Abstract
Post-natal growth and regeneration of skeletal muscle is highly dependent on a population of resident myogenic precursors known as satellite cells. Transcription factors from the Pax gene family, Pax3 and Pax7, are critical for satellite cell biogenesis, survival and potentially self-renewal; however, the underlying molecular mechanisms remain unsolved. This is particularly true in the case of Pax7, which appears to regulate myogenesis at multiple levels. Accordingly, recent data have highlighted the importance of a functional relationship between Pax7 and the MyoD family of muscle regulatory transcription factors during normal muscle formation and disease. Here we will critically review key findings suggesting that Pax7 may play a dual role by promoting resident muscle progenitors to commit to the skeletal muscle lineage while preventing terminal differentiation, thus keeping muscle progenitors poised to differentiate upon environmental cues. In addition, potential regulatory mechanisms for the control of Pax7 activity will be proposed.
Keywords: Pax7, satellite cells, pan genes, MyoD
Introduction
The ability to repair adult damaged tissues is critical for organ function and maintenance. In vertebrates, adult skeletal muscle possesses a tremendous capacity for regeneration in response to acute injury or chronic disease. Because the nuclei within skeletal muscle fibres are terminally differentiated and thus incapable of participating in muscle repair, these responses are largely attributed to a distinct and small population (1–6% of total muscle nuclei) of resident myogenic progenitors referred to as satellite cells [1-4]. This cell population is physically distinct from the adult myofibre as they reside between the sarcolemma and the basal lamina in a non-proliferative, low metabolic state [1, 4, 5]. Quiescent satellite cells do not express Muscle Regulatory transcription Factors (MRFs) [3, 6], which are required for myogenic lineage commitment and terminal differentiation. However, upon stimuli such as muscle injury, satellite cells become activated, proliferate extensively and induce the expression of MRFs such as MyoD and Myf-5. Proliferating satellite cells are referred to as adult myoblasts. Ultimately, adult myoblasts become committed to terminal differentiation by inducing the expression of the MRF myogenin, withdraw from the cell cycle and either fuse with existing fibres or fuse one to another to form new myofibres [7]. During muscle regeneration, the quiescent satellite cell pool must be replenished to ensure muscle maintenance and additional muscle regeneration throughout adulthood [8, 9] (Fig. 1). Maintenance of the satellite cell population occurs via self-renewal [10-18], for which satellite cells are considered tissue-specific adult stem cells [19-21].
Fig 1.

Satellite cells and skeletal muscle regeneration. In resting and uninjured muscles, satellite cells reside quiescent (green) sandwiched between the muscle fibre (multi-nucleated cell) plasma membrane and the basal lamina (not depicted). Upon external stimuli such as muscle injury (blue flash symbol), satellite cells become activated (yellow/green), re-enter the cell cycle and commit to the muscle lineage. After a few rounds of proliferation, myoblasts (yellow/ orange) undergo terminal differentiation (red cells) and fuse one to another to form new myotubes (red syncytia) or to pre-existing damaged fibres. During the process of myofibre repair, the pool of quiescent satellite cells is renewed.
Although important advances have been made toward understanding the molecular regulation of embryonic muscle formation and adult myoblast differentiation, the mechanisms that control satellite cell fate decisions and lead to maintenance and renewal of the progenitor population remain to be elucidated. Quiescent satellite cells express a number of molecular markers including cell surface receptors and adhesion molecules such as Syndecan-3, syndecan-4, c-Met, calcitonin receptor, p75 NTR/BDNF, α7-integrin, CD34 and m-cadherin [6, 22-24] and transcription factors such as Sox8/9 and Pax7 [25, 26]. However, the relationship between expression of these markers and the establishment and maintenance of the quiescent state is not clear.
Since the beginning of the present decade, a number of reports have suggested a critical role for Pax7 function in satellite cell biogenesis, perinatal satellite cell survival and regulation of myogenic progression [13, 26-32]. Nevertheless, the exact nature of Pax7’s role in these processes remains veiled behind contradictory results concerning Pax7 function and the lack of detailed analysis regarding Pax7 protein regulation. In this review we will discuss how the available data may be reconciled and suggest that Pax7 plays a bivalent role in satellite cells: while promoting myogenic commitment by inducing MRF expression, Pax7 prevents differentiation by repressing MRF function. Finally, we will discuss putative regulatory mechanisms for Pax7 activity in satellite cells and the potential for the extrinsic regulation of muscle stem cells in cell-based treatment of acute and/or chronic muscle loss.
Pax proteins in muscle formation
The Pax gene family defines an evolutionary conserved group of transcription factors that play critical roles during organogenesis and tissue homeostasis [33-35]. Nine Pax proteins have been described in mammals (Table 1), where presence of the paired box DNA-binding domain is a common feature. The family is further sub-grouped by the presence of an octapeptide motif and the presence, absence or truncation of a homeodomain, which can mediate both DNA–protein and protein–protein interactions. Spontaneous mutations in Pax genes can lead to developmental defects both in mice and humans [36]. Perturbation in Pax function can also lead to cancer [35, 37].
Table 1.
Pax family of transcription factors in mammals
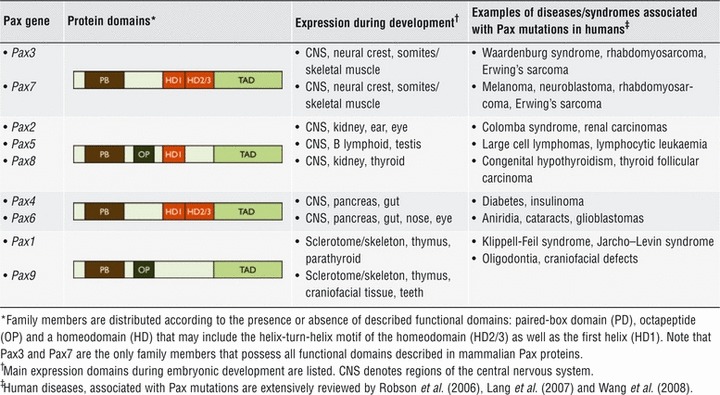 |
Pax3 and Pax7 are two closely related family members involved in the specification and maintenance of skeletal muscle progenitors [38-40]. In contrast, Pax7’s role in muscle development may not be critical because Pax7−/− mice have no gross defects in muscle formation. However, in the absence of Pax7 adult skeletal muscles appear devoid of satellite cells [26]. Accordingly, Pax7 null mice exhibit reduced muscle growth, marked muscle wasting and an extreme deficit in muscle regeneration after acute injury [26, 27]. Both Pax3 and Pax7 mark a population of muscle progenitors in the embryonic somites (Pax3+/Pax7+ cells) [41-44]. Pax3+/Pax7+ cells proliferate and persist throughout embryonic and foetal development and are proposed to give rise to postnatal satellite cells. Pax3 expression is down-regulated before birth, and in the adult is confined to a subpopulation of satellite cells in specific muscle groups [28, 43], whereas Pax7 expression is considered a universal satellite cell marker. Inducible post-natal inactivation of PAX7 [32] has recently challenged the significance of Pax7 expression in satellite cells. In this model, early post-natal and juvenile (P0–P20) PAX7 inactivation severely impaired myogenesis, whereas Pax7 inactivation later after birth (over P20) did not compromise adult myogenesis (i.e. muscle regeneration) [32]. The authors of that study concluded that Pax7 is critical for maintenance of satellite cell progenitors, but after the establishment of the quiescent phenotype (∼P20), Pax7 function is dispensable for muscle repair. It is worth noting that these studies do not rule out the participation of infiltrating cells, such as bone marrow–derived cells [45] compensating for Pax7 depletion during muscle repair. The effect of PAX7 post-natal inactivation was not explored during muscle aging, although a correlation between Pax7 expression, regeneration potential and self-renewal in aged satellite cells has been suggested [46]. Similarly, Pax7 function during regeneration may be partially redundant with other pathways, which may compensate for Pax7 loss in adult muscle progenitors [47]. Thus, the absence of dramatic in vivo phenotypes cannot directly rule out a Pax7 function in adult myogenesis.
Pax7 persists in recently activated, proliferating satellite cells and is rapidly down-regulated in cells that commit to terminal differentiation [13, 15]. In primary adult, myoblast cultures a small population of Pax7+/MyoD+ cells down-regulate MyoD expression while retaining or up-regulating Pax7 (Pax7+/MyoD− cells). This subpopulation remains undifferentiated and mitotically inactive, resembling a quiescent satellite cell [13, 15, 48]. In this context, we have previously shown that transient Pax7 overexpression in primary adult myoblasts and satellite cell–derived cell lines results in: (i) down-regulation of MyoD expression, (ii) inhibition of myogenesis and (iii) reduction in BrdU incorporation in transfected cells [13, 29]. Thus, expression pattern analyses and gain of function experiments were among the first indications that Pax7 could functionally interact with the MRFs to regulate satellite cell fate decisions.
