Abstract
T cell receptor (TCR)α and β chains cooperatively recognize peptide-MHC (pMHC) complexes. It has been shown that a ‘chain-centric’ TCR hemichain can, by itself, dictate MHC-restricted antigen specificity without requiring major contributions from the paired TCR counterchain. Little is known, however, regarding the relative contributions and roles of chain-centric and its counter, non-chain-centric hemichains in determining T cell avidity. We comprehensively analyzed a thymically unselected T cell repertoire generated by transducing the α chain-centric HLA-A*02:01(A2)/MART127–35 TCRα, clone SIG35α, into A2-matched and unmatched post-thymic T cells. Regardless of their HLA-A2 positivity, a substantial subset of peripheral T cells transduced with SIG35α gained reactivity for A2/MART127–35. While the generated A2/MART127–35-specific T cells used various TRBV genes, TRBV27 predominated with >102 highly diverse and unique clonotypic CDR3β sequences. T cells individually reconstituted with various A2/MART127–35 TRBV27 TCRβ genes along with SIG35α possessed a wide range (>2 log orders) of avidity. Approximately half possessed avidity higher than T cells expressing clone DMF5, a naturally occurring A2/MART127–35 TCR with one of the highest affinities. Importantly, similar findings were recapitulated with other self-antigens. Our results indicate that, although a chain-centric TCR hemichain determines antigen specificity, the paired counterchain can regulate avidity over a broad range (>2 log orders) without compromising antigen specificity. TCR chain centricity can be exploited to generate a thymically unselected antigen-specific T cell repertoire, which can be used to isolate high avidity antitumor T cells and their uniquely encoded TCRs rarely found in the periphery due to tolerance.
Introduction
Conventional αβ T cell receptors (TCRs), which recognize peptide-MHC (pMHC) complexes, are comprised of TCRα and β chains, which both possess three complementarity-determining region (CDR) loops. The variable TCRα or β CDR1 and 2 regions are encoded within the germline Vα or β segment, and the hypervariable CDR3 region is determined by the junction of spliced VJα or VDJβ gene segment accompanied by random insertion and deletion of nucleotides (1). The heterogeneity in these 6 TCRα and β chain CDR regions coordinately determines the breadth of target antigens and the affinity of a given TCR. Thus, the TCR CDR sequence diversity defines a repertoire of T cells, whose mission is to recognize and target a large array of foreign antigens as adaptive lymphocytes. The repertoire of naïve T cells is vast, if not infinite, and contains millions of unique TCR structures resulting from CDR sequence diversity. In the face of such diversity, expansion out of this gigantic repertoire of clonotypic T cells with antigen specificity and defined affinity was believed to be a largely stochastic and random process that results in a highly individualized response to an antigen.
However, accumulating evidence suggests that T cell responses exist where multiple individuals generate T cells with identical or near-identical TCRs in response to the same antigenic epitope. These shared or public TCRs have been observed to occur in many types of immune responses in multiple species across many facets of immunology including infectious diseases, malignancy, autoimmunity and allergy (2, 3). It is believed that public TCRs result from a mixture of recombinatorial bias in the thymus and antigen-driven selection in the periphery. Public TCRα or β chains can promiscuously pair with multiple clonotypic counterchains with various CDR3 sequences while preserving antigen specificity. For example, public clonotypic HLA-B*07:02 (B7)-restricted HSV-2 VP2249–57-specific TRAV1-1 TCRα chain forms functional heterodimers with TRBV5-1, 6-1, 9, and 12-3 TCRβ chains (4). In this example, the TCRα chain appears dominant and contributes more to the overall strength of the TCR:pMHC interaction compared with paired TCRβ chains. In contrast, CD8+ T cell responses to an HLA-B7-restricted pp65265–275 epitope of human CMV was highly biased and frequently dominated by a public TRBV4-3 TCRβ chain (5). The presence of these public TCR hemichains which form antigen-specific heterodimers in conjunction with multiple clonotypic TCR counterchains suggests that either TCRα or β chain can play a dominant role in binding pMHC complexes requiring minimal contributions from the counterchain.
