SUMMARY
The C-C chemokine receptor type 5 (CCR5) is a receptor expressed by T-cells and macrophages that serves as a co-receptor for macrophage-tropic HIV-1. Loss of CCR5 is associated with resistance to HIV-1. Here we combine the live cell-based SELEX with high throughput sequencing technology to generate CCR5 RNA aptamers capable of specifically targeting HIV-1 susceptible cells (as siRNA delivery agent) and inhibiting HIV-1 infectivity (as antiviral agent) via block of the CCR5 required for HIV-1 to enter cells. One of the best candidates, G-3, efficiently bound and was internalized into human CCR5 expressing cells. The G-3 specifically neutralized R5 virus infection in primary peripheral blood mononuclear cells, and in vivo generated human CD4+ T cells with a nanomolar IC50. G-3 was also capable of transferring functional siRNAs to CCR5 expressing cells. Collectively, the cell-specific, internalizing, CCR5-targeted aptamers and aptamer-siRNA conjugates offer promise for overcoming some of the current challenges of drug resistance in HIV-1 by providing cell-type- or tissue-specific delivery of various therapeutic moieties.
INTRODUCTION
Nucleic acid-based therapeutics are quickly emerging as a strong alternative or co-therapy to the chemical antiviral agents currently used to treat HIV-1/AIDS. The combinatorial use of various antiviral nucleic acids could be more efficacious in blocking viral replication and preventing the emergence of resistant HIV-1 variants (Joshi, et al., 2003, Scherer, et al., 2007). Additionally, highly specific nucleic acid-based aptamers and aptamer-functionalized agents have been used extensively for targeted diseases therapy (Li, et al., 2013,Nimjee, et al., 2005,Sundaram, et al., 2013,Thiel and Giangrande, 2009,Zhang, et al., 2004). These aptamers often have favorable characteristics, such as: small size, high stability (dehydrated form), lack of immunogenicity, facile chemical synthesis, adaptable modification and cell-free evolution. To date, many nucleic acid aptamers have been shown to be specific to various regions of the HIV-1 genome and to HIV-1 dependent proteins, including: HIV-1 reverse transcriptase (RT), integrase (IN), nucleocapsid (NC), group-specific antigen (Gag), trans-activation response element (TAR), Regulator of Expression of Virion Proteins (Rev), Trans-Activator of Transcription (Tat), envelope glycoprotein 120 (gp120) and cluster of differentiation 4 (CD4) protein (Shum, et al., 2013). These aptamers have been raised through purified protein-based SELEX (Systematic Evolution of Ligand EXponential enrichment) method and shown to effectively suppress viral replication (Held, et al., 2006, Shum, et al., 2013, Zhang, et al., 2004). Importantly, a number of cell-specific aptamers targeting cell surface proteins have been adapted as promising delivery vehicles for the targeted cell-type specific delivery of small interfering RNA (siRNA) (Mallikaratchy, et al., 2009, Zhou and Rossi, 2011). Moreover, the combined use of siRNAs and aptamers could effectively block viral replication and prevent the emergence of resistant variants (Zhou and Rossi, 2012).
In our previous efforts, anti-HIV gp120 aptamers were combined with anti-HIV siRNAs to achieve a dual-inhibitory drug capable of delivering siRNAs selectively to HIV-infected cells as well as inhibiting viral entry via blocking of the envelope interaction with the CD4 (Neff, et al., 2011,Zhou, et al., 2013). Human CCR5 (C-C chemokine receptor type 5), a protein expressed by T-cells and macrophages, is an important co-receptor for macrophage-tropic virus, including HIV-1 R5 isolates (Berger, et al., 1999,Pelchen-Matthews, et al., 1999). Variations in CCR5 are associated with resistance or susceptibility to HIV-1. As an essential factor for viral entry, CCR5 has represented an attractive cellular target for the treatment of HIV-1 (Meanwell and Kadow, 2003,Ugolini, et al., 1999). We therefore sought to develop CCR5 directed RNA aptamers to target HIV-1 susceptible cells, and specifically regulate both gene silencing of HIV-1 and the blockage of CCR5 which is required for HIV-1 to enter cells.
By combining the “live cell-based SELEX” strategy (Cerchia, et al., 2009,Fang and Tan, 2009) (Figure 1A) with high throughput sequencing (HTS) and bioinformatics analysis, we have successfully identified several 2′-Fluoropyrimidine modified RNA aptamers directed to human CCR5. One of the best candidates (G-3) efficiently bound to CCR5 and was internalized into human CCR5 expressing Magi-U373-CCR5E cells, CEM-NKr-CCR5 cells and primary PBMCs. The G-3 aptamer specifically neutralized R5-tropic virus infection in primary PBMCs and in vivo generated human CD4+ T cells with about 50~350 nM of IC50. Moreover, the G-3 aptamer was capable of delivering functional anti-HIV siRNAs to CCR5 expressing cells in a receptor-targeted manner, thereby resulting in a dual inhibitory effect on HIV-1 replication. Collectively, we describe the derivation and mechanistic characterization of new CCR5 targeted aptamers, which may prove useful in several applications, including use as a novel antiviral therapy.
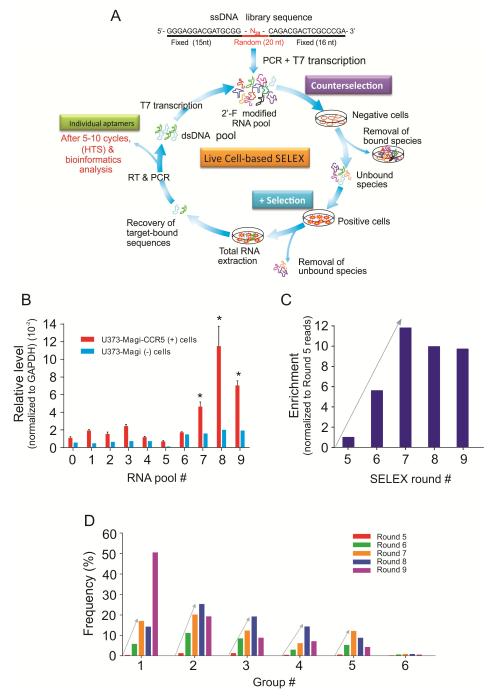
Figure 1. Live cell-based SELEX and bioinformatics analysis of high throughput sequence data from selection rounds.
A) Schematic of live cell-based SELEX procedure for evolution of RNA aptamers. It consists of four main steps: 1) counter selection by incubating library with negative cells that do not express the target protein; 2) positive selection by incubating recovered unbound sequences with positive cells expressing the target protein; 3) recovery of target-bound sequences; and finally 4) re-amplification of recovered species and make new RNA pool for next selection round. Individual aptamer sequences are identified through barcode-based high throughput, Illumina Deep Sequencing (HTS) and bioinformatics analysis. B) The progress of SELEX. Nine rounds of live cell-based SELEX were performed to enrich for RNA aptamers. Cell-type specific binding/internalization of ten RNA pools was evaluated by qRT-PCR. Error bars represent standard deviation (SD). C) The molecular enrichment at each round. The molecular enrichment at each round was calculated by the formula (total reads of top 1000 sequences in round X / round 5). D) The frequency of each group at each selection round. After alignment of top 40 sequences, six groups of aptamers were identified. The percent frequency of each group at each selection round was calculated by the formula (the reads of each group / the useful reads at each round).
RESULTS
Live cell-based SELEX of CCR5 aptamers
RNA aptamers, 2′-Fluoropyrimidine modified, were selected using the live cell-based SELEX strategy (Cerchia, et al., 2005,Cerchia, et al., 2009,Thiel, et al., 2012,Thiel, et al., 2012) (Figure 1A). Cellular surface CCR5 expression in target U373-Magi-CCR5E cells was first verified using flow cytometric analysis (Supplementary Figure S1) and a counter-selection step with U373-Magi cells (CCR5 negative). The detailed selection conditions are summarized in Figure 1A and Supplementary Table S1. Nine selection rounds containing both positive and negative selection were performed with cell-specific binding and internalization of the RNA pools monitored using quantitative RT-PCR (qRTPCR). A significant enrichment was observed between the 7th (7-RNA pool) to the 9th round RNA pool (9-RNA pool) (Figure 1B). No further increase could be detected after the 8th selection round, suggesting that maximal binding/internalization of the RNA pool may have been reached.
