Abstract
Background
T-cell targeted peptide epitope tolerogens from grass pollen allergens may be useful in treating seasonal allergic rhinitis, but there is urgent need for optimisation of approaches from improved understanding of mechanism.
Objective
We sought to identify human leukocyte antigen (HLA)-DR1-restricted epitopes from the Timothy grass pollen allergen, Phleum pratense, and characterise T-cell immune regulation following intranasal administration of a single, immunodominant epitope.
Methods
T-cell epitopes within P pratense were identified using HLA-DR1 transgenic mice and tetramer-guided epitope mapping (TGEM) in HLA-DR1-positive individuals with grass allergy. An immunodominant epitope was tested in HLA-DR1 transgenics for impact on responses to whole Phl p5 b or peptide. Microarrays and quantitative PCR were used to characterise T-cell immunity.
Results
Peptide 26 ( p26) was identified in HLA-DR1 transgenic mice and by TGEM analysis of HLA-DR1-positive individuals with grass allergy. p26 shows promiscuous binding to a wide range of HLA class II alleles, making it of relevance across immunogenetically diverse patients. The epitope is conserved in rye and velvet grass, making it applicable across a spectrum of grass pollen allergy. Intranasal pretreatment of mice with p26 results in significantly reduced T-cell responses. Transcriptomic array analysis in mice showed T-cell regulation in the intranasal treatment group associated with increased expression of members of the Cbl-b and Itch E3 ubiquitin ligase pathway.
Conclusions
We defined an immunodominant P pratense epitope, p26, with broad binding across multiple HLA class II alleles. Intranasal treatment of mice with p26 results in T-cell regulation to whole allergen, involving the Cbl-b and Itch regulatory pathway.
INTRODUCTION
Allergic rhinitis and asthma triggered by grass pollen is an important cause of morbidity.1 Many patients with these conditions express dissatisfaction with pharmacotherapy. For these patients, specific grass pollen immunotherapy, given as crude extract either subcutaneously or sublingually, is effective and associated with long-term benefits persisting after treatment is discontinued.2,3 However, subcutaneous immunotherapy is associated with a risk of IgE-mediated systemic side effects. Sublingual grass pollen vaccines, while safe, require large quantities of allergen daily, usually for several years.4
An alternative immunotherapy involves targeting allergen-specific T cells with peptides containing immunodominant epitopes. This approach has shown promise in cat5,6 and bee venom7 allergy and avoids the immediate reactions associated with IgE crosslinking that limit the amount of native allergen that can be given subcutaneously. We have shown, using a human leukocyte antigen (HLA)-DR1 transgenic model of allergic asthma, that treatment with a peptide derived from the cat allergen Fel d 1 inhibited allergic airway inflammation.8 Data from patients who received the vaccine also suggested that T-cell responses were suppressed to epitopes that were not administered, through ‘linked suppression’. The Allervax trial using Fel d 1 peptides showed improved pulmonary function in patients with reduced baseline FEV1 and global evaluation of the subjects’ ability to tolerate cats demonstrated significant improvement.9 The recent trials with short-course Cat-PAD found persisting improvement in the ocular and nasal components of rhinoconjunctivitis symptoms in allergic subjects.10
In comparison to cat and bee venom, grass pollen contains a relatively large number of major allergens.11 Furthermore, numerous isoallergen variants have been described within allergen families.12 The viability of peptide vaccines for grass allergy is therefore likely to depend on identifying a relatively limited selection of immunodominant epitopes and establishing the potential for these to induce regulatory responses extending to T cells specific for other epitopes within the same and other major allergens. The need is to define immunodominant T-cell epitopes within common allergens, basing design of therapeutics around epitopes widely recognised by different individuals through the ability to bind multiple HLA class II molecules.
We investigated the HLA-DR1-restricted CD4T-cell responses to a major group five Timothy grass pollen allergen in transgenic HLA-DR1 mice and HLA-DR1-positive donors with grass allergy. The work builds on previous studies in which efforts have been made to map pollen epitope/HLA class II patterns of immune recognition using ex vivo analysis of T-cell lines in allergic individuals.13–16 The novelty of our approach is in the combination of immunisation of HLA-DR1 transgenic mice to define immunodominance and characterisation of human responses by tetramer-guided epitope mapping (TGEM),17,18 allowing definition of immunodominant allergen epitopes and of epitopes binding across diverse HLA polymorphisms.
METHODS
Allergens and peptides
Phleum pratense extract and purified natural Phl p 5b (nPhl p 5b) were supplied by ALK Abello (Horsholm, Denmark). Recombinant Phl p 5b (rPhl p 5b) was supplied by Allergopharma (Germany).19,20 A Phl p 5b peptide library comprising 20-mer peptides overlapping by 12 amino acids was synthesised from the sequence of Phl p 5.0201 (Proimmune, Oxford, UK) (table 1).
Table 1.
Amino acid sequence of Phl p 5b 20-mer overlapping peptide sequences used for epitope mapping studies
| Phl p 5b peptide | Sequence |
|---|---|
| 1 | DAGYAPATPAAAGAAAGKAT |
| 2 | PAAAGAAAGKATTEEQKLIE |
| 3 | GKATTEEQKLIEDINVGFKA |
| 4 | KLIEDINVGFKAAVAAAASV |
| 5 | GFKAAVAAAASVPAADKFKT |
| 6 | AASVPAADKFKTFEAAFTSS |
| 7 | KFKTFEAAFTSSSKAAAAKA |
| 8 | FTSSSKAAAAKAPGLVPKLD |
| 9 | AAKAPGLVPKLDAAYSVAYK |
| 10 | PKLDAAYSVAYKAAVGATPE |
| 11 | VAYKAAVGATPEAKFDSFVA |
| 12 | ATPEAKFDSFVASLTEALRV |
| 13 | SFVASLTEALRVIAGALEVH |
| 14 | ALRVIAGALEVHAVKPVTEE |
| 15 | LEVHAVKPVTEEPGMAKIPA |
| 16 | VTEEPGMAKIPAGELQIIDK |
| 17 | KIPAGELQIIDKIDAAFKVA |
| 18 | IIDKIDAAFKVAATAAATAP |
| 19 | FKVAATAAATAPADDKFTVF |
| 20 | ATAPADDKFTVFEAAFNKAI |
| 21 | FTVFEAAFNKAIKESTGGAY |
| 22 | NKAIKESTGGAYDTYKCIPS |
| 23 | GGAYDTYKCIPSLEAAVKQA |
| 24 | CIPSLEAAVKQAYAATVAAA |
| 25 | VKQAYAATVAAAPQVKYAVF |
| 26 | VAAAPQVKYAVFEAALTKAI |
| 27 | YAVFEAALTKAITAMSEVQK |
| 28 | TKAITAMSEVQKVSQPATGA |
| 29 | EVQKVSQPATGAATVAAGAA |
| 30 | ATGAATVAAGAATTAAGAAS |
| 31 | AGAATTAAGAASGAATVAAG |
| 32 | GAASGAATVAAGGYKV |
Each peptide overlaps by 12 amino acids.
Recruitment and characterisation of study subjects
Subjects with a history of seasonal allergic rhinitis21 for a minimum of 2 years during the UK grass pollen season were recruited from the Royal Brompton Hospital Allergy Clinic or through advertisement (table 2). All subjects had skin prick tests to common inhalant allergens: Dermatophagoides pteronyssinus, 6-grass pollen mix, Timothy grass, Alternaria alternata, three tree mix, Betula verrucosa, cat dander, dog dander, horse dander, and mugwort in the presence of positive (histamine) and negative (diluent) controls (Soluprick, ALK Abello, Horsholm). Subjects showed sensitisation to Timothy grass (P pratense), evidenced by a positive skin test response (wheal diameter >3 mm larger than elicited by diluent control) and elevated P pratense specific IgE. Non-allergic controls were asymptomatic with negative skin prick tests to common aeroallergens including P pratense and normal levels of total and allergen-specific IgE. All subjects and controls were HLA-DRB1*0101 positive. The study was approved by the Royal Brompton and Harefield NHS Trust Ethics Committee (06/ Q0404/43) and informed written consent was obtained (table 2).
