Abstract
How Darwin’s “endless forms most beautiful” have evolved remains one of the most exciting questions in biology. The significant variety of bacterial shapes is most likely due to the specific advantages they confer with respect to the diverse environments they occupy. While our understanding of the mechanisms generating relatively simple shapes has improved tremendously in the last few years, the molecular mechanisms underlying the generation of complex shapes and the evolution of shape diversity are largely unknown. The emerging field of bacterial evolutionary cell biology provides a novel strategy to answer this question in a comparative phylogenetic framework. This relatively novel approach provides hypotheses and insights into cell biological mechanisms, such as morphogenesis, and their evolution that would have been difficult to obtain by studying only model organisms. We discuss the necessary steps, challenges, and impact of integrating “evolutionary thinking” into bacterial cell biology in the genomic era.
Keywords: bacterial shape, evolutionary cell biology, morphological transitions, evolutionary developmental biology, co-option, tree-thinking, non-model organisms
Introduction
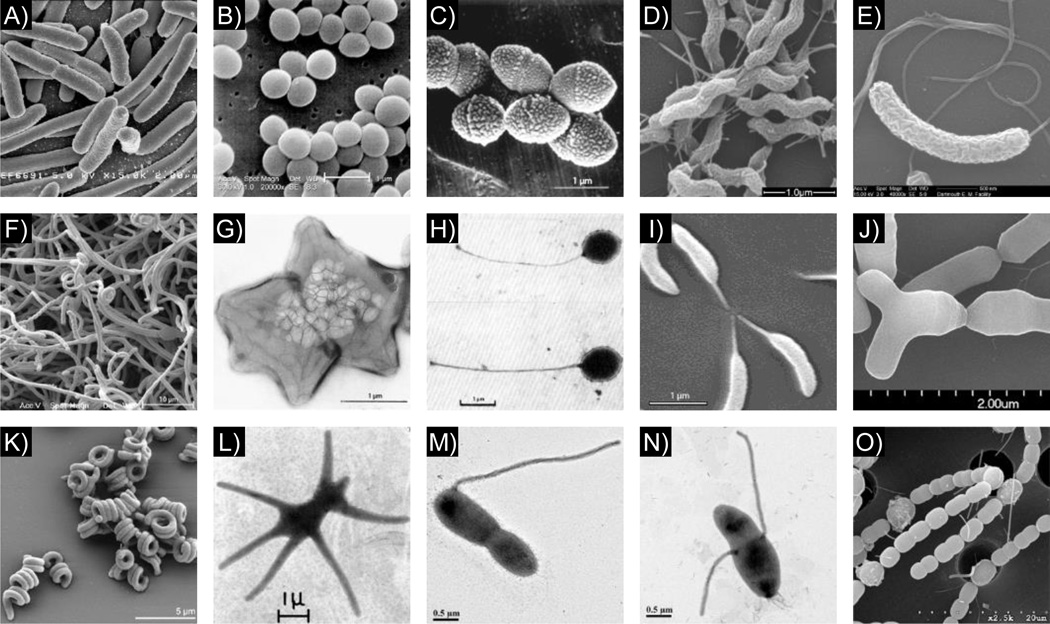
It is difficult not to marvel at the amazing diversity of shapes in the living world: we witness it every day when we encounter plants and animals of different shapes and sizes. How the diversity of organismal forms evolved remains one of the most fundamental and fascinating questions in biology. Since the dawn of microbiology, shape, in particular the classic rod, sphere, and spiral forms, has served as an important descriptor of bacterial species. A simple, but often overlooked, fact is that there is significant morphological diversity in the microbial world, hidden to the naked eye. Under the microscope, bacteria can be found in multiple shapes and sizes, from simple spheres, rods, and spirals to unconventional chains, coils, stars, and more complex shapes such as branching filaments or bacteria that radiate cell envelope extensions from the cell body [1] (Fig. 1).
Figure 1.
The diversity of bacterial shapes. For each shape, a brief description and the name of one representative species is provided, followed by the image source in parenthesis. A: Rod, Escherichia coli (NIAID); B: Sphere, Staphylococcus aureus (Janice Haney Carr, CDC); C: Ovococcoid, Streptococcus pneumoniae [105]; D: Spiral, Campylobacter jejuni [106]; E: Crescent, Vibrio cholerae (Louisa Howard, Dartmouth College); F: Branched filaments, Streptomyces coelicolor (Paul Hoskisson, University of Strathclyde); G: Star, Stella vacuolata [107]; H: Stalked, Planctomyces maris [108]; I: Stalked and crescent, Caulobacter crescentus (Ellen Quardokus, Indiana University); J: Bifid/Y-shaped, Bifidobacterium breve (Daria Zhurina and Paul Walther, University of Ulm); K: Coil, Spirosoma linguale [109]; L: Multi-stalked, Ancalomicrobium adetum [110]; M: Stalked, Asticcacaulis excentricus (Chao Jiang, Stanford University); N: Stalked, Asticcacaulis biprosthecum (Chao Jiang, Stanford University); O: Chain and Heterocysts, Anabaena variabilis (Jinshun Zhong, University of Missouri-St. Louis) [111]. All images are reproduced with permission.
It is intuitive that the different shapes observed in the macroscopic world, for example fins, wings, or a long neck, confer specific advantages. Why a particular bacterium has a given shape is a difficult question to answer, one confounded by the fact that a single shape rarely dominates a given environment and simple shapes such as spheres, ovoids, and rods can be found in a variety of environments. Ultimately, shape will be influenced by a combination of factors, including, but not limited to, nutrient availability, attachment and dispersal strategies, motility requirements, and predation, and therefore more than one shape may provide advantages in a given environment [1]. While the advantages of most bacterial shapes are yet to be determined [1], the high fidelity of bacterial species morphology and the conservation of shapes spanning distant taxa (and hence long periods of time) suggest that their shapes confer specific advantages. For example, Helicobacter pylori is hypothesized to use its corkscrew shape to traverse the thick mucus layer that covers and protects the epithelial lining of the stomach mucosa, and shape mutants that have lost this characteristic helical twist exhibit attenuated stomach colonization [2–4]. Other examples come from aquatic bacteria living in oligotrophic environments. Oligotrophy is often connected with small coccoid bacteria, simply because this shape increases the surface/volume ratio [1]. Another morphological feature found in oligotrophic bacteria, albeit less frequently than the small coccoid shape, is known as the stalk, a thin cylindrical extension of the cell envelope that protrudes from the cell body and serves as a nutrient scavenging antenna thought to improve the efficiency of nutrient uptake [5] (Figs. 1 and 2). In addition, some bacterial species can vary their shape in order to optimize their ability to survive and reproduce in different environmental conditions or as a natural part of their life cycle, a process known as morphological plasticity [6]. When the filamentous soil bacterium Streptomyces coelicolor finds itself in a favorable environment, it forms a branched vegetative mycelium that allows it to spread and burrow deep into the surrounding substrate (Fig. 1F). However, when the environment becomes unfavorable, the branches extend upwards from the surface to form aerial hyphae, which differentiate further into a series of spores that are released into the environment to facilitate cell dispersal [7]. Morphological plasticity is also a hallmark of a number of pathogens. For example, uropathogenic Escherichia coli (UPEC) switches from non-motile rods to cocci, then to motile rods, and ultimately to a filamentous form whose size is thought to prevent phagocytosis during the course of infection [6]. H. pylori and Campylobacter jejuni have been shown to assume coccoid forms after starvation and, although these coccid-shaped cells are non-cultivable, research has shown that these cells are still able to infect hosts [8,9].
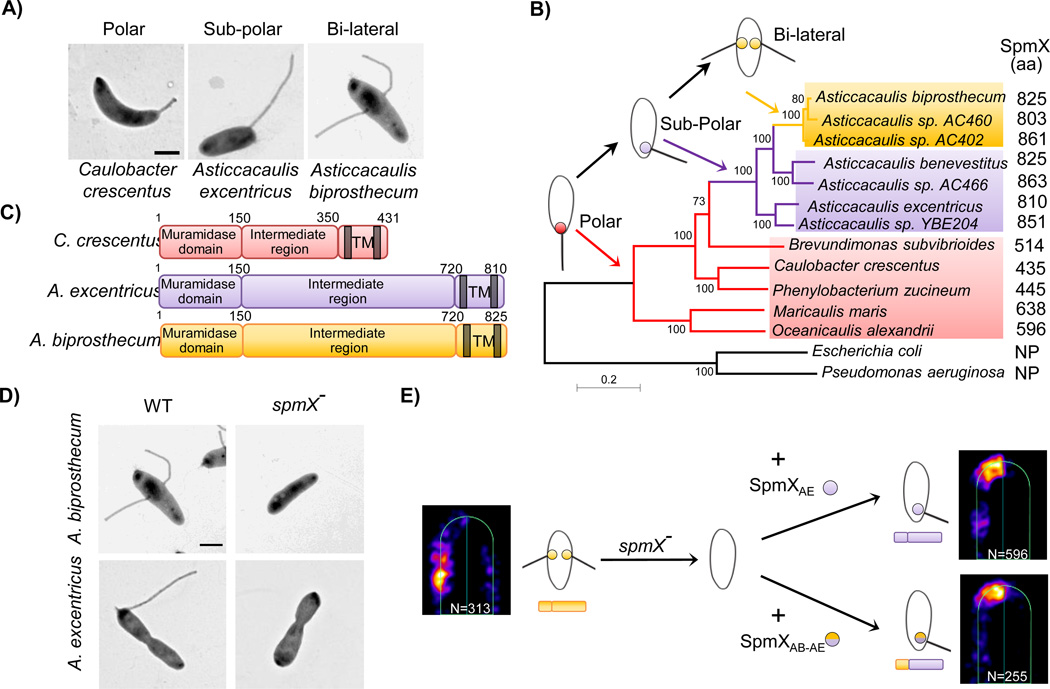
Figure 2.