Defining the relative contributions of TCRα or β chain in pMHC binding has been a topic of great interest. According to crystallographic studies, either TCRα or β hemichain can be dominant depending on the particular target pMHC complexes that is recognized (6, 7). The existence of dominant TCR hemichains has also been demonstrated using other approaches. Yokosuka et al. reported that, when coexpressed with H-2Dd-restricted HIVgp160315–329-specific TRAV16N/J32 TCRα chain, clone RT-1, one-third of TRBV13-3 TCRβ chains randomly chosen from naive mouse T cells were able to generate antigen-specific TCRαβ dimers (8). Interestingly, Jβ usage affected the functional avidity of reconstituted TCRs. Using mice transgenic for the Db-restricted H-Y738–746-specific TCRβ chain, Bouneaud et al. found that this β chain was able to pair with multiple TCRα chains with various CDR3α sequences and that the TCRα structure correlated with T cell avidity (9).
MART1, a melanocyte differentiation antigen, was identified as a target of HLA-A2-restricted cytotoxic T lymphocytes (CTLs) isolated from patients with malignant melanoma (10, 11). Since MART1 is expressed by the majority of melanoma tumors but not by normal tissues except for normal melanocytes, a number of immunotherapy clinical trials have utilized MART1 as a target (12–20). It is well known that the frequency of precursor CTLs specific for A2/MART127–35 (hereafter A2/MART1) is unusually high in HLA-A2+ healthy individuals (21, 22). TCR sequencing analysis of A2/MART1 CD8+ T cell clones isolated from tumor-infiltrating lymphocytes and peripheral blood lymphocytes demonstrated a striking bias in the usage of TRAV12-2 across different individuals (23, 24). Cole et al. suggested this bias could be due to the interaction between the TRAV12-2 CDR1α loop and the peptide, describing it as “innate-like” recognition of the pMHC complex (25). Both A2/MART1 TCR, clone MEL5, and A2/HTLV-I TAX12–19 TCR, clone A6, bear TRAV12-2 TCRα chains but their CDR3α sequences are different: their TCRβ chains utilize different TRBV genes and encode distinct CDR3β sequences. Interestingly, crystallographic studies revealed that MEL5 and A6 align in nearly identical positions and orientations over the cognate pMHC complex (25, 26). Based on this, it was suggested that the Vα segments, i.e. CDR1/2 regions, of TRAV12-2 TCRα chains play a dominant role in TCR:A2 docking with minimal contributions from heterogeneous TCRβ chains, allowing TRAV12-2 TCRα chains to bind A2/peptide complexes in an ‘α-centric’ manner (25, 27, 28).
These studies suggest that a dominant TCR hemichain or TCR hemichain with chain centricity alone can largely dictate its MHC-restricted antigen specificity. However, virtually all studies analyzed peripheral T cells, which have undergone thymic selection that results in the substantial depletion of a subset of antigen-specific T cell precursors, especially those with high avidity. Accordingly, these studies may have underestimated the magnitude of heterogeneity and avidity of T cells which express a dominant TCR hemichain. Therefore, it remains to be determined, in the absence of constraints by thymic selection, how permissive a dominant TCR can be in selecting TCR counterchains while preserving antigen specificity, and how broad the range of TCR avidity can be for the cognate antigen complex.
To address these questions, we generated thymically unselected A2/MART1 TCR repertoires by transducing a public A2/MART1 TCRα chain into human peripheral T cells from HLA-A2+ and A2− donors. By utilizing an artificial antigen-presenting cell-based system, which can deliver a controlled level of T cell stimulation (29), we isolated highly polyclonal A2/MART1 T cells from these de novo A2/MART1 T cell repertoires and cloned their TCRβ chains. T cells reconstituted with a single public A2/MART1 TCRα chain along with various clonotypic TCRβ chains possessed a wide range avidities spanning 2 log orders that is solely dependent on the primary CDR3β structures. Importantly, similar findings held true with other self-antigens in addition to A2/MART1.
