hep3B (Fig. 3A, B; Supporting Fig. S3B) or PLC/5 (Supporting Fig. S3C) sensitized the cells to energy starvation (glucose and glutamine double deprivation) induced cell death. Similarly, the C/EBPα-deficient HepG2 and HCC-M cells, but not the expressing Hep3B and Huh7 cells, were sensitive to energy starvation (Fig. 3C, D). More importantly, this sensitization effect could be replicated even in a hypoxic environment (Supporting Fig. S3D). On the other hand, overexpression of C/EBPα using a metallothionein-inducible promoter system13 in the C/EBPα-deficient HCC-M cells resulted in partial protection against starvation-induced cell death (Fig. 3E).
Figure 3.
Hepatocarcinoma cells were protected from energy starvation–induced cell death by C/EBPα. (A) The stable C/EBPα-expressing shNC control cells and C/EBPα–silenced cells (sh4 and sh7) were starved in glucose- and glutamine-free Dulbecco's modified Eagle's medium (Glu+Gln starvation) for 2 days. Cell images are shown in the upper panel, followed by cell cycle profiles with the proportion of sub-G1 dead cells indicated as mean ± standard deviation. Western blotting in the lower panel shows the expression of C/EBPα. (B) Cells were starved in glucose- and glutamine-free Dulbecco's modified Eagle's medium, fetal bovine serum–free Dulbecco's modified Eagle's medium, or fetal bovine serum– and amino acid–double free Earle's Balanced Salt Solution medium for 2 days. (C,D) The C/EBPα-expressing Hep3B and Huh7 and C/EBPα-deficient HepG2 and HCC-M cells were starved as above. (E) Overexpression of C/EBPα using a metallothionein inducible promoter system (induced by 100 µM zinc chloride) in C/EBPα-deficient HCC-M cells resulted in partial protection against starvation-induced cell death. *P < 0.05 compared with cells cultured in full medium. Abbreviations: FBS, fetal bovine serum; EBSS, Earle's Balanced Salt Solution; MT, metallothionein.
Lipid Catabolism Was Essential for C/EBPα-Mediated Protection
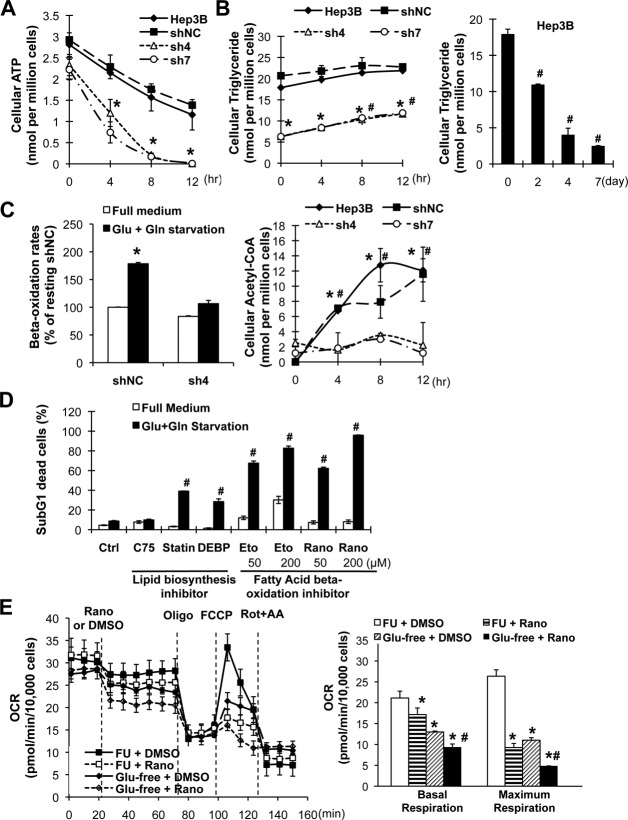
To find out how C/EBPα-expressing cells survived during energy starvation, we examined the time-dependent changes of key energy metabolites in the two different cell types. The C/EBPα-silenced sh4 and sh7 cells were depleted of adenosine triphosphate after 12 hours starvation (Fig. 4A). In contrast, the C/EBPα-expressing Hep3B and shNC cells maintained adenosine triphosphate concentrations at up to 50% of the basal level at 12 hours. Pyruvate, the final product of glycolysis, was also decreased at a higher rate in C/EBPα-silenced cells (Supporting Fig. S4A). Glycogen, the carbohydrate energy reserve, was consumed at the same rate in both cell types during the first 4 hours but depleted in C/EBPα-silenced cells at later time points (Supporting Fig. S4A), possibly due to lower basal levels. Strikingly, the kinetics by which C/EBPα-expressing and C/EBPα-silenced cells utilized triglyceride (the lipid form of energy reserve) was different. In the C/EBPα-expressing cells, triglyceride was stored at a higher basal level and maintained unchanged after 12-hour starvation but decreased in a time-dependent manner from day 2 to day 7 (Fig. 4B). In contrast, the C/EBPα-silenced (sh4 and sh7) and C/EBPα-deficient (HepG2) cells increased triglyceride two-fold despite cell death occurring (Fig. 4B; Supporting Fig. S4B). Lastly, C/EBPα-expressing cells had higher starvation-induced lipid catabolism (as evidenced by a higher fatty acid beta-oxidation rate and an increased acetyl-coenzyme A level) compared with silenced cells (Fig. 4C). The lipid mobilization kinetics suggests that lipid biosynthesis was terminated and lipid catabolism switched on in C/EBPα-expressing cells, but not in C/EBPα-silenced cells, during starvation.