Table 2.
Clinical characteristics of study participants
| Volunteer | M/F | Age (years) | HLA-DRB1 alleles | SPT to Phleum pretense (mm)* | SPT to other allergens | IgE to Phleum pretense (IU/mL) | Total IgE (IU/mL) |
|---|---|---|---|---|---|---|---|
| Allergic | 7:4 | ||||||
| URM 018 | M | 29 | 1:15 | 6 | Nil | 1.49 | 3.0 |
| URM 022 | F | 26 | 1:10 | 10 | D | 100.0 | 760.0 |
| URM 028 | M | 34 | 1:4 | 6 | D, M | 9.66 | 26.0 |
| URM 031 | M | 49 | 1:4 | 9 | C, D, H | 5.70 | 229.0 |
| URM 039† | F | 42 | 1:14 | 13 | Nil | 9.21 | 15.0 |
| URM 044 | M | 45 | 1:4 | 9 | A, C, D | 2.98 | 23.0 |
| URM 067 | M | 41 | 1:13 | 6 | B, C, D, M | 11.8 | 36.0 |
| URM 068 | F | 36 | 1:3 | 11 | B, HDM | 9.13 | 107.0 |
| URM 069 | M | 58 | 1:15 | 13 | B, C, D, HDM | 7.98 | 89.0 |
| URM 074 | F | 45 | 1:3 | 10 | B, D | 23.4 | 73.0 |
| URM 087 | M | 38 | 1:1502 | 7 | B, M | 15.4 | 123.0 |
| Non-allergic | 6:4 | ||||||
| URM 095 | M | 38 | 1:3 | 0 | Nil | <0.35 | 53.0 |
| URM 213 | M | 41 | 1:7 | 0 | Nil | <0.35 | 5.0 |
| URM 222 | F | 48 | 1:14 | 0 | Nil | <0.35 | 25.0 |
| URM 302 | M | 25 | 1:7 | 0 | Nil | <0.35 | 7.0 |
| URM 304 | M | 37 | 1:3 | 0 | Nil | <0.35 | 35.0 |
| URM 306 | M | 47 | 1:7 | 0 | Nil | <0.35 | 26.0 |
| URM 308 | M | 27 | 1:15 | 0 | Nil | <0.35 | 15.0 |
| URM 318 | F | 24 | 1:4 | 0 | Nil | <0.35 | 4.0 |
| URM 321 | F | 29 | 1:3 | 0 | Nil | <0.35 | 18.0 |
| URM 322 | F | 29 | 1:12 | 0 | Nil | <0.35 | 3.0 |
Size of wheal defined as (Dl/dp)/2, where Dl is the longest diameter and dp is the diameter mid-orthogonal to Dl.
Currently undergoing immunotherapy.
There is no significant difference in age between individuals with and without allergy in the study groups; mean age (±SEM), allergic 40 (3) years; non-allergic 34.5 (3) years (age: allergic vs non-allergic; p=0.1597). An unpaired t test was used to determine significant differences between groups. Statistically significant differences were defined as a p value of less than 0.05. The HLA-DRB1 genotype was determined by PCR-based HLA genotyping; HLA-DRB1*0101 patients were selected as a study group on the grounds that this is a common allele in the UK population and facilitated matched studies in the HLA-DRB1*0101 transgenic mouse model. Entries against each patient indicate that they are DRB1*0101, with the other allele carried by them indicated after the colon.
A, Alternaria alternata; B, Betula verrucosa; C, cat hair; D, dog hair; H, horse dander; HLA, human leukocyte antigen; HDM, house dust mite, Dermatophagoides pteronyssinus; M, mugwort; SPT, skin prick test.
Transgenic mice
HLA-DR1 (DRA*0101/DRB1*0101) Aβ° transgenic mice have been described previously22; 8–14-week-old mice were used. Animal procedures were approved by the UK Home Office. UK Home Office regulations for animal welfare, based on the Animals (Scientific Procedures) Act 1986, were strictly observed.
HLA-DR1-restricted T-cell responses to Phl p 5b protein and overlapping peptide panel
HLA-DR1-restricted T-cell lines were made against Phl p 5 protein. HLA-DR1-tg/Aβ° mice (n=6) were footpad immunised with 25 μg nPhl p 5b in Complete Freund’s Adjuvant (CFA). Popliteal draining lymph nodes (DLNs) were harvested at day 10 and T-cell lines generated through two restimulations against nPhl p 5 protein (25 μg/mL) in the presence of irradiated mouse splenocytes as antigen presenting cells; this was repeated on three occasions and representative data from one of these are shown. T-cell proliferation in response to nPhl p 5 and 20-mer overlapping peptides (table 1) spanning Phl p 5b at 50 μg/mL were assessed by addition of 1 μCi/well3H-thymidine during the final 18 h of a 72 h culture. Data are presented as the mean from triplicate wells minus the mean from triplicate wells containing cells plus medium.
Tetramer-guided epitope mapping
TGEM17,18 was used to identify epitopes within Phl p 5b. The 32 overlapping peptides in pools were used to stimulate T-cell cultures from two individuals with grass allergy (table 2, URM018, URM031). Cells were cultured with peptide pools for 14 days prior to tetramer staining. Purified soluble DRA1*0101/DRB1*0101 molecules were produced from Schneider cells and loaded with peptide pools to generate tetramers. Tetramer-positive cultures staining with these tetramers were restained using HLA-DR1 tetramers loaded with each peptide from the positive pool.
Isolation and culture of PBMCs for tetramer analysis
Freshly isolated peripheral blood mononuclear cells (PBMCs) from DRB1*0101-allergic subjects and controls were cultured at 2×106 cells/mL in RPMI-1640 (Invitrogen, Life Technologies, UK) containing, L-glutamine (Sigma, UK), penicillin/streptomycin (Sigma) and 10% human AB serum (PAA Laboratories GmbH, Pasching, Austria) with 20 μg/mL Phl p 5b peptide 26 (p26) for 14 days. Recombinant human interleukin (IL)-2 (1 ng/mL) was added at day 7 (Proleukin, Novartis, UK).
Purified soluble DRA1*0101/DRB1*0101 molecules were loaded to generate HLA-DR1 tetramer with Phl p 5 p26 or an irrelevant peptide (Phl p 1, p26) as a control tetramer.
At day 14, T cells were stained with anti-CD3-FITC, anti-CD4-PE-Cy5 (BD Pharmingen, Oxford, UK), PE-conjugated-DRB1*0101 tetramers and analysed by flow cytometry. Cultured Tcells were incubated with 10 μg/mL tetramer at 37°C for 1 h and costained with anti-CD3-FITC and anti-CD4-PE-Cy5 (BD Pharmingen) for 30 min. Cells were gated on live lymphocytes according to forward and side scatter and the CD3 CD4 positive population to detect the p26-specific T-cell population and data acquired on a FACSCalibur (BD Biosciences, Oxford, UK).
For ex vivo phenotyping of tetramer-positive PBMCs, cells were gated on the tetramer-positive CD4 population, then analysed within this gate for expression of CD45RA (Clone L48, BD Pharmingen), CCR4 (Clone 205410, R&D Systems), CCR3 (Clone 61828.111, R&D Systems), CXCR1 (Clone 5A12 BD, Pharmingen), CCR5 (Clone 45523, R&D Systems), CXCR3 (Clone 49801, R&D Systems), CCR7 (Clone 3D12, eBiosciences), CD62L (Clone DREG-56, eBiosciences) and CD38 (Clone 240742, R&D Systems).
