Background: Transcriptional regulation of smooth muscle cells is an understudied component of intestinal development and physiology.
Results: Foxf2 deletion from smooth muscle causes intestinal malformations and colon remodeling.
Conclusion: Foxf2 regulation of PDGF and myocardin/SRF signaling is essential for intestinal development and homeostasis.
Significance: Better understanding of transcriptional mechanisms regulating postnatal intestine development and homeostasis may provide therapeutic approaches for congenital and acquired gastrointestinal diseases.
Keywords: Gene Knockout, Intestine, Myocardin, Smooth Muscle, Transcription Factor, Transgenic Mice, Forkhead Protein, Foxf2
Abstract
Alterations in the forkhead box F2 gene expression have been reported in numerous pathologies, and Foxf2−/− mice are perinatal lethal with multiple malformations; however, molecular mechanisms pertaining to Foxf2 signaling are severely lacking. In this study, Foxf2 requirements in murine smooth muscle cells were examined using a conditional knock-out approach. We generated novel Foxf2-floxed mice, which we bred to smMHC-Cre-eGFP mice to generate a mouse line with Foxf2 deleted specifically from smooth muscle. These mice exhibited growth retardation due to reduced intestinal length as well as inflammation and remodeling of the small intestine. Colons of Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice had expansion of the myenteric nerve plexus and increased proliferation of smooth muscle cells leading to thickening of the longitudinal smooth muscle layer. Foxf2 deficiency in colonic smooth muscle was associated with increased expression of Foxf1, PDGFa, PDGFb, PDGF receptor α, and myocardin. FOXF2 bound to promoter regions of these genes indicating direct transcriptional regulation. Foxf2 repressed Foxf1 promoter activity in co-transfection experiments. We also show that knockdown of Foxf2 in colonic smooth muscle cells in vitro and in transgenic mice increased myocardin/serum response factor signaling and increased expression of contractile proteins. Foxf2 attenuated myocardin/serum response factor signaling in smooth muscle cells through direct binding to the N-terminal region of myocardin. Our results indicate that Foxf2 signaling in smooth muscle cells is essential for intestinal development and serum response factor signaling.
Introduction
The mammalian gut is derived from the endodermal and mesodermal germ layers, which give rise to gut epithelium, mesenchyme, and smooth muscle (1). The enteric nervous system is derived from invading cells from the neural crest (1). Digestive diseases affect 60–70 million Americans resulting in 13.5 million hospitalizations annually (2). Digestive diseases run a gamut from viral infections to irritable bowel syndrome and hemorrhoids, and they inflict all tissue layers (2). To this point, studies of gut development and diseases have primarily focused on the epithelial layer due to its obvious importance in digestion and nutrient absorption. However, smooth muscle is also of critical importance to gut anatomy and physiology, maintaining shape and allowing for motility of luminal content. Despite the great breadth of knowledge pertaining to vascular smooth muscle development and pathology, surprisingly little is known about molecular and transcriptional mechanisms critical for development of visceral smooth muscle lining the gut.
The development of visceral smooth muscle requires controlled cross-talk between epithelial, neuronal, and mesenchymal cell layers (3–5). The muscularis of the gastrointestinal (GI)3 tract contains two distinct smooth muscle layers separated by the myenteric plexus, an inner circular layer and an outer longitudinal layer (1, 6, 7). The muscularis mucosa is also of mesenchymal origin and demonstrates features similar to smooth muscle (1). Numerous signaling pathways have been identified that play important roles in regulating visceral smooth muscle development during embryogenesis, including the TGF-β and bone morphogenetic protein pathways (5, 8, 9), Wnt signaling (9, 10), the Hedgehog pathway (3, 4, 9), and a number of growth factor-mediated pathways (i.e. PDGF, fibroblast growth factor (FGF), and insulin-like growth factor (IGF)) (5, 7, 11, 12). However, knowledge pertaining to the regulation of postnatal visceral smooth muscle development remains scarce.
The forkhead box (Fox) family of transcription factors has been shown to mediate a wide variety of cellular activities, including embryonic and postnatal cell growth, tissue repair after injury, cell migration, and tumor formation (13). The Foxf subgroup contains two members, Foxf1 and Foxf2 (1, 14). Lessons from knock-out mice indicate that both Foxf members are critical for embryonic development as Foxf1−/− mice die in utero (15, 16), and Foxf2−/− as well as compound heterozygotic (Foxf1+/−/Foxf2+/−) mice die shortly after birth (17). Several Fox proteins, including Foxo4, Foxq1, and Foxf1, have been shown to be critical for smooth muscle cell development via regulation of the myocardin/serum response factor (SRF) axis (18–20). Using in situ hybridization, embryonic Foxf2 expression has been found in the mesenchyme of the oral cavity, limb buds, genitalia, CNS, eyes, lung, prostate, ear, and placenta as well as the lamina propria region and smooth muscle of the developing GI tract (21, 22). During embryonic development, Hedgehog signaling from the epithelium induces Foxf2 expression (17); however, postnatal Foxf2 expression and signaling remain poorly characterized. Studies have shown alterations in Foxf2 expression in prostate cancer (23) and that Foxf2 decreases the size and frequency of colonic polyps in colon adenoma (24). Foxf2 has been further shown to mediate cardiac metabolism (25), and a W174R amino acid substitution has been shown to cause anterior segment mesenchymal dysgenesis in the eye (Foxf2W174R) (26). However, molecular mechanisms for Foxf2 signaling in smooth muscle cells have not been previously elucidated due to lack of mouse models allowing conditional inactivation of Foxf2.
In this study, we generated a novel Foxf2-floxed (Foxf2fl) mouse line to investigate the importance of Foxf2 in smooth muscle cells. By breeding these mice with smooth muscle myosin heavy chain-Cre mice (smMHC-Cre-eGFP) (27), we were able to efficiently delete Foxf2 from mature smooth muscle in the gut. Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice were viable and fertile. Juvenile mice showed no difference in size or morphology; however, adult Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice were significantly smaller in stature and had a significant decrease in the length of the GI tract (both large and small intestine). Adult mice had focal inflammatory regions in the small intestine and a significant increase in thickness of the smooth muscle and enteric neuron layers in the colon. We used colon smooth muscle from Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice and cultured Foxf2−/− smooth muscle cells to demonstrate that Foxf2 regulates the PDGF pathway and physically interacts with myocardin to inhibit SRF signaling in visceral smooth muscle cells.
EXPERIMENTAL PROCEDURES
Generation of Foxf2-floxed Mice and Deletion of Foxf2 from Smooth Muscle Myocytes
A LoxP site was inserted into the Foxf2 promoter via the Foxf2-targeting vector and the PGK-gb2 LoxP/FRT-flanked neomycin (neo) cassette was placed into the first intron (Fig. 1D). Electroporation of mouse ES cells (C57Bl/6 × 129/SVEV) with the Foxf2fl-targeting vector, following neo (G418) selection, was performed at the inGenious Targeting Laboratory (Stony Brook, NY). PCR analysis with multiple primer sets identified ES cells with the appropriate Foxf2fl-targeted locus. Foxf2fl ES cells were subsequently used to generate chimeric mice by injection into mouse blastocysts. Mice containing the Foxf2fl-targeted allele were determined by PCR amplification with primers flanking the LoxP sequence located in the Foxf2 promoter (P1 and P2) (Table 1) and primers located in the 3′ region of the Foxf2fl allele (P3 and P4). Chimeric mice were bred with C57Bl/6 mice in the animal facility at Cincinnati Children's Research Foundation to produce Foxf2fl/+ mice. The neo cassette was deleted by breeding Foxf2fl/+ mice with ACT-FLP1 mice (The Jackson Laboratory) (Fig. 1D). The loss of neo in Foxf2fl/+ mice was confirmed by PCR using P5 and P6 primers (Table 1) and sequencing of the Foxf2 locus using mouse tail DNA. Foxf2fl/+ mice were backcrossed to generate viable Foxf2fl/fl mice that were bred into the C57Bl/6 background for 10 generations. Deletion of the Foxf2fl alleles from visceral smooth muscle was accomplished through breeding with smooth muscle myosin heavy chain-Cre-eGFP (smMHC-Cre-eGFP) transgenic mice (C57Bl/6) (27). Animal studies were approved by the Animal Care and Use Committee of Cincinnati Children's Research Foundation.
FIGURE 1.
