Abstract
Predicting the impact of genetic variation on human health remains an important and difficult challenge. Often, algorithmic classifiers are tasked with predicting binary traits (e.g. positive or negative for a disease) from missense variation. Though useful, this arrangement is limiting and contrived, because human diseases often comprise a spectrum of severities, rather than a discrete partitioning of patient populations. Furthermore, labeling variants as causal or benign can be error prone, which is problematic for training supervised learning algorithms (the so-called garbage in, garbage out phenomenon). We explore the potential value of training classifiers using continuous-valued quantitative measurements, rather than binary traits. Using 20 variants from cystic fibrosis transmembrane conductance regulator (CFTR) nucleotide-binding domains and six quantitative measures of cystic fibrosis (CF) severity, we trained classifiers to predict CF severity from CFTR variants. Employing cross validation, classifier prediction and measured clinical/functional values were significantly correlated for four of six quantitative traits (correlation P-values from 1.35 × 10−4 to 4.15 × 10−3). Classifiers were also able to stratify variants by three clinically relevant risk categories with 85–100% accuracy, depending on which of the six quantitative traits was used for training. Finally, we characterized 11 additional CFTR variants using clinical sweat chloride testing, two functional assays, or all three diagnostics, and validated our classifier using blind prediction. Predictions were within the measured sweat chloride range for seven of eight variants, and captured the differential impact of specific variants on the two functional assays. This work demonstrates a promising and novel framework for assessing the impact of genetic variation.
Introduction
The ability to accurately diagnose and optimally manage disease from a patients unique molecular profile is paramount to the realization of individualized medicine (1). The accumulation of databases that catalog mutations putatively causal of disease provide the opportunity to develop general principles for interpreting genetic variation (2,3). These principles can be encoded in algorithms capable of utilizing the large variant databases for training and/or testing, and the resulting classifiers employed for predicting the disease liability of previously unclassified variants (2–4). When assessed independently across multiple variant databases, popular contemporary methods typically achieve classification accuracies of ∼60 to ∼80% (5–7). Unfortunately, the performance of individual methods can vary significantly between genes and databases (5–8). In addition, there is often a significant imbalance in prediction sensitivity and specificity (e.g. high sensitivity, but unacceptably low specificity). These large changes in predictive performance in different settings significantly limit the clinical utility of current methods for assessing the consequence of genetic variation.
The inability of contemporary methods to achieve consistent and clinically relevant performance highlights methodological limitations. And in many cases, simplifying assumptions about complex clinical phenotypes could directly contribute to misclassification (or perceived misclassification). Even though it is increasingly acknowledged that many human diseases fall on a spectrum of severity (9–15), classifiers are commonly tasked with partitioning patients into discrete populations as positive or negative for a given phenotype (5–8,16). Similarly, supervised learning algorithms are often trained with genetic variants that have been partitioned into two classes (e.g. disease causing and neutral), ignoring the relative severity within the classes (3,17). Also confounding is the presence of variants associated with mild-to-moderate phenotypes or incomplete penetrance (18). When labeled as disease causing or neutral for the purposes of algorithmic training, these “borderline” variants could have a negative impact on the performance of the resulting classifier.
One improvement to supervised learning algorithms might be to utilize quantitative correlates of phenotype, known as endophenotypes (19–21). Initially formulated in entomology studies and popularized in psychiatric fields, the concept of the endophenotype is now being considered in other disease areas (19,21,22). As an example, in a population dichotomized as having coronary heart disease (CHD) or not, each patient will have continuous-valued risk factors of disease such as blood pressure, cholesterol level or coronary calcium score (19). Labeling a patient as positive for the CHD phenotype does not explicitly indicate disease severity. This could be problematic for classifier training because variants with significantly different impact on disease could be in the same class. Additionally, diagnosis of borderline cases can be subjective and varied. For instance, a variant associated with 30–60% narrowing of a major coronary artery (stenosis) could reasonably be considered neutral, borderline or CHD causing (23), and could be differentially classified across multiple clinical studies. Conversely, an endophenotype such as blood pressure can influence CHD severity, and is a quantitative measurement rather than an arbitrary clinical interpretation. Therefore, endophenotypic data could limit contamination introduced by human interpretation or the information loss associated with dichotomization, and might have benefits with respect to training algorithmic classifiers.
The cystic fibrosis transmembrane conductance regulator (CFTR) protein is a 1480 residue chloride channel encoded for by the CFTR gene (24); the disease cystic fibrosis (CF) is caused by variants in the CFTR gene. The recently established clinical and functional translation of CFTR (CFTR2) database catalogs data from ∼40 000 individuals with CF (25). Among these individuals, 1044 distinct genetic CFTR variations were found, with 159 of these variants having an allele frequency of 0.01% or greater and accounting for 96.4% of all variants observed. These 159 variants include 64 missense variants, for which the CFTR2 database includes endophenotypic data for up to six parameters, including clinical traits in patients carrying those variants and in vitro functional assays. Of these 64 missense variants, there are 20 variants that reside in CFTR nucleotide-binding domains (NBD) and have data available for all six endophenotypic measurements.
We recently developed a supervised learning algorithm called phenotype-optimized sequence ensemble (POSE) (26). Provided a multiple sequence alignment (MSA) and variants of known phenotypic impact, POSE isolates an optimal set of sequences for predicting the phenotype. Once this optimized MSA is created, POSE can use the alignment to assess the impact of other variants in the target gene. When tasked with predicting CF disease from mutation in the CFTR protein, the POSE method had significantly higher prediction accuracy than other popular methods tested using the same variants. POSE-derived MSAs also improved the accuracy of other methods, relative to using their default MSAs (26).
For this study, we extended the utility of POSE to account for quantitative disease risk factors, by facilitating the use of continuous-valued endophenotypic data for training. The expanded algorithm also now includes the option of using 3D protein structure for training and prediction. Here, we explore the potential value of using these continuous-valued quantitative traits for the purposes of classifier training, and predict CF disease liability as a function of CFTR variation using endophenotypic data from six clinical and functional assays. Training and prediction from a leave-one-out cross-validation strategy applied to 20 CFTR variants results in high predictive performance of both continuous-valued endophenotypes and annotated CF phenotypes. Finally, we clinically and functionally characterized 11 additional CFTR variants to validate our classifier using blind prediction; predicted and measured endophenotypes were in broad agreement. This novel approach, of training a supervised learning algorithm with disease endophenotypes for the subsequent prediction of both endophenotype and phenotype, could be of immediate utility for prioritizing functional assays, further elucidating pathogenesis, and as a complement to existing CF diagnostics.