Peptide binding to HLA-DR and HLA-DQ molecules
HLA-DR and HLA-DQ molecules were purified from homozygous Epstein–Barr virus cell lines by affinity chromatography with L24323–25 (HLA-DR) and SPVL3 (HLA-DQ).23,24 Binding to HLA-DR and HLA-DQ was assessed by competitive ELISA,23–25 evaluating the peptide concentration preventing binding of 50% of the labelled peptide (IC50). Data were expressed as relative affinity: ratio of the half maximal inhibitory concentration (IC50) of the peptide to the IC50 of the reference peptide, which is a high binder to the HLA class II molecule. Unlabelled forms of the biotinylated peptides were used as reference peptides. Their sequences and IC50 values were: HA 306–318 (PKYVKQNTLKLAT) for DRB1*0101 (2 nM), DRB1*0401 (14 nM), DRB1*1101 (72 nM); YKL (AAYAAAKAAALAA) for DRB1*0701 (5 nM), A3 152–166 (EAEQLRAYLDGTGVE) for DRB1*1501 (41 nM), MT 2–16 (AKTIAYDEEARRGLE) for DRB1*0301 (71 nM), B1 21– 36 (TERVRLVTRHIYNREE) for DRB1*1301 (46 nM), CTP 427–441 (VHGFYNPAVSRIVEA) for DRB1*0901 (23 nM), TFR141–155 (TGTIKLLNENSYVPR) (363 nM) for DRB1*1202, TFR607–620 (LNLDYERYNSQLLS) for DRB1*1502 (4 nM), B7150–164 (LNEDLRSWTAADTAA) for DQ6 (DQA1*0103/ DQB1*0603) (37 nM) and DQB45–57 (ADVEVYRAVTPLGPPD) for DQ8 (DQA1*0301/DQB1*0302) (98 nM).
Intranasal peptide treatment and T-cell assays
HLA-DR1-tg/Aβ° mice (seven per group) were treated with intranasal phosphate-buffered saline (PBS) or Phl p 5 p26 (100 μg) in 20 μL on days 1, 2 and 3. On day 21 mice were footpad primed with 25 μg of whole rPhl p 5b protein, Phl p 5b p26, or whole rPhl p 1 protein as an emulsion with CFA (Sigma-Aldrich, UK). At day 31, single-cell suspensions were prepared from DLNs. Cells were cultured in triplicate with doses of whole rPhl p 5b protein, p26, or whole rPhl p 1 protein for 3 days. 1 μCi/well of 3H-thymidine was added 18 h before termination of 72 h cultures. Data are represented as the mean from triplicate wells minus the mean from triplicate wells containing cells plus medium without antigen.
Microarray and real-time PCR analysis
HLA-DR1-tg/Aβ° mice received intranasal challenge with PBS or rPhl p 5b p26 (100 μg) on days 1, 2 and 3. On day 21 mice were primed with 25 μg of p26/CFA. At day 31, DLN suspensions were established as cultures with p26 (50 μg/mL). At 72 h cells were harvested and lysed for RNA analysis. cDNA was synthesised from 700 ng RNA (RT2 First strand kit, Qiagen, UK) and sample quality assessed by Mouse RT2 RNA QC PCR Array (PAMM-999A, Qiagen). cDNA samples were assayed by PCR array (RT2 Profiler PCR array, PAMM-074Z, Qiagen). Plates were run on a Stratagene Mx3000p RT PCR machine for 10 min at 95°C followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Data were analysed using Partek Genomics Suite V.6.6 and significance determined using analysis of variance.
For real-time PCR, cDNA was synthesised from 500 ng RNA using SuperScript III reverse transcriptase (Invitrogen, Life Technologies). Real-time PCR was carried out in 20 μL containing 0.5 μL cDNA, 10μL 2XTaqMan Universal PCR Master Mix (Applied Biosystems, USA) and gene-specific probe and primers (Applied Biosystems: transforming growth factor β (TGFβ) inducible early gene 1 (TIEG-1) Mm00449812_m1, Itch Mm00492683_m1, Cblb Mm01343092_m1, Egr3 Mm00516979_m1; Sigma: Foxp3 Forward 5′-ATAGTTCCTTCCCAGAGTTCTTCC, Reverse 5′-ATGGTAGATTTCATTGAGTGTCCTC, Probe 5′-[FAM] CACCTATGCCACCCTTATCCGATGG[TAM]). PCR reactions were run in triplicate and CT values obtained using a MX3000P real-time PCR machine (Stratagene, USA) normalising to glyceraldehyde-3-phosphate dehydrogenase. Student’s t test was used to determine significance between groups. Statistically significant differences were defined as p values of less than 0.05.
RESULTS
HLA-DR1 restricted T-cell response to a Phl p 5b peptide panel identifies immunodominant epitopes of Phl p 5b
Since most humans are heterozygous with respect to the HLA region and also will present peptide to CD4 T cells via multiple HLA class II isotypes, definition of common peptide–major histocompatibility complex combinations can be challenging. We addressed this through a combination of HLA transgenic, HLA-binding studies, TGEM and patient T-cell responses. HLA-DR1-tg/Aβ° mice were immunised with nPhl p 5b and T-cell lines made against nPhl p 5b protein. T-cell responses of these lines were tested against 20-mer overlapping peptides encompassing the Phl p 5b sequence (table 1). Peptides p18, p23 and p26 together with whole allergen elicited strong T-cell responses (figure 1). p26 shows sequence conservation with allergens from other grass species, including rye grass and velvet grass, making it of applicability across a spectrum of grass pollen allergy (table 3).
Figure 1.
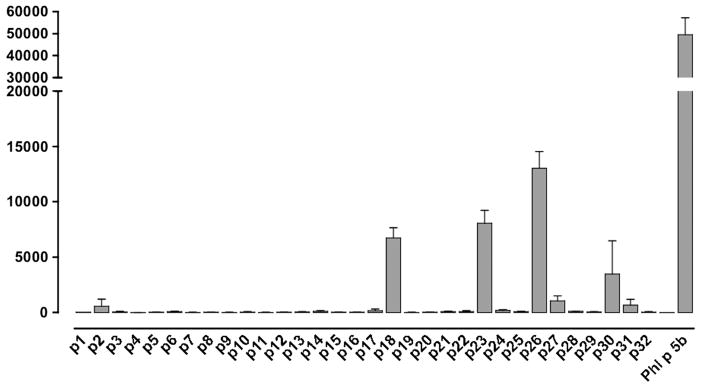
Human leukocyte antigen (HLA)-DR1 restricted T-cell responses to Phl p 5b protein and Phl p 5b 20-mer overlapping peptide panel. HLA-DR1-restricted T-cell lines were made against nPhl p 5 protein. HLA-DR1-Tg/Aβ° mice (n=6) expressing human HLA-DR1 were immunised with nPhl p 5b emulsified in Complete Freund’s Adjuvant. Draining lymph node cells were harvested at day 10 and T-cell lines generated against Phl p 5 protein. T-cell responses of lines were assessed after 72 h in a 3H-thymidine incorporation assay against Phl p 5 protein and 20-mer overlapping peptides spanning Phl p 5b, each at 50 μg/mL. The y-axis indicates T-cell proliferation in response to peptide, measured in Δcpm of 3H-thymidine incorporation (ie, the difference between the mean count from triplicate wells in the presence or absence of peptide). The x-axis indicates the identity of the tested Phl p 5b peptides that were assayed for the ability to elicit a T-cell response. This assay was repeated on three separate occasions. Epitopes p18, p23 and p26 were confirmed by priming with the specific peptides (data not shown).
Table 3.