Identification of CCR5 aptamers using high-throughput sequencing (HTS)
By using barcode based-Illumina deep sequencing technology, high throughput sequencing (HTS) was performed for the RNA pools from selection round 5 to 9 with approximate 30~50 million total reads obtained from each sequenced round (Table 1A). After removal of constant region and sequencing adapter sequences, the raw reads were filtered based on the length of RNA library random region sequence, and the most frequent 1,000 unique sequences identified (Table 1A and Supplementary Figure S2A). A significant increase in the frequencies of the top 1,000 unique sequences was observed after the 7th round of selection, suggesting a decrease in library sequence diversity and an increase in library sequence enrichment at this step. Supporting this notion are the observations that after round 7 the molecular diversity was dramatically converged, thereby suggesting some specific sequences have been successfully enriched during the selection (Figure 1C). These observations are consistent with SELEX progress (Figure 1B).
Table 1. Bioinformatics analysis of high throughput sequence data from selection rounds.
A) The total reads and useful reads were defined as follows. The most frequent 1,000 unique sequences were identified and listed for the clarity. The molecular enrichment at each round was calculated by the formula (total reads of top 1000 unique sequences at round X / round 5). B) Bioinformatics analysis of RNA aptamers to identify related sequence and structure groups. After alignment of top 40 sequences, six groups of RNA aptamers were identified. The representative RNA aptamers and the reads of each group are listed here. Only the random sequences of the aptamer core regions (5′-3′) are indicated.
| A) | |||||
|---|---|---|---|---|---|
| Round 5 | Round 6 | Round 7 | Round 8 | Round 9 | |
| Total Reads | 48,175,338 | 37,973,185 | 36,503,548 | 27,876,693 | 30,723,210 |
| Useful Reads | 35,003,576 | 24,043,982 | 25,650,155 | 17,994,328 | 16,273,964 |
| Total Reads of top 1000 unique sequences | 1,553,154 | 8,877,699 | 18,721,730 | 15,831,299 | 15,473,124 |
| Molecular Enrichment (fold) | 1.00 | 5.72 | 12.05 | 10.19 | 9.96 |
| B) | |||||||
|---|---|---|---|---|---|---|---|
| Groups | RNA | 20-nt random sequence | Reads of each group | ||||
| Round 5 | Round 6 | Round 7 | Round 8 | Round 9 | |||
| 1 | G-1 | AUCGUCUAUUAGUCGCUGGC | 144,560 | 1,405,945 | 4,385,441 | 2,572,509 | 8,228,896 |
| 2 | G-2 | UCCUUGGCUUUUCGUCUGUG | 447,271 | 2,700,796 | 5,191,611 | 4,572,814 | 3,151,265 |
| 3 | G-3 | GCCUUCGUUUGUUUCGUCCA | 409,975 | 2,033,703 | 3,150,589 | 3,452,540 | 1,427,892 |
| 4 | G-4 | UCCCGGCUCGUUCGUCUGUG | 129,752 | 695,110 | 1,564,701 | 2,568,638 | 1,148,903 |
| 5 | G-5 | UUCGUCAU UUUUCGUCUGGG | 208,652 | 1,282,740 | 3,133,297 | 1,591,731 | 701,763 |
| 6 | G-6 | CCUUUCGUCUGUUUCUGCGC | 69,760 | 158,855 | 233,840 | 159,454 | 94,378 |
| Others | Orphan sequences | 20,151 | 42,315 | 0 | 0 | 0 | |
| Total Reads of all groups | 1,430,121 | 8,319,464 | 17,659,479 | 14,917,686 | 14,753,097 | ||
The distribution of each nucleotide (A, T, C and G) at the 20-nt random region within the RNA sequence of the top 1000 candidates from each round was identified (Supplementary Figure S2B). The individual sequences were classified into six major groups (Group 1-6) based on the alignments of the top 1000 unique aptamer sequences (Table 1B). One representative sequence from each group (G-1, G-2, G-3, G-4, G-5 and G-6) was listed for further characterization because of their relative abundance within their group. Theoretical secondary structures were predicted by using RNA folding algorithm Mfold (Supplementary Figure S2C). Group 2, 4 and 5 shared a conserved sequence, which is comprised of 10 nucleotides UUCGUCUG(U/G)G (Supplementary Figure S2C). Furthermore, we calculated the percent frequency to determine the evolution of each sequence group from the round 5 to 9 (Figure 1D). A progressive evolution of all the groups was observed from the round 5 to round 7, where the maximal selection convergence or binding/internalization of the RNA pool has been achieved (Table 1 and Figure 1B). Importantly, a significant increase of group 1 sequence observed at round 9 compromised a population of other groups, implying further selection may adversely affect the enrichment of the candidate aptamers.
CCR5 RNA aptamers bind to cell-surface CCR5 and are internalized into CCR5-expressing cells
To evaluate the binding affinity and internalization potential of these individual RNA aptamers, one representative sequence from each group (G-1, G-2, G-3, G-4, G-5 and G-6) was synthesized for further characterization. U373-Magi-CCR5E cells stably expressing the CCR5 protein and its parental U373-Magi (CCR5 negative) control cells were used to test for binding and internalization of the candidate aptamers. Cell-type specific binding and internalization was firstly detected by using quantitative RT-PCR (qRT-PCR). These RNA aptamers were able to selectively bind and internalize into U373-Magi-CCR5E cells (Figure 2A). Next, these RNA aptamers were labeled with Cy3 dye and their binding and cellular uptake was determined. Flow cytometric analysis revealed that the aptamers specifically bound to the U373-Magi CCR5E cells but did not bind to the control U373-Magi cells (CCR5 negative) (Figure 2B and Supplementary Figure S3A, B).
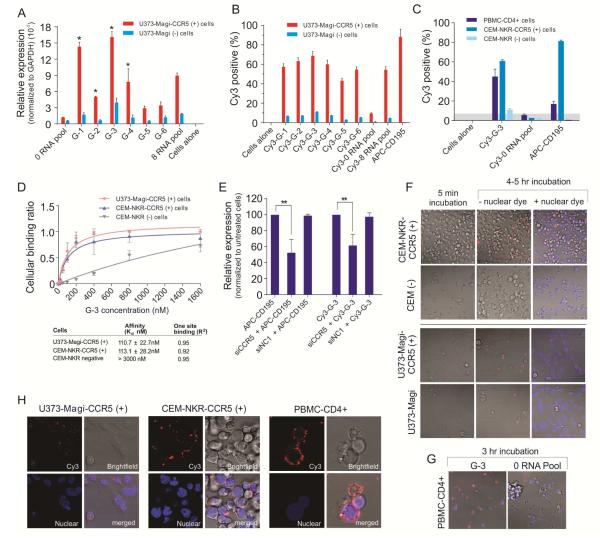
Figure 2. Cell-type specific binding and internalization studies of individual RNA aptamers.
A) Cell-type specific binding/internalization of individual aptamer was evaluated by qRT-PCR. The 0-RNA pool was used as negative control. Data represent the average of three replicates. Cell surface binding of Cy3-labeled RNAs was assessed by flow cytometry. B) Cy3-labeled RNAs were tested for binding to U373-Magi-CCR5 positive cells and U373-Magi negative cells. APC-CD195 antibody was used to stain cellular surface CCR5. C) G-3 aptamer was chosen for further binding affinity test with PBMC-CD4+ cells, CEM-NKr-CCR5 positive cells, CEM negative cells. D) Cell surface binding constant (Kd) of G-3 was evaluated by flow cytometry. E) Knockdown of CCR5 reduced binding affinity of aptamers. A scrambled siRNA (NC1) was used as negative control. Asterisk indicates a significant difference compared with control (P < 0.01, student’s t-test). Data represent the average of three replicates. Internalization analysis. F) U373-Magi-CCR5 positive cells, U373-Magi negative cells, CEM-NKr-CCR5 positive cells, CEM negative cells, or G) PBMC-CD4+ cells were grown in 35 mm plates treated with polylysine and incubated with a 67 nM concentration of Cy3-labeled G-3 aptamer in complete culture media for real-time live-cell confocal microscopy analysis. The images were collected using 40×magnification. H) Localization analysis.