Results
For this work, we further developed our POSE algorithm, such that continuous-valued quantitative phenotypes (endophenotypes) could be used to train the classifier. We tested this expanded functionality by predicting CF disease liability from CFTR amino acid substitution using six different clinical and functional data types for training. For each of these six endophenotypes, individually, the POSE algorithm trained using all but one of the variants, and prediction was made on that remaining variant; this process was repeated for each variant (i.e. a leave-one-out strategy). Importantly, the residue position for the variant being predicted on was never present in the training set. For example, when predictions were made for R560T, R560K was absent from the training set, and vice versa, because both variants occur at residue 560. Our evaluation is performed in three phases. First, we assess the correlation between prediction (POSE score) and measured endophenotype. Second, we threshold the continuous-valued POSE scores to compare predictions with annotated CF phenotypes. Finally, we perform blind validation by clinically or functionally evaluating 11 additional CFTR variants and comparing those measurements with POSE predictions.
Correlation between prediction and endophenotype varied considerably among the different CFTR domains. Calculations considering all variants, or variants from either the NBDs or transmembrane domains (TMDs) separately revealed significantly better performance in the NBDs. Correlation between POSE prediction and endophenotype was low for CFTR TMD variants, where predictions were not significant for any of the six endophenotypes (P-values < 0.05; see Supplementary Material, Table S1). While predictions for some endophenotypes were statistically significant when all variants were considered, correlations were generally low (Supplementary Material, Table S2). Conversely, correlation was generally high for CFTR NBD variants, and predictions were statistically significant for four of six endophenotypes (see below). Because current algorithmic performance is too low to reliably aid in the interpretation of TMD variants, we restrict further analysis and additional validation to variants in the NBDs.
Table 1 shows variant-specific endophenotype data for the NBD variants used in this study, with the corresponding POSE scores obtained from leave-one-out cross validation. This includes all 20 NBD variants from the CFTR2 database that had data available across all six endophenotypes. Additionally, the annotated phenotype is shown in Table 1. See Materials and Methods for a complete description of all endophenotypic data and phenotype definitions.
Table 1.
Twenty CFTR nucleotide-binding domain mutations from the CFTR2 database with corresponding endophenotypic measurements, POSE scores and annotated phenotype
| Variant | Sweat [Cl−] | PI | P.a. infection | [Cl−] cond. | C/(C + B) | FEV1%pred | Phenotype |
|---|---|---|---|---|---|---|---|
| A455E | 83 (0.86) | 53.5 (0.70) | 46.2 (0.88) | 6.8 (0.74) | 0 (1.12) | 75.7 (0.52) | CF-causing |
| L467P | 97 (1.53) | 100 (1.54) | 18.2 (1.62) | 0 (1.61) | 0 (1.61) | 83.7 (1.62) | CF-causing |
| S492F | 72 (0.26) | 20 (0.49) | 60 (0.40) | 0 (0.34) | 0 (0.37) | 67.9 (0.23) | CF-causing |
| V520F | 108 (1.13) | 100 (1.31) | 43.6 (1.49) | 0.2 (1.15) | 0 (1.43) | 78.2 (1.06) | CF-causing |
| S549N | 101 (0.89) | 93.9 (0.89) | 53.2 (0.89) | 1.6 (0.88) | 95 (0.83) | 72.7 (0.89) | CF-causing |
| S549R | 109 (0.89) | 95.5 (0.89) | 54.5 (0.88) | 0.1 (0.88) | 75.3 (0.83) | 72 (0.89) | CF-causing |
| G551D | 104 (0.99) | 97.8 (1.30) | 58 (1.31) | 1.3 (1.24) | 99.4 (0.70) | 74.4 (1.24) | CF-causing |
| A559T | 99 (1.55) | 96.4 (1.56) | 55.6 (1.56) | 0 (1.43) | 0 (1.56) | 72.3 (1.56) | CF-causing |
| R560K | 112 (0.86) | 100 (0.82) | 40 (0.84) | 0 (0.71) | 0 (0.85) | 71.5 (1.00) | CF-causing |
| R560T | 103 (0.86) | 99.2 (0.82) | 66.1 (0.84) | 0.1 (0.73) | 0 (0.87) | 76.2 (1.00) | CF-causing |
| G1244E | 99 (1.37) | 81.8 (1.50) | 54.2 (1.50) | 1 (1.36) | 98.8 (0.99) | 73.6 (1.50) | CF-causing |
| S1251N | 87 (0.86) | 88.7 (0.89) | 51.9 (0.81) | 5.2 (0.87) | 98.8 (0.60) | 61.5 (0.62) | CF-causing |
| N1303K | 103 (1.10) | 95.7 (1.07) | 54.4 (1.09) | 0.5 (1.03) | 10.5 (1.10) | 72.5 (1.10) | CF-causing |
| D579G | 75 (1.0) | 72.7 (1.00) | 30 (1.00) | 13.9 (0.93) | 76.3 (0.80) | 82.7 (1.00) | Indeterminate |
| D614G | 72 (0.76) | 0 (0.77) | 20 (0.53) | 18 (0.63) | 40.9 (0.65) | 59.2 (0.56) | Indeterminate |
| I1234V | 99 (0.32) | 40 (0.79) | 50 (0.82) | 39.9 (0.36) | 98.8 (0.68) | 79.8 (0.46) | Indeterminate |
| D1270N | 65 (0.50) | 25 (0.5) | 11.1 (0.53) | 53.2 (0.50) | 97.4 (0.39) | 92 (0.52) | Indeterminate |
| M470V | 81 (−0.01) | 57.1 (0.01) | 21.4 (0.00) | 95.7 (0.00) | 93 (0.17) | 83.3 (0.02) | Not CF-causing |
| G576A | 50 (−0.01) | 30 (−0.06) | 8.3 (−0.09) | 147 (0.03) | 98.5 (0.31) | 91.7 (−0.01) | Not CF-causing |
| S1235R | 54 (0.08) | 57.9 (0.05) | 25 (0.08) | 78.7 (0.09) | 95.9 (0.21) | 87.4 (−0.22) | Not CF-causing |
Sweat [Cl−] is the mean sweat chloride for patients with the variant; PI is the percentage of patients displaying pancreatic insufficiency; P.a. infection is the percentage of patients with P. aeruginosa infection; [Cl−] cond. is the mean chloride conductance for cells expressing the CFTR variant; C/(C + B) estimates the fraction of properly processed (“mature”) CFTR protein; FEV1%pred is the mean lung function as a percent of wild type. Increasing sweat [Cl−], PI and psuedo are each associated with increasing CF severity. For [Cl−] cond., C/(C + B) and FEV1%pred, decreasing values are associated with increasing CF severity. For POSE scores, shown in parentheses, increasing values always correspond to increasing disease severity. See Endophenotypic data and Annotated phenotype for a detailed description of functional–clinical endophenotypes and phenotype definitions.
