Abstract
Many infectious diseases arise from co-infections or re-infections with more than one genotype of the same pathogen. These mixed infections could alter host fitness, the severity of symptoms, success in pathogen transmission and the epidemiology of the disease. Trypanosoma cruzi, the etiological agent of Chagas disease, exhibits a high biological variability often correlated with its genetic diversity. Here, we developed an experimental approach in order to evaluate biological interaction between three T. cruzi isolates belonging to different Discrete Typing Units (DTUs TcIII, TcV and TcVI). These isolates were obtained from a restricted geographical area in the Chaco Region. Different mixed infections involving combinations of two isolates (TcIII + TcV, TcIII + TcVI and TcV + TcVI) were studied in a mouse model. The parameters evaluated were number of parasites circulating in peripheral blood, histopathology and genetic characterization of each DTU in different tissues by DNA hybridization probes. We found a predominance of TcVI isolate in blood and tissues respect to TcIII and TcV; and a decrease of the inflammatory response in heart when the damage of mice infected with TcVI and TcIII + TcVI mixture were compared. In addition, simultaneous presence of two isolates in the same tissue was not detected. Our results show that biological interactions between isolates with different biological behaviors lead to changes in their biological properties. The occurrence of interactions among different genotypes of T. cruzi observed in our mouse model suggests that these phenomena could also occur in natural cycles in the Chaco Region.
Background
Advances in molecular typing techniques show that many infectious diseases may arise from co-infections or re-infections with more than one genotype of the same pathogen. In these mixed infections the co-infecting parasites may be interacting among each other within the same host determining host fitness, severity of disease symptoms, parasite transmission successful rate and epidemiology of the disease [1]. Various mechanisms can cause interactions between parasite species or among different genotypes of the same species within an individual host. For example, parasites can infect the same target site within a host and directly interact among each other by interference competition, or indirectly by resources competition or via the host immune system [2].
In general, biological interactions between protozoan parasites have been divided into two main groups: those who involved parasites belonging to the same species and the ones that occur between closely to distantly related different species [3]. In this sense, several research studies have reported mixed infections in Leishmania spp [4,5,6], Plasmodium spp [7,8,9], Trypanosoma brucei and Trypanosoma congolense [10,11,12].
Trypanosoma cruzi is the etiologic agent of Chagas disease, an illness that affects several million people in Latin America and still remains an important public health problem in certain endemic areas of Argentina. This parasite shows a high genetic variability which has been the basis to classify it into six Discrete Typing Units (DTUs), TcI to TcVI [13]. In addition to this genetic diversity, in vitro and in vivo T. cruzi infection models showed a high biological variability among different genotypes of T. cruzi [14,15,16,17,18,19]. Although it is supposed that genetic and biological diversities of the parasite are essential to determine the clinical course of Chagas disease, specific associations between particular clinical manifestations and a determined lineage have not been clearly demonstrated [20]. Furthermore, the host genetics and its ability to establish an immune response to control the infection are very important in the outcome of the disease [21].
The consequences of mixed infections by different T. cruzi DTUs have been studied in animal models using different laboratory strains. It has been demonstrated in vivo that the tissue tropism of one T. cruzi genotype could change in the presence of another genotype of a different DTU [22,23]. Even more, the histopathological damage and the intensity of the inflammatory process resulting of these co-infections also present remarkable variations [24,25]. Other studies involving T. cruzi mixed infections showed that the parasite load in peripheral blood could be altered either increasing or decreasing according to the co-infecting strains [26,27,28]. Even the outcome of specific chemotherapy has been proven to be altered by these events of concomitant infection by T. cruzi [26,29].
In several geographical areas of the Southern Cone of America, the occurrence of natural mixed infections by different genotypes of T. cruzi have been widely reported in humans [20,30,31,32,33], in wild and domestic animals [34,35] and in the vector Triatoma infestans [34,36].
In a previous work we described the different biological properties displayed by three selected isolates obtained from the Chaco region of Argentina and belonging to DTUs TcIII, TcV and TcVI [17]. These isolates have the particularity of circulating sympatrically in a restricted geographical area; therefore, here we describe the biological outcome resulting of in vivo experimental dual-mixed infections with these T. cruzi strains. Our working hypothesis is the existence of biological interactions among different T. cruzi isolates in the vertebrate host.
Methods
Ethics statement
All animal protocols adhered to the National Institutes of Health (NIH) ‘‘Guide for the care and use of laboratory animals” and were approved by the Animal Ethics Committee of the School of Health Sciences, National University of Salta (Nu 014–2011) [37].
Trypanosoma cruzi isolates
Different Trypanosoma cruzi isolates were examined in the present work. These isolates were obtained from Las Leonas settlement (W 61° 39’ 8.7”, S 27° 01’ 49”), located in the South-west of Chaco Province, Argentina. The protocol of obtaining samples was approved by the Bioethics Committee of the Faculty of Health Sciences of the National University of Salta, Argentina (Resolution N°052–10). The inhabitants signed an informed consent form before sampling at each house. This study did not involve endangered or protected species.
Parasites were recovered from the feces of either, naturally infected Triatoma infestans or insects used for xenodiagnosis of mammalian hosts. The isolates were identified as DTUs TcIII (LL051-P24), TcV (LL014–1) and TcVI (LL040–1) by Multilocus Sequence Typing (MLST) technique, using a typing scheme proposed by Lauthier and cols, [38]. These parasites were maintained in a vector transmission model developed by Ragone and cols, (2012). Hereafter, each isolates will be named according to the corresponding DTU.