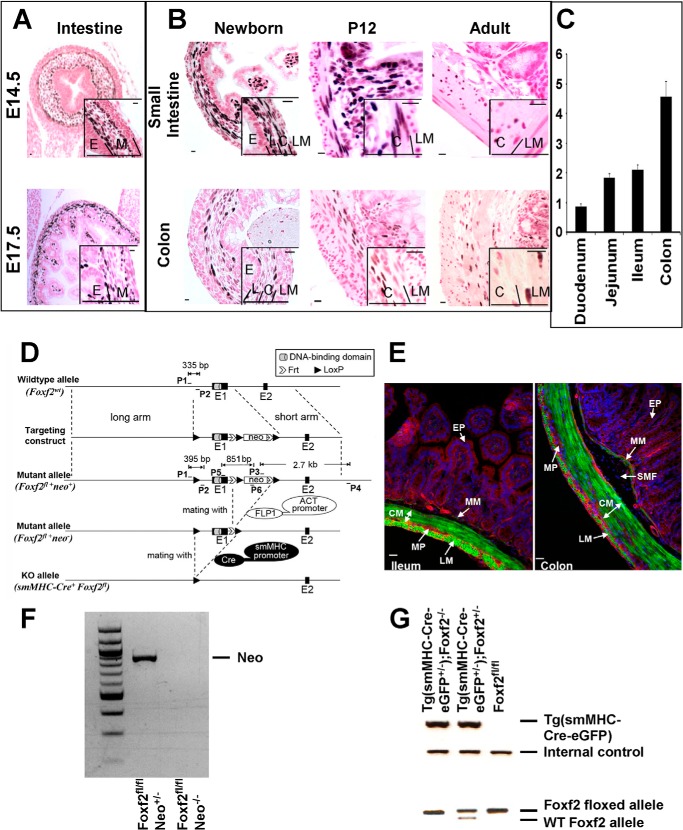
Deletion of Foxf2 from GI smooth muscle. Foxf2 is expressed in the mouse GI tract from embryonic time points through adulthood. A and B, immunostaining with anti-FOXF2 antibodies showed restriction of FOXF2 to mesenchymal layers of the developing GI tract at E14.5 and E17.5. Postnatally, Foxf2 was expressed in lamina propria (L) and circular muscle layer (C) but was absent from the longitudinal muscle layer (LM) and epithelium (E). C, qRT-PCR analysis of adult whole organ mRNA showed that Foxf2 is more highly expressed in the jejunum and ileum than the duodenum of the small intestine and more highly expressed in the colon than small intestine. Expression levels were normalized to β-actin (n = 3). D, schematic shows Foxf2-floxed targeting construct. LoxP sites (solid triangles) were inserted to flank the first exon (E1) of the Foxf2 gene. Locations of primers (P) and sizes of PCR products are indicated. Primers used are listed in Table 1. Foxf2fl/fl mice were bred with ACT-FLP1 mice to remove neo and later with smMHC-Cre-eGFP mice to delete Foxf2 from smooth muscle cells. E, visualization of immunofluorescence in frozen-fixed cross-sections of adult ileum and colon from smMHC-Cre-eGFP mice crossed with the Cre-dependent mT/mG reporter strain. Membrane-bound enhanced GFP is detected in the smooth muscle layers as well as the muscularis mucosa but not lamina propria. eGFP is expressed stronger in the muscularis mucosa of colon compared with ileum. The abbreviations used are as follows: CM, circular muscle; LM, longitudinal muscle; MP, myenteric nerve plexus; MM, muscularis mucosa; SMF, submucosal fibroblasts; EP, epithelium. F and G, PCR analysis of mouse tail DNA shows the presence of the neo cassette, Cre transgene, as well as wild type and Foxf2-floxed alleles. Scale bar, 10 μm.
TABLE 1.
Foxf2-floxed allele mouse primers
Primer sets in table were used for genotyping from mouse tail DNA.
| Primer name | Sequence |
|---|---|
| P1 | TGGAGGAGTGTTCTCGAATGGAG |
| P2 | CACTGGACGCCCTTGAGCTG |
| P3 | CCAGAGGCCACTTGTGTAGC |
| P4 | AAAGCTTGTGTCTAGCGGTTGTCCAC |
| P5 | AGCATGTCTTCCTACTCGTTGGAG |
| P6 | GCTCCTGCCGAGAAAGTATCC |
Tissue Collection
Intestinal tracts were collected from Foxf2fl/fl and Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice and placed in ice-cold, calcium-free, sterile PBS. Colons and small intestines were separated at this time. Luminal contents were manually forced out by gentle manipulation with forceps. Tissues destined for immunohistochemistry were placed in 4% paraformaldehyde at this time. Tissues for whole organ RNA were placed in RNA-STAT-60. To enrich the smooth muscle layer of the colon, colons were split along their length and laid flat with the smooth muscle side down. A scalpel was then used to scrape away the epithelium and other layers leaving behind a smooth muscle enriched segment that was then placed in RNA-STAT-60 for RNA isolation or used for enzymatic digestion to prepare a single cell suspension as described previously (18).
Immunohistochemistry
To characterize FOXF2 expression throughout the development of the GI tract, intestinal tissues were collected from wild type mice at embryonic day 14.5 (E14.5), E17.5, postnatal day 1 (P1), as well as Tg(smMHC-Cre-eGFP+/−);Foxf2−/− and Foxf2fl/fl mice at E16.5, E18.5, P12, and in the adult (8–12 weeks). Intestines and abdomens were fixed in 4% paraformaldehyde overnight and embedded into paraffin blocks. Paraffin sections of 5 μm were immunostained with antibodies against FOXF2 (1:1000; Santa Cruz Biotechnology), CGRP (1:4000; Sigma), PDGF receptor α (1:200; Santa Cruz Biotechnology), α-smooth muscle actin (αSMA; 1:10,000; Sigma), γSMA (1:1000; 7 Hills Biotech), PECAM-1 (1:500; Pharmingen), FOXA2 (1:300; 7 Hills Biotech), FOXF1 (1:1000 (29)), pan-cytokeratin (1:500; Sigma), Ki-67 (1:10,000; Dako), or cyclin D1 (1:250; Abcam). Antibody-antigen complexes were detected using biotinylated secondary antibody followed by the avidin-HRP complex and 3,3′-diaminobenzidine substrate (Vector Laboratories, Burlingame, CA) as described previously (30–34). Sections were counterstained with nuclear fast red (Vector Laboratories). Intestinal sections were also stained with hematoxylin and eosin (H&E) to evaluate morphology, Masson's Trichrome to detect fibrosis, or wheat germ agglutinin (Sigma) to measure myocyte size. Size of myocytes was measured from wheat germ agglutinin-stained slides as described previously (35, 36). Slides were photographed using a Zeiss Axioplan2 microscope and AxioVision Rel 4.8 software.
To detect Cre-dependent reporter activity, smMHC-Cre-eGFP mice were crossed with the Cre-dependent reporter strain, mT/mG (B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J (The Jackson Laboratory). Tissues were harvested from adult double heterozygous mice, fixed in 4% paraformaldehyde, and frozen in OCT tissue freezing media (Tissue-Tech). Frozen sections were washed in 100 mm Tris, pH 7.6, 150 mm NaCl, stained with Hoechst, mounted in Prolong Gold (Invitrogen), and visualized by confocal microscopy (Olympus Fluoview FV1000). Under these conditions cytoplasmic enhanced GFP encoded by the smMHC-Cre-eGFP transgene was washed out of the tissue sections.
Quantitative Real Time RT-PCR (qRT-PCR)
Whole organ RNA was prepared from small intestine and P12 colon. Smooth muscle-enriched RNA was prepared as described (18) from adult colon of individual Tg(smMHC-Cre-eGFP+/−);Foxf2−/− and control Foxf2fl/fl mice using RNA-STAT-60 (Tel-Test “B” Inc. Friendswood, TX). cDNA was generated using the Applied Biosystems High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). Evaluation of expression levels of specific genes was performed by qRT-PCR using inventoried TaqMan probes (Table 2) and the StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA) as described previously (37–42).
TABLE 2.
TaqMan probes
Table lists inventoried TaqMan probes used for qRT-PCR analysis of gene expression.
| Gene name | Assay no. |
|---|---|
| acta2 | Mm00725412_s1 |
| Actb | Mm00607939_s1 |
| actg2 | Mm00656102_m1 |
| bmp4 | Mm01321704_m1 |
| ccnb1 | Mm00838401_g1 |
| ccnd1 | Mm00432359_m1 |
| cdc25b | Mm00499136_m1 |
| Foxf1 | Mm00487497_m1 |
| Foxf2 | Mm00515793_m1 |
| Foxm1 | Mm00514924_m1 |
| gli1 | Mm01160468_g1 |
| gli2 | Mm01293116_m1 |
| myh11 | Mm00443013_m1 |
| Myocd | Mm00455051_m1 |
| Pdgfa | Mm01205760_m1 |
| Pdgfb | Mm01298578_m1 |
| Pdgfra | Mm01211694_m1 |
| ptch1 | Mm00436026_m1 |
| sox9 | Mm00448840_m1 |
| Srf | Mm00491032_m1 |
| Tagln | Mm00441661_g1 |
| wnt5a | Mm00437347_m1 |
Chromatin Immunoprecipitation (ChIP) Assays
Stably transfected mouse rhabdomyosarcoma cells with a dual His/FLAG-tagged Foxf2 construct were generated and used for ChIP analysis. The pMIEG3 retroviral vector was used for dual-tagged protein expression in mammalian cells and has been described previously (43). To generate a double-tagged construct, Foxf2 ORF was PCR-amplified using high fidelity Pfx polymerase with N-terminal FLAG and C-terminal His6 affinity epitopes according to manufacturer's protocol (Invitrogen). Nuclear extracts from transfected mouse rhabdomyosarcoma cells were cross-linked by addition of formaldehyde, sonicated, and used for immunoprecipitation with anti-His6-tagged rabbit polyclonal antibodies (Abcam) as described previously (44–48). DNA fragments were ∼500 bp as verified by agarose gel. Reverse cross-linked ChIP DNA samples were subjected to PCR, using oligonucleotides specific to promoter regions of mouse PDGFa, PDGFb, PDGFRα, Foxf1, and myocardin genes (Table 3). Potential FOXF2-binding sites were identified using the MacVector program and the previously published FOXF2 consensus binding sequence TA(G)TTTA(G)T (44). DNA sites with the highest homology to the published FOXF2 consensus binding sequence were investigated via ChIP. DNA binding was normalized to control ChIP DNA samples, which were immunoprecipitated using control rabbit IgG.
TABLE 3.
