Summary
Bile acid (BA) biosynthesis is tightly controlled by intrahepatic negative feedback signaling elicited by BA binding to farnesoid X receptor (FXR), and also by enterohepatic communication involving ileal BA reabsorption and FGF15/19 secretion. However, how these pathways are coordinated is poorly understood. We show here that non-receptor tyrosine phosphatase Shp2 is a critical player that couples and regulates the intrahepatic and enterohepatic signals for repression of BA synthesis. Ablating Shp2 in hepatocytes suppressed signal relay from FGFR4, receptor for FGF15/19, and attenuated BA activation of FXR signaling, resulting in elevation of systemic BA levels and chronic hepatobiliary disorders in mice. Acting immediately downstream of FGFR4, Shp2 associates with FRS2α and promotes the receptor activation and signal relay to several pathways. These results elucidate a molecular mechanism for the control of BA homeostasis by Shp2 through orchestration of multiple signals in hepatocytes.
Introduction
The biosynthesis of bile acids (BAs) in hepatocytes is a primary pathway for cholesterol catabolism and removal of excess cholesterol via fecal disposal (Chiang, 2002; de Aguiar Vallim et al., 2013; Russell, 2003; Thomas et al., 2008). When secreted into duodenum postprandially, BAs act as “physiological detergent” to emulsify food lipids and facilitate their absorption by intestine. Recently, BAs are also viewed as signaling molecules in several metabolic processes (Houten et al., 2006; Vallim and Edwards, 2009).
Because of its toxicity in excess amounts, BA synthesis is tightly controlled by a negative feedback mechanism. BAs bind farnesoid X receptor (FXR) in hepatocytes (Makishima et al., 1999; Parks et al., 1999), and transactivate small heterodimer partner (SHP) to repress the expression of Cyp7a1 that encodes cholesterol 7a-hydroxylase, the rate-limiting enzyme for BA synthesis (Lu et al., 2000). FXR knockout (KO) mice displayed increased BA levels, higher plasma cholesterol, phospholipids and triglycerides, and were more susceptible to cholesterol-induced hepatic steatosis (Anakk et al., 2011; Sinal et al., 2000). However, SHP deletion rendered only a mild increase of the BA pool size in mice (Kerr et al., 2002), and SHP KO mice were protected from liver damage induced by cholesterol and BA diet (Wang et al., 2003). These observations suggest SHP-independent pathways in control of Cyp7a1 expression. Consistently, FXR and SHP double knockout (DKO) mice displayed early onset cholestasis, more severe liver damage and higher BA synthesis than mice with loss of either gene alone (Anakk et al., 2011).
Ileum is the major site for BA reabsorption in the intestine (Baker and Searle, 1960; Buchwald and Gebhard, 1968; Thomas et al., 2008). BA/FXR signaling induces intestinal production of FGF15 (FGF19 in humans), which also inhibits Cyp7a1 expression in hepatocytes by activating FGFR4 signaling (Fon Tacer et al., 2010; Inagaki et al., 2005). Selective FXR deletion or transgenic expression of an activated FXR in the intestine abolished or enhanced ileal FGF15 expression (Modica et al., 2012; Stroeve et al., 2010). Recently, Diet1 was shown to be required for FGF15/19 expression in enterocytes (Vergnes et al., 2013). Gut microbiota, which metabolize primary BAs into secondary BAs, also regulate intestinal FGF15 production in an FXR-dependent manner (Sayin et al., 2013). Both FGF15 and FGFR4 KO mice exhibited elevated BA levels and enhanced Cyp7a1 expression (Inagaki et al., 2005; Yu et al., 2000). Further, FXR agonist feeding failed to inhibit Cyp7a1 expression in FGFR4 or FGF15 KO mice (Inagaki et al., 2005; Kong et al., 2012), suggesting FGFR4 signaling is necessary for FXR-mediated repression of BA biosynthesis. Experimental data also showed that SHP was required for repression of Cyp7a1 by exogenous FGF15/19 (Inagaki et al., 2005; Kir et al., 2012). SHP suppresses Cyp7a1 expression via interaction with HNF4α and LRH-1 on Cyp7a1 promoter (Kir et al., 2012), both of which are regulators of Cyp7a1 transcription (Inoue et al., 2006; Lu et al., 2000). Despite the dependence on SHP for transcriptional suppression of Cyp7a1 by FGF19, no altered affinity to Cyp7a1 promoter was detected for HNF4α, LRH-1 and SHP after FGF19 treatment (Kir et al., 2012). How activated FGFR4 signaling impacts on BA biosynthesis remains elusive.
Shp2 is a non-receptor tyrosine phosphatase with two Src-homology 2 domains, which promotes signaling through the Ras-Erk pathway (Chan and Feng, 2007; Neel et al., 2003). Mice with Shp2/Ptpn11 ablated in hepatocytes (Shp2hep−/−) displayed impaired hepatocyte proliferation and liver regeneration after partial hepatectomy (Bard-Chapeau et al., 2006). Shp2hep−/− animals suffered chronic hepatic injury and inflammation, and were more susceptible to carcinogen-induced liver tumorigenesis (Bard-Chapeau et al., 2011). Here, we show that Shp2 loss in hepatocytes disrupts BA homeostasis and causes hepatobiliary damage. Our results identify Shp2 as a crucial factor that orchestrates the FGF15/19-FGFR4 and BA-FXR signaling pathways for control of BA biosynthesis.
Results
Early Onset Hepatobiliary Defects in Mice Deficient for Shp2 in Hepatocytes
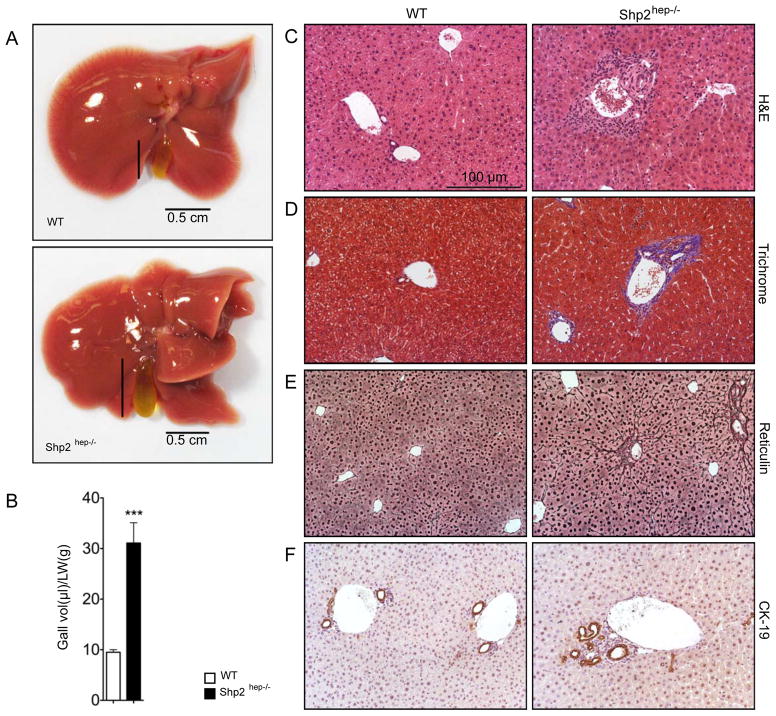
In previous experiments, we generated a mouse line (Shp2hep−/−, Albumin-Cre +:Shp2fl/fl) with Shp2 deleted in hepatocytes (Bard-Chapeau et al., 2011; Bard-Chapeau et al., 2006). We observed hepatic necrosis, inflammatory infiltration and peri-portal fibrosis, dented lobe edges and significantly enlarged gallbladders in Shp2hep−/− mice at age of 2 months (Figures 1A–1D). These hepatic disorders are similar to those in rats fed with sodium cholate, a bile acid detergent (Jeong et al., 2005) or mice after bile duct ligation (BDL, Figure S1) (Georgiev et al., 2008). Liver sections from Shp2hep−/− mice displayed evident biliary fibrosis around portal triad, with positive collagen staining (blue) around bile duct (Figure 1D). Stronger reticulin fiber staining (Figure 1E) also indicates hepatic damage in Shp2hep−/− mice. Around the portal triad, sporadic ductal cell proliferation was consistently observed in Shp2hep−/− livers, as revealed by cytokeratin-19 (CK-19) staining (Figure 1F). Together, these results demonstrate that ablating Shp2 in hepatocytes induces multiple hepatobiliary defects.
Figure 1. Hepatobilliary defects in Shp2hep−/− mice.
(A) Macroscopic view of the whole livers from two-month-old WT (Alb-cre−:Shp2fl/fl) and Shp2hep−/− (Alb-cre+:Shp2fl/fl) mice.
