Abstract
Scl/Tal1 confers hemogenic competence and prevents ectopic cardiomyogenesis in embryonic endothelium by unknown mechanisms. We discovered that Scl binds to hematopoietic and cardiac enhancers that become epigenetically primed in multipotent cardiovascular mesoderm, to regulate the divergence of hematopoietic and cardiac lineages. Scl does not act as a pioneer factor but rather exploits a pre-established epigenetic landscape. As the blood lineage emerges, Scl binding and active epigenetic modifications are sustained in hematopoietic enhancers, whereas cardiac enhancers are decommissioned by removal of active epigenetic marks. Our data suggest that, rather than recruiting corepressors to enhancers, Scl prevents ectopic cardiogenesis by occupying enhancers that cardiac factors, such as Gata4 and Hand1, use for gene activation. Although hematopoietic Gata factors bind with Scl to both activated and repressed genes, they are dispensable for cardiac repression, but necessary for activating genes that enable hematopoietic stem/progenitor cell development. These results suggest that a unique subset of enhancers in lineage-specific genes that are accessible for regulators of opposing fates during the time of the fate decision provide a platform where the divergence of mutually exclusive fates is orchestrated.
Keywords: cardiac specification, enhancer, hematopoiesis, mesoderm diversification, transcriptional regulation
Introduction
Specification of cell types is dictated by few master regulators that activate lineage-specific transcriptional networks. This concept is underscored by studies in which overexpression of a small set of transcription factors can reprogram fibroblasts (or other cells) into a variety of cell types that closely resemble pluripotent cells, neurons, pancreatic beta cells, cardiomyocytes or hematopoietic cells (Takahashi & Yamanaka, 2006; Ieda et al, 2010; Huang et al, 2011; Kim et al, 2011; Pereira et al, 2013). Hematopoietic and cardiovascular systems are important targets for cell-based therapies due to the high morbidity and mortality associated with blood and heart diseases; however, so far, in vitro generation of transplantable cells for treating these diseases has not been successful. Blood cells, vasculature and the heart share not only an intimate functional relationship, but also a common origin in Flk1+ mesoderm (Fehling et al, 2003; Huber et al, 2004; Iida et al, 2005; Kattman et al, 2006). Although many regulators of the blood and circulatory system have been discovered, it is unclear how the divergence of these lineages in multipotent cardiovascular mesoderm is orchestrated and how their cell identity is solidified. Although several specific epigenetic modifications have been associated with activation or repression of the regulatory regions of genes (Cui et al, 2009; Creyghton et al, 2010; Zentner et al, 2011), to what degree they cause, or are the consequence of, gene activation/repression by cell type-specific transcription factors is still unclear.
The embryonic hematopoietic system is established in multiple waves, starting with the generation of lineage-restricted progenitors in the yolk sac and culminating in the emergence of multipotent hematopoietic stem/progenitor cells (HS/PC) in the major vessels in the yolk sac (Li et al, 2005; Yoshimoto et al, 2011), aorta-gonad mesonephros (AGM) region (North et al, 1999; De Bruijn et al, 2000; Zovein et al, 2008; Chen et al, 2009; Bertrand et al, 2010; Boisset et al, 2010; Kissa & Herbomel, 2010) and the placenta (Rhodes et al, 2008). The Ets factor Etv2/ER71/Etsrp first distinguishes the hemato-vascular lineages from cardiac mesoderm by specifying endothelium (Lee et al, 2008; Sumanas et al, 2008; Kataoka et al, 2011) and activates the bHLH factor Scl/Tal1, which confers hemogenic identity to endothelium (Van Handel et al, 2012). Scl also has a less well understood but important function in repressing the cardiac fate (Schoenebeck et al, 2007; Ismailoglu et al, 2008; Van Handel et al, 2012). Inducible overexpression of Scl during ES cell differentiation resulted in the expansion of hematopoietic lineage at the expense of cardiac and paraxial mesoderm (Ismailoglu et al, 2008), while the analysis of Scl knockout (SclKO) embryos revealed robust generation of ectopic cardiomyocytes in yolk sac vasculature, verifying a physiological role for Scl in cardiac repression (Van Handel et al, 2012). The requirement for Scl to repress cardiogenesis in hemogenic tissues was restricted to early midgestation of mouse development, suggesting the presence of epigenetic or other contributing mechanisms that solidify the fate choice. Scl also prevented misspecification of endocardium and cushion mesenchymal cells to cardiomyocytes. Although these studies pointed to a critical requirement for Scl in cardiac repression, it remained unknown whether Scl or its downstream targets repress cardiogenesis.
Scl functions in a multi-factor complex with Gata factors 1 or 2, E2A, Ldb1 and Lmo2 (Wadman et al, 1997) that promotes gene activation in hematopoietic cells (Tripic et al, 2009; Yu et al, 2009; Xu et al, 2012). Gata2 functions in HS/PC generation and survival (De Pater et al, 2013; Gao et al, 2013) whereas Gata1 regulates the maturation of erythroid, megakaryocytic and eosinophilic lineages (Bresnick et al, 2012), although there is redundancy between these factors (Takahashi et al, 2000). Embryos deficient for both Gata1 and Gata2 lack all blood cells, similar to Scl knockout embryos (Fujiwara et al, 2004). However, it is unknown whether hematopoietic Gata factors cooperate with Scl to repress cardiomyogenesis.
Here we show that Scl-dependent activation of hematopoietic fate and repression of cardiac fate during mesoderm diversification is accompanied by Scl binding to hematopoietic and cardiac enhancers that were epigenetically primed for activation in mesoderm. Our findings suggest that the accessibility and decommissioning of Scl-regulated enhancers is determined by modulation of active rather than repressive epigenetic modifications and is a major determinant of available fate choices and developmental stage specificity of target genes. We discovered a distinct subset of Scl-regulated enhancers that are also accessible for cardiac transcription factors in mesoderm, and propose that these enhancers serve as a platform where the fate choice between mutually exclusive fates is determined. Although hematopoietic Gata factors can bind with Scl to both activated and repressed enhancers, their function becomes critical only for activating genes required for HS/PC emergence from hemogenic endothelium. Elucidation of the stepwise process of how master regulators exploit the epigenetic landscape to establish hematopoietic or cardiovascular cell types from multipotent mesoderm provides new insights into improving directed differentiation and lineage reprogramming of stem/progenitor cells of these lineages for regenerative medicine.
Results
Scl specifies mesoderm by binding to genes encoding hematopoietic and cardiac regulators
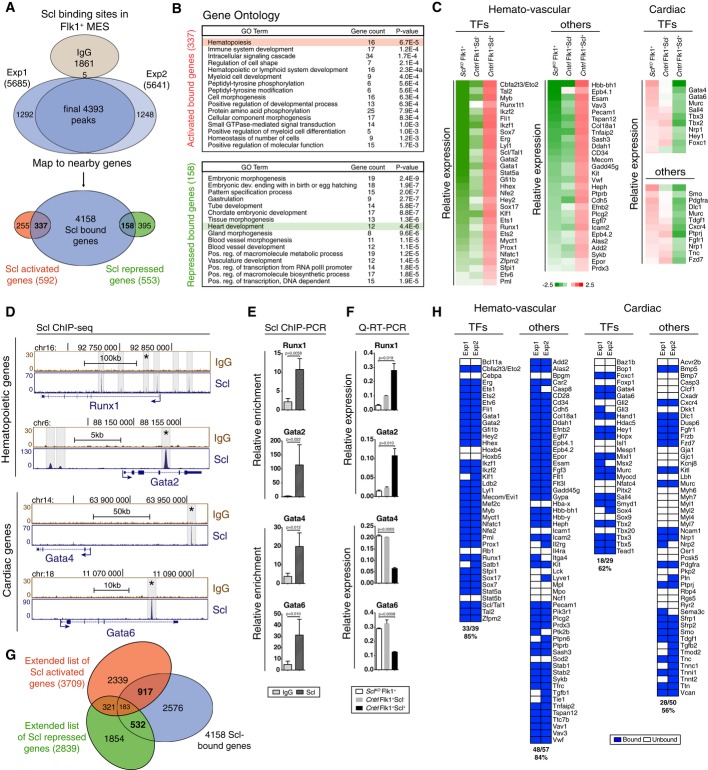
To understand how Scl specifies hemogenic endothelium while simultaneously repressing the cardiac fate, we defined Scl target genes by combining gene expression and chromatin immunoprecipitation sequencing (ChIP-seq) analysis on Flk1+ mesodermal cells differentiated from mouse ES cells (Supplementary Fig S1A). Scl binding associated genes (4,393 binding sites associated with 4,158 genes within 200 kb from transcriptional start sites) (Supplementary Table S1A) were intersected with Scl-dependent genes (592 and 553 genes that were significantly up- or down-regulated in mesoderm upon Scl expression (P-value < 0.05, fold change < 1.5, see details in Supplementary Fig S1A–C, Supplementary Table S1B and C, and Supplementary Materials and Methods). Integrated analysis showed Scl binding to 57% of the activated and 29% of the repressed genes in mesoderm (Fig1A, Supplementary Table S1B and C), which is significantly higher than would be expected by random chance (P-values 4.08 × 10−103 and 3.91 × 10−10, respectively). The genes in activated/bound group were enriched for GO term hematopoiesis, including key hematopoietic transcription factors Runx1, Gata2, cMyb and Cfba2t3/Eto2 (Fig1B–D). The genes in repressed/bound group were enriched for GO term heart development, including cardiac transcription factors Gata4, Gata6 and Tbx3 (Fig1B–D). Quantitative RT–PCR and ChIP–PCR verified Scl-dependent expression and Scl binding with both activated and repressed genes (Fig1E and F).
Figure 1.
- Venn diagram showing the number of Scl binding sites and overlap with Scl activated and repressed genes in Flk1+ MES (mesoderm) documents that Scl binds to both Scl-dependent activated and repressed genes.
- DAVID (Huang et al, 2007) GO enrichment analysis for Scl-bound and activated or Scl-bound and repressed genes shows enrichment of hematopoietic and heart-related terms, respectively.
- Gene expression heatmaps of selected genes from the bound and activated and bound and repressed groups show activation of key hematopoietic target genes and repression of key cardiac target genes in Scl-expressing mesoderm as compared to Scl-deficient mesoderm. Control [Scl+/hCD4, divided into Scl-expressing (Scl+) and non-expressing cells (Scl−)] and SclKO EBs are shown.
- Scl and control IgG ChIP-seq tracks show examples of Scl binding sites near hematopoietic (Runx1 and Gata2) and cardiac (Gata4 and Gata6) genes in Flk1+ mesoderm (MES).
- Verification of Scl binding sites marked with asterisks in (D) using ChIP-PCR, average enrichment of at least three independent biological experiments over negative control region (chr16: 92230219–92230338) with SEM are shown.
- Verification of Scl-dependent gene expression using qRT–PCR. Average of three biological replicates with SD are shown.
- Venn diagrams show the number of potential ‘extended activated’ and ‘extended repressed’ Scl target genes by intersecting genes associated with Scl binding in Flk1+ MES with Scl activated and repressed genes from day 4 EBs, E9.5 yolk sac, placenta and endocardium (Van Handel et al, 2012).