SpmX is the evolving morphogen of stalk synthesis. A: Transmission Electron micrographs of three species with distinct stalk positioning. B: Phylogenetic tree and inferred evolutionary trajectory of stalk positioning. Colors of shading, branches, and SpmX (filled circles) denote the polar (red), sub-polar (purple), and bi-lateral (yellow) stalk positioning, respectively. Arrows point to the origin of respective morphologies. The size of SpmX is indicated in amino acids (aa). NP, orthologs not present. Scale bar, number of substitutions per site. C: Domain organization of SpmX. Transmembrane domains (TM) are shown as grey bars. All versions of SpmX share a conserved N-terminal putative muramidase domain and two C-terminal transmembrane domains. However, the intermediate region is highly variable in both length and sequence. D: SpmX is required for stalk synthesis in Asticcacaulis. Transmission electron microscopy images of Asticcacaulis species and their respective spmX− stalkless mutants. E: Heat maps of SpmX localization in the A. biprosthecum spmX− mutant expressing SpmXAB-EGFP (left), SpmXAE-EGFP (Right, top) or the chimeric SpmXAB-AE-EGFP, with the N-terminal A. biprosthecum muramidase domain fused to the C-terminal A. excentricus intermediate and TM domains (Right, bottom). Notice that both SpmXAE-EGFP and the chimeric SpmXAB-AE-EGFP are able to drive morphological transitions from bi-lateral to sub-polar predominantly. “N” indicates the number of foci analyzed. Figure adapted from Jiang et al (2014) with permission.
Phylogenetic analysis suggests that the last common ancestor of bacteria was probably rod shaped, giving rise at various times to cocci/ovococci or other shapes [10]. It has been suggested that there is a significant correlation between cell shape and the arrangement of the dcw cluster of genes involved in cell division and cell wall synthesis [11], but it remains to be seen if this correlation still holds, given the ever growing amount of available genomic data. Morphological variations are often found in closely related bacterial species: the diversity in the number and positioning of stalks or flagella in several phyla [12–16], the variation in the number and shape of endospores in the Firmicutes [17], the structural diversity of fruiting bodies of Myxobacteria [18], and the diverse helical shapes within the Helicobacter and Campylobacter genera [19,20] to name but a few. The mechanisms that control shape changes within a bacterial species are beginning to be understood, but the mechanisms by which new morphologies evolved from ancestral ones mostly remain to be described. However, while several studies have pinpointed the molecular mechanisms behind morphological transitions in multicellular eukaryotes, highlighting the importance of regulatory and functional sequence evolution in the plant and animal kingdoms [21–25], the mechanisms underlying the transitions leading to the morphological diversity of bacteria remain unknown. In this review, we describe the mechanisms of bacterial cell shape generation and evolution, and we discuss the importance of conducting future bacterial cell and developmental biology studies in a comparative phylogenetic framework.
The nature of bacterial shape
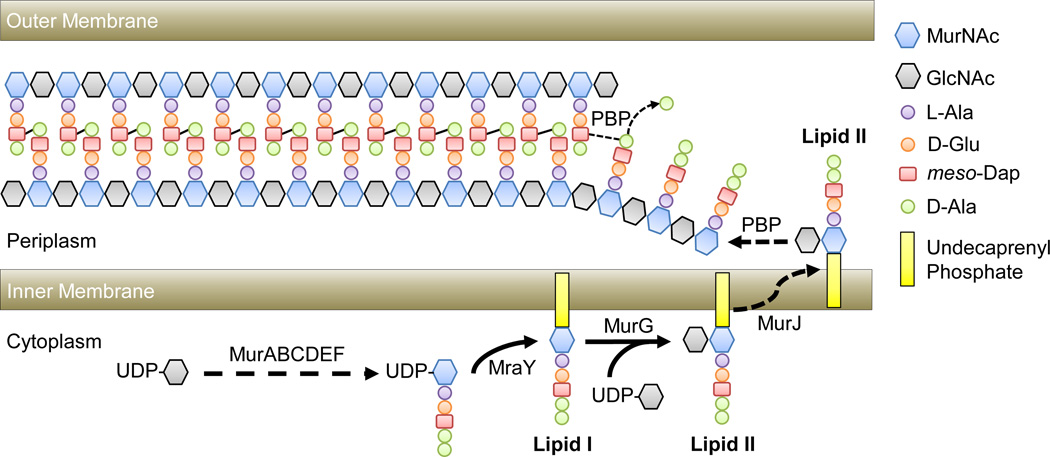
The bacterial cell wall plays a pivotal role in maintaining the shape of bacterial cells [26]. The most prevalent form of bacterial cell wall is the peptidoglycan (PG), a structure composed of glycan strands made of repeating disaccharide subunits composed of N-Acetylmuramic acid (MurNAc) and N-Acetylglucosamine (GlcNAc), which are further crosslinked by pentapeptide bridges attached to the MurNAc units (For details please see Fig. 3). The resulting mesh-like structure is rigid enough to maintain bacterial shapes, yet also is elastic and can be dynamically modified [26,27]. Indeed, disruption of PG structure or synthesis in E. coli and Bacillus subtilis can quickly lead to a round-shaped cell called a “spheroplast” [26,28,29].
Figure 3.
A simplified model of peptidoglycan synthesis in E. coli. PG synthesis is a complex process coordinated by a number of proteins that are conserved in almost all bacterial species. Due to the scope of this review, we will only briefly cover the biochemical fundamentals of PG synthesis using E. coli as an example (for detailed reviews see [26,27]). PG synthesis begins with the synthesis of PG precursors in the cytoplasm. The nucleotide sugar uridine diphosphate N-Acetylglucosamine (UDP-GlcNAc) is converted to uridine diphosphate N-Acetylmuramic acid (UDP-MurNAc) by MurAB. MurCEDF then catalyze the addition of an amino acid side chain to UDP-MurNAc through the sequential addition of L-alanine (L-Ala), D-glutamic acid (D-Glu), meso-diaminopimelic acid (meso-Dap; a derivative of lysine), and two D-alanines (D-Ala). This UDP-MurNAc-pentapeptide is then anchored to the inner membrane via the transport lipid undecaprenyl phosphate by MraY to form Lipid I. A UDP-GlcNAc moiety is attached to Lipid I via glycosidic bond by MurG to form the disaccharide-pentapeptide precursor known as Lipid II, the basic building block of the PG. The disaccharide-pentapeptide is flipped across the inner membrane to the periplasmic space by a flippase (MurJ) where it is incorporated into the nascent PG chain by penicillin-binding protein (PBP) transglycoslylase activity. Once incorporated, PBP transpeptidases crosslink meso-Dap of one pentapeptide to D-Ala of an opposing pentapeptide, concomitant with the cleavage of the terminal D-Ala, thus incorporating a new chain into the PG sacculus. Due to the crucial roles of these enzymatic activities in PG synthesis, PG transpeptidases, also known as Penicillin-Binding Proteins (PBPs), remain the best targets for antibiotics. In addition to the synthesis machinery, numerous enzymes also exist to remodel the existing PG structure (for a review, please see [27]). It is important to note that the PG is not essential for organisms to form distinct shapes, as seen in some intracellular parasitic bacterial species that lack PG, such as Mycoplasma and Spiroplasma in Tenericutes (Mollicutes), and free-living bacteria like the Planctomycetes [114,115]. Interestingly, recent work has shown that in the Chlamydia group, where the presence of a PG was uncertain, at least two species have a detectable PG structure [64,116]. These findings suggest that rigorous re-examination of other presumably PG-deficient bacterial species, such as the Planctomycetes, is crucial to our understanding of the evolution of cell wall synthesis in bacteria.
The not-so-simple ways of generating simple sphere, rod, or spiral shapes
The morphogenesis of different bacterial shapes requires the spatiotemporal modulation of the PG synthesis machinery, the exact mechanisms of which remain largely unknown. Most studies have focused on a few model organisms, revealing some of the principles of how the basic sphere/ovoid, rod, and spiral shapes are generated.