(B) Gallbladder volumes were adjusted by liver weight from WT and Shp2hep−/− mice (n = 6–7). Data is shown as mean ± s.e.m. *** p < 0.001.
(C–F) Liver sections were stained with H&E (C), Manson’s Trichrome (D), and Reticulin (E) and CK-19 (F).
Scale bars in (D, E and F) were same as in (C).
Shp2hep−/− mice Are More Susceptible to BDL
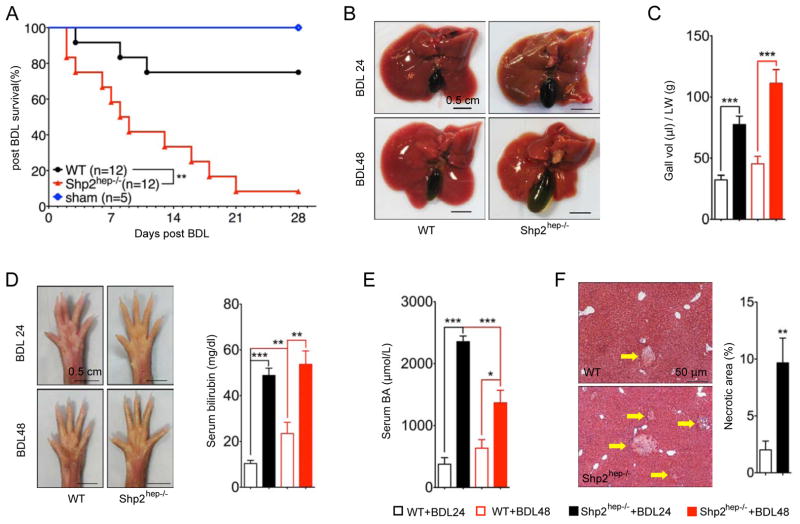
The spontaneous hepatobiliary defects strongly suggest biliary dysfunction in Shp2hep−/− mice. To this end, we performed a BDL experiment, a well-characterized cholestasis model (Georgiev et al., 2008). Strikingly, almost all Shp2hep−/− mice (11/12) died within 4 weeks after BDL, while 75% of WT animals survived the experiment (Figure 2A). Shp2hep−/− mice displayed larger gallbladders 24 and 48 hrs after surgery (Figures 2B and 2C), and more severe jaundice, with darker yellowish color seen on the palms (Figure 2D). Consistently, higher serum bilirubin and BA levels were detected in Shp2hep−/− than WT mice at these time points (Figures 2D and 2E). However, Shp2hep−/− mice also exhibited decreasing serum BA levels from 24 to 48 hrs after BDL (Figure 2E), and BDL induced larger areas of infarction in Shp2hep−/− mice as examined at 24 hrs (Figure 2F). The more extensive necrosis and deteriorating liver function may explain the higher mortality and the drop in serum BA levels in Shp2hep−/− mice. Thus, the Shp2hep−/− mice were more vulnerable than WT controls to biliary obstruction, characterized by higher mortality rate, more severe liver damage and jaundice.
Figure 2. Severe hepatobiliary damages in Shp2hep−/− mice following bile duct ligation.
(A) Kaplan-Meier survival analysis of WT and Shp2hep−/− mice after BDL. ** p = 0.0014, as determined by Log-rank (Mantel-Cox) Test.
(B) Macroscopic views of WT and Shp2hep−/− livers were taken 24 and 48 hrs after BDL.
(C) Gallbladder volumes were adjusted to liver weight after BDL (n = 4–10).
(D) Macroscopic view of palms was shown 24 and 48 hrs after BDL. Serum bilirubin levels were measured (n = 6–12).
(E) Serum BA levels were measured after BDL (n = 6–12)
(F) Liver sections were stained with H&E (left), and statistical analysis (n = 5–7) of necrotic areas (right).
Data in (E, D, E and F) are shown as the mean ± s.e.m. ** p < 0.01 and *** p < 0.001, as determined by Student’s t test.
Shp2 Deficiency in Hepatocytes Led to Increase of Systemic BA Levels
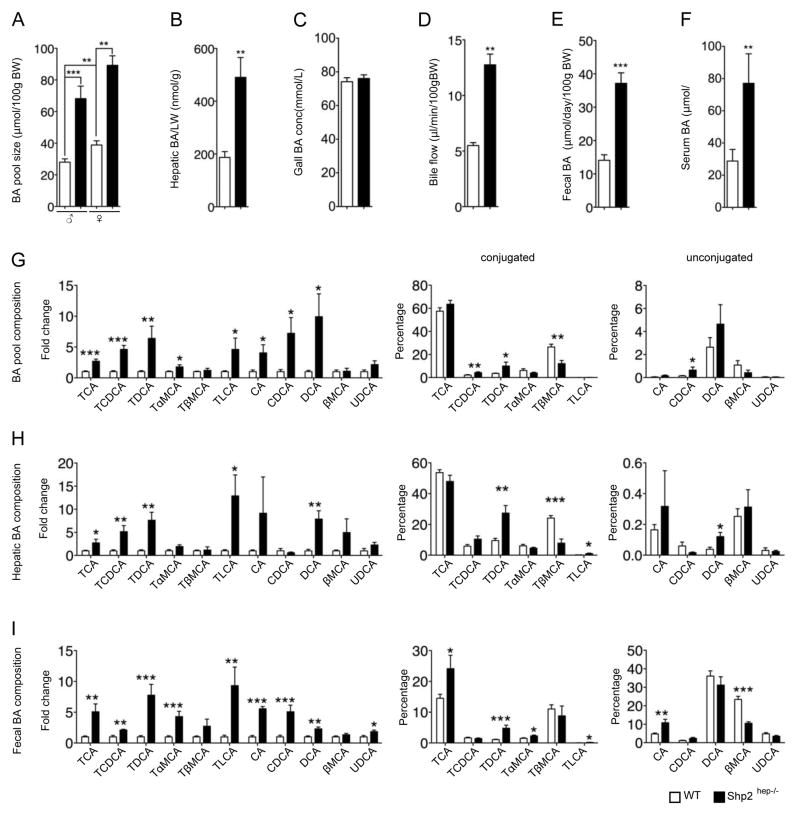
We then measured BA levels in different ways. Consistent with the literature (Rao et al., 2008), WT female mice exhibited larger BA pool sizes than male (Figure 3A). The BA pool size increased significantly in both male and female Shp2hep−/− mice compared to controls (Figure 3A). Since an increase in BA pool size could be due to obstruction of bile flow or BA overproduction, we measured BA levels in serum, liver, gallbladder and feces, as well as bile flow rate. Both hepatic and serum BA levels were elevated in Shp2hep−/− mice, compared to controls (Figures 3B and 3F). Although the gallbladder BA concentrations were similar (Figure 3C), the total BA amounts in gallbladder were significantly elevated in Shp2hep−/− mice, due to the larger size (Figures 1A and 1B). The bile flow rate in Shp2hep−/− mice increased significantly (Figure 3D), ruling out intrahepatic biliary obstruction. With similar daily excretion of feces excretion weight (Figure S2A), the daily fecal BA excretion was significantly higher in Shp2hep−/− than control animals (Figure 3E). All these results indicate elevation of systemic BA levels in Shp2hep−/− mice, which was evidently not caused by biliary hindrance.
Figure 3. Elevation of systemic BA levels in Shp2hep−/− mice.
(A) BA pool (liver, gallbladder and intestine) sizes were measured in both genders of the two genotypes (n = 6–10).
(B–E) BA levels in liver (B), gallbladder (C), feces (D), serum (E) were measured (n = 5–11). All data were collected in males, hepatic BA concentration was adjusted to every gram liver weight, and fecal BA excretion was adjusted to 100 g body weight/day.
(F) Bile flow rate was adjusted to 100 g body weight/min (n = 3).
(G–I) BA composition in BA pool (n=6–9), liver (n=6–7) and feces (n=6–9) was analyzed by liquid chromatography/mass spectrometry. The fold changes of BA species in Shp2hep−/− mice were calibrated to WT (the average value was designated as 1, left panels in G–I). The percentile representations of each conjugated and unconjugated BA species are shown in two panels separately in the right.
Data in (A–I) are shown as the mean ± s.e.m. * p<0.05, ** p < 0.01 and *** p < 0.001, as determined by Student’s t test.