Sequence alignment of the in silico defined epitope sequence (YAVFEAALT) within Phl p 5b peptide 26 (p26), from Phleum pratense and other commonly occurring grass species allergens in the UK
| Grass allergen | p26 epitope sequence | Percentage sequence alignment |
|---|---|---|
| Phl p 5.0201 | YAVFEAALT | – |
| Dac g 5 isoallergen 2 | YAVFEAALT | 100 |
| Dac g 5 isoallergen 1 | YAVFEAALT | 100 |
| Dac g 5 | YAVFEAALT | 100 |
| Lol p VA | YAVFEAALT | 100 |
| Lol p 5C | YAVFEAALT | 100 |
| Hol l 5.0201 | YAVFEAALT | 100 |
| Hol l 5b | FAVFKAALT | 77 |
| Poa p 5.0201 | FAVFKAALT | 77 |
| Phl p 5.0101 | YTVFETALK | 66 |
| Hol l 5.0101 | YTVFETALK | 66 |
| Lol p 5.0101 | YTVSETALK | 55 |
The TEPITOPE-predicted HLA-DR1-restricted epitope within Phl p 5b-derived p26 (YAVFEAALT) was identified in other grass species. Sequences were aligned using the CLUSTAL 2.0.12 multiple sequence alignment programme available via the European Bioinformatics Institute server: (http://www.ebi.ac.uk/Tools/services/web/toolform.ebi?tool=clustalw2). The Phl p isoallergen investigated in this study is shown in bold. The table highlights the percentage identity of the epitope sequences following calculation of pairwise sequence alignment scores when compared with Phl p 5.0201. Dac g, Dactylis glomerata; HLA, human leukocyte antigen; Hol l, Holcus lanatus; Lol p, Lolium perenne; Phl p, Phleum pratense; Poa p, Poa pratensis.
Immunodominant epitopes of Phl p 5b bind diverse HLA class II alleles
Epitopes, p18, p23 and p26 were analysed for binding to common HLA-DR and DQ alleles (table 4). p26 is unusual in its ability to bind with moderate to high affinity to extremely diverse HLA class II sequences, irrespective of isotype with the exception of HLA-DQ8. Note, however, that HLA-DQ8 would generally be carried on an HLA-DR4 haplotype, so that these individuals could in any case present the peptide through HLA-DR. The epitope also binds HLA-DRB1*1502, the more common DR15 variant in Asian populations, suggesting that the relevance of the epitope would extend beyond Caucasian patients.
Table 4.
Affinity of p26, 18 and 23 binding to HLA-DR and HLA-DQ molecules
| DR1 | DR3 | DR4 | DR7 | DR9 | DR11 | DR13 | DR1501 | DR1502 | DQ6 | DQ8 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 14% | 30% | 20% | 25% | 1% | 14% | 16% | 27% | 2% | 16% | 21% | |
| p26 | 1 | 83 | 5 | 1 | 0.3 | 14 | 38 | 9 | 1 | 16 | 267 |
| p18 | 11 | nd | 0.3 | 32 | nd | 3 | >456 | 42 | 2000 | nd | nd |
| p23 | 1 | nd | 0.4 | 8 | nd | 2 | >456 | 37 | 4 | nd | nd |
Results are expressed as a relative binding ratio obtained by dividing the IC50 of allergen peptide by that of a reference peptide that binds strongly to the HLA molecule. Lower numbers correspond to a higher binding affinity. Numbers in bold (ratio of 20 or less) = high affinity binding; numbers in italic (ratio 20–100) = moderate binding affinity. Each peptide—major histocompatibility complex combination was evaluated in two independent experiments. Percentages are the frequency of the HLA-DR or HLA-DQ alleles in a Caucasian population calculated from allele frequency net (http://www.allelefrequencies.net): a database and online repository for immune gene frequencies in worldwide populations.
HLA, human leukocyte antigen; nd, not done.
TGEM confirms Phl p 5b p26 as an HLA-DR1-restricted T-cell epitope
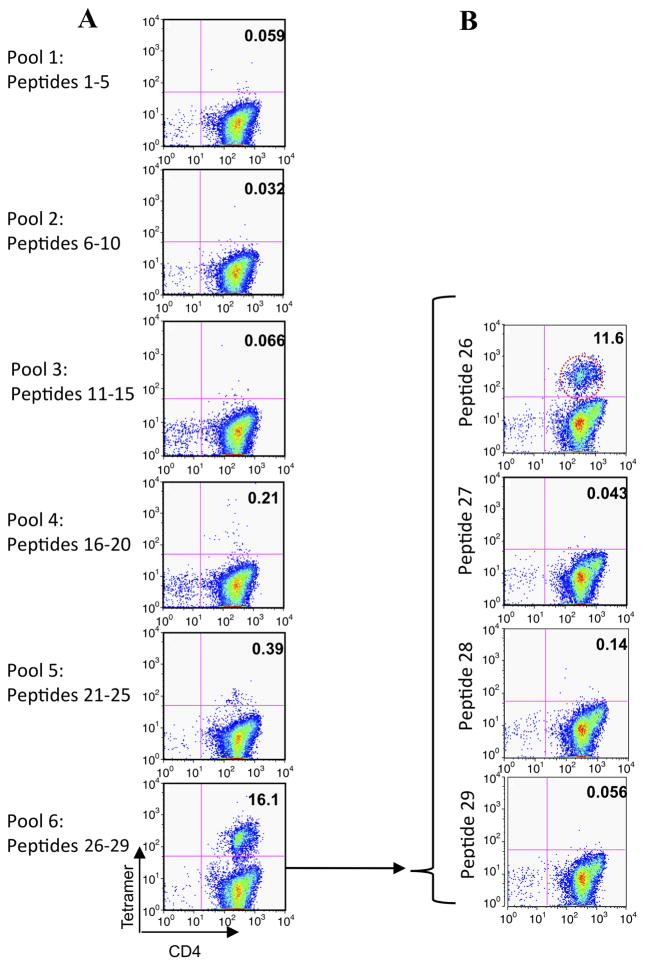
Seven pools of peptides were used to stimulate CD4 T cells from two HLA-DR1-positive subjects with grass allergy (table 2, URM018, URM031). Twelve days later, cells were analysed using HLA-DR1 tetramers loaded with each peptide pool; representative data are shown from donor URM031. Cells stimulated with pool 6 (p26–29) gave positive staining, with a weaker signal for pool 5 (p21–25) (figure 2A). On day 14, HLA-DR1 tetramers loaded with individual tetramers from pool six were used to stain the cells, identifying HLA-DR1-restricted p26 (pool 6) (figure 2B).
Figure 2.
Tetramer-guided epitope mapping identifies peptide 26 (p26) as a human leukocyte antigen (HLA)-DR1-restricted CD4T-cell epitope of Phl p 5b. A panel of overlapping 20-mer peptides spanning Phl p 5b was synthesised. Peptides were divided into pools and loaded onto PE-conjugated HLA-DR1 tetramers. Isolated CD4 T cells were cultured with adherent cells in the presence of the pooled peptides for 7 days before adding interleukin 2. Between days 12 and 14 the cells were stained with anti-CD4 and the pooled peptide tetramers (A). Positive tetramer staining was identified by flow cytometry. Individual peptide tetramers were generated and staining repeated (B). In this representative example, positive labelling is seen with HLA-DR1 tetramers loaded with p26 of Phl p 5b in patient URM031.