ChIP primers
Primer sets in table were used in ChIP analysis of Foxf2 binding to gene promoter regions (listed in parentheses).
| Gene (promoter region) | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
|---|---|---|
| Foxf1 + enhancer (248/255) | CCCCACCCCTAACGGATTATTTG | TCCCCTGCTTCTAAAAACTGTCCC |
| Foxf1 3′ region (negative control) | ACCACCATCTCCATCACCAGCC | CCTCCCGTCCAGAATGTCAAGTC |
| Myocardin (−4532/−4525 and −4523/−4516) | CCTGGGACCACACAAACAAC | AGGCTCCGTCTCAGCGTG |
| PDGFa (−6421/−6414) | CCCATCATCCTCTCCTCTTTGC | GGTAGCCTTTAGAAATGTGTGCCAG |
| PDGFb (−1435/−1428) | TAGATGAGTTCTGGGACTGGACT | AGACATAACCGGAGGAGAAGAAG |
| PDGFRα (−531/−524) | TGTGCCTCCCAATGGATGAAATC | CCTCTGTGAGACGGAAGCCTGC |
Luciferase and Mammalian Two-hybrid Assays
Foxf2 was amplified by RT-PCR from mRNA isolated from mouse intestine. The encoded protein of 446 amino acids is identical to that encoded by NM_010225.2. A Foxf1 mammalian expression construct was amplified by PCR from an HFH8 clone obtained from Robert Costa (44). This resulted in expression of a FOXF1 protein of 353 amino acids identical to that encoded by NM_010426.1. Fox expression plasmids were transfected together with SRF expression plasmid, telokin promoter luciferase reporter gene (−256 to +147), and telokin-Renilla luciferase internal control into 10T1/2 cells, and luciferase assays were performed as described previously (46). The Foxf1 luciferase reporter (−5.3 kb Foxf1 + 3′ response element) and Foxf2 luciferase reporter (six repeats of Foxf2-binding sequence) genes were transfected into U2OS cells, and luciferase assays were performed as described previously (46, 49). Mammalian two-hybrid assays utilizing SRF fused to the GAL4 DNA binding domain and myocardin fused to the GAL4 activation domain were performed as described previously (50).
GST Pulldown Assays
GST-SRF and GST-myocardin bacterial expression plasmids were described previously (50–53). GST-NT myocardin transcription factor A (MRTFA) (encoding amino acids 1–628) and GST-CT MRTFA (encoding amino acids 618–929) were generated by PCR amplification of MRTFA fragments from the MRTFA mammalian expression vector (52). All expression constructs were confirmed by DNA sequencing. Full-length and fragments of Foxf2 were PCR-amplified and cloned into pET vectors for expression in bacteria. GST pulldown assays were performed as described previously (53).
Statistical Analysis
Student's t test was used to determine statistical significance. p values <0.05 were considered significant. Values for all measurements were expressed as means ± S.E.
RESULTS
Generation of Mice with Foxf2 Deletion from Smooth Muscle Cells
As Foxf2 expression has been poorly studied and the emergence of reliable commercial antibodies fairly recent, we performed immunohistochemistry on embryonic (E14.5 and E17.5), newborn, juvenile (P12), and adult (8–12 weeks) wild type mice to investigate FOXF2 expression patterns in the GI tract during development. As early as E14.5 and at E17.5, FOXF2-positive nuclei were detected in the intestinal mesenchyme located adjacent to the epithelial layer (Fig. 1A). In the newborn, P12, and adult, FOXF2 was detected in the lamina propria and inner circular smooth muscle layer of the muscularis externa in both the small intestine and colon (Fig. 1B). FOXF2 was not detected in the outer longitudinal smooth muscle layer (Fig. 1B). FOXF2 was not expressed in vascular smooth muscle cells (data not shown). qRT-PCR analysis indicated that Foxf2 expression increases along the cranial to caudal axis in the adult gut, with little expression in the duodenum and highest expression in the colon (Fig. 1C).
To assess Foxf2 requirements in smooth muscle cells, we created a PGK-Neo-Foxf2 targeting construct and used it for electroporation of ES cells (Fig. 1D and “Experimental Procedures”). A Foxf2-floxed (Foxf2fl) mouse line was generated and confirmed by PCR amplification of tail genomic DNA (Fig. 1, F and G). The neo cassette was deleted by crossing the mice with ACT-FLP1 transgenic mice (Fig. 1D). Foxf2fl/fl mice contain LoxP sites flanking exon one of the Foxf2 gene, which contains the DNA binding domain and one of two transcriptional activation domains of the FOXF2 protein. Foxf2fl/fl mice were bred with smMHC-Cre-eGFP mice to delete Foxf2 from intestinal smooth muscle. Analysis of Cre-dependent reporter mice demonstrated that the smMHC-Cre-eGFP transgene directs robust Cre activity in the smooth muscle layers as well as muscularis mucosa but not lamina propria of small intestine and colon (Fig. 1E).
Growth Retardation and Diminished Intestinal Length in Tg(smMHC-Cre-eGFP+/−);Foxf2−/− Mice
Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice were born in normal Mendelian ratio and were viable and fertile. As early as E16.5, Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice had a noticeable decrease in FOXF2 staining in smooth muscle cells of the circular smooth muscle layer, without loss of FOXF2 from the lamina propria (Fig. 2A), a finding consistent with GFP reporter studies (Fig. 1E). Similar results were obtained from adult Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice. Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice exhibited loss of FOXF2 from the inner circular muscle layer (Fig. 2A) and decreased Foxf2 mRNA in the colon and small intestine (Fig. 2B). Although intestine structure and size were unchanged in juvenile Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice at P12, 8–12-week-old adult Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice had a truncated GI tract with shorter small and large intestines (Fig. 2, D and E). Adult Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice also had lower body weight than control littermates (19.8 ± 1.2 g for Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice compared with 26.4 ± 1.4 g for control mice) and less fecal production (1.3 ± 0.3 g/day for Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice compared with 2 ± 0.1 g/day for control mice), indicating diminished intestinal function. No intestinal abnormalities were observed in mice containing only the smMHC-Cre-eGFP transgene (39).
FIGURE 2.
Decreased Foxf2 and length of intestinal tract in Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice. A and B, FOXF2 staining and Foxf2 mRNA were significantly decreased in Tg(smMHC-Cre-eGFP+/−);Foxf2−/− embryonic intestines at E16.5 as well as adult small intestine and colon smooth muscle. Expression levels were analyzed by qRT-PCR and normalized to β-actin (n = 4). *, p < 0.05 versus control. C, representative images of H&E-stained adult small intestine and colon from Foxf2fl/fl and Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice are shown. D, gross view of intestines from adult Tg(smMHC-Cre-eGFP+/−);Foxf2−/− and control Foxf2fl/fl mice indicated shorter intestinal tract in Foxf2-deficient mice. E, measurements of intestinal tract confirmed it was significantly shorter in Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice due to decreased length of small and large intestines. (n = 4). Scale bar, 10 μm in A, 100 μm in C. The abbreviations used are as follows: CM, circular muscle; LM, longitudinal muscle; M, muscularis; E, epithelium; L, lamina propria.
Focal Inflammation in Foxf2-deficient Small Intestine
Decreased Foxf2 protein and mRNA were found in Tg(smMHC-Cre-eGFP+/−);Foxf2−/− small intestines, a finding consistent with efficient deletion of Foxf2 by the smMHC-Cre-eGFP transgene (Fig. 2, A and B). Adult Tg(smMHC-Cre-eGFP+/−);Foxf2−/− small intestines were mostly normal in morphology but exhibited ulcer-like patches with loss of villus structure and inflammatory cell infiltrates (Fig. 3, A and B). Morphological appearance of smooth muscle layers was normal (Fig. 3A). Interestingly, the small intestines of juvenile (P12) and embryonic (E17.5) Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice were morphologically normal (Fig. 3C and data not shown), indicating that Foxf2 is important for maintaining intestinal structure in adults.
FIGURE 3.
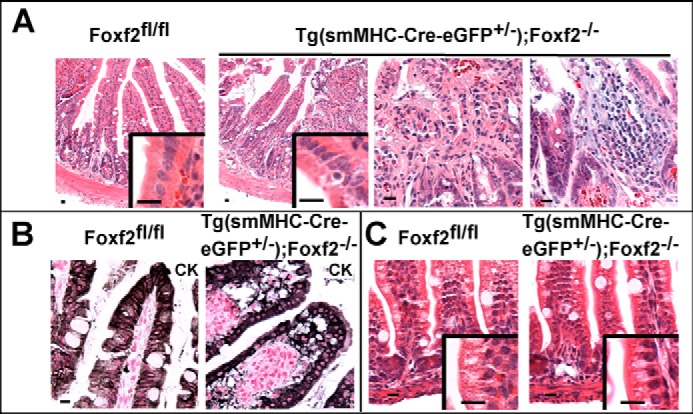
Focal inflammation in Tg(smMHC-Cre-eGFP+/−);Foxf2−/− small intestine. A, adult Tg(smMHC-Cre-eGFP+/−);Foxf2−/− small intestines had mostly normal morphology with focal regions of inflammatory infiltration leading to disruption of villus structure, although muscle morphology remained normal. B, cytokeratin (CK) staining shows disruption of villus epithelium in Tg(smMHC-Cre-eGFP+/−);Foxf2−/− small intestines. C, H&E staining of control and Foxf2-deficient small intestines showed normal morphology at P12. Scale bar, 10 μm.