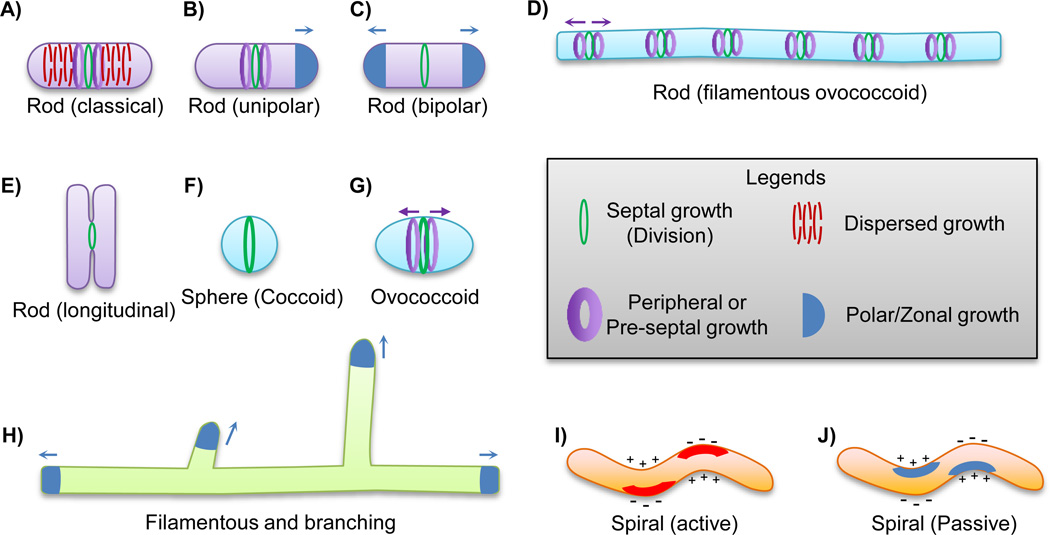
Conceptually, there are two major PG synthesis modes whose combination likely leads to the majority of bacterial shapes: growth and cytokinesis, which may have a common ancestry [30]. 1) Growth. Growth can occur by PG synthesis evenly distributed throughout the cell (dispersed growth) or from one or many spatially restricted zones, leading to zonal growth. As explained below, zonal growth can be specified spatially by various molecular mechanisms to yield different shapes. 2) Cytokinesis (often called septation in bacteria). Cell division requires directing PG synthesis inwards, usually at the midcell, perpendicular to the long axis of the cell. Cell division is mostly governed by the tubulin homolog FtsZ [31], which assembles into filaments to form a ring-like structure (Z-ring) around the division plane (Fig. 4) [32–34]. FtsZ recruits, directly or indirectly, a large number of proteins involved in PG synthesis, as shown in various species [30]. It is still not clear if the Z-ring is the major driving force during cell division, or if the Z-ring simply serves as a scaffold for proteins that provide constrictive force [33–35]. FtsZ is conserved in all bacterial phyla except for the Tenericutes (Mollicutes), Planctomycetes and the Chlamydia group [36,37].
Figure 4.
Mechanisms underlying the synthesis of various bacterial shapes. Arrows indicate the direction of the various zonal growth mechanisms. A–E: The various ways of making a rod-shaped cell body. Septation is required to resolve two or more daughter cells in each case (Green ring). A: The dispersed elongation model in which new material is uniformly incorporated into the side wall (Red dashed rings). In E. coli and C. crescentus, a specialized type of growth called pre-septal growth also contributes to elongation (Purple bands). B: Unipolar growth elongates the cell body in a “budding” fashion (Blue cap). In A. tumefaciens, pre-septal elongation occurs and defines the future sites of active polar growth (Purple bands). C: Some Actinobacteria species elongate the cell body in a bi-polar fashion (Blue caps), driven by DivIVA as detailed in the text. D: Filamentous ovococcoid cells may also achieve a rod-shaped cell body by a combination of inhibition of cell division (Green rings) and persistent peripheral growth around the septal region (Purple bands), as seen in Lactococcus lactis. E: Strikingly, one ectosymbiotic Gammaproteobacterium that attaches to the surface of the marine nematode Laxus oneistus grows in width and divides longitudinally. The Z-ring is also positioned longitudinally to divide the cell (Green ring). F–G: The sphere (coccoid) and oval shape (ovococcoid) utilize septal growth (Green ring) to synthesize the hemispheres of two respective daughter cells. In addition, regions of peripheral growth (Purple bands) can elongate the sphere-shaped cell body to sculpt an oval-shaped cell. H: The long branched filaments of Streptomyces coelicolor are achieved by tip growth at discrete positions directed by the protein DivIVA, (Blue caps), as detailed in the main text. I–J: The spiral shape can be achieved in at least two different ways. An “active” mechanism in which proteins localize to one side of the cell cylinder and induce negative curvature formation by relaxing cross-links of glycan strands (I, red stripes). Alternatively, cytoskeleton proteins can induce the formation of positive curvature by physically molding one side of the cell body (J, blue stripes). The positive and negative curvature of the cell body are indicated by + and − signs. The Z-ring positioning is imprecise in the only studied spiral-shaped organism Helicobacter pylori, hence it is not depicted in the schematics.
How spheres are made: the perfect symmetry
Synthesizing a spherical cell body is intuitive, as it is physically the perfect shape for a membrane bound structure under osmotic pressure. Spherical cells are generated by uniformly growing inwards from the septum of the dividing cell. This mechanism ensures that both daughter cells are equally spherical (Fig. 4F) [38]. In addition, peripheral growth regions around the septum can elongate the sphere-shaped cell body, sculpting an oval-shaped cell (Fig. 4G) [38]. This so-called peripheral growth is coordinated by DivIVA and FtsZ in Streptococcus pneumoniae [38–40]. DivIVA binds preferentially to negatively curved regions of cells and drives different types of zonal growth (see below). Interestingly, most spherical or oval-shaped bacteria lack the actin homolog mreB gene (Fig. 5) that is required for lateral PG synthesis in many rod-shaped species (see below) [41]. Whether this is a secondary loss or the cause in the evolution from rod to sphere/ovococcus remains to be investigated.
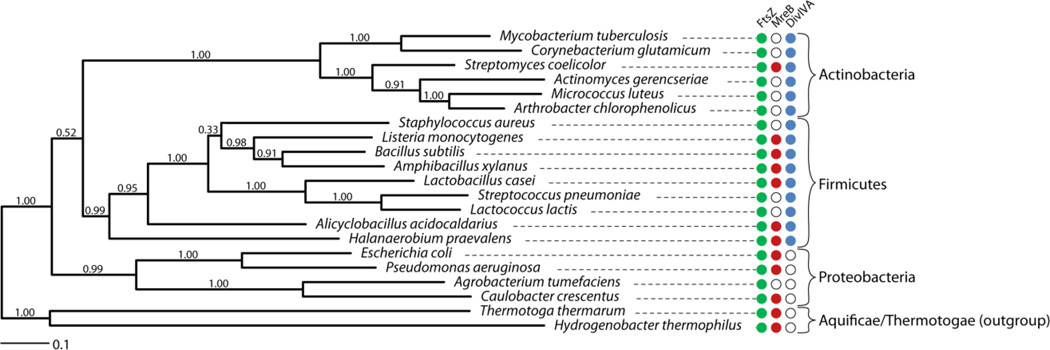
Figure 5.
The distribution of FtsZ, MreB, and DivIVA in selected Gram-positive bacteria. We randomly chose 138 genera across different orders and then chose 1–3 species in each genus, preferably with finished genomes. The Aquificae/Thermotogae outgroup is a deep-branched bacterial group usually found in extreme environments. Overall, the phylogenetic tree is representative of the result of 186 sampled genomes: 1. FtsZ is found in all species tested. 2. MreB is missing in most Actinobacteria and coccoid species. However, we note exceptions to this “rule” as several coccoid species in the Actinobacteria and Firmicutes have identifiable MreB. 3. DivIVA is almost strictly restricted to Gram-positive bacteria (Actinobacteria and Firmicutes). The phylogenetic tree of representative species (selected based on the scope of this review) belonging to Actinobacteria, Firmicutes, Proteobacteria and the outgroup deep branching extremophiles was calculated based on the alignment of the GyrA protein using the maximum likelihood method based on the LG model in MEGA 6 [117], supported by 100 bootstrap replicates. A discrete Gamma distribution with invariant positions was used to model evolutionary rate differences among sites. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The low support value for grouping Actinobacteria and Firmicutes together agrees with a previous report [118]. Filled or open circles indicate the genomic presence or absence of FtsZ (Green), MreB (Red), and DivIVA (Blue) detected by the Bi-directional Best Hit method (BBH) [119], respectively. Note that DivIVA is only present in Gram-positive bacterial species (Actinobacteria and Firmicutes), although its function diverges significantly in different species. Specifically, DivIVA is required for polar elongation in Actinobacteria but not in Firmicutes. MreB is absent in characterized coccoid/ovococcoid species [38]. In addition, MreB is also absent in polarly growing species (including A. tumefaciens in Proteobacteria), except for S. coelicolor in which MreB is required for sporulation, but not elongation [73].
How rods are made: many solutions to the same problem
Synthesizing a rod shape can be achieved through a number of mechanisms, including some variations on zonal growth (Fig. 4A–E). The actin-homolog MreB is required for rod-shaped cell elongation by dispersed PG synthesis along the cell body in a number of species, including the major experimental models E. coli, B. subtilis, and C. crescentus (Fig. 4A). MreB has been postulated to form membrane-associated filaments that rotate circumferentially inside the cell body [42–44]. Although it is still debated whether MreB can form extended filaments or simply local, discrete patches [45,46], lateral PG synthesis is clearly associated with MreB [47]. When MreB synthesis is disrupted, the localization of PG synthesis proteins is also disrupted, and cells display several growth defects leading to a rounded cell morphology [48]. Conversely, when PG synthesis is disrupted, either through the depletion of PG precursors or the addition of antibiotics, the rotation of MreB filaments is halted [42–44]. Simulations have predicted that the rotation of MreB may be critical to the morphogenesis of the rod shape by ensuring appropriately distributed PG incorporation throughout the cell body [49–51]. Feedback between cell geometry and MreB localization targets PG synthesis to regions of negative PG curvature to maintain the rod shape [51].
In addition to the dispersed mode of growth described above, some rod-shaped species, such as E. coli and C. crescentus, also elongate partly from the midcell using so-called pre-septal PG synthesis (Fig. 4A, purple bands). This FtsZ-dependent mode of PG synthesis occurs just prior to septation [52–54] and likely involves an interaction with MreB. These two proteins co-localize in C. crescentus [55,56] and E. coli [57], and they have recently been shown to interact directly in E. coli to transfer PG synthesis enzymes from the cell elongation machinery to the midcell for pre-septal and/or septal PG synthesis [57]. Therefore, the coordinated action of MreB and FtsZ apparently mediates a shift from dispersed to zonal PG synthesis in many species.