Pax7/MRF cross-regulation and the control of satellite cell fate
MyoD is considered the myogenic master gene as its activity can trigger the entire myogenic program when ectopically expressed in non–muscle cell types [49, 50]. Interestingly, ectopic expression of Pax7 can efficiently repress the MyoD-dependent conversion of C3H10T1/2 mesenchymal cells to the muscle lineage [13] and myogenic progression in C2C12 myoblasts [17, 30, 51, 52]. Interestingly, Pax3 overexpression also inhibits myogenesis in MyoD-expressing fibroblasts and C2C12 myoblasts [53]. Analysis of MyoD transcriptional activity upon Pax7 co-expression and identification of potential Pax7 transcriptional targets indicate that Pax7 inhibits early events in the molecular cascade leading to muscle differentiation [13, 29, 52], possibly by repressing MyoD transcriptional activity.
In apparent contradiction with a role for Pax7 repressing muscle differentiation, genetic interactions and transcriptional profile analyses indicate that Pax7 could participate in the induction of the myogenic program during development and in cell culture models [30, 31, 41-44], possibly through induction of MYOD and/or MYF-5 expression [28, 30, 54]. In addition, different cell populations isolated from skeletal muscle tissue (distinct from satellite cells) that have shown myogenic capacity in vivo, such as Pw1+ interstitial cells and CD45+/Sca1+ cells, also require the induction of Pax7 expression to commit to the skeletal muscle lineage [55, 56].
Could it be possible that a single transcription factor can trigger lineage commitment while preventing further progression towards terminal differentiation? It is usually argued that different experimental models (i.e. in vivo versus tissue culture) may account for the apparent contradictory reports analysing Pax7 function. Despite many technological advances, satellite cells are still difficult to isolate in sufficient numbers for biochemical analysis without expanding the purified population in vitro, which also precludes a fine biochemical characterization of truly quiescent satellite cells. In addition, the adult regenerating tissue environment is difficult to manipulate and to ‘decode’ on a molecular scale. Thus, in vitro studies on primary adult myoblasts and/or myogenic cell lines have been largely exploited as valuable methods to examine the biochemical and genetic pathways that direct satellite cell function. Aside from the variability associated to different experimental models, there is increasing evidence suggesting a stem cell-specific molecular mechanism underlying the two-faced behaviour of Pax7 in muscle progenitors.
Lang et al. elegantly showed that in skin stem cells, Pax3 activates expression of Mitf, a transcription factor critical for melanogenesis [57], while competing with Mitf to occupy an enhancer element of a key target gene required for melanin synthesis [58]. Thus, Pax3-expressing melanoblasts are committed yet undifferentiated. Activated β-catenin can then relieve Pax3-dependent repression leading to rapid melanocyte differentiation [58]. Based on these results, Lang et al. suggested the existence of a heterogeneous class of stem cell transcription factors that can both determine cell fate and simultaneously maintain an undifferentiated state, keeping a cell poised to differentiate upon external stimuli.
A similar model has been described for alveolar rhabdomyosarcoma (ARMS) development. The most common cytogenetic feature of this soft tissue sarcoma is represented by the chromosomal translocations leading to fusion between either PAX3 or PAX7 and the FKHR (FOXO1) gene. The Pax-FKHR fusion protein contains intact Pax3 or Pax7 DNA binding domains fused to the FKHR transcriptional activation domain [59]. Pax-FKHR proteins exhibit increased abundance and transcriptional activity compared to their wild-type counterparts [60-63]. Thus, it has been suggested that deregulation of Pax3/Pax7 downstream target genes contributes to tumourigenesis. Although ARMS exhibits a muscle lineage phenotype, the cells evade terminal differentiation despite expressing the potent myogenic transcriptional regulator MyoD. Intriguingly, Pax-FKHR proteins can regulate myogenesis by inducing expression of MyoD but preventing terminal differentiation, via a mechanism that involves repression of MyoD activity [64-68]. Thus, by differentially regulating MyoD expression and function, Pax7-FKHR may promote retention of muscle progenitor characteristics and ARMS development.
Additional examples for transcription factors with similar dual functions have been described including Sox1 that maintains neural crest lineage commitment while inhibiting neuronal differentiation [69] and other two members of the Pax family: Pax5 during B-lymphocyte lineage maintenance and differentiation [70, 71] and Pax6 during retina development [72, 73].
Thus, it is likely that also Pax7 plays a dual role in the regulation of myogenesis by activating commitment to the myogenic program and simultaneously preventing terminal differentiation (Fig. 2). In the short term, such mechanism would allow muscle progenitor expansion and/or maintenance preventing precautious differentiation. Similarly, Pax7 expression in quiescent satellite cells may be associated with quick entry into the myogenic program upon satellite cell activation. In the context of tissue regeneration, it seems advantageous to have a ‘designated driver’ in charge of both the gas and the brake pedal regulating adult stem cell function. The evolution of this strategy may be related to a transition from a regeneration strategy characterized by cell de-differentiation followed by de novo tissue formation, which is present in organisms such as urodeles [74, 75], to the regeneration plan that relies on tissue-specific stem cells with specific differentiation repertoires present in mammals [76, 77]. A centralized mechanism that can direct adult stem cell fate may allow for maintaining their regenerative potential throughout the lifespan of an organism in the form of quiescent precursors (therefore, minimizing the potential for transformation and tumour formation), but at the same time assuring prompt and efficient response upon specific activation.
Fig 2.

Proposed model for Pax7 function as a Pan gene. Pax7 (and possibly Pax7/Pax3 during embryonic development) are required for acquisition of muscle commitment (yellow/orange cell) by inducing the expression of MyoD (and/or Myf-5). MyoD will eventually activate the expression of key targets such as myogenin (red cells), directing progression trough myogenesis and terminal differentiation (pink multi-nucleated cell). Pax7 may act downstream the induction of MyoD expression regulating MyoD activity, preventing myogenin induction and thus, muscle differentiation. This dual role of Pax7 may allow for the expression of key myogenic factors in an undifferentiated state, keeping a cell poised to rapidly differentiate on external stimuli.
Different lines of evidence indicate that regulation of MyoD activity by Pax7 represents a nodal point for the regulation of myogenic progression. Indeed, once myogenin expression is induced, Pax7 is unable to repress muscle differentiation [13, 51]. Accordingly, gain and loss of function experiments suggest that myogenin up-regulation results in rapid loss of Pax7, while preventing myogenin induction may be necessary to maintain Pax7 expression. Thus, a model has been proposed where an inhibitory circuit between Pax7, MyoD and myogenin may regulate the cell fate decisions in muscle progenitors [29]. In this model, the Pax7:MyoD ratio plays a critical role in cell fate determination (Fig. 3). From this model, it is possible to predict that even transient changes in this ratio can likely introduce variability in cellular phenotypes when manipulating Pax7 and/or MRFs levels in muscle progenitors.
Fig 3.
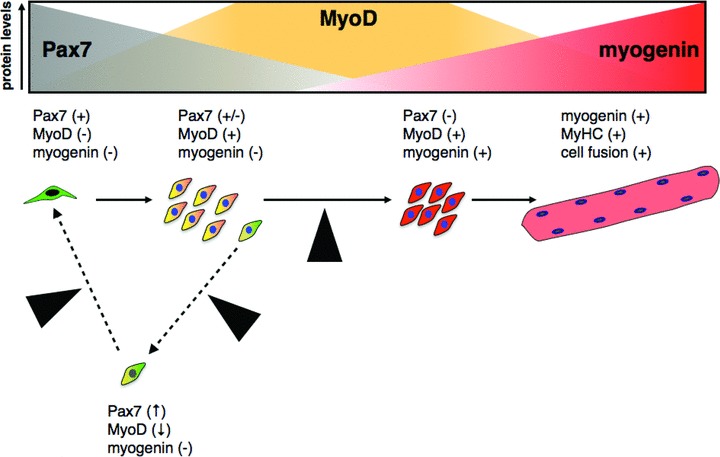
Proposed model for the significance of Pax7:MRF protein ratios and satellite cell fate decisions. Modified from Olguin et al. (2007). Satellite cells (Pax7+/MyoD−/myogenin−) must commit to proliferate, differentiate or renew the progenitor population to maintain muscle function. Extrinsic signalling activates satellite cells and up-regulate MyoD (orange) with a concomitant decline in Pax7 expression (grey). Upon commitment to terminal differentiation, up-regulation of myogenin (red) directly or indirectly down-regulates Pax7. In a small subset, Pax7 down-regulation is prevented by unknown mechanisms resulting in MyoD down-regulation, blocking myogenin induction and eventually leading to the commitment to a quiescent, undifferentiated phenotype (yellow cell, black nucleus). In this model, the Pax7:MyoD expression ratio is regulated primarily by post-translational regulation of Pax7 and MyoD. Black arrowheads indicate putative control points (discussed in the text) where regulation of Pax7 activity/stability may participate in the control of satellite cell fate decisions.