Materials and Methods
Cells
Peripheral blood samples were obtained from healthy donors following institutional review board approval. All donors were identified to be positive or negative for HLA-A*02:01 (A2) by high resolution HLA DNA typing (American Red Cross). Mononuclear cells were obtained by density gradient centrifugation (Ficoll-Paque PLUS; GE Healthcare). K562 is an erythroleukemic cell line defective for HLA expression. T2 is an HLA-A2 positive T cell leukemia/B-LCL hybrid cell line. SupT1 is a preTCRα+/β+ pre-T cell leukemia cell line. Jurkat 76 is a T cell leukemic cell line lacking TCR and CD8 expression (a gift from Dr. Heemskerk) (30). A375 (A2+ MART1−), Malme-3M (A2+ MART1+), SK-MEL-37 (A2+ MART1−), Me275 (A2+ NY-ESO-1+), SK-MEL-28 (A2− MART1+ NY-ESO-1−), and SK-MEL-21 (A2+ NY-ESO-1−) are melanoma cell lines. All cell lines except for melanoma cell lines were cultured in RPMI1640 supplemented with 10% FCS and gentamicin (Invitrogen) as reported previously (20, 30–34). Melanoma cell lines were grown in DMEM medium supplemented with 10% FCS and gentamicin.
cDNAs
Codon-optimized A2/MART1 TCR gene (clone SIG35α) was produced by Life Technologies (Burlingame, CA) according to the published sequence (23, 24). Except for DMF5β, 1G4α, 1G4β and 1G4LYα genes, each TCRα or β chain gene of interest was fused with ΔNGFR gene via an optimized intervening sequence consisting of a furin cleavage site, an SGSG spacer sequence, and an F2A sequence (35). Mutagenesis was conducted using standard molecular biology techniques. A2/MART1 TCR (clone DMF5) and A2/NY-ESO-1157–165 (hereafter named A2/NY-ESO-1) TCR (clone 1G4) genes were kindly provided by Dr. Rosenberg (NIH/NCI, Bethesda, MD). To clone TCR TRBV27 genes, RT-PCR was performed using TRBV27-specific primer, 5′-TRBV27 (5′-ATCCCAGTGTGGTGGTACGGGAATTCTGCCATGGGCCCCCAGCTCCTTGGC-3′), and β constant region specific reverse primers, 3′-Cβ-1 (5′-ATCGTCGACCACTGTGCTGGCGGCCGCTCGAGTTCCAGGGCTGCCTTCAGAAATCC-3′) or 3′-Cβ-2 primer (5′-GACCACTGTGCTGGCGGCCGCTCGAGCTAGCCTCTGGAATCCTTTCTCTTGACCATTGC-3′). Full-length MART1 and NY-ESO-1 cDNAs were cloned from Malme-3M and Me275 cells by RT-PCR according to published sequences, respectively. cDNAs were cloned into pMX vector and utilized to transduce all cell lines and primary human T cells (36). Nucleotide sequencing was performed at the Centre for Applied Genomics, The Hospital for Sick Children (Toronto, Canada). TCRα and β gene allele names are in accordance with IMGT unique gene nomenclatures (http://www.imgt.org/).
Peptides
Peptides used were A2-restricted wild-type MART127–35 (27AAGIGILTV35), heteroclitic NY-ESO-1157–165 (157SLLMWITQV165), and HIV pol476–484 (A2/HIV) (476ILKEPVHGV484) peptides. Synthetic peptides were obtained from ProImmune. A2/HIV pol476–484 peptide was always used as control peptide. Throughout the study, wild-type but not heteroclitic A2/MART1 peptide was utilized for expansion and functional analysis of T cells.