Figure 4.
Lipid catabolism was essential for C/EBPα-mediated protection against energy starvation. Cells expressing C/EBPα (Hep3B and shNC) and C/EBPα-silenced stable cells (sh4 and sh7) were starved in glucose- and glutamine-free medium. The intracellular levels of adenosine triphosphate (A) and triglyceride (B) were determined in a time course. (C) Fatty acid beta-oxidation rates in Hep3B and sh4 cells were determined after 2-hour starvation. Intracellular levels of acetyl-coenzyme A were determined as in A. (D) The Hep3B cells were either pretreated with the liposynthesis inhibitor C75 (5 µg/mL), simvastatin (statin, 10 µM), and diethylum-belliferyl phosphate (50 µM) for 2 weeks before starvation or treated with the fatty acid beta-oxidation inhibitor etomoxir and ranolazine in starvation medium for 3 days. Dead cells were determined by sub-G1 assay as above. *P < 0.05 compared with the corresponding Hep3B and shNC cells of the same time point, #P < 0.05 compared with unstarved cells. (E) Oxygen consumption rates in Hep3B shNC cells were determined by Seahorse mitochondrial stress analysis, under basal conditions and in response to the indicated inhibitors (n = 4). Basal and maximum respiration rates are summarized in the right panel. *P < 0.05 compared with resting cells, #P < 0.05 compared with the corresponding starved cells. Abbreviations: ATP, adenosine triphosphate; CoA, coenzyme A; DEBP, diethylum-belliferyl phosphate; Eto, etomoxir; Rano, ranolazine; DMSO, dimethyl sulfoxide; FCCP, fluorocarbonyl cyanide phenylhydrazone; Rot+AA, rotenone and antimycin A; FU, full medium; OCR, oxygen consumption rate.
To further investigate the role of lipid metabolism in cell survival, we used pharmacological inhibitors to decrease either lipid anabolism or lipid catabolism. Pretreatment of Hep3B cells with C75 (fatty acid synthase inhibitor), simvastatin (3-hydroxy-3-methylglutaryl-coenzyme A reductase inhibitor), or diethylum-belliferyl phosphate (cholesterol esterase inhibitor) decreased (but did not deplete) intracellular lipid content (Supporting Fig. S4C). Simvastatin and diethylum-belliferyl phosphate, but not C75, mildly increased starvation-induced cell death (around 30%; Fig. 4D). In contrast, specific inhibition of fatty acid beta-oxidation by etomoxir or ranolazine significantly sensitized Hep3B cells to cell death in a dose-dependent manner (60%–90%; Fig. 4C). To confirm the role of beta-oxidation, we further determined the changes of mitochondrial oxygen respiration. Ranolazine treatment, especially in the absence of glucose, significantly impaired basal oxygen consumption and maximum spare respiratory capacity (Fig. 4E), as described.16 These results collectively indicate that lipid catabolism is important for C/EBPα-involved cell survival.