Transcriptional and non-transcriptional Pax7 functions in myogenic progenitors
Unlike Pax3, Pax7 is a poor transcriptional activator. Moreover, Pax7 contains two cis-acting repressor domains at the N-terminus and the homeodomain, respectively, which may correlate with poor Pax7-dependent reporter gene activation in vitro [78]. Interestingly, Pax7 represses myogenesis in the absence of either its paired-box or the transactivation domains, both thought to be critical for its transcriptional activity. Deletion of the homeodomain, however, completely abrogates the ability of Pax7 to inhibit myogenesis [29]. In light of these observations, it is possible that Pax7 could act to indirectly regulate transcription of additional gene(s) required for MyoD function and/or via a non-transcriptional mechanism, such as post-translational control of MyoD. Both mechanisms have been experimentally supported: Pax7 activates the expression of Id2 and Id3, two inhibitory porteins from the helix-loop-helix (HLH) superfamily, that directly regulate MyoD function and expression in muscle precursors [52]. On the other hand, Pax7 can induce proteasome-dependent degradation of MyoD [29]. Proteasome-dependent MyoD degradation is inhibited by MyoD binding to DNA in vitro [79], thus decreased MyoD protein stability could be a consequence of Pax7-dependent disruption of MyoD transcriptional complexes.
Replacement of the Pax7 transcriptional activation domain (TAD) by the Engrailed repressor domain (Pax7:EnR) converts Pax7 into a dominant-repressor of transcription and can be used to analyse the effect of negative regulation of putative Pax7 target genes. Ectopic expression of Pax7:EnR in primary myoblasts and myogenic cell lines in culture indicates that Pax7 transcriptional activity also is involved in the regulation of cell survival, proliferation and morphology [28, 31, 54]. So it is likely that additional Pax7 functions require its transcriptional activity. However, is worth noting that manipulation of the Pax7 TAD may have secondary effects besides inducing or repressing expression of putative Pax7 targets. It has been shown that substitution of the TAD in Pax3 alters its homeodomain function, changing Pax3 transcriptional activity and DNA-binding specificity [80]. Normal Pax3 regulatory protein–protein interactions are also affected by replacement of its TAD [81]. Similar effects may occur upon replacement of Pax7 TAD by the Engrailed repressor domain. Supporting this concept, we have recently shown that MyoD is required for Pax7-FKHR to activate transcription in cell cultures. Interestingly, the MyoD domains sensitive to repression by Pax7 and those that cooperate with Pax7-FKHR are distinct [68]. Thus, it is likely that chimeric proteins such as Pax7:EnR or Pax7-FKHR affect muscle progenitor cell fate via additional mechanisms not directly related to activation or repression of endogenous Pax7 pathways.
Protein interactions, post-translational modifications and the regulation of Pax7
Protein–protein interactions provide an attractive mechanism for regulating Pax7 function in satellite cells. Moreover, this appears to be a conserved theme in the Pax family [33]. Pax proteins interact with many cofactors, resulting in a wide range of biological effects including: (i) regulation of their transcriptional activity [82, 83], (ii) limiting access to regulatory kinases [84], (iii) changes in subcellular localization [85] and (iv) establishment of new protein interactions [86].
Co-immunoprecipitation experiments indicate that Pax7 and MyoD co-exist in a protein complex through indirect interactions [29]. The importance of protein–protein interactions on the regulation of Pax7 function is underscored by recent studies indicating that Pax7 associates with the Wdr5–Ash2L–MLL2 histone methyltransferase complex at the MYF-5 proximal promoter. Thus, Pax7 may regulate entry into the developmental myogenic program by inducing specific chromatin modifications at the MYF-5 promoter [30]. Intriguingly, MYF-5 induction by Pax7 requires the paired box domain but not the homeodomain [30]. These results indicate that the ability of Pax7 to either induce or repress myogenesis depends on differential contribution of both transcriptional and non-transcriptional mechanisms, which reside in different Pax7 functional domains. In addition, attempts to identify direct Pax7 transcriptional targets have returned a low number of putative targets and only a fraction of them have been further confirmed [52, 87], suggesting that an indirect control of transcription by Pax7 could be more prevalent than previously anticipated.
Currently, several groups are involved in the identification of new Pax7 interacting proteins. Indeed, the most recent data from our laboratory suggest that Pax7 may be part of additional protein complexes (Olguín et al., in preparation). Differential protein–protein interactions may be critical to sustain Pax7’s dual role in myogenesis.
Pax proteins undergo post-translational modifications, including phosphorylation [88], sumoylation [89], ubiquitination [90, 91] and glutathionylation [92]. No specific post-translational modifications have been described for Pax7 during myogenesis, although our research suggests that Pax7 undergoes proteasome-mediated degradation in cells that up-regulate myogenin and commit to terminal differentiation [29]. Although recent reports from Boutet et al. suggest that Pax3, but not Pax7, is subject to ubiquitination and proteasome-dependent degradation [90, 91], work in progress from our group may suggest that Pax7 stability is also tightly regulated (Bustos et al., in preparation). Alternatively, there is increasing evidence for the importance of regulated ubiquitin-independent proteasomal degradation of proteins including c-Fos, p53 and retinoblastoma protein (reviewed in Ref. [93]). Similar mechanisms could explain the absence of Pax7 ubiquitination in previously reported in vitro ubiquitination-degradation assays [90, 91]. Control of Pax7 levels during myogenic progression is likely to be complex and to involve additional mechanisms. Accordingly, recent reports indicate that Pax7 expression is negatively regulated by microRNAs [94, 95]. miR-1, miR-206 and miR-486 have been shown to target elements in the Pax7 3’-untranslated region [94, 95]. Loss of function experiments and the expression of a microRNA-resistant form of Pax7, result in delay of myoblast differentiation and persistent Pax7 expression. Interestingly, MyoD induces miR-1, miR-206 and miR-486 expression during differentiation [94, 95]. Because MyoD and Pax7 are generally co-expressed in myoblasts, it is possible to envision an expression level balance maintained by mutual inhibition. In this context, disruption of Pax7:MyoD ratio (Fig. 3) triggered by decrease in Pax7 protein stability (e.g. via proteasome degradation) would disrupt such balance, allowing microRNA-dependent down-regulation to further suppress Pax7 expression in differentiating cells.
In silico analysis of the Pax7 protein sequence reveals several regions with high probability of being post-translationally modified. Such modifications include Ser/Thr phosphorylation, ubiquitination and caspase-mediated cleavage (Olguín, unpublished results). Interestingly, it has been shown that phosphorylation can directly regulate degradation of several key players in the control of myogenesis progression, including MyoD [41, 96-101]. Pax7 contains potential sites for phosphorylation by PKCs, CSK2 and CaMKII (Olguín, unpublished results). Although whether Pax7 represents a direct substrate for these candidate kinases still needs to be determined, differential phosphorylation of Pax7 represents a potential molecular switch regulating Pax7 dual role in muscle progenitors.
Extracellular signalling and the control of Pax7 in muscle progenitors
In its anatomical niche, the satellite cell interacts directly with the myofibre and its basal lamina, however other cell types present in the muscle tissue in close proximity with satellite cells have been shown to play important roles in regulating satellite cell function through paracrine signalling. These cells include endothelial cells [102], fibro-adipogenic progenitors [103] and macrophages that infiltrate the muscle tissue during injury-induced regeneration [104]. In this section, we summarize some of the major extracellular signalling pathways triggered by the satellite cell niche that control satellite cell fate decisions via mechanisms that potentially involve regulation of Pax7 protein (Fig. 4).
Fig 4.
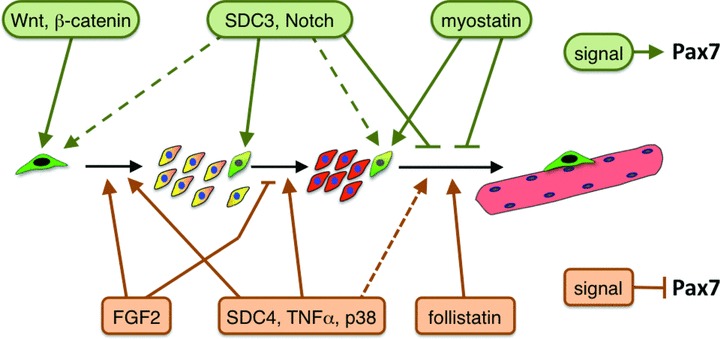
Extracellular regulation of satellite cell fate. Extracellular signalling pathways that participate in the regulation of satellite cell fate and are associated with positive regulation of Pax7 expression/function are depicted in green. Signalling pathways exhibiting negative regulation of Pax7 expression/function are depicted in orange. Molecules that function in the same pathway (e.g. Syndecan-3 and Notch; Wnt and β-catenin) or leading to equivalent downstream effects in muscle progenitors (such as Syndecan-4 and p38), are depicted in common boxes. Signalling pathways that affect satellite cell fate but have no clearly defined effects on Pax7 expression/function are not depicted. Dotted lines indicate effects on satellite cell fate that are currently less defined or controversial. Extracellular signalling can direct the transition from one status to another (black arrows), or can stabilize a specific status (coloured cells). Green cells: uncommitted progenitor (i.e. recently activated and/or quiescent satellite cells); green/yellow cells: self-renewing satellite cells; yellow/orange cells: proliferating, MyoD+ muscle precursors (i.e. adult myoblasts); red cells: myogenin+, differentiating myocytes; pink multi-nucleated cell: myofibre.