Transfectants
SupT1 cells reconstituted with TCRs were purified using CD3 Microbeads (Miltenyi Biotec) according to the manufacturer’s instruction. Jurkat 76 was transduced with CD8α and CD8β cDNAs to generate Jurkat 76/CD8αβ as reported previously (37). Jurkat 76 or Jurkat 76/CD8αβ transfectants were further transduced with individual TCRβ genes along with SIG35α and the transfectants were purified using CD3 Microbeads. K562-based aAPCs expressing HLA-A2 (wt-aAPC) and mutated HLA-A2 (mut-aAPC) in conjunction with CD80 and CD83 were reported elsewhere (37). Mutated HLA-A2 molecules bear two amino acid substitutions at positions 227 and 228 which abrogate the interaction with A2 (38). Mut-aAPC was engineered to constitutively secrete IL-21 to enable T cell expansion (37). 293GPG-derived retrovirus supernatants were used to introduce TCR genes into SupT1 as reported previously (37). PG13-derived retrovirus supernatants were utilized to transduce TCR genes into Jurkat 76, Jurkat 76/CD8αβ, and human primary T cells. TransIT293 (Mirus Bio) was used to transfect TCR genes into packaging cell lines. A retroviral vector encoding ΔNGFR alone was employed as a control vector. MART1-negative A375 was retrovirally transduced with full-length MART1 cDNA to generate A375/MART1. Similarly, NY-ESO-1-negative SK-MEL-21 and SK-MEL-28 were infected with retrovirus encoding full-length NY-ESO-1 cDNA to produce SK-MEL-21/NY-ESO-1 and SK-MEL-28/NY-ESO-1, respectively. HLA-A2 negative SK-MEL-28 was retrovirally transduced with wild-type HLA-A2 to generate SK-MEL-28/A2. To knockdown the MART1 gene, target cells were retrovirally infected with small-interfering RNAs against MART1 (siMART1) as reported previously (39). The target sequences of siMART1 were as follows: 5′-GAGAAGATGCTCACTTCATCT-3′, 5′-CACTCTTACACCACGGCTGAA-3′, 5′-GGCACTCAATGTGCCTTAACA-3′ and 5′-AAGACGAAATGGATACAGAGC-3′. Malme-3M was transduced with the siMART1 using retrovirus system to generate Malme-3M/siMART1 with suppressed MART1 expression. 293GPG-derived retrovirus supernatants for retroviral transduction as reported previously (32, 37). The expression of MART1 and NY-ESO-1 in the transduced cells was evaluated by western blot analysis with anti-MART1 (clone A103; Santa Crus Biotechnology) and anti-NY-ESO-1 (clone E978; Santa Crus Biotechnology), respectively. HLA-A2 expression in SK-MEL-28/A2 cells was analyzed by flow cytometry following staining with anti-HLA-A2 (clone BB7.2; Biolegend) as reported previously (32).
Expansion of TCR gene-modified CD8+ T cells in an HLA-A2-restricted peptide-specific manner
Peptide-specific CD8+ T cells were expanded using an aAPC as described previously (31, 32, 40–42). PBMCs were isolated from healthy volunteers and stimulated with 50 ng/ml anti-CD3 mAb (clone OKT3) in the presence of 100 IU/ml human IL-2 (Novartis) 3 days before transduction. Activated T cells were retrovirally transduced with TCR genes by centrifuging 1 hour at 1,000 g at 32°C. Following transduction, CD8+ T cells were purified and plated at 2 × 106 cells/well in RPMI 1640 supplemented with 10% human AB serum. The stimulator wt-aAPC or mut-aAPC was pulsed with 10 μg/ml A2-restricted wild-type MART127–35 or heteroclitic NY-ESO-1157–165 peptide for 6 hours at room temperature. The aAPC was then irradiated at 200 Gy, washed, and added to the responder T cells at a responder to stimulator ratio of 20:1. Starting the next day, 10 IU/ml IL-2 (Novartis) and 10 ng/ml IL-15 (Peprotech) were added to the cultures every three days. T cells were harvested, counted, and restimulated every week. T cell analysis was performed one day prior to or on the day of restimulation. A2/HIV pol476–484 peptide was used as a control.
Flow cytometry analysis
Cell surface molecules on transfectants were counterstained with PC5-conjugated anti-CD8 mAb (clone B9.11, Beckman Coulter), FITC-conjugated anti-NGFR (clone ME20.4; Biolegend), and FITC-conjugated anti-CD3 (clone UCHT1; Biolegend). Assessment of TCR Vβ subfamily usage was performed using TCR Vβ mAbs (Beta Mark, Coulter, CA) as reported previously (31). Stained cells were analyzed with flow cytometry (BD Biosciences) and data analysis was performed using FlowJo (TreeStar) as published previously (40–43).