Autophagy Was Required for C/EBPα-Mediated Lipid Catabolism and Protection
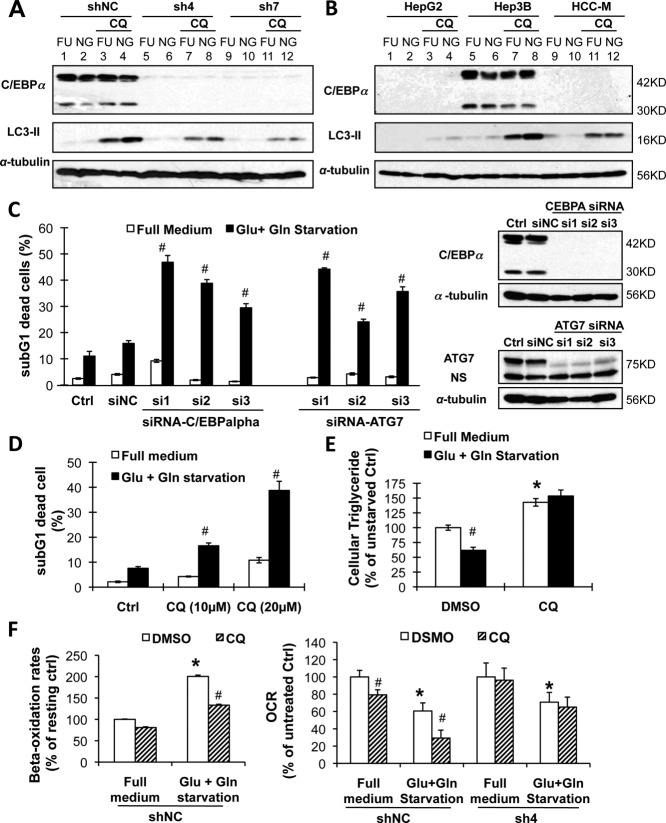
It has been reported that autophagy is essential for lipid catabolism during nutrient deprivation.17 We thus investigated the possible role of autophagy in C/EBPα-mediated lipid catabolism and cell survival. The C/EBPα-expressing cells had higher basal levels of LC3-II than the C/EBPα-silenced or -deficient cells (compare lanes 3 versus 7 and 11 in Fig. 5A and lanes 7 versus 3 and 11 in Fig. 5B). More importantly, cotreatment of CQ (a lysosomotropic agent that prevents endosomal acidification and lysosomal enzymatic activity) further increased starvation-induced LC3-II expression, a hallmark of autophagy induction (also called LC3 flux;18 compare lanes 4 versus 3 in Fig. 5A and lanes 8 versus 7 in Fig. 5B) in C/EBPα-expressing cells. These results suggest that autophagy was induced in C/EBPα-expressing cells but not in C/EBPα-silenced or -deficient cells.
Figure 5.
Autophagy was essential for C/EBPα-mediated lipid catabolism and protection against starvation. (A) Cells expressing C/EBPα (shNC) and C/EBPα-silenced stable cells (sh4 and sh7) were treated with or without 25 µM of the lysosomal inhibitor CQ in glucose- and glutamine-free medium for 3 hours. Flux of LC3-II was determined by western blotting. (B) Cells expressing C/EBPα (Hep3B) and C/EBPα-deficient HepG2 and HCC-M cells were treated as in A. (C) The Hep3B cells were silenced of ATG7 or C/EBPα by specific siRNAs for 2 days before starvation treatment (siNC for nonspecific siRNA) for 3 days. Dead cells were determined by sub-G1 assay (left panel) and the proteins by western blotting (right panel). (D,E) The Hep3B cells were starved with or without CQ for 2 days. Dead cells were determined (D), and the intracellular triglyceride level was calculated as in Fig. 3B. (F) The shNC and sh4 cells were starved with or without CQ (100 µM). Fatty acid beta-oxidation rates were determined (left panel), and oxygen consumption rates were separately determined by mitochondrial stress assay (n = 4, right panel). Abbreviations: FU, full medium; NG, glucose- and glutamine-free medium; NS, nonspecific staining; DMSO, dimethyl sulfoxide.