Alternatively, the rod shape can be achieved by cell elongation from one or both poles. For example, Agrobacterium tumefaciens grows unipolarly (Fig. 4B) and the actinobacteria Corynebacterium glutamicum and Mycobacterium tuberculosis grow bipolarly (Fig. 4C) [58–62]. Polar growth in C. glutamicum [58] and M. tuberculosis [62] requires DivIVA, the disruption of which leads to a rounded cell shape, similar to when dispersed elongation is disrupted in rod-shaped E. coli or B. subtilis cells. Interestingly, A. tumefaciens, C. glutamicum, and M. tuberculosis all lack MreB, suggesting that the polar elongation mechanism is functionally equivalent to MreB-directed dispersed elongation (Fig. 5). It remains to be determined if the evolution of polar growth machineries made MreB dispensable in these clades. Interestingly, in the case of A. tumefaciens, cells that have roughly doubled in length switch their zonal growth from a polar to a pre-septal mode analogous to the pre-septal mode described above for E. coli and C. crescentus (Fig. 4B, purple bands), which positions the elongation machinery at the new poles following division [59]. It is possible that polar growth arose in rod-shaped cells that lost the dispersed mode of growth but instead maintained pre-septal growth and repurposed it for growth at the poles. However, it remains to be determined which growth mode is ancestral. Since growth modes can now be readily detected with recently developed fluorescent probes for PG synthesis [63,64], the evolution of growth modes can be experimentally analyzed in a proper phylogenetic context to determine ancestral and derived states.
Intriguingly, the rod shape can also be generated by yet more mechanisms. Lactococcus lactis, an ovococcoid species, can form long rod-shaped filamentous cells in a synthetic medium (Fig. 4D). L. lactis also lacks MreB (Fig. 5), achieving its filamentous rod shape by forming a series of peripheral growth zones at Z-rings that are inhibited for cytokinesis (Fig. 4D, purple bands) [65]. Finally, perhaps the most eccentric way of synthesizing a rod is found in one ectosymbiotic gammaproteobacterium (not formally named) that attaches to the surface of the marine nematode Laxus oneistus [66]. This ectosymbiotic bacterium attaches to its host polarly and forms a monolayer biofilm that expands as the nematode grows in size. This bacterium grows in width and divides longitudinally, defying what is known in all previously studied rod-shaped bacterial species (Fig. 4E). This growth mechanism is well suited ecologically to maintain coverage of the nematode surface. Not surprisingly, the Z-ring is also positioned longitudinally to coordinate the cytokinesis of two daughter cells, bringing up the fascinating question of how FtsZ can localize in this fashion [66].
How spirals are made: the art of twisting
The spiral shape is a bit trickier to generate, and mechanistic studies are relatively scarce. The twisted spiral shape can be viewed as a summation of at least three distinct growth modes: 1) elongation, 2) curvature, and 3) twist [3,67]. There are at least two distinct mechanisms for generating a spiral, both of which utilize differential growth of the PG to induce either positive or negative curvature on one side of the cylindrical cell body. The first is an “active” mechanism in which genes are directly associated with generating the helical shape. H. pylori, a pathogenic species well known for its iconic helical shape, has been the major model organism for studying this mechanism. A number of genes have been shown to affect the helical nature of the cell body to varying degrees. It was suggested that the helical shape is achieved by local modification of PG crosslinks for every twist to create flexible regions that introduce the negative curvature, thereby forming the spiral shape (Fig. 4I) [3,49,68]. The alternative “passive” mechanism can be mediated by protein filaments “molding” the cell shape. In C. crescentus, the intermediate filament (IF)-like cytoskeleton protein CreS forms a protein bundle on only one side of the cell body [69]. By limiting the local lateral growth, possibly via MreB, this mechanism introduces positive curvature, resulting a crescent-shaped cell body [70]. However, the C. crescentus curved cell body is actually also twisted (Fig. 4J). This becomes obvious during prolonged growth in stationary phase, resulting in filamentous helices [71], indicating that a similar passive mechanism can also facilitate the synthesis of a spiral-shaped cell body.
Zonal growth is used to generate complex shapes
In the previous section, we briefly summarized decades of research on simple bacterial cell morphologies. Specifically, we have shown that simple rods can be achieved by at least five different mechanisms, some of which may be used to generate other shapes. However, the metaphorical “elephant in the room” question remains: what mechanisms are required to generate more complex or eccentric cell shapes? Unfortunately, we know little about how these shapes are generated at the molecular or even cellular level. Conceptually, these complex morphologies (Fig. 1) can be achieved by specifying zones of PG synthesis to target growth at specific subcellular locations [19,72].
A classic example of how the positioning of zonal growth can generate complex morphologies comes from studies of branch formation in Streptomyces. As is the case for most, if not all, Actinomycetales, Streptomyces species grow polarly. In addition, Streptomyces species often initiate lateral growth to form long branched filaments, resulting in a complex network of branched mycelium (Fig. 1F). In S. coelicolor, the formation of new branches occurs behind the tip of growing hyphae [7,73]. The negative curvature binding protein DivIVA localizes to the poles of growing hyphae and forms a structure called the polarisome, which recruits the PG synthesis machinery. Phosphorylation of DivIVA by the kinase AfsK causes the disassembly of part of the apical polarisome. The resulting DivIVA foci left behind the growing tip initiate the formation of new polarisomes and therefore the formation of a new zone of growth (Fig. 4H) [7,74,75]. In Firmicutes, DivIVA is required to prevent Z-ring formation at the new cell poles after division in B. subtilis, and to coordinate midcell elongation in S. pneumoniae [76,77]. Interestingly, only DivIVA from Actinobacteria species (either S. coelicolor or M. tuberculosis), but not Firmicutes (B. subtilis or S. pneumoniae), can rescue the elongation defect of a divIVA mutant in the Actinobacterium C. glutamicum [58]. These results indicate that the function of DivIVA is different in Actinobacteria compared to the Firmicutes (Fig. 5), but the mode of growth has only been studied in a few species in these groups. It may be that the role of DivIVA has shifted from regulating septal PG synthesis in Firmicutes to coordinating polar growth and lateral growth for branching in Actinobacteria (Fig. 5). A robust phylogenetic study of the mode of growth and DivIVA function in this group will be required to resolve the ancestral state of DivIVA (see section 3).
The knowledge gained from studying branch formation in S. coelicolor provides a glimpse into the mechanisms that generate complex morphologies: nature extends and builds upon basic shapes. Compared to rod-shaped cells, S. coelicolor branching can be viewed as a result of controlled zonal growth at discrete localized positions along the cell body. This simple strategy is most efficient with a highly modular mechanism, in which a master regulator controls the activity and/or localization of the whole complex/pathway. Here, simply changing the location of DivIVA is both necessary and sufficient to recruit the PG synthesis machinery to new positions to from branches. However, DivIVA is not found in Gram-negative bacteria, which possess equal, if not more, morphological diversity. Therefore, a great variety of such modular mechanisms must exist, as demonstrated in the next example.
The mechanism and evolution of morphogenesis: the study of stalk synthesis and localization provides a model
The stalk is a thin, appendage-like extension of all three layers of the cell envelope (inner membrane, peptidoglycan, and outer membrane) found in phylogenetically diverse groups of bacteria [14] (Figs. 1 and 2). Not to be confused with the hypha structure in Streptomyces, the stalk is much narrower than the cell body and hence a morphologically distinct organelle, analogous to the cilium of the eukaryotic cells [78]. Stalk structure, synthesis, and function have been mostly studied in C. crescentus, a model organism for bacterial development and adhesion [79–81]. The stalk is synthesized from its cell-proximal region and is compartmentalized from the cell body by proteinaceous structures called crossbands [81,82]. The stalk increases cell buoyancy and can be used as a nutrient scavenging organelle [5,83,84]. In C. crescentus, the stalk grows precisely from the polar location bearing the adhesive holdfast, and therefore pushes the attached cell away from the surface. Because there is little to no flow at a surface, pushing the cell away provides access to flowing nutrients: increasing the distance from the surface from 1 to 10 µm would provide an ~10% increase in nutrient flux [5,83,85]. The stalk can even serve as a "birth canal" through which budding bacteria produce daughter cells in the families Hyphomonadaceae and Hyphomicrobiaceae [86]. Although studies have shown that certain genes involved in PG synthesis and its modulation play a role in the synthesis of the stalk, the exact molecular mechanism for stalk synthesis and positioning remains undetermined [87].
Recently, an evolutionary cell biology study has provided novel insights into the mechanisms of stalk synthesis and positioning. In the closely related Asticcacaulis genus, the number and location of the stalks drastically differs from that of C. crescentus, yet its structure appears identical in the two genera [13] (Figs. 1 and 2). In C. crescentus, the stalk is positioned at a single cell pole; in Asticcacaulis excentricus, the stalk is made at a subpolar position off-center of a cell pole; and finally, in Asticcacaulis biprosthecum, two stalks are positioned bilaterally on the cell body [12–14] (Figs. 1 and 2). Stalks are synthesized from their cell body-proximal region in all species, suggesting that a common molecular mechanism may exist to account for the positioning and growth of stalks [14]. To identify potential stalk morphogens, the localization of proteins known to localize polarly in C. crescentus was determined in A. biprosthecum, leading to the identification of two proteins that localize at the base of its bi-lateral stalks rather than the cell pole [14]. One of the proteins, SpmX, was shown to be required for stalk synthesis in the Asticcacaulis genus (Fig. 2D), whereas it is not required in C. crescentus [88]. Expression of SpmX in an exogenous species could drive stalk synthesis at alternative positions (Fig. 2E). These results show that SpmX is necessary and sufficient to drive stalk synthesis to specific positions, indicating that it functions in a modular manner, much like DivIVA in the localization of the required PG synthesis machinery for branch formation in S. coelicolor. Therefore, SpmX serves as a morphogen for stalk synthesis in Asticcacaulis, responsible for coordinating zonal growth at species-dependent locations to produce the stalk(s), ultimately contributing to the diversity of cell shape [14].