HLA/peptide multimer staining
Biotinylated HLA-A2/peptide monomers were purchased from ProImmune, multimerized in-house using SA-PE and SA-APC, and utilized to stain antigen-specific T cells as described previously (20, 37, 44, 45). A2/HIV multimer was always used as a control. Structural avidity was determined by staining with graded concentrations of A2/MART1 multimer.
Cytokine ELISPOT analysis
IL-2 and IFN-γ ELISPOT assays were conducted as described elsewhere (37, 43–45). Briefly, PVDF plates (Millipore) were coated with capture mAb. T cells were incubated with 2 × 104 per well of T2 cells in the presence of wild-type A2/MART127–35 peptide for 20–24 hours at 37°C. Plates were washed and incubated with biotin-conjugated detection mAb. Functional avidity was tested using T2 cells pulsed with graded concentrations of wild-type A2/MART127–35 peptide as stimulators in ELISPOT assays as reported previously (37).
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 5.0e. To determine whether two groups were statistically different for a given variable, analysis was performed using the Welch’s t test (two-sided). P values of < 0.05 were considered significant.
Results
When paired with the endogenous irrelevant SupT1 TCRβ chain, SIG35α, but not DMF5α, recognizes A2/MART1
The A2/MART1 TCRα gene, clone SIG35α, (hereafter called SIG35α) utilizes TRAV12-2/J35. Although this TRAV-J usage does not match with the previously described public A2/MART1 TCR, TRAV12-2/J34 or J45, SIG35α has been repeatedly isolated from A2/MART1 CTLs by many groups including us (3, 20, 23, 24, 46). SIG35α has been shown to pair with TRBV5-1 and TRBV27 TCRβ chains with diverse CDR3β sequences, suggesting that recognition of A2/MART1 by SIG35α containing TCRs is α chain centric (Table I). SupT1 is a human pre-T cell leukemia cell line, which expresses preTCRα and TRBV9/J2-1 TCRβ chains but not a mature TCRα chain (47). This suggests that the SupT1 was derived from T cells, which had yet to experience HLA-restricted selection in the thymus. When transduced with SIG35α, SupT1 cells were successfully stained by A2/MART1 multimer but not control A2/HIV multimer (Fig. 1, left). In contrast, SupT1 cells transfected with the TCRα gene from the high affinity A2/MART1 TCR, clone DMF5 (called DMF5 hereafter), which harbors TRAV12-2/J23, was not stained by A2/MART1 multimer (48). Surface CD3 expression on both transfectants was similarly upregulated confirming the successful transduction and surface expression of both TCRα genes (Fig. 1, left). Supra-transduction of DMF5β into SupT1 transduced with DMF5α rendered the tranfectant positive for A2/MART1 multimer staining, further confirming the successful transduction of DMF5α. These results indicate that, compared to DMF5α, SIG35α plays a dominant role in the recognition of A2/MART1 and requires minor contributions from TCRβ chains to determine its A2-restricted MART127–35 epitope specificity.
Table I.
CDR3 sequences of TCRβ chains paired with SIG35α in A2/MART1 T cells
Fig. 1. SIG35α but not DMF5α expressing SupT1 cells are stained by A2/MART1 multimers when paired with the endogenous irrelevant TCRβ chain of SupT1 cells.