To determine the ability of the candidate aptamers to selectively bind to different cells expressing human CCR5, the aptamers were applied to human T-lymphoblastoid cell line (CEM-NKr-CCR5 cell) and primary peripheral blood mononuclear cells (PBMCs) isolated from different donors. The top candidate aptamer, G-3, was selected for further analysis as it was able to bind CCR5 in the various CCR5 expressing cells. Moreover, this aptamer was refractory to binding the non-CCR5 expressing control cells (Figure 2C and Supplementary Figure S3C, D). The cell surface-binding constant (Kd) of the G-3 aptamer was evaluated by flow cytometry. G-3 demonstrated good binding affinity to CCR5 expressing U373-Magi-CCR5E and CEM-NKr-CCR5 cells (Figure 2D) with an apparent Kd values of ~110 nM. In the CCR5 negative CEM cells, only very higher concentration (>800 nM) of G-3 caused nonspecific cellular binding (Kd > 3000 nM) (Figure 2D).
We next wanted to determine whether the selected G-3 aptamer requires CCR5 expression to target CCR5 expressing cells. The specific knockdown of CCR5 using a previously validated CCR5 siRNA (Wheeler, et al., 2011) demonstrated a loss of CCR5 at both the mRNA and cell surface (Supplementary Figures S3E, F). The suppression of CCR5 using siRNAs resulted in a loss of the Cy3-labeled G-3 aptamer and control APC-labeled CCR5 antibody (APC-CD195) binding to the CCR5 expressing cells (Figure 2E and Supplementary Figure S3G).
In addition, to examine whether or not the G-3 aptamer internalized in target cells, we carried out real-time live-cell Z-axis confocal microscopy. The Cy3-labeled G-3 aptamer appeared to be selectively internalized within the CCR5 expressing U373-Magi-CCR5E cells, CEM-NKr-CCR5 cells and primary PBMCs, but not the U373-Magi and CEM control cells (Figure 2F, G) and appeared to be preferentially retained in the cytoplasm (Figure 2H and Supplementary Figure S3H).
CCR5 aptamer suppresses HIV-1 infectivity of R5-tropic HIV-1 in primary PBMCs and in vivo generated human CD4+ T cells
HIV-1 commonly uses CCR5 or CXCR4 as coreceptors along with CD4 to enter target cells (Berger, et al., 1999). A number of new experimental CCR5 receptor antagonists have been designed to interfere with the interaction between CCR5 and HIV-1 (Signoret, et al., 2000,Ugolini, et al., 1999,Vila-Coro, et al., 2000). We therefore conducted a “prophylactic” HIV-1 experiment to determine whether the CCR5 aptamer would also block HIV infectivity of R5 viruses in cell culture. In this assay, primary PBMCs were first incubated with G-3 aptamer for 4-6 hours followed by an infection with various HIV-1 stains (R5 viruses: JR-FL and Bal; X4 viruses: IIIB and NL4-3). The G-3 aptamer efficiently neutralized HIV-1 infectivity of R5 strains with about 170~350 nM of IC50 (Table 2 and Supplementary Figures S4A, B). Notably, the G3-aptamer had no observable suppression of those cells infected with X4 strains (Supplementary Figures S4C, D).
Table 2. The IC50 value was calculated based on HIV-1 protection assay.
Human PBMC-CD4+ cells or in vivo generated human CD4+ T cells were pre-treated with experimental RNAs before exposure to HIV-1 viruses.
| Virus | Day-3 post-treatment | Day-5 post-treatment | Day-7 post-treatment | R2 | ||
|---|---|---|---|---|---|---|
|
|
||||||
| IC50 (nM) | ||||||
| R5 | JR-FL | 219.7 ± 55.6 | 232.6 ± 38.3 | 170.4 ± 47.0 | >0.9 | |
| Primary PBMCs | R5 | Bal | 349.4 ± 77.24 | 354.3 ± 113.3 | >1000 | >0.9 |
| (G-3 aptamer) | X4 | IIIB | >1000 | Not converged | Not converged | <0.2 |
| X4 | NL4-3 | Not converged | Not converged | Not converged | <0.5 | |
|
| ||||||
| Primary PBMCs | R5 | JR-FL | 23.2 ± 6.2 | 142.6 ± 41.3 | 79.3 ± 21.6 | >0.9 |
| (G-3-TNPO3 OVH Chimera) | ||||||
|
| ||||||
| in vivo generated human | R5 | JR-FL | 47.98 ± 8.57 | 58.51 ± 11.22 | 17.09 ± 5.90 | >0.9 |
| CD4+ T cells | R5 | Bal | 147.6 ± 45.69 | 245.5 ± 35.76 | 243.2 ± 46.16 | >0.9 |
| (G-3 aptamer) | X4 | NL4-3 | Not converged | Not converged | Not converged | ---- |
Furthermore, the capacity of CCR5 aptamers to inhibit HIV-1 replication was evaluated on in vivo generated human CD4+ T-cells. We isolated mature human CD4+ T-cells from humanized mice. At sacrifice, splenocytes were harvested and human CD4+ T-cells were collected by cell sorting. The cellular surface CCR5 expression was detected by flow cytometry analysis (Supplementary Figure S4E, F). The sorted CD4+ T-cells were pre-treated with G-3 and subsequently were challenged ex vivo by R5 stains (JR-FL or Bal) or X4 strains (NL4-3) as described above. Similarly, HIV-1 JR-FL or Bal replication was inhibited in human CD4+ T-cells from humanized mice (Supplementary Figures S4G, H, and I) which showed about 50~250 nM of IC50 as listed in Table 2. Taken together, these data indicate that the selected CCR5 aptamer (G-3) inhibits HIV-1 p24 production and provided protection from HIV infection by R5 viruses.
Design of CCR5 aptamer-siRNA chimera delivery systems that can bind to CCR5 and are internalized by CCR5 expressing cells
We have previously delivered anti-HIV siRNAs specifically to HIV-1 infected cells using RNA aptamer against HIV-1 envelop gp120 protein (Zhou, et al., 2009). In this study, we utilized a similar strategy for cell-specific targeting of TNPO3 siRNA into cells expressing CCR5. TNPO3 (Transportin-3) is a cellular factor that is involved in facilitating cytoplasmic - nuclear trafficking of the HIV-1 pre-integration complex and was previously shown by an siRNA screen to block HIV-1 infection at the afferent stage (Brass, et al., 2008,Shah, et al., 2013). As shown in Figure 3A, B, two CCR5 aptamer-siRNA chimeras (G-3-TNPO3 OVH chimera and G-3-TNPO3 Blunt chimera) were designed and prepared as previously described.
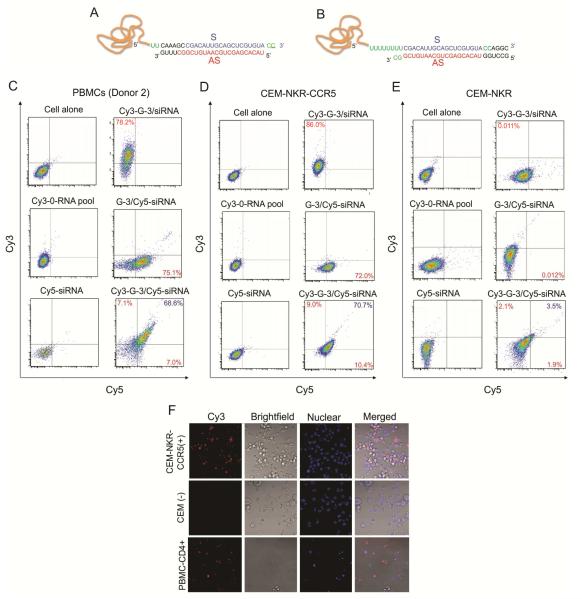
Figure 3. The design and binding affinity of aptamer-siRNA chimeras.