Figure 1A–E shows the correlation between prediction (POSE score) and each of the six endophenotypes. Each data point is the measured endophenotype versus POSE score for a given variant, where each POSE score resulted from leave-one-out cross validation. For POSE, increasing score indicates a prediction of increasing disease severity (possible range is −3.0 to 3.0, where zero is equivalent to wild type and negative values indicate a protective variant). For each of the six endophenotypes, the measured values and POSE scores showed the expected, general correlation. For instance, increasing sweat chloride, pancreatic insufficiency and Pseudomonas infection are each associated with increasing CF disease severity; POSE predictions captured this positive correlation. Chloride-conductance, fraction of mature-to-total protein and lung function endophenotypes are all inversely associated with CF disease severity; for these three endophenotypes, POSE predictions captured this inverse correlation.
Figure 1.
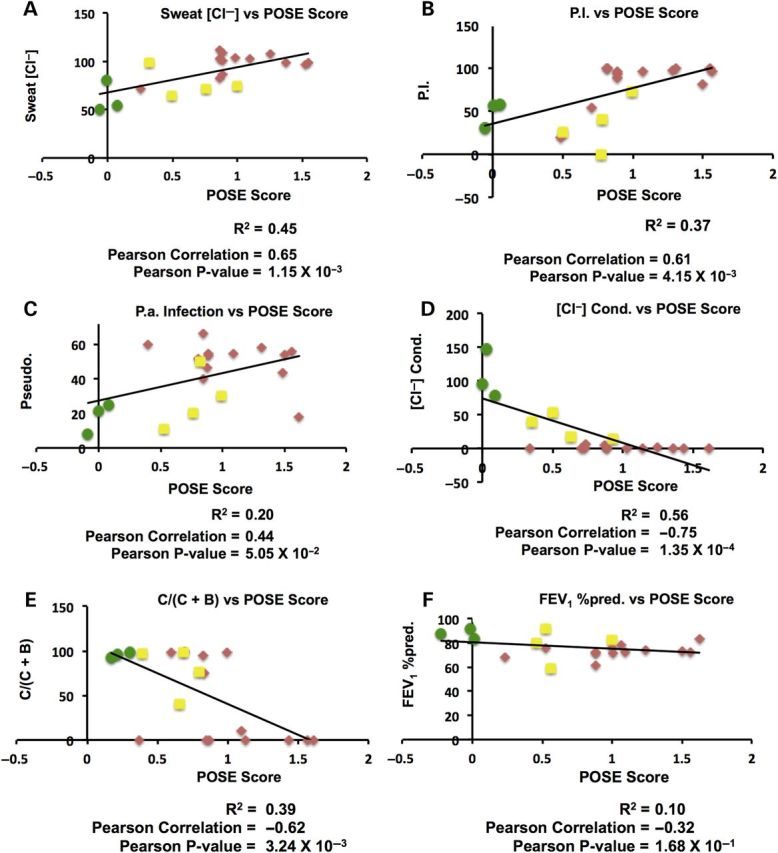
Correlation of POSE score with six individual CFTR endophenotypes. Each plot shows the results of 20 leave-one-out POSE calculations (i.e. one data point for each mutation), trained using the corresponding endophenotypic data. In each plot, the X-axis is the predicted impact (POSE score) for each of 20 CFTR amino acid substitutions, and the Y-axis is the experimentally or clinically determined endophenotype. Green circles, yellow squares, and red diamonds denote the not CF-causing, indeterminate and CF-causing annotated phenotypes, respectively. See Table 1 for a description of each endophenotypic measurement type and annotated phenotypes.
Across all 20 variants, POSE scores were highly predictive of mean patient sweat chloride, achieving an R2 and Pearson correlation of 0.45 and 0.65, respectively, and a Pearson P-value of 1.15 × 10−3 (Fig. 1A). POSE score was also highly correlated with pancreatic insufficiency (Fig. 1B), having correlation statistics similar with that of the sweat chloride predictions. POSE score was only moderately correlated with the percentage of patients with Pseudomonas aeruginosa infection, achieving an R2 of 0.20, a Pearson correlation coefficient of 0.44 and a Pearson correlation P-value of 0.05 (Fig. 1C). Overall correlation of predicting variant-specific chloride conductance in Fisher rat thyroid cells was high, achieving an R2 of 0.56, Pearson correlation of −0.75 and a Pearson P-value of 1.35 × 10−4 (Fig. 1D); training with this endophenotype resulted in the highest overall correlation among the six endophenotypes. POSE scores did reasonably well at predicting the ratio of mature-to-total CFTR protein in HeLa cells, as a function of mutation, across the 20 leave-one-out calculations (Fig. 1E). Patient lung function, measured as the forced expiratory volume in 1 s (FEV1%pred), was the most difficult endophenotype to predict using the POSE algorithm (Fig. 1F). While POSE scores did show the expected negative correlation with FEV1%pred measurements, the associated P-value was 0.168 and the R2 and Pearson correlation was only 0.10 and −0.32, respectively.