- Heatmaps show Scl binding to majority of activated hematopoietic and repressed cardiac transcription factors and other proteins. See also Supplementary Fig S1 and Supplementary Table S1A–E.
As the expression analysis in day 4 EB Flk1+ cells was limited to genes that were immediately activated or repressed upon Scl induction in mesoderm, we extended the analysis of Scl-dependent genes to those that became differentially expressed in endothelium in the yolk sac or the placenta, or endocardium in the heart in SclKO embryos (Van Handel et al, 2012). This analysis yielded an extended list of 3,709 Scl-dependent activated and 2,839 Scl-dependent repressed genes, of which 1,100 (29.6%) and 715 (25.2%) (Supplementary Fig S1A, Supplementary Table S1D and E) were bound by Scl in Flk1+ mesoderm (Fig1G), which is significantly higher than would be randomly expected (P-values 2.75 × 10−83 and 4.44 × 10−25, respectively). Analysis of the ‘extended activated’ genes showed that Scl binds to regulatory regions of the majority (33/39) of known hematopoietic transcription factors, many (48/57) surface markers and other proteins characteristic of nascent HS/PCs and hemogenic endothelium such as CD41 (Itga2b) and Cdh5 (VE-cadherin) (Fig1H). Scl binding was also associated with many (18/29) cardiac transcription factors (Gata4, Gata6, Hand1 and Tbx5), PDGFRα, a marker for cardiogenic mesoderm and ectopic cardiomyogenic precursors in SclKO embryos (Van Handel et al, 2012), and cardiomyocyte contractile proteins (Tnnc1, Tnnt2, Tnni1 and Ttn) (Fig1H). These data showed that during mesoderm diversification, Scl binds to a broad network of genes that govern hematopoietic and cardiac development, including the master transcription factors of each lineage.
To verify that the ES cell in vitro differentiation system recapitulates ectopic cardiomyogenesis in Scl-deficient endothelial precursors, SclKO and wild-type ES cells were differentiated toward mesodermal lineages and assayed for differentiation potential and gene expression. SclKO ES cells with doxycycline-inducible Scl overexpression (SclKOiScl) were included to assess whether reintroduction of Scl is sufficient to reverse the phenotype. As expected, both wild-type and SclKOiScl EBs, but not SclKO EBs, could generate CD41+c-Kit+ hematopoietic progenitors by day 7 (Supplementary Fig S1D). Likewise, Tie2+CD31+ endothelial precursors isolated from day 4.75 EBs from wild-type and SclKOiScl cells, but not SclKO cells, robustly generated CD45+CD11b+/− hematopoietic cells on OP9 stroma (Supplementary Fig S1E). SclKO Tie2+CD31+ endothelial precursors readily differentiated to troponin T-expressing cardiomyocytes, whereas re-expression of Scl abolished the ectopic cardiogenic potential in ES cell-derived endothelial cells (Supplementary Fig S1E). These data were reinforced by qRT–PCR analysis that verified the lack of expression of hematopoietic transcription factors and ectopic induction of cardiac factors in Scl-deficient endothelium, and rescue of these molecular defects by Scl overexpression (Supplementary Fig S1F). These data validate the ES cell in vitro differentiation system as a suitable model to study Scl-dependent cardiac repression and ectopic cardiogenesis from endothelial precursors.
Scl regulates mesodermal fate diversification via pre-established enhancers
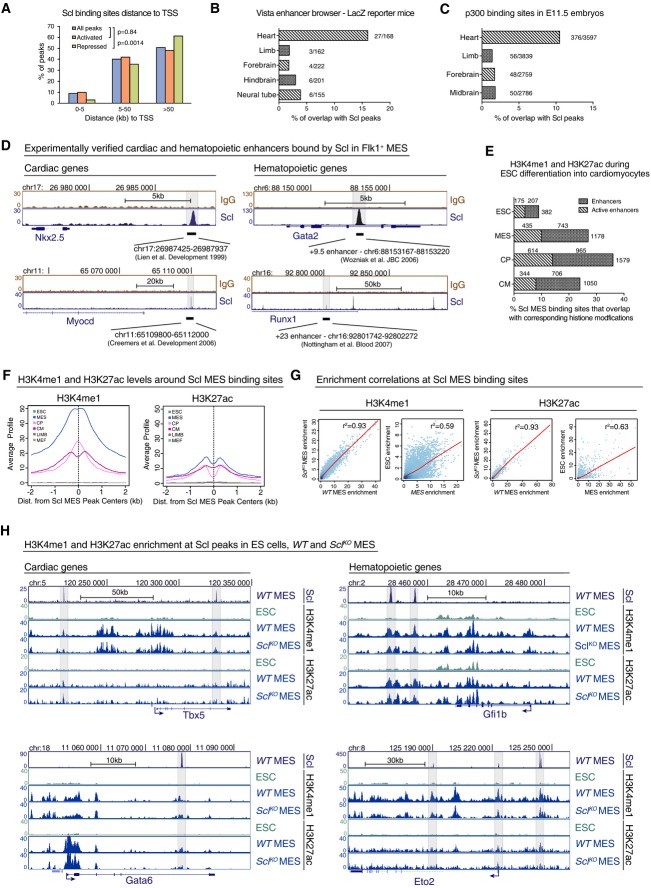
Analysis of genomic locations of Scl binding showed that the majority of Scl binding sites in Flk1+ mesoderm reside away from transcriptional start site (TSS) (Fig2A), suggesting that Scl functions through enhancer elements. This was most pronounced in the repressed genes, where only 3% of Scl binding sites were found within 5 kb of TSS. We thus correlated Scl mesodermal binding sites with published datasets for cardiac enhancers, including the Vista Enhancer database that contains in vivo experimentally verified enhancers (Visel et al, 2007) and ChIP-seq analysis for co-activator p300, a marker of enhancers (Visel et al, 2009), in E11.5 mouse hearts (Blow et al, 2010). Intersecting these data with Scl binding sites in Flk1+ mesoderm revealed Scl binding in 10–16% of all putative cardiac enhancers, which is significantly (P = 8.6E−5) higher than the overlap with enhancers in other tissues (Fig2B and C). Analysis of well-established enhancers of cardiac transcription factors Myocardin and Nkx2.5 (Fig2D) (Lien et al, 1999; Creemers et al, 2006) and hematopoietic regulators Runx1 (+23 enhancer) (Nottingham et al, 2007) and Gata2 (+9.5 enhancer) (Wozniak et al, 2007; Gao et al, 2013) (Supplementary Fig S2A) showed clear overlap with Scl binding.
Figure 2.
- Distribution of Scl binding sites relative to TSS in Scl-bound activated or repressed genes shows that majority of Scl binding sites locate away from TSS. Chi-square test was used to assess differences in the distribution.
- Percent of tissue-specific enhancers validated by LacZ reporter mice (Vista enhancer browser) that overlap with Scl MES binding sites shows that Scl binds to experimentally verified heart enhancers more often compared to enhancers from other tissues.
- Percent of Scl MES binding sites that overlap with heart enhancers in E11.5 embryos defined by p300 binding (Blow et al, 2010) is higher than overlap with enhancers from other tissues.
- Scl MES binding sites overlap with experimentally verified heart-specific enhancers upstream of Myocardin and Nkx2.5 genes and with hematopoietic enhancers within Runx1 and Gata2 genes.
- Percent of enhancers from different cardiac developmental stages identified by H3K4me1 (enhancers) and H3K4me1 combined with H3k27ac (active enhancers) (Wamstad et al, 2012) that overlap with Scl MES binding sites.
- Average H3K4me1 (left) and H3K27ac (right) profiles around all 4,393 Scl binding sites show tissue-specific enrichment in Flk1+ mesoderm (MES), cardiac precursors (CP) and cardiomyocytes (CM) but not in ESC (mouse ES cells) mouse embryonic fibroblasts (MEF) and embryonic limbs (LIMB).
- Correlation analysis of H3K4me1 (left) and H3K27ac (right) levels between WT and SclKO MES and ES cells and MES (Wamstad et al, 2012) around 4,393 Scl binding sites shows that establishment of these marks occurs independently of Scl.
- Scl, H3K4me1 and H3K27ac ChIP-seq tracks show comparable levels of H3K4me1 and H3K27ac in WT and SclKO mesoderm around cardiac (Gata6, Tbx5) and hematopoietic (Eto2, Gfi1b) genes.
To assess the epigenetic state of Scl-bound enhancers, 4,393 Scl mesodermal binding sites were assessed for the average levels of histone modifications associated with enhancers (H3K4me1) and active enhancers (H3K4me1 and H3K27ac) (Creyghton et al, 2010; Wamstad et al, 2012) during ES cell differentiation to cardiomyocytes. In ES cells, Scl mesodermal binding sites were largely devoid of H3K4me1 and H3K27ac; however, in Flk1+ mesoderm they had acquired high levels of H3K4me1 and some H3K27ac, which were in part retained during differentiation into cardiac progenitors and cardiomyocytes (Fig2E and F). Analysis of the average H3K4me1 and H3K27ac levels around Scl binding sites in other mesodermal tissues (limb, fibroblasts) showed no enrichment (Fig2F). This indicates that the regulatory regions to which Scl binds become epigenetically primed for activation specifically in Flk1+ mesoderm.
We next asked whether Scl is required for depositing active histone marks at hematopoietic and/or cardiac enhancers or whether these enhancers have been epigenetically pre-established prior to Scl binding. ChIP-seq analysis for histone H3K4me1 and H3K27ac in WT and SclKO Flk1+ mesoderm evidenced co-localization of the active marks across all Scl binding sites (Fig2G) including cardiac and hematopoietic genes (Fig2H) in both cell lines. Although H3K27ac levels were generally lower and this mark was not present in all hematopoietic or cardiac enhancers, there was no significant difference in H3K27ac levels at Scl mesodermal binding sites in WT and SclKO Flk1+ mesoderm (Fig2G and H). These results show that Scl is not required for the establishment of active enhancer marks in mesoderm, suggesting that these enhancers have become primed for activation in multipotent cardiovascular mesoderm to pave the way for Scl action.
To investigate whether the epigenetically primed status of enhancers is necessary for Scl binding, we induced Scl expression ectopically in SclKOiScl ES cells and compared Scl binding in these cells to that in Flk1+ mesoderm in wild-type and SclKOiScl cells. While inducible Scl was faithfully bound to the hematopoietic and cardiac enhancers in mesoderm, minimal binding was observed in ES cells (Supplementary Fig S2). Notably, the few binding sites where ectopically expressed Scl was able to bind in ES cells also harbored H3K4me1, whereas the key hematopoietic and cardiac enhancers were devoid of both Scl binding and H3K4me1 (Supplementary Fig S2). These data imply that Scl binding is tightly developmentally controlled, and at least in part determined by epigenetic status of the enhancers.