Since different BA species may act as either FXR agonists or antagonists (Makishima et al., 1999; Parks et al., 1999; Sayin et al., 2013), we analyzed BA compositions in BA pool, liver and feces. Shp2hep−/− mice exhibited significant increase in relative fold (Figures 3G–3I, left panels) or absolute amounts (Figure S2B) for almost all BA species. The majority of BAs were conjugated in BA pool and liver (Figures 3G and 3H, middle and left panels), while most fecal BAs were unconjugated (Figure 3I, middle and left panels). The general representation of each species was similar in the BA pool and liver (Figures 3G and 3H, middle and left panels). Of note, the amount of FXR antagonist species tauro-β-muricholic acid (TβMCA) was unchanged in Shp2hep−/− liver, with decrease in its representation in BA pools and feces (Figures 3G–3I). Further, the FXR agonist species, such as tauro-chenodeoxycholic acid (TCDCA), taurodeoxycholic acid (TDCA), taurolithocholic acid (TLCA) and taurocholic acid (TCA), increased significantly in the liver, and TLCA and TDCA even showed an increased representation in hepatic BA composition (Figure 3H).
BA Sequestration Ameliorates Hepatobiliary Defects in Shp2hep−/− Mice
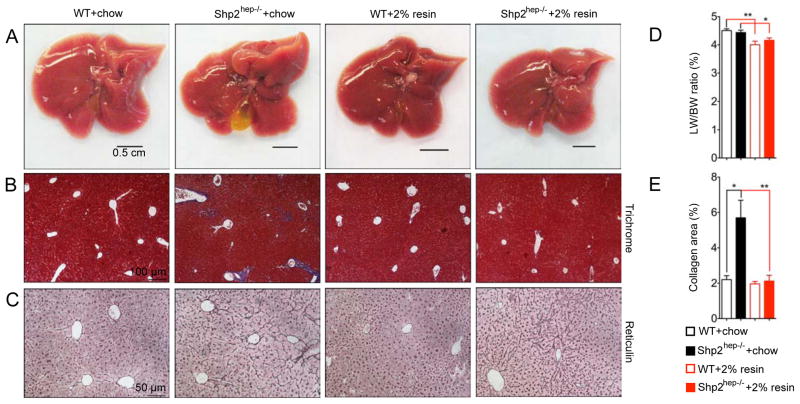
Next, we asked whether the excess BAs are responsible for the hepatobiliary defects in Shp2hep−/− mice. We fed the mice with chow diet supplemented with 2% cholestyramine from weaning to 2-month. Cholestyramine is a BA sequestrant that binds BAs to prevent its ileal reabsorption and to increase its fecal discharge, and therefore it lowers BA pool size in mice (Huang et al., 2006; Kong et al., 2012). The hepatobiliary defects including enlarged gallbladder and dented edges were greatly improved in Shp2hep−/− mice treated with cholestyramine (Figure 4A). Trichrome staining showed significant decrease of portal fibrosis in Shp2hep−/− livers, down to the WT level (Figures 4B and 4E), with no obvious difference in reticulum staining (Figure 4C). Cholestyramine treatment reduced the liver/body weight ratios in WT and Shp2hep−/− mice (Figure 4D). Thus, these results demonstrated that the hepatobiliary defects in Shp2hep−/− mice are at least in part due to the excess BAs.
Figure 4. Lowering BA levels in Shp2hep−/− mice alleviates hepatobilliary defects.
(A) Macroscopic views of WT and Shp2hep−/− livers fed with chow without or with 2% cholestyramine from age of 3 weeks to 2 months.
(B) Liver sections were stained with Manson’s Trichrome.
(C) Liver sections were stained with reticulin.
(D) The ratios of liver/body weight were determined for each group (n = 5–8).
(E) Collagen areas (blue) were measured from images in (B) (n = 4–7).
BA Biosynthesis Is Dramatically Increased in Shp2hep−/− Liver
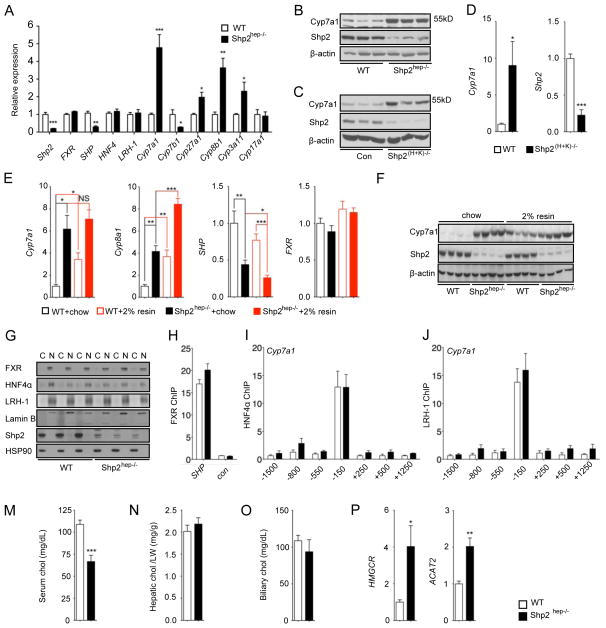
Without biliary obstruction, the augmented fecal BA excretion indicated higher hepatic BA synthesis rate. Indeed, qRT-PCR analysis revealed increased expression of key genes involved in both classical and alternative BA synthetic pathways (Chiang, 2002; de Aguiar Vallim et al., 2013), including Cyp7a1, Cyp8b1 and Cyp27a1, in Shp2hep−/− livers (Figure 5A). The increased Cyp8b1 expression also explained the elevated TDCA levels in BA composition in Shp2hep−/− mice (Figures 3G–I). Expression of the BA-intoxication gene Cyp3a11 was also increased in Shp2hep−/− livers (Figure 5A), likely due to increased hepatic BA levels. Similar to mRNA expression, elevated Cyp7a1 protein levels were detected in Shp2hep−/− livers (Figures 5B, S3A and S3B). To determine if the elevated Cyp7a1 expression was caused by Shp2 ablation directly, we used another mouse model Mx1-Cre+:Shp2fl/fl (referred as Shp(H+K−/−) hereafter), in which Shp2 is acutely deleted in hepatocytes and non-parenchymal cells in adult mice following injection of polyinosinic:polycytidylic acid (poly-I:C) (Zhu et al., 2011). Consistently, acute removal of Shp2 also led to enhanced Cyp7a1 expression significantly at both mRNA and protein levels (Figures 5C, 5D and S3C). Therefore, the deregulated BA biosynthesis is a direct effect of Shp2 loss in hepatocytes. Consistent with previous reports (Huang et al., 2006; Kong et al., 2012), we found that the expression of both Cyp7a1 and Cyp8b1 markedly increased in WT mice after cholestyramine treatment (Figure 5E). Notably, cholestyramine feeding did not further increase Cyp7a1 expression in Shp2hep−/− livers, albeit with an enhancing effect on Cyp8b1 (Figure 5E). Immunoblot analysis confirmed the qRT-PCR result on Cyp7a1 expression in WT and Shp2hep−/− livers (Figures 5F and S3D). Cholestyramine treatment induced modest reduction of SHP expression in WT, and further decreased SHP expression in Shp2hep−/− livers (Figure 5E).
Figure 5. BA synthesis-related genes are significantly up-regulated in Shp2hep−/− liver.
(A) The expression of genes as indicated was determined by qRT-PCR in 2-month-old WT or Shp2hep−/− livers (n = 4–5).
(B) Cyp7a1, Shp2 and β-actin protein levels were determined by immunoblotting of liver lysates from WT and Shp2hep−/− mice. Each lane represents each mouse.
(C) Cyp7a1, Shp2 and β-actin protein levels were determined by immunoblotting of liver lysates from Control and Shp2(H+K)−/− mice. Each lane represents each mouse.
(D) Relative expression of Shp2 and Cyp7a1 was measured by qRT-PCR in liver extracts of WT and Shp2(H+K)−/− mice following poly-I:C injection (n = 5).
(E) Hepatic expression of Cyp7a1, Cyp8b1,SHP and FXR mRNAs was determined by qRT-PCR in mice fed with chow without or with 2% cholestyramine from 3 weeks to 2 months. (n = 4–8).
(F) Cyp7a1, Shp2 and β-actin protein levels were determined by immunoblotting of liver lysates as in (F). Each lane represents each mouse.
(G) Cytoplasmic (C) and nuclear (N) fractions were prepared from freshly-isolated liver samples. FXR, HNF4α, LRH-1, Lamin B (nuclear marker), Shp2 and Hsp90 (cytoplasmic marker) protein levels were determined by immunoblot analysis. Each pair of (C) and (N) samples was prepared from the same mouse.
(H) Chromatin Immunoprecipitation (ChIP) was performed with liver samples (n=3) using FXR antibody. qPCR was performed with FXR binding region on SHP promoter (SHP) and coding region (con). Data are shown as fold enrichment.
(I–J) ChIP assay was performed with HNF4α or LRH-1 antibodies, and different DNA sequences in Cyp7a1 promoter and proximal regions (n=4). Data are shown as fold enrichment.
(M–O) Cholesterol (chol) levels of serum (h), liver (i) and gallbladder (j) were measured. Hepatic cholesterol was adjusted to mg/liver weight (g).