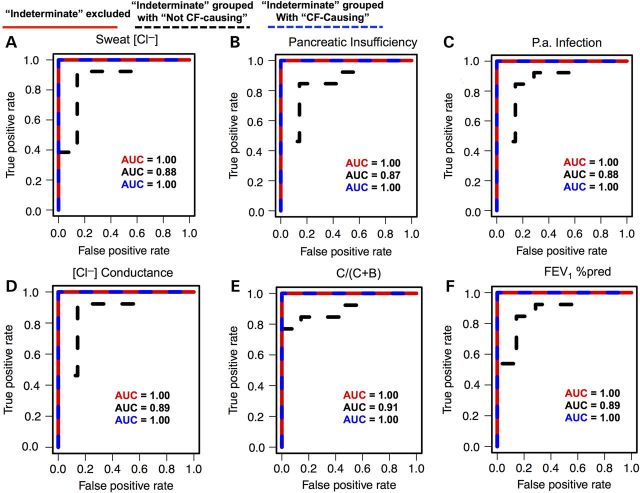
Next, we assessed the agreement between POSE scores and the annotated phenotype associated with each of the 20 variants (see Table 1). This is useful because an endophenotype-trained POSE classifier could be used to help inform clinical diagnoses in the same way the functional and clinical assays currently help inform clinical diagnoses. And for the abovementioned reasons, endophenotypic data can limit contamination and information loss relative to phenotypic data, which could make endophenotype-trained classifiers more reliable than phenotype-trained classifiers. To do this, we performed area under the curve (AUC), receiver–operator characteristic (ROC) analysis using POSE scores as the continuous variable and annotated phenotypes as the binary variable (Fig. 2A–F). Table 2 shows the corresponding sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV). Dichotomization of POSE scores to calculate these predictive performance statistics was achieved by choosing the cutoff that maximized balanced accuracy (arithmetic mean of sensitivity and specificity). The 20 variants were previously annotated as being associated with three distinct CF phenotypes (25), classified as not CF-causing, indeterminate and CF-causing. Dichotomization of the tripartite data was achieved by either removing the indeterminate group (“indeterminate excluded”), grouping the indeterminate and not CF-causing variants (“indeterminate + not CF-causing”) or grouping the indeterminate and CF-causing variants (“indeterminate + CF-causing”). As an example, for binarization using the indeterminate + CF-causing scheme, the negative class comprises only variants labeled not CF-causing, and the positive class comprises variants labeled indeterminate and CF-causing.
Figure 2.
ROC analysis for prediction of CF phenotypes from POSEs trained using six individual CFTR endophenotypes. Clinical phenotypes were defined as not CF-causing, indeterminate or CF-causing. To achieve dichotomization, variants defined as indeterminate were either excluded, grouped with variants defined as CF causing or grouped with the not CF-causing class (i.e. three individual ROC curves per chart). Continuous variables are POSE scores resulting from 20 leave-one-out calculations trained on each of six individual endophenotypes. AUC is the area under the curve. See Table 1 for a description of each endophenotypic measurement.
Table 2.
Predictive performance for classifying CF phenotypes from POSEs trained using six individual CFTR endophenotypes
| Endophenotype | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |
|---|---|---|---|---|---|
| “Indeterminate” excluded | Sweat [Cl−] | 100 | 100 | 100 | 100 |
| PI | 100 | 100 | 100 | 100 | |
| P.a. infection | 100 | 100 | 100 | 100 | |
| [Cl−] cond. | 100 | 100 | 100 | 100 | |
| C/(C + B) | 100 | 100 | 100 | 100 | |
| FEV1%pred. | 100 | 100 | 100 | 100 | |
| “Indeterminate” grouped with “not CF-causing” | Sweat [Cl−] | 92 | 86 | 86 | 92 |
| PI | 85 | 86 | 75 | 92 | |
| P.a. infection | 85 | 86 | 75 | 92 | |
| [Cl−] cond. | 92 | 86 | 86 | 92 | |
| C/(C + B) | 77 | 100 | 70 | 100 | |
| FEV1%pred. | 85 | 86 | 75 | 92 | |
| “Indeterminate” grouped with “CF-causing” | Sweat [Cl−] | 100 | 100 | 100 | 100 |
| PI | 100 | 100 | 100 | 100 | |
| P.a. infection | 100 | 100 | 100 | 100 | |
| [Cl−] cond. | 100 | 100 | 100 | 100 | |
| C/(C + B) | 100 | 100 | 100 | 100 | |
| FEV1%pred. | 100 | 100 | 100 | 100 |
Comparing Figure 2A–E and Table 2, endophenotype-trained POSE classifiers were good predictors of annotated phenotype using any of the six endophenotypes for training. In addition, predictive performance metrics (AUC, sensitivity, specificity, PPV and NPV) were generally good for any dichotomization of the tripartite phenotype annotations (Fig. 2A–E and Table 2). For the indeterminate excluded and indeterminate + CF-causing dichotomizations, five of the six endophenotype-trained POSE classifiers were perfect in all predictive performance metrics; POSE classifiers trained using patient P. aeruginosa infection show slightly reduced predictive performance for these phenotype groupings. For all six POSE classifiers, POSE scores were worse predictors of phenotype for the indeterminate + not CF-causing dichotomization, relative to the other two groupings. This result suggests that the POSE score function scores variants from the indeterminate group with a severity closer to the CF-causing group than the not CF-causing group. However, even for the indeterminate + not CF-causing dichotomization, POSE scores result in good separation of the classes.
POSE scores and endophenotypic measurements comparably partitioned the variants by annotated phenotype. For instance, along the X-axis in Figure 1A the not CF-causing variants (green circles) cluster around a POSE score of 0.0, indeterminate variants (red squares) have POSE scores from 0.32 to 1.0, and CF-causing variants (blue diamonds) have POSE scores partially overlapping with indeterminate variants and extending to 1.55 (see also Table 1). Looking at the Y-axis on the same plot, mean patient sweat chloride partitioned the indeterminate and CF-causing variants similar to the sweat chloride-trained POSE classifier. In addition, the sweat chloride-trained POSE classifier more distinctly separated the not CF-causing and indeterminate variants than do the actual sweat chloride measurements. Indeed, this trend is clear in almost all plots in Figure 1. Endophenotype-trained POSE classifiers separate the not CF-causing and indeterminate classes more distinctly than the corresponding endophenotypic measurements. And, separation of the indeterminate and CF-causing classes is comparable from the endophenotypes and the corresponding POSE scores. The exception to these observations occurs for the in vivo chloride-conductance measurements (Fig. 1D), which almost perfectly separates all three annotated phenotypes; notably, this endophenotypic data type also produced the best correlation with POSE score among all endophenotypes.