Repression of cardiac enhancers by Scl occurs transiently during mesoderm diversification
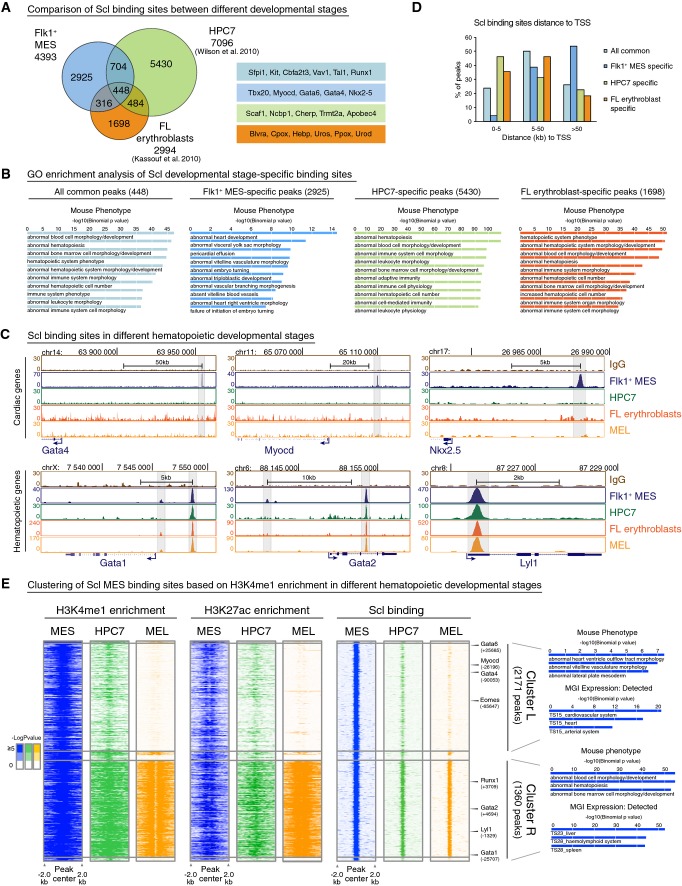
Since Scl is required only during a brief developmental window in midgestation to repress cardiogenesis in hemogenic tissues (Van Handel et al, 2012), we assessed whether Scl binding to cardiac enhancers is maintained in hematopoietic cells later in development. Intersecting Scl binding sites in Flk1+ mesoderm with previously published Scl binding sites in ES cell-derived hematopoietic progenitor cell line (HPC7) (Wilson et al, 2010) or fetal liver erythroblasts (Kassouf et al, 2010) showed that many Scl binding sites are developmental stage specific (Fig3A). GO enrichment analysis revealed enrichment of heart-related terms only with Flk1+ mesoderm-specific peaks (Fig3B) and genes (Supplementary Fig S3A). Analysis of Scl ChIP-seq tracks for cardiac regulators Gata4, Nkx2-5 and Myocardin verified Scl binding only in Flk1+ mesoderm (Fig3C). To confirm that different methods for ChIP-seq analysis between labs did not account for the difference, we performed Scl ChIP-seq on MEL cells (mouse erythroleukemia line) and verified the absence of Scl binding in cardiac genes (Fig3C, Supplementary Fig S3B). In comparison, Scl binding sites shared between different stages were enriched for hematopoiesis-related terms, and included major hematopoietic transcription factors (e.g. Gata1, Gata2, Lyl1, Gif1b, Runx1 and Myb) (Fig3C, Supplementary Fig S3A and B). While Flk1+ mesoderm-specific peaks were rarely found within 5 kb of TSS (Fig3D), 46% of the HPC7 or 34% of the erythroid cell-specific peaks were located within 5 kb to TSS. These data suggest that Scl binding extends from distant enhancers to promoters as hematopoiesis progresses.
Figure 3.
- Venn diagram showing overlaps of Scl binding sites in different developmental stages—Flk1+ MES, fetal liver (FL) erythroblast cells (Kassouf et al, 2010) and hematopoietic progenitors (HPC7) (Wilson et al, 2010) (left)—shows that majority of Scl binding sites are developmental stage specific. Examples of developmental stage-specific Scl-bound genes are shown (right).
- GREAT (McLean et al, 2010) analysis of significantly enriched GO terms in Mouse Phenotype category for all common, Flk1+ MES-specific, HPC7-specific and FL erythroblast-specific peaks shows enrichment of heart-related terms only among Flk1+ MES-specific peaks.
- ChIP-seq tracks with Scl binding in Flk1+ MES, HPC7, FL erythroblasts and MEL (mouse erythroleukemia) cells around cardiac (Gata4, Myocd, Nkx2.5) and hematopoietic (Gata1, Gata2 and Lyl1) genes show that binding to cardiac genes occurs only in mesoderm, while binding to hematopoietic genes is maintained throughout hematopoietic development.
- Comparison of the location of developmental stage-specific Scl binding sites relative to TSS shows that Scl binding near TSS occurs more often in later hematopoietic development compared to mesoderm.
- Heatmaps of combinatorial clustering of H3K4me1 around Scl mesodermal binding sites in MES, HPC7 and MEL reveal two major clusters: Cluster L loses H3K4me1, H3K27ac and Scl binding during hematopoietic differentiation and is enriched for genes involved in heart development; cluster R retains H3K4me1, H3K27ac and Scl binding during hematopoietic differentiation and is enriched for hematopoiesis-related genes. Selected GO terms among top 10 most enriched are shown.
Since Scl was no longer bound to cardiac enhancers in hematopoietic cells, we assessed whether the epigenetic landscape in Scl binding sites was modified upon differentiation of mesoderm to hematopoietic cells. Combinatorial clustering of Scl mesodermal binding sites based on H3K4me1 patterns in mesoderm, hematopoietic progenitors (HPC7) and erythroid cells (MEL) revealed two major clusters. Although the majority of Scl binding sites harbored H3K4me1 in mesoderm, this enhancer mark was lost in Scl binding sites in cluster L (= Loss of H3K4me1) but retained in cluster R (= Retention of H3K4me1) as hematopoietic development progressed. The gradual loss of H3K4me1 enrichment from HPC7 to MEL in cluster L strongly correlated with the loss of H3K27ac and Scl binding, while Scl binding sites in cluster R retained H3K4me1, H3K27ac and Scl binding (Fig3E). The peaks in cluster L were enriched for cardiac GO terms and included key cardiac regulators Gata4, Gata6 and Myocardin (Fig3E, Supplementary Fig S3B), while Scl peaks in cluster R were enriched for hematopoietic-related GO terms and included hematopoietic regulators Gata1, Gata2, Lyl1 and Runx1 (Fig3E, Supplementary Fig S3B). The opposite was observed in the cardiomyocyte cell line HL1, in which the enhancers of key hematopoietic regulators lost H3K4me1 and H3K27ac, while the enhancers of regulators of cardiomyocyte differentiation maintained these active marks (Supplementary Fig S3B). These results imply that the epigenetic landscape becomes dynamically remodeled in unused enhancers upon mesodermal fate diversification to blood and heart.
Decommissioning of Scl-regulated cardiac enhancers is associated with loss of active epigenetic marks rather than gain of repressive marks
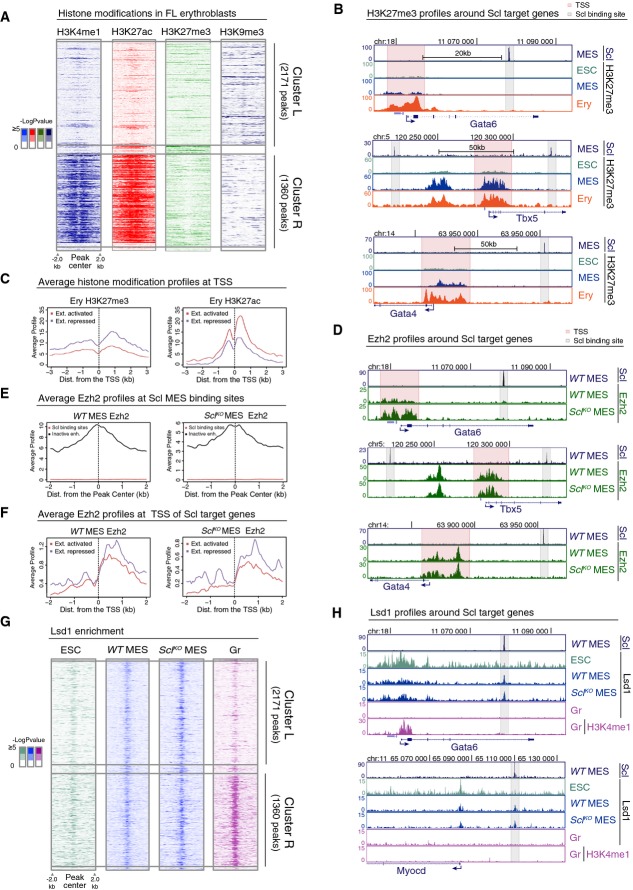
We next investigated whether the active epigenetic marks in Scl-regulated cardiac enhancers are replaced by repressive marks in hematopoietic cells to solidify the fate choice. Analysis of Ter119 erythroid cells for several histone marks (ENCODE project and Kowalczyk et al, 2012) in Scl mesodermal binding sites confirmed a strong correlation between Scl binding and retention of the active histone marks H3K4me1 and H3K27ac in Cluster R, and loss of these marks in Cluster L (Fig4A), also in erythroid cells in vivo. A candidate for a repression mechanism was Polycomb-mediated deposition of H3K27me3, which has been associated with silencing of both promoters and enhancers (Harmston & Lenhard, 2013). Moreover, Scl has been shown to interact with Ezh2, a core component of the Polycomb repressive complex 2 (PRC2) (Pinello et al, 2014). Analysis of H3K27me3 ChIP-seq data provided no evidence of H3K27me3 acquisition at Scl binding sites in erythroid cells in either Cluster R or L (Fig4A). ChIP-seq tracks around cardiac genes Gata4, Gata6 and Tbx5 confirmed the lack of H3K27me3 at Scl binding sites in ES cells, mesoderm and erythroid cells (Fig4B). However, H3K27me3 was observed at the TSS of some Scl-regulated genes (Fig4B). Comparison of H3K27me3 levels in Scl-bound extended activated and extended repressed genes (see Fig1, Supplementary Table S1D and E) in erythroid cells revealed an increase in H3K27me3 at the TSS of repressed genes, while H3K27ac was more enriched at the TSS of activated genes (Fig4C). These data suggested that Polycomb may contribute to the silencing of Scl-regulated genes at promoters, and raised the question whether Scl binding at enhancers is required for bringing PRC2 complex to promoters via chromosomal looping. ChIP-seq analysis for Ezh2 in mesoderm showed strong correlation between Ezh2 binding and H3K27me3 at TSS; however, there was no enrichment of Ezh2 at Scl binding sites (Fig4E). Moreover, there was no difference in the ability to recruit Ezh2 to the TSS of Scl-regulated genes in WT versus SclKO Flk1+ mesoderm (Fig4D and F). Altogether, these data imply that although Polycomb-mediated repression may contribute to the silencing of cardiac genes in the blood lineage, it is not a major factor governing accessibility to Scl-bound enhancers in mesoderm or their derivatives, and the failure to repress cardiac genes in Scl-deficient mesoderm is not caused by lack of Ezh2 recruitment to these genes.
Figure 4.
- Heatmaps of H3K4me1, H3K27ac, H3K27me3 and H3K9me3 histone modifications in FL erythroblasts around Scl MES binding sites in cluster L and cluster R show no gain of common repressive histone marks in cluster L.