Experimental infection in mice
Six groups of 4 male C57BL/6J mice (one month old) were inoculated by intraperitoneal (i.p.) route with parasites recovered from the feces of infected insects and each infected group was followed during 30 days after infection. Prior infection, the feces were visualized microscopically, in order to distinguish epimastigotes, metacyclic and intermediate forms (parasites whose morphology is intermediate between epimastigotes and tripomastigotes); according to Kollien and cols. [39]. The final inoculation dose was adjusted according to the amount of metacyclic and intermediate forms. For single infections with TcIII, TcV or TcVI, 104 parasites were inoculated per mouse. Instead, 5x103 parasites from each isolate were inoculated for dual mixed infections (TcIII + TcV, TcIII + TcVI and TcV + TcVI). We decided to maintain constant the final dose used in both, single and dual-mixed infections, in order to avoid possible differences due to the final number of inoculated parasites. For this reason, we examined in previous assays if the two doses, 104 and 5x103 parasite/mouse of each isolate show differences in the biological parameters to be studied. No statistical differences were observed between these doses (data not shown). Control groups inoculated with Phosphate Buffer Saline (PBS) were included in the experiment. Animal care guidelines adopted by the Health Sciences Faculty, National University of Salta, Argentina, were strictly followed.
Biological parameters evaluated
Parasitemia
Fresh blood from inoculated animals was collected in heparinized glass capillary pipettes by sectioning the tail tip under slight anesthesia. Ten microliters (μl) of blood were placed between slide and cover slip and the number of parasites per 100 fields was recorded microscopically (400X) at different time points.
Histopathology
Animals were sacrificed at 30 days post infection (dpi) by exposure to halothane, and cardiac and skeletal muscle samples were collected. Tissue samples were divided into two parts; one was stored at -80°C for DNA extractions and the other part was fixed in 10% formalin and processed using routine histological techniques. Serial histological sections (3 to 5 μm thick), were stained with hematoxylin-eosin and observed under microscope (50, 200 and 400X). Quantification of the inflammatory response (IR) was assessed taking into account the presence and intensity of inflammatory foci. Criteria were set according to the size and number of foci in order to quantify the inflammatory process in different organs. Thus, IR was blindly quantified as null, mild, moderate and severe as reported in Ragone and cols. [17]. In some cases intermediate values for the inflammatory response were admitted: mild to moderate and moderate to severe.
Detection of T. cruzi DNA in blood and tissue samples
Peripheral blood was obtained at 30 dpi from each animal (350 μl) and mixed with 700 μl of guanidine buffer. DNA extractions were performed from 100μl of the mixture blood-guanidine buffer by using the phenol-chloroform method. DNA extractions from a skeletal and cardiac muscle sample (obtained at 30 dpi) were performed using a kit (PureLink Genomic DNA Kit, Invitrogen). Subsequently, PCR amplification of 330 bp corresponding to minicircle hypervariable regions (mHVR) was performed using primers 121 (5’-AATAATGTACGGG(T/G)GAGATGCATGA-3’) and 122 (5’-GGTTCGATTGGGGTTGGTGTAATATA-3’) [40]. Amplification was performed in a MJR PTC-100 thermocycler (MJ Research, Watertown, MA, USA). The reaction products were visualized in a 2% agarose gel stained with ethidium bromide.
Identification of T. cruzi Discrete Typing Units in biological samples
Detection of each isolate was carried out by hybridization with specific mHVR-kDNA non-radioactive probes in blood and tissues samples taken at 30 dpi. The probes were constructed using DNA from isolates LL051-P24 (TcIII), LL055R3cl2 (TcV) and CL-Brener (TcVI). The primers for probe construction were CV1 (5’-GATTGGGGTTGGAGTACTAT-3’) and CV2 (5’-TTGAACGGCCCTCCGAAAAC-3’) which produced a 290-bp fragment. Restriction sites for Sau96I and ScaI which allow elimination of the minicircle constant region of these PCR fragments were included in the sequence of these primers [41]. PCR fragments were further digested with the restriction endonucleases obtaining a 250-bp final product. The specificity of each generated DNA probe was evaluated in our laboratory by Southern blot analysis against different 330-bp mHVR PCR products of several T. cruzi isolates belonging to the different DTUs [42]. Briefly, Southern blot analysis was performed using 10 μl of each 121–122 PCR product. Samples were subjected to electrophoresis, transferred to Hybond N + nylon membranes (Roche Diagnostics) and cross-linked with UV light to fix the DNA. The membranes were pre-hybridized for at least 30 minute at 42°C and individually hybridized with the generated probes. Labeling of the probe and DNA hybridization were performed according to the protocol supplied with the PCR-DIG DNA-labeling and detection kit (Roche Applied Science, USA).
For assessing the limit of detection of the technique, T. cruzi DNA of the different isolates involved in the study was analyzed individually and combined. The limit of the technique in detecting single infections was 0.1 fg/μl for TcIII and TcV DTUs, and 0.5 fg/μl for TcVI DTU. Whereas that in the mixed infections we evaluated different proportions of one isolate combined with other isolate, from 0.1 fg/μl to 0.9 fg/μl. As a result of this experiment, the detection limit for TcV was 0.1 fg/μl in the TcIII + TcV and TcV + TcVI mixtures; while the detection limit for TcIII was 0.5 fg/μl in TcIII + TcV and TcIII+TcVI. Finally the detection limit for TcVI in the mixtures TcV+TcVI and TcIII+TcVI was 0.5 fg/μl.
Statistical analysis
Differences in the number of parasites in peripheral blood and inflammatory response among groups were evaluated using one-way variance analysis (ANOVA). To analyze inflammatory response, numeric values for the different levels were assigned: null: 0, mild: 1, mild to moderate: 2, moderate: 3, moderate to severe: 4, and severe: 5. Statistical analysis was performed using the software GraphPad Prism V5.00.