Finally, we sought to test our algorithm using a blinded validation scheme. We evaluated 11 CFTR variants that had sweat chloride, chloride-conductance and/or CFTR processing measurements. Authors responsible for deriving POSE predictions were not provided the measured data until after the predictions were shared. Table 3 shows measured and predicted endophenotypes, and POSE scores for this 11-variant validation set. POSE “predictions” were derived using the new POSE scores and the linear equations resulting from the initial leave-one-out cross validations, for each relevant endophenotype (see Fig. 1).
Table 3.
Measured and predicted endophenotypes for 11 newly characterized CFTR variants
| Variant | Sweat [Cl−] |
[Cl−] cond. |
C/(C + B) |
|||
|---|---|---|---|---|---|---|
| Measured | Predicted (score) | Measured | Predicted (score) | Measured | Predicted (score) | |
| Y569D | NA | NA | 0 | 5.4 (1.0) | 8 ± 7 | 11.4 (1.42) |
| A561E | NA | NA | 0 | 0 (1.6) | 8 ± 5 | 3.6 (1.53) |
| G1349D | NA | NA | 2.9 ± 0.9 | 19.5 (0.83) | 85 ± 6 | 76.4 (0.5) |
| G551S | 79.7 ± 28.9 | 93.4 (1.0) | 8.1 ± 3.4 | 0 (1.2) | 86 ± 10 | 67 (0.64) |
| Y563N | 97.0 ± 15.3 | 95.8 (1.1) | NA | NA | NA | NA |
| L1335P | 81.4 ± 31.6 | 65.4 (0) | NA | NA | NA | NA |
| T1246I | 63.9 ± 19.1 | 94.8 (1.1) | NA | NA | NA | NA |
| V456A | 62.6 ± 14.4 | 82.8 (0.6) | NA | NA | NA | NA |
| G622D | 78.2 ± 28.2 | 104.5 (1.4) | NA | NA | NA | NA |
| I502T | 96.5 ± 4.8 | 84.3 (0.7) | NA | NA | NA | NA |
| I1269N | 94.2 ± 10.6 | 95.3 (1.1) | NA | NA | NA | NA |
Measured endophenotype and corresponding standard deviation, POSE-predicted endophenotype and POSE score (parentheses) for each of 11 newly characterized CFTR variants. NA indicates that the specific measurement was not made for the corresponding mutation.
Measured and POSE-predicted endophenotypes were in broad agreement for the 11-variant validation set (Table 3). The predicted sweat chloride was within 1 SD of the mean for five of eight CFTR variants (sweat chloride measurements were not available for three of the 11 variants). Predicted and measured sweat chlorides were almost identical for the Y563N and I1269N variants (see Table 3). For I502T, the predicted sweat chloride level (84.3) is >1 SD from the measured sweat chloride (96.5 ± 4.8); however, both the measured and predicted values are indicative of a CF-causing variant. For variants T1246I and V456A, the comparison between prediction and clinical measurement is less clear. First, there is conflicting evidence for the role V456A in CF disease pathogenesis (27,28). One group reported that T1246 variants can significantly impact CFTR function (29). In this study, these variants have mean sweat chloride values associated with the indeterminate phenotype, but have individual measurements that span large ranges of disease liability (33.0–98.0; see the Supplementary Material, supplemental spreadsheet). POSE predictions suggest that both of these variants are CF-causing.
Functional data were available for four variants from the 11-variant validation set (Table 3). Cells expressing either the Y549D or A561E variants were predicted to exhibit severely reduced chloride-conductance and CFTR processing, as confirmed by the corresponding functional measurements. For G1349D and G551S, POSE predicts that the majority of CFTR protein is properly processed, but that cells expressing these variants are still poorly conductive. Remarkably, this mechanism of disease pathogenesis is supported by the functional measurements, for both the G1349D and G551S variants. A potential discrepancy between predicted and measured functional endophenotypes occurred for impact of G1349D on chloride conductance. For this variant, the prediction was indicative of the indeterminate phenotype, whereas the measured value clearly suggests a CF-causing phenotype (see Table 1).
Discussion
In this study, we explored the potential value of using continuous-valued quantitative traits, rather than binary traits, to inform the classification of missense variants. When available, these continuous-valued endophenotypes might provide some advantages for classifier training, relative to training with data that has been labeled as either positive or negative for a given trait. Labeling can be subjective and is a potential source of contamination, and there is information loss associated with ignoring relative severity within the classes. These sources of error can propagate downstream to algorithmic training, in turn causing error in subsequent predictions.
For a dataset of 20 CF-associated variants in CFTR NBDs, endophenotype-trained POSE classifiers show promise. Employing a leave-one-out cross-validation strategy, these classifiers were generally well correlated with the corresponding endophenotype. And in many cases, POSE scores more distinctly separated three annotated phenotypes than did the corresponding clinical/experimental measurements. This suggests that endophenotype-trained POSE classifiers might be worthy of consideration, along with existing clinical diagnostics, for assessing the disease liability of CFTR variants. POSE predictions might also be useful for prioritizing in vivo assays of CFTR function. For example, chloride conductance in Fisher rat thyroid cells is a good predictor of variant-specific disease liability, but it is far too expensive and time consuming to experimentally test all possible CFTR variants. In this report, we showed that POSE scores are correlated with chloride conductance, providing a tractable way to rank NBD missense CFTR variants and prioritize ongoing experiments.
Blind prediction on an additional 11 CF-associated CFTR variants also showed promising results. Particularly encouraging was the ability of endophenotype-trained POSEs to elucidate the different molecular mechanisms contributing to disease pathogenesis. POSE accurately predicted that variants G1349D and G551S impact CF disease via channel gating, rather than improper CFTR processing. For Y569D and A561E, POSE predicted an almost complete abrogation of CFTR processing, consistent with the C/(C + B) measurements. Because these variants result in a severe reduction of mature CFTR protein, gating is also expected to be low, consistent with both the measured and predicted chloride conductance.
One shortcoming of this work is the low correlation between POSE score and endophenotype for variants in CFTR TMDs. This result is consistent with that of a previous study, using an earlier version of POSE to study CF-causing CFTR variants (26); that study did not consider endophenotypes. The POSE score function relies entirely on sequences homologous to the target protein, and 3D structural data, when available. CFTR is a member of the large superfamily of ABC transporters, which bear high sequence similarity among the NBDs and very low sequence similarity among the TMDs (30,31). Similarly, high-resolution X-ray crystals structures are available for human CFTR NDBs, but not for human CFTR TMDs. Furthermore, high-resolution structural studies of bacterial ABC transporters show structurally conserved NBDs, but TMDs that lack significant structural homology (32). Also, CFTR is unique among ABC transporters in that CFTR is an ion channel, rather than a transporter. Given the lack of sequence and structural homology among ABC TMDs, it is not surprising that a method relying on sequence and structure would suffer in these domains. It is possible, however, that a different set of sequences in the initial MSA could improve overall prediction.