(P) Hepatic expression of HMGCR and ACAT2 mRNA was determined by qRT-PCR (n = 4–5).
All PCR data was normalized against β-actin, and fold change was calibrated to WT group. Data (A, D, F, H, I, J and K) are shown as the means ± s.e.m. * p < 0.05, ** p < 0.01 and *** p < 0.001, as determined by Student’s t test.
The basal SHP expression was significantly down-regulated in Shp2hep−/− livers (Figures 5A and 5E). Given the elevated FXR agonist BA species and unchanged antagonist TβMCA in Shp2hep−/− livers, decreased SHP expression suggests defective FXR signaling. To address this, we first examined FXR protein expression and subcellular distribution. Cytoplasmic and nuclear fractions were prepared, and immunoblotting showed clean separation of the two fractions using HSP90 as the cytoplasmic marker and lamin B1 for nucleus (Figure 5G). Deletion of Shp2 did not alter FXR protein level or its nuclear localization (Figure 5G). Of note, Shp2 was almost exclusively located in the cytoplasm (Figure 5G), and we failed to detect physical association of Shp2 with FXR even with over-expressed tagged-FXR by co-immunoprecipitation (data not shown). By chromatin-immunoprecipitation (ChIP) assay, we found that binding of FXR to the SHP promoter was unchanged in Shp2hep−/− livers (Figure 5H).
To further determine the FXR activation status, we fed both WT and Shp2hep−/− mice with synthetic FXR agonist GW4064 by oral gavage. Hepatic SHP expression was significantly induced in WT mice, but no SHP induction was observed in Shp2hep−/− mice (Figure S3E). However, GW4064 induced SHP expression in the ileums of both WT and Shp2hep−/− mice, where Shp2 expression was intact (Figure S3E). This result strongly suggests defective FXR activation in Shp2-deficient hepatocytes. We also examined HNF4α abd LRH-1, two nuclear receptors that bind and activate Cyp7a1 promoter (Kir et al., 2012; Lu et al., 2000). Both mRNA and protein levels of HNF4α and LRH-1 remained unchanged in Shp2hep−/− liver (Figures 5A and 5G). Similar binding of HNF4α anf LRH-1 to Cyp7a1 promoter was detected by ChIP in WT and Shp2hep−/− livers (Figures 5I and 5J).
To interrogate if the BA overproduction is fueled by excess cholesterol in mutant mice, we measured cholesterol levels in serum, liver and gallbladder. Serum cholesterol was even lower in Shp2hep−/− than WT mice (Figure 5M), while the cholesterol concentrations in liver (Figure 5N) and gallbladder (Figure 5O) were similar. Of note, the expression of cholesterol synthesis-related genes, such as HMGCR and ACAT2, was enhanced in Shp2hep−/− livers (Figure 5P). Thus, aberrantly increased BA synthesis in Shp2hep−/− livers lowered circulating cholesterol levels, resulting in compensatory increase of hepatic cholesterol synthesis.
Shp2 Mediates both FGF15/19 and BA Signals to Suppress BA Synthesis
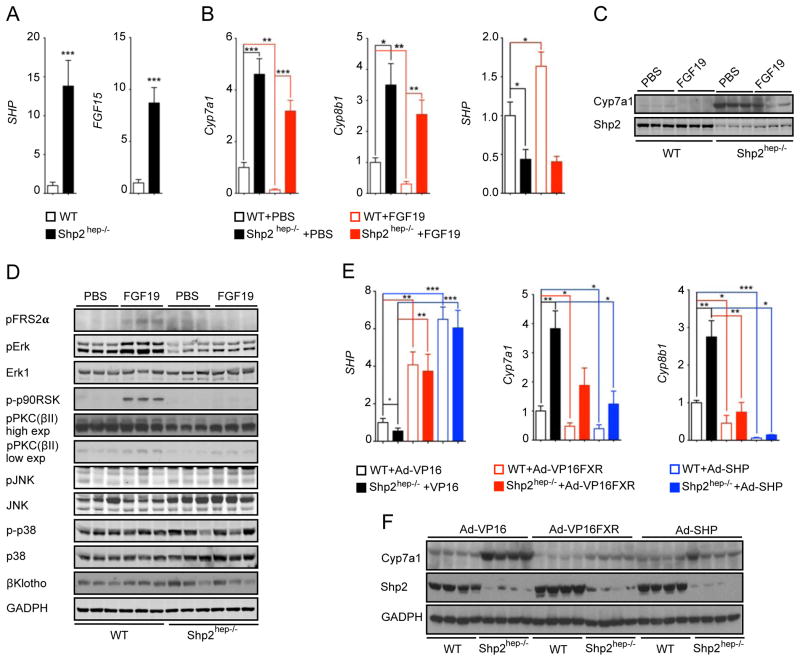
Given the elevated bile flow and enhanced fecal excretion of BAs in Shp2hep−/− animals (Figure 3), we measured ileal expression of FGF15 and SHP, both of which are induced by ileal BA-FXR signaling (Kong et al., 2012; Modica et al., 2012; Stroeve et al., 2010). The mRNA levels of FGF15 and SHP were markedly increased in the ileum of mutant mice (Figure 6A), where Shp2 expression was normal (Figure S4A). The failed repression of BA synthesis in the mutant liver and the drastically elevated ileal FGF15 expression suggests insensitivity of Shp2-deficient hepatocytes to this gut hormone. To test this, we injected recombinant hFGF19 intraperitoneally (IP), and measured gene expression 6 hrs later. Exogenous hFGF19 exerted a strong inhibition of Cyp7a1 and Cyp8b1 expression in WT controls (Figures 6B, 6C and S4B). However, the response of Shp2hep−/− livers to hFGF19 was significantly diminished (though not completely blocked), as evaluated by Cyp7a1 mRNA and protein levels (Figures 6B, 6C and S4B). Furthermore, hFGF19 failed to up-regulate SHP expression in Shp2hep−/− livers (Figure 6B).
Figure 6. Shp2hep−/− mice are refractory to FGF15/19 repression of BA synthesis.
(A) Relative expression of SHP and FGF15 mRNA was determined by qRT-PCR in ileum samples (n = 5–7).
(B) Relative expression of Cyp7a1, Cyp8b1 and SHP mRNAs in liver samples was determined by qRT-PCR. The animals (n = 5–10) were injected with PBS or hFGF19 (1 mg/kg body weight) and fasted for 6 hrs before sample collection.
(C) Cyp7a1 and Shp2 protein levels were determined by immunoblot analysis of liver lysates from mice as in (B). Each lane represents one mouse.
(D) Immunoblotting of liver lysates was performed with antibodies against pFRS2α(Y196), pErk, Erk1, p-p90RSK, p-PKC(pan) (βII Ser660), pJNK, JNK, p-p38, p38, β-Klotho and GAPDH. Two-month-old WT or Shp2hep−/− mice were fasted for 5.5 hours before IP injection of PBS or hFGF19 (1 mg/kg body weight). The animals were sacrificed 30 min after injection.
(E) Relative expression of SHP, Cyp7a1 and Cyp8b1 mRNA was determined by qRT-PCR in liver samples. The mice (n = 4–5) were injected with 2×109 virions of VP16, VP16-FXR or SHP adenoviruses through tail vein and liver samples were collected 5 days later.
(F) Cyp7a1, V5, Shp2 and GAPDH protein levels were determined by immunoblotting of liver samples collected as in (E). Each lane represents one mouse.
Relative gene expression was normalized to β-actin, and fold change was calibrated to WT group. Data are shown as the means ± s.e.m. * p < 0.05, ** p < 0.01 and *** p < 0.001, as determined by Student’s t test.
To identify Shp2-modulated signaling events downstream of FGFR4, receptor for FGF15/19, we prepared liver lysates 30 min after hFGF19 injection. hFGF19 potently stimulated Erk1/2 phosphorylation in WT but not in Shp2hep−/− livers (Figures 6D and S4C), and similarly defective activation was also observed for ribosomal S6 kinase (p90RSK) (Figure 6D). The specific effect on Erk1/2 activation by hFGF19 was ascertained by the similar p-p38 MAPK levels detected in both lysates (Figures 6D and S4C). In contrast, FGF19-induced p-Jnk signals were higher in Shp2hep−/− liver (Figures 6D and S4C), consistent with our previous observations (Bard-Chapeau et al., 2006; Shi et al., 1998). Using an antibody that recognizes several protein kinase C (PKC) family members, we detected hFGF19-induced phosphorylation of PKCs in controls, which was attenuated in Shp2hep−/− livers (Figures 6D and S4C). To rule out the effect by the chronic liver damages in Shp2hep−/− mice, we injected hFGF19 into Shp2(H+K)−/− mice, and obtained similar results in these mice (Figure S4D). All these data suggest that Shp2 deletion suppressed hFGF19-stimulated Erk and PKC activation in hepatocytes.