How did stalk positioning evolve?
Stalks are faithfully reproduced at defined positions by a number of diverse species across multiple phyla, but the positions can vary between species. The evolutionary progression of stalk positioning, inferred from phylogeny, places the polar stalk of C. crescentus ancestral to sub-polar and bi-lateral stalks (Fig. 2B) [14]. This intuitive progression is in agreement with the evolutionary principle that more complex structures are typically built from simpler, yet similar ones [89]. SpmX regulates development in C. crescentus, but it is not required for stalk synthesis in this species, making it a surprising candidate for a morphogen for stalk positioning in Asticcacaulis [88]. These observations indicate that certain changes have occurred for SpmX to evolve new functions. The process of repurposing an existing biological unit (gene, pathway, organ, etc.) for a new function is referred to as co-option [90]. Based on similar evolutionary developmental biology (evo-devo) studies of eukaryotic multicellular organisms, such changes could be regulatory and/or functional. Coincidentally, the Asticcacaulis SpmX has expanded by as many as 400 amino acids to over 800, compared to the 435 amino acids of Caulobacter SpmX (Fig. 2C). Furthermore, this expansion is limited to a highly divergent intermediate region, as the N-terminal muramidase domain and C-terminal transmembrane domains remain conserved (Fig. 2C). To test the hypothesis that the expansion of this domain is the key to SpmX’s role in stalk synthesis, a series of chimeric proteins, in which separate domains of SpmX from different species are fused together, were constructed to test their function in stalk synthesis and positioning in different species. The results indicated that through progressive changes in its divergent C-terminal domain, SpmX evolved the ability to synthesize, and then target, stalk synthesis at specific positions (Fig. 2E). Last but not least, it was shown that over-expression of SpmX in A. excentricus leads to the formation of multiple sub-polar stalks, hinting that an increase in the expression level of SpmX might be a prerequisite for bi-lateral stalk synthesis in A. biprosthecum [14].
In summary, functional evolution of a specific domain of SpmX is the key to the evolution of cell morphology in Asticcacaulis and Caulobacter. Although SpmX is not required for stalk synthesis in C. crescentus, it is certainly reasonable to assume that a protein analogous to SpmX exists in C. crescentus to coordinate stalk synthesis, and that the actual downstream stalk synthesis machineries are likely homologous in Asticcacaulis and Caulobacter, resulting in almost identical stalk ultrastructure. Furthermore, to generate complex cell morphologies, as is observed in some of the Rhizobiales (Fig. 1L) [59], which lack spmX orthologs, a morphogen analogous to SpmX may exist to coordinate the zonal growth that specifies distinct morphologies.
The emerging field of bacterial evolutionary cell biology
We have briefly covered how basic sphere, rod, and spiral shapes can be generated in bacteria. Furthermore, we described the mechanisms underlying the synthesis of two distinct complex morphologies, cell branching and stalks. While it is clear that our knowledge of how bacterial shapes are generated is still very limited, we observe an emerging pattern in which complex shapes can evolve from basic shapes using a similar evolutionary mechanism – controlled zonal growth. Such mechanisms are only identifiable when we investigate multiple species that are morphologically distinct yet closely related. The importance of using evolutionary principles and phylogenetically informed comparative biology in the study of complex processes is nicely illustrated in the evo-devo (evolution of development) field [91]. Three important mechanisms have emerged from studies of morphological transitions in eukaryotes: 1) changes in cis-regulatory elements or protein sequences are frequently associated with morphological transitions; 2) modularity is usually present in the morphogenesis pathways (morphogen); 3) genes with existing functions can acquire new roles through evolution (co-option) [91–93].
Although evo-devo studies have historically focused on the shape of multicellular eukaryotes, their findings are potentially applicable to any evolutionary process. Conversely, any cell biological process can be studied using the approaches of evo-devo with minor modifications, as exemplified in the emerging field of evolutionary cell biology where these principles are beginning to be applied to the evolution of subcellular organization [94].
Why evolutionary cell biology in bacteria?
Two examples illustrate the potential benefits of studying evolutionary cell biology in bacteria: 1) The role of SpmX in stalk synthesis could not be inferred from its role in the much studied model C. crescentus and instead its discovery required its study in the closely related Asticcacaulis genus. In addition, the machinery for stalk synthesis remains to be identified in any genus. Now, SpmX can be used as a starting point to identify the downstream stalk synthesis machinery in Asticcacaulis and the acquired knowledge can be applied back to C. crescentus, where most of the stalk synthesis machinery is expected to be the same given the common ultrastructure of stalks in the two genera. 2) The rod-shaped A. tumefaciens had been assumed to elongate by incorporating new PG material in a dispersed manner along the side wall, similar to E. coli [95]. However, many phylogenetically closely related Rhizobiales species were known to grow polarly. In addition, the A. tumefaciens genome lacks mreB, which is essential for dispersed cell elongation in E. coli and B. subtilis. These evolutionary observations led to testing the mode of growth of A. tumefaciens, leading to the discovery that A. tumefaciens, and likely most species in the Rhizobiales order, grow polarly [59]. A more detailed study of growth modes in the Alphaproteobacteria will be required to answer this question.
Next generation sequencing paves the way for bacterial evolutionary cell biology studies
Recent advances in next generation sequencing (NGS) technologies have enabled the affordable sequencing of a large number of genomes [96]. At the time of this writing, approximately 22,000 bacterial genomes had been sequenced and deposited in public databases. However, most of the sequencing efforts have focused on pathogenic strains and a few selected model organisms, such as E. coli and B. subtilis, although efforts are underway to compile a phylogeny-driven genomic encyclopedia of bacteria [97]. The availability of genomic data provides an opportunity to study different organisms without experimental manipulation. For example, the presence or absence of metabolic pathways can be used to define the lifestyle of species of interest [98]. Alternatively, the evolutionary conservation of genes helps to predict whether a protein of interest may have broadly important or species-specific functions. For example, FtsZ and MreB are widely conserved in a majority of bacterial phyla, supporting their crucial roles as described above; whereas SpmX is only found in Caulobacter-related species, indicating that its role must be specific to this clade of bacteria, such as developmental regulation and/or stalk synthesis. Finally, genomic sequences enable the construction of rigorous phylogenies, which is essential in bacterial evolutionary studies, as we will discuss next.
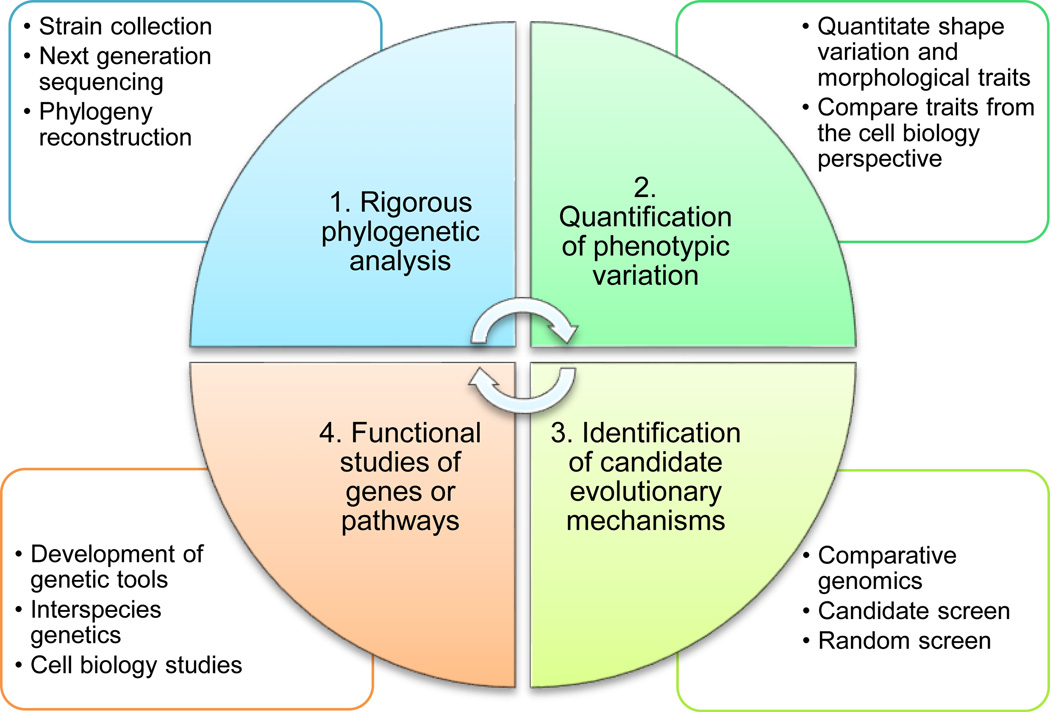
The four key steps in bacterial evolutionary cell biology research
Mallarino et al. (2012) have summarized a research approach for evo-devo studies of animal morphological evolution, which includes three key steps: 1) quantification of morphological variation, 2) identification of candidate developmental mechanism, and 3) functional analysis of genes and pathways. Here we adapt these three steps for the bacterial evolutionary cell biology field, with one key ingredient added as the first step: rigorous phylogenetic analysis to identify closely related species with variations of interest (Fig. 6):
-
Rigorous phylogenetic analysis of bacterial species
In eukaryotes, studies focusing on morphological evolution are always performed within phylogenetically closely related species. However, animals have numerous visible morphological features that can be used to reliably predict phylogeny most of the time (only birds look like birds). In bacteria, phylogeny cannot be reliably predicted based on morphological traits alone since there is insufficient character state variation in bacterial morphology, especially in the most common round, oval, and rod-shaped species. Furthermore, convergent evolution in bacterial shape complicates analysis (the coccoid form potentially arose multiple times). However, systematic analysis of cell size and shape in various bacterial species using recently developed automated quantitative image analysis tools could provide finer resolution to alleviate this problem [14,50,99,100]. Therefore, to reliably evaluate the phylogeny of bacteria, relevant molecular data are essential. Traditionally, the 16S rRNA sequence has been used to infer the phylogeny of bacteria, but in cases where the phylogeny is difficult to resolve based on a single gene, the best strategy is to sequence the genomes of interest, which is becoming increasingly affordable [101]. For example, to study stalk positioning, several closely related stalked and non-stalked species were sequenced [102] so that rigorous phylogenetic analysis could be performed to provide insights into the generation of morphological variation [14].