For the autosomal recessive disease phenylketonuria, there is evidence that phenylalanine hydroxylase variant severity, assessed by in vitro assays, does not have a linear correlation with disease severity (33). Considering that example, there is no reason to assume a priori that the relationship between experimental data and POSE score should be linear. One potential improvement to the POSE algorithm might be to include nonlinear regression during the sequence optimization (training) phase. Here, CFTR homologs were selected that maximized R2 for the linear regression of endophenotypes onto POSE scores. But, among the POSE score–endophenotype relationships shown in Figure 1A–E, the chloride conductance (Fig. 1D) and ratio of mature-to-total protein (Fig. 1E) appear exponential and sigmoidal, respectively. Figure 3 shows chloride conductance versus POSE score fit using a polynomial (Fig. 3A) and linear (Fig. 3B) regression; R2 is 30% greater when fit with a third-degree polynomial regression, relative to linear regression. Algorithmic performance might benefit from the addition of second- and third-degree polynomial and sigmoidal fitting during training, with the appropriate penalty to prevent over fitting.
Figure 3.
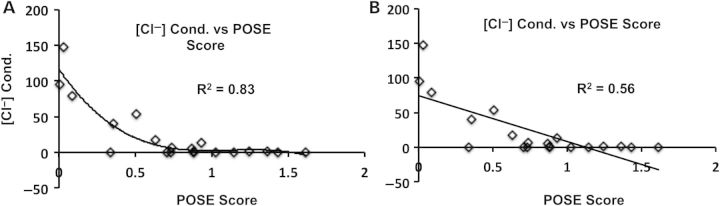
Correlation of chloride-conductance and POSE score using either a linear or exponential relationship. Variant-specific chloride-conductance ([Cl−] cond.) versus POSE score fit using a third-degree polynomial regression (A). For comparison, (B) shows the same data fit using a linear regression (same as Fig. 1D).
It is tempting to conclude that an apparent disagreement between POSE prediction and annotated phenotype indicates algorithmic misclassification. However, there is also disagreement across the measured endophenotypes, which might be instructive. For instance, S492F has an annotated phenotype of CF-causing, but the POSE classifier consistently classified this variant in the indeterminate regime. The lung function (FEV1%pred), mature-to-total protein, chloride-conductance and Pseudomonas infection endophenotypes are all indicative of a CF-causing classification for this variant. However, patient sweat chloride and pancreatic insufficiency rates suggest that this variant was milder. In reality, this variant could be differentially affecting different components of disease, could be associated with alternative disease mechanisms (deleterious to the airway in a manner that was disruptive to chloride conductance) or could be associated with less than complete penetrance. In this way, POSE classifiers might be informative for identifying variants deserving more study.
The continued improvement of methods for predicting the impact of genetic variation appears to be a recognized priority for the near future. It is also worth acknowledging the upper limits of these methods, such that efforts are allocated to achieve maximum benefit. Even for “textbook” monogenetic disorders such as CF, there are likely other endogenous factors that influence disease severity (34,35). These factors include expression levels, modifying mutations in other genes, small molecule or ion concentrations, etc. And of course, other exogenous or environmental factors can have significant influence on human disease (36). Indeed, highly accurate individualized medicine will likely require multi-scale approaches that consider everything from molecular-level phenomena to lifestyle circumstances (36). Methods such as POSE should be a necessary component of a larger algorithmic framework, utilizing diverse clinical and laboratory inputs.
Materials and Methods
Previously, we developed the POSE algorithm to predict the impact of protein amino acid substitution on phenotype. (26) The algorithm is executed in two phases: (i) a supervised learning (training) phase where the algorithm isolates subsets of sequences, from a pre-computed MSA, that facilitates an optimal classification of variants of known phenotypic impact. (ii) The POSE that results from training is then used to predict the phenotypic impact of variants not present in the initial training phase. In this report, we extend the functionality of our algorithm to utilize continuous-valued, quantitative disease risk data (endophenotypic data) during the supervised learning stage. POSE is written in python and is freely available for nonprofit use at http://karchinlab.org/apps/appPose.html. See the accompanying Supplementary Material, Materials and Methods for a detailed description of POSE and the implementation used for this work.
CFTR sequences
CFTR is member 7 of subfamily C of the large family of evolutionarily related ABC transporters. For this study, we constructed the MSA's from all CFTR (ABCC7) orthologs, and CFTR paralogs (ABCC1–6 and 8–12) from the UCSC 46-vertebrate genome alignments; this resulted in a total of 547 CFTR homologs. Sequences were aligned using ClustalW in default mode (37).
CFTR homology model
POSE calculates relative residue burial from protein 3D structure, if available, for scoring variants. For this study, we supplied POSE with a homology model of CFTR in the so-called inward-facing conformation (26). There is no crystal structure of full-length CFTR available in the protein databank (PDB). However, crystal structures for both CFTR NBD1 and NDB2, individually, were available at the time of this writing. The added utility of using the homology model, relative to isolated NBD crystal structures, is that the homology model provides an estimation of the interface formed between the interacting NBDs. Importantly, the backbone RMSD is low between the homology model and each of the NDB crystal structures (1.70Å for NBD1, PDB ID 2BB0; 3.0Å for NBD2, PDB ID 3GD7). Unfortunately, no crystal structure for CFTR TMD1 and TMD2 is currently available in the PDB.
CFTR variants
There were a total of 59 variants that had endophenotypic data available in the CFTR2 database for each of six unique data types (25). This included 20 variants in CFTR NBDs, 37 in the TMDs and 2 in the regulatory domain. We performed leave-one-out cross-validation calculations using three different groupings of the variants: (i) including only the 20 NBD variants. (ii) Including only the 37 TMD variants. (iii) All 57 variants included in the cross-validation calculation. Considering membrane and cytosolic protein regions separately was useful to determine differences in performance between the domains.