The essential role of autophagy in C/EBPα-mediated lipid catabolism and cell survival during energy starvation was further investigated by the following experiments. Firstly, knocking down the autophagy-essential gene ATG7 by specific siRNAs sensitized Hep3B cells to starvation-induced cell death, replicating the effect of C/EBPα-specific siRNA silencing (Fig. 5C). Similarly, inhibition of autolysosomal activity by CQ treatment dose-dependently sensitized Hep3B cells to starvation-induced cell death (Fig. 5D). Secondly, CQ treatment also increased the basal levels of intracellular triglyceride and prevented triglyceride utilization during energy starvation (Fig. 5E), indicating the essential role of autophagy in lipid metabolism in both resting and starvation states. More importantly, CQ treatment metabolically decreased the basal and starvation-induced fatty acid beta-oxidation in the C/EBPα-expressing Hep3B cells (Fig. 5F, left panel), which is consistent with a previous report.17 This result was further supported by changes in the mitochondrial OCR. In the C/EBPα-expressing shNC cells, CQ pretreatment (100 µM for 2 hours) decreased the OCR in the resting state and further decreased it when the cells were energy-deprived (Fig. 5F, right panel). In contrast, in the C/EBPα-silenced sh4 cells, CQ pretreatment failed to decrease the OCR in both states.
TMEM166 Was Responsible for C/EBPα-Mediated Autophagy Induction
Autophagy induction involves two common upstream signaling pathways: adenosine monophosphate–activated protein kinase (AMPK) and mammalian target of rapamycin. We showed that glucose and glutamine starvation caused AMPK activation (as shown by increased AMPK phosphorylation at thr172) and mammalian target of rapamycin deactivation (decreased S6 phosphorylation at ser235/236) in both C/EBPα-expressing and C/EBPα-silenced cells (Supporting Fig. S5A). This suggests that the alterations of these two pathways are independent of C/EBPα expression in the cells.
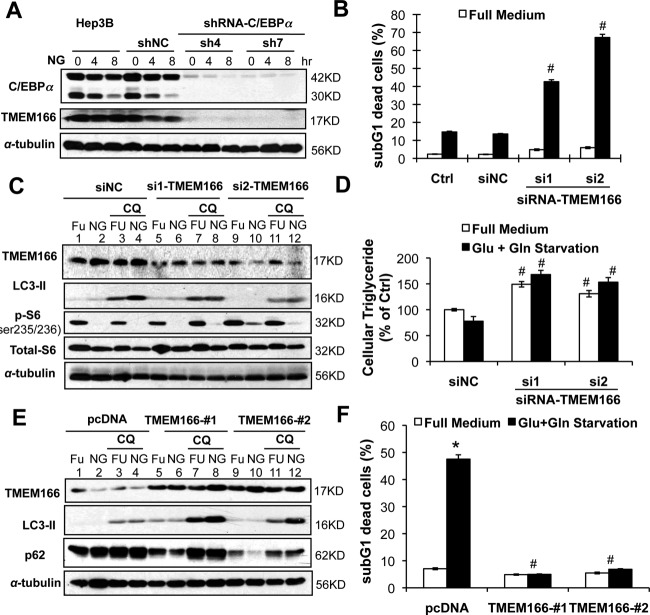
To identify the critical genes involved in autophagy that may be regulated by C/EBPα, we compared the transcriptomes obtained from C/EBPα-expressing cells versus C/EBPα-silenced or -deficient cells (Supporting Fig. S6A). Among 84 genes differentially expressed at least 10-fold, TMEM166 (EVA1A) has been reported to be associated with autophagy induction.14 We confirmed that both TMEM166 mRNA and protein were up-regulated in C/EBPα-expressing cells (Fig. 6A; Supporting Fig. S6B). Knocking down TMEM166 by specific siRNAs inhibited starvation-induced autophagy (compare lanes 8 versus 7 and lanes 12 versus 11 in Fig. 6C) and triglyceride catabolism (Fig. 6B; Supporting Fig. S6C) and subsequently sensitized significant cell death (Fig. 6D). On the other hand, forced overexpression of TMEM166 in the low-expressing HCC-M cells restored autophagy induction (compare lanes 8 versus 7 and lanes 12 versus 11 in Fig. 6E) and decreased p62 stability (another hallmark of autophagy induction). Overexpression of TMEM166 protected HCC-M cells from starvation-induced cell death (Fig. 6F). Although TMEM overexpression increased beta-oxidation in HCC-M cells under resting and starvation conditions (Supporting Fig. S6D), it was not able to mediate the starvation-induced increase in beta-oxidation observed in the C/EBPα-expressing cells.
Figure 6.