Wnt signalling
Wnts are soluble ligands involved in several developmental pathways as well as regulation of a number of stem cell populations. Wnts can signal either through a canonical pathway or through non-canonical pathways [105]. The canonical pathway involves β-catenin dephosphorylation and translocation into the nucleus, where β-catenin binding to Tcf/Lef transcription factors induces Wnt-responsive genes [106]. In contrast, the non-canonical pathways affect cell function independently of β-catenin status [105, 107]. Both canonical and non-canonical Wnt signalling have been reported to regulate embryonic and adult myogenesis [108]. However, is yet to be determined how Wnt signalling affects critical effectors of cell fate specification, such as Pax7 and the MRFs [109-111]. In post-natal and adult myogenesis, Wnt signalling has been reported to induce Pax7 expression and myogenic specification of muscle-derived CD45+ cells [112] and rat bone marrow mesenchymal stem cells [113]. In satellite cells, β-catenin promotes self-renewal and represses satellite cell commitment to myogenesis by inducing Pax7 expression while repressing MyoD expression [114]. Interestingly, Wnt signalling (by means of Wnt7a and Fzd7) has been shown to play a role in asymmetric satellite cell division by promoting symmetric expansion of Pax7+/Myf5− satellite stem cells, therefore assuring maintenance of the Pax+ undifferentiated population throughout life [115]. It is unknown if unequal Pax7 expression during asymmetric cell division is driven mainly by differential transcriptional regulation, changes in protein stability or both.
Notch signalling
Notch is a family of transmembrane receptors involved in several developmental pathways and in adult stem cell fate specification [116, 117]. In embryonic myogenesis, Notch target genes (mainly members of the Hes and Hey family of bHLH transcription factors) have been shown to repress terminal myogenic differentiation by directly binding MyoD and repressing its activity [118]. This could possibly be the mechanism by which Pax7+ muscle progenitors are allowed to expand and support both the generation of a myogenic lineage in the embryo and the generation of satellite cells at later stages of embryonic development [119, 120]. Although a direct role for Notch signalling in controlling Pax7 expression and function has not been demonstrated yet, activation of Notch signalling has been shown to increase Pax7 protein levels and decrease MyoD and myogenin protein levels in adult myoblast and satellite cell-derived cell lines in culture [121], whereas inhibition of Notch signalling leads to down-regulation of Pax7, loss of reserve cells and concomitant enhancement of terminal differentiation [122, 123]. Thus, an attractive possibility considers the Notch signalling pathway as a key controller of the Pax7:MyoD ratio to regulate satellite cell fate, as proposed in Figure 2.
The transforming growth factor-β (TGF-β) superfamily
The TGF-β superfamily comprises soluble ligands, which have been shown to regulate myogenesis, including TGF-β [124-126], BMPs [127], myostatin [133-135] as well as by repressing cell cycle progression [125]. Conversely, it has been shown that expression of TGFβ-type I receptor in rat myotubes is negatively regulated by electrical activity [136], whereas TGF-β bio-availability and signalling is negatively regulated by proteoglycans present in muscle extracellular matrix (such as decorin, biglycan and betaglycan) [137]. Despite regulating myogenesis, no direct effect of TGFβ signalling on Pax7 activity or expression has been reported yet.
Myostatin and follistatin have been shown to play a role in controlling Pax7 expression and/or function, although the molecular mechanisms involved remain largely uncharacterized [17, 130]. It is known that myostatin increases Pax7 levels through an extracellular signal-regulated kinase (ERK), isoforms 1 and 2-dependent pathway and that Pax7 is the ultimate mediator of myostatin-dependent control of terminal differentiation [17]. Pax7 overexpression in myostatin-treated differentiating C2C12 restores the balance between differentiation and self-renewal to non-treated levels [17]. By contrast follistatin, which binds to the same receptors that interact with myostatin, has been shown to promote myoblast terminal differentiation and fusion leading to myotube hypertrophy [131, 132]. Whether the downstream effects of follistatin are entirely dependent on inhibition of myostatin [138] or, at least in part, myostatin independent [139] requires further investigation.
Syndecans
Syndecans are transmembrane heparan sulphate proteoglycans expressed in several tissues where they are involved in mediating autocrine and paracrine signalling, cell–cell adhesion and cell–matrix anchorage [140-142]. Of the four mammalian syndecans, two are expressed in satellite cells, Syndecan-3 (SDC3) and syndecan-4 (SDC4), which play key roles in satellite cell fate specification. In sdc4−/− injured muscles satellite cells fail to activate, to up-regulate MyoD and to enter the cell cycle [143]. Conversely, in sdc3−/− injured muscles activated satellite cells fail to return to quiescence and remain in a transit-amplifying status eventually leading to myofibre hypertrophy, myofibre hyperplasia and partial loss of the undifferentiated Pax7+ pool [144]. Interestingly, injection of Syndecan-3 null C2C12 myoblasts into normal regenerating muscle, fail to fuse into new myofibres [145], highlighting a cell-autonomous requirement for Syndecan-3 function during myoblast differentiation. Syndecan-3 is required for Notch processing and signalling in activated satellite cells as well as for promoting FGF2 signalling [146-148]. Whether Syndecan-3–mediated signalling directly affects Pax7 expression and/or function is yet to be determined. As discuss previously, regulation of Notch and/or FGF signalling through Syndecan-3 may control muscle progenitor cell fate by modulation of Pax7:MyoD protein ratio.
Tumour necrosis factor-α(TNF-α) and p38
The role played by TNF-α in satellite cells has been long debated [149-151]. However, the elegant work produced by Palacio et al. [153] and by Mozzetta et al. [152] show how p38-mediated TNF-α–signalling is transduced into the nucleus of satellite cells and accounts for epigenetic control of Pax7 expression. In activated satellite cells, the Pax7 gene is responsive to p38 signalling due to a bivalent chromatin signature that features the coexistence of H3-K27(3me) and H3-K4(3me) marks, which results in repression of Pax7 expression. In the presence of active p38, H3-K4(3me) is removed resulting in repression of Pax7 expression [152] and cell cycle re-entry [153, 154]. Conversely, p38 blockade promotes Pax7 expression by preventing H3-K27(3me) [152] and satellite cell quiescence [154].
Concluding remarks
In several forms of muscular dystrophy, muscle damage is triggered by genetic defects in proteins present in the dystrophin protein complex [155, 156]. Although the biochemical and mechanical events that lead to continuous myofibre damage in dystrophic muscles are known and well characterized [155, 157], it is not understood why dystrophic muscles fail to regenerate leading to extensive fibrosis and eventually muscle loss. It is believed, however, that loss of muscle regenerative capacity arises from exhaustion of the satellite cell pool [158, 159]. Consistent with this hypothesis, analysis of telomere length in satellite cells suggests that when clinical symptoms appear in Duchenne muscular dystrophy (DMD) patients, satellite cells have already undergone extensive cell proliferation by participating in repeated cycles of regeneration [160, 161]. Recently, telomerase inactivation in the mdx mouse model of DMD has given strong support to this hypothesis [159]. Thus, transplantation of muscle progenitors as been proposed as a viable therapy to remodel dystrophic muscle [20, 162, 163]. However, the success of this approach has been highly variable, mainly due to low homing and survival of implanted cells (up to 98–99% die less than a week after injection) and their minor contribution to re-populate the satellite cell niche. Interestingly, recent studies suggest that myogenic commitment (i.e. MyoD expression) directly correlates with the poor survival of cell transplants [164]. As discussed in this review, Pax7 appears to act as a dual regulator of muscle progenitor cell fate, inducing lineage commitment but stabilizing an undifferentiated state. As part of a regulatory nodal point, understanding the molecular control of Pax7 function and expression represents an attractive target for therapeutic manipulation of muscle progenitors, potentially allowing (i) in vitro expansion without loss of myogenic potential, (ii) enhanced cell survival and engraftment after implantation and (iii) higher contribution of engrafted cells to the self-renewing satellite cell compartment. These could be applicable not only to treat muscle dystrophies but also to recover extensive muscle loss upon acute traumatic injury.
Acknowledgments
This work was supported by Internal Grant from P. Catholic University of Chile (VRAID 20/2009 and 17/2010; to H.O.). H.O. designed manuscript structure; A.P. and H.O. wrote the paper.
Conflict of interest
The authors confirm that there are no conflicts of interest.