We constructed a final validation dataset of 11 NBD variants. Clinical endophenotype data on sweat chloride concentration was derived from a new data collection for the CFTR2 project that includes a larger number of CF patients. By using this new, larger dataset we were able to evaluate data for 11 additional variants that did not have enough patients in the first dataset. All patients included in analysis of the 11 additional variants had a known CF-causing mutation on the other allele and were identified from patients in the CFTR2 database and occurred at a frequency of at least 0.004%. Sweat chloride means were derived from a minimum of five patients. Detailed information, including variant-specific sweat chloride ranges, is included in Supplementary Material, supplemental spreadsheet.
Annotated phenotype
The variants in the CFTR2 database were reported in CF patients collected in national registries and large clinics (25). To confirm that these variants were causing disease, the CFTR2 project used the clinical severity of patients that carried the variant, laboratory based functional testing of the variants in experimental cell lines, and a test of penetrance that looked at the non-transmitted allele in fathers of CF offspring (25). This allowed the CFTR variants observed in CF patients to be characterized as CF-causing if clinical parameters were consistent with the diagnostic criteria for CF, functional evaluation demonstrated the variant would disrupt protein expression or function, and there was no evidence that the variant was not fully penetrant to cause infertility in fathers. One hundred and twenty-seven variants of the 159 evaluated met all these criteria. The remaining variants were assigned an “indeterminate” phenotype (20 of 159), unless there was evidence the variant was not penetrant (12 of 159) and could therefore be classified as not CF-causing.
Endophenotypic data
In addition to the annotated phenotypes (CF, not-CF, indeterminate) described above, we utilized specific endophenotypes that allowed variants to be placed on a spectrum of severity (25). Two of these phenotypes refer to functional testing of experimentally generated cell lines that express a single copy of the mutated protein. These cell lines were tested for CFTR maturation, and for CFTR-specific chloride current. A variant was defined as more severe if there was greater disruption of wild-type CFTR processing, or if the chloride conductance was low. Clinical endophenotypes were derived from patients in the CFTR2 database that carried a given variant on one chromosome, with a variant known to be associated with zero CFTR function on the other chromosome. More severe CFTR variants would be expected to have higher sweat chloride concentration, lower lung function (measured as forced expiratory volume in 1 s, FEV1%pred), to be associated with infection with a common pathogen seen in CF (P. aeruginosa), and to be associated with exocrine pancreatic dysfunction.
Performance metrics
POSE natively calculates the sensitivity, specificity, PPV, NPV, ROC AUC, and R2 for the linear regression of POSE scores versus endophenotypic measurements. Pearson correlation coefficients and P-values are calculated using the R cor.test module, which interfaces with POSE via RPy2. All ROC plots were generated in R.
Sensitivity is defined as number of true positives (TP) divided by the sum of TP and false negatives (FN). Specificity is defined as the number of true negatives (TN) divided by the sum of TN and false positives (FP). The PPV and NPV are defined as TP/(TP + FP) and TN/(TN + FN), respectively. These predictive performance statistics are calculated by dichotomizing POSE scores such that the balanced accuracy of phenotype prediction is maximized (i.e. POSE scores are dichotomized and compared with the binary annotated phenotypes).
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
Funding
This work supported by the US CF Foundation (KARCHI1210 and CUTTIN11A0).
Supplementary Material
References
- 1.Hamburg M.A., Collins F.S. The path to personalized medicine. N. Engl. J. Med. 2010;363:301–304. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 2.Stitziel N., Kiezun A., Sunyaev S. Computational and statistical approaches to analyzing variants identified by exome sequencing. Genome Biol. 2011;12:227. doi: 10.1186/gb-2011-12-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cline M.S., Karchin R. Using bioinformatics to predict the functional impact of SNVs. Bioinformatics. 2011;27:441–448. doi: 10.1093/bioinformatics/btq695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mah J.T.L., Low E.S.H., Lee E. In silico SNP analysis and bioinformatics tools: a review of the state of the art to aid drug discovery. Drug Discov. Today. 2011;16:800–809. doi: 10.1016/j.drudis.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Thusberg J., Olatubosun A., Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum. Mutat. 2011;32:358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- 6.Chan P.A., Duraisamy S., Miller P.J., Newell J.A., McBride C., Bond J.P., Raevaara T., Ollila S., Nyström M., Grimm A.J., et al. Interpreting missense variants: comparing computational methods in human disease genes CDKN2A, MLH1, MSH2, MECP2, and tyrosinase (TYR) Hum. Mutat. 2007;28:683–693. doi: 10.1002/humu.20492. [DOI] [PubMed] [Google Scholar]
- 7.Shihab H.A., Gough J., Cooper D.N., Stenson P.D., Barker G.L.A., Edwards K.J., Day I.N.M., Gaunt T.R. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hicks S., Wheeler D.A., Plon S.E., Kimmel M. Prediction of missense mutation functionality depends on both the algorithm and sequence alignment employed. Hum. Mutat. 2011;32:661–668. doi: 10.1002/humu.21490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wieckowska A., Zein N.N., Yerian L.M., Lopez A.R., McCullough A.J., Feldstein A.E. In vivo assessment of liver cell apoptosis as a novel biomarker of disease severity in nonalcoholic fatty liver disease. Hepatology. 2006;44:27–33. doi: 10.1002/hep.21223. [DOI] [PubMed] [Google Scholar]