With the reduced response to FGF15/19 signal, the elevated BA levels failed to activate FXR to up-regulate SHP expression in Shp2hep−/− livers, suggesting a role of Shp2 upstream of FXR. To test this, we investigated if expression of a constitutively active FXR can rescue Shp2 deficiency and therefore inhibit Cyp7a1 expression. Adenoviruses expressing VP16-FXR, SHP or VP16 were injected into WT and Shp2hep−/− mice through tail vein. Similarly increased SHP expression was observed in both WT and Shp2hep−/− livers after VP16-FXR injection (Figure 6E). Cyp7a1 mRNA and protein levels were decreased in WT livers, and were also down-regulated by lesser extent in Shp2hep−/− livers (Figures 6E, 6F and S4E). SHP overexpression also suppressed Cyp7a1 expression in Shp2hep−/− livers, though not to the WT level (Figures 6E, 6F and S4E). These results argue that FXR and SHP do not operate in a simple linear relationship, and also suggest that Shp2 modulates signaling to both independently. Although Cyp8b1 expression was significantly elevated in Shp2hep−/− livers (Figure 5A, 6E), overexpression of VP16-FXR or SHP caused similar suppression of Cyp8b1 in control and mutant mice (Figure 6E). Thus, the expression of Cyp7a1 and Cyp8b1 is likely controlled by common and distinct pathways.
Shp2 Is Required for Hepatic FGFR4 Activation by FGF15/19
As described above, Shp2 is required for hepatic response to ileal FGF15/19 signal and also for intrahepatic FXR activation by BAs. To gain a broad view on Shp2 function, we performed microarray analysis of gene expression in 2-month-old Shp2hep−/− and WT livers, and compared the results with two published datasets. One was on FGF15/19-treated livers (Potthoff et al., 2011), which showed induction of Erk pathway and inhibition of BA synthesis. Another was on FXR/SHP DKO livers, which exhibited increased BA synthesis (Anakk et al., 2011). Overall, opposite gene expression patterns were observed between Shp2hep−/− and FGF15/19-treated livers (Figure S5A). Only one group of up-regulated genes is enriched between Shp2hep−/− and FXR/SHP DKO mice (Figure S5A). Gene ontology (GO) analysis showed significant enrichment of BA metabolism-related processes, such as steroid synthesis and primary BA biosynthesis, in the group of up-regulated genes in Shp2hep−/− livers, which were down-regulated in FGF15/19-treated livers (Figure S5B). Furthermore, GO analysis revealed a group of genes that were up-regulated in both Shp2hep−/− and FXR/SHP DKO livers and also a set of genes that were oppositely regulated in FGF15/19-treated and FXR/SHP DKO livers (Figure S5B). Therefore, the large-scale data analysis suggests that Shp2 is a positive regulator of FGF15/19 signal and acts cooperatively with FXR and SHP in hepatic control of BA synthesis.
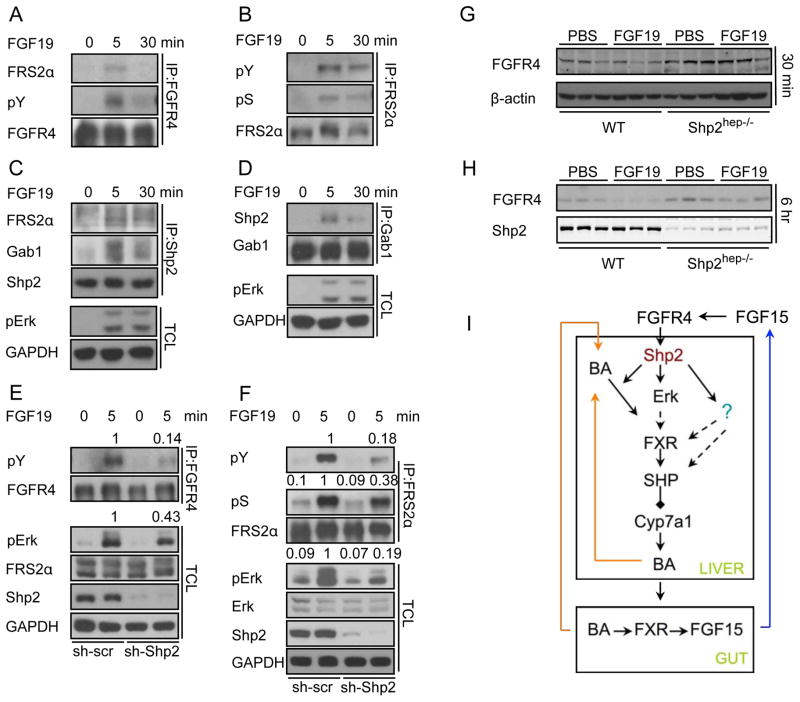
We further dissected the effect of Shp2 deficiency on signaling events proximal to FGFR4. Treatment of Hep3B cells with hFGF19 induced robust tyrosine-phosphorylation of FGFR4 and its immediate target FRS2α (Figures 7A and 7B). Consistent with a previous report (Zhou et al., 2009), FRS2α was also highly phosphorylated at serine/threonine residues (Figure 7B). hFGF19 stimulation induced physical association of FGFR4 with FRS2α (Figure 7A), and assembly of Shp2/FRS2α and Shp2/Gab1 complexes (Figures 7C and 7D). shRNA-mediated Shp2 knockdown (KD) decreased tyrosyl phosphorylation of FGFR4 (Figure 7E) and reduced FRS2α phosphorylation on tyrosine and serine (Figure 7F), resulting in impaired Erk activation (Figures 7E and 7F).
Figure 7. Shp2 is required for FGF15/19-stimulated FGFR4 activation.
(A–F) Serum-starved Hep3B cells were stimulated with 100 ng/ml hFGF19 as indicated. Immunoblotting was performed with indicated antibodies for total cell lysate (TCL) or immunoprecipitates. In E–F, the cells were treated with lenti-viruses expressing either scrambled (sh-scr) or Shp2-specific (sh-Shp2) shRNAs for 72 hrs before starvation.
(G) The same samples as in Figure 6D were blotted with antibodies to FGFR4 and β-actin.
(H) The same samples as in Figure 6C were immunoblotted for FGFR4 and Shp2.
(I) A model shows how Shp2 orchestrates BA and FGF15/19 signaling in control of BA biosynthesis.
Consistent with the observations in Hep3B cells, FRS2α was not phosphorylated in Shp2hep−/− liver following hFGF19 injection, suggesting defective FGFR4 activation (Figure 6D). However, FGFR4 mRNA levels remained unchanged in mutant livers and were not affected by hFGF19 injection (Figure S5D). Treatment with hFGF19 for 6 hrs induced downregulation of FGFR4 in WT livers (Figures 7H and S5C), suggesting that activation of FGFR4 is followed by endocytosis and degradation after ligand binding, similar to other FGFRs reported previously (Beenken and Mohammadi, 2009; Haugsten et al., 2008). Notably, this process was attenuated in Shp2hep−/− livers, as evidenced by more steady FGFR4 protein contents after hFGF19 treatment (Figures 7G, 7H and S5C). These results indicate a requirement of Shp2 for FGF15/19 activation of FGFR4 and its downstream signaling pathways in hepatocytes.
Discussion
Tight control of BA homeostasis is essential, given the critical roles of BAs in lipid digestion and cholesterol metabolism but also the toxic effect of excess BAs. Numerous data suggested cross-talk of FGF15/19-FGFR4 and BA-FXR signaling events, although the underlying mechanism is unclear. This report presents physiological and biochemical data proving that Shp2 acts to coordinate the signals elicited by FGF15/19 and BAs in the liver (Figure 7I).
The Shp2hep−/− mice exhibited early onset hepatobiliary defects, including enlarged gallbladder, elevation of systemic BA levels, and ductal cell proliferation (Figures 1 and 3). Furthermore, Shp2hep−/− animals were more susceptible to biliary obstruction (Figure 2). BA sequestration by cholestyramine improved the hepatobiliary phenotypes (Figure 4), suggesting that excess BAs account for the liver damages. The increased fecal BA discharge together with elevated bile flow (Figures 3D and 3E) directly points to unrestrained BA synthesis in Shp2hep−/− mice. Indeed, several lines of evidence highlight an indispensable role of Shp2 in repression of BA synthesis. First, basal levels of Cyp7a1 mRNA and protein were markedly increased in Shp2hep−/− livers (Figure 5). Second, increased intrahepatic BAs did not suppress BA synthesis in Shp2−/− hepatocytes (Figures 5 and 6). Third, increased ileal FGF15 expression, and even IP injection of hFGF19, did not effectively inhibit BA synthesis in mutant mice (Figures 6A-D). These observations indicate that Shp2 is positively required for hepatic response to both intrahepatic and ileal inhibitory signals.