Increased Smooth Muscle Cell Proliferation in Foxf2-deficient Colons
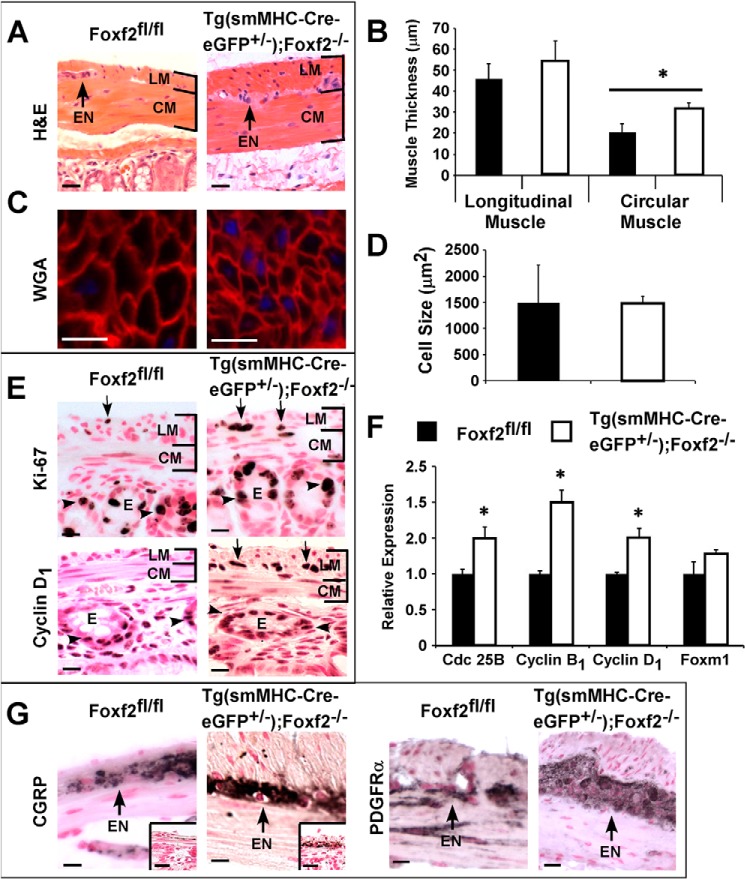
The outer longitudinal smooth muscle layer was significantly thicker in adult Tg(smMHC-Cre-eGFP+/−);Foxf2−/− colons compared with control littermates (Figs. 2C and 4, A and B). Thicker muscle in the colon of Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice was not due to pronounced hypertrophy as there was no detectable difference in myocyte size between control and Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice (Fig. 4, C and D). This would suggest that increased muscle thickness was due to hyperplasia. Although no myocyte proliferation was observed in adult colons, neonatal P12 Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice exhibited increased Ki-67 and cyclin D1 staining in myocytes from the outer longitudinal smooth muscle layer (Fig. 4E). qRT-PCR analysis confirmed increased expression of the cell cycle regulators Cdc 25B, cyclin B1, and cyclin D1 in P12 Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice compared with age-matched controls (Fig. 4F). Therefore, increased smooth muscle thickness in Tg(smMHC-Cre-eGFP+/−); Foxf2−/− colons likely results from increased or prolonged myocyte proliferation during postnatal development and not from hypertrophy of individual myocytes.
FIGURE 4.
Increased muscle thickness and expanded myenteric plexus in Tg(smMHC-Cre-eGFP+/−);Foxf2−/− colons. A, H&E staining indicated increased muscle thickness in adult Tg(smMHC-Cre-eGFP+/−);Foxf2−/− colons. Enteric neurons (EN) are shown with arrows, and longitudinal (LM) and circular (CM) smooth muscle layers are indicated by brackets. B, measurement of muscle thickness was performed in 12 random colon sections with n = 4 mice per group. The longitudinal muscle layer was significantly thicker (*, p < 0.05 versus control) in Tg(smMHC-Cre-eGFP+/−);Foxf2−/− colons. C, wheat germ agglutinin (WGA) staining; D, measurement of cell size in adult mice eliminated hypertrophy as the cause of muscle thickening. E, immunostaining with Ki-67 and cyclin D1 showed increased myocyte proliferation (arrows) in Tg(smMHC-Cre-eGFP+/−);Foxf2−/− colons at P12. Longitudinal and circular smooth muscle layers are shown with brackets, and epithelial layer is indicated as E (arrowheads show epithelial proliferation). F, mRNA levels of Cdc 25B, cyclin B1, and cyclin D1 were increased, but Foxm1 mRNA was unaltered in P12 Tg(smMHC-Cre-eGFP+/−);Foxf2−/− colons. Expression levels were determined by qRT-PCR and normalized to β-actin mRNA (n = 4). G, CGRP and PDGF receptor α immunostaining showed expansion of myenteric plexus (arrows) in adult Tg(smMHC-Cre-eGFP+/−);Foxf2−/− colons. Insets show expanded CGRP staining in P12 Tg(smMHC-Cre-eGFP+/−);Foxf2−/− colons. Scale bar, 10 μm.
Expansion of the Myenteric Plexus in Tg(smMHC-Cre-eGFP+/−);Foxf2−/− Colons
Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice also had an expanded myenteric nerve plexus. Immunohistochemical staining using the CGRP, which marks serotonergic neurons, showed a clearly enlarged network of enteric neurons in adult Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice (Fig. 4G). Greater CGRP staining of the myenteric plexus was also observed at P12, prior to thickening of the longitudinal muscle layer (Fig. 4G, inset). Expansion of the myenteric plexus in Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice was confirmed by immunostaining with antibodies against the platelet-derived growth factor receptor α (PDGF receptor α) (Fig. 4G) that is highly expressed in neurons (54–56) and weakly expressed in smooth muscle cells (57). Consistent with increased PDGF receptor α staining, qRT-PCR analysis showed increased expression of PDGF receptor α, as well as its ligands PDGFa and PDGFb, in Tg(smMHC-Cre-eGFP+/−);Foxf2−/− colons (Fig. 5A).
FIGURE 5.
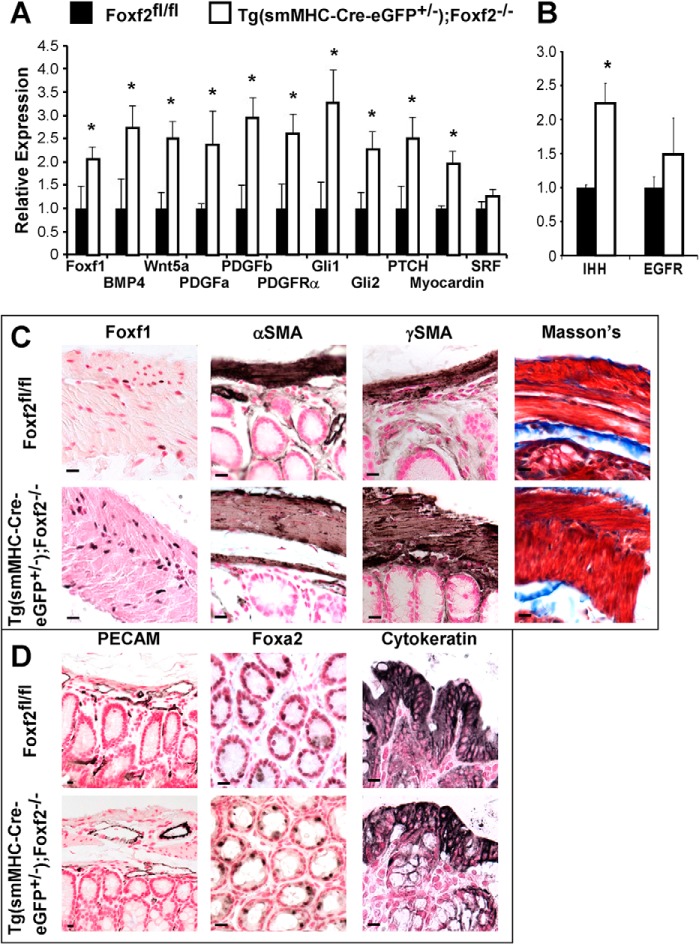
Regulation of gene transcription by Foxf2. A, increased expression of Foxf1, Wnt, bone morphogenetic protein, as well as members of the PDGF, Hedgehog, and SRF pathways in smooth muscle isolated from adult Tg(smMHC-Cre-eGFP+/−);Foxf2−/− colons as determined by qRT-PCR. B, Indian Hedgehog (IHH) expression was increased in whole colon RNA from adult Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice. Epidermal growth factor receptor (EGFR) expression was unaltered. Expression levels were normalized to β-actin (n = 4). *, p < 0.05 versus control. C, FOXF1-positive myocytes were more abundant in Tg(smMHC-Cre-eGFP+/−);Foxf2−/− smooth muscle than control myocytes as shown by immunostaining of the adult colon. Despite prolonged proliferation and altered gene expression, muscle markers αSMA and γSMA were observed in Tg(smMHC-Cre-eGFP+/−);Foxf2−/− colon. Fibrosis was not observed in adult Tg(smMHC-Cre-eGFP+/−);Foxf2−/− colons as determined by Masson's Trichrome staining of intestinal paraffin sections. D, vasculature was unaltered in adult Tg(smMHC-Cre-eGFP+/−);Foxf2−/− colons as visualized by PECAM-1 staining. Epithelial staining for FOXA2 and cytokeratin was normal in adult colons. Scale bar, 10 μm.