References
- 1.Mauro A. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol. 1961;9:493–5. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuang S, Rudnicki MA. The emerging biology of satellite cells and their therapeutic potential. Trends Mol Med. 2008;14:82–91. doi: 10.1016/j.molmed.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Hawke TJ, Garry DJ. Myogenic satellite cells: physiology to molecular biology. J Appl Physiol. 2001;91:534–51. doi: 10.1152/jappl.2001.91.2.534. [DOI] [PubMed] [Google Scholar]
- 4.Schultz E, McCormick KM. Skeletal muscle satellite cells. Rev Physiol Biochem Pharmacol. 1994;123:213–57. doi: 10.1007/BFb0030904. [DOI] [PubMed] [Google Scholar]
- 5.Schultz E, Gibson MC, Champion T. Satellite cells are mitotically quiescent in mature mouse muscle: an EM and radioautographic study. J Exp Zool. 1978;206:451–6. doi: 10.1002/jez.1402060314. [DOI] [PubMed] [Google Scholar]
- 6.Cornelison DD, Wold BJ. Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev Biol. 1997;191:270–83. doi: 10.1006/dbio.1997.8721. [DOI] [PubMed] [Google Scholar]
- 7.Ciciliot S, Schiaffino S. Regeneration of mammalian skeletal muscle. Basic mechanisms and clinical implications. Curr Pharm Des. 2010;16:906–14. doi: 10.2174/138161210790883453. [DOI] [PubMed] [Google Scholar]
- 8.Chargé SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209–38. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 9.Shea KL, Xiang W, Laporta VS, et al. Sprouty1 regulates reversible quiescence of a self-renewing adult muscle stem cell pool during regeneration. Cell Stem Cell. 2010;6:117–29. doi: 10.1016/j.stem.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Angello JC, Hauschka SD. Skeletal muscle satellite cells: timelapse videomicroscopic evidence that renewal is stochastic. BAM. 1996;6:491–502. [Google Scholar]
- 11.Baroffio A, Hamann M, Bernheim L, et al. Identification of self-renewing myoblasts in the progeny of single human muscle satellite cells. Differentiation. 1996;60:47–57. doi: 10.1046/j.1432-0436.1996.6010047.x. [DOI] [PubMed] [Google Scholar]
- 12.Collins CA, Olsen I, Zammit PS, et al. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 13.Olguin HC, Olwin BB. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: a potential mechanism for self-renewal. Dev Biol. 2004;275:375–88. doi: 10.1016/j.ydbio.2004.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schultz E, Jaryszak DL, Gibson MC, et al. Absence of exogenous satellite cell contribution to regeneration of frozen skeletal muscle. J Muscle Res Cell Motil. 1986;7:361–7. doi: 10.1007/BF01753657. [DOI] [PubMed] [Google Scholar]
- 15.Zammit PS, Golding JP, Nagata Y, et al. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J Cell Biol. 2004;166:347–57. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuang S, Kuroda K, Le Grand F, et al. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McFarlane C, Hennebry A, Thomas M, et al. Myostatin signals through Pax7 to regulate satellite cell self-renewal. Exp Cell Res. 2008;314:317–29. doi: 10.1016/j.yexcr.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 18.Sacco A, Doyonnas R, Kraft P, et al. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–6. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rudnicki MA, Le Grand F, McKinnell I, et al. The molecular regulation of muscle stem cell function. Cold Spring Harb Symp Quant Biol. 2008;73:323–31. doi: 10.1101/sqb.2008.73.064. [DOI] [PubMed] [Google Scholar]
- 20.Peault B, Rudnicki M, Torrente Y, et al. Stem and progenitor cells in skeletal muscle development, maintenance, and therapy. Mol Ther. 2007;15:867–77. doi: 10.1038/mt.sj.6300145. [DOI] [PubMed] [Google Scholar]
- 21.Shi X, Garry DJ. Muscle stem cells in development, regeneration, and disease. Genes Dev. 2006;20:1692–708. doi: 10.1101/gad.1419406. [DOI] [PubMed] [Google Scholar]
- 22.Cornelison DD, Filla MS, Stanley HM, et al. Syndecan-3 and syndecan-4 specifically mark skeletal muscle satellite cells and are implicated in satellite cell maintenance and muscle regeneration. Dev Biol. 2001;239:79–94. doi: 10.1006/dbio.2001.0416. [DOI] [PubMed] [Google Scholar]
- 23.Fukada S, Uezumi A, Ikemoto M, et al. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells. 2007;25:2448–59. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- 24.Mousavi K, Jasmin BJ. BDNF is expressed in skeletal muscle satellite cells and inhibits myogenic differentiation. J Neurosci. 2006;26:5739–49. doi: 10.1523/JNEUROSCI.5398-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt K, Glaser G, Wernig A, et al. Sox8 is a specific marker for muscle satellite cells and inhibits myogenesis. J Biol Chem. 2003;278:29769–75. doi: 10.1074/jbc.M301539200. [DOI] [PubMed] [Google Scholar]
- 26.Seale P, Sabourin LA, Girgis-Gabardo A, et al. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–86. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 27.Kuang S, Chargé SB, Seale P, et al. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J Cell Biol. 2006;172:103–13. doi: 10.1083/jcb.200508001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Relaix F, Montarras D, Zaffran S, et al. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J Cell Biol. 2006;172:91–102. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olguin HC, Yang Z, Tapscott SJ, et al. Reciprocal inhibition between Pax7 and muscle regulatory factors modulates myogenic cell fate determination. J Cell Biol. 2007;177:769–79. doi: 10.1083/jcb.200608122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKinnell IW, Ishibashi J, Le Grand F, et al. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat Cell Biol. 2008;10:77–84. doi: 10.1038/ncb1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins CA, Gnocchi VF, White RB, et al. Integrated functions of Pax3 and Pax7 in the regulation of proliferation, cell size and myogenic differentiation. PLoS One. 2009 doi: 10.1371/journal.pone.0004475. ; doi: 10.1371/journal.pone.0004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lepper C, Conway SJ, Fan CM. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460:627–31. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buckingham M, Relaix F. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu Rev Cell Dev Biol. 2007;23:645–73. doi: 10.1146/annurev.cellbio.23.090506.123438. [DOI] [PubMed] [Google Scholar]
- 34.Chi N, Epstein JA. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 2002;18:41–7. doi: 10.1016/s0168-9525(01)02594-x. [DOI] [PubMed] [Google Scholar]
- 35.Robson EJ, He SJ, Eccles MR. A PANorama of PAX genes in cancer and development. Nat Rev Cancer. 2006;6:52–62. doi: 10.1038/nrc1778. [DOI] [PubMed] [Google Scholar]
- 36.Lang D, Powell SK, Plummer RS, et al. PAX genes: roles in development, pathophysiology, and cancer. Biochem Pharmacol. 2007;73:1–14. doi: 10.1016/j.bcp.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Fang WH, Krupinski J, et al. Pax genes in embryogenesis and oncogenesis. J Cell Mol Med. 2008;12:2281–94. doi: 10.1111/j.1582-4934.2008.00427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bober E, Franz T, Arnold HH, et al. Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development. 1994;120:603–12. doi: 10.1242/dev.120.3.603. [DOI] [PubMed] [Google Scholar]
- 39.Goulding M, Lumsden A, Paquette AJ. Regulation of Pax-3 expression in the dermomyotome and its role in muscle development. Development. 1994;120:957–71. doi: 10.1242/dev.120.4.957. [DOI] [PubMed] [Google Scholar]
- 40.Tajbakhsh S, Rocancourt D, Cossu G, et al. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell. 1997;89:127–38. doi: 10.1016/s0092-8674(00)80189-0. [DOI] [PubMed] [Google Scholar]
- 41.Ben-Yair R, Kalcheim C. Lineage analysis of the avian dermomyotome sheet reveals the existence of single cells with both dermal and muscle progenitor fates. Development. 2005;132:689–701. doi: 10.1242/dev.01617. [DOI] [PubMed] [Google Scholar]
- 42.Gros J, Manceau M, Thomé V, et al. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature. 2005;435:954–8. doi: 10.1038/nature03572. [DOI] [PubMed] [Google Scholar]
- 43.Kassar-Duchossoy L, Giacone E, Gayraud-Morel B, et al. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes Dev. 2005;19:1426–31. doi: 10.1101/gad.345505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Relaix F, Rocancourt D, Mansouri A, et al. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–53. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- 45.LaBarge MA, Blau HM. Biological progression from adult bone marrow to mononucleate muscle stem cell to multinucleate muscle fiber in response to injury. Cell. 2002;111:589–601. doi: 10.1016/s0092-8674(02)01078-4. [DOI] [PubMed] [Google Scholar]