A, B) Schematic of CCR5 aptamer– siRNA chimeras. The region of the aptamer is responsible for binding to CCR5, and the siRNA is targeting TNPO3 gene. A linker (2 or 8 Us) between the aptamer and siRNA is indicated in green. Two versions, G-3-TNPO3 OVH chimera (A) and G-3-TNPO3 Blunt chimera (B), were designed. C, D, E) Cell surface binding of fluorescent dye –labeled RNAs was assessed by flow cytometry. The Cy3-labeled 0-RNA pool and Cy5-labeld siRNA were used as negative control. G-3-TNPO3 OVH chimera was chosen for binding affinity test with C) PBMC-CD4+ cells, D) CEM-NKr-CCR5 positive cells, E) CEM negative cells. The aptamer-sense strand and antisense strand of the chimera were labeled by Cy3 and Cy5 dye, respectively. And then they were annealed to form aptamer-siRNA chimera. F) Internalization analysis. The images were collected using 40×magnification as described previously.
To determine whether the designed chimeras are able to specifically bind and be internalized in CCR5 expressing cells, we performed flow cytometry and live cells confocal microscopy experiment. G-3-TNPO3 OVH chimera was chosen for binding affinity test with PBMC-CD4+ cells, CEM-NKr-CCR5 positive cells, and CEM negative cells. The aptamer-sense strand and antisense strand of the chimera were labeled by Cy3 and Cy5 dye, respectively. Subsequently, sense and antisense RNAs were annealed to form aptamer-siRNA chimera containing either single color or dual color labeling. Flow cytometric results showed that G-3 (either non-labeled or Cy3-labeled) delivered Cy5 labeled siRNA to about 70% ~ 80%of CCR5 expressing PBMCs and CEM-NKr-CCR5 cells (Figure 3C-E and Supplementary Figure S5). Similar with the parental G-3 aptamer, the Cy3-labeled aptamer-siRNA chimeras selectively bound and were internalized into cells expressing CCR5 after 5 h post-treatment (Figure 3F).
CCR5 aptamer-siRNA chimeras specifically knockdown TNPO3 expression via RNAi pathway and do not trigger a type I interferon response
To determine the functionality of the aptamer delivered siRNAs, after internalization, we evaluated the relative levels of inhibition of TNPO3 expression. Specially, CEM-NKr-CCR5 cells or CEM-NKr control cells were directly incubated with the aptamer-siRNA chimeras, G-3 aptamer, or a non-functional control aptamer-scrambled siRNA chimera. In parallel, cells were transfected with the experimental RNAs by using a commercial transfection agent (Trans IT-TKO). Silencing of TNPO3 was assessed by the degree of TNPO3 mRNA knockdown using qRT-PCR (Figure 4A). In the presence of transfection agent, TNPO3 gene silencing was observed in both CEM-NKr-CCR5 and CEM-NKr control cells after the treatment of G-3-TNPO3 chimeras. However, in the absence of transfection agent, the G-3-TNPO3 chimeras, but not the G-3-scrambled siRNA chimera or non-targeting aptamer-TNPO3 siRNA conjugate (gp120 aptamer A-1-stick-TNPO3), reduced TNPO3 mRNA levels. Importantly, the reduction was specific to CCR5 positive cells, as control CEM-NKr negative cells treated with either of the G-3-TNPO3 chimeras exhibited no TNPO3 mRNA reduction (Figure 4A). Similarly, the G-3 aptamer delivered TNPOs DsiRNA also resulted in a decrease in target mRNA levels 48 hours post-treatment in human PBMC-CD4+ cells, with efficiency comparable to a previous validated transfection agent (G5 dendrimer) (Zhou, et al., 2011) (Figure 4B).
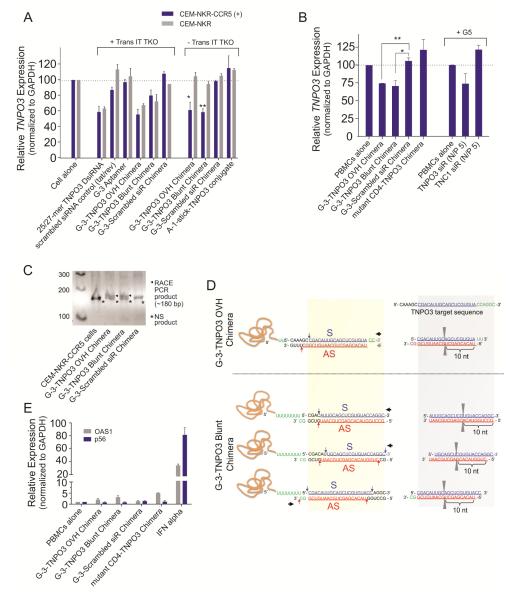
Figure 4. CCR5 aptamer delivered siRNAs specifically knockdown TNPO3 expression via RNAi pathway.
Relative TNPO3 mRNA expression was detected by real-time PCR in A) CEM-NKr-CCR5 and CEM-NKr negative cells, and B) human PBMC-CD4+ cells. As negative control, unrelated aptamer-siRNA chimera (gp120 aptamer A-1 or mutant CD4 aptamer) and G-3 aptamer-scrambled siRNA chimera were used. Asterisk indicates a significant difference compared with control (P < 0.01, student’s t-test). 5′-RACE PCR analysis of TNPO3 DsiRNA delivered by CCR5 aptamer – siRNA chimeras. C) Nested PCR products were resolved in an agarose gel and specific siRNA-mediated RACE PCR cleavage mRNA products are marked by a bold arrow. D) The positions of the siRNA directed cleavage sites in the TNPO3 target RNA are indicated with a pair of grey triangle. According to mRNA cleavage, these predicated siRNA species also are showed with blue and red arrow. The proposed directions of Dicer entry are indicated by a bold black arrow. E) IFN gene activation assays in human PBMCs. The interferon response genes encoding P56 (CDKL2) and OAS1, were measured by quantitative RT-PCR. These data represent the average of three replicate measurements.
In order to determine whether the siRNAs released from the chimeras were actually triggering RNAi, we next investigated siRNA-directed mRNA cleavage using a modified 5′-RACE (Rapid amplification of cDNA ends) PCR assay. It has been established that Ago2 mediates cleavage between bases 10 and 11 relative to the 5′ end of each siRNA (Matranga, et al., 2005). Thus the RACE PCR product sequence analyses of the target should reveal a 3′ linker addition at the base positioned 11 nucleotides from the 5′ end of the siRNA guide strand. PCR bands of the predicted lengths were detected in the total RNAs from CEM-NKr-CCR5 cells treated with the chimeras after two nested PCR reactions (Figure 4C). The individual clones were sequenced to verify the expected PCR products. Several cleavage sites were found in the samples treated with the two chimeras. Figure 4D indicated the Ago2 cleavage sites and proposed direction of Dicing. For the OVH chimera, one major cleavage was observed, suggesting that Dicer preferentially entered the DsiRNA from 3′-overhang of the sense strand. The short linker (2Us) may limit the Dicer entry from 5′-end of the sense strand. In the case of the Blunt chimera, several different cleavage sites were generated, suggesting that Dicer might bi-directionally enter the DsiRNA to generate different 21-mer siRNA species. Different from the observation in the OVH chimeras, the longer linker (8Us) of the Blunt chimera may allow the Dicer entry from 5′-end of the sense strand. Taken together, these results provide strong evidence that the chimera-delivered siRNAs are processed intracellularly and trigger sequence-specific degradation of the TNPO3 target mRNA.
As a final test for nonspecific inhibitory activity, the aptamer mediated siRNA delivery system was monitored for induction of type I interferon response (IFN). The levels of two different type I IFN-stimulated gene expressions (mRNAs) were quantified by quantitative RT-PCR (Figure 4E). We used IFN-α as a positive control to confirm up-regulation of p56 (CDKL2, Cyclin-dependent kinase-like 2) and OAS1 (2′-5′-oligoadenylate synthetase 1) gene expression. Our results indicated that the treatment of PBMC-CD4+ cells with these chimeras did not activate the type I IFN pathway.
CCR5 aptamer-siRNA chimeras inhibit HIV-1 infection in primary human PBMCs
It has been reported previously that as one of HIV-1 host dependency factors (HDFs), TNPO3 is a karyopherin required for viral integration, suggesting it is potential therapeutic target. Therefore, we conducted two different experiments to assess the anti-HIV-1 activities of the CCR5 aptamer-mediated TNPO3 siRNA delivery system.