It has been well recognized that the BA-FXR-SHP axis plays a central role in repression of Cyp7a1 expression. However, phenotypic analyses of FXR and SHP KO or DKO mice argued against a simple linear relationship of the FXR-SHP-Cyp7a1 pathway (Anakk et al., 2011; Sinal et al., 2000). Our results indicate defective FXR activation in Shp2hep−/− livers. First, the BA composition analysis showed increase of most FXR agonist species with similar levels of antagonist in Shp2hep−/− livers (Figures 3G–I). However, the basal SHP expression was reduced in the mutant livers (Figure 5A). Second, synthetic FXR agonist GW4064 failed to up-regulate SHP expression in Shp2hep−/− livers (Figure S3E). Third, exogenous expression of an activated FXR or SHP partially repressed Cyp7a1 expression (Figure 6), placing Shp2 upstream of FXR. However, with distinct subcellular localization (Figure 5G), Shp2 does not form a physical complex with and regulate FXR activity directly.
The defective response to hFGF19 in Shp2hep−/− livers is very similar to FGFR4 and FGF15 KO mice, indicating a critical role of Shp2 in this pathway. With normal expression of Shp2 in the ileum (Figure S4A), the intestinal BA-FXR signaling remained intact in Shp2hep−/− animals. In fact, ileal FGF15 and SHP expression was increased (Figure 6A) due to enhanced bile flow. However, this data may have also revealed a compensatory mechanism for the insensitivity of Shp2−/− hepatocytes to FGF15. Indeed, IP injection of hFGF19 suppressed Cyp7a1 and Cyp8b1 expression in WT livers, but this response was diminished in Shp2hep−/− livers (Figure 6). Tyrosyl phosphorylation of FGFR4 and FRS2α was reduced in Shp2 KD cells following hFGF19 stimulation (Figure 7E). The ligand-stimulated FGFR4 activation/down-regulation was also attenuated in Shp2hep−/− livers (Figures 7G and 7H). These biochemical data suggest a requirement for Shp2 in FGFR4 activation by hFGF19, which involves its association with FRS2α.
Consistently, we detected multiple signaling defects downstream of FGFR4 in Shp2hep−/− livers and Shp2 KD cells. hFGF19-stimulated p-Erk1/2 and p90RSK activation was almost blocked in Shp2hep−/− livers (Figure 6D). Consistently, several groups reported that pharmaceutical or siRNA-mediated inhibition of Erk alleviated repression of Cyp7a1 expression in human hepatocytes or mouse livers (Henkel et al., 2011; Li et al., 2012b; Song et al., 2009). Therefore, defective Erk activation may account for deregulated BA synthesis in Shp2−/− hepatocytes. Several molecules have been proposed as potential Shp2 targets in promoting the Erk pathway, including PAG/Cbp, RasGAP, Gab1 and Sprouty (Chan and Feng, 2007; Neel et al., 2003). Previous data also suggested BA activation of PKCα, β and δ (Gineste et al., 2008; Rao et al., 1997), and a recent report showed PKCζ activation by FGF19 (Seok et al., 2013). These studies suggested a mechanism for FXR regulation via phosphorylation by PKCs, which can be stimulated by FGF15/19. We observed that Shp2-deficiency resulted in reduced PKC phosphorylation in control and hFGF19-treated livers. Further studies are needed to elucidate distinct roles of specific PKC isoforms in FGFR4 signaling and FXR activation. Together, our results show that Shp2 is a critical player immediately downstream of FGFR4 to regulate BA synthesis. The biochemical data was further supported by comparative analysis of global gene expression profiles in Shp2hep−/−, FXR/SHP DKO and FGF15/19-treated livers (Figures S5A and S5B).
BAs are also considered as carcinogens due to their amphipathic nature (Wang et al., 2013). Both FXR and SHP KO animals developed liver cancers spontaneously (Yang et al., 2007; Zhang et al., 2008). FXR/SHP DKO mice suffered from accelerated liver tumorigenesis due to BA activation of Hippo signaling (Anakk et al., 2013). Shp2hep−/− mice developed hepatocellular adenomas spontaneously and were more susceptible to chemical carcinogen (Bard-Chapeau et al., 2011; Li et al., 2012a). Lowering BAs by cholestyramine significantly improved hepatobiliary damages in mutant animals, suggesting that persistent elevation of hepatic BA contents is a contributing factor to oncogenesis in Shp2-deficient livers. Recent studies showed FGF19 and FGFR4 are deregulated in several human cancers (Desnoyers et al., 2008; French et al., 2012). In Shp2hep−/− mice, up-regulated ileal FGF15 expression (Figure 6A) may contribute to enhanced liver tumorigenesis.
BA biosynthesis is a primary route for disposal of excess cholesterol, and the intricate balance between BAs and cholesterol is exemplified by the cholesterol-lowering effect of BA sequestration. Similar to Cyp7a1 transgenic mice (Li et al., 2011), Shp2hep−/− animals also showed lower plasma cholesterol levels and increased hepatic cholesterol synthesis (Figures 5M and 5P), indicating that the enhanced BA synthesis is not driven by cholesterol accumulation but rather is due to uncontrolled expression of Cyp7a1 and other BA-synthetic genes. All this supports Shp2 as a bona fide regulator of BA biosynthesis.
EXPERIMENTAL PROCEDURES
Animal Procedures
Generation of hepatocyte-specific Shp2 KO mice (Shp2hep−/−) were described previously (Bard-Chapeau et al., 2011; Bard-Chapeau et al., 2006). The animal protocols (S09108) with all used procedures were approved by UCSD Institutional Animal Care and Use Committee. BDL was performed as previously reported (Georgiev et al., 2008). For BA sequestration, mice were fed with chow diet (Cat. # 7012, Harlan Laboratories, Madison, WI) supplemented with 2% cholestyramine-resin (Cat. # C4650 Sigma-Aldrich) from weaning to 2-month. All experimental data were collected from male animals at age of 8–10 weeks, except that BDL and measurement of BA pool size and bile flow were done on both male and female mice. All samples were collected from WT and mutant animals between 3:00–5:00 pm during the day.
Histology Staining and Image Acquisition
Liver samples were prepared as reported (Bard-Chapeau et al., 2011), embedded, sectioned and stained with H&E at a UCSD core facility. Manson’s trichrome staining (Cat. No. KTMTRPT American MasterTech) and reticulum staining (Cat. No. KTCPRPT American MasterTech) were performed following manufacturer’s instructions. Necrotic areas were counted using ImageJ and normalized with parenchymal areas. The images were acquired with Olympus IX71 microscope and CellSense Software.
qRT-PCR and Immunoblot Analyses
Liver or ileum samples were lysed in TRIzol® reagent (Cat. No. 15596, Invitrogen) using MagNA Lyser (Roche). RNA was extracted and reverse transcribed with a kit (Cat. No. 4374966, invitrogen). Quantitative Real-time PCR (qRT-PCR) was performed with commercial master mix (Cat. No 600882, Agilent Technologies) using Mx3000P QPCR system (Agilent). A list of PCR primers is provided in the Supplemental Information. Immunoblot analysis was performed with standard protocols and visualized with ECL or ECL-plus. Some blots were visualized by Lico-Odyssey system. The list of primary antibodies is provided in the Supplemental Information. Freshly isolated liver lysates were separated into cytoplasmic and nuclear fractions using a kit (Pierce, Cat. # 78835).
Measurement of BAs, Bilirubin and Cholesterol
Bile flow rate was measured as described previously (Modica et al., 2011). Levels of total bile acids (Cat. No. DZ042A-K, Diazyme), total bilirubin (Cat. No. B577, Teco Diagnostics) and total cholesterol (Cat. No.439-17501 Wako Diagnostics) were measured according to manufacturer’s instructions. BA composition in BA pool, liver and feces was analyzed as previously reported (Li et al., 2012b).
Cell Culture, Treatment and Immunoprecipitation
Hep3B cells (ATCC® HB-8064) were starved in DMEM with 1% FBS for 16 hrs and stimulated with 100 ng/ml hFGF19 in DMEM for indicated time periods. Immunoprecitation was performed as reported (Shi et al., 2000). Tyrosyl-phosphorylated proteins were detected with three anti-pY antibodies combined (Supplemental information).
Adeno-virus and Lenti-virus Generation and Purification
PCR fragments of VP16, VP16-FXR and SHP were cloned into pENTR™/D-TOPO®, then shuttled into pAd/CMV/V5-DEST™. VP16-FXR fragment was amplified with VP16-ad-F and FXR-ad-R primers. The virus stocks were generated according to manufacturer’s instructions. The purification and titration of viruses were performed as previously described (Qiao et al., 2006). Lenti-virus constructs with scrambled or Shp2-specific shRNAs were generated as previously reported (Lu et al., 2011).