Results
Predominance of the more virulent isolate in peripheral blood and DTU detection by mHVR-kDNA hybridization
As in a previous study Ragone and cols. [17] in this work a marked difference was well established respecting the parasitemia between the three different isolates. The TcV isolate presented non-detectable parasitemia in fresh blood mounts. However, PCR assays corroborated infection by this isolate. On the other hand, circulating parasites in peripheral blood were detected in mice inoculated with TcIII and TcVI, being the parasitemia of TcVI significantly higher than the one obtained by TcIII (p = 0,013). When co-infection models (TcIII + TcVI, TcIII + TcV and TcV + TcVI) were considered, the pattern of parasitemia was the one corresponding to the more virulent DTU in all cases (Figs. 1A-C). In the co-infection involving TcIII + TcVI and TcV + TcVI, the parasitemia was the one described for TcVI alone. In addition, in the mixture TcIII + TcV, a behavior equal to TcIII was observed. As expected, there were non-significant differences between TcVI vs. TcIII + TcVI and TcVI vs. TcV + TcVI; neither between TcIII vs. TcIII + TcV. No statistical comparison between TcV vs. TcIII + TcV or TcV + TcVI was carried out since no circulating parasites were detected for TcV isolate.
Fig 1. Parasitemia in peripheral blood of singles and mixed infections.
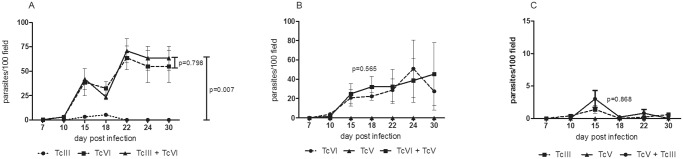
This variable was measured by microscopic observation, of animals inoculated with single and mixed isolates. (A) Shows difference between TcIII vs TcVI vs TcIII + TcVI, (B) TcVI vs TcV + TcVI and (C) TcIII vs TcIII + TcV. The parasitemia of TcV isolate was sub-patent.
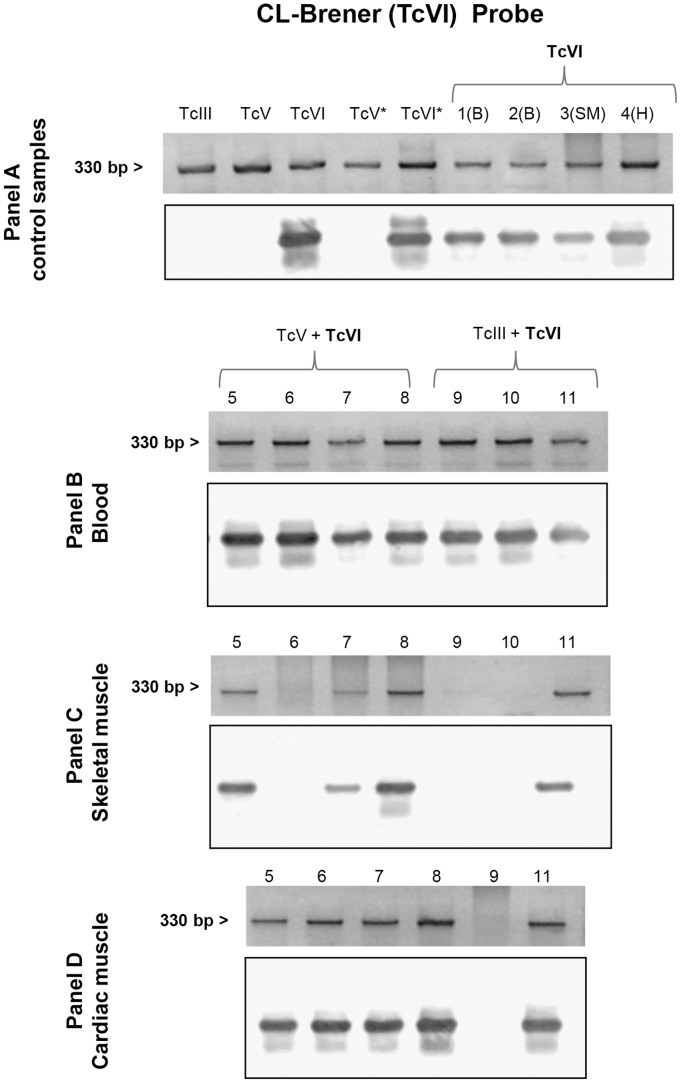
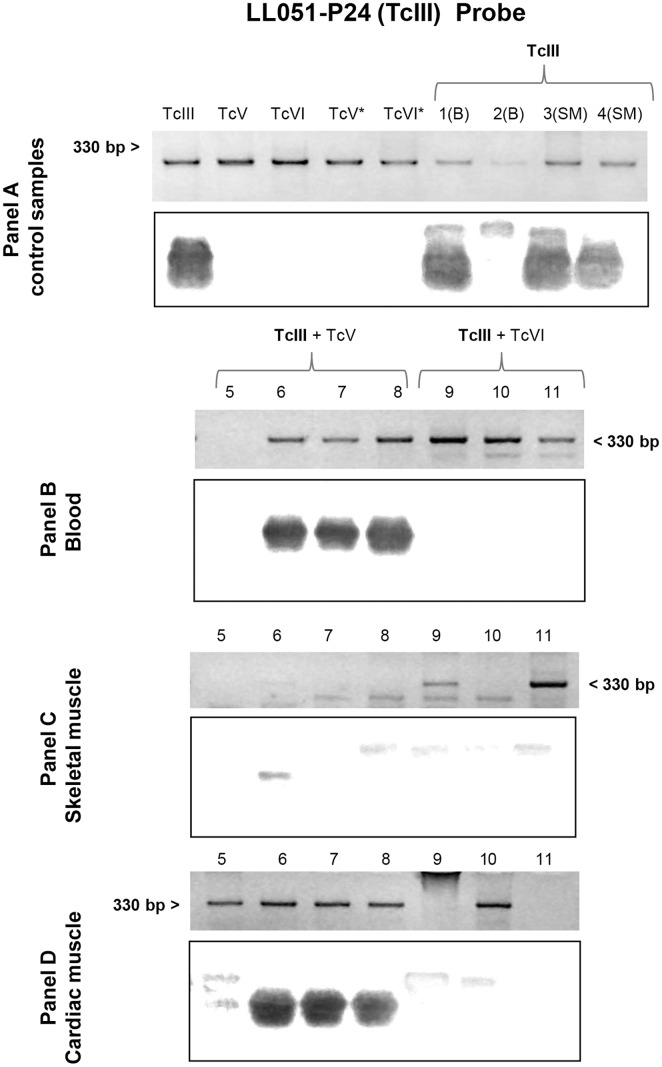
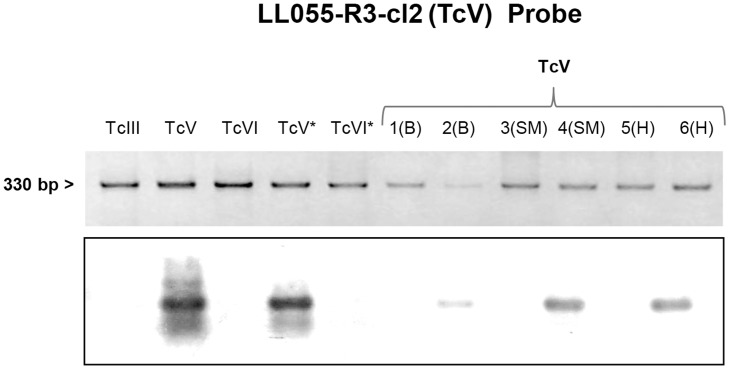
Finally, in blood samples collected at 30 days post-infection we applied specific mHVR-kDNA hybridizations to determine the circulating DTU in single and dual infections. TcVI was identified in all the infection formulas involved (TcIII + TcVI and TcV + TcVI mixtures) (Fig. 2B), while TcIII was only detected in the mixture TcIII + TcV (Fig. 3B). Even though no visible parasites were detected for TcV, positive PCR could be obtained (Fig. 4); however, no hybridization signal for TcV was detected in the mixture TcV + TcVI. In addition, for the blend TcV + TcIII, only TcIII could be detected (Fig. 3B). Table 1 shows the number of mice with positive signal for each specific probe.
Fig 2. Southern blot analyses using the TcVI (CL-Brener) probe.