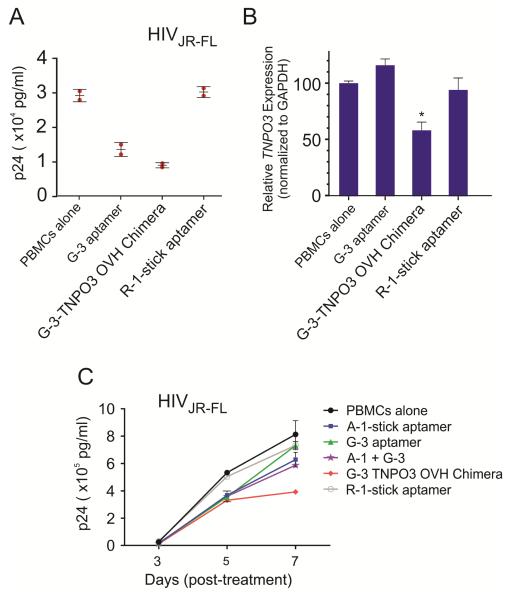
The first assay is a “prophylactic” HIV-1 experiment performed as described above. The experimental RNAs were incubated with human PBMC-CD4+ cells. After 4-6 hours treatment, the cells were challenged by the R5 virus (HIV-1 JR-FL). The culture supernatants were collected at five days after treatment for HIV-1 p24 antigen ELISA assay. Results presented in Figure 5A showed that both of the aptamers and chimera inhibited p24 production, but the strongest inhibition was observed with the G-3-TNPO3 OVH chimera treatment which showed a lower IC50 value (Table 2). To confirm that the siRNA component was functioning along with the aptamer, following internalization of the chimera in infected cells, we also evaluated the relative levels of inhibition of TNPO3 gene expression by qRTPCR assay (Figure 5B). Specific down regulation of the TNPO3 mRNA was observed, a direct consequence of the CCR5 aptamer-mediated DsiRNA delivery. In contrast, treatment of these cells with the CCR5 aptamer alone or unrelated RNA had no effect on TNPO3 levels.
Figure 5. Dual inhibition on HIV-1 infection mediated by aptamer-based siRNA delivery system.
A) HIV-1 protection assay. Human PBMC-CD4+ cells were pre-treated with experimental RNAs and then infected with JR-FL virus. B) The siRNA delivered by aptamers knocked down TNPO3 gene expression in human PBMCs. Relative TNPO3 mRNA expression were detected by real-time PCR, with GAPDH as internal control. Asterisk indicates a significant difference compared with control (P < 0.01, student’s t-test). C) HIV-1 challenge assay. Human PBMC-CD4+ cells were infected with JR-FL and then incubated with experimental RNAs. A gp120 aptamer (A-1-stick) and an unrelated aptamer (R-1-stick) were used as positive and negative controls, respectively. Data represent the average of triplicate measurements of p24.
In the second experiment, human PBMC-CD4+ cells were infected with JR-FL virus for 5 days and then incubated with CCR5 aptamer and aptamer-siRNA chimeras. At different days post treatment, aliquots of the media were assayed for viral p24 antigen levels. An anti-gp120 aptamer (A-1-stick) was used here as a positive control. The results of these analyses (Figure 5C and Supplementary Figure S6) indicated that both CCR5 aptamer and aptamer-siRNA chimera were also able to suppress HIV-1 replication at day 3 and day 5 post-treatment in which cells were infected with HIV-1 virus prior to the treatment. Although gp120 aptamer showed a better inhibition than CCR5 aptamer, the strongest and prolonged suppression was observed with G-3-TNPO3 OVH chimera treatment. Collectively, these results provide further evidence that the aptamer delivered siRNA triggers RNAi, thereby resulting in dual inhibitory effect on HIV-1 replication.
DISCUSSION
The concept of nucleic acid-based therapeutic (ribozyme, decoy, antisense oligonucleotide, mRNA, aptamer, siRNA, microRNA) has been extensively exploited for the treatment of various human diseases (Stull and Szoka, 1995). Although they each could be employed as a stand-alone inhibitor, the combinatorial use of various nucleic acids has shown more potential and advantages, including synergistic, prolonged suppression, and especially targeted therapy when a cell-type specific nucleic acid aptamer was adopted (Thiel and Giangrande, 2009). In previous studies, investigators have demonstrated that HIV-1 genome and host genes can be targeted in combination with various antiviral nucleic acids (ribozyme, siRNA, decoy, etc.) in a tailored manner, thereby providing more efficacies in blocking viral replication and preventing the emergence of resistant variants (Shum, et al., 2013,Zhou and Rossi, 2011,Zhou and Rossi, 2011). By functionalizing the cell-specific aptamers with therapeutic agents, the cellular uptake is enhanced, thereby improving the therapeutic efficacy. We also have capitalized on the exquisite specificity of an anti-gp120 aptamer to delivery anti-HIV siRNAs into HIV infected cells with the net result that the replication and spread of HIV is strongly inhibited by the combined action of the aptamer and the siRNAs against HIV-1 tat/rev or host dependency factors (HDFs). In this case, the aptamers can function both as targeted delivery reagents and antiviral agents. In the present study, we wish to generate anti-CCR5 RNA aptamers capable of specifically targeting HIV-1 susceptible cells (as a therapeutic siRNA delivery agent) and inhibiting HIV-1 infectivity (as antiviral agent) via block of the CCR5 required for HIV-1 to enter cells.
Considering the limited resource and the risk of changed conformation of purified CCR5 membrane protein after purification, we therefore utilized live cell-based SELEX methodology for generating cell-type specific RNA aptamers. High throughput sequencing (HTS) technology and bioinformatics analysis were also combined here to facilitate the rapid identification of individual RNA aptamers and show the library evolution. We obtained and processed millions of sequence reads from each round, which provide comprehensive and in-depth information, such as the basic sequences, total reads, the complexity of the library, the enrichment factor, frequency of each unique sequences, and distribution of each nucleotide at random region, thereby helping us to better understand the selection progression and the molecular evolution. In comparison, conventional approach for aptamer identification in which the final SELEX library is generally cloned into a plasmid vector and dozens to a few hundred individual clones are picked up for sequencing analysis cannot provide such thorough insight into the selection progression and aptamer evolution. Our HTS analysis revealed that maximal molecular enrichment was reached at the 7th or the 8th selection round which suggested a stop time point. Subsequently additional selection rounds did not further improve the enrichment, probably implying that an increase of one moderate sequence probably occupied the proportion of some better aptamers. Furthermore, the evolution of a particular sequence or group revealed that some aptamer candidates have already been identified by HTS at much earlier selection round (e.g., the 5th or 6th round) which cannot be done by traditional cloning and sequencing approach due to their limited sequences. The use of the minimum number of selection cycles is able to significantly improve the SELEX efficiency and the aptamer efficacy. Taken together, we demonstrate that the power of high throughput sequencing technique and bioinformatics analysis in the rapid identification of aptamers.
Human immunodeficiency virus (HIV) replicates primarily in T-lymphocytes and cells of the macrophage lineage. CD4 is required for virus binding to the cell surface, and coreceptors (CCR5, CXCR4) are required for viral fusion with the cell membrane (Berger, et al., 1999,Murphy, et al., 2000,Pelchen-Matthews, et al., 1999). M-tropic HIV-1 stains (R5 viruses) use CCR5 for the coreceptor, and T-tropic strains (X4 viruses) use CXCR4. Successes in discovering new classes of CCR5 inhibitors targeted to the step of HIV-1 entry have been reported in recent years. Previous studies have demonstrated that the chemokine RANTES (regulated on activation, normal T cell expressed and secreted), a natural CCR5 ligand, and an NH2-terminal modified form of RANTES (AOP-RANTES) protect cells from HIV infection by R5 viruses (Sabbe, et al., 2001,Signoret, et al., 2000). This inhibition of virus infection may be explained by either occupancy of CCR5 and blocking of interaction with the CD4-gp120 complex, or receptor sequestration following internalization (Mack, et al., 1998,Mariani, et al., 1999,Vila-Coro, et al., 1999). Therefore, we designed a “prophylactic” HIV-1 experiment to determine whether or not CCR5 aptamer can specifically suppress R5-tropic virus infection. Our results indicated that G-3 aptamer specifically neutralized R5 virus infection in primary PBMCs and in vivo generated human CD4+ T cells with a nanomolar IC50 value, but no obvious anti-viral effect was observed in X4 virus infected cell, thereby suggesting a HIV-1 tropic-selective inhibition. Therefore, CCR5 aptamers show potential as a therapeutic by itself for HIV-1 infection.