In C/EBPα-expressing Hep3B cells TMEM166 was required for the induction of autophagy. (A) The TMEM166 protein was determined by western blotting in stable cell lines. (B-D) In Hep3B cells TMEM166 was silenced by specific siRNA before starvation. The sub-G1 cell deaths were determined (B), western blotting is shown (C), and intracellular triglyceride levels were measured (D). (E,F) The TMEM166 protein was overexpressed in the low-expressing HCC-M cells and then starved. Abbreviations: FU, full medium; NG, glucose- and glutamine-free medium.
Discussion
In this article, we have demonstrated that C/EBPα overexpression is correlated with poorer HCC overall survival (Fig. 1 and Table1). Expression of C/EBPα is also helpful for subdividing patients with different survival outcomes by combining with serum AFP or vascular invasion (Fig. 1; Supporting Fig. S1). The results indicate that C/EBPα may be a useful prognosticator in HCC, either independently or in combination with serum AFP and vascular invasion. Although neither C/EBPα overexpression nor its level in tumor tissues was associated with recurrence (Supporting Fig. S7), we found that C/EBPα expression by HCC may be a stable trait of the tumor. In nine patients who had a second surgery to remove a recurrent tumor, C/EBPα levels in the recurrent tumors were not different from those in the primary tumors (P = 0.800; Supporting Table S3), regardless of the interval between resection of primary tumors and diagnosis of recurrence. Consistent with the primary HCC (Fig. 1), patients with recurrent tumors and who had elevated C/EBPα expression in the primary tumor were more likely to have poorer overall survival (P = 0.047; Supporting Fig. S7C).
The negative correlation between C/EBPα expression and HCC survival is consistent with the better survival outcome observed in AML patients who had double-allele CEBPA mutations19, 20 or decreased expression due to DNA hypermethylation.21, 22 Strikingly, C/EBPα is reported as a tumor suppressor in AML because human CEBPA mutation predisposes one to familial AML23 and mice carrying the engineered CEBPA alleles that only express p30 developed AML with complete penetrance.24 The opposing roles of C/EBPα in oncogenesis in different types of cancer may be determined by a posttranslational phosphorylation switch. Phosphorylation of C/EBPα at ser190 (ser193 in mice homologue) is essential for its binding with cyclin-dependent kinases and growth inhibitory activity.25 However, dephosphorylation by the PI3K/AKT-protein phosphatase 2 pathway switches C/EBPα to sequester Rb protein and releases E2F to promote cell growth.25 We and others have shown that inhibition of C/EBPα dephosphorylation by treatment with AKT or phosphatase inhibitors could reverse this change and inhibit cell growth in HCC12, 26 and prostate cancer cells.27 However, the role of C/EBPα phosphorylation in human cancers is as yet undetermined.
During solid tumor development, cancer cells may experience limited nutrient and oxygen availability as a result of the distance to the vasculature and lack of neoangiogenesis.4 We have convincingly showed that C/EBPα promoted cell survival in both in vivo tumor development (Fig. 2 for mice xenograft and Table2 for human HCC) and in vitro cell studies (Fig. 3). In contrast, the C/EBPα-silenced and C/EBPα-deficient cells were sensitive to glucose and glutamine double starvation under normoxia or hypoxia (Fig. 3; Supporting Fig. S3). We found that the C/EBPα-dependent sensitization effect was specific to energy starvation because neither FBS nor amino acid deprivation could consistently kill the C/EBPα-deficient cells (Fig. 3B, D). Similar to our findings, it has been reported that leukemia cells expressing C/EBPα or its mutant were resistant to Fas ligand–induced apoptosis by transcriptionally induced antiapoptotic Bcl-2 and FLIP.13, 28 However, we did not observe increased expression of these two antiapoptotic molecules in the HCC cells, indicating that different molecular mechanisms against cell death are involved in HCC and leukemia.