Each panel shows the electrophoretic pattern of minicircle regions and kDNA transferred to a nylon membrane. Panel A, lanes TcIII, TcV, TcVI, TcV* and TcVI* correspond to DNA of parasite culture from LL051-P24 (DTU TcIII), LL055R3cl2 (DTU TcV), CL-Brener (DTU TcVI), LL014–1 (DTU TcV*) and LL040–1 (DTU TcVI*) respectively; lane 1–4: blood (B), skeletal muscle (SM) and heart (H) samples of mouse infected with TcVI isolate. The asterisk as superscript of the DTU indicates DNA sample from culture of the same inoculated isolate. Panel B, C and D: blood, skeletal muscle and cardiac muscle, respectively, of animals infected with TcV + TcVI (Lane 5–8) and TcIII + TcVI (Lane 9–11).
Fig 3. Southern blot analyses using the TcIII (LL051-P24) probe.
Each panel shows the electrophoretic pattern of minicircle regions and kDNA transferred to a nylon membrane. Panel A: lane TcIII, TcV, TcVI, TcV* and TcVI* correspond to DNA of parasite culture from LL051-P24 (DTU TcIII), LL055R3cl2 (DTU TcV), CL-Brener (DTU TcVI), LL014–1 (DTU TcV*) and LL040–1 (DTU TcVI*) respectively; and lane 1–4: blood (B) and skeletal muscle (SM) samples of mouse infected with TcIII isolate. The asterisk as superscript of the DTU indicates DNA sample from culture of the same inoculated isolate. Panel B, C and D: blood, skeletal muscle and cardiac muscle, respectively, of animals infected with TcIII + TcV (Lane 5–8) and TcIII + TcVI (Lane 9–11).
Fig 4. Southern blot analyses using the TcV (LL055R3cl2) probe.
Electrophoretic pattern of minicircle regions and kDNA transferred to a nylon membrane. Lane TcIII, TcV, TcVI, TcV* and TcVI* correspond to DNA of parasite culture from LL051-P24 (DTU TcIII), LL055R3cl2 (DTU TcV), CL-Brener (DTU TcVI), LL014–1 (DTU TcV) and LL040–1 (DTU TcVI), respectively; and lane: 1–6 blood (B) skeletal muscle (SM) and cardiac muscle (H) of mouse infected with TcV isolate.
Table 1. mHVR-kDNA hybridization results for the detection of individual DTUs in mixed infections.
| MIXTURES | SAMPLES | Probe TcIII | Probe TcV | Probe TcVI |
|---|---|---|---|---|
| TcIII + TcV | Blood | 3/4 | 0/4 | - |
| Skeletal muscle | 1/4 | 0/4 | - | |
| Heart | 4/4 | 0/4 | - | |
| TcIII + TcVI | Blood | 0/3* | - | 3/3 |
| Skeletal muscle | 0/3* | - | 2/3* | |
| Heart | 0/2* | - | 1/2* | |
| TcV + TcVI | Blood | - | 0/4 | 4/4 |
| Skeletal muscle | - | 0/4 | 3/4 | |
| Heart | - | 0/4 | 4/4 |
The values correspond to mice with positive hybridization for a specific probe in relation to the total of mice inoculated with the mixture.
* indicate one or two sample lost.