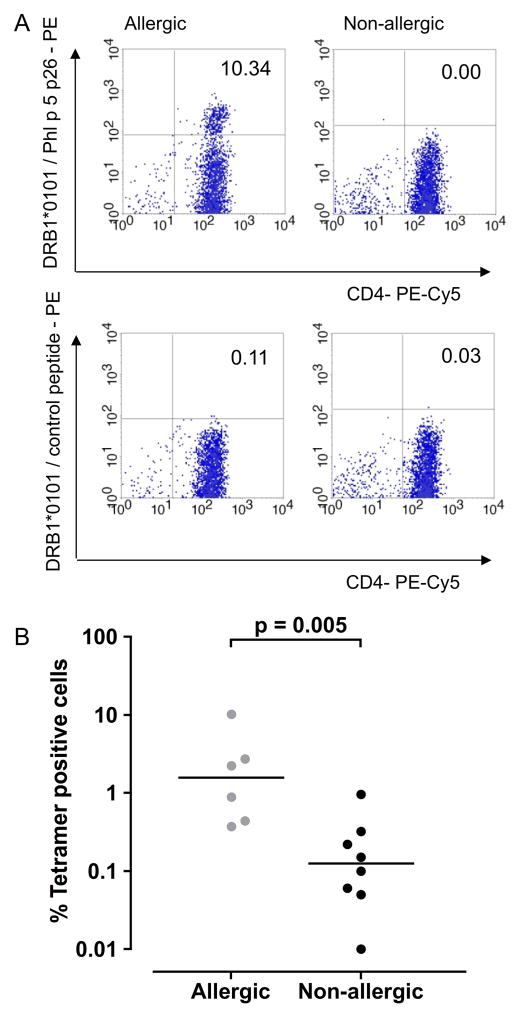
Phl p 5b p26 HLA-DR1 tetramer-positive populations identified individuals with grass allergy
HLA-DR1-restricted CD4 T-cell responses to p26 were examined in six of the subjects with grass allergy and eight subjects without grass allergy, described in table 2 (figure 3A,B). Subjects with grass allergy (n=6) showed higher numbers of HLA-DR1 p26 tetramer positive CD4 T cells than controls without allergy (n=8) (p<0.01).
Figure 3.
Phl p 5b peptide 26 (p26) human leukocyte antigen (HLA)-DR1 tetramer-positive populations in study participants with and without allergy. Whole peripheral blood mononuclear cells isolated from HLA-DR1-positive individuals with grass allergy and seasonal allergic rhinitis, and healthy controls without allergy were cultured with Phl p 5b p26. At day 14, cells were labelled with anti-CD3-FITC, anti-CD4-PE-Cy5 antibodies and PE-conjugated-HLA-DR1 tetramer loaded with p26 (A, top panel) or control tetramer loaded with irrelevant peptide (A, bottom panel) and analysed by flow cytometry. Cells were gated on live lymphocytes according to forward and side scatter and the CD3 CD4 positive population to detect the p26-specific T-cell population. The percentage of p26 tetramer-positive cells for subjects with and without allergy is shown (B). Statistical analysis was performed using the Mann– Whitney U test (bars indicate median values). Samples were processed during the grass pollen season, 2010 (subjects with allergy, n=6; healthy controls without allergy, n=8).
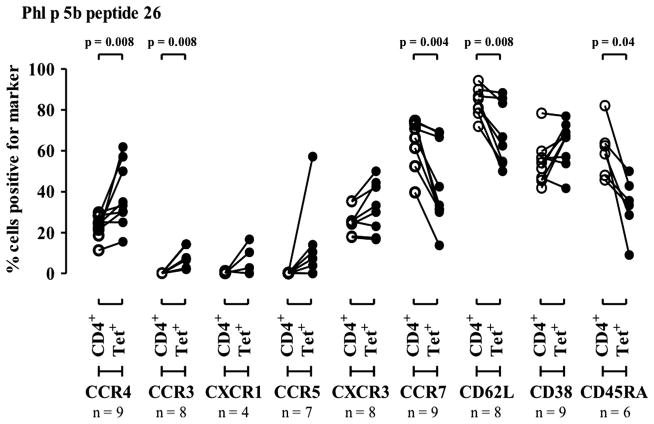
To assess the phenotype of p26-reactive donor cells, PBMCs from subjects with allergy were labelled ex vivo with p26/DR1 tetramer and costained for CD4, CXCR1, CCR3, CCR4, CXCR5, CCR7, CD62L, CD38 and CD45RA (figure 4). Tetramer-positive cells are of an effector memory type: CD45RAl°, CD62Ll°, CCR4hi, CCR3hi. In line with this, patient responses to p26 encompass an IL-5 response, but no detectable interferon γ (data not shown).
Figure 4.
Effector memory phenotype of Phl p 5 peptide 26 (p26) tetramer-positive CD4 T cells from subjects with allergy. Peripheral blood mononuclear cells isolated from subjects with allergy were labelled immediately ex vivo with Phl p 5b peptide 26 tetramer. This was followed by cell surface staining with FL-1 conjugated cell surface maker antibodies and an anti-CD4PE-Cy5. The expression of a FL-1 conjugated cell surface maker antibody was compared on total CD4high and CD4 human leukocyte antigen (HLA)-DR1 Phl p 5 p26 tetramer-positive cells from HLA-DR1-positive individuals with grass allergy. The reduction in mean fluorescence intensity for CD62L and CD45RA staining of tetramer-positive cells is indicated in online supplementary figure S1. Statistical analysis was performed using a Wilcoxon signed rank test. Samples were processed during the grass pollen season, summer 2010.
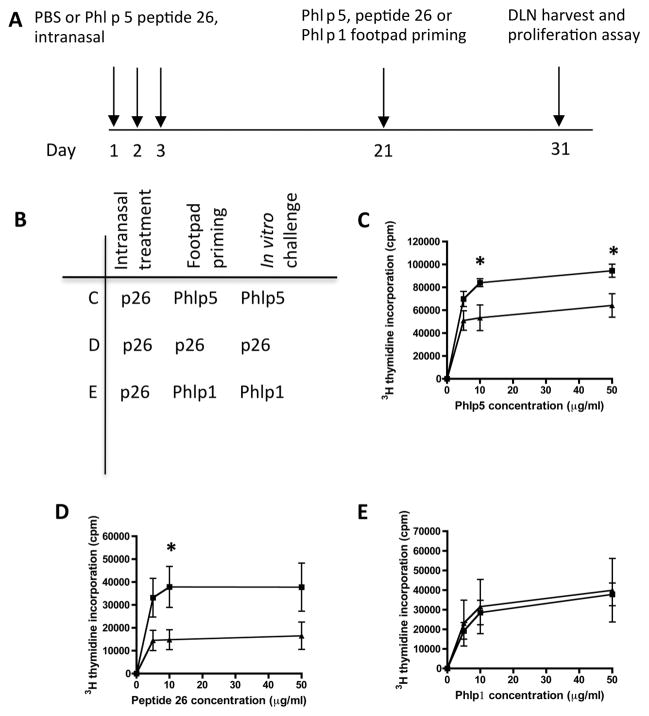
Intranasal treatment of mice with Phl p 5b p26 results in reduced response to p26 and whole rPhl p 5b protein
Intranasal administration of p26 substantially reduces the subsequent T-cell response to immunogenic challenge with p26 (figure 5). Intranasal treatment with just p26 is sufficient to significantly reduce the recall response to immune challenge with whole protein (figure 5C) or with p26 itself (figure 5D). The induction of immune regulation is antigen specific since intranasal administration of p26 has no effect on priming and challenge with another Timothy antigen, Phl p 1 (figure 5E).
Figure 5.
Intranasal treatment with peptide 26 (p26) results in reduced T-cell proliferation to rPhl p 5b and p26. (A, B) Mice were treated intranasally with phosphate-buffered saline (PBS, filled squares) or p26 (filled triangles) at days 1, 2 and 3 and footpad primed at day 21 with (C) rPhl p 5 protein (each group, n=7), (D) p26 (each group, n=7) or (E) whole Phl p 1 protein (each group, n=5). Draining lymph nodes were harvested at day 31 and 3Hthymidine incorporation measured for protein or peptide as shown. An unpaired t test was used to determine significant differences between groups. Statistically significant differences were defined as p values of less than 0.05 (*p<0.05).