Similar to other novel oncogenic signaling pathways uncovered for HCC,7 we have shown that overexpressed C/EBPα endowed HCC cells with a metabolic advantage against energy deprivation, where glucose and glutamine had been removed. Our findings reinforce the results that C/EBPα is involved in glucose and lipid metabolism.9–11 The higher baseline level of triglyceride in C/EBPα-expressing cells (Fig. 4A; Supporting Fig. S4B) could be explained by the increased expression of several lipogenic enzymes (Supporting Fig. S5B, C), including long chain fatty acid coenzyme A 1, 3, and 4.29, 30 However, the energetic switch from lipid biosynthesis to utilization (Fig. 4) may result from enhanced phosphorylation of acetyl-coenzyme A carboxylase. Acetyl-coenzyme A carboxylase is essential for lipid biosynthesis; however, its phosphorylation at ser79 by the AMPK pathway in response to starvation switches off its bioactivity.31 Strong acetyl-coenzyme A carboxylase phosphorylation was observed in the C/EBPα-expressing cells, but it became weaker at 4 hours and disappeared at 8 hours in the C/EBPα-silenced cells (Supporting Fig. S4B), despite higher AMPK phosphorylation observed at both time points (Supporting Fig. S4A). More importantly, it appears that the ability to mobilize triglyceride was the key factor in protecting cells from starvation-induced cell death. An increasing number of reports have shown that lipids are degraded by autophagy during starvation to maintain lipid homeostasis and energy balance.17 We confirmed the importance of autophagy in C/EBPα-mediated lipid catabolism and cell survival by silencing the essential autophagy gene, ATG7, or by treating with CQ (Fig. 5). From the differential genes regulated by C/EBPα, we identified that TMEM166 was involved in C/EBPα-mediated autophagy induction and protection of cells from energy deprivation (Fig. 6). However, it appears that TMEM166 alone was not able to fully reconstitute the C/EBPα-mediated increase in beta-oxidation during energy deprivation in non-C/EBPα-expressing HCCM cells. It may be that TMEM166 facilitates autophagosome formation;14 however, the exact mechanism is as yet unknown. Although we cannot exclude the possibility of other C/EBPα-regulated autophagic genes involved in responding to starvation, the results indicate that TMEM166 may, at least in part, explain the survival advantage of C/EBPα-expressing cells under starvation.
The molecular mechanisms involved in C/EBPα overexpression in HCC are as yet unclear. However, silencing C/EBPα (Fig. 3) or inhibiting its downstream pathways (e.g., autophagy by CQ [clinically approved for the treatment and prevention of malaria, Fig. 5]; beta-oxidation by etomoxir or ranolazine [clinically approved for the treatment of angina, Fig. 4]) could significantly sensitize resistant HCC cells to energy starvation–induced cell death, suggesting C/EBPα as a potential therapeutic target in HCC. It is tempting to speculate that these findings could be applied to in vivo HCC therapy, such as antineoangiogenesis treatment which inhibits de novo tumor blood vessel formation to starve cancer cells and control cancer growth. Several antineoangiogenesis drugs (e.g., brivanib, ABT-869) are currently under HCC phase II or III clinical trials.32, 33 Furthermore, these findings may also be applied to enhance the current use of embolization therapy. The blood supply of HCC tumors comes mainly (90%-100%) from the hepatic artery, while a normal liver has a dual blood supply, with 75%-85% from the portal vein and 20%-25% from the hepatic artery.34 Hence, embolization of the hepatic artery deprives the blood supply predominantly to the tumor, while maintaining adequate blood supply to the normal liver. Embolization treatment, especially transarterial chemoembolization, proved to be effective in extending HCC patient survival.8 How HCC patients respond differently to transarterial chemoembolization is as yet unknown. Thus, it will be interesting to learn whether targeting C/EBPα or its downstream autophagy-involved lipolysis pathway could further improve transarterial chemoembolization.
Taken together, C/EBPα is an independent prognosticator of HCC patient survival and a novel molecular target for HCC cell survival in HCC development and therapy. These findings may pave the way for a better understanding and management of HCC.
Author names in bold designate shared co-first authorship.
Acknowledgments
We thank Dr. A.D. Friedman (Johns Hopkins University, Baltimore, MD) for providing the inducible C/EBPα expression plasmid and Dr. Yingyu Chen (Peking University, Beijing, China) for the human TMEM166 plasmid.
Glossary
- AFP
α-fetoprotein
- AML
acute myeloid leukemia
- AMPK
adenosine monophosphate–activated protein kinase
- C/EBPα
CCAAT enhancer binding protein α
- CQ
chloroquine
- HCC
hepatocellular carcinoma
- OCR
oxygen consumption rate
- siRNA
small interfering RNA
Footnotes
Additional Supporting Information may be found at onlinelibrary.wiley.com/doi/10.1002/hep.27593/suppinfo.