Altered Expression of Genes Critical for Gut Morphogenesis in Tg(smMHC-Cre-eGFP+/−);Foxf2−/− Colons
To determine molecular mechanisms underlying increased proliferation in Foxf2-deficient colons, we examined expression of genes critical for the Hedgehog pathway, which has been shown to be important for proliferation of smooth muscle cells and their mesenchymal precursors (3, 4, 9). Indian Hedgehog mRNA was increased in adult Tg(smMHC-Cre-eGFP+/−);Foxf2−/− colons but was not changed at P12 (Fig. 5B and data not shown). In agreement with increased Hedgehog signaling, there was increased mRNA expression of Hedgehog target genes Foxf1, Gli1, Gli2, and the Hedgehog receptor Patched in smooth muscle-enriched tissue from Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice (Fig. 5A). Considerably more FOXF1-positive myocytes were observed in the colon of Tg(smMHC-Cre-eGFP+/−);Foxf2−/− compared with control mice (Fig. 5C), further indicating increased Hedgehog signaling. qRT-PCR analysis showed elevated mRNA levels of Wnt5a, BMP4, and myocardin (Fig. 5A), factors critical for gut development. Despite significant alterations in smooth muscle regulatory genes in Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice, muscle markers αSMA and γSMA were unaltered following Foxf2 deletion (Fig. 5C), indicating that Foxf2-deficient smooth muscle cells are fully differentiated. No fibrosis was detected in Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice as visualized by Masson's trichrome stain (Fig. 5C). In addition, there was no observed difference in vascularization of the muscle layers of the colon as indicated by PECAM-1 immunostaining (Fig. 5D). Furthermore, neither FOXA2 nor cytokeratin immunostaining demonstrated any difference in the structure of colon epithelium (Fig. 5D). Altogether, our data indicate that Foxf2 deletion from smooth muscle cells alters expression of genes critical for intestinal morphogenesis.
Foxf2 Binds to Promoter Regions of PDGFa, PDGFb, PDGF Receptor α, Myocardin, and Foxf1 Genes
Because expression of genes critical for the PDGF and Hedgehog signaling pathways was altered in the smooth muscle of Tg(smMHC-Cre-eGFP+/−);Foxf2−/− colons (Fig. 5A), we examined whether FOXF2 could directly bind to promoter regions of these genes. Potential FOXF2-binding sites (TGTTTAT, TATTTAT, or TATTTGT (44)) were identified in the promoter regions of PDGFa, PDGFb, PDGFRα, and Foxf1 genes. Subsequently, a mouse myosarcoma cell line was created that stably expressed a His- and FLAG-tagged Foxf2 construct. ChIP with anti-His antibodies demonstrated that His-FOXF2 bound to the promoter regions of the PDGFa, PDGFb, and PDGFRα genes (Fig. 6A). His-FOXF2 protein also bound to promoter regions of myocardin, a critical co-activator of SRF signaling, and Foxf1 transcription factor, a downstream target of Hedgehog signaling (Fig. 6B). In co-transfection experiments, FOXF2 repressed Foxf1 promoter activity in a concentration-dependent manner as shown by luciferase assay (Fig. 6C). Thus, Foxf1 is a direct transcriptional target of Foxf2.
FIGURE 6.
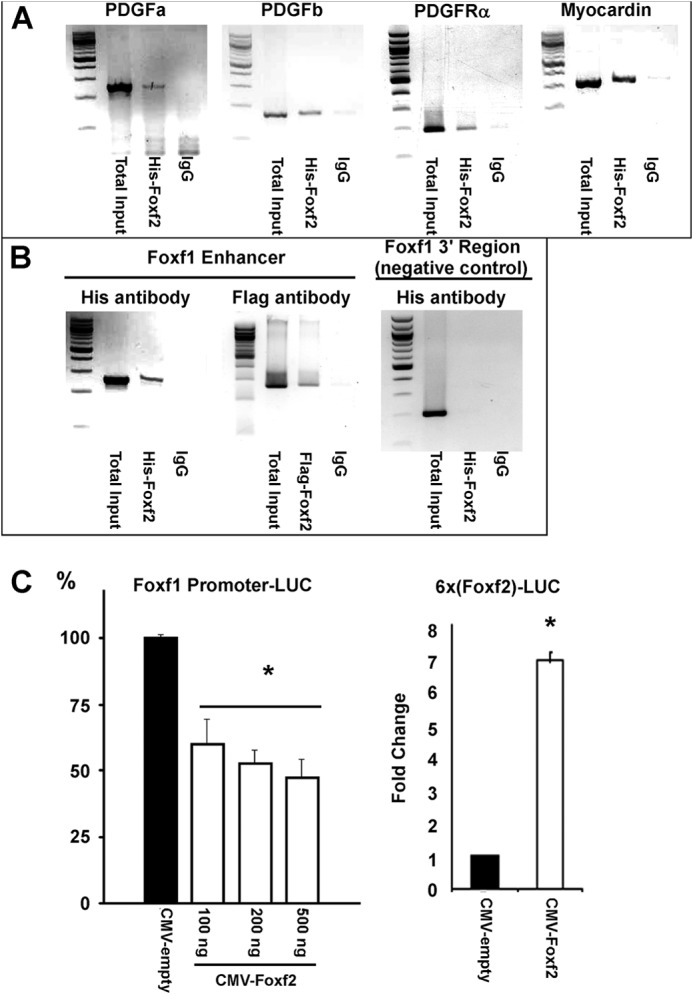
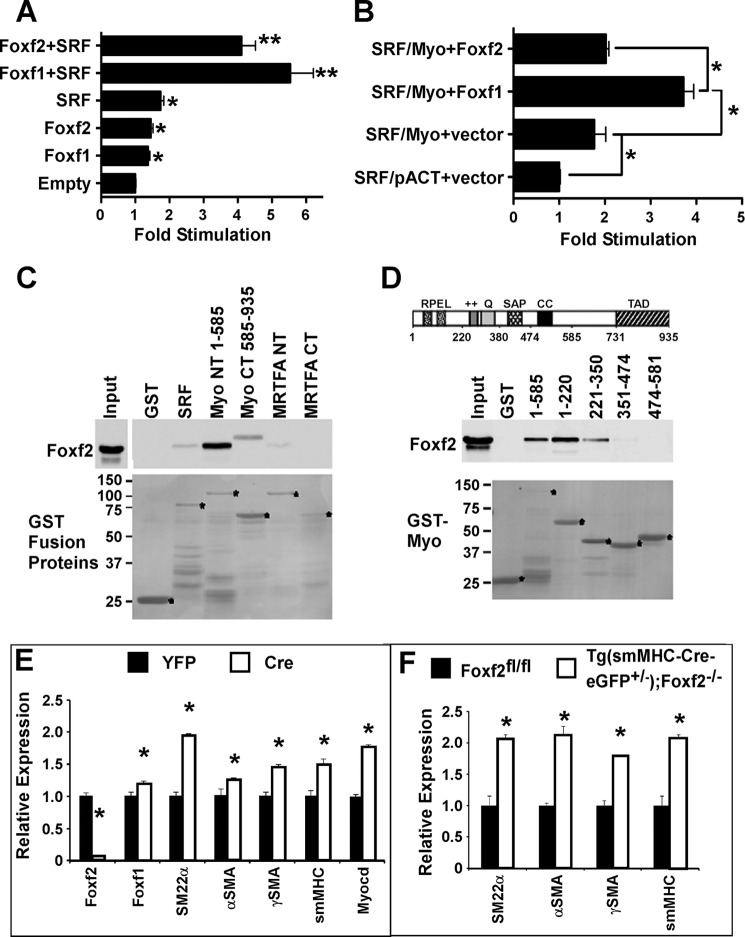
FOXF2 binds to PDGF, myocardin, and Foxf1 promoter regions. A and B, FOXF2 binds to the promoter regions of PDGFa, PDGFb, PDGFRα, myocardin, and Foxf1 genes as determined by ChIP analysis. ChIP was performed using a rhabdomyosarcoma cell line that stably expressed His- and FLAG-tagged FOXF2 protein. Anti-His antibodies or control IgG were used for ChIP. (n = 3.) Negative control for ChIP included a DNA region 1.7 kb 3′ to the Foxf1 regulatory element (Foxf1 3′ Region). Anti-FLAG antibody was used for ChIP of Foxf1 to verify results obtained with anti-His antibody. C, luciferase reporter gene assays show that FOXF2 inhibits transcriptional activity of the Foxf1 promoter (−5.3 kb Foxf1 + 3′ response element) in a dose-dependent manner (left panel). Conversely, FOXF2 activated a reporter gene driven by multiple copies of a FOXF2-binding site (right panel). (n = 3.) *, p < 0.05 versus CMV-empty.
Foxf2 Directly Interacts with Myocardin and Influences Myocardin/SRF Signaling
Gene expression studies in Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice suggest that Foxf2 may regulate multiple signaling pathways during gut morphogenesis. Next, we focused our studies on the myocardin/SRF signaling pathway. SRF and myocardin are known to be essential for smooth muscle development and function (18). Having shown that FOXF2 was able to bind to the myocardin promoter, we investigated the effects of the FOXF proteins on myocardin/SRF signaling in vitro. Co-transfection experiments with an SRF-dependent luciferase reporter (telokin promoter) showed that both FOXF proteins, FOXF1 and FOXF2, can increase SRF-dependent stimulation of the telokin reporter gene in the absence of myocardin, although FOXF2 to a lesser extent (Fig. 7A). A mammalian two-hybrid assay, examining the binding of SRF to myocardin also showed that FOXF1 promotes the interaction of SRF with myocardin, whereas FOXF2 does not (Fig. 7B). Utilizing a glutathione pulldown assay, we showed that FOXF2 protein has a direct physical interaction with myocardin protein (Fig. 7C). FOXF2 was found to bind strongly to the N terminus of myocardin (Myo NT(1–585)). FOXF2 weakly bound to SRF, the C terminus of myocardin (Myo CT(585–935)) and the N terminus of the myocardin family member, MRTFA (Fig. 7C). Further analysis of myocardin deletion mutants determined that FOXF2 binds to a region within the first 350 amino acids of myocardin (Fig. 7D), which contains two RPEL motifs (58). Interestingly, this is distinct from FOXF1 which binds to the myocardin SAP domain (18).