- H3K27me3 ChIP-seq tracks in ES cells (ESC), Flk1+ mesoderm (MES) and FL erythroblasts (Ery) show that cardiac genes Gata6, Tbx5 and Gata4 harbor H3K27me3 at the TSS (pink) but not at Scl binding sites (gray).
- Average H3K27me3 (left) and H3K27ac (right) levels in FL erythroblasts around the TSS of extended list of Scl activated and repressed genes show that Scl repressed genes have on average more H3K27me3 and less H3K27ac as compared to Scl activated genes.
- Ezh2 ChIP-seq tracks in WT and SclKO mesoderm showing that Ezh2 recruitment to the TSS of cardiac genes Gata6, Tbx5 and Gata4 is not Scl dependent.
- Average Ezh2 enrichment in WT (left) and SclKO mesoderm (right) for inactive enhancers (defined by having H3K4me1 and H3K27me3 modifications (Wamstad et al, 2012) and distance greater than 5 kb from TSS) and for Scl binding sites shows no Ezh2 enrichment at Scl binding sites.
- Average Ezh2 enrichment at the TSS of extended list of Scl activated and repressed genes is similar in WT (left) and SclKO mesoderm (right).
- Heatmaps of Lsd1 enrichment in ES cells, WT and SclKO mesoderm and in granulocytes (Gr) (Kerenyi et al, 2013) in cluster L and cluster R show co-localization of Lsd1 binding with Scl binding sites and reveal that recruitment of Lsd1 can happen independently of Scl.
- Lsd1 ChIP-seq tracks (ES cells, WT and SclKO Flk1+ mesoderm and granulocytes) and H3K4me1 ChIP-seq track (granulocytes) show that Lsd1 enrichment coincides with Scl MES binding sites nearby cardiac genes Gata6 and Myocd in WT and SclKO Flk1+ mesoderm.
We next assessed whether the repressive histone modification H3K9me3 is acquired at the decommissioned enhancers during hematopoietic differentiation. The average levels of H3K9me3 were slightly higher in cluster L binding sites than in cluster R binding sites in three hematopoietic cell types (Ter119 erythroid cells, Fig4A, Supplementary Fig S4B, and G1E embryonic erythroid cell line and Ch12 B lymphoid cells, Supplementary Fig S4A and B). However, analysis of H3K9me3 around key cardiac genes did not show specific enrichment for H3K9me3 at Scl-bound enhancers (Supplementary Fig S4C). These data imply that although regions around some Scl mesodermal binding sites acquire H3K9me3 during hematopoietic differentiation, deposition of H3K9me3 is unlikely the predominant mechanism that initiates Scl-mediated cardiac repression.
As DNA methylation can restrict transcription factor binding to its target DNA (Blattler & Farnham, 2013), we asked whether changes in DNA methylation mediate the inactivation of Scl-regulated cardiac enhancers in hematopoietic cells. Comparison of the average DNA methylation levels at Scl mesodermal binding sites associated with extended list of Scl activated or repressed genes in genome-wide methylation datasets from mouse ES cells (Habibi et al, 2013), bone marrow, heart, pancreas and skin (Hon et al, 2013), showed that Scl binding sites are on average hypomethylated compared to the surrounding regions (Supplementary Fig S4D). Nevertheless, both Scl's extended activated and repressed target genes showed a general increase in DNA methylation within 200 kb of Scl binding sites from ES cells to all adult cell types analyzed (Supplementary Fig S4E). The methylation levels in Scl extended activated genes were lower in the bone marrow compared to other tissues, while the extended repressed genes showed lower methylation in the heart (Supplementary Fig S4E).
To investigate whether Scl initiates differential methylation in mesoderm, we performed RRBS (reduced representation bisulfite sequencing) in ES cells, WT and SclKO Flk1+ mesodermal cells, and MEL cells. Global covariance analysis showed that major changes in DNA methylation occur later in hematopoietic development rather than in mesoderm (Supplementary Fig S4F). Intersection of RRBS data with Scl mesodermal binding sites showed no difference in methylation levels in ES cells and WT and SclKO Flk1+ mesoderm; only MEL cells had higher DNA methylation (Supplementary Fig S4G). Thus, while DNA methylation may contribute to the silencing of Scl target genes in differentiated cells, this occurs later in development and is not centered at Scl-bound enhancers. These data imply that DNA methylation is not a primary mechanism that silences Scl-regulated cardiac enhancers during hematopoietic specification.
Our analysis of known repressive histone marks did not provide evidence that Scl-regulated enhancers would be actively silenced during mesodermal lineage diversification, but rather implied that gain and loss of active epigenetic marks is the key determinant that governs tissue-specific access to these enhancers. We thus assessed whether the recruitment of Lsd1, a lysine-specific demethylase 1 that decommissions enhancers by removing H3K4me1 (Whyte et al, 2012), to cardiac enhancers is interrupted in the absence of Scl. Comparison of ChIP-seq data of Lsd1 mesodermal binding to published Lsd1 binding datasets in ES cells and myeloid cells (Whyte et al, 2012; Kerenyi et al, 2013) showed robust co-localization of Lsd1 at Scl-bound enhancers (Fig4G and H) in both cluster L and R in Flk1+ mesoderm, which is a heterogenous population of cells committed to hemato-vascular or cardiac fates. In myeloid cells, cardiac enhancers were devoid of both Lsd1 and H3K4me1 (Fig4H), documenting a short temporal window for Lsd1 action. Nevertheless, analysis of Lsd1 binding in Scl-deficient mesodermal cells did not show evidence of impaired Lsd1 binding in cardiac enhancers in Cluster L (Fig4G and H). These results imply that, although Lsd1 is likely involved in the decommissioning of unused enhancers upon mesodermal lineage diversification, defective cardiac repression in Scl-deficient mesodermal derivatives is not caused by an inability to recruit Lsd1 to primed cardiac enhancers.
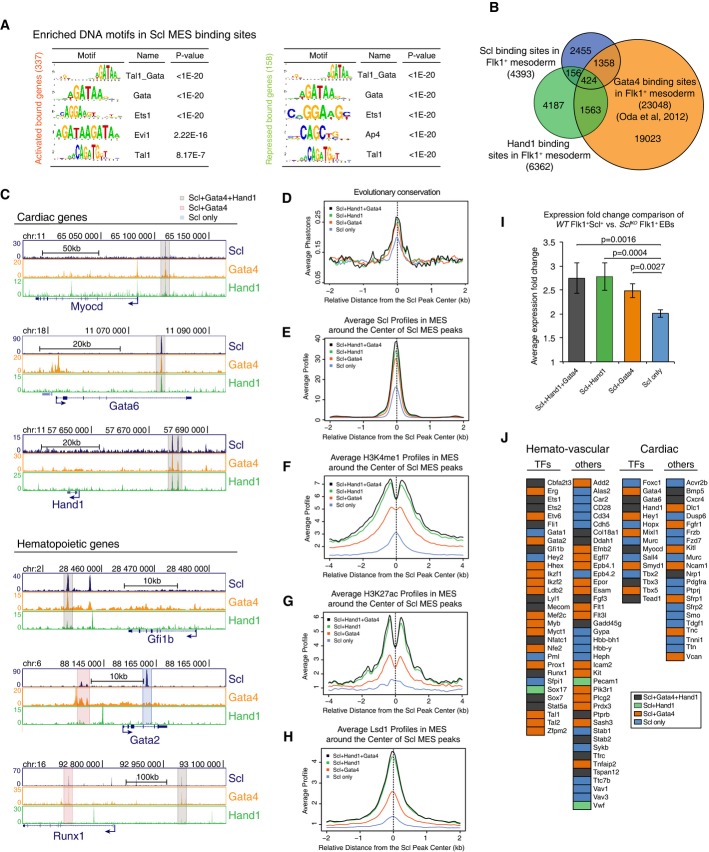
Binding by Scl and cardiac Gata4 and/or Hand1 define enhancer subgroups that may determine fate choice between opposing mesodermal fates
As the analysis of repressive marks and corepressors provided no evidence for direct Scl-dependent corepressor recruitment to enhancers or promoters in mesoderm, we considered the possibility that Scl inhibits cardiac fate by interfering with the activation of these genes by cardiac regulators. To identify candidate factors that could also bind to Scl-regulated enhancers, we performed DNA motif enrichment analysis for known transcription factor binding sites in Transfac and Jaspar databases. This analysis identified the composite Tal1(Scl)_Gata motif and Gata motifs as the most prevalent motifs in both activated and repressed genes (Fig5A).
Figure 5.
- A Scl DNA binding motif analysis shows enrichment of Tal1_Gata, Gata and Ets motifs in both Scl activated and repressed genes.
- B Venn diagram showing overlaps between Scl, Hand1 and Gata4 (Oda et al, 2013) binding sites in Flk1+ mesoderm documents that some of Scl MES binding sites are also bound by cardiac TFs.
- C Gata4, Hand1 and Scl ChIP-seq tracks with cardiac and hematopoietic gene regions show examples of three subgroups of enhancers—those bound by Scl, Hand1 and Gata4 (gray), Scl and Gata4 (orange) and those bound only by Scl (blue).
- D-H Scl binding sites that overlap with both Hand1 and Gata4 (gray) or with Hand1 (green) or Gata 4 (orange) alone show higher evolutionary conservation (D) and higher average enrichment of Scl (E), H3K4me1 (F), H3K27ac (G) and Lsd1 (H) in mesoderm compared to sites bound by Scl alone (blue).
- I Average expression changes between Scl-expressing and Scl-deficient mesoderm shows that genes that are regulated by enhancers bound by all three (Scl, Hand1 and Gata4) factors or two factors (Scl and Hand1 or Scl and Gata4) show higher absolute expression changes compared to those bound by Scl alone.
- J Analysis of Scl-regulated key hematopoietic and cardiac genes shows that many of them can be bound also by cardiac factors, Scl, Gata4 and Hand1 (orange); Scl and Hand1 binding (green); Scl and Gata4 binding (orange); and Scl binding alone (blue).
There are six known Gata factors, of which Gata1, 2 and 3 act in hematopoietic cells and Gata4, 5 and 6 regulate cardiac development (Chlon & Crispino, 2012). Gata4 can regulate cardiac development both indirectly via endoderm and directly by binding to cardiac genes in Flk1+ mesoderm (Holtzinger et al, 2010; Oda et al, 2013). Analysis of a published dataset for Gata4 binding in Flk1+ mesoderm revealed that Gata4 binding overlaps with a large fraction (41%) of Scl binding sites (Fig5B). Moreover, as other bHLH factors can also bind to E-box motifs similar to Scl, we also performed ChIP-seq for Hand1, a key cardiac bHLH factor that can interact with cardiac Gata factors. 13.2% all Scl-bound enhancers were co-occupied by Scl and Hand1 in Flk1+ mesoderm (9.7% Scl, Hand1 and Gata4 altogether, and 3.5% Scl and Hand1 alone). Analysis of the regulatory regions for key cardiac (Gata6, Tbx5 and Myocardin) and hematopoietic (Runx1, Gfi1b and Gata2) transcription factors identified at least one enhancer in each gene bound by Scl, Hand1 and/or Gata4 (Fig5C). The sites that can be occupied by all three factors (424 sites) included both that remain active (cluster R, 167 sites) and sites that become decommissioned during hematopoietic development (cluster L, 201 sites), and they associated with both Scl-dependent activated (128) and repressed (79) genes. Similar distribution was observed for Scl binding sites that were shared with Gata4 or Hand1 alone. These data suggest that a distinct subset of Scl-bound enhancers may be used by both hematopoietic and cardiac bHLH and Gata transcription factors to activate their own lineage or repress a competing lineage.