Supporting Information
Additional Supporting Information may be found at onlinelibrary.wiley.com/doi/10.1002/hep.27593/suppinfo.
Supplementary Information
References
- Villanueva A, Hernandez-Gea V, Llovet JM. Medical therapies for hepatocellular carcinoma: a critical view of the evidence. Nat Rev Gastroenterol Hepatol. 2013;10:34–42. doi: 10.1038/nrgastro.2012.199. [DOI] [PubMed] [Google Scholar]
- Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25–34. doi: 10.1016/S1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491:364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- Huang Q, Tan Y, Yin P, Ye G, Gao P, Lu X, et al. Metabolic characterization of hepatocellular carcinoma using nontargeted tissue metabolomics. Cancer Res. 2013;73:4992–5002. doi: 10.1158/0008-5472.CAN-13-0308. [DOI] [PubMed] [Google Scholar]
- Beyoğlu D, Imbeaud S, Maurhofer O, Bioulac-Sage P, Zucman-Rossi J, Dufour JF, et al. Tissue metabolomics of hepatocellular carcinoma: tumor energy metabolism and the role of transcriptomic classification. Hepatology. 2013;58:229–238. doi: 10.1002/hep.26350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhu A, Roessler S, Zhao X, Yu Z, Forgues M, Ji J, et al. Integrated metabolite and gene expression profiles identify lipid biomarkers associated with progression of hepatocellular carcinoma and patient outcomes. Gastroenterology. 2013;144:1066–1075, e1. doi: 10.1053/j.gastro.2013.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llovet JM, Bruix J. Systematic review of randomized trials for unresectable hepatocellular carcinoma: chemoembolization improves survival. Hepatology. 2003;37:429–442. doi: 10.1053/jhep.2003.50047. [DOI] [PubMed] [Google Scholar]
- Koschmieder S, Halmos B, Levantini E, Tenen DG. Dysregulation of the C/EBPα differentiation pathway in human cancer. J Clin Oncol. 2009;27:619–628. doi: 10.1200/JCO.2008.17.9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ND, Finegold MJ, Bradley A, Ou CN, Abdelsayed SV, Wilde MD, et al. Impaired energy homeostasis in C/EBP α knockout mice. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- Matsusue K, Gavrilova O, Lambert G, Brewer HB, Jr, Ward JM, Inoue Y, et al. Hepatic CCAAT/enhancer binding protein α mediates induction of lipogenesis and regulation of glucose homeostasis in leptin-deficient mice. Mol Endocrinol. 2004;18:2751–2764. doi: 10.1210/me.2004-0213. [DOI] [PubMed] [Google Scholar]
- Lu GD, Leung CH, Yan B, Tan CM, Low SY, Aung MO, et al. C/EBPα is up-regulated in a subset of hepatocellular carcinomas and plays a role in cell growth and proliferation. Gastroenterology. 2010;139:632–643, 643. doi: 10.1053/j.gastro.2010.03.051. e1-4. [DOI] [PubMed] [Google Scholar]
- Paz-Priel I, Ghosal AK, Kowalski J, Friedman AD. C/EBPα or C/EBPα oncoproteins regulate the intrinsic and extrinsic apoptotic pathways by direct interaction with NF-kappaB p50 bound to the bcl-2 and FLIP gene promoters. Leukemia. 2009;23:365–374. doi: 10.1038/leu.2008.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yu C, Lu Y, He P, Guo J, Zhang C, et al. TMEM166, a novel transmembrane protein, regulates cell autophagy and apoptosis. Apoptosis. 2007;12:1489–1502. doi: 10.1007/s10495-007-0073-9. [DOI] [PubMed] [Google Scholar]
- Miranda DA, Koves TR, Gross DA, Chadt A, Al-Hasani H, Cline GW, et al. Re-patterning of skeletal muscle energy metabolism by fat-storage-inducing transmembreane protein 2. J Biol Chem. 2011;286:42188–42199. doi: 10.1074/jbc.M111.297127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Windt GJ, Everts B, Chang CH, Curtis JD, Freitas TC, Amiel E, et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour A, Schneider F, Metzeler KH, Hoster E, Schneider S, Zellmeier E, et al. Acute myeloid leukemia with biallelic CEBPA gene mutations and normal karyotype represents a distinct genetic entity associated with a favorable clinical outcome. J Clin Oncol. 2010;28:570–577. doi: 10.1200/JCO.2008.21.6010. [DOI] [PubMed] [Google Scholar]