- 46.Collins CA, Zammit PS, Ruiz AP, et al. A population of myogenic stem cells that survives skeletal muscle aging. Stem Cells. 2007;25:885–94. doi: 10.1634/stemcells.2006-0372. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Conboy I. Embryonic versus adult myogenesis: challenging the ‘regeneration recapitulates development’ paradigm. J Mol Cell Biol. 2010;2:1–4. doi: 10.1093/jmcb/mjp027. [DOI] [PubMed] [Google Scholar]
- 48.Reimann J, Brimah K, Schröder R, et al. Pax7 distribution in human skeletal muscle biopsies and myogenic tissue cultures. Cell Tissue Res. 2004;315:233–42. doi: 10.1007/s00441-003-0833-y. [DOI] [PubMed] [Google Scholar]
- 49.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 50.Weintraub H, Tapscott SJ, Davis RL, et al. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci USA. 1989;86:5434–8. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zammit PS, Relaix F, Nagata Y, et al. Pax7 and myogenic progression in skeletal muscle satellite cells. J Cell Sci. 2006;119:1824–32. doi: 10.1242/jcs.02908. [DOI] [PubMed] [Google Scholar]
- 52.Kumar D, Shadrach JL, Wagers AJ, et al. Id3 is a direct transcriptional target of Pax7 in quiescent satellite cells. Mol Biol Cell. 2009;20:3170–7. doi: 10.1091/mbc.E08-12-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Epstein JA, Lam P, Jepeal L, et al. Pax3 inhibits myogenic differentiation of cultured myoblast cells. J Biol Chem. 1995;270:11719–22. doi: 10.1074/jbc.270.20.11719. [DOI] [PubMed] [Google Scholar]
- 54.Chen Y, Lin G, Slack JM. Control of muscle regeneration in the Xenopus tadpole tail by Pax7. Development. 2006;133:2303–13. doi: 10.1242/dev.02397. [DOI] [PubMed] [Google Scholar]
- 55.Mitchell KJ, Pannérec A, Cadot B, et al. Identification and characterization of a non-satellite cell muscle resident progenitor during postnatal development. Nat Cell Biol. 2010;12:257–66. doi: 10.1038/ncb2025. [DOI] [PubMed] [Google Scholar]
- 56.Seale P, Ishibashi J, Scimé A, et al. Pax7 is necessary and sufficient for the myogenic specification of CD45+:Sca1+ stem cells from injured muscle. PLoS Biol. 2004;2:664–72. doi: 10.1371/journal.pbio.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Potterf SB, Furumura M, Dunn KJ, et al. Transcription factor hierarchy in Waardenburg syndrome: regulation of MITF expression by SOX10 and PAX3. Hum Genet. 2000;107:1–6. doi: 10.1007/s004390000328. [DOI] [PubMed] [Google Scholar]
- 58.Lang D, Lu MM, Huang L, et al. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature. 2005;433:884–7. doi: 10.1038/nature03292. [DOI] [PubMed] [Google Scholar]
- 59.Barr FG. Gene fusions involving PAX and FOX family members in alveolar rhabdomyosarcoma. Oncogene. 2001;20:5736–46. doi: 10.1038/sj.onc.1204599. [DOI] [PubMed] [Google Scholar]
- 60.Davis RJ, Barr FG. Fusion genes resulting from alternative chromosomal translocations are overexpressed by gene-specific mechanisms in alveolar rhabdomyosarcoma. Proc Natl Acad Sci USA. 1997;94:8047–51. doi: 10.1073/pnas.94.15.8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fredericks WJ, Galili N, Mukhopadhyay S, et al. The PAX3-FKHR fusion protein created by the t(2;13) translocation in alveolar rhabdomyosarcomas is a more potent transcriptional activator than PAX3. Mol Cell Biol. 1995;15:1522–35. doi: 10.1128/mcb.15.3.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bennicelli JL, Fredericks WJ, Wilson RB, et al. Wild type PAX3 protein and the PAX3-FKHR fusion protein of alveolar rhabdomyosarcoma contain potent, structurally distinct transcriptional activation domains. Oncogene. 1995;11:119–30. [PubMed] [Google Scholar]
- 63.Bennicelli JL, Edwards RH, Barr FG. Mechanism for transcriptional gain of function resulting from chromosomal translocation in alveolar rhabdomyosarcoma. Proc Natl Acad Sci USA. 1996;93:5455–9. doi: 10.1073/pnas.93.11.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davicioni E, Finckenstein FG, Shahbazian V, et al. Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res. 2006;66:6936–46. doi: 10.1158/0008-5472.CAN-05-4578. [DOI] [PubMed] [Google Scholar]
- 65.Cao L, Yu Y, Bilke S, et al. Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer. Cancer Res. 2010;70:6497–508. doi: 10.1158/0008-5472.CAN-10-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Graf Finckenstein F, Shahbazian V, Davicioni E, et al. PAX-FKHR function as pangenes by simultaneously inducing and inhibiting myogenesis. Oncogene. 2008;27:2004–14. doi: 10.1038/sj.onc.1210835. [DOI] [PubMed] [Google Scholar]
- 67.Khan J, Bittner ML, Saal LH, et al. cDNA microarrays detect activation of a myogenic transcription program by the PAX3-FKHR fusion oncogene. Proc Natl Acad Sci USA. 1999;96:13264–9. doi: 10.1073/pnas.96.23.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Olguín HC, Patzlaf NE, Olwin BB. PAX7-FKHR transcriptional activity is enhanced by transcriptionally repressed MyoD. J Cell Biochem. 2011;112:1410–17. doi: 10.1002/jcb.23057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim J, Lo L, Dormand E, et al. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron. 2003;38:17–31. doi: 10.1016/s0896-6273(03)00163-6. [DOI] [PubMed] [Google Scholar]
- 70.Nutt SL, Heavey B, Rolink AG, et al. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–62. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 71.Mikkola I, Heavey B, Horcher M, et al. Reversion of B cell commitment upon loss of Pax5 expression. Science. 2002;297:110–13. doi: 10.1126/science.1067518. [DOI] [PubMed] [Google Scholar]
- 72.Marquardt T, Ashery-Padan R, Andrejewski N, et al. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 73.Oron-Karni V, Farhy C, Elgart M, et al. Dual requirement for Pax6 in retinal progenitor cells. Development. 2008;135:4037–47. doi: 10.1242/dev.028308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yin VP, Poss KD. New regulators of vertebrate appendage regeneration. Curr Opin Genet Dev. 2008;18:381–6. doi: 10.1016/j.gde.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tanaka EM. Cell differentiation and cell fate during urodele tail and limb regeneration. Curr Opin Genet Dev. 2003;13:497–501. doi: 10.1016/j.gde.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 76.Stoick-Cooper CL, Moon RT, Weidinger G. Advances in signaling in vertebrate regeneration as a prelude to regenerative medicine. Genes Dev. 2007;21:1292–315. doi: 10.1101/gad.1540507. [DOI] [PubMed] [Google Scholar]
- 77.Gurtner GC, Werner S, Barrandon Y, et al. Wound repair and regeneration. Nature. 2008;453:314–21. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- 78.Bennicelli JL, Advani S, Schäfer BW, et al. PAX3 and PAX7 exhibit conserved cis-acting transcription repression domains and utilize a common gain of function mechanism in alveolar rhabdomyosarcoma. Oncogene. 1999;18:4348–56. doi: 10.1038/sj.onc.1202812. [DOI] [PubMed] [Google Scholar]
- 79.Abu Hatoum O, Gross-Mesilaty S, Breitschopf K, et al. Degradation of myogenic transcription factor MyoD by the ubiquitin pathway in vivo and in vitro: regulation by specific DNA binding. Mol Cell Biol. 1998;18:5670–7. doi: 10.1128/mcb.18.10.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cao Y, Wang C. The COOH-terminal transactivation domain plays a key role in regulating the in vitro and in vivo function of Pax3 homeodomain. J Biol Chem. 2000;275:9854–62. doi: 10.1074/jbc.275.13.9854. [DOI] [PubMed] [Google Scholar]
- 81.Hollenbach AD, Sublett JE, McPherson CJ, et al. The Pax3-FKHR oncoprotein is unresponsive to the Pax3-associated repressor hDaxx. EMBO J. 1999;18:3702–11. doi: 10.1093/emboj/18.13.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Muhr J, Andersson E, Persson M, et al. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell. 2001;104:861–73. doi: 10.1016/s0092-8674(01)00283-5. [DOI] [PubMed] [Google Scholar]
- 83.Eberhard D, Jiménez G, Heavey B, et al. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J. 2000;19:2292–303. doi: 10.1093/emboj/19.10.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cai Y, Brophy PD, Levitan I, et al. Groucho suppresses Pax2 transactivation by inhibition of JNK-mediated phosphorylation. EMBO J. 2003;22:5522–9. doi: 10.1093/emboj/cdg536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sato H, Wang D, Kudo A. Dissociation of Pax-5 from KI and KII sites during kappa-chain gene rearrangement correlates with its association with the underphosphorylated form of retinoblastoma. J Immunol. 2001;166:6704–10. doi: 10.4049/jimmunol.166.11.6704. [DOI] [PubMed] [Google Scholar]