Microarray and Bioinformatic Data Analysis
Total RNA from Shp2hep−/− and WT mice liver was prepared with RNeasy mini kit (Qiagen Cat # 74104). Labeled cRNA was prepared from 500 ng RNA using the Illumina® RNA Amplification Kit from Ambion (Austin, TX, USA). The labeled cRNA (750 ng) was hybridized overnight at 58°C to the Sentrix Mouse -8 Expression BeadChip (>23,000 gene transcripts; Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. BeadChips were subsequently washed and developed with fluorolink streptavidin-Cy3 (GE Healthcare). BeadChips were scanned with an Illumina BeadArray Reader. The microarray data have been deposited in the Gene Expression Omnibus (GEO) under the accession number of GSE51860. The gene expression data (GSE20599) for FXR−/−/SHP−/− DKO mice at 5 weeks of age were downloaded from the GEO (Anakk et al., 2011), processed with BeadStudio software and quantile normalized. The data (GSE29426) for FGF15/19-treated mice were downloaded from GEO (Potthoff et al., 2011) and processed with MAS5 algorithm (Affymetrix). Probes were filtered with detection p-value > 0.01 (for Shp2hep−/− and FXR−/−/SHP−/− DKO data) or with ABS call (for FGF15/19 data) before further analysis. Transcripts shared between datasets were used for K-means clustering with Cluster 3.0 software. Heat maps were generated with Java TreeView. GO analysis was performed with DAVID v.6.7 program.
Statistical Analyses
Data analysis was performed using a two-tailed unpaired Student’s t test. Values are expressed as mean ± SEM (*p < 0.05; **p < 0.01; ***p < 0.001).
Supplementary Material
Highlights.
Shp2/Ptpn11 is required to repress bile acid (BA) biosynthesis in hepatocytes.
Shp2 coordinates hepatic responses to BA and FGF15/19 signals.
FGFR4 activation by FGF15/19 requires Shp2 activity.
Hepatic Shp2 is essential to maintain systemic BA and hepatobiliary homeostasis.
Acknowledgments
We thank R Evans (Salk Inst.) for the vp16-FXR construct, J Luo (Peking Uni.) for the Shp2 knockdown lenti-viruses, Z Chen (Shanghai Inst of Biochem. & Cell Bio.) for anti-FRS2α antibody and J Qi (Uni. of Maryland) and G Hon (UCSD) for helpful discussion and protocols. This research was supported by NIH grants R01HL096125 and R01CA176012 (GSF), R01DK066202 (HEX), R01HD069634 (JS), and P20GM103549 (TL), and ADA grant 1-13-BS-048 (GSF).
Footnotes
AUTHOR CONTRIBUTIONS
SL and GSF conceived the project, analyzed the data and wrote the paper. GSF provided the reagents. SL designed and performed most of the experiments. DH did the BA measurements and made FXR and VP-16 FXR virus stocks. XL provided some animals and microarray data. SL, NA and LQ purified the adenoviruses. NA did liver fraction and some western blots. JL made the SHP adenovirus stock. LM helped set up the ChIP assay. HZ provided liver samples from Shp2H+K−/− mice, JS and ZH helped design the experiments and analyzed the data. TL performed the BA composition analysis. BL performed the bioinformatics analysis. KSP and HEX prepared recombinant hFGF19, KJ did the histology analysis.
The authors declare no competing financial interests.
Supplemental information contains three supplemental tables, four supplemental figures and references.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anakk S, Bhosale M, Schmidt VA, Johnson RL, Finegold MJ, Moore DD. Bile Acids Activate YAP to Promote Liver Carcinogenesis. Cell reports. 2013 doi: 10.1016/j.celrep.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anakk S, Watanabe M, Ochsner SA, McKenna NJ, Finegold MJ, Moore DD. Combined deletion of Fxr and Shp in mice induces Cyp17a1 and results in juvenile onset cholestasis. J Clin Invest. 2011;121:86–95. doi: 10.1172/JCI42846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RD, Searle GW. Bile salt absorption at various levels of rat small intenstine. Proc Soc Exp Biol Med. 1960;105:521–523. doi: 10.3181/00379727-105-26163. [DOI] [PubMed] [Google Scholar]
- Bard-Chapeau EA, Li S, Ding J, Zhang SS, Zhu HH, Princen F, Fang DD, Han T, Bailly-Maitre B, Poli V, et al. Ptpn11/Shp2 acts as a tumor suppressor in hepatocellular carcinogenesis. Cancer Cell. 2011;19:629–639. doi: 10.1016/j.ccr.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard-Chapeau EA, Yuan J, Droin N, Long S, Zhang EE, Nguyen TV, Feng GS. Concerted functions of Gab1 and Shp2 in liver regeneration and hepatoprotection. Mol Cell Biol. 2006;26:4664–4674. doi: 10.1128/MCB.02253-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwald H, Gebhard RL. Localization of bile salt absorption in vivo in the rabbit. Ann Surg. 1968;167:191–198. doi: 10.1097/00000658-196802000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RJ, Feng GS. PTPN11 is the first identified proto-oncogene that encodes a tyrosine phosphatase. Blood. 2007;109:862–867. doi: 10.1182/blood-2006-07-028829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang JY. Bile acid regulation of gene expression: roles of nuclear hormone receptors. Endocr Rev. 2002;23:443–463. doi: 10.1210/er.2000-0035. [DOI] [PubMed] [Google Scholar]
- de Aguiar Vallim TQ, Tarling EJ, Edwards PA. Pleiotropic roles of bile acids in metabolism. Cell Metab. 2013;17:657–669. doi: 10.1016/j.cmet.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoyers LR, Pai R, Ferrando RE, Hotzel K, Le T, Ross J, Carano R, D’Souza A, Qing J, Mohtashemi I, et al. Targeting FGF19 inhibits tumor growth in colon cancer xenograft and FGF19 transgenic hepatocellular carcinoma models. Oncogene. 2008;27:85–97. doi: 10.1038/sj.onc.1210623. [DOI] [PubMed] [Google Scholar]
- Fon Tacer K, Bookout AL, Ding X, Kurosu H, John GB, Wang L, Goetz R, Mohammadi M, Kuro-o M, Mangelsdorf DJ, et al. Research resource: Comprehensive expression atlas of the fibroblast growth factor system in adult mouse. Mol Endocrinol. 2010;24:2050–2064. doi: 10.1210/me.2010-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French DM, Lin BC, Wang M, Adams C, Shek T, Hotzel K, Bolon B, Ferrando R, Blackmore C, Schroeder K, et al. Targeting FGFR4 inhibits hepatocellular carcinoma in preclinical mouse models. PLoS One. 2012;7:e36713. doi: 10.1371/journal.pone.0036713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev P, Jochum W, Heinrich S, Jang JH, Nocito A, Dahm F, Clavien PA. Characterization of time-related changes after experimental bile duct ligation. Br J Surg. 2008;95:646–656. doi: 10.1002/bjs.6050. [DOI] [PubMed] [Google Scholar]
- Gineste R, Sirvent A, Paumelle R, Helleboid S, Aquilina A, Darteil R, Hum DW, Fruchart JC, Staels B. Phosphorylation of farnesoid X receptor by protein kinase C promotes its transcriptional activity. Mol Endocrinol. 2008;22:2433–2447. doi: 10.1210/me.2008-0092. [DOI] [PubMed] [Google Scholar]
- Haugsten EM, Malecki J, Bjorklund SM, Olsnes S, Wesche J. Ubiquitination of fibroblast growth factor receptor 1 is required for its intracellular sorting but not for its endocytosis. Mol Biol Cell. 2008;19:3390–3403. doi: 10.1091/mbc.E07-12-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel AS, Anderson KA, Dewey AM, Kavesh MH, Green RM. A chronic high-cholesterol diet paradoxically suppresses hepatic CYP7A1 expression in FVB/NJ mice. J Lipid Res. 2011;52:289–298. doi: 10.1194/jlr.M012781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houten SM, Watanabe M, Auwerx J. Endocrine functions of bile acids. EMBO J. 2006;25:1419–1425. doi: 10.1038/sj.emboj.7601049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Ma K, Zhang J, Qatanani M, Cuvillier J, Liu J, Dong B, Huang X, Moore DD. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. 2006;312:233–236. doi: 10.1126/science.1121435. [DOI] [PubMed] [Google Scholar]
- Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Inoue Y, Yu AM, Yim SH, Ma X, Krausz KW, Inoue J, Xiang CC, Brownstein MJ, Eggertsen G, Bjorkhem I, et al. Regulation of bile acid biosynthesis by hepatocyte nuclear factor 4alpha. J Lipid Res. 2006;47:215–227. doi: 10.1194/jlr.M500430-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong WI, Jeong DH, Do SH, Kim YK, Park HY, Kwon OD, Kim TH, Jeong KS. Mild hepatic fibrosis in cholesterol and sodium cholate diet-fed rats. J Vet Med Sci. 2005;67:235–242. doi: 10.1292/jvms.67.235. [DOI] [PubMed] [Google Scholar]