FIGURE 7.
FOXF2 regulates myocardin/SRF signaling in smooth muscle cells. A, FOXF1 and, to a lesser extent, FOXF2 stimulate a SRF-dependent telokin reporter in the absence of myocardin. SRF, Foxf1, and Foxf2 expression plasmids were co-transfected with the SRF-dependent telokin reporter gene into 10TI1/2 cells. **, p < 0.01. B, mammalian two-hybrid assay examining the binding between SRF and myocardin in the presence or absence of FOXF1 or FOXF2. In these assays, SRF was fused to the GAL4 DNA binding domain and myocardin to the GAL4 activation domain. Interaction between the SRF binding domain and myocardin activation domain results in activation of a GAL4-dependent reporter gene (SRF/Myo+vector compared with SRF/pACT+vector: pACT− empty GAL4 activation domain vector). FOXF1 promotes myocardin binding to SRF, although FOXF2 has no effect on myocardin-SRF binding. C, GST pulldown assays in which bacterially expressed FOXF2 was incubated with SRF, myocardin, or MRTFA-GST fusion proteins. Following extensive washing, glutathione-bound fusion proteins were analyzed by Western blot (upper panel). Input GST-fusion proteins visualized by Ponceau staining are shown in the lower panel. FOXF2 protein physically binds to SRF, the N-terminal half of myocardin (Myo NT 1–585), more weakly to the C-terminal half of myocardin (Myo CT 585–935), and weakly with the N-terminal half of MRTFA but not the C-terminal half of MRTFA. D, upper panel, schematic representation of myocardin structural domains. Middle panel, GST pulldown assays using various fragments of myocardin fused to GST. The GST fusion protein-bound FOXF2 is shown in the middle panel and GST fusion protein inputs in the lower panel. FOXF2 specifically bound to the first 220 amino acids of the myocardin protein, which contains RPEL motifs. Weak binding was also observed for amino acids 221–350 of myocardin. E, adenoviral Cre-mediated deletion of Foxf2fl alleles from primary colon smooth muscle cells increased mRNA levels of Foxf1 as well as expression of myocardin/SRF target genes SM22α, αSMA, γSMA, smMHC, and myocardin. mRNA expression levels were quantitated by qRT-PCR and normalized to β-actin (n = 6). *, p < 0.05 versus control. F, mRNA levels of myocardin/SRF target genes SM22α, αSMA, γSMA, and smMHC were increased in adult Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mouse colons (n = 4).
To investigate functional consequences of FOXF2 binding to myocardin on expression of SRF target genes, adenoviral Cre-mediated deletion of Foxf2-floxed alleles was performed in a primary culture of colon smooth muscle cells purified from Foxf2fl/fl mice. Foxf2 mRNA was nearly completely ablated indicating efficient Foxf2 deletion from primary smooth muscle cells (Fig. 7E). Foxf2 deletion significantly increased Foxf1 mRNA (Fig. 7E), a finding consistent with increased Foxf1 expression in Foxf2-deficient colons (Fig. 5A). Foxf2 knockdown increased expression of the SRF target gene smooth muscle 22α (sm22α), αSMA, γSMA, smMHC, and myocardin in vitro (Fig. 7E). Furthermore, increased mRNA levels of SRF target genes were observed in colon tissue obtained from adult Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice (Fig. 7F). Thus, Foxf2 deficiency in colonic smooth muscle cells causes increased expression of genes regulated by the SRF signaling pathway. Altogether, our results indicate that FOXF2 directly binds to myocardin and inhibits myocardin/SRF signaling in smooth muscle cells.
DISCUSSION
To circumvent perinatal lethality in Foxf2−/− mice, we generated a novel mouse model in which Foxf2 is selectively deleted from smooth muscle cells beginning at mid-gestation and throughout the life of the mouse. Loss of Foxf2 from smooth muscle decreased the length of the intestinal tract, truncating both the small intestine and colon. Foxf2 deletion resulted in increased thickness of longitudinal smooth muscle and expansion of the myenteric nerve plexus. PDGF receptor α-positive nerves have been previously observed in the GI tract (54), and staining of control and Tg(smMHC-Cre-eGFP+/−);Foxf2−/− colons showed PDGF receptor α clearly localized to the myenteric plexus with increased staining in Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice. Tg(smMHC-Cre-eGFP+/−);Foxf2−/− colons also had elevated expression of PDGFa and PDGFb. Increased PDGF signaling can contribute to expansion of the myenteric plexus in Foxf2-deficient colons as PDGF signaling has been previously shown to induce neural mitosis and hyperplasia (55, 59, 60). Interestingly, Foxf2−/− mice have been reported to lack ganglia in the colon (17), which is in stark contrast to the increased myenteric plexus observed in our Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice. The reason for this discrepancy could be due to the contribution of Foxf2-regulated signals from the lamina propria, which is not targeted by smMHC-Cre-eGFP and thus not Foxf2-deficient in our mouse model. Foxf2 may regulate the expression of genes essential for neural crest cell migration, proliferation, or survival. Alternatively, the difference may be due to the timing in which Foxf2 is deleted, as smMHC-Cre-eGFP is not detected prior to E12.5 (27), which is after the initiation of neural crest infiltration of the colon (61).
In addition to expression in enteric neurons, PDGF receptor α, and PDGF ligands are expressed in colon smooth muscle (1, 57). PDGF has been shown to be a potent mitogenic agent in smooth muscle cells (62), and a cell autonomous role for PDGF signaling in smooth muscle proliferation has been previously demonstrated (63). Although we did not observe any smooth muscle cell proliferation in the colon of adult Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice, we did observe increased proliferation during early postnatal development. Unlike vascular smooth muscle, smooth muscle of the GI tract continues to proliferate for the first few days after birth. By 12 days after birth, proliferation of colon smooth muscle cells in control mice appears to have ceased. In contrast, in Tg(smMHC-Cre-eGFP+/−);Foxf2−/− mice there was still detectable proliferation in the circular muscle layer of the colon and marked proliferation in the longitudinal muscle layer. The increased expression of PDGF ligands and receptor in Foxf2-deficient colons could thus be stimulating proliferation of the neonatal colonic smooth muscle and/or delaying cell cycle exit. It has been previously shown that PDGF ligands originate from the circular muscle layer and myenteric plexus of the gut, and PDGF receptor α is more highly expressed in the longitudinal muscle than the circular smooth muscle layer (57). This expression pattern could explain why the circular muscle layer is less affected by Foxf2 deletion, because it lacks the receptor to respond to PDGF ligands. Increased expression of PDGF proteins, however, does not appear to prevent colonic smooth muscle cells from ultimately exiting the cell cycle and differentiating. Cell cycle exit coupled with the direct increase in myocardin expression seen following loss of Foxf2 likely contributes to increased expression of contractile proteins in the adult colon.
The experiments performed in this study provide novel insight into the in vivo signaling of Foxf2 as well as regulatory pathways for postnatal visceral smooth muscle development, particularly in the intestines. It was previously shown that FOXF1 and FOXF2 bind to the same consensus cis-acting regulatory element sequence (28). This together with analysis of global knock-out and compound Foxf1/f2 heterozygous mice suggests that Foxf1 and Foxf2 may be partially redundant (17). In contrast, our studies have clearly shown that Foxf1 and Foxf2 have quite distinct roles in GI smooth muscle cells. We have previously shown that loss of Foxf1 from smooth muscle cells results in decreased contractile protein expression (18); however, in this study we found that loss of Foxf2 increases expression of these genes. Binding experiments show that FOXF1 and FOXF2 bind to different regions of myocardin (this study and Ref. 18), and the effects of this binding are quite distinct. FOXF2 is a weak activator of SRF-regulated genes as compared with FOXF1, and binding of FOXF1 to myocardin promotes its interaction with SRF, whereas the binding of FOXF2 to myocardin does not. Our data indicate that Foxf2 may act as a functional decoy for Foxf1, having only minimal co-stimulatory activity, at least as pertains to myocardin/SRF signaling. In addition, FOXF2 inhibits Foxf1 promoter activity, which can further exacerbate diminished myocardin/SRF signaling.
In addition to regulating the activity of myocardin/SRF complexes through protein-protein interactions, FOXF2 also directly affects the expression of myocardin through binding to a forkhead site in the myocardin promoter. ChIP analysis showed that FOXF2 can bind to the promoter region of myocardin, implicating Foxf2 in transcriptional regulation of the myocardin gene. Our data indicate that Foxf2 influences GI tract development via regulation of PDGF, SRF and Hedgehog pathways, all of which have been shown to be critical for GI tract development and physiology. Foxf2 may directly or indirectly regulate expression of genes critical for all three pathways and through this regulation mediate postnatal intestinal development and maintenance.
In summary, we generated a novel mouse line containing Foxf2-floxed alleles and used it to create smooth muscle-specific Foxf2 knock-out mice. Utilizing this model, we demonstrated a distinct phenotype for smooth muscle Foxf2 deletion compared with Foxf2−/− mice. We also showed that FOXF2 bound to the promoter region of the myocardin gene as well as several members of the PDGF and Hedgehog signaling pathways. FOXF2 directly binds to and inhibits Foxf1 promoter activity and regulates SRF signaling through a protein-protein interaction with myocardin. By its capacity to regulate PDGF, Hedgehog and SRF signaling, Foxf2 is a critical regulator of GI tract development and maintenance.