-
Quantification of phenotypic variation
After rigorous phylogenetic analysis, it is essential to quantitatively characterize the morphological traits. In the case of stalk positioning, the difference may be obvious, but within the stalked species, there are also variations in the length or even the diameter of the stalks, which would require more thorough characterization. Alternatively, the spiral shape of diverse Helicobacter and Campylobacter species are inherently different and also require rigorous characterizations. Finally, from the perspective of cell biology, phenotypic variation can be observed in different forms: variation in function and localization of proteins of interest, changes in the arrangement of intracellular organelles, and even divergence of gene function after duplication, all of which require rigorous quantification.
-
Identification of candidate evolutionary mechanisms
The identification of candidate evolutionary mechanisms usually requires a systematic and creative approach. One strategy is to take advantage of existing knowledge by branching out from established model organisms. For example, in the study of stalk synthesis in the Caulobacter clade, knowledge about the localization of developmental regulators was exploited to identify proteins that localize at the base of stalks in Asticcacaulis. Similarly, the mechanisms by which branches form in Streptomyces may have evolved from polar growth, as the branching morphogen, DivIVA, is also required for polar growth in the closely related rod-shaped Corynebacterium and Mycobacterium (Fig. 5). It would be interesting to sample more species in Actinobacteria to see if the function of DivIVA in branch formation is a derived state.
-
Functional studies of genes or pathways
Once candidate genes or pathways are identified, the next critical step is to study the candidate genes in the model organism and closely related non-model organisms. For example, after SpmX was identified as a candidate morphogen for stalk synthesis, spmX mutants were constructed in both Asticcacaulis species and were found to be stalkless, confirming the prediction (Fig. 2D). Next, cross-complementation genetics provides a powerful test as to whether changes to the genes of interest generate phenotypic variation in closely related species. For example, different alleles of SpmX were expressed in different strains from the same xylose-inducible promoter, ensuring that the only variable was the SpmX protein. The results showed that exogenous SpmX could still drive stalk synthesis, albeit at a different position, indicating that SpmX evolution is the mechanism underlying the evolution of stalk positioning (Fig. 2E). This study also demonstrated yet another advantage of branching out from a model organism, since related genetic tools are much more likely to work in closely related non-model organisms.
Figure 6.
Steps of evolutionary cell biology research in bacteria. Each of the four steps is indispensable to identify the evolutionary mechanisms underlying variation in different species from either a cell or developmental biology perspective. Construction of a rigorous phylogeny is the foundation of evolutionary cell biology studies, in that an unreliable phylogeny will lead to misinterpretations of the evolutionary history of species/traits and mistakes in experimental design and implementation. Development of genetic tools in non-model organisms serves as the other technical barrier for this type of studies; however, certain biochemical approaches, such as the use of antibiotics, universal or specific molecular probes, or specific antibodies, can be exploited to study organisms where genetic experiments are not feasible.
It is important to point out that the four steps need not be ordered in a flow-chart fashion (Fig. 6). For example, following quantitation of morphological traits, sequenced strains might not encompass the needed variation at either the cellular or the molecular level, which would lead to further strain collection and sequencing efforts. Alternatively, after functional studies, the candidate genes or pathways may not prove sufficient to account for the variation observed in the chosen organisms, which would require either additional strain collection and/or a revised strategy to identify candidate genes or pathways. Therefore, it is more appropriate to perceive the four steps in a dynamic fashion, in which each step may require revisions based on the other step(s).
Conclusion and outlook
Through billions of years of evolution, bacteria have achieved their dominant success in today’s world [1]. As Theodosius Dobzhansky elegantly stated, “Nothing in biology makes sense except in the light of evolution” [103]. The benefits of understanding the mechanisms underlying the evolution of bacterial shape and other cellular processes include: 1) Confirmation of the knowledge acquired from model systems, often limited to studies of a single species, and expansion of what we cannot learn from model organisms; 2) Understanding how old genes can be co-opted for a new purpose by means of the evolution of regulatory and/or protein sequences; 3) Potential implications in medical science; as described earlier, many pathogenic species use shape to their advantage when invading hosts, some are even capable of transforming on the fly; 4) The ability to control the shape or metabolic processes of bacterial cells can be potentially useful in synthetic biology to maximize the efficiency of industrial applications such as fermentation, because the ability to take up nutrients at the micro-scale is limited by diffusion [5]; 5) Finally, studying the mechanisms underlying Darwin’s “endless forms most beautiful” [104] provides an excellent opportunity for researchers to convey the beauty of evolution and science to the public and to inspire future generations of scientists.
Acknowledgements
We thank the Brun laboratory for critical reading of the manuscript. We thank Charles Daghlian, Paul Hoskisson, Hans-Peter Klenk, Nikos Kyrpides, Ellen Quardokus, Christian Riedel, and Teresa Thiel for generously providing electron micrographs to be used in this review. Our work on evolutionary bacterial cell biology has been supported by National Institutes of Health Grant GM051986 and National Science Foundation Grant MCB0731950 to YVB. YVB was supported by a Fulbright Scholar Award during the writing of this article.
Abbreviations
- PG
peptidoglycan
- Evo-Devo
evolution of development
Footnotes
The authors declare no competing financial interests.
References
- 1.Young KD. The selective value of bacterial shape. Microbiol Mol Biol Rev. 2006;70:660–703. doi: 10.1128/MMBR.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montecucco C, Rappuoli R. Living dangerously: how Helicobacter pylori survives in the human stomach. Nature reviews. Molecular cell biology. 2001;2:457–466. doi: 10.1038/35073084. [DOI] [PubMed] [Google Scholar]
- 3.Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, et al. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori's helical shape and stomach colonization. Cell. 2010;141:822–833. doi: 10.1016/j.cell.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sycuro LK, Wyckoff TJ, Biboy J, Born P, Pincus Z, et al. Multiple peptidoglycan modification networks modulate Helicobacter pylori's cell shape, motility, and colonization potential. PLoS pathogens. 2012;8:e1002603. doi: 10.1371/journal.ppat.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner JK, Brun YV. Out on a limb: how the Caulobacter stalk can boost the study of bacterial cell shape. Molecular microbiology. 2007;64:28–33. doi: 10.1111/j.1365-2958.2007.05633.x. [DOI] [PubMed] [Google Scholar]
- 6.Horvath DJ, Jr, Li B, Casper T, Partida-Sanchez S, Hunstad DA, et al. Morphological plasticity promotes resistance to phagocyte killing of uropathogenic Escherichia coli. Microbes and infection / Institut Pasteur. 2011;13:426–437. doi: 10.1016/j.micinf.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flardh K, Buttner MJ. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nature reviews. Microbiology. 2009;7:36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- 8.Andersen LP, Rasmussen L. Helicobacter pylori-coccoid forms and biofilm formation. FEMS immunology and medical microbiology. 2009;56:112–115. doi: 10.1111/j.1574-695X.2009.00556.x. [DOI] [PubMed] [Google Scholar]
- 9.Ikeda N, Karlyshev AV. Putative mechanisms and biological role of coccoid form formation in Campylobacter jejuni. European journal of microbiology & immunology. 2012;2:41–49. doi: 10.1556/EuJMI.2.2012.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siefert JL, Fox GE. Phylogenetic mapping of bacterial morphology. Microbiology. 1998;144(10):2803–2808. doi: 10.1099/00221287-144-10-2803. [DOI] [PubMed] [Google Scholar]
- 11.Tamames J, Gonzalez-Moreno M, Mingorance J, Valencia A, Vicente M. Bringing gene order into bacterial shape. Trends in genetics : TIG. 2001;17:124–126. doi: 10.1016/s0168-9525(00)02212-5. [DOI] [PubMed] [Google Scholar]
- 12.Poindexter JS. Biological Properties and Classification of the Caulobacter Group. Bacteriol Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pate JL, Ordal EJ. The fine structure of two unusual stalked bacteria. J Cell Biol. 1965;27:133–150. doi: 10.1083/jcb.27.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang C, Brown PJ, Ducret A, Brun YV. Sequential evolution of bacterial morphology by co-option of a developmental regulator. Nature. 2014;506:489–493. doi: 10.1038/nature12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kojima M, Nishioka N, Kusumoto A, Yagasaki J, Fukuda T, et al. Conversion of mono-polar to peritrichous flagellation in Vibrio alginolyticus. Microbiology and immunology. 2011;55:76–83. doi: 10.1111/j.1348-0421.2010.00290.x. [DOI] [PubMed] [Google Scholar]
- 16.Guttenplan SB, Shaw S, Kearns DB. The cell biology of peritrichous flagella in Bacillus subtilis. Molecular microbiology. 2013;87:211–229. doi: 10.1111/mmi.12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Angert ER. Alternatives to binary fission in bacteria. Nature reviews. Microbiology. 2005;3:214–224. doi: 10.1038/nrmicro1096. [DOI] [PubMed] [Google Scholar]