- Kerr TA, Saeki S, Schneider M, Schaefer K, Berdy S, Redder T, Shan B, Russell DW, Schwarz M. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev Cell. 2002;2:713–720. doi: 10.1016/s1534-5807(02)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kir S, Zhang Y, Gerard RD, Kliewer SA, Mangelsdorf DJ. Nuclear receptors HNF4alpha and LRH-1 cooperate in regulating Cyp7a1 in vivo. J Biol Chem. 2012;287:41334–41341. doi: 10.1074/jbc.M112.421834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong B, Wang L, Chiang JY, Zhang Y, Klaassen CD, Guo GL. Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology. 2012;56:1034–1043. doi: 10.1002/hep.25740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Hsu DD, Wang H, Feng GS. Dual faces of SH2-containing protein-tyrosine phosphatase Shp2/PTPN11 in tumorigenesis. Front Med. 2012a;6:275–279. doi: 10.1007/s11684-012-0216-4. [DOI] [PubMed] [Google Scholar]
- Li T, Francl JM, Boehme S, Ochoa A, Zhang Y, Klaassen CD, Erickson SK, Chiang JY. Glucose and insulin induction of bile acid synthesis: mechanisms and implication in diabetes and obesity. J Biol Chem. 2012b;287:1861–1873. doi: 10.1074/jbc.M111.305789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Matozel M, Boehme S, Kong B, Nilsson LM, Guo G, Ellis E, Chiang JY. Overexpression of cholesterol 7alpha-hydroxylase promotes hepatic bile acid synthesis and secretion and maintains cholesterol homeostasis. Hepatology. 2011;53:996–1006. doi: 10.1002/hep.24107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- Lu Y, Xiong Y, Huo Y, Han J, Yang X, Zhang R, Zhu DS, Klein-Hessling S, Li J, Zhang X, et al. Grb-2-associated binder 1 (Gab1) regulates postnatal ischemic and VEGF-induced angiogenesis through the protein kinase A-endothelial NOS pathway. Proc Natl Acad Sci U S A. 2011;108:2957–2962. doi: 10.1073/pnas.1009395108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- Modica S, Murzilli S, Moschetta A. In Current Protocols in Mouse Biology. John Wiley & Sons, Inc; 2011. Characterizing Bile Acid and Lipid Metabolism in the Liver and Gastrointestinal Tract of Mice. [DOI] [PubMed] [Google Scholar]
- Modica S, Petruzzelli M, Bellafante E, Murzilli S, Salvatore L, Celli N, Di Tullio G, Palasciano G, Moustafa T, Halilbasic E, et al. Selective Activation of Nuclear Bile Acid Receptor FXR in the Intestine Protects Mice Against Cholestasis. Gastroenterology. 2012;142:355–365. e354. doi: 10.1053/j.gastro.2011.10.028. [DOI] [PubMed] [Google Scholar]
- Neel BG, Gu H, Pao L. The ‘Shp’ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem Sci. 2003;28:284–293. doi: 10.1016/S0968-0004(03)00091-4. [DOI] [PubMed] [Google Scholar]
- Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, et al. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Boney-Montoya J, Choi M, He T, Sunny NE, Satapati S, Suino-Powell K, Xu HE, Gerard RD, Finck BN, et al. FGF15/19 regulates hepatic glucose metabolism by inhibiting the CREB-PGC-1alpha pathway. Cell Metab. 2011;13:729–738. doi: 10.1016/j.cmet.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao L, MacLean PS, You H, Schaack J, Shao J. knocking down liver ccaat/enhancer-binding protein alpha by adenovirus-transduced silent interfering ribonucleic acid improves hepatic gluconeogenesis and lipid homeostasis in db/db mice. Endocrinology. 2006;147:3060–3069. doi: 10.1210/en.2005-1507. [DOI] [PubMed] [Google Scholar]
- Rao A, Haywood J, Craddock AL, Belinsky MG, Kruh GD, Dawson PA. The organic solute transporter alpha-beta, Ostalpha-Ostbeta, is essential for intestinal bile acid transport and homeostasis. Proc Natl Acad Sci U S A. 2008;105:3891–3896. doi: 10.1073/pnas.0712328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao YP, Stravitz RT, Vlahcevic ZR, Gurley EC, Sando JJ, Hylemon PB. Activation of protein kinase C alpha and delta by bile acids: correlation with bile acid structure and diacylglycerol formation. J Lipid Res. 1997;38:2446–2454. [PubMed] [Google Scholar]
- Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, Angelin B, Hyotylainen T, Oresic M, Backhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Seok S, Kanamaluru D, Xiao Z, Ryerson D, Choi SE, Suino-Powell K, Xu HE, Veenstra TD, Kemper JK. Bile acid signal-induced phosphorylation of small heterodimer partner by protein kinase Czeta is critical for epigenomic regulation of liver metabolic genes. J Biol Chem. 2013;288:23252–23263. doi: 10.1074/jbc.M113.452037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi ZQ, Lu W, Feng GS. The Shp-2 tyrosine phosphatase has opposite effects in mediating the activation of extracellular signal-regulated and c-Jun NH2-terminal mitogen-activated protein kinases. J Biol Chem. 1998;273:4904–4908. doi: 10.1074/jbc.273.9.4904. [DOI] [PubMed] [Google Scholar]
- Shi ZQ, Yu DH, Park M, Marshall M, Feng GS. Molecular mechanism for the Shp-2 tyrosine phosphatase function in promoting growth factor stimulation of Erk activity. Mol Cell Biol. 2000;20:1526–1536. doi: 10.1128/mcb.20.5.1526-1536.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- Song KH, Li T, Owsley E, Strom S, Chiang JY. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7alpha-hydroxylase gene expression. Hepatology. 2009;49:297–305. doi: 10.1002/hep.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroeve JH, Brufau G, Stellaard F, Gonzalez FJ, Staels B, Kuipers F. Intestinal FXR-mediated FGF15 production contributes to diurnal control of hepatic bile acid synthesis in mice. Lab Invest. 2010;90:1457–1467. doi: 10.1038/labinvest.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- Vallim TQdA, Edwards PA. Bile Acids Have the Gall to Function as Hormones. Cell metabolism. 2009;10:162–164. doi: 10.1016/j.cmet.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Vergnes L, Lee JM, Chin RG, Auwerx J, Reue K. Diet1 functions in the FGF15/19 enterohepatic signaling axis to modulate bile acid and lipid levels. Cell Metab. 2013;17:916–928. doi: 10.1016/j.cmet.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Han Y, Kim CS, Lee YK, Moore DD. Resistance of SHP-null mice to bile acid-induced liver damage. J Biol Chem. 2003;278:44475–44481. doi: 10.1074/jbc.M305258200. [DOI] [PubMed] [Google Scholar]
- Wang X, Fu X, Van Ness C, Meng Z, Ma X, Huang W. Bile Acid Receptors and Liver Cancer. Curr Pathobiol Rep. 2013;1:29–35. doi: 10.1007/s40139-012-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Huang X, Yi T, Yen Y, Moore DD, Huang W. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 2007;67:863–867. doi: 10.1158/0008-5472.CAN-06-1078. [DOI] [PubMed] [Google Scholar]
- Yu C, Wang F, Kan M, Jin C, Jones RB, Weinstein M, Deng CX, McKeehan WL. Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor FGFR4. J Biol Chem. 2000;275:15482–15489. doi: 10.1074/jbc.275.20.15482. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xu P, Park K, Choi Y, Moore DD, Wang L. Orphan receptor small heterodimer partner suppresses tumorigenesis by modulating cyclin D1 expression and cellular proliferation. Hepatology. 2008;48:289–298. doi: 10.1002/hep.22342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou WC, Feng XJ, Wu YJ, Benge J, Zhang Z, Chen ZJ. FGF-receptor substrate 2 functions as a molecular sensor integrating external regulatory signals into the FGF pathway. Cell Res. 2009;19:1165–1177. doi: 10.1038/cr.2009.95. [DOI] [PubMed] [Google Scholar]
- Zhu HH, Ji K, Alderson N, He Z, Li S, Liu W, Zhang DE, Li L, Feng GS. Kit-Shp2-Kit signaling acts to maintain a functional hematopoietic stem and progenitor cell pool. Blood. 2011;117:5350–5361. doi: 10.1182/blood-2011-01-333476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.