Increased expression of regulatory genes and decreased expression of Th2 inflammatory genes in p26-induced immune regulation
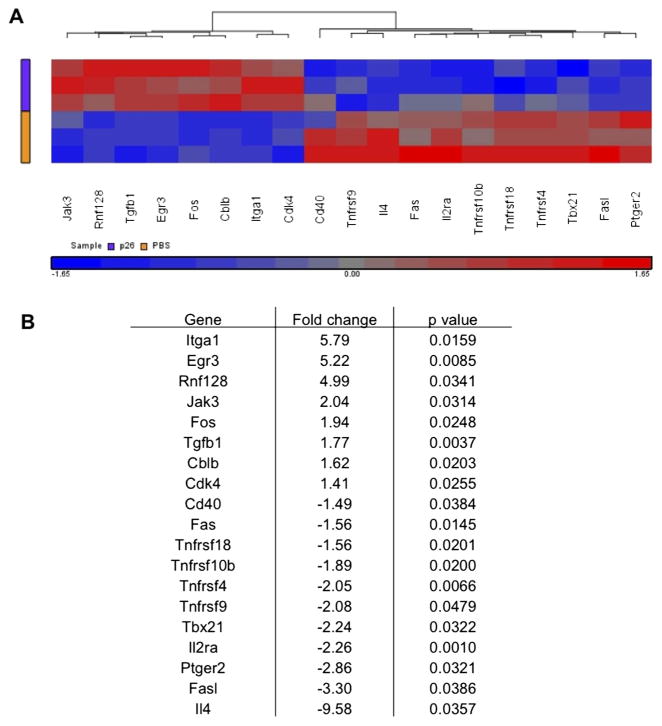
We then explored the transcriptomic pathways underlying p26-induced immune regulation by cDNA microarray analysis of peptide re-stimulated DLN cultures from peptide-treated or control HLA-DR1 transgenics (figure 6). The heatmap shows key genes differentially expressed following in vitro p26 re-stimulation of DLN cells. Intranasal peptide treatment prevents the full development of effector function: il4, il2ra are not upregulated, nor are Cd40, Fas and FasL. This is accompanied by an increase in transcripts that define a more regulatory response, including upregulation of Jak3, Tgfb1, Cbl-b, Egr3 and Rnf128.26,27 The most highly upregulated transcript in the dataset, Itga-1 (CD49a), is the α 1 chain of VLA-1. It is likely to have a role in regulation of cell trafficking to sites of allergen exposure.
Figure 6.
Intranasal treatment of peptide 26 (p26) is associated with increased expression of regulatory genes and decreased expression of T helper 2 inflammatory genes. Mice were treated intranasally with phosphate-buffered saline (PBS) (n=6) or p26 (n=6) before footpad immunisation and in vitro p26 stimulation of draining lymph node (DLN) cells. (A) A heat map shows the key genes differentially expressed following in vitro p26 stimulation of DLN cells where fold change is greater than ±1.4 with a p value of <0.05 by analysis of variance. (B) Fold change values and p values for each of these differentially expressed genes are shown.
Cbl-b and Itch are in a pathway mediating p26-induced T-cell regulation
Changes in regulation-associated transcripts were confirmed by quantitative PCR (qPCR) (figure 7). Transcriptional changes in Itch and Cbl-b were noteworthy since they are considered gatekeepers regulating a key peripheral tolerance pathway in the anergy and autoimmunity.26,27 We therefore investigated further the transcriptional changes to these genes and other changes potentially feeding through this pathway into changes at the Foxp3 promoter, including those to TIEG-1.27 Individual qPCR assays on DLN cells from p26-treated mice confirmed significant upregulation of Cbl-b, Itch, TIEG-1, Egr-3 and Foxp3.
Figure 7.
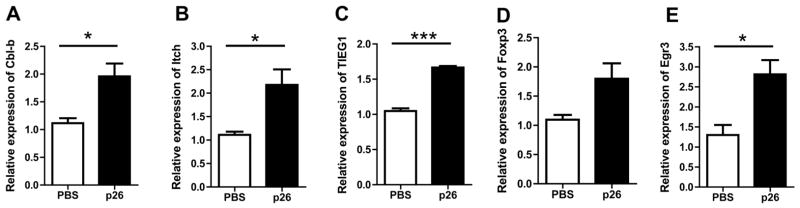
Intranasal treatment of peptide 26 (p26) results in increased expression of key targets in the Itch-Foxp3-transforming growth factor β inducible early gene 1 (TIEG-1) pathway. (A) Cbl-b, (B) Itch, (C) TIEG-1, (D) Foxp3 and (E) Egr3 are upregulated in p26-treated (n=3) compared with phosphate-buffered saline (PBS)-treated (n=3) mouse draining lymph node cells by real-time PCR. Student’s t test was used to determine significant differences between groups. Statistically significant differences were defined as p values less than 0.05 (*p<0.05, ***p<0.0005).
DISCUSSION
There may be therapeutic value in the wider use of allergen peptide immunotherapy to regulate CD4 T cells without the risk of IgE binding.28 Grass pollens encompass the large numbers of allergens and isoallergens, making it important to identify T-cell grass pollen epitopes conserved across diverse grass species as a step towards defining the minimal antigenic requirements for clinical tolerance to whole grass pollen. Previously, an HLA-DR4-restricted epitope was identified in rye grass Lol p 1.29 Identification of T-cell reactive regions within group five allergens of P pratense has been performed using Phl p 5 Tcell clones.8,30 A drawback of the latter studies is that they included small numbers of subjects of various HLA-DR haplotypes. As a result it is not clear if and how the identified T-cell reactive regions can or should be extrapolated to larger subject populations.
Screening T-cell lines from Phl p 5b immunised mice, we identified responses to p18, p23 and p26. Of these, p26 is conserved across diverse grass pollens, including rye grass and velvet grass. A US national survey of individuals on prevalence of 19 allergen-specific IgEs showed the response to rye grass to be the most common of allergens tested.31 While evidence exists to support the importance of Phl p 5, as we have used here, it is possible that group I are also clinically important and that mixed peptide cocktails may be used for therapy.
The reductionism of HLA class II transgenics to identify the presenting class II heterodimer with TGEM analysis of human PBMCs offers a powerful approach to epitope characterisation. Tetramers loaded with the individual peptide revealed a population responding to p26. Further comparison showed a difference in the number of p26 tetramer-positive cells expanded in individuals with allergy compared with those without allergy. While this study was based on cells expanded in culture ex vivo, our directly ex vivo TGEM data on individuals with allergy suggests frequencies in the range 1/60 000–1/300 000 T cells (data not shown). Whereas it can be hard to generalise from analysis of CD4 epitopes in a limited sample of individuals to applicability in a large outbred population expressing diverse HLA class II alleles, we show that p26 is likely to be of broad relevance due to its ability to bind multiple HLA class II heterodimers. Sette and colleagues characterised antigen-dominant regions of Phl p antigens predicted to bind diverse HLA alleles, a prediction we confirm empirically.32
Our microarray analysis pinpoints pathways defining a transcriptomic signature of regulation. At the top of this cascade stand changes to the transcription factor Egr-3, upregulated in peptide-treated mice. Overexpression of Egr-3 has been associated with upregulation of Cbl-b, inhibition of T-cell activation and consequent tolerance.33 Conversely, Egr-3-null mice show downregulation of Cbl-b and are resistant to peptide-induced tolerance. The E3 ligase, Itch, named for the itchy skin allergic phenotype with autoimmunity and interstitial lung disease in knockout mice, is a regulator of peripheral tolerance.27 Cbl-b, another ubiquitin ligase controlling tolerance, was also upregulated in this biosignature (figures 6 and 7). This ubiquitin ligase is proposed to impact on the immunity/anergy switch point at several levels through interactions with different target substrates. Itch-mediated ubiquitination re-routes PLC-γ1 and PKC-θ to the lysosome for degradation, with the effect of curtailing the immune synapse.34 In general, the enhanced re-sorting and degradation of immune signalling proteins mediated by E3 ubiquitin ligases is thought to cause an inability to maintain an intact immune synapse in stimulated T cells. Furthermore, there may be a more direct effect on TGFβ-induced Foxp3 expression as Itch knockouts show impairment of this pathway, presumably via its targeting by monoubiquitination of TIEG-1.27 The resulting response is a discrete effector rather than null response, evidenced by upregulation of Jak-3, which associates with the common γ chain, γc, pairing with ligand-specific subunits to supply receptors for IL-2, IL-4, IL-7, IL-9, IL-15 and IL-21.35
Itga-1 (CD49a), the α 1 chain of VLA-1, was very highly upregulated in mice treated with intranasal p26. This is noteworthy as a gene expression profiling study of human T regulatory type 1 (Tr1) cells by the Roncarolo group pinpoints CD49b and LAG-3 coexpression as a defining signature, allowing therapeutic Tr1 cells to be tracked in vivo.36
It should be tractable to use these approaches demonstrated here for high-throughput mapping of immunodominant epitopes from allergens and the design of optimal tolerogenic regimens. We acknowledge that the present article does not extend from exploring the mechanism of immune regulation to in vivo evaluation of therapeutic efficacy. Further studies will be needed to determine if it is possible to manipulate tolerance using therapeutic drug agonists of the E3 ubiquitin ligases, as under consideration in other settings including oncogenesis.37,38
Supplementary Material
Key messages.