In the present study, we used TNPO3 siRNA as proof of principle to show successful delivery as well as specific mRNA cleavage. Through dual-color flow cytometry assay, we found that at least 80% siRNA could be delivered by CCR5 aptamer into various CCR5 expression cells. We also observed that the aptamer or aptamer-siRNA chimeras were mainly located within the cytoplasm by real-time confocal microscopy, suggesting that the CCR5 aptamer may be internalized probably by CCR5 receptor-mediated endocytosis. Additionally, we conducted both “prophylactic” HIV-1 experiment and HIV-1 challenge experiment to access the anti-HIV-1 activity of chimera. When the CCR5 aptamer-siRNA chimera was applied on human PBMCs either before exposure to the virus or after HIV-1 infection, it resulted in stronger and prolonged suppression on HIV-1 replication in comparison of CCR5 aptamer alone. This is likely due to the contribution of siRNA component of the chimera in silencing TNPO3 expression in the treated cells.
Several major challenges still remain that hinder the rapid transition of aptamer-mediated technology from the research laboratory to the clinic. As single-stranded nucleic acid molecules, aptamers are subject to nuclease-mediated degradation and rapid renal filtration / clearance. The lower or moderate affinities of aptamers often compromise the therapeutic efficacy. Proper modifications of aptamers can help to improve their pharmacokinetics. For example, aptamers have been conjugated to PEG to increase their bioavailability and circulating half-life in vivo. Incorporating protective functional groups (e.g. thiol-phosphate, 2′-Fluoro, 2′-amino, etc.) in the phosphate backbone or 2′-position of the ribose sugar, can improve their nuclease resistance, even their binding affinity. Additionally, multiple aptamers and therapeutic agents can be combined to various nanocarrier system, probably further enhancing binding affinity of the aptamer and loading capacity of drug, eventually improving therapeutic index.
In summary, we have demonstrated that the combinatorial use of live cell-based SELEX with HTS and bioinformatics analysis not only represents a powerful and rapid method for generating cell-type specific, internalizing aptamers that are able to recognize a particular target membrane protein under native condition, but also provides comprehensive information that could help us to better understand the selection progression and improve the evolution efficiency. Additionally, the CCR5 aptamer-based siRNA delivery system serves as dual functional inhibitors and therefore provides better efficacy than either the aptamer or siRNA applied alone. Therefore, the cell-specific, internalizing CCR5 aptamersfunctionalized agents may offer great promise for cell-type- or tissue-specific delivery of various therapeutic drugs for targeted HIV-1 therapy. Notably, these targeted delivery approaches can be utilized in disease models beyond HIV-1 such as cancer.
EXPERIMENTAL PROCEDURES
Live Cell-Based SELEX
The starting DNA library contained 20 nt of random sequences and was synthesized by Integrated DNA Technologies (Coralville, IA, USA). The random region is flanked by constant regions, which include the T7 promoter (underlined) for in vitro transcription and a 3′-tag for reverse transcription– polymerase chain reaction (RT-PCR). 51-mer ssDNA oligo library for RNA Library is 5′- GGG AGG ACG ATG CGG - N20- CAG ACG ACT CGC CCG A - 3′ (51 nt). The 5′ and 3′constant sequences are 5′- TAA TAC GAC TCA CTA TAG GGA GGA CGA TGC GG −3′ (32 mer) and 5′- TCG GGC GAG TCG TCT G −3′ (16 mer), respectively. The DNA random library (0.4 μM) was amplified by PCR using 3 μM each of 5′- and 3′- primers, along with 2 mM MgCl2 and 200 μM of each dNTP. To preserve the abundance of the original DNA library, PCR was limited to 10 cycles. The PCR amplification protocol is as follows: 93 °C for 3 min, followed by 10 cycl es of heating to 93 °C for 1 min, 63 °C for 1 min a nd 72 °C 1 min. A final extension step was performed f or 7 min at 72 °C. After the PCR reactions (10 reactions, 100 μl per reaction), the amplified dsDNA pool was recovered using a QIAquick Gel purification Kit. The resulting dsDNA was converted to an RNA library using the DuraScription Kit (Epicentre, Madison, WI, USA) according to the manufacturer′s instructions. In the transcription reaction mixture, CTP and UTP were replaced with 2′-F-CTP and 2′-F-UTP to produce ribonuclease resistant RNA. The reactions were incubated at 37°C for 6 h, and subsequently the template DNA was removed by DNase I digestion. The transcribed RNA pool was purified in an 8% polyacrylamide/7 M urea gel. The purified RNA library was quantified by ultraviolet spectrophotometry.
The SELEX was performed principally as described by Tuerk and Gold (Tuerk and Gold, 1990) and here we applied a modified cell-based SELEX described by Thiel et al (Cerchia, et al., 2005,Cerchia, et al., 2009,Thiel, et al., 2012,Thiel, et al., 2012). Generally, in each round, the desired amount of RNA pools were refolded in 3 mL of refolding buffer, heated to 65°C for 5 min and then slowly cooled to 37°C. Incubation was continued at 37°C for 10 min. The refolding or washing buffer contained DPBS (pH 7.0 ~7.4) and Ca2+ and Mg2+, 1 mM CaCl2, 2.7 mM KCl, 1.47 mM KH2PO4, 1 mM MgCl2, 136.9 mM NaCl, 2.13 mM Na2HPO4. The binding buffer used during the selection was prepared by adding yeast tRNA to washing buffer to reduce nonspecific binding. To avoid nonspecific interaction between nucleic acids and the cell surface, the tRNA (100 μg/mL) as a competitor was first incubated with non-targeted cells or targeted cells at 37 °C for 25 min and then ready for selection step. Counter-selection step was performed per cycle to minimize nonspecific binding with the non-targeted cells. Subsequently, the unbound RNA pool was transferred to the targeted cells for positive selection.
For the first cycle of selection, 24 hours before selection, U373-Magi cells (CCR5 negative) and U373-Magi-CCR5E cells (CCR5 positive) were seeded at equal density (5*106 cells per plate) on 150 mm tissue culture dish with 25 mL complete culture medium. On the day of selection, U373-Magi negative cells were washed three times with 15 mL per-warmed washing buffer to remove dead cells and then added 15 mL pre-warmed binding buffer supplemented with 100 μg/mL yeast tRNAs. After 25 min incubation at 37 °C, the buffer was removed and the refolded RNA pool (4 nmol 0-RNA pool in 15 mL refolding buffer) was added to the U373-Magi negative cells for 30 min at 37 °C. The pre-cleared 0-RNA pool (supernatants with unbound sequences from negative cells plate) was ready for positive selection. Meanwhile, as described above, U373-Magi CCR5 positive cells were also washed and incubated with 15 mL pre-warmed binding buffer supplemented with 100 μg/mL yeast tRNAs. After 25 min incubation at 37 °C, the buffer was re moved and the pre-cleared 0-RNA pool was subsequently transferred to the U373-Magi CCR5 positive cells for 30 min at 37 °C. Following incubation of the pre-clean RNA pool, the U373-Magi CCR5 positive cells were washed twice with 12 mL pre-warm washing buffer to remove unbound sequences and cell-surface RNAs with weak binding. Cell-surface bound RNA with strong binding affinity and internalized RNA sequences were recovered by TRIzol (Invitrogen) extraction by following the manufacturer’s instructions.