We next assessed whether there are any unique features that sets the ‘dually accessible’ enhancers apart from enhancers bound by Scl alone. The enhancers bound by both Scl and Gata4 and/or Hand1 were more evolutionarily conserved as compared to other Scl mesodermal binding sites (Fig5D) and showed higher levels of Scl binding (Fig5E), H3K4me1, H3K27ac (Fig5F and G) and Lsd1 enrichment (Fig5H). Moreover, Scl target genes that harbor an enhancer that can be bound by Scl and Gata4 and/or Hand1 showed higher average gene expression change in Scl-expressing WT versus SclKO Flk1+ mesoderm as compared to Scl target genes that do not have a dually accessible enhancer (Fig5I). The genes that contain a dually accessible enhancer included many hematopoietic (27/31, 87%) and cardiac (10/15, 67%) transcription factors (Fig5J).
We next assessed whether similar developmental stage-specific binding observed with Scl is also seen with Gata4, for which several published datasets from fetal and adult stages are available (He et al, 2014). Analysis of Gata4 binding in different stages of heart development showed gradual loss of overlap between Scl mesodermal binding sites and Gata4 binding from Flk1+ mesoderm to E12.5 hearts to adult hearts (Supplementary Fig S5A and B). While Gata4 binding was observed in both hematopoietic (Gfi1b and Myb) and cardiac enhancers (Tbx5 and Gata6) in mesoderm, hematopoietic enhancers had lost Gata4 binding in fetal hearts by E12.5. Many cardiac enhancers also lost Gata4 binding in adult hearts (Supplementary Fig S5C). These data show that the key genes regulating cardiovascular fate determination harbor dually accessible enhancers that can be bound by both hematopoietic and cardiac factors in mesoderm, but are dynamically remodeled during lineage diversification and maturation. Our data propose that this subset of enhancers may be critical for initiating the fate choice between competing cardiovascular lineages.
Gata1 and/or Gata2 are dispensable for cardiac repression but are essential for emergence of HS/PCs from hemogenic endothelium
Since Scl and Gata4 share common binding sites in both hematopoietic and cardiac enhancers and Scl acts together with hematopoietic Gata factors 1 and 2 in blood forming cells, we investigated whether hematopoietic Gata factors are required for Scl-induced gene activation and/or repression during mesoderm diversification. ChIP–PCR for Gata2 showed that, similar to Scl, Hand1 and Gata4, Gata2 binds to dually accessible enhancers in both hematopoietic and cardiac transcription factors that are activated or repressed by Scl (Supplementary Fig S5D).
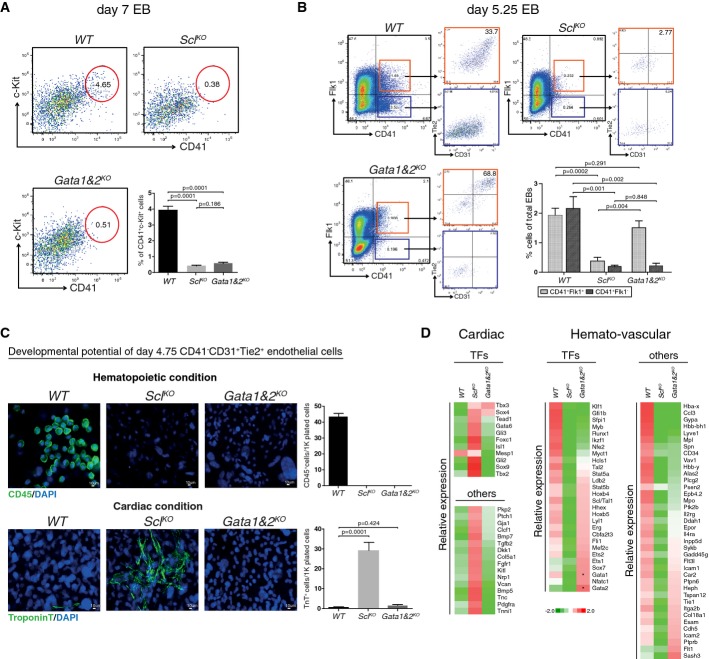
To investigate whether hematopoietic Gata factors function in Scl-mediated gene activation and repression, we assessed the developmental potential of Gata1 and 2 double knockout (Gata1&2KO) ES cells, which eliminates possible redundancy between Gata1 and 2. No c-Kit+CD41+ hematopoietic progenitors were detected in day 7 Gata1&2KO EBs or SclKO EBs (Fig6A). However, FACS analysis on day 5.25 EBs identified a subpopulation of cells in Gata1&2KO EBs that expressed the early HS/PC marker CD41, while SclKO EBs were devoid of this population. Nevertheless, all CD41+ cells in Gata1&2KO EBs co-expressed Flk1 and the majority also expressed endothelial markers CD31 and Tie2, raising the hypothesis that HS/PC development becomes stalled at hemogenic endothelium in the absence of Gata1 and 2 (Fig6B).
Figure 6.
- FACS analysis of day 7 EBs with markers CD41 and c-Kit shows efficient generation of HS/PCs from WT, but not SclKO or Gata1&2KO cells. Average of six independent experiments with SEM is shown.
- FACS analysis of day 5.25 EBs with markers CD41, Flk1, Tie2 and CD31 shows the generation of hemogenic endothelial cells from WT and Gata1&2KO cells, but not SclKO cells. Average of five independent biological experiments with SEM is shown.
- Assessment of the developmental potential of day 4.75 EB CD41−CD31+Tie2+ endothelial cells on OP9 for 14 days shows the generation of CD45+ hematopoietic cells from WT cells but not Gata1&2KO or SclKO cells, and troponin T+ cardiomyocytes from SclKO cells, but not WT or Gata1&2KO cells. Average of four independent experiments with SEM is shown.
- Heatmaps show gene expression differences in subsets of hematopoietic genes between Gata1&2KO and WT or SclKO day 4.75 CD41−CD31+Tie2+ endothelial cells. Cardiac derepression is observed only in SclKO cells. *designates non-functional transcripts.
To assess the differentiation potential of Gata1- and 2-deficient endothelial cells, EBs were differentiated for 4.75 days, after which CD41−CD31+Tie2+ endothelial cells were sorted and cultured for 14 days in hematopoietic and cardiac conditions on OP9 stroma. Neither Gata1&2KO nor SclKO endothelial cells generated CD45+ hematopoietic cells (Fig6C). In contrast, only SclKO, but not Gata1&2KO or WT endothelial cells, robustly differentiated into troponin T+ cardiomyocytes. These data show that Gata1 and/or 2 is required for hematopoiesis, but not for the repression of cardiogenesis in endothelial precursors.
To investigate the effect of Gata1 and 2 on endothelial gene expression, CD41−CD31+Tie2+ endothelial cells from day 4.75 EBs were sorted for RNA sequencing. No significant derepression of cardiac regulators was observed in Gata1&2KO endothelial cells, unlike SclKO endothelial cells which showed induction of several cardiac genes (Fig6D, Supplementary Table S2). Notably, expression of many hemato-vascular transcription factors that were down-regulated in SclKO endothelial cells (e.g. Lyl1, Hhex, Cbfa2t3/Eto2, Erg, Fli1, Sox7) was not significantly decreased in Gata1&2KO endothelial cells. Likewise, many surface markers of hemogenic endothelium and HS/PCs (e.g. CD41/Itga2b, VE-cadherin/Cdh5, Esam, Icam2) were expressed in Gata1&2KO endothelial cells, further supporting the notion that Gata1&2KO cells can specify hemogenic endothelium. However, the expression of key HS/PC transcription factors Runx1, Myb, Gfi1b, Pu.1/Sfpi1 and Klf1 was down-regulated in Gata1&2KO endothelial cells (Fig6D), implying that Gata1 and/or 2 is required for the induction of the hematopoietic transcription factor network that enables the emergence of HS/PCs from hemogenic endothelium.
To exclude the possibility that Gata3, a hematopoietic Gata factor that functions in T cells (Hosoya et al, 2010) and HSC maintenance (Ku et al, 2012) compensates for Gata1 and 2 in cardiac repression, Gata1-, 2- and 3-deficient (Gata1&2KOGata3KD) ES cells were generated using lentiviral knockdown of Gata3. Similar to Gata1&2KO, Gata1&2KOGata3KD endothelial cells showed neither up-regulation of cardiac program nor generation of ectopic cardiomyocytes in culture, and the hematopoietic program was similarly stalled at hemogenic endothelium with intact expression of many Scl-dependent hemato-vascular genes (Hhex, Ets2, Lyl1, Itga2b/CD41, Cadh5/Ve-Cad etc) and defective expression of key HS/PC transcription factors (e.g. Runx1 and Myb) (Supplementary Fig S6). Collectively, these data show that hematopoietic Gata factors are critical for full activation of Scl-dependent hematopoietic transcriptional network, but not for cardiac repression.
Gata factors 1 and 2 recruit Scl to specific binding sites in key hematopoietic transcription factors
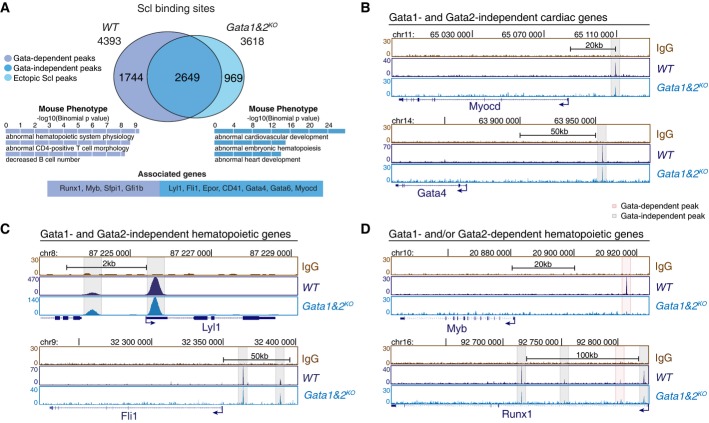
To investigate whether loss of Gata1 and Gata2 compromises Scl binding to its targets during mesoderm specification, we determined genome-wide Scl binding sites in Flk1+ mesoderm from Gata1&2KO EBs using ChIP-seq (Fig7A, Supplementary Table S3). The GO terms enriched among the genes associated with overlapping Scl binding sites included both hematopoiesis and cardiogenesis, while the peaks lost in Gata1&2KO cells were associated with genes related to hematopoiesis (Fig7A). Analysis of the individual genes confirmed intact Scl binding to key cardiac enhancers (Myocardin, Gata4 and Gata6) in the absence of Gata1 and 2 (Fig7B, Supplementary Fig S7B). Likewise, a majority of hemato-vascular transcription factors (Lyl1, Fli1) that were expressed in Gata1&2KO endothelium retained Scl binding (Fig7C). The subset of genes that lost an Scl binding site in Gata1&2KO Flk1+ cells included transcription factors Runx1, Myb, Pu.1/Sfpi1 and Gfi1b, whose expression in endothelium was Gata1 and 2 dependent (Fig7D, Supplementary Fig S7B). Remarkably, while several other Scl binding sites were preserved near Runx1 gene, the binding site that was lost in Gata1&2KO cells corresponds to +23 enhancer that has been shown to be essential for Runx1 regulation during HSC emergence (Nottingham et al, 2007) and becomes the strongest Scl binding site in HPC7 hematopoietic progenitor cells (Supplementary Fig S3B). These results imply that although Gata1 and 2 are not essential for Scl binding to the majority of its activated or repressed target genes, they are required for the recruitment of Scl to specific regulatory regions that govern the expression of key hematopoietic transcription factors. ChIP-PCR for Gata1&2KOGata3KD mesoderm confirmed similar binding for Scl in key hematopoietic and cardiac enhancers in the absence of all three hematopoietic Gata factors (Supplementary Fig S7A).