- Marcucci G, Maharry K, Radmacher MD, Mrózek K, Vukosavljevic T, Paschka P, et al. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B Study. J Clin Oncol. 2008;26:5078–5087. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackanson B, Bennett KL, Brena RM, Jiang J, Claus R, Chen SS, et al. Epigenetic modification of CCAAT/enhancer binding protein α expression in acute myeloid leukemia. Cancer Res. 2008;68:3142–3151. doi: 10.1158/0008-5472.CAN-08-0483. [DOI] [PubMed] [Google Scholar]
- Jost E, do ON, Wilop S, Herman JG, Osieka R, Galm O. Aberrant DNA methylation of the transcription factor C/EBPα in acute myelogenous leukemia. Leuk Res. 2009;33:443–449. doi: 10.1016/j.leukres.2008.07.027. [DOI] [PubMed] [Google Scholar]
- Pabst T, Eyholzer M, Haefliger S, Schardt J, Mueller BU. Somatic CEBPA mutations are a frequent second event in families with germline CEBPA mutations and familial acute myeloid leukemia. J Clin Oncol. 2008;26:5088–5093. doi: 10.1200/JCO.2008.16.5563. [DOI] [PubMed] [Google Scholar]
- Kirstetter P, Schuster MB, Bereshchenko O, Moore S, Dvinge H, Kurz E, et al. Modeling of C/EBPα mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 2008;13:299–310. doi: 10.1016/j.ccr.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Wang GL, Iakova P, Wilde M, Awad S, Timchenko NA. Liver tumors escape negative control of proliferation via PI3K/Akt-mediated block of C/EBP α growth inhibitory activity. Genes Dev. 2004;18:912–925. doi: 10.1101/gad.1183304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Timchenko NA. Dephosphorylated C/EBPα accelerates cell proliferation through sequestering retinoblastoma protein. Mol Cell Biol. 2005;25:1325–1338. doi: 10.1128/MCB.25.4.1325-1338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Lowery M, Glass J. In prostate cancer C/EBPα promotes cell growth by the loss of interactions with CDK2, CDK4, and E2F and by activation of AKT. Prostate. 2009;69:1001–1016. doi: 10.1002/pros.20947. [DOI] [PubMed] [Google Scholar]
- Paz-Priel I, Houng S, Dooher J, Friedman AD. C/EBPα and C/EBPα oncoproteins regulate nfkb1 and displace histone deacetylases from NF-kappaB p50 homodimers to induce NF-kappaB target genes. Blood. 2011;117:4085–4094. doi: 10.1182/blood-2010-07-294470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu SY, Mashek MT, Mashek DG. Suppression of long chain acyl-CoA synthetase 3 decreases hepatic de novo fatty acid synthesis through decreased transcriptional activity. J Biol Chem. 2009;284:30474–30483. doi: 10.1074/jbc.M109.036665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LO, Ellis JM, Paich HA, Wang S, Gong N, Altshuller G, et al. Liver-specific loss of long chain acyl-CoA synthetase-1 decreases triacylglycerol synthesis and beta-oxidation and alters phospholipid fatty acid composition. J Biol Chem. 2009;284:27816–27826. doi: 10.1074/jbc.M109.022467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha J, Daniel S, Broyles SS, Kim KH. Critical phosphorylation sites for acetyl-CoA carboxylase activity. J Biol Chem. 1994;269:22162–22168. [PubMed] [Google Scholar]
- Park JW, Finn RS, Kim JS, Karwal M, Li RK, Ismail F, et al. Phase II, open-label study of brivanib as first-line therapy in patients with advanced hepatocellular carcinoma. Clin Cancer Res. 2011;17:1973–1983. doi: 10.1158/1078-0432.CCR-10-2011. [DOI] [PubMed] [Google Scholar]
- Toh HC, Chen PJ, Carr BI, Knox JJ, Gill S, Ansell P, et al. Phase 2 trial of linifanib (ABT-869) in patients with unresectable or metastatic hepatocellular carcinoma. Cancer. 2013;119:380–387. doi: 10.1002/cncr.27758. [DOI] [PubMed] [Google Scholar]
- Soulen MC. Chemoembolization of hepatic malignancies. Oncology (Williston Park) 1994;8:77–84. discussion 84, 89-90. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information