The recovered RNA pool was reversed transcribed using the ThermoScript RT-PCR system (Invitrogen) and amplified for 15 cycles of PCR. The PCR amplification protocol is as follows: 95 °C for 5 min, followed by 15 cycles of heating to 95 °C for 1 min, 63 °C for 1 min and 72 °C 1 min. A fin al extension step was performed for 7 min at 72 °C. Af ter the amplified dsDNA template was purified a QIAquick Gel purification Kit, it was transcribed to new RNA pool as described above for the next round of selection. With the SELEX progress, the cells number, density, volume, RNA amount, washing times, tRNA competitor amount, and incubation time were progressively adjusted in order to increase the pressure of aptamer selection. The selection conditions are summarized in Supplementary Table S1.
Inhibit HIV-1 replication in human PBMCs
Assay 1: HIV-1 protection assay
HIV-1 protection assay was performed in 24-well tissue culture plates. PBMCs were washed once with pre-warmed PBS, and 4×105 PBMCs were seeded to each well of assay plates. Subsequently, experimental RNAs (800 nM final concentration) were added. Plates were incubated for 4 - 6 hours at 37 °C in a humidified 5% CO 2 incubator. HIV-1 R5 strains (JR-FL, MOI=0.01) were added into each well. After 24 hours incubation, the cells were gently washed with pre-warmed PBS to eliminate free viruses and were incubated sequentially at 37 °C in a humidified 5% CO2 incubator. The culture supernatants were collected at different time points after infection (3, 5 and 7 days). The HIV-1 p24 antigen analyses were performed using a Coulter HIV-1 p24 antigen assay (Beckman Coulter, Fullerton, CA) in accordance with the manufacturer’s instructions. The total RNA was isolated for qRT-PCR analysis.
Assay 2: HIV-1 challenge assay
Experimental RNAs were prepared and refolded in refolding buffer. Human PBMCs were infected with HIV-1 JR-FL virus for 5 days (MOI=0.01). Prior to RNA treatments the infected cells were gently washed with PBS three times to remove free virus. Next 1×105 infected cells and 1×105 uninfected cells were incubated with experimental RNAs at 800 nM final concentration in 24-well plates at 37°C. The culture supernatants were collected at different times (3 d, 5 d and 7 d). The p24 antigen analyses were performed as described above.
Supplementary Material
Significance.
Although there is currently no cure for HIV/AIDS, a variety of antiretroviral drugs that act on different stages of the HIV lifecycle can be used in combination to control the virus. Highly active antiretroviral treatment (HAART) has proven to be the standard approach for delaying the progression to AIDS and reducing the risk of death and disease complication. However, severe side effects, compliance problems and drug resistances complicate the use of this therapy. Nucleic acid-based therapeutics has been actively developed as alternative or adjuvant agents for those chemical antiviral drugs in order to surmount those drawbacks.
RNA interference-based therapeutics has unique therapeutic potential for the treatment of HIV-1 infection since the entire genome of the virus is target. Another type of nucleic acid – aptamers – shows promise as a potent class of anti-HIV agent and can additionally function as a cell-type specific delivery vehicle for targeted RNAi. We reasoned that a combinatorial use of various antiviral nucleic acids such as siRNAs and aptamers could be more efficacious in blocking viral replication and preventing the emergence of resistant variants.
The C-C chemokine receptor type 5 (CCR5) is a 7 pass transmembrane receptor expressed by T-cells and macrophages which serves as a co-receptor for macrophage-tropic HIV-1. Loss of CCR5 is associated with resistance to HIV-1. We therefore generated CCR5 RNA aptamers capable of specifically targeting HIV-1 susceptible cells (as siRNA delivery agent) and inhibiting HIV-1 infectivity (as antiviral agent) via block of the CCR5 required for HIV-1 to enter cells. The importance of such a strategy for the treatment of HIV-1 cannot be overstated as viral variants resistant to current therapies are on the rise in developed nations and portend a new pandemic of resistant virus. Notably, these delivery approaches can be utilized in disease models beyond HIV-1 such as cancer.
Highlights.
Combinatorial use of various antiviral nucleic acids for HIV-1 therapy
Cell-type specific delivery for targeted RNA interference
Dual inhibitory function aptamer-siRNA conjugates
ACKNOWLEDGEMENT
We thank James O. McNamara II and Paloma H. Giangrande for helpful discussions. We thank Mayumi Takahashi for providing primary PBMCs. The authors would like to thank City of Hope DNA sequencing core and Bioinformatics Core facility (Harry Gao, Xiwei Wu and Jinhui Wang) for Solexa Deep sequencing and data processing. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: the U373-Magi and U373-Magi-CCR5E cell lines were obtained from Dr. Michael Emerman (Vodicka, et al., 1997); CEM-NKr and CEM-NKr-CCR5 cell lines were obtained from Dr. Peter Cresswell (Howell, et al., 1985,Lyerly, et al., 1987); the HIV-1JR-FL and HIV-1BaL virus.
FUNDING This work was supported by National Institutes of Health grants [grant numbers R01AI29329, R01AI42552, and R01HL07470 to J.J.R, and grant number P01AI099783 to K.V. M]. M.S.W. is supported by grants from the South African National Research Foundation (NRF) and Medical Research Council (MRC). Funding for open access charge: National Institutes of Health.
Footnotes
Conflict of Interest declaration: J. J. R. and J. Z. have an issued patent entitled “Cell-type specific aptamer-siRNA delivery system for HIV-1 therapy”. USPTO, No. US 8, 222, 226 B2, issued date: July 17, 2012. J.J.R., J. Z., M.S.W and K.V.M. have a patent pending on “Cell-specific internalizing RNA aptamers against human CCR5 and used therefore”, The United States Patent, and application number: 62/025,368, filed on July 16, 2014.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annual review of immunology. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Cerchia L, Duconge F, Pestourie C, Boulay J, Aissouni Y, Gombert K, Tavitian B, de Franciscis V, Libri D. Neutralizing aptamers from whole-cell SELEX inhibit the RET receptor tyrosine kinase. PLoS Biol. 2005;3:e123. doi: 10.1371/journal.pbio.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cerchia L, Esposito CL, Jacobs AH, Tavitian B, de Franciscis V. Differential SELEX in human glioma cell lines. PloS one. 2009;4:e7971. doi: 10.1371/journal.pone.0007971. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cerchia L, Giangrande PH, McNamara JO, de Franciscis V. Cell-specific aptamers for targeted therapies. Methods Mol Biol. 2009;535:59–78. doi: 10.1007/978-1-59745-557-2_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Tan W. Aptamers Generated from Cell-SELEX for Molecular Medicine: A Chemical Biology Approach. Acc Chem Res. 2009 doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held DM, Kissel JD, Patterson JT, Nickens DG, Burke DH. HIV-1 inactivation by nucleic acid aptamers. Front Biosci. 2006;11:89–112. doi: 10.2741/1782. [DOI] [PubMed] [Google Scholar]
- Howell DN, Andreotti PE, Dawson JR, Cresswell P. Natural killing target antigens as inducers of interferon: studies with an immunoselected, natural killing-resistant human T lymphoblastoid cell line. Journal of immunology. 1985;134:971–976. [PubMed] [Google Scholar]
- Joshi PJ, Fisher TS, Prasad VR. Anti-HIV inhibitors based on nucleic acids: emergence of aptamers as potent antivirals. Curr Drug Targets Infect Disord. 2003;3:383–400. doi: 10.2174/1568005033481060. [DOI] [PubMed] [Google Scholar]