- 18.Kaiser D, Robinson M, Kroos L. Myxobacteria, polarity, and multicellular morphogenesis. Cold Spring Harbor perspectives in biology. 2010;2:a000380. doi: 10.1101/cshperspect.a000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown PJ, Kysela DT, Brun YV. Polarity and the diversity of growth mechanisms in bacteria. Seminars in cell & developmental biology. 2011;22:790–798. doi: 10.1016/j.semcdb.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ochman H, Elwyn S, Moran NA. Calibrating bacterial evolution. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:12638–12643. doi: 10.1073/pnas.96.22.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan YF, Marks ME, Jones FC, Villarreal G, Jr, Shapiro MD, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loehlin DW, Werren JH. Evolution of shape by multiple regulatory changes to a growth gene. Science. 2012;335:943–947. doi: 10.1126/science.1215193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mallarino R, Grant PR, Grant BR, Herrel A, Kuo WP, et al. Two developmental modules establish 3D beak-shape variation in Darwin's finches. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4057–4062. doi: 10.1073/pnas.1011480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ronshaugen M, McGinnis N, McGinnis W. Hox protein mutation and macroevolution of the insect body plan. Nature. 2002;415:914–917. doi: 10.1038/nature716. [DOI] [PubMed] [Google Scholar]
- 25.Werner T, Koshikawa S, Williams TM, Carroll SB. Generation of a novel wing colour pattern by the Wingless morphogen. Nature. 2010;464:1143–1148. doi: 10.1038/nature08896. [DOI] [PubMed] [Google Scholar]
- 26.Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS microbiology reviews. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 27.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nature reviews. Microbiology. 2012;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allan EJ, Hoischen C, Gumpert J. Bacterial L-forms. Advances in applied microbiology. 2009;68:1–39. doi: 10.1016/S0065-2164(09)01201-5. [DOI] [PubMed] [Google Scholar]
- 29.Errington J. L-form bacteria, cell walls and the origins of life. Open biology. 2013;3:120143. doi: 10.1098/rsob.120143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szwedziak P, Lowe J. Do the divisome and elongasome share a common evolutionary past? Current opinion in microbiology. 2013;16:745–751. doi: 10.1016/j.mib.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Erickson HP, Anderson DE, Osawa M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev. 2010;74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loose M, Mitchison TJ. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nature cell biology. 2014;16:38–46. doi: 10.1038/ncb2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. The EMBO journal. 2007;26:4694–4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pilhofer M, Ladinsky MS, McDowall AW, Petroni G, Jensen GJ. Microtubules in bacteria: Ancient tubulins build a five-protofilament homolog of the eukaryotic cytoskeleton. PLoS biology. 2011;9:e1001213. doi: 10.1371/journal.pbio.1001213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Hsin J, Zhao L, Cheng Y, Shang W, et al. FtsZ protofilaments use a hinge-opening mechanism for constrictive force generation. Science. 2013;341:392–395. doi: 10.1126/science.1239248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Margolin W. FtsZ and the division of prokaryotic cells and organelles. Nature reviews. Molecular cell biology. 2005;6:862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erickson HP, Osawa M. Cell division without FtsZ--a variety of redundant mechanisms. Molecular microbiology. 2010;78:267–270. doi: 10.1111/j.1365-2958.2010.07321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinho MG, Kjos M, Veening JW. How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. Nature reviews. Microbiology. 2013;11:601–614. doi: 10.1038/nrmicro3088. [DOI] [PubMed] [Google Scholar]
- 39.Fleurie A, Manuse S, Zhao C, Campo N, Cluzel C, et al. Interplay of the Serine/Threonine-Kinase StkP and the Paralogs DivIVA and GpsB in Pneumococcal Cell Elongation and Division. PLoS genetics. 2014;10:e1004275. doi: 10.1371/journal.pgen.1004275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sham LT, Tsui HC, Land AD, Barendt SM, Winkler ME. Recent advances in pneumococcal peptidoglycan biosynthesis suggest new vaccine and antimicrobial targets. Current opinion in microbiology. 2012;15:194–203. doi: 10.1016/j.mib.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daniel RA, Errington J. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 42.van Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS, et al. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15822–15827. doi: 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dominguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Soldner R, et al. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 44.Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, et al. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reimold C, Defeu Soufo HJ, Dempwolff F, Graumann PL. Motion of variable-length MreB filaments at the bacterial cell membrane influences cell morphology. Molecular biology of the cell. 2013;24:2340–2349. doi: 10.1091/mbc.E12-10-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swulius MT, Chen S, Jane Ding H, Li Z, Briegel A, et al. Long helical filaments are not seen encircling cells in electron cryotomograms of rod-shaped bacteria. Biochemical and biophysical research communications. 2011;407:650–655. doi: 10.1016/j.bbrc.2011.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.White CL, Gober JW. MreB: pilot or passenger of cell wall synthesis? Trends in microbiology. 2012;20:74–79. doi: 10.1016/j.tim.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 48.Jones LJ, Carballido-Lopez R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 49.Huang KC, Mukhopadhyay R, Wen B, Gitai Z, Wingreen NS. Cell shape and cell-wall organization in Gram-negative bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19282–19287. doi: 10.1073/pnas.0805309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Furchtgott L, Wingreen NS, Huang KC. Mechanisms for maintaining cell shape in rod-shaped Gram-negative bacteria. Molecular microbiology. 2011;81:340–353. doi: 10.1111/j.1365-2958.2011.07616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ursell TS, Nguyen J, Monds RD, Colavin A, Billings G, et al. Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E1025–E1034. doi: 10.1073/pnas.1317174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Pedro MA, Quintela JC, Holtje JV, Schwarz H. Murein segregation in Escherichia coli. Journal of bacteriology. 1997;179:2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Varma A, de Pedro MA, Young KD. FtsZ directs a second mode of peptidoglycan synthesis in Escherichia coli. Journal of bacteriology. 2007;189:5692–5704. doi: 10.1128/JB.00455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, et al. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Molecular microbiology. 2007;64:938–952. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- 55.Figge RM, Divakaruni AV, Gober JW. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Molecular microbiology. 2004;51:1321–1332. doi: 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- 56.Gitai Z, Dye N, Shapiro L. An actin-like gene can determine cell polarity in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8643–8648. doi: 10.1073/pnas.0402638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fenton AK, Gerdes K. Direct interaction of FtsZ and MreB is required for septum synthesis and cell division in Escherichia coli. The EMBO journal. 2013;32:1953–1965. doi: 10.1038/emboj.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Letek M, Ordonez E, Vaquera J, Margolin W, Flardh K, et al. DivIVA is required for polar growth in the MreB-lacking rod-shaped actinomycete Corynebacterium glutamicum. Journal of bacteriology. 2008;190:3283–3292. doi: 10.1128/JB.01934-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brown PJ, de Pedro MA, Kysela DT, Van der Henst C, Kim J, et al. Polar growth in the Alphaproteobacterial order Rhizobiales. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1697–1701. doi: 10.1073/pnas.1114476109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kysela DT, Brown PJ, Huang KC, Brun YV. Biological consequences and advantages of asymmetric bacterial growth. Annual review of microbiology. 2013;67:417–435. doi: 10.1146/annurev-micro-092412-155622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zupan JR, Cameron TA, Anderson-Furgeson J, Zambryski PC. Dynamic FtsA and FtsZ localization and outer membrane alterations during polar growth and cell division in Agrobacterium tumefaciens. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9060–9065. doi: 10.1073/pnas.1307241110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kang CM, Nyayapathy S, Lee JY, Suh JW, Husson RN. Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology. 2008;154:725–735. doi: 10.1099/mic.0.2007/014076-0. [DOI] [PubMed] [Google Scholar]
- 63.Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, et al. In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angewandte Chemie. 2012;51:12519–12523. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liechti GW, Kuru E, Hall E, Kalinda A, Brun YV, et al. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature. 2014;506:507–510. doi: 10.1038/nature12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perez-Nunez D, Briandet R, David B, Gautier C, Renault P, et al. A new morphogenesis pathway in bacteria: unbalanced activity of cell wall synthesis machineries leads to coccus-to-rod transition and filamentation in ovococci. Molecular microbiology. 2011;79:759–771. doi: 10.1111/j.1365-2958.2010.07483.x. [DOI] [PubMed] [Google Scholar]
- 66.Leisch N, Verheul J, Heindl NR, Gruber-Vodicka HR, Pende N, et al. Growth in width and FtsZ ring longitudinal positioning in a gammaproteobacterial symbiont. Current biology : CB. 2012;22:R831–R832. doi: 10.1016/j.cub.2012.08.033. [DOI] [PubMed] [Google Scholar]