What is the key question?
To identify important immunodominant epitopes from the Timothy grass pollen allergen, Phleum pratense, and study T cell immune regulation following intranasal treatment using a single immunodominant epitope.
What is the bottom line?
We describe an immunodominant Phleum pratense epitope, p26, with binding across multiple Human Leukocyte Antigen (HLA) class II alleles and sequence conservation with allergens from other grass species including rye grass and velvet grass making it an epitope of potential applicability across a spectrum of grass pollen allergy in the majority of individuals; intranasal treatment with this peptide results in T cell regulation to whole allergen mediated through the Cbl-b and Itch pathway.
Why read on?
This study exploits an approach combining HLA transgenic mice, tetramer guided epitope mapping, HLA binding studies and therapeutic studies in mice to define immune targets for the development of therapeutics in pollen allergy and identify a novel molecular pathway for the induced tolerance, offering the potential for additional future drug targets.
Acknowledgments
The authors would like to thank the Anthony Nolan Histocompatibility Laboratories and the Department of Clinical Immunology, Hammersmith Hospital, Imperial College Healthcare NHS Trust for HLA typing subjects; Allergopharma, Germany for supplying rPhl p 5; ALK Abello, Denmark for supplying nPhl p 5. The authors acknowledge the support of the NIHR Imperial BRC multi-parameter flow cytometry and confocal imaging facility, Hammersmith Campus.
Funding This study was funded by Asthma UK (RJB, SJT, SRD); Medical Research Council and Asthma UK Centre in Allergic Mechanisms of Asthma, UK PhD studentships (EJR, KJQ); The Health Foundation and Academy of Medical Sciences Clinician Scientist Fellowship (SJT); The Welton Foundation (RJB); Royal Brompton and Harefield Charitable Trust (RJB, SJT, SRD); WWK is supported by NIH NIAID. The NIHR Imperial BRC multi-parameter flow cytometry and confocal imaging facility, Hammersmith Campus supported this work.
Footnotes
Additional material is published online only. To view please visit the journal online (http://dx.doi.org/10.1136/thoraxjnl-2013-204324).
Contributors Conception, design, acquisition of data, analysis and interpretation of data: RJB, SJT. Drafting article and/or revising article critically for important intellectual content: RJB, SJT, DMA, SRD. All the authors contributed to either the acquisition of data and/or analysis and interpretation of data. Recruitment and characterisation of study subjects: AG-K, EJR, SJT, SRD. HLA-DR1 (DRA*0101/ DRB1*0101) Aβ° mice transgenics: DMA. TGEM analysis: WWK. T-cell epitope mapping in HLA-DR1 transgenics: EJR, CJR, RJB. Isolation and culture of human PBMCs and tetramer analysis: EJR, LRS, SJT, RJB. Peptide binding studies: BM. Murine peptide treatment and T-cell assays: CJR, RJB. Microarray and real-time PCR analysis: KJQ, CJR, RJB. HLA-DR1 genotyping and breeding: AG-K, KJQ, CJR, RJB. All authors were involved in writing and revising the final article prior to submission.
Competing interests SRD reports grants and personal fees from ALK-Abello, grants and personal fees from Merck, personal fees from Circassia, outside the submitted work; Dr Till reports grants from ALK-Abello, outside the submitted work; the rest of the authors declare that they have no conflict of interest.
Ethical approval The study was approved by the Royal Brompton and Harefield NHS Trust Ethics Committee (06/Q0404/43) and all participants gave written informed consent to participate.
Provenance and peer review Not commissioned; externally peer reviewed.
References
- 1.Meltzer EO. The prevalence and medical and economic impact of allergic rhinitis in the United States. J Allergy Clin Immunol. 1997;99(6 Pt 2):S805–28. [PubMed] [Google Scholar]
- 2.Durham SR, Walker SM, Varga EM, et al. Long-term clinical efficacy of grass-pollen immunotherapy. N Engl J Med. 1999;341:468–75. doi: 10.1056/NEJM199908123410702. [DOI] [PubMed] [Google Scholar]
- 3.Durham SR, Emminger W, Kapp A, et al. SQ-standardized sublingual grass immunotherapy: confirmation of disease modification 2 years after 3 years of treatment in a randomized trial. J Allergy Clin Immunol. 2012;129:717–25. e5. doi: 10.1016/j.jaci.2011.12.973. [DOI] [PubMed] [Google Scholar]
- 4.Walker SM, Durham SR, Till SJ, et al. Immunotherapy for allergic rhinitis. Clin Exp Allergy. 2011;41:1177–200. doi: 10.1111/j.1365-2222.2011.03794.x. [DOI] [PubMed] [Google Scholar]
- 5.Oldfield WL, Larche M, Kay AB. Effect of T-cell peptides derived from Fel d 1 on allergic reactions and cytokine production in patients sensitive to cats: a randomised controlled trial. Lancet. 2002;360:47–53. doi: 10.1016/s0140-6736(02)09332-7. [DOI] [PubMed] [Google Scholar]
- 6.Worm M, Lee HH, Kleine-Tebbe J, et al. Development and preliminary clinical evaluation of a peptide immunotherapy vaccine for cat allergy. J Allergy Clin Immunol. 2011;127:89–97. e1–14. doi: 10.1016/j.jaci.2010.11.029. [DOI] [PubMed] [Google Scholar]
- 7.Muller U, Akdis CA, Fricker M, et al. Successful immunotherapy with T-cell epitope peptides of bee venom phospholipase A2 induces specific T-cell anergy in patients allergic to bee venom. J Allergy Clin Immunol. 1998;101(6 Pt 1):747–54. doi: 10.1016/S0091-6749(98)70402-6. [DOI] [PubMed] [Google Scholar]
- 8.Campbell JD, Buckland KF, McMillan SJ, et al. Peptide immunotherapy in allergic asthma generates IL-10-dependent immunological tolerance associated with linked epitope suppression. J Exp Med. 2009;206:1535–47. doi: 10.1084/jem.20082901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maguire P, Nicodemus C, Robinson D, et al. The safety and efficacy of ALLERVAX CAT in cat allergic patients. Clin Immunol. 1999;93:222–31. doi: 10.1006/clim.1999.4795. [DOI] [PubMed] [Google Scholar]
- 10.Patel D, Couroux P, Hickey P, et al. Fel d 1-derived peptide antigen desensitization shows a persistent treatment effect 1 year after the start of dosing: a randomized, placebo-controlled study. J Allergy Clin Immunol. 2013;131:103–9. e1–7. doi: 10.1016/j.jaci.2012.07.028. [DOI] [PubMed] [Google Scholar]
- 11.Schulten V, Greenbaum JA, Hauser M, et al. Previously undescribed grass pollen antigens are the major inducers of T helper 2 cytokine-producing T cells in allergic individuals. Proc Natl Acad Sci U S A. 2013;110:3459–64. doi: 10.1073/pnas.1300512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chabre H, Gouyon B, Huet A, et al. Molecular variability of group 1 and 5 grass pollen allergens between Pooideae species: implications for immunotherapy. Clin Exp Allergy. 2010;40:505–19. doi: 10.1111/j.1365-2222.2009.03380.x. [DOI] [PubMed] [Google Scholar]