Histopathological damage and its association with the different infecting DTUs
When single infections were considered, different patterns of tissue damage were observed. TcVI isolate induced significantly more histological lesions in skeletal muscle (p = 0.0026) and heart (p<0.0001) than TcIII and TcV. In addition, TcIII produced significantly more damage in heart than TcV (p<0.0001). Briefly, TcVI induced severe damage in heart and moderate damage in skeletal muscle; additionally amastigote nests and fiber homogenizations were found in both tissues in animals infected with this isolate. On the other hand, TcIII induced mild-moderate lesions in heart and skeletal muscle and no amastigote nests or cellular alteration were found; while TcV induced only mild lesions in the analyzed tissues.
However, in our co-infecting models a moderate damage in heart and skeletal muscle was found in the mixtures TcIII + TcVI and TcV + TcVI, while in mice infected with TcIII + TcV, mild histological lesions were observed in both tissues. In addition, amastigote nests were found in heart and skeletal muscle of animals infected with TcVI + (TcIII or TcV).
In cardiac muscle, no differences were detected between the damage induced by TcIII or by TcIII + TcVI co-infection; however, the intensity of the lesions induced by this mixture were significantly milder respect to the damage induced by TcVI (p = 0,0003) (Fig. 5A). Instead, the histological lesions in heart of animals infected with TcV + TcVI were statistically different from the ones detected in animals infected with TcV alone (p = 0.002) but not to the produced by TcVI (Fig. 5B). The same results were observed when the damage in heart of mice infected with TcIII + TcV was compared to the one of mice infected with TcIII; in this case, no statistical differences between single and mixed infections were detected; however, the lesions induce by the mixture were significantly different than the induce by TcV alone (p = 0,001) (Fig. 5C).
Fig 5. Histological damage in mice infected with single and mixed infections.
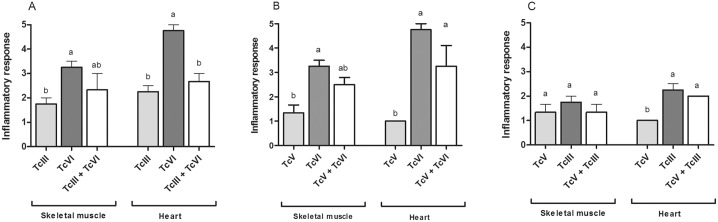
Inflammatory response observed in skeletal muscle and heart samples of experimental groups inoculated with: (A) TcIII, TcVI or TcIII + TcVI, (B) TcV, TcVI or TcV + TcVI and (C) TcIII, TcV or TcIII + TcV. Different letters above error bars indicate statistical difference (p<0.05).
In skeletal muscle, the intensity of the lesions found in mice infected with TcIII + TcVI and TcV + TcVI mixtures was intermediate respect to each single infection; however, no statistical differences were detected (Fig. 5A and 5B). In TcIII + TcV co-infection the damage in skeletal muscle was equal to the one detected in single infections with TcIII or TcV (Fig. 5C).
Heart and muscle tissues were analyzed by PCR and posterior DTU specific hybridization assays. T. cruzi DNA was detected in both, skeletal and cardiac muscle, in all experimental groups. In addition, we found the presence of the isolates TcVI or TcIII in the mixed infections (Fig. 2C and D and Fig. 3C and D). Again, although TcV probe worked correctly (Fig. 4), no hybridization against positive PCR samples of animals infected with TcV + TcIII or TcV + TcVI mixtures was observed. DTU detection by hybridization tests is summarized in Table 1.
Discussion
Here, we developed an experimental approach in order to evaluate if mixed infections involving T. cruzi isolates, belonging to different DTUs and collected from a restricted area in the Chaco Region of Argentina, display evidence of biological interaction in a mouse model. In this work we analyzed the parasitemia and histological damage in heart and skeletal muscle of C57BL/6J mice infected with a combination of the different isolates analyzed. The intrinsic properties of the isolates under study were as follow: TcVI isolate induces high parasitemia, severe histological damage in heart and moderate lesions in skeletal muscle. On the other hand, the TcIII isolate induces less parasitemia than TcVI, and mild-moderate histological damage in heart and skeletal muscle. Finally TcV isolate induces sub-patent parasitemia and mild lesions in the analyzed tissues. When TcVI was combined, either with TcIII or TcV, only the parasitemia pattern of TcVI was observed, suggesting that TcVI isolate predominates over TcIII and TcV isolates. This result was supported by the fact that only DNA of TcVI was detected in the co-infection models involving this isolate and TcIII or TcV. Similar results were obtained for the mixture TcIII + TcV, where the observed parasitemia pattern corresponded to the TcIII pattern and only DNA of TcIII was detected by the probes. This result could be due to several factors: the survival of one isolate in peripheral blood could be related to different mechanisms associated with its ability to escape from the host immune system [28], or due to a selective process within the host cells in favor of a given DTU [43]. On the other hand, in a previous work we reported that the isolates TcIII and TcVI induce a higher serological response than TcV [17]; in consequence we believe that in a mixed infection event between TcV + (TcIII or TcVI), the isolate TcV may be susceptible to the immunological response induced by TcIII or TcVI. Therefore, TcV availability in the host would be reduced, or even eliminated from the host, and not detected at least by the technique herein applied.
When cardiac muscle samples of mice infected with the mixtures TcIII + TcVI and TcV + TcVI were analyzed; only presence of TcVI isolate was confirmed by hybridizations assays. However, cardiac lesions induced by TcIII + TcVI mixture were the same that the induced by TcIII isolate alone, suggesting that the presence of TcIII, perhaps in an early infection, modifies the alterations that TcVI alone could cause in the co-infected mice, in spite of the TcIII isolate is not detected in this mixture. On the other hand, skeletal muscle samples of animals co-infected with TcVI + (TcV or TcIII) showed intermediate values of damage compared to single infections, indicating that the combination of two isolates could modify the expected lesions in this tissue. These results could be due to the ability of each isolate to infect cells since it is known that T. cruzi is capable of infecting a wide variety of host cells, but the persistence of this parasite in cardiac and skeletal muscles depends on its ability to enter in the host cell and in the interaction with this [44]. On the other hand, it has been reported that the co-infections between different strains of T. cruzi trigger both protective inflammatory immunity and regulatory immune mechanisms that attenuate the damage in heart in a mouse model [25].