- 67.Specht M, Dempwolff F, Schatzle S, Thomann R, Waidner B. Localization of FtsZ in Helicobacter pylori and consequences for cell division. Journal of bacteriology. 2013;195:1411–1420. doi: 10.1128/JB.01490-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Waidner B, Specht M, Dempwolff F, Haeberer K, Schaetzle S, et al. A novel system of cytoskeletal elements in the human pathogen Helicobacter pylori. PLoS pathogens. 2009;5:e1000669. doi: 10.1371/journal.ppat.1000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ausmees N, Kuhn JR, Jacobs-Wagner C. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell. 2003;115:705–713. doi: 10.1016/s0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- 70.Cabeen MT, Charbon G, Vollmer W, Born P, Ausmees N, et al. Bacterial cell curvature through mechanical control of cell growth. The EMBO journal. 2009;28:1208–1219. doi: 10.1038/emboj.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wortinger MA, Quardokus EM, Brun YV. Morphological adaptation and inhibition of cell division during stationary phase in Caulobacter crescentus. Molecular microbiology. 1998;29:963–973. doi: 10.1046/j.1365-2958.1998.00959.x. [DOI] [PubMed] [Google Scholar]
- 72.Margolin W. Sculpting the bacterial cell. Current biology : CB. 2009;19:R812–R822. doi: 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Flardh K, Richards DM, Hempel AM, Howard M, Buttner MJ. Regulation of apical growth and hyphal branching in Streptomyces. Current opinion in microbiology. 2012;15:737–743. doi: 10.1016/j.mib.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 74.Hempel AM, Wang SB, Letek M, Gil JA, Flardh K. Assemblies of DivIVA mark sites for hyphal branching and can establish new zones of cell wall growth in Streptomyces coelicolor. Journal of bacteriology. 2008;190:7579–7583. doi: 10.1128/JB.00839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hempel AM, Cantlay S, Molle V, Wang SB, Naldrett MJ, et al. The Ser/Thr protein kinase AfsK regulates polar growth and hyphal branching in the filamentous bacteria Streptomyces. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2371–E2379. doi: 10.1073/pnas.1207409109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Massidda O, Novakova L, Vollmer W. From models to pathogens: how much have we learned about Streptococcus pneumoniae cell division? Environmental microbiology. 2013;15:3133–3157. doi: 10.1111/1462-2920.12189. [DOI] [PubMed] [Google Scholar]
- 77.Rowlett VW, Margolin W. The bacterial Min system. Current biology : CB. 2013;23:R553–R556. doi: 10.1016/j.cub.2013.05.024. [DOI] [PubMed] [Google Scholar]
- 78.Kim S, Dynlacht BD. Assembling a primary cilium. Current opinion in cell biology. 2013;25:506–511. doi: 10.1016/j.ceb.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Curtis PD, Brun YV. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol Mol Biol Rev. 2010;74:13–41. doi: 10.1128/MMBR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brown PJ, Hardy GG, Trimble MJ, Brun YV. Complex regulatory pathways coordinate cell-cycle progression and development in Caulobacter crescentus. Adv Microb Physiol. 2009;54:1–101. doi: 10.1016/S0065-2911(08)00001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schlimpert S, Klein EA, Briegel A, Hughes V, Kahnt J, et al. General protein diffusion barriers create compartments within bacterial cells. Cell. 2012;151:1270–1282. doi: 10.1016/j.cell.2012.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hughes HV, Huitema E, Pritchard S, Keiler KC, Brun YV, et al. Protein localization and dynamics within a bacterial organelle. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5599–5604. doi: 10.1073/pnas.0909119107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wagner JK, Setayeshgar S, Sharon LA, Reilly JP, Brun YV. A nutrient uptake role for bacterial cell envelope extensions. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11772–11777. doi: 10.1073/pnas.0602047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Poindexter JS. Selection for nonbuoyant morphological mutants of Caulobacter crescentus. Journal of bacteriology. 1978;135:1141–1145. doi: 10.1128/jb.135.3.1141-1145.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Klein EA, Schlimpert S, Hughes V, Brun YV, Thanbichler M, et al. Physiological role of stalk lengthening in Caulobacter crescentus. Communicative & integrative biology. 2013;6:e24561. doi: 10.4161/cib.24561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hirsch P. Budding bacteria. Annual review of microbiology. 1974;28:391–444. doi: 10.1146/annurev.mi.28.100174.002135. [DOI] [PubMed] [Google Scholar]
- 87.Wagner JK, Galvani CD, Brun YV. Caulobacter crescentus requires RodA and MreB for stalk synthesis and prevention of ectopic pole formation. Journal of bacteriology. 2005;187:544–553. doi: 10.1128/JB.187.2.544-553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Radhakrishnan SK, Thanbichler M, Viollier PH. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev. 2008;22:212–225. doi: 10.1101/gad.1601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Atchley WR, Hall BK. A model for development and evolution of complex morphological structures. Biological reviews of the Cambridge Philosophical Society. 1991;66:101–157. doi: 10.1111/j.1469-185x.1991.tb01138.x. [DOI] [PubMed] [Google Scholar]
- 90.True JR, Carroll SB. Gene co-option in physiological and morphological evolution. Annual review of cell and developmental biology. 2002;18:53–80. doi: 10.1146/annurev.cellbio.18.020402.140619. [DOI] [PubMed] [Google Scholar]
- 91.Mallarino R, Abzhanov A. Paths less traveled: evo-devo approaches to investigating animal morphological evolution. Annual review of cell and developmental biology. 2012;28:743–763. doi: 10.1146/annurev-cellbio-101011-155732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 93.Nunes MD, Arif S, Schlotterer C, McGregor AP. A perspective on micro-evo-devo: progress and potential. Genetics. 2013;195:625–634. doi: 10.1534/genetics.113.156463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brodsky FM, Thattai M, Mayor S. Evolutionary cell biology: Lessons from diversity. Nature cell biology. 2012;14:651. doi: 10.1038/ncb2539. [DOI] [PubMed] [Google Scholar]
- 95.Parte A. Bergey's manual of systematic bacteriology. New York: Springer; 2012. [Google Scholar]
- 96.Shendure J, Lieberman Aiden E. The expanding scope of DNA sequencing. Nature biotechnology. 2012;30:1084–1094. doi: 10.1038/nbt.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wisniewski-Dye F, Borziak K, Khalsa-Moyers G, Alexandre G, Sukharnikov LO, et al. Azospirillum genomes reveal transition of bacteria from aquatic to terrestrial environments. PLoS genetics. 2011;7:e1002430. doi: 10.1371/journal.pgen.1002430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guberman JM, Fay A, Dworkin J, Wingreen NS, Gitai Z. PSICIC: noise and asymmetry in bacterial division revealed by computational image analysis at sub-pixel resolution. PLoS computational biology. 2008;4:e1000233. doi: 10.1371/journal.pcbi.1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol. 2011;80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Williams KP, Sobral BW, Dickerman AW. A robust species tree for the alphaproteobacteria. Journal of bacteriology. 2007;189:4578–4586. doi: 10.1128/JB.00269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brown PJ, Kysela DT, Buechlein A, Hemmerich C, Brun YV. Genome sequences of eight morphologically diverse Alphaproteobacteria. Journal of bacteriology. 2011;193:4567–4568. doi: 10.1128/JB.05453-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dobzhansky T. Nothing in Biology Makes Sense except in the Light of Evolution. The American Biology Teacher. 1973 Mar.35:125–129. [Google Scholar]
- 104.Darwin C. On the origin of species by means of natural selection. 1. ix. London: J. Murray; 1859. p. 502. [Google Scholar]
- 105.Kobayashi R, Konomi M, Hasegawa K, Morozumi M, Sunakawa K, et al. In vitro activity of tebipenem, a new oral carbapenem antibiotic, against penicillin-nonsusceptible Streptococcus pneumoniae. Antimicrobial agents and chemotherapy. 2005;49:889–894. doi: 10.1128/AAC.49.3.889-894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xie Y, He Y, Irwin PL, Jin T, Shi X. Antibacterial activity and mechanism of action of zinc oxide nanoparticles against Campylobacter jejuni. Applied and environmental microbiology. 2011;77:2325–2331. doi: 10.1128/AEM.02149-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Garrity GM, Brenner DJ, Krieg NR, Staley JT. The Alpha-, Beta-, Delta-, and Epsilonproteobacteria. New York: Springer; 2005. Bergey's Manual of Systematic Bacteriology Vol. 2., Part C. [Google Scholar]
- 108.Bauld J, Staley JT. Planctomyces maris sp. nov.: a Marine Isolate of the Planctomyces-Blastocaulis Group of Budding Bacteria. J Gen Microbiol. 1976;97:45–55. [Google Scholar]
- 109.Lail K, Sikorski J, Saunders E, Lapidus A, Glavina Del Rio T, et al. Complete genome sequence of Spirosoma linguale type strain (1) Standards in genomic sciences. 2010;2:176–185. doi: 10.4056/sigs.741334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Staley JT. Prosthecomicrobium and Ancalomicrobium: new prosthecate freshwater bacteria. Journal of bacteriology. 1968;95:1921–1942. doi: 10.1128/jb.95.5.1921-1942.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Thiel T, Pratte BS, Zhong J, Goodwin L, Copeland A, et al. Complete genome sequence of Anabaena variabilis ATCC 29413. Standards in genomic sciences. 2014;9:562–573. doi: 10.4056/sigs.3899418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ruiz N. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15553–15557. doi: 10.1073/pnas.0808352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sham LT, Butler EK, Lebar MD, Kahne D, Bernhardt TG, et al. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science. 2014;345:220–222. doi: 10.1126/science.1254522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fuerst JA, Sagulenko E. Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nature reviews. Microbiology. 2011;9:403–413. doi: 10.1038/nrmicro2578. [DOI] [PubMed] [Google Scholar]
- 115.Razin S, Hayflick L. Highlights of mycoplasma research--an historical perspective. Biologicals : journal of the International Association of Biological Standardization. 2010;38:183–190. doi: 10.1016/j.biologicals.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 116.Pilhofer M, Aistleitner K, Biboy J, Gray J, Kuru E, et al. Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nature communications. 2013;4:2856. doi: 10.1038/ncomms3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular biology and evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lang JM, Darling AE, Eisen JA. Phylogeny of bacterial and archaeal genomes using conserved genes: supertrees and supermatrices. PloS one. 2013;8:e62510. doi: 10.1371/journal.pone.0062510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]