Acknowledgment
We thank Yufang Zhang for excellent technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants HL84151 (to V. V. K.), HL123490 (to V. V. K.), CA142724 (to T. V. K.), and DK061130 (to B. P. H.).
- GI
- gastrointestinal
- Fox
- Forkhead box
- smMHC
- smooth muscle myosin heavy chain
- P
- postnatal day
- E
- embryonic day
- qRT-PCR
- quantitative real time RT-PCR
- CGRP
- calcitonin gene-related peptide
- PDGFR
- platelet-derived growth factor receptor
- SRF
- serum response factor
- neo
- neomycin
- SMA
- smooth muscle actin
- MRTFA
- myocardin-related transcription factor A.
REFERENCES
- 1. McLin V. A., Henning S. J., Jamrich M. (2009) The role of the visceral mesoderm in the development of the gastrointestinal tract. Gastroenterology 136, 2074–2091 [DOI] [PubMed] [Google Scholar]
- 2. Stoll B. J., Hansen N. I., Bell E. F., Shankaran S., Laptook A. R., Walsh M. C., Hale E. C., Newman N. S., Schibler K., Carlo W. A., Kennedy K. A., Poindexter B. B., Finer N. N., Ehrenkranz R. A., Duara S., et al. (2010) Neonatal outcomes of extremely preterm infants from the NICHD Neonatal Research Network. Pediatrics 126, 443–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Apelqvist A., Ahlgren U., Edlund H. (1997) Sonic hedgehog directs specialised mesoderm differentiation in the intestine and pancreas. Curr. Biol. 7, 801–804 [DOI] [PubMed] [Google Scholar]
- 4. Theodosiou N. A., Tabin C. J. (2003) Wnt signaling during development of the gastrointestinal tract. Dev. Biol. 259, 258–271 [DOI] [PubMed] [Google Scholar]
- 5. Torihashi S., Hattori T., Hasegawa H., Kurahashi M., Ogaeri T., Fujimoto T. (2009) The expression and crucial roles of BMP signaling in development of smooth muscle progenitor cells in the mouse embryonic gut. Differentiation 77, 277–289 [DOI] [PubMed] [Google Scholar]
- 6. Kedinger M., Simon-Assmann P., Bouziges F., Arnold C., Alexandre E., Haffen K. (1990) Smooth muscle actin expression during rat gut development and induction in fetal skin fibroblastic cells associated with intestinal embryonic epithelium. Differentiation 43, 87–97 [DOI] [PubMed] [Google Scholar]
- 7. McCulloch D. R., Le Goff C., Bhatt S., Dixon L. J., Sandy J. D., Apte S. S. (2009) Adamts5, the gene encoding a proteoglycan-degrading metalloprotease, is expressed by specific cell lineages during mouse embryonic development and in adult tissues. Gene Expr. Patterns 9, 314–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Santa Barbara P., Williams J., Goldstein A. M., Doyle A. M., Nielsen C., Winfield S., Faure S., Roberts D. J. (2005) Bone morphogenetic protein signaling pathway plays multiple roles during gastrointestinal tract development. Dev. Dyn. 234, 312–322 [DOI] [PubMed] [Google Scholar]
- 9. Wallace K. N., Akhter S., Smith E. M., Lorent K., Pack M. (2005) Intestinal growth and differentiation in zebrafish. Mech. Dev. 122, 157–173 [DOI] [PubMed] [Google Scholar]
- 10. Spence J. R., Lauf R., Shroyer N. F. (2011) Vertebrate intestinal endoderm development. Dev. Dyn. 240, 501–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Flynn R. S., Mahavadi S., Murthy K. S., Kellum J. M., Kuemmerle J. F. (2009) Insulin-like growth factor-binding protein-5 stimulates growth of human intestinal muscle cells by activation of Gαi3. Am. J. Physiol. Gastrointest. Liver Physiol. 297, G1232–G1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hazelgrove K. B., Flynn R. S., Qiao L. Y., Grider J. R., Kuemmerle J. F. (2009) Endogenous IGF-I and αvβ3 integrin ligands regulate increased smooth muscle growth in TNBS-induced colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G1230–G1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kalin T. V., Ustiyan V., Kalinichenko V. V. (2011) Multiple faces of FoxM1 transcription factor: lessons from transgenic mouse models. Cell Cycle 10, 396–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ormestad M., Astorga J., Carlsson P. (2004) Differences in the embryonic expression patterns of mouse Foxf1 and -2 match their distinct mutant phenotypes. Dev. Dyn. 229, 328–333 [DOI] [PubMed] [Google Scholar]
- 15. Kalinichenko V. V., Lim L., Stolz D. B., Shin B., Rausa F. M., Clark J., Whitsett J. A., Watkins S. C., Costa R. H. (2001) Defects in pulmonary vasculature and perinatal lung hemorrhage in mice heterozygous null for the Forkhead Box f1 transcription factor. Dev. Biol. 235, 489–506 [DOI] [PubMed] [Google Scholar]
- 16. Mahlapuu M., Ormestad M., Enerbäck S., Carlsson P. (2001) The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development 128, 155–166 [DOI] [PubMed] [Google Scholar]
- 17. Ormestad M., Astorga J., Landgren H., Wang T., Johansson B. R., Miura N., Carlsson P. (2006) Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development 133, 833–843 [DOI] [PubMed] [Google Scholar]
- 18. Hoggatt A. M., Kim J. R., Ustiyan V., Ren X., Kalin T. V., Kalinichenko V. V., Herring B. P. (2013) The transcription factor Foxf1 binds to serum response factor and myocardin to regulate gene transcription in visceral smooth muscle cells. J. Biol. Chem. 288, 28477–28487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoggatt A. M., Kriegel A. M., Smith A. F., Herring B. P. (2000) Hepatocyte nuclear factor-3 homologue 1 (HFH-1) represses transcription of smooth muscle-specific genes. J. Biol. Chem. 275, 31162–31170 [DOI] [PubMed] [Google Scholar]
- 20. Liu Z. P., Wang Z., Yanagisawa H., Olson E. N. (2005) Phenotypic modulation of smooth muscle cells through interaction of Foxo4 and myocardin. Dev. Cell 9, 261–270 [DOI] [PubMed] [Google Scholar]
- 21. Aitola M., Carlsson P., Mahlapuu M., Enerbäck S., Pelto-Huikko M. (2000) Forkhead transcription factor FoxF2 is expressed in mesodermal tissues involved in epithelio-mesenchymal interactions. Dev. Dyn. 218, 136–149 [DOI] [PubMed] [Google Scholar]
- 22. McLin V. A., Shah R., Desai N. P., Jamrich M. (2010) Identification and gastrointestinal expression of Xenopus laevis FoxF2. Int. J. Dev. Biol. 54, 919–924 [DOI] [PubMed] [Google Scholar]
- 23. van der Heul-Nieuwenhuijsen L., Dits N., Van Ijcken W., de Lange D., Jenster G. (2009) The FOXF2 pathway in the human prostate stroma. Prostate 69, 1538–1547 [DOI] [PubMed] [Google Scholar]
- 24. Nik A. M., Reyahi A., Pontén F., Carlsson P. (2013) Foxf2 in intestinal fibroblasts reduces numbers of Lgr5(+) stem cells and adenoma formation by inhibiting Wnt signaling. Gastroenterology 144, 1001–1011 [DOI] [PubMed] [Google Scholar]
- 25. Philip-Couderc P., Tavares N. I., Roatti A., Lerch R., Montessuit C., Baertschi A. J. (2008) Forkhead transcription factors coordinate expression of myocardial KATP channel subunits and energy metabolism. Circ. Res. 102, e20–e35 [DOI] [PubMed] [Google Scholar]
- 26. McKeone R., Vieira H., Gregory-Evans K., Gregory-Evans C. Y., Denny P. (2011) Foxf2: a novel locus for anterior segment dysgenesis adjacent to the Foxc1 gene. PLoS One 6, e25489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xin H. B., Deng K. Y., Rishniw M., Ji G., Kotlikoff M. I. (2002) Smooth muscle expression of Cre recombinase and eGFP in transgenic mice. Physiol. Genomics 10, 211–215 [DOI] [PubMed] [Google Scholar]
- 28. Hellqvist M., Mahlapuu M., Samuelsson L., Enerbäck S., Carlsson P. (1996) Differential activation of lung-specific genes by two forkhead proteins, FREAC-1 and FREAC-2. J. Biol. Chem. 271, 4482–4490 [DOI] [PubMed] [Google Scholar]
- 29. Malin D., Kim I. M., Boetticher E., Kalin T. V., Ramakrishna S., Meliton L., Ustiyan V., Zhu X., Kalinichenko V. V. (2007) Forkhead box F1 is essential for migration of mesenchymal cells and directly induces integrin-β3 expression. Mol. Cell. Biol. 27, 2486–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kalin T. V., Wang I. C., Meliton L., Zhang Y., Wert S. E., Ren X., Snyder J., Bell S. M., Graf L., Jr., Whitsett J. A., Kalinichenko V. V. (2008) Forkhead Box m1 transcription factor is required for perinatal lung function. Proc. Natl. Acad. Sci. U.S.A. 105, 19330–19335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim I. M., Ramakrishna S., Gusarova G. A., Yoder H. M., Costa R. H., Kalinichenko V. V. (2005) The forkhead box m1 transcription factor is essential for embryonic development of pulmonary vasculature. J. Biol. Chem. 280, 22278–22286 [DOI] [PubMed] [Google Scholar]
- 32. Kim I. M., Zhou Y., Ramakrishna S., Hughes D. E., Solway J., Costa R. H., Kalinichenko V. V. (2005) Functional characterization of evolutionarily conserved DNA regions in forkhead box f1 gene locus. J. Biol. Chem. 280, 37908–37916 [DOI] [PubMed] [Google Scholar]