Based on these observations we evidence biological interaction in our murine co-infection model. However, it is important to note that the prevalence of either one isolate or the another could vary according to the infection period in which blood samples are taken [25,28,45] as well as the analyzed tissue [22].
In spite of several studies demonstrating the presence of biological interactions within mammalian hosts [23,25,27,28,46], these works analyzed strains of T. cruzi from very different geographical regions. In the present work, the three T. cruzi isolates studied were obtained in the same temporal period, in the same geographical area and simultaneously introduced into the animal model.
Is important to note that the DTUs studied in this work have epidemiological relevance, since the TcV and TcVI are more prevalent in domestic cycles [34,42,47,48], whereas the TcIII DTU was found in domestic animals [34]. It is clear that the extrapolation of co-infection results obtained from animal models to the natural occurrence of the phenomena in other hosts should be carefully considered. We emphasize the predominance of TcVI in the mixture TcV + TcVI since this mixture was found in human blood samples in endemics areas of Argentina [42,49].
Many studies have shown that the biological differences between closely related DTUs are smaller than the biological differences among genetically divergent DTUs [14,15,19,27]. However, an interesting question that emerges from this work is why two genetically close strains of T. cruzi, as TcV and TcVI, exhibit opposite biological properties. Even the biological interaction between them could lead to the predominance of one isolate (TcVI) over the other (TcV), at least during the acute phase and in our specific experimental model. In this sense we believe that the interaction with the host immune response, as well as the mechanisms related to the regulation of the acute inflammatory response and the proteomic expression of different DTUs could also be contributing to the pathology of each isolate and its interactions.
Conclusions
The results presented in this work show that biological interactions among different genotypes of T. cruzi from the Chaco Region do occur in our experimental model. Consequently, our results demonstrate that the biological interaction between T. cruzi strains in a mammal host is phenomenon that could be occurring in natural cycles. However, to examine this hypothesis field studies involving natural hosts should be performed.
Acknowledgments
We are grateful to Marcela Portelli for technical help in the histological preparations, María Celia Mora for assistance in animal care and Dr. Julio Nasser for contributions to this study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This manuscript was supported by funding agency Argentina: Agencia Nacional de Promoción Científica y Tecnológica (http://www.agencia.mincyt.gob.ar), through a grant given to Dr. Patricio Diosque (PICT-2012-2174). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pedersen AB, Fenton A (2007) Emphasizing the ecology in parasite community ecology. Trends Ecol Evol 22: 133–139. [DOI] [PubMed] [Google Scholar]
- 2. Telfer S, Birtles R, Bennett M, Lambin X, Paterson S, et al. (2008) Parasite interactions in natural populations: insights from longitudinal data. Parasitology 135: 767–781. 10.1017/S0031182008000395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cox FE (2001) Concomitant infections, parasites and immune responses. Parasitology 122 Suppl: S23–38. [DOI] [PubMed] [Google Scholar]
- 4. Bastrenta B, Mita N, Buitrago R, Vargas F, Flores M, et al. (2003) Human mixed infections of Leishmania spp. and Leishmania-Trypanosoma cruzi in a sub Andean Bolivian area: identification by polymerase chain reaction/hybridization and isoenzyme. Mem Inst Oswaldo Cruz 98: 255–264. [DOI] [PubMed] [Google Scholar]
- 5. Mendes DG, Lauria-Pires L, Nitz N, Lozzi SP, Nascimento RJ, et al. (2007) Exposure to mixed asymptomatic infections with Trypanosoma cruzi, Leishmania braziliensis and Leishmania chagasi in the human population of the greater Amazon. Trop Med Int Health 12: 629–636. [DOI] [PubMed] [Google Scholar]
- 6. Veland N, Valencia BM, Alba M, Adaui V, Llanos-Cuentas A, et al. (2013) Simultaneous infection with Leishmania (Viannia) braziliensis and L. (V.) lainsoni in a Peruvian patient with cutaneous leishmaniasis. Am J Trop Med Hyg 88: 774–777. 10.4269/ajtmh.12-0594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bell AS, de Roode JC, Sim D, Read AF (2006) Within-host competition in genetically diverse malaria infections: parasite virulence and competitive success. Evolution 60: 1358–1371. [PubMed] [Google Scholar]
- 8. Dinko B, Oguike MC, Larbi JA, Bousema T, Sutherland CJ (2013) Persistent detection of Plasmodium falciparum, P. malariae, P. ovale curtisi and P. ovale wallikeri after ACT treatment of asymptomatic Ghanaian school-children. Int J Parasitol Drugs Drug Resist 3: 45–50. 10.1016/j.ijpddr.2013.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Flores-Mendoza C, Fernandez R, Escobedo-Vargas KS, Vela-Perez Q, Schoeler GB (2004) Natural Plasmodium infections in Anopheles darlingi and Anopheles benarrochi (Diptera: Culicidae) from eastern Peru. J Med Entomol 41: 489–494. [DOI] [PubMed] [Google Scholar]