Figure 7.
- A Intersection of Scl binding sites in WT and Gata1&2KO Flk1+ MES shows Gata1- and/or Gata2-dependent redistribution of Scl binding sites in a subset of target genes (two independent experiments). Selected GO terms among top 10 most enriched for Gata-independent (blue) and Gata-dependent (purple) Scl binding sites together with example genes reveal Gata1- and/or Gata2-dependent functions for Scl-mediated hematopoietic regulation.
- B-D Scl ChIP-seq tracks in WT and Gata1&2KO Flk1+ MES show Gata1- and 2-independent Scl binding to cardiac regulators (Myocardin, Gata4) (B), Gata1- and 2-independent Scl binding to key hemato-vascular regulators (Lyl1, Fli1) (C) and Gata1- and/or 2-dependent Scl binding to key HS/PC transcription factors (Myb, Runx1) (D).
We also assessed whether the hematopoietic Gata factors function by modifying the epigenetic landscape to enable access of Scl complex to key hematopoietic enhancers. Analysis of H3K4me1 and H3K27ac in Gata1- and 2-deficient mesodermal cells did not reveal differences in the establishment of primed state in key hematopoietic and cardiac enhancers, including those that could not be activated in Gata1&2KO cells (Supplementary Fig S7B). Likewise, there was a strong correlation of global H3K4me1 and H3K27ac levels between WT and Gata1&2KO Flk1+ mesoderm (Supplementary Fig S7C). Collectively, these data show that Scl and hematopoietic Gata factors bind to enhancers that have been epigenetically primed for activation in mesoderm prior to their binding. While the repression of the cardiac fate by Scl can occur in the absence of hematopoietic Gata factors, they help recruit Scl to new regulatory regions to enable activation of key hematopoietic transcription factors that secure the emergence of HS/PCs from hemogenic endothelium.
Discussion
The ability to execute developmental fate decisions with proper temporal and spatial control is critical for embryonic development, as well as for harnessing the power of stem cell and reprogramming technologies for therapeutic applications. The bHLH factor Scl has emerged as a true master regulator of mesoderm fate as it not only governs the establishment of the entire hematopoietic system, but it also is critical for preventing ectopic cardiomyocyte development in hemogenic tissues (Van Handel et al, 2012). Here we show that Scl directly binds to enhancers regulating a broad network of hematopoietic and cardiac transcription factors to specify the hematopoietic lineage and override the cardiac fate and thus has a dual function in coordinating the segregation of multipotent cardiovascular mesoderm to downstream cell fates.
We discovered that Scl regulates mesodermal fates by binding to enhancers that have been primed for activation specifically in Flk1+ mesoderm. A previous study with DNA binding mutant of Scl showed that 80% of Scl peaks in erythroid cells were lost in the mutant and could neither be bound by Scl's interaction partner Gata1, raising the question whether Scl acts as a pioneer factor at these sites (Kassouf et al, 2010). Our finding that the establishment of the active histone modifications, H3K4me1 and H3K27ac, at the enhancers of Scl's hematopoietic and cardiac target genes occurs independent of Scl and its complex partners Gata1 and 2 implies that these factors do not act as a pioneer factors but rather exploit pre-established enhancers in mesoderm. These results suggest that yet another factor(s) is responsible for establishing primed enhancers that can attract Scl and Gata factors to their cell type-specific binding sites. Our data are consistent with a model that the acquisition of a primed, or provisionally activated, state at hematopoietic and cardiac enhancers precedes Scl and Gata1 and 2 binding and determines the available fate options in mesoderm. Our finding that overexpression of Scl in ES cells did not lead to Scl binding to hematopoietic or cardiac enhancers also supports this model.
Several recent studies have documented highly cell type-specific, functional Scl binding during later stages of hematopoietic development (Calero-Nieto et al, 2014; Pimkin et al, 2014; Wu et al, 2014). Our study documents the dynamic nature of Scl binding already in mesoderm. In contrast to the key hematopoietic enhancers that retained Scl binding and active epigenetic marks throughout hematopoietic development, Scl binding to cardiac enhancers was restricted to Flk1+ mesoderm and correlated with the transient presence of active histone marks. These data imply that Scl represses the cardiac fate during a brief window of developmental plasticity before mesodermal fates become solidified, providing a molecular basis for the inducible loss-of-function studies that narrowed down the temporal requirement for Scl function in cardiac repression to mesoderm/early angioblasts (Van Handel et al, 2012). Moreover, these data suggest enhancer decommissioning as a key mechanism that extinguishes the cardiac fate option in hematopoietic cells.
Our studies propose a model that Scl suppresses cardiogenesis by interfering with the activation of key cardiac genes until the ‘window of opportunity’ for their activation is closed, rather than directly recruiting corepressors. Analyses correlating Scl binding with major corepressors and repressive epigenetic marks (H3K27me3, H3K9me3 and DNA methylation) did not provide a mechanism by which transient Scl binding to these enhancers results in the repression of cardiac genes, but rather suggests that these repression mechanisms contribute to the overall gene silencing later, once the Scl-driven lineage choice decision has already been made.
Lsd1 emerged as a key candidate for modulating enhancer activity during mesoderm diversification as it decommissions cell type-specific enhancers by demethylating H3K4me1 in several developmental contexts including ES cells and myeloid cells (Whyte et al, 2012; Kerenyi et al, 2013), and interaction of Scl with Lsd1 has been implicated in erythroid progenitors and T-ALL (Hu et al, 2009a,b; Li et al, 2012). Our analysis verified a strong correlation between Lsd1 and Scl binding sites and H3K4me1 in mesoderm. However, Lsd1 binding to cardiac enhancers was not impaired in Scl-deficient cells. Thus, although Lsd1 is involved in decommissioning of unused enhancers during mesoderm diversification, its recruitment to cardiac enhancers is not Scl dependent. These data do not, however, exclude the possibility that the function of Lsd1, or the COREST complex Lsd1 is part of, may be influenced by Scl. As acetylation can impede Lsd1 function (Amente et al, 2013), it is plausible that Scl binding at hematopoietic enhancers reinforces a fully active state, possibly by chromosomal looping to promoter, and thereby indirectly impairs Lsd1 function. On the other hand, Scl binding at cardiac enhancers may interfere with chromosomal looping and/or the establishment of a proper cardiac activation complex, enabling Lsd1 to decommission cardiac enhancers. Future studies are required to directly test these hypotheses.
Our findings that there is substantial overlap with Scl and Hand1 and/or Gata4 mesodermal binding sites, and essentially all major hematopoietic and cardiac transcription factors that are regulated by Scl contain at least one enhancer that can also be bound by Hand1 and/or Gata4, support the model that repression of an alternative fate choice may be the result of preventing activation of these genes by competing lineage-specific regulators. The subset of Scl-bound enhancers that can also be bound by Hand1 and/or Gata4 showed higher evolutionary conservation, higher H3K4me1 and H3K27ac enrichment and Lsd1 binding in mesoderm, and stronger correlation with Scl-dependent gene expression change. We propose that these dually accessible enhancers that can be regulated by factors of two opposing lineages act as a platform where the fate choice is determined. Future studies will determine which cardiac factor(s) is most critical for activating these cardiac enhancers and whether they are also critical for suppressing the hematopoietic program.
We found that Gata2 can bind together with Scl to both hematopoietic and cardiac enhancers; however, we discovered that hematopoietic Gata factors (1, 2 and 3) are dispensable for Scl-mediated cardiac repression, and only necessary for recruiting Scl to specific binding sites to activate hematopoietic factors required for HS/PC emergence and differentiation (Runx1, Pu.1/Sfpi1, Klf1, Gfi1b). These data suggest that although the core Gata/Scl complex can bind to both activated and repressed genes, Gata/Scl interaction becomes critical principally in gene activation. Similar to hematopoietic Gata factors, Runx1 can redistribute Scl to new binding sites as development progresses from endothelium to HS/PCs (Lichtinger et al, 2012). Thus, Scl establishes the blood lineage by binding to a broad set of hematopoietic enhancers; it then induces downstream transcriptional network with the help of its target genes, which enable Scl to relocate to new binding sites to build a functional hematopoietic system.
The findings that Scl targets Gata1, Gata2 and Runx1 (Van Handel et al, 2012), which are critical for modulating Scl activity in hematopoietic genes, are not essential for cardiac repression underscore the unique role of Scl as a dual regulator of hematopoietic versus cardiac fate choice. Apart from the studies with Scl, ectopic cardiogenesis in hemato-vascular mesoderm has only been observed upon ablation of Etv2, the upstream regulator for Scl (Ren et al, 2010; Palencia-Desai et al, 2011; Rasmussen et al, 2011; Liu et al, 2012; Wareing et al, 2012). Future studies will be required to determine whether Etv2 has a specific function in silencing cardiac fate or whether it contributes indirectly by inducing Scl expression.
Our finding that Scl directs mesoderm diversification via enhancers is concordant with the data that transcription factor binding at enhancers is a key determinant of tissue-specific gene expression (Heintzman et al, 2009; Visel et al, 2009; Xu et al, 2012). Our discovery that Scl directly binds to enhancers of key transcription factors of two alternative fates suggests a mechanism by which lineage-specific transcription factors secure a mutually exclusive lineage choice by both activating their own lineage and preventing the establishment of alternative fates. Coordination of both processes by the same factor enables accurate fate specification and prevents the generation of a cell with mixed characteristics. As reprogramming of induced pluripotent stem cells is also initiated at enhancers (Soufi et al, 2012), understanding the repressive function of master regulators may have broader implications in regenerative medicine as the presence of a factor that could block lineage-specific enhancers during lineage reprogramming may influence the efficiency and outcome of reprogramming. Thus, better understanding of the pre-requisites for gene activation and repression by master regulators, and how the epigenetic boundaries are created between cell types, will help develop more effective protocols for directing cell fates for therapeutic applications.
Materials and Methods
ES cell culture and differentiation
Standard ES cell culture conditions were used to maintain SclhCD4 (Chung et al, 2002), SclKO (Porcher et al, 1996), SclKOiScl, WT, Gata1&2KO and Gata1&2KOGata3KD ES cells. SclKOiScl cell line was generated by transduction of SclKO ES cells with rtTA and PNL1-Scl-GFP lentiviral vectors followed by clonal selection. Gata1&2 KOGata3KD ES cells were generated by transducing Gata1&2KO ES cells with Gata3 shRNA lentiviral vector (Santa Cruz). Flk1+ mesoderm and hemato-vascular derivatives were generated using EB differentiation as described (Mikkola et al, 2003). EBs were digested enzymatically and analyzed by FACS or enriched for desired populations by MACS or FACS sorting. For further details about cell culture, see Supplementary Materials and Methods.