- 33. Ramakrishna S., Kim I. M., Petrovic V., Malin D., Wang I. C., Kalin T. V., Meliton L., Zhao Y. Y., Ackerson T., Qin Y., Malik A. B., Costa R. H., Kalinichenko V. V. (2007) Myocardium defects and ventricular hypoplasia in mice homozygous null for the Forkhead Box M1 transcription factor. Dev. Dyn. 236, 1000–1013 [DOI] [PubMed] [Google Scholar]
- 34. Balli D., Zhang Y., Snyder J., Kalinichenko V. V., Kalin T. V. (2011) Endothelial cell-specific deletion of transcription factor FoxM1 increases urethane-induced lung carcinogenesis. Cancer Res. 71, 40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bolte C., Newman G., Schultz Jel J. (2009) Hypertensive state, independent of hypertrophy, exhibits an attenuated decrease in systolic function on cardiac κ-opioid receptor stimulation. Am. J. Physiol. Heart Circ. Physiol. 296, H967–H975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bolte C., Zhang Y., Wang I. C., Kalin T. V., Molkentin J. D., Kalinichenko V. V. (2011) Expression of Foxm1 transcription factor in cardiomyocytes is required for myocardial development. PLoS One 6, e22217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ren X., Shah T. A., Ustiyan V., Zhang Y., Shinn J., Chen G., Whitsett J. A., Kalin T. V., Kalinichenko V. V. (2013) FOXM1 promotes allergen-induced goblet cell metaplasia and pulmonary inflammation. Mol. Cell. Biol. 33, 371–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ren X., Zhang Y., Snyder J., Cross E. R., Shah T. A., Kalin T. V., Kalinichenko V. V. (2010) Forkhead box M1 transcription factor is required for macrophage recruitment during liver repair. Mol. Cell. Biol. 30, 5381–5393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ustiyan V., Wang I. C., Ren X., Zhang Y., Snyder J., Xu Y., Wert S. E., Lessard J. L., Kalin T. V., Kalinichenko V. V. (2009) Forkhead box M1 transcriptional factor is required for smooth muscle cells during embryonic development of blood vessels and esophagus. Dev. Biol. 336, 266–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang I. C., Meliton L., Ren X., Zhang Y., Balli D., Snyder J., Whitsett J. A., Kalinichenko V. V., Kalin T. V. (2009) Deletion of Forkhead Box M1 transcription factor from respiratory epithelial cells inhibits pulmonary tumorigenesis. PLoS One 4, e6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Balli D., Ustiyan V., Zhang Y., Wang I. C., Masino A. J., Ren X., Whitsett J. A., Kalinichenko V. V., Kalin T. V. (2013) Foxm1 transcription factor is required for lung fibrosis and epithelial-to-mesenchymal transition. EMBO J. 32, 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cheng X. H., Black M., Ustiyan V., Le T., Fulford L., Sridharan A., Medvedovic M., Kalinichenko V. V., Whitsett J. A., Kalin T. V. (2014) SPDEF inhibits prostate carcinogenesis by disrupting a positive feedback loop in regulation of the Foxm1 oncogene. PLoS Genet. 10, e1004656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Singh T. R., Saro D., Ali A. M., Zheng X. F., Du C. H., Killen M. W., Sachpatzidis A., Wahengbam K., Pierce A. J., Xiong Y., Sung P., Meetei A. R. (2010) MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol. Cell 37, 879–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peterson R. S., Lim L., Ye H., Zhou H., Overdier D. G., Costa R. H. (1997) The winged helix transcriptional activator HFH-8 is expressed in the mesoderm of the primitive streak stage of mouse embryos and its cellular derivatives. Mech. Dev. 69, 53–69 [DOI] [PubMed] [Google Scholar]
- 45. Wang I. C., Snyder J., Zhang Y., Lander J., Nakafuku Y., Lin J., Chen G., Kalin T. V., Whitsett J. A., Kalinichenko V. V. (2012) Foxm1 mediates cross-talk between Kras/mitogen-activated protein kinase and canonical Wnt pathways during development of respiratory epithelium. Mol. Cell. Biol. 32, 3838–3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhou J., Hoggatt A. M., Herring B. P. (2004) Activation of the smooth muscle-specific telokin gene by thyrotroph embryonic factor (TEF). J. Biol. Chem. 279, 15929–15937 [DOI] [PubMed] [Google Scholar]
- 47. Ren X., Ustiyan V., Pradhan A., Cai Y., Havrilak J. A., Bolte C. S., Shannon J. M., Kalin T. V., Kalinichenko V. V. (2014) FOXF1 transcription factor is required for formation of embryonic vasculature by regulating VEGF signaling in endothelial cells. Circ. Res. 115, 709–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang I. C., Ustiyan V., Zhang Y., Cai Y., Kalin T. V., Kalinichenko V. V. (2014) Foxm1 transcription factor is required for the initiation of lung tumorigenesis by oncogenic Kras(G12D.). Oncogene 33, 5391–5396 [DOI] [PubMed] [Google Scholar]
- 49. Cai Y., Balli D., Ustiyan V., Fulford L., Hiller A., Misetic V., Zhang Y., Paluch A. M., Waltz S. E., Kasper S., Kalin T. V. (2013) Foxm1 expression in prostate epithelial cells is essential for prostate carcinogenesis. J. Biol. Chem. 288, 22527–22541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou J., Hu G., Herring B. P. (2005) Smooth muscle-specific genes are differentially sensitive to inhibition by Elk-1. Mol. Cell. Biol. 25, 9874–9885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Herring B. P., Kriegel A. M., Hoggatt A. M. (2001) Identification of Barx2b, a serum response factor-associated homeodomain protein. J. Biol. Chem. 276, 14482–14489 [DOI] [PubMed] [Google Scholar]
- 52. Zhang M., Fang H., Zhou J., Herring B. P. (2007) A novel role of Brg1 in the regulation of SRF/MRTFA-dependent smooth muscle-specific gene expression. J. Biol. Chem. 282, 25708–25716 [DOI] [PubMed] [Google Scholar]
- 53. Zhou J., Zhang M., Fang H., El-Mounayri O., Rodenberg J. M., Imbalzano A. N., Herring B. P. (2009) The SWI/SNF chromatin remodeling complex regulates myocardin-induced smooth muscle-specific gene expression. Arterioscler. Thromb. Vasc. Biol. 29, 921–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Blair P. J., Bayguinov Y., Sanders K. M., Ward S. M. (2012) Relationship between enteric neurons and interstitial cells in the primate gastrointestinal tract. Neurogastroenterol. Motil. 24, e437–e449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Eccleston P. A., Funa K., Heldin C. H. (1993) Expression of platelet-derived growth factor (PDGF) and PDGF α- and β-receptors in the peripheral nervous system: an analysis of sciatic nerve and dorsal root ganglia. Dev. Biol. 155, 459–470 [DOI] [PubMed] [Google Scholar]
- 56. Lobsiger C. S., Schweitzer B., Taylor V., Suter U. (2000) Platelet-derived growth factor-BB supports the survival of cultured rat Schwann cell precursors in synergy with neurotrophin-3. Glia 30, 290–300 [DOI] [PubMed] [Google Scholar]
- 57. Kurahashi M., Niwa Y., Cheng J., Ohsaki Y., Fujita A., Goto H., Fujimoto T., Torihashi S. (2008) Platelet-derived growth factor signals play critical roles in differentiation of longitudinal smooth muscle cells in mouse embryonic gut. Neurogastroenterol. Motil. 20, 521–531 [DOI] [PubMed] [Google Scholar]
- 58. Guettler S., Vartiainen M. K., Miralles F., Larijani B., Treisman R. (2008) RPEL motifs link the serum response factor cofactor MAL but not myocardin to Rho signaling via actin binding. Mol. Cell. Biol. 28, 732–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fruttiger M., Calver A. R., Richardson W. D. (2000) Platelet-derived growth factor is constitutively secreted from neuronal cell bodies but not from axons. Curr. Biol. 10, 1283–1286 [DOI] [PubMed] [Google Scholar]
- 60. Reneker L. W., Overbeek P. A. (1996) Lens-specific expression of PDGF-A in transgenic mice results in retinal astrocytic hamartomas. Invest. Ophthalmol. Vis. Sci. 37, 2455–2466 [PubMed] [Google Scholar]
- 61. Young H. M., Turner K. N., Bergner A. J. (2005) The location and phenotype of proliferating neural-crest-derived cells in the developing mouse gut. Cell Tissue Res. 320, 1–9 [DOI] [PubMed] [Google Scholar]
- 62. Stanzel R. D., Lourenssen S., Nair D. G., Blennerhassett M. G. (2010) Mitogenic factors promoting intestinal smooth muscle cell proliferation. Am. J. Physiol. Cell Physiol. 299, C805–C817 [DOI] [PubMed] [Google Scholar]
- 63. Graham M. F., Willey A., Adams J., Yager D., Diegelmann R. F. (1996) Interleukin 1β down-regulates collagen and augments collagenase expression in human intestinal smooth muscle cells. Gastroenterology 110, 344–350 [DOI] [PubMed] [Google Scholar]