- Li X, Zhao Q, Qiu L. Smart ligand: aptamer-mediated targeted delivery of chemotherapeutic drugs and siRNA for cancer therapy. J Control Release. 2013;171:152–162. doi: 10.1016/j.jconrel.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Lyerly HK, Reed DL, Matthews TJ, Langlois AJ, Ahearne PA, Petteway SR, Jr., Weinhold KJ. Anti-GP 120 antibodies from HIV seropositive individuals mediate broadly reactive anti-HIV ADCC. AIDS research and human retroviruses. 1987;3:409–422. doi: 10.1089/aid.1987.3.409. [DOI] [PubMed] [Google Scholar]
- Mack M, Luckow B, Nelson PJ, Cihak J, Simmons G, Clapham PR, Signoret N, Marsh M, Stangassinger M, Borlat F, et al. Aminooxypentane-RANTES induces CCR5 internalization but inhibits recycling: a novel inhibitory mechanism of HIV infectivity. The Journal of experimental medicine. 1998;187:1215–1224. doi: 10.1084/jem.187.8.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallikaratchy P, Liu H, Huang YF, Wang H, Lopez-Colon D, Tan W. Using aptamers evolved from cell-SELEX to engineer a molecular delivery platform. Chem Commun (Camb) 2009:3056–3058. doi: 10.1039/b823258j. [DOI] [PubMed] [Google Scholar]
- Mariani R, Wong S, Mulder LC, Wilkinson DA, Reinhart AL, LaRosa G, Nibbs R, O’Brien TR, Michael NL, Connor RI, et al. CCR2-64I polymorphism is not associated with altered CCR5 expression or coreceptor function. Journal of virology. 1999;73:2450–2459. doi: 10.1128/jvi.73.3.2450-2459.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Meanwell NA, Kadow JF. Inhibitors of the entry of HIV into host cells. Curr Opin Drug Discov Devel. 2003;6:451–461. [PubMed] [Google Scholar]
- Murphy PM, Baggiolini M, Charo IF, Hebert CA, Horuk R, Matsushima K, Miller LH, Oppenheim JJ, Power CA. International union of pharmacology. XXII. Nomenclature for chemokine receptors. Pharmacological reviews. 2000;52:145–176. [PubMed] [Google Scholar]
- Neff CP, Zhou J, Remling L, Kuruvilla J, Zhang J, Li H, Smith DD, Swiderski P, Rossi JJ, Akkina R. An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4(+) T cell decline in humanized mice. Sci Transl Med. 2011;3:66ra66. doi: 10.1126/scitranslmed.3001581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: an emerging class of therapeutics. Annu Rev Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- Pelchen-Matthews A, Signoret N, Klasse PJ, Fraile-Ramos A, Marsh M. Chemokine receptor trafficking and viral replication. Immunological reviews. 1999;168:33–49. doi: 10.1111/j.1600-065x.1999.tb01281.x. [DOI] [PubMed] [Google Scholar]
- Sabbe R, Picchio GR, Pastore C, Chaloin O, Hartley O, Offord R, Mosier DE. Donor- and ligand-dependent differences in C-C chemokine receptor 5 reexpression. Journal of virology. 2001;75:661–671. doi: 10.1128/JVI.75.2.661-671.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer L, Rossi JJ, Weinberg MS. Progress and prospects: RNA-based therapies for treatment of HIV infection. Gene Ther. 2007;14:1057–1064. doi: 10.1038/sj.gt.3302977. [DOI] [PubMed] [Google Scholar]
- Shah VB, Shi J, Hout DR, Oztop I, Krishnan L, Ahn J, Shotwell MS, Engelman A, Aiken C. The host proteins transportin SR2/TNPO3 and cyclophilin A exert opposing effects on HIV-1 uncoating. Journal of virology. 2013;87:422–432. doi: 10.1128/JVI.07177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum KT, Zhou J, Rossi JJ. Aptamer-based therapeutics: new approaches to combat human viral diseases. Pharmaceuticals. 2013;6:1507–1542. doi: 10.3390/ph6121507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signoret N, Pelchen-Matthews A, Mack M, Proudfoot AE, Marsh M. Endocytosis and recycling of the HIV coreceptor CCR5. The Journal of cell biology. 2000;151:1281–1294. doi: 10.1083/jcb.151.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stull RA, Szoka FC., Jr. Antigene, ribozyme and aptamer nucleic acid drugs: progress and prospects. Pharm Res. 1995;12:465–483. doi: 10.1023/a:1016281324761. [DOI] [PubMed] [Google Scholar]
- Sundaram P, Kurniawan H, Byrne ME, Wower J. Therapeutic RNA aptamers in clinical trials. Eur J Pharm Sci. 2013;48:259–271. doi: 10.1016/j.ejps.2012.10.014. [DOI] [PubMed] [Google Scholar]
- Thiel KW, Giangrande PH. Therapeutic applications of DNA and RNA aptamers. Oligonucleotides. 2009;19:209–222. doi: 10.1089/oli.2009.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel KW, Hernandez LI, Dassie JP, Thiel WH, Liu X, Stockdale KR, Rothman AM, Hernandez FJ, McNamara JO, 2nd, Giangrande PH. Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res. 2012;40:6319–6337. doi: 10.1093/nar/gks294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel WH, Bair T, Peek AS, Liu X, Dassie J, Stockdale KR, Behlke MA, Miller FJ, Jr., Giangrande PH. Rapid identification of cell-specific, internalizing RNA aptamers with bioinformatics analyses of a cell-based aptamer selection. PloS one. 2012;7:e43836. doi: 10.1371/journal.pone.0043836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Ugolini S, Mondor I, Sattentau QJ. HIV-1 attachment: another look. Trends Microbiol. 1999;7:144–149. doi: 10.1016/s0966-842x(99)01474-2. [DOI] [PubMed] [Google Scholar]
- Vila-Coro AJ, Mellado M, Martin de Ana A, Lucas P, del Real G, Martinez AC, Rodriguez-Frade JM. HIV-1 infection through the CCR5 receptor is blocked by receptor dimerization. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3388–3393. doi: 10.1073/pnas.050457797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila-Coro AJ, Mellado M, Martin de Ana A, Martinez AC, Rodriguez-Frade JM. Characterization of RANTES- and aminooxypentane-RANTES-triggered desensitization signals reveals differences in recruitment of the G protein-coupled receptor complex. Journal of immunology. 1999;163:3037–3044. [PubMed] [Google Scholar]
- Vodicka MA, Goh WC, Wu LI, Rogel ME, Bartz SR, Schweickart VL, Raport CJ, Emerman M. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology. 1997;233:193–198. doi: 10.1006/viro.1997.8606. [DOI] [PubMed] [Google Scholar]
- Wheeler LA, Trifonova R, Vrbanac V, Basar E, McKernan S, Xu Z, Seung E, Deruaz M, Dudek T, Einarsson JI, et al. Inhibition of HIV transmission in human cervicovaginal explants and humanized mice using CD4 aptamer-siRNA chimeras. J Clin Invest. 2011;121:2401–2412. doi: 10.1172/JCI45876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Blank M, Schluesener HJ. Nucleic acid aptamers in human viral disease. Arch Immunol Ther Exp (Warsz) 2004;52:307–315. [PubMed] [Google Scholar]
- Zhou J, Neff CP, Liu X, Zhang J, Li H, Smith DD, Swiderski P, Aboellail T, Huang Y, Du Q, et al. Systemic administration of combinatorial dsiRNAs via nanoparticles efficiently suppresses HIV-1 infection in humanized mice. Molecular therapy : the journal of the American Society of Gene Therapy. 2011;19:2228–2238. doi: 10.1038/mt.2011.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Neff CP, Swiderski P, Li H, Smith DD, Aboellail T, Remling-Mulder L, Akkina R, Rossi JJ. Functional In Vivo Delivery of Multiplexed Anti-HIV-1 siRNAs via a Chemically Synthesized Aptamer With a Sticky Bridge. Molecular therapy : the journal of the American Society of Gene Therapy. 2013;21:192–200. doi: 10.1038/mt.2012.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Rossi JJ. Aptamer-targeted RNAi for HIV-1 therapy. Methods Mol Biol. 2011;721:355–371. doi: 10.1007/978-1-61779-037-9_22. [DOI] [PubMed] [Google Scholar]
- Zhou J, Rossi JJ. Cell-specific aptamer-mediated targeted drug delivery. Oligonucleotides. 2011;21:1–10. doi: 10.1089/oli.2010.0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Rossi JJ. Progress in RNAi-based antiviral therapeutics. Methods Mol Biol. 2011;721:67–75. doi: 10.1007/978-1-61779-037-9_4. [DOI] [PubMed] [Google Scholar]
- Zhou J, Rossi JJ. Therapeutic Potential of Aptamer-siRNA Conjugates for Treatment of HIV-1. BioDrugs : clinical immunotherapeutics, biopharmaceuticals and gene therapy. 2012;26:393–400. doi: 10.2165/11635350-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Swiderski P, Li H, Zhang J, Neff CP, Akkina R, Rossi JJ. Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res. 2009;37:3094–3109. doi: 10.1093/nar/gkp185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.