- 10.Bozza F., Salluh J., Japiassu A., Soares M., Assis E., Gomes R., Bozza M., Castro-Faria-Neto H., Bozza P. Cytokine profiles as markers of disease severity in sepsis: a multiplex analysis. Crit. Care. 2007;11:R49. doi: 10.1186/cc5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakuntabhai A., Turbpaiboon C., Casademont I., Chuansumrit A., Lowhnoo T., Kajaste-Rudnitski A., Kalayanarooj S.M., Tangnararatchakit K., Tangthawornchaikul N., Vasanawathana S., et al. A variant in the CD209 promoter is associated with severity of dengue disease. Nat. Genet. 2005;37:507–513. doi: 10.1038/ng1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stahl E., Lindberg A., Jansson S.-A., Ronmark E., Svensson K., Andersson F., Lofdahl C., Lundback B. Health-related quality of life is related to COPD disease severity. Health Qual. Life Outcomes. 2005;3:56. doi: 10.1186/1477-7525-3-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson P.T., Jicha G.A., Schmitt F.A., Liu H., Davis D.G., Mendiondo M.S., Abner E.L., Markesbery W.R. Clinicopathologic correlations in a large Alzheimer disease center autopsy cohort: neuritic plaques and neurofibrillary tangles “do count” when staging disease severity. J. Neuropathol. Exp. Neurol. 2007;66:1136. doi: 10.1097/nen.0b013e31815c5efb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drumm M.L., Konstan M.W., Schluchter M.D., Handler A., Pace R., Zou F., Zariwala M., Fargo D., Xu A., Dunn J.M., et al. Genetic modifiers of lung disease in cystic fibrosis. N. Engl. J. Med. 2005;353:1443–1453. doi: 10.1056/NEJMoa051469. [DOI] [PubMed] [Google Scholar]
- 15.Dorfman R., Sandford A., Taylor C., Huang B., Frangolias D., Wang Y., Sang R., Pereira L., Sun L., Berthiaume Y., et al. Complex two-gene modulation of lung disease severity in children with cystic fibrosis. J. Clin. Invest. 2008;118:1040–1049. doi: 10.1172/JCI33754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stone E.A., Sidow A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 2005;15:978–986. doi: 10.1101/gr.3804205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tavtigian S.V., Greenblatt M.S., Lesueur F., Byrnes G.B. In silico analysis of missense substitutions using sequence-alignment based methods. Hum. Mutat. 2008;29:1327–1336. doi: 10.1002/humu.20892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper D., Krawczak M., Polychronakos C., Tyler-Smith C., Kehrer-Sawatzki H. Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum. Genet. 2013;132:1077–1130. doi: 10.1007/s00439-013-1331-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almasy L., Blangero J. Endophenotypes as quantitative risk factors for psychiatric disease: rationale and study design. Am. J. Med. Genet. 2001;105:42–44. [PubMed] [Google Scholar]
- 20.Waddington J.L., Corvin A.P., Donohoe G., O'Tuathaigh C.M.P., Mitchell K.J., Gill M. Functional Genomics and Schizophrenia: endophenotypes and mutant models. Psychiatr. Clin. North Am. 2007;30:365–399. doi: 10.1016/j.psc.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Cannon T.D., Keller M.C. Endophenotypes in the genetic analyses of mental disorders. Annu. Rev. Clin. Psychol. 2006;2:267–290. doi: 10.1146/annurev.clinpsy.2.022305.095232. [DOI] [PubMed] [Google Scholar]
- 22.Thaker G. Psychosis endophenotypes in schizophrenia and bipolar disorder. Schizophr. Bull. 2008;34:720–721. doi: 10.1093/schbul/sbn055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson R.D., Pepine C.J. Coronary angiography: is it time to reassess? Circulation. 2013;127:1760–1762. doi: 10.1161/CIRCULATIONAHA.113.002566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riordan J.R. CFTR function and prospects for therapy. Annu. Rev. Biochem. 2008;77:701–726. doi: 10.1146/annurev.biochem.75.103004.142532. [DOI] [PubMed] [Google Scholar]
- 25.Sosnay P.R., Siklosi K.R., Van Goor F., Kaniecki K., Yu H., Sharma N., Ramalho A.S., Amaral M.D., Dorfman R., Zielenski J. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat. Genet. 2013;45:1160–1167. doi: 10.1038/ng.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masica D.L., Sosnay P.R., Cutting G.R., Karchin R. Phenotype-optimized sequence ensembles substantially improve prediction of disease-causing mutation in cystic fibrosis. Hum. Mutat. 2012;33:1267–1274. doi: 10.1002/humu.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strom C.M., Huang D., Chen C., Buller A., Peng M., Quan F., Redman J., Sun W. Extensive sequencing of the cystic fibrosis transmembrane regulator gene: assay validation and unexpected benefits of developing a comprehensive test. Genet. Med. 2003;5:9–14. doi: 10.1097/00125817-200301000-00002. [DOI] [PubMed] [Google Scholar]
- 28.Uppaluri L., England S.J., Scanlin T.F. Clinical evidence that V456A is a cystic fibrosis causing mutation in South Asians. J. Cyst. Fibrosis. 2012;11:312–315. doi: 10.1016/j.jcf.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Vergani P., Lockless S.W., Nairn A.C., Gadsby D.C. CFTR channel opening by ATP-driven tight dimerization of its nucleotide-binding domains. Nature. 2005;433:876–880. doi: 10.1038/nature03313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zolnerciks J.K., Andress E.J., Nicolaou M., Linton K.J. Structure of ABC transporters. Essays Biochem. 2011;50:43–61. doi: 10.1042/bse0500043. [DOI] [PubMed] [Google Scholar]
- 31.Rahman K.S., Cui G., Harvey S.C., McCarty N.A. Modeling the conformational changes underlying channel opening in CFTR. PLoS ONE. 2013;8:e74574. doi: 10.1371/journal.pone.0074574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenberg M.F., Kamis A.B., Aleksandrov L.A., Ford R.C., Riordan J.R. Purification and crystallization of the cystic fibrosis transmembrane conductance regulator (CFTR) J. Biol. Chem. 2004;279:39051–39057. doi: 10.1074/jbc.M407434200. [DOI] [PubMed] [Google Scholar]
- 33.Kayaalp E., Treacy E., Waters P.J., Byck S., Nowacki P., Scriver C.R. Human phenylalanine hydroxylase mutations and hyperphenylalaninemia phenotypes: a metanalysis of genotype-phenotype correlations. Am. J. Hum. Genet. 1997;61:1309–1317. doi: 10.1086/301638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hull J., Thomson A.H. Contribution of genetic factors other than CFTR to disease severity in cystic fibrosis. Thorax. 1998;53:1018–1021. doi: 10.1136/thx.53.12.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garred P., Pressler T., Madsen H.O., Frederiksen B., Svejgaard A., Høiby N., Schwartz M., Koch C. Association of mannose-binding lectin gene heterogeneity with severity of lung disease and survival in cystic fibrosis. J. Clin. Invest. 1999;104:431–437. doi: 10.1172/JCI6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehner B. Genotype to phenotype: lessons from model organisms for human genetics. Nat. Rev. Genet. 2013;14:168–178. doi: 10.1038/nrg3404. [DOI] [PubMed] [Google Scholar]
- 37.Thompson J.D., Gibson T.J., Higgins D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics. 2002;chapter 2:unit 2.3. doi: 10.1002/0471250953.bi0203s00. doi:10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.