- 86.Nitsch R, Di Palma T, Mascia A, et al. WBP-2, a WW domain binding protein, interacts with the thyroid-specific transcription factor Pax8. Biochem J. 2004;377:553–60. doi: 10.1042/BJ20031233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.White RB, Ziman MR. Genome-wide discovery of Pax7 target genes during development. Physiol Genomics. 2008;33:41–9. doi: 10.1152/physiolgenomics.00256.2007. [DOI] [PubMed] [Google Scholar]
- 88.Cai Y, Lechner MS, Nihalani D, et al. Phosphorylation of Pax2 by the c-Jun N-terminal kinase and enhanced Pax2-dependent transcription activation. J Biol Chem. 2002;277:1217–22. doi: 10.1074/jbc.M109663200. [DOI] [PubMed] [Google Scholar]
- 89.Yan Q, Gong L, Deng M, et al. Sumoylation activates the transcriptional activity of Pax-6, an important transcription factor for eye and brain development. Proc Natl Acad Sci USA. 2010;107:21034–9. doi: 10.1073/pnas.1007866107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boutet SC, Disatnik MH, Chan LS, et al. Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell. 2007;130:349–62. doi: 10.1016/j.cell.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 91.Boutet SC, Biressi S, Iori K, et al. Taf1 regulates Pax3 protein by monoubiquitination in skeletal muscle progenitors. Mol Cell. 2010;40:749–61. doi: 10.1016/j.molcel.2010.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cao X, Kambe F, Lu X, et al. Glutathionylation of two cysteine residues in paired domain regulates DNA binding activity of Pax-8. J Biol Chem. 2005;280:25901–6. doi: 10.1074/jbc.M411443200. [DOI] [PubMed] [Google Scholar]
- 93.Jariel-Encontre I, Bossis G, Piechaczyk M. Ubiquitin-independent degradation of proteins by the proteasome. Biochim Biophys Acta. 2008;1786:153–77. doi: 10.1016/j.bbcan.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 94.Chen JF, Tao Y, Li J, et al. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol. 2010;190:867–79. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dey BK, Gagan J, Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol. 2011;31:203–14. doi: 10.1128/MCB.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lin DI, Barbash O, Kumar KG, et al. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Mol Cell. 2006;24:355–66. doi: 10.1016/j.molcel.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu Y, Hedvat CV, Mao S, et al. The ETS protein MEF is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCFSkp2. Mol Cell Biol. 2006;26:3114–23. doi: 10.1128/MCB.26.8.3114-3123.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mouravlev A, Young D, During MJ. Phosphorylation-dependent degradation of transgenic CREB protein initiated by heterodimerization. Brain Res. 2007;1130:31–7. doi: 10.1016/j.brainres.2006.10.076. [DOI] [PubMed] [Google Scholar]
- 99.Poizat C, Puri PL, Bai Y, et al. Phosphorylation-dependent degradation of p300 by doxorubicin-activated p38 mitogen-activated protein kinase in cardiac cells. Mol Cell Biol. 2005;25:2673–87. doi: 10.1128/MCB.25.7.2673-2687.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Spengler ML, Guo LW, Brattain MG. Phosphorylation mediates Sp1 coupled activities of proteolytic processing, desumoylation and degradation. Cell Cycle. 2008;7:623–30. doi: 10.4161/cc.7.5.5402. [DOI] [PubMed] [Google Scholar]
- 101.Wang Y, Liao M, Hoe N, et al. A role for protein phosphorylation in cytochrome P450 3A4 ubiquitin-dependent proteasomal degradation. J Biol Chem. 2009;284:5671–84. doi: 10.1074/jbc.M806104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Christov C, Chrétien F, Abou-Khalil R, et al. Muscle satellite cells and endothelial cells: close neighbors and privileged partners. Mol Biol Cell. 2007;18:1397–409. doi: 10.1091/mbc.E06-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Joe AW, Yi L, Natarajan A, et al. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol. 2010;12:153–63. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Arnold L, Henry A, Poron F, et al. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med. 2007;204:1057–69. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rao TP, Kühl M. An updated overview on Wnt signaling pathways: a prelude for more. Circ Res. 2010;106:1798–806. doi: 10.1161/CIRCRESAHA.110.219840. [DOI] [PubMed] [Google Scholar]
- 106.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4:68–75. doi: 10.4161/org.4.2.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Snider L, Tapscott SJ. Emerging parallels in the generation and regeneration of skeletal muscle. Cell. 2003;113:811–12. doi: 10.1016/s0092-8674(03)00474-4. [DOI] [PubMed] [Google Scholar]
- 109.Takata H, Terada K, Oka H, et al. Involvement of Wnt4 signaling during myogenic proliferation and differentiation of skeletal muscle. Dev Dyn. 2007;236:2800–7. doi: 10.1002/dvdy.21327. [DOI] [PubMed] [Google Scholar]
- 110.Han XH, Jin YR, Seto M, et al. A WNT/{beta}-catenin signaling activator, R-spondin, plays positive regulatory roles during skeletal myogenesis. J Biol Chem. 2011;286:10649–59. doi: 10.1074/jbc.M110.169391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim CH, Neiswender H, Baik EJ, et al. Beta-catenin interacts with MyoD and regulates its transcription activity. Mol Cell Biol. 2008;28:2941–51. doi: 10.1128/MCB.01682-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Polesskaya A, Seale P, Rudnicki MA. Wnt signaling induces the myogenic specification of resident CD45+ adult stem cells during muscle regeneration. Cell. 2003;113:841–52. doi: 10.1016/s0092-8674(03)00437-9. [DOI] [PubMed] [Google Scholar]
- 113.Shang YC, Wang SH, Xiong F, et al. Wnt3a signaling promotes proliferation, myogenic differentiation, and migration of rat bone marrow mesenchymal stem cells. Acta Pharmacol Sin. 2007;28:1761–74. doi: 10.1111/j.1745-7254.2007.00671.x. [DOI] [PubMed] [Google Scholar]
- 114.Perez-Ruiz A, Ono Y, Gnocchi VF, et al. Beta-Catenin promotes self-renewal of skeletal-muscle satellite cells. J Cell Sci. 2008;121:1373–82. doi: 10.1242/jcs.024885. [DOI] [PubMed] [Google Scholar]
- 115.Le Grand F, Jones AE, Seale V, et al. Wnt7a activates the planar cell polarity pathway to drive the symmetric expansion of satellite stem cells. Cell Stem Cell. 2009;4:535–47. doi: 10.1016/j.stem.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–6. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 117.Chiba S. Notch signaling in stem cell systems. Stem Cells. 2006;24:2437–47. doi: 10.1634/stemcells.2005-0661. [DOI] [PubMed] [Google Scholar]
- 118.Kopan R, Nye JS, Weintraub H. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development. 1994;120:2385–96. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- 119.Vasyutina E, Lenhard DC, Wende H, et al. RBP-J (Rbpsuh) is essential to maintain muscle progenitor cells and to generate satellite cells. Proc Natl Acad Sci USA. 2007;104:4443–8. doi: 10.1073/pnas.0610647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schuster-Gossler K, Cordes R, Gossler A. Premature myogenic differentiation and depletion of progenitor cells cause severe muscle hypotrophy in Delta1 mutants. Proc Natl Acad Sci USA. 2007;104:537–42. doi: 10.1073/pnas.0608281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sun D, Li H, Zolkiewska A. The role of Delta-like 1 shedding in muscle cell self-renewal and differentiation. J Cell Sci. 2008;121:3815–23. doi: 10.1242/jcs.035493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Conboy IM, Rando TA. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- 123.Kitzmann M, Bonnieu A, Duret C, et al. Inhibition of Notch signaling induces myotube hypertrophy by recruiting a subpopulation of reserve cells. J Cell Physiol. 2006;208:538–48. doi: 10.1002/jcp.20688. [DOI] [PubMed] [Google Scholar]
- 124.Furutani Y, Umemoto T, Murakami M, et al. Role of endogenous TGF-β family in myogenic differentiation of C2C12 cells. J Cell Biochem. 2011;112:614–24. doi: 10.1002/jcb.22953. [DOI] [PubMed] [Google Scholar]
- 125.Carlson ME, Hsu M, Conboy IM. Imbalance between pSmad3 and Notch induces CDK inhibitors in old muscle stem cells. Nature. 2008;454:528–32. doi: 10.1038/nature07034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Droguett R, Cabello-Verrugio C, Santander C, et al. TGF-beta receptors, in a Smad-independent manner, are required for terminal skeletal muscle differentiation. Exp Cell Res. 2010;316:2487–503. doi: 10.1016/j.yexcr.2010.04.031. [DOI] [PubMed] [Google Scholar]
- 127.Patterson SE, Bird NC, Devoto SH. BMP regulation of myogenesis in zebrafish. Dev Dyn. 2010;239:806–17. doi: 10.1002/dvdy.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Amthor H, Otto A, Macharia R, et al. Myostatin imposes reversible quiescence on embryonic muscle precursors. Dev Dyn. 2006;235:672–80. doi: 10.1002/dvdy.20680. [DOI] [PubMed] [Google Scholar]