Gene expression analysis
RNA preparation, hybridization to Mouse Genome 430 2.0 arrays, and analysis was conducted as described previously (Van Handel et al, 2012). Libraries for RNA sequencing were constructed using Encore Complete RNA-Seq DR Multiplex System 1-8 (Nugen). Libraries were sequenced using HIseq-2000 (Illumina). Reads were mapped to mouse genome (mm9) using TopHat v2.0.4 (Trapnell et al, 2009). Abundance estimations (FPKMs) were performed with Cufflinks v2.0.1 (Trapnell et al, 2010). Assemblies for all samples were merged using Cuffmerge, and differential expression (P < 0.01) was determined using Cuffdiff. Quantitative reverse-transcriptase polymerase chain reaction (qRT–PCR) was performed with LightCycler 480 SYBR Green I Master (Roche) using LightCycler 480 (Roche). Primer sequences are listed in Supplementary Table S4. For further details, see Supplementary Materials and Methods.
Chromatin immunoprecipitation sequencing analysis
ChIP with ES, Flk1+ mesodermal (MES), HPC7, MEL or HL1 cells were performed as described previously (Ferrari et al, 2012; Kerenyi et al, 2013). Libraries were generated using Nugen Ovation ultralow kit and sequenced using HIseq-2000 (Illumina). Mapping was performed using Bowtie (Langmead et al, 2009) as described (Ferrari et al, 2012). Peak identification was performed with MACS v1.3.7.1 (Zhang et al, 2008). For published ChIP-seq datasets used and more detailed methods, see Supplementary Materials and Methods.
Reduced representation bisulfite sequencing
RRBS libraries were generated from genomic DNA of mouse ES cells, WT and SclKO Flk1+ mesoderm (MES) and MEL cells as described previously (Meissner et al, 2005) with minor modifications. RRBS data were aligned with BS-Seeker2 (Guo et al, 2013). Differentially methylated cytosines were calculated with methylKit (Akalin et al, 2012). For detailed methods and the published genome-wide methylation datasets used in this study, see Supplementary Materials and Methods.
Analysis of endothelial differentiation potential
After 4.75 days of EB induction, CD41−CD31+Tie2+cells were sorted from WT, SclKO, SclKOiScl, Gata1&2KO and Gata1&2KO Gata3KD EBs and 20,000 cells were plated on irradiated OP9 stroma in 8-well chamber slides (354118 BD Falcon™). 1 mg/ml doxycycline (1:1,000) is added to SclKOiScl culture and is kept on since day 2 of EB induction. The media contained α-MEM, 20% FBS, 1% penicillin, streptomycin and 1% glutamine. For hematopoietic differentiation, 50 ng/ml SCF, 5 ng/ml IL3, 5 ng/ml IL6, 5 ng/ml TPO and 10 ng/ml Flt3L were added. For cardiac differentiation, 5 ng/ml hVEGF, 30 ng/ml mFGF, 50 ng/ml hBMP4 and 1 μM Wnt/Beta-Catenin Inhibitor XAV939 (Sigma) were added. After 14 days, FACS staining or immunostaining was performed as described (Van Handel et al, 2012). See Supplementary Materials and Methods for details.
Accession numbers
Microarray, ChIP-seq, RNA-seq and RRBS datasets have been deposited to the NCBI GEO database under the accession number GSE47085. All datasets used in this study are listed in Supplementary Table S5.
Acknowledgments
The authors thank the Broad Stem Cell Research Center (BSCRC) Flow Cytometry Core for FACS sorting and BSCRC Sequencing core for next-generation sequencing. This work was funded by the California Institute for Regenerative Medicine (CIRM) New Faculty Award (RN1-00557), the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at UCLA Research Award, American Heart Association (#14GRNT20480340), and Leukemia Lymphoma Society Scholar Award to HKAM (20103778). TO was supported by Leukemia Lymphoma Society postdoctoral fellowship (57537-13) and by the European Union through the European Social Fund (Mobilitas Grant No. MJD284). DD was supported by the government of P.R.C through the State Scholarship Fund (File No. 2011624028). AMH was supported by a fellowship from HFSP and American Society of Hematology. BVH was supported by the NIH/NHLBI T32 HL69766. Contributions of YF and SHO were supported by P30DK049216.
Author contributions
TO and DD were involved in project design, experimental work, data analysis and interpretation and manuscript preparation. RF and RS participated in data analysis and interpretation, and manuscript editing. AM-H, BVH and MAK were involved in experimental work and manuscript editing. LR was involved in experimental work. YF and SHO provided essential reagents and edited the manuscript. MP and SKK interpreted the data and edited the manuscript. HKAM was involved in project design, data analysis and interpretation, and manuscript preparation.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Supplementary Table S5
Supplementary Figure Legends
Supplementary Materials and Methods
Review Process File
References
- Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amente S, Lania L, Majello B. The histone LSD1 demethylase in stemness and cancer transcription programs. Biochim Biophys Acta. 2013;1829:981–986. doi: 10.1016/j.bbagrm.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DYR, Traver D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464:108–111. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattler A, Farnham PJ. Cross-talk between site-specific transcription factors and DNA methylation states. J Biol Chem. 2013;288:34287–34294. doi: 10.1074/jbc.R113.512517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Bristow J, Ren B, Black BL, Rubin EM, Visel A, Pennacchio LA. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset J-C, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- Bresnick EH, Katsumura KR, Lee H-Y, Johnson KD, Perkins AS. Master regulatory GATA transcription factors: mechanistic principles and emerging links to hematologic malignancies. Nucleic Acids Res. 2012;40:5819–5831. doi: 10.1093/nar/gks281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero-Nieto FJ, Ng FS, Wilson NK, Hannah R, Moignard V, Leal-Cervantes AI, Jimenez-Madrid I, Diamanti E, Wernisch L, Göttgens B. Key regulators control distinct transcriptional programmes in blood progenitor and mast cells. EMBO J. 2014;33:1212–1226. doi: 10.1002/embj.201386825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlon TM, Crispino JD. Combinatorial regulation of tissue specification by GATA and FOG factors. Development. 2012;139:3905–3916. doi: 10.1242/dev.080440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung YS, Zhang WJ, Arentson E, Kingsley PD, Palis J, Choi K. Lineage analysis of the hemangioblast as defined by FLK1 and SCL expression. Development. 2002;129:5511–5520. doi: 10.1242/dev.00149. [DOI] [PubMed] [Google Scholar]
- Creemers EE, Sutherland LB, McAnally J, Richardson JA, Olson EN. Myocardin is a direct transcriptional target of Mef2, Tead and Foxo proteins during cardiovascular development. Development. 2006;133:4245–4256. doi: 10.1242/dev.02610. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh T-Y, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bruijn MF, Speck NA, Peeters MC, Dzierzak E. Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J. 2000;19:2465–2474. doi: 10.1093/emboj/19.11.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pater E, Kaimakis P, Vink CS, Yokomizo T, Yamada-Inagawa T, van der Linden R, Kartalaei PS, Camper SA, Speck N, Dzierzak E. Gata2 is required for HSC generation and survival. J Exp Med. 2013;210:2843–2850. doi: 10.1084/jem.20130751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehling HJ, Lacaud G, Kubo A, Kennedy M, Robertson S, Keller G, Kouskoff V. Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development. 2003;130:4217–4227. doi: 10.1242/dev.00589. [DOI] [PubMed] [Google Scholar]
- Ferrari R, Su T, Li B, Bonora G, Oberai A, Chan Y, Sasidharan R, Berk AJ, Pellegrini M, Kurdistani SK. Reorganization of the host epigenome by a viral oncogene. Genome Res. 2012;22:1212–1221. doi: 10.1101/gr.132308.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y, Chang AN, Williams AM, Orkin SH. Functional overlap of GATA-1 and GATA-2 in primitive hematopoietic development. Blood. 2004;103:583–585. doi: 10.1182/blood-2003-08-2870. [DOI] [PubMed] [Google Scholar]
- Gao X, Johnson KD, Chang Y-I, Boyer ME, Dewey CN, Zhang J, Bresnick EH. Gata2 cis-element is required for hematopoietic stem cell generation in the mammalian embryo. J Exp Med. 2013;210:2833–2842. doi: 10.1084/jem.20130733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Fiziev P, Yan W, Cokus S, Sun X, Zhang MQ, Chen P-Y, Pellegrini M. BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genom. 2013;14:774. doi: 10.1186/1471-2164-14-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi E, Brinkman AB, Arand J, Kroeze LI, Kerstens HHD, Matarese F, Lepikhov K, Gut M, Brun-Heath I, Hubner NC, Benedetti R, Altucci L, Jansen JH, Walter J, Gut IG, Marks H, Stunnenberg HG. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell. 2013;13:360–369. doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Harmston N, Lenhard B. Chromatin and epigenetic features of long-range gene regulation. Nucleic Acids Res. 2013;41:7185–7199. doi: 10.1093/nar/gkt499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A, Gu F, Hu Y, Ma Q, Yi Ye L, Akiyama JA, Visel A, Pennacchio LA, Pu WT. Dynamic GATA4 enhancers shape the chromatin landscape central to heart development and disease. Nat Commun. 2014;5:4907. doi: 10.1038/ncomms5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzinger A, Rosenfeld GE, Evans T. Gata4 directs development of cardiac-inducing endoderm from ES cells. Dev Biol. 2010;337:63–73. doi: 10.1016/j.ydbio.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon GC, Rajagopal N, Shen Y, McCleary DF, Yue F, Dang MD, Ren B. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet. 2013;45:1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya T, Maillard I, Engel JD. From the cradle to the grave: activities of GATA-3 throughout T-cell development and differentiation. Immunol Rev. 2010;238:110–125. doi: 10.1111/j.1600-065X.2010.00954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Li X, Valverde K, Fu X, Noguchi C, Qiu Y, Huang S. LSD1-mediated epigenetic modification is required for TAL1 function and hematopoiesis. Proc Natl Acad Sci USA. 2009a;106:10141–10146. doi: 10.1073/pnas.0900437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Ybarra R, Qiu Y, Bungert J, Huang S. Transcriptional regulation by TAL1: a link between epigenetic modifications and erythropoiesis. Epigenetics. 2009b;4:357–361. doi: 10.4161/epi.4.6.9711. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- Huber TL, Kouskoff V, Fehling HJ, Palis J, Keller G. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature. 2004;432:625–630. doi: 10.1038/nature03122. [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu J-D, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida M, Heike T, Yoshimoto M, Baba S, Doi H, Nakahata T. T Identification of cardiac stem cells with FLK1, CD31, and VE-cadherin expression during embryonic stem cell differentiation. FASEB J. 2005;19:371–378. doi: 10.1096/fj.04-1998com. [DOI] [PubMed] [Google Scholar]