- 13.Van Neerven RJJ, Arnved J, Ipsen H. Phleum pratense-specific T cells of allergic rhinitis patients display a broader recognition pattern than Phleum pratense-specific serum immunoglobulin E. Clin Exp Allergy. 2000;30:242–54. doi: 10.1046/j.1365-2222.2000.00707.x. [DOI] [PubMed] [Google Scholar]
- 14.Jahn-Schmid B, Fischer GF, Bohle B, et al. Antigen presentation to the immunodominant T-cell epitope of the major mugwort pollen allergen, Art v 1, is associated with the expression of HLA-DRB1*01. J Allergy Clin Immunol. 2005;115:399–404. doi: 10.1016/j.jaci.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Etto T, de Boer C, Prickett S, et al. Unique and cross-reactive T cell epitope peptides of the major Bahia grass pollen allergen, pas n1. Int Arch All Appl Immunol. 2012;159:355–66. doi: 10.1159/000338290. [DOI] [PubMed] [Google Scholar]
- 16.Knapp B, Fischer G, Van Hemelen D, et al. Association of HLA-DR1 with the allergic response to the major mugwort pollen allergen: molecular background. BMC Immunol. 2012;13:43. doi: 10.1186/1471-2172-13-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Novak EJ, Liu AW, Gebe JA, et al. Tetramer-guided epitope mapping: rapid identification and characterization of immunodominant CD4+ T cell epitopes from complex antigens. J Immunol. 2001;166:6665–70. doi: 10.4049/jimmunol.166.11.6665. [DOI] [PubMed] [Google Scholar]
- 18.Yang J, James EA, Huston L, et al. Multiplex mapping of CD4T cell epitopes using class II tetramers. Clin Immunol. 2006;120:21–32. doi: 10.1016/j.clim.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Cromwell O, Suck R, Kahlert H, et al. Transition of recombinant allergens from bench to clinical application. Methods. 2004;32:300–12. doi: 10.1016/j.ymeth.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 20.Jutel M, Jaeger L, Suck R, et al. Allergen-specific immunotherapy with recombinant grass pollen allergens. J Allergy Clin Immunol. 2005;116:608–13. doi: 10.1016/j.jaci.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Bousquet J, Khaltaev N, Cruz AA, et al. Allergic Rhinitis and its Impact on Asthma (ARIA) 2008 update (in collaboration with the World Health Organization, GA(2) LEN and AllerGen) Allergy. 2008;63(Suppl 86):8–160. doi: 10.1111/j.1398-9995.2007.01620.x. [DOI] [PubMed] [Google Scholar]
- 22.Altmann DM, Douek DC, Frater AJ, et al. The T cell response of HLA-DR transgenic mice to human myelin basic protein and other antigens in the presence and absence of human CD4. J Exp Med. 1995;181:867–75. doi: 10.1084/jem.181.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Texier C, Pouvelle S, Busson M, et al. HLA-DR restricted peptide candidates for bee venom immunotherapy. J Immunol. 2000;164:3177–84. doi: 10.4049/jimmunol.164.6.3177. [DOI] [PubMed] [Google Scholar]
- 24.Texier C, Pouvelle-Moratille S, Busson M, et al. Complementarity and redundancy of the binding specificity of HLA-DRB1, -DRB3, -DRB4 and -DRB5 molecules. Eur J Immunol. 2001;31:1837–46. doi: 10.1002/1521-4141(200106)31:6<1837::aid-immu1837>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 25.Pancré V, Georges B, Angyalosi G, et al. Novel promiscuous HLA-DQ HIV Nef peptide that induces IFN-gamma-producing memory CD4+ T cells. Clin Exp Immunol. 2002;129:429–37. doi: 10.1046/j.1365-2249.2002.01934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venuprasad K. Cbl-b and itch: key regulators of peripheral T-cell tolerance. Cancer Res. 2010;15:70, 3009–12. doi: 10.1158/0008-5472.CAN-09-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venuprasad K, Huang H, Harada Y, et al. The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1. Nat Immunol. 2008;9:245–53. doi: 10.1038/niXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haselden BM, Kay AB, Larché M. Immunoglobulin E-independent major histocompatibility complex-restricted T cell peptide epitope-induced late asthmatic reactions. J Exp Med. 1999;189:1885–94. doi: 10.1084/jem.189.12.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macaubas C, Wahlstrom J, Galvao da Silva AP, et al. Allergen-specific MHC class II tetramer+ cells are detectable in allergic, but not in nonallergic, individuals. J Immunol. 2006;176:5069–77. doi: 10.4049/jimmunol.176.8.5069. [DOI] [PubMed] [Google Scholar]
- 30.Schramm G, Kahlert H, Suck R, et al. ‘Allergen engineering’: variants of the Timothy grass pollen allergen Phl p 5b with reduced IgE-binding capacity but conserved T cell reactivity. J Immunol. 1999;162:2406–14. [PubMed] [Google Scholar]
- 31.Salo PM, Calatroni A, Gergen PJ, et al. Allergy-related outcomes in relation to serum IgE: results from the National Health and Nutrition Examination Survey 2005–2006. J Allergy Clin Immunol. 2011;127:1226–35. doi: 10.1016/j.jaci.2010.12.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oseroff C, Sidney J, Kotturi MF, et al. Molecular determinants of T cell epitope recognition to the common Timothy grass allergen. J Immunol. 2010;185:943–55. doi: 10.4049/jimmunol.1000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Safford M, Collins S, Lutz MA, et al. Egr-2 and Egr-3 are negative regulators of T cell activation. Nat Immunol. 2005;6:472–80. doi: 10.1038/ni1193. [DOI] [PubMed] [Google Scholar]
- 34.Jeon M, Atfield A, Venuprasad K, et al. Essential role of the E3 ubiquitin ligase Cbl-b in T cell anergy induction. Immunity. 2004;21:167–77. doi: 10.1016/j.immuni.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 35.Ghoreschi K, Laurence A, O’Shea JJ. Janus kinases in immune cell signaling. Immunol Rev. 2009;228:273–87. doi: 10.1111/j.1600-065X.2008.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gagliani N, Magnani CF, Huber S, et al. Coexpression of CD49b and LAG-3 identifies human and mouse T regulatory type 1 cells. Nat Med. 2013;19:739–46. doi: 10.1038/nm.3179. [DOI] [PubMed] [Google Scholar]
- 37.Ernst A, Avvakumov G, Tong J, et al. A strategy for modulation of enzymes in the ubiquitin system. Science. 2013;339:590–5. doi: 10.1126/science.1230161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Micel LN, Tentler JJ, Smith PG, et al. Role of ubiquitin ligases and the proteasome in oncogenesis: novel targets for anticancer therapies. J Clin Oncol. 2013;31:1231–8. doi: 10.1200/JCO.2012.44.0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.