- 129.Siriett V, Salerno MS, Berry C, et al. Antagonism of myostatin enhances muscle regeneration during sarcopenia. Mol Ther. 2007;15:1463–70. doi: 10.1038/sj.mt.6300182. [DOI] [PubMed] [Google Scholar]
- 130.Li X, Nie F, Yin Z, et al. Enhanced hyperplasia in muscles of transgenic zebrafish expressing Follistatin1. Sci China Life Sci. 2011;54:159–65. doi: 10.1007/s11427-010-4121-2. [DOI] [PubMed] [Google Scholar]
- 131.Iezzi S, Di Padova M, Serra C, et al. Deacetylase inhibitors increase muscle cell size by promoting myoblast recruitment and fusion through induction of follistatin. Dev Cell. 2004;6:673–84. doi: 10.1016/s1534-5807(04)00107-8. [DOI] [PubMed] [Google Scholar]
- 132.Pisconti A, Brunelli S, Di Padova M, et al. Follistatin induction by nitric oxide through cyclic GMP: a tightly regulated signaling pathway that controls myoblast fusion. J Cell Biol. 2006;172:233–44. doi: 10.1083/jcb.200507083. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 133.Vaidya TB, Rhodes SJ, Taparowski EJ, et al. Fibroblast growth factor and transforming growth factor fl repress transcription of the myogenic regulatory gene MyoD1. Mol Cell Biol. 1989;9:3576–9. doi: 10.1128/mcb.9.8.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Martin JF, Li L, Olson EN. Repression of myogenin function by TGF-beta 1 is targeted at the basic helix-loop-helix motif and is independent of E2A products. J Biol Chem. 1992;267:10956–60. [PubMed] [Google Scholar]
- 135.Li X, McFarland DC, Velleman SG. Transforming growth factor-beta1-induced satellite cell apoptosis in chickens is associated with beta1 integrin-mediated focal adhesion kinase activation. Poult Sci. 2009;88:1725–34. doi: 10.3382/ps.2008-00534. [DOI] [PubMed] [Google Scholar]
- 136.Ugarte G, Brandan E. Transforming growth factor beta (TGF-beta) signaling is regulated by electrical activity in skeletal muscle cells. TGF-beta type I receptor is transcriptionally regulated by myotube excitability. J Biol Chem. 2006;281:18473–81. doi: 10.1074/jbc.M600918200. [DOI] [PubMed] [Google Scholar]
- 137.Droguett R, Cabello-Verrugio C, Riquelme C, et al. Extracellular proteoglycans modify TGF-beta bio-availability attenuating its signaling during skeletal muscle differentiation. Matrix Biol. 2006;25:332–41. doi: 10.1016/j.matbio.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 138.Amthor H, Nicholas G, McKinnell I, et al. Follistatin complexes Myostatin and antagonises Myostatin-mediated inhibition of myogenesis. Dev Biol. 2004;270:19–30. doi: 10.1016/j.ydbio.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 139.Lee SJ, Lee YS, Zimmers TA, et al. Regulation of muscle mass by follistatin and activins. Mol Endocrinol. 2010;24:1998–2008. doi: 10.1210/me.2010-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tkachenko E, Rhodes JM, Simons M. Syndecans: new kids on the signaling block. Circ Res. 2005;96:488–500. doi: 10.1161/01.RES.0000159708.71142.c8. [DOI] [PubMed] [Google Scholar]
- 141.Couchman JR. Syndecans: proteoglycan regulators of cell-surface microdomains? Nat Rev Mol Cell Biol. 2003;4:926–37. doi: 10.1038/nrm1257. [DOI] [PubMed] [Google Scholar]
- 142.Rapraeger AC. Syndecan-regulated receptor signaling. J Cell Biol. 2000;149:995–8. doi: 10.1083/jcb.149.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cornelison DD, Wilcox-Adelman SA, Goetinck PF, et al. Essential and separable roles for Syndecan-3 and Syndecan-4 in skeletal muscle development and regeneration. Genes Dev. 2004;18:2231–6. doi: 10.1101/gad.1214204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pisconti A, Cornelison DD, Olguín HC, et al. Syndecan-3 and Notch cooperate in regulating adult myogenesis. J Cell Biol. 2010;190:427–41. doi: 10.1083/jcb.201003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Casar JC, Cabello-Verrugio C, Olguin H, et al. Heparan sulfate proteoglycans are increased during skeletal muscle regeneration: requirement of Syndecan-3 for successful fiber formation. J Cell Sci. 2004;117:73–84. doi: 10.1242/jcs.00828. [DOI] [PubMed] [Google Scholar]
- 146.Jones NC, Fedorov YV, Rosenthal RS, et al. ERK1/2 is required for myoblast proliferation but is dispensable for muscle gene expression and cell fusion. J Cell Physiol. 2001;186:104–15. doi: 10.1002/1097-4652(200101)186:1<104::AID-JCP1015>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 147.Fuentealba L, Carey DJ, Brandan E. Antisense inhibition of Syndecan-3 expression during skeletal muscle differentiation accelerates myogenesis through a basic fibroblast growth factor-dependent mechanism. J Biol Chem. 1999;274:37876–84. doi: 10.1074/jbc.274.53.37876. [DOI] [PubMed] [Google Scholar]
- 148.Olwin BB, Hauschka SD. Identification of the fibroblast growth factor receptor of Swiss 3T3 cells and mouse skeletal muscle myoblasts. Biochemistry. 1986;25:3487–92. doi: 10.1021/bi00360a001. [DOI] [PubMed] [Google Scholar]
- 149.Acharyya S, Sharma SM, Cheng AS, et al. TNF inhibits Notch-1 in skeletal muscle cells by Ezh2 and DNA methylation mediated repression: implications in Duchenne muscular dystrophy. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012479. : e12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Valerio A, Cardile A, Cozzi V, et al. TNF-alpha downregulates eNOS expression and mitochondrial biogenesis in fat and muscle of obese rodents. J Clin Invest. 2006;116:2791–8. doi: 10.1172/JCI28570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Li YP. TNF-alpha is a mitogen in skeletal muscle. Am J Physiol Cell Physiol. 2003;285 doi: 10.1152/ajpcell.00453.2002. : C370–6. [DOI] [PubMed] [Google Scholar]
- 152.Mozzetta C, Consalvi S, Saccone V, et al. Selective control of Pax7 expression by TNF-activated p38α/polycomb repressive complex 2 (PRC2) signaling during muscle satellite cell differentiation. Cell Cycle. 2011;10:191–8. doi: 10.4161/cc.10.2.14441. [DOI] [PubMed] [Google Scholar]
- 153.Palacios D, Mozzetta C, Consalvi S, et al. TNF/p38α/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell. 2010;7:455–69. doi: 10.1016/j.stem.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jones NC, Tyner KJ, Nibarger L, et al. The p38alpha/beta MAPK functions as a molecular switch to activate the quiescent satellite cell. J Cell Biol. 2005;169:105–16. doi: 10.1083/jcb.200408066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Cohn RD, Campbell KP. Molecular basis of muscular dystrophies. Muscle Nerve. 2000;23:1456–71. doi: 10.1002/1097-4598(200010)23:10<1456::aid-mus2>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 156.Emery AE. The muscular dystrophies. Lancet. 2002;359:687–95. doi: 10.1016/S0140-6736(02)07815-7. [DOI] [PubMed] [Google Scholar]
- 157.Rüegg MA, Glass DJ. Molecular mechanisms and treatment options for muscle wasting diseases. Annu Rev Pharmacol Toxicol. 2011;51:373–95. doi: 10.1146/annurev-pharmtox-010510-100537. [DOI] [PubMed] [Google Scholar]
- 158.Scimè A, Rudnicki MA. Anabolic potential and regulation of the skeletal muscle satellite cell populations. Curr Opin Clin Nutr Metab Care. 2006;9:214–19. doi: 10.1097/01.mco.0000222102.21385.7d. [DOI] [PubMed] [Google Scholar]
- 159.Sacco A, Mourkioti F, Tran R, et al. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell. 2010;143:1059–71. doi: 10.1016/j.cell.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Decary S, Hamida CB, Mouly V, et al. Shorter telomeres in dystrophic muscle consistent with extensive regeneration in young children. Neuromuscul Disord. 2000;10:113–20. doi: 10.1016/s0960-8966(99)00093-0. [DOI] [PubMed] [Google Scholar]
- 161.Decary S, Mouly V, Hamida CB, et al. Replicative potential and telomere length in human skeletal muscle: implications for satellite cell-mediated gene therapy. Hum Gene Ther. 1997;8:1429–38. doi: 10.1089/hum.1997.8.12-1429. [DOI] [PubMed] [Google Scholar]
- 162.Cossu G, Mavilio F. Myogenic stem cells for the therapy of primary myopathies: wishful thinking or therapeutic perspective. J Clin Invest. 2000;105:1669–74. doi: 10.1172/JCI10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kapsa R, Kornberg AJ, Byrne E. Novel therapies for Duchenne muscular dystrophy. Lancet Neurol. 2003;2:299–310. doi: 10.1016/s1474-4422(03)00382-x. [DOI] [PubMed] [Google Scholar]
- 164.Asakura A, Hirai H, Kablar B, et al. Increased survival of muscle stem cells lacking the MyoD gene after transplantation into regenerating skeletal muscle. Proc Natl Acad Sci USA. 2007;104:16552–7. doi: 10.1073/pnas.0708145104. [DOI] [PMC free article] [PubMed] [Google Scholar]