- 10. Balmer O, Tostado C (2006) New fluorescence markers to distinguish co-infecting Trypanosoma brucei strains in experimental multiple infections. Acta Trop 97: 94–101. [DOI] [PubMed] [Google Scholar]
- 11. Ezeokonkwo RC, Ezeh IO, Onunkwo JI, Obi PO, Onyenwe IW, et al. (2010) Comparative haematological study of single and mixed infections of mongrel dogs with Trypanosoma congolense and Trypanosoma brucei brucei . Vet Parasitol 173: 48–54. 10.1016/j.vetpar.2010.06.020 [DOI] [PubMed] [Google Scholar]
- 12. Omeje JN, Anene BM (2012) Comparative serum biochemical changes induced by experimental infection of T. brucei and T. congolense in pigs. Vet Parasitol 190: 368–374. 10.1016/j.vetpar.2012.07.008 [DOI] [PubMed] [Google Scholar]
- 13. Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM, et al. (2012) The revised Trypanosoma cruzi subspecific nomenclature: rationale, epidemiological relevance and research applications. Infect Genet Evol 12: 240–253. 10.1016/j.meegid.2011.12.009 [DOI] [PubMed] [Google Scholar]
- 14. Dos Reis D, Monteiro WM, Bossolani GD, Teston AP, Gomes ML, et al. (2012) Biological behaviour in mice of Trypanosoma cruzi isolates from Amazonas and Parana, Brazil. Exp Parasitol 130: 321–329. 10.1016/j.exppara.2012.02.016 [DOI] [PubMed] [Google Scholar]
- 15. dos Santos DM, Talvani A, Guedes PM, Machado-Coelho GL, de Lana M, et al. (2009) Trypanosoma cruzi: Genetic diversity influences the profile of immunoglobulins during experimental infection. Exp Parasitol 121: 8–14. 10.1016/j.exppara.2008.09.012 [DOI] [PubMed] [Google Scholar]
- 16. Lisboa CV, Pinho AP, Monteiro RV, Jansen AM (2007) Trypanosoma cruzi (kinetoplastida Trypanosomatidae): biological heterogeneity in the isolates derived from wild hosts. Exp Parasitol 116: 150–155. [DOI] [PubMed] [Google Scholar]
- 17. Ragone PG, Perez Brandan C, Padilla AM, Monje Rumi M, Lauthier JJ, et al. (2012) Biological behavior of different Trypanosoma cruzi isolates circulating in an endemic area for Chagas disease in the Gran Chaco region of Argentina. Acta Trop 123: 196–201. 10.1016/j.actatropica.2012.05.003 [DOI] [PubMed] [Google Scholar]
- 18. Revollo S, Oury B, Laurent JP, Barnabe C, Quesney V, et al. (1998) Trypanosoma cruzi: impact of clonal evolution of the parasite on its biological and medical properties. Exp Parasitol 89: 30–39. [DOI] [PubMed] [Google Scholar]
- 19. Toledo MJ, de Lana M, Carneiro CM, Bahia MT, Machado-Coelho GL, et al. (2002) Impact of Trypanosoma cruzi clonal evolution on its biological properties in mice. Exp Parasitol 100: 161–172. [DOI] [PubMed] [Google Scholar]
- 20. del Puerto R, Nishizawa JE, Kikuchi M, Iihoshi N, Roca Y, et al. (2010) Lineage analysis of circulating Trypanosoma cruzi parasites and their association with clinical forms of Chagas disease in Bolivia. PLoS Negl Trop Dis 4: e687 10.1371/journal.pntd.0000687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Macedo AM, Machado CR, Oliveira RP, Pena SD (2004) Trypanosoma cruzi: genetic structure of populations and relevance of genetic variability to the pathogenesis of chagas disease. Mem Inst Oswaldo Cruz 99: 1–12. [DOI] [PubMed] [Google Scholar]
- 22. Andrade LO, Machado CR, Chiari E, Pena SD, Macedo AM (1999) Differential tissue distribution of diverse clones of Trypanosoma cruzi in infected mice. Mol Biochem Parasitol 100: 163–172. [DOI] [PubMed] [Google Scholar]
- 23. Franco DJ, Vago AR, Chiari E, Meira FC, Galvao LM, et al. (2003) Trypanosoma cruzi: mixture of two populations can modify virulence and tissue tropism in rat. Exp Parasitol 104: 54–61. [DOI] [PubMed] [Google Scholar]
- 24. Andrade SG, Campos RF, Sobral KS, Magalhaes JB, Guedes RS, et al. (2006) Reinfections with strains of Trypanosoma cruzi, of different biodemes as a factor of aggravation of myocarditis and myositis in mice. Rev Soc Bras Med Trop 39: 1–8. [DOI] [PubMed] [Google Scholar]
- 25. Rodrigues CM, Valadares HM, Francisco AF, Arantes JM, Campos CF, et al. (2010) Coinfection with different Trypanosoma cruzi strains interferes with the host immune response to infection. PLoS Negl Trop Dis 4: e846 10.1371/journal.pntd.0000846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martins HR, Silva RM, Valadares HM, Toledo MJ, Veloso VM, et al. (2007) Impact of dual infections on chemotherapeutic efficacy in BALB/c mice infected with major genotypes of Trypanosoma cruzi . Antimicrob Agents Chemother 51: 3282–3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martins HR, Toledo MJ, Veloso VM, Carneiro CM, Machado-Coelho GL, et al. (2006) Trypanosoma cruzi: Impact of dual-clone infections on parasite biological properties in BALB/c mice. Exp Parasitol 112: 237–246. [DOI] [PubMed] [Google Scholar]
- 28. Sales-Campos H, Kappel HB, Andrade CP, Lima TP, Mattos ME Jr., et al. (2014) A DTU-dependent blood parasitism and a DTU-independent tissue parasitism during mixed infection of Trypanosoma cruzi in immunosuppressed mice. Parasitol Res 113: 375–385. 10.1007/s00436-013-3665-z [DOI] [PubMed] [Google Scholar]