- Ismailoglu I, Yeamans G, Daley GQ, Perlingeiro RCR, Kyba M. Mesodermal patterning activity of SCL. Exp Hematol. 2008;36:1593–1603. doi: 10.1016/j.exphem.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Kassouf MT, Hughes JR, Taylor S, McGowan SJ, Soneji S, Green AL, Vyas P, Porcher C. Genome-wide identification of TAL1's functional targets: insights into its mechanisms of action in primary erythroid cells. Genome Res. 2010;20:1064–1083. doi: 10.1101/gr.104935.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka H, Hayashi M, Nakagawa R, Tanaka Y, Izumi N, Nishikawa S, Jakt ML, Tarui H, Nishikawa S-I. Etv2/ER71 induces vascular mesoderm from Flk1+PDGFRα+ primitive mesoderm. Blood. 2011;118:6975–6986. doi: 10.1182/blood-2011-05-352658. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Kerenyi MA, Shao Z, Hsu Y-J, Guo G, Luc S, O'Brien K, Fujiwara Y, Peng C, Nguyen M, Orkin SH. Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation. Elife. 2013;2:e00633. doi: 10.7554/eLife.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Efe JA, Zhu S, Talantova M, Yuan X, Wang S, Lipton SA, Zhang K, Ding S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci USA. 2011;108:7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 2010;464:112–115. doi: 10.1038/nature08761. [DOI] [PubMed] [Google Scholar]
- Kowalczyk MS, Hughes JR, Garrick D, Lynch MD, Sharpe JA, Sloane-Stanley JA, McGowan SJ, De Gobbi M, Hosseini M, Vernimmen D, Brown JM, Gray NE, Collavin L, Gibbons RJ, Flint J, Taylor S, Buckle VJ, Milne TA, Wood WG, Higgs DR. Intragenic enhancers act as alternative promoters. Mol Cell. 2012;45:447–458. doi: 10.1016/j.molcel.2011.12.021. [DOI] [PubMed] [Google Scholar]
- Ku C-J, Hosoya T, Maillard I, Engel JD. GATA-3 regulates hematopoietic stem cell maintenance and cell-cycle entry. Blood. 2012;119:2242–2251. doi: 10.1182/blood-2011-07-366070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Park C, Lee H, Lugus JJ, Kim SH, Arentson E, Chung YS, Gomez G, Kyba M, Lin S, Janknecht R, Lim D-S, Choi K. ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell. 2008;2:497–507. doi: 10.1016/j.stem.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ferkowicz MJ, Johnson SA, Shelley WC, Yoder MC. Endothelial cells in the early murine yolk sac give rise to CD41-expressing hematopoietic cells. Stem Cells Dev. 2005;14:44–54. doi: 10.1089/scd.2005.14.44. [DOI] [PubMed] [Google Scholar]
- Li Y, Deng C, Hu X, Patel B, Fu X, Qiu Y, Brand M, Zhao K, Huang S. Dynamic interaction between TAL1 oncoprotein and LSD1 regulates TAL1 function in hematopoiesis and leukemogenesis. Oncogene. 2012;31:5007–5018. doi: 10.1038/onc.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtinger M, Ingram R, Hannah R, Müller D, Clarke D, Assi SA, Lie-A-Ling M, Noailles L, Vijayabaskar MS, Wu M, Tenen DG, Westhead DR, Kouskoff V, Lacaud G, Göttgens B, Bonifer C. RUNX1 reshapes the epigenetic landscape at the onset of haematopoiesis. EMBO J. 2012;31:4318–4333. doi: 10.1038/emboj.2012.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien CL, Wu C, Mercer B, Webb R, Richardson JA, Olson EN. Control of early cardiac-specific transcription of Nk2–5 by a GATA-dependent enhancer. Development. 1999;126:75–84. doi: 10.1242/dev.126.1.75. [DOI] [PubMed] [Google Scholar]
- Liu F, Kang I, Park C, Chang L-W, Wang W, Lee D, Lim D-S, Vittet D, Nerbonne JM, Choi K. ER71 specifies Flk-1+ hemangiogenic mesoderm by inhibiting cardiac mesoderm and Wnt signaling. Blood. 2012;119:3295–3305. doi: 10.1182/blood-2012-01-403766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola HKA, Fujiwara Y, Schlaeger TM, Traver D, Orkin SH. Expression of CD41 marks the initiation of definitive hematopoiesis in the mouse embryo. Blood. 2003;101:508–516. doi: 10.1182/blood-2002-06-1699. [DOI] [PubMed] [Google Scholar]
- North T, Gu TL, Stacy T, Wang Q, Howard L, Binder M, Marín-Padilla M, Speck NA. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- Nottingham WT, Jarratt A, Burgess M, Speck CL, Cheng J-F, Prabhakar S, Rubin EM, Li P-S, Sloane-Stanley J, Kong-A-San J, de Bruijn MFTR. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood. 2007;110:4188–4197. doi: 10.1182/blood-2007-07-100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda M, Kumaki Y, Shigeta M, Jakt LM, Matsuoka C, Yamagiwa A, Niwa H, Okano M. DNA methylation restricts lineage-specific functions of transcription factor Gata4 during embryonic stem cell differentiation. PLoS Genet. 2013;9:e1003574. doi: 10.1371/journal.pgen.1003574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palencia-Desai S, Kohli V, Kang J, Chi NC, Black BL, Sumanas S. Vascular endothelial and endocardial progenitors differentiate as cardiomyocytes in the absence of Etsrp/Etv2 function. Development. 2011;138:4721–4732. doi: 10.1242/dev.064998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira C-F, Chang B, Qiu J, Niu X, Papatsenko D, Hendry CE, Clark NR, Nomura-Kitabayashi A, Kovacic JC, Ma'ayan A, Schaniel C, Lemischka IR, Moore K. Induction of a hemogenic program in mouse fibroblasts. Cell Stem Cell. 2013;13:205–218. doi: 10.1016/j.stem.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimkin M, Kossenkov A V, Mishra T, Morrissey CS, Wu W, Keller CA, Blobel GA, Lee D, Beer MA, Hardison RC, Weiss MJ. Divergent functions of hematopoietic transcription factors in lineage priming and differentiation during erythro-megakaryopoiesis. Genome Res. 2014;24:1932–1944. doi: 10.1101/gr.164178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinello L, Xu J, Orkin SH, Yuan G-C. Analysis of chromatin-state plasticity identifies cell-type-specific regulators of H3K27me3 patterns. Proc Natl Acad Sci USA. 2014;111:E344–E353. doi: 10.1073/pnas.1322570111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt FW, Orkin SH. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- Rasmussen TL, Kweon J, Diekmann MA, Belema-Bedada F, Song Q, Bowlin K, Shi X, Ferdous A, Li T, Kyba M, Metzger JM, Koyano-Nakagawa N, Garry DJ. ER71 directs mesodermal fate decisions during embryogenesis. Development. 2011;138:4801–4812. doi: 10.1242/dev.070912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Gomez GA, Zhang B, Lin S. Scl isoforms act downstream of etsrp to specify angioblasts and definitive hematopoietic stem cells. Blood. 2010;115:5338–5346. doi: 10.1182/blood-2009-09-244640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes KE, Gekas C, Wang Y, Lux CT, Francis CS, Chan DN, Conway S, Orkin SH, Yoder MC, Mikkola HKA. The emergence of hematopoietic stem cells is initiated in the placental vasculature in the absence of circulation. Cell Stem Cell. 2008;2:252–263. doi: 10.1016/j.stem.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenebeck JJ, Keegan BR, Yelon D. Vessel and blood specification override cardiac potential in anterior mesoderm. Dev Cell. 2007;13:254–267. doi: 10.1016/j.devcel.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumanas S, Gomez G, Zhao Y, Park C, Choi K, Lin S. Interplay among Etsrp/ER71, Scl, and Alk8 signaling controls endothelial and myeloid cell formation. Blood. 2008;111:4500–4510. doi: 10.1182/blood-2007-09-110569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Shimizu R, Suwabe N, Kuroha T, Yoh K, Ohta J, Nishimura S, Lim KC, Engel JD, Yamamoto M. GATA factor transgenes under GATA-1 locus control rescue germline GATA-1 mutant deficiencies. Blood. 2000;96:910–916. [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripic T, Deng W, Cheng Y, Zhang Y, Vakoc CR, Gregory GD, Hardison RC, Blobel GA. SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood. 2009;113:2191–2201. doi: 10.1182/blood-2008-07-169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Handel B, Montel-Hagen A, Sasidharan R, Nakano H, Ferrari R, Boogerd CJ, Schredelseker J, Wang Y, Hunter S, Org T, Zhou J, Li X, Pellegrini M, Chen J-N, Orkin SH, Kurdistani SK, Evans SM, Nakano A, Mikkola HKA. Scl represses cardiomyogenesis in prospective hemogenic endothelium and endocardium. Cell. 2012;150:590–605. doi: 10.1016/j.cell.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA enhancer browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman IA, Osada H, Grütz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, Erwin G, Kattman SJ, Keller GM, Srivastava D, Levine SS, Pollard KS, Holloway AK, Boyer LA, Bruneau BG. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wareing S, Mazan A, Pearson S, Göttgens B, Lacaud G, Kouskoff V. The Flk1-Cre-mediated deletion of ETV2 defines its narrow temporal requirement during embryonic hematopoietic development. Stem Cells. 2012;30:1521–1531. doi: 10.1002/stem.1115. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schütte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, Pimanda JE, de Bruijn MFTR, Göttgens B. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Wozniak RJ, Boyer ME, Grass JA, Lee Y, Bresnick EH. Context-dependent GATA factor function: combinatorial requirements for transcriptional control in hematopoietic and endothelial cells. J Biol Chem. 2007;282:14665–14674. doi: 10.1074/jbc.M700792200. [DOI] [PubMed] [Google Scholar]
- Wu W, Morrissey CS, Keller CA, Mishra T, Pimkin M, Blobel GA, Weiss MJ, Hardison RC. Dynamic shifts in occupancy by TAL1 are guided by GATA factors and drive large-scale reprogramming of gene expression during hematopoiesis. Genome Res. 2014;24:1945–1962. doi: 10.1101/gr.164830.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Shao Z, Glass K, Bauer DE, Pinello L, Van Handel B, Hou S, Stamatoyannopoulos JA, Mikkola HKA, Yuan G-C, Orkin SH. Combinatorial assembly of developmental stage-specific enhancers controls gene expression programs during human erythropoiesis. Dev Cell. 2012;23:796–811. doi: 10.1016/j.devcel.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto M, Montecino-Rodriguez E, Ferkowicz MJ, Porayette P, Shelley WC, Conway SJ, Dorshkind K, Yoder MC. Embryonic day 9 yolk sac and intra-embryonic hemogenic endothelium independently generate a B-1 and marginal zone progenitor lacking B-2 potential. Proc Natl Acad Sci USA. 2011;108:1468–1473. doi: 10.1073/pnas.1015841108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, Bernstein BE, Fraenkel E, Cantor AB. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell. 2009;36:682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovein AC, Hofmann JJ, Lynch M, French WJ, Turlo KA, Yang Y, Becker MS, Zanetta L, Dejana E, Gasson JC, Tallquist MD, Iruela-Arispe ML. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3:625–636. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Supplementary Table S5
Supplementary Figure Legends
Supplementary Materials and Methods
Review Process File