- 29. Toledo MJ, Bahia MT, Carneiro CM, Martins-Filho OA, Tibayrenc M, et al. (2003) Chemotherapy with benznidazole and itraconazole for mice infected with different Trypanosoma cruzi clonal genotypes. Antimicrob Agents Chemother 47: 223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Burgos JM, Diez M, Vigliano C, Bisio M, Risso M, et al. (2010) Molecular identification of Trypanosoma cruzi discrete typing units in end-stage chronic Chagas heart disease and reactivation after heart transplantation. Clin Infect Dis 51: 485–495. 10.1086/655680 [DOI] [PubMed] [Google Scholar]
- 31. Coronado X, Zulantay I, Albrecht H, Rozas M, Apt W, et al. (2006) Variation in Trypanosoma cruzi clonal composition detected in blood patients and xenodiagnosis triatomines: implications in the molecular epidemiology of Chile. Am J Trop Med Hyg 74: 1008–1012. [PubMed] [Google Scholar]
- 32. Diez C, Lorenz V, Ortiz S, Gonzalez V, Racca A, et al. (2010) Genotyping of Trypanosoma cruzi sublineage in human samples from a North-East Argentina area by hybridization with DNA probes and specific polymerase chain reaction (PCR). Am J Trop Med Hyg 82: 67–73. 10.4269/ajtmh.2010.09-0391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Garcia A, Ortiz S, Iribarren C, Bahamonde MI, Solari A (2014) Congenital co-infection with different Trypanosoma cruzi lineages. Parasitol Int 63: 138–139. [PubMed] [Google Scholar]
- 34. Cardinal MV, Lauricella MA, Ceballos LA, Lanati L, Marcet PL, et al. (2008) Molecular epidemiology of domestic and sylvatic Trypanosoma cruzi infection in rural northwestern Argentina. Int J Parasitol 38: 1533–1543. 10.1016/j.ijpara.2008.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rozas M, Botto-Mahan C, Coronado X, Ortiz S, Cattan PE, et al. (2007) Coexistence of Trypanosoma cruzi genotypes in wild and periodomestic mammals in Chile. Am J Trop Med Hyg 77: 647–653. [PubMed] [Google Scholar]
- 36. Bosseno MF, Yacsik N, Vargas F, Breniere SF (2000) Selection of Trypanosoma cruzi clonal genotypes (clonet 20 and 39) isolated from Bolivian triatomines following subculture in liquid medium. Mem Inst Oswaldo Cruz 95: 601–607. [DOI] [PubMed] [Google Scholar]
- 37. Grossblatt A (1996) Guide for the use and care of laboratory animals. National Academy Press. [Google Scholar]
- 38. Lauthier JJ, Tomasini N, Barnabe C, Rumi MM, D'Amato AM, et al. (2012) Candidate targets for Multilocus Sequence Typing of Trypanosoma cruzi: validation using parasite stocks from the Chaco Region and a set of reference strains. Infect Genet Evol 12: 350–358. 10.1016/j.meegid.2011.12.008 [DOI] [PubMed] [Google Scholar]
- 39. Kollien AH, Schaub GA (2000) The development of Trypanosoma cruzi in triatominae. Parasitol Today 16: 381–387. [DOI] [PubMed] [Google Scholar]
- 40. Gomes ML, Macedo AM, Vago AR, Pena SD, Galvao LM, et al. (1998) Trypanosoma cruzi: optimization of polymerase chain reaction for detection in human blood. Exp Parasitol 88: 28–33. [DOI] [PubMed] [Google Scholar]
- 41. Veas F, Breniere SF, Cuny G, Brengues C, Solari A, et al. (1991) General procedure to construct highly specific kDNA probes for clones of Trypanosoma cruzi for sensitive detection by polymerase chain reaction. Cell Mol Biol 37: 73–84. [PubMed] [Google Scholar]
- 42. Rumi MM, Perez Brandan C, Gil JF, D'Amato AM, Ragone PG, et al. (2013) Benznidazole treatment in chronic children infected with Trypanosoma cruzi: serological and molecular follow-up of patients and identification of Discrete Typing Units. Acta Trop 128: 130–136. 10.1016/j.actatropica.2013.07.003 [DOI] [PubMed] [Google Scholar]
- 43. Pena DA, Eger I, Nogueira L, Heck N, Menin A, et al. (2011) Selection of TcII Trypanosoma cruzi population following macrophage infection. J Infect Dis 204: 478–486. 10.1093/infdis/jir292 [DOI] [PubMed] [Google Scholar]
- 44. Andrade LO, Andrews NW (2005) The Trypanosoma cruzi-host-cell interplay: location, invasion, retention. Nat Rev Microbiol 3: 819–823. [DOI] [PubMed] [Google Scholar]
- 45. D'Avila DA, Macedo AM, Valadares HM, Gontijo ED, de Castro AM, et al. (2009) Probing population dynamics of Trypanosoma cruzi during progression of the chronic phase in chagasic patients. J Clin Microbiol 47: 1718–1725. 10.1128/JCM.01658-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lo Presti MS, Esteves BH, Moya D, Bazan PC, Strauss M, et al. (2014) Circulating Trypanosoma cruzi populations differ from those found in the tissues of the same host during acute experimental infection. Acta Trop 133: 98–109. 10.1016/j.actatropica.2014.02.010 [DOI] [PubMed] [Google Scholar]
- 47. Corrales RM, Mora MC, Negrette OS, Diosque P, Lacunza D, et al. (2009) Congenital Chagas disease involves Trypanosoma cruzi sub-lineage IId in the northwestern province of Salta, Argentina. Infect Genet Evol 9: 278–282. 10.1016/j.meegid.2008.12.008 [DOI] [PubMed] [Google Scholar]
- 48. Diosque P, Barnabe C, Padilla AM, Marco JD, Cardozo RM, et al. (2003) Multilocus enzyme electrophoresis analysis of Trypanosoma cruzi isolates from a geographically restricted endemic area for Chagas' disease in Argentina. Int J Parasitol 33: 997–1003. [DOI] [PubMed] [Google Scholar]
- 49. Monje-Rumi MM, Brandan CP, Ragone PG, Tomasini N, Lauthier JJ, et al. (2014) Trypanosoma cruzi diversity in the Gran Chaco: Mixed infections and differential host distribution of TcV and TcVI. Infect Genet Evol 29C: 53–59. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.