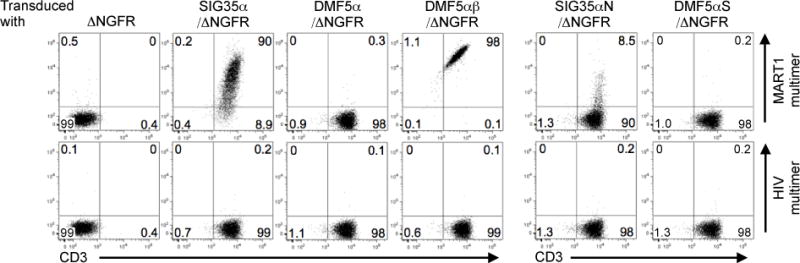
The human TCRα−/β+ pre-T cell leukemia cell line, SupT1 was transduced with 5 different clonotypic A2/MART1-specific TCRα chains, SIG35α/ΔNGFR, DMF5α/ΔNGFR, SIG35αN/ΔNGFR, or DMF5αS/ΔNGFR, or TCRαβ chains, DMF5αβ/ΔNGFR. SIG35αN is a SIG35α-derived mutant encoding Asn instead of Ser at the V-J junction. DMF5 is a high affinity A2/MART1 TCR (48). DMF5αS is a DMF5α-derived mutant coding for Ser instead of Asn at the V-J junction. All TCRα genes were fused with ΔNGFR gene via an optimized intervening sequence consisting of a furin cleavage site, an SGSG spacer sequence, and an F2A sequence (35). ΔNGFR alone was employed as a control. The transduction efficiency of SupT1 transfectants was approximately 90% as determined by the percentage of ΔNGFR+ cells (data not shown). All SupT1 transfectants were stained with A2/MART1 or A2/HIV multimer along with anti-CD3 mAb. Data shown are gated on ΔNGFR+ cells and a representative of two independent experiments.
The CDR3α regions of SIG35α and DMF5α encode CAVSIGFGNVL and CAVNFGGGKLIF, respectively. SIG35α but not DMF5α harbors a flexible amino acid, Ser at the V-J junction as underlined. When SIG35αN, which is a SIG35α-derived mutant encoding Asn in lieu of Ser, was transduced into SupT1 cells, positive A2/MART1 multimer staining was largely lost, suggesting that the Ser residue was critical for the chain centricity of SIG35α (Fig. 1, right). The DMF5α mutant, DMF5αS, which carries a Ser residue instead of Asn at the V-J junction, was not able to acquire stronger chain centricity compared to parental DMF5α. This indicates that the mere existence of a flexible amino acid, Ser, at the V-J junction is not sufficient to confer chain centricity to A2/MART1 TCRα genes.
Both HLA-A2+ and A2− peripheral T cells recognize A2/MART1 when transduced with chain-centric SIG35α
Peripheral T cells from 4 donors, 2 each for HLA-A2+ and A2− individuals, were transduced with SIG35α alone and stained by A2/MART1 multimer (Fig. 2A). To distinguish A2/MART1 T cells derived from untransduced and transduced T cells, the SIG35α gene was fused to the ΔNGFR gene by the F2A sequence as in Fig. 1. The overall transduction efficiency of peripheral T cells was approximately 50–85% as determined by the percentage of ΔNGFR+ cells (Supplementary Fig. 1A). ΔNGFR and A2/MART1 multimer double positive cells were detectable in all donors tested regardless of their HLA-A2 positivity. Previously, we reported a series of human cell-based artificial antigen-presenting cells (aAPCs), which can expand in vitro antigen-specific CD4+ and CD8+ T cells, and polyclonal CD3+ T cells (31, 32, 37, 40–45). When exogenously pulsed with wild-type A2/MART1 peptide, aAPCs expressing wild-type HLA-A2 (wt-aAPC) or mutated HLA-A2 (mut-aAPC) successfully expanded A2/MART1 T cells (Fig. 2B). Mut-aAPCs express mutated A2 molecules which cannot engage CD8 coreceptors so that they specifically expand a subset of A2-restricted antigen-specific T cells with higher avidity (see below) (37). Importantly, when not exogenously pulsed with wild-type A2/MART1 peptide, both aAPCs failed to grow A2/MART1 T cells suggesting that the observed expansion of A2/MART1 T cells is dependent on pulsed A2/MART1 peptide (data not shown). The T cells expressing SIG35α recognized A2+ target cells pulsed with wild-type A2/MART1 but not A2/HIV control peptide (Fig. 2C). Furthermore, they were capable of targeting unpulsed A2+ MART1+ Malme-3M tumor cells but not A2+ MART1− A375 tumor cells, suggesting that the SIG35α-transduced T cells possessed functional avidity sufficient to recognize endogenously processed and presented A2/MART1 peptide.
Fig. 2. Both HLA-A2+ and A2− peripheral T cells can recognize A2/MART1 when transduced with chain-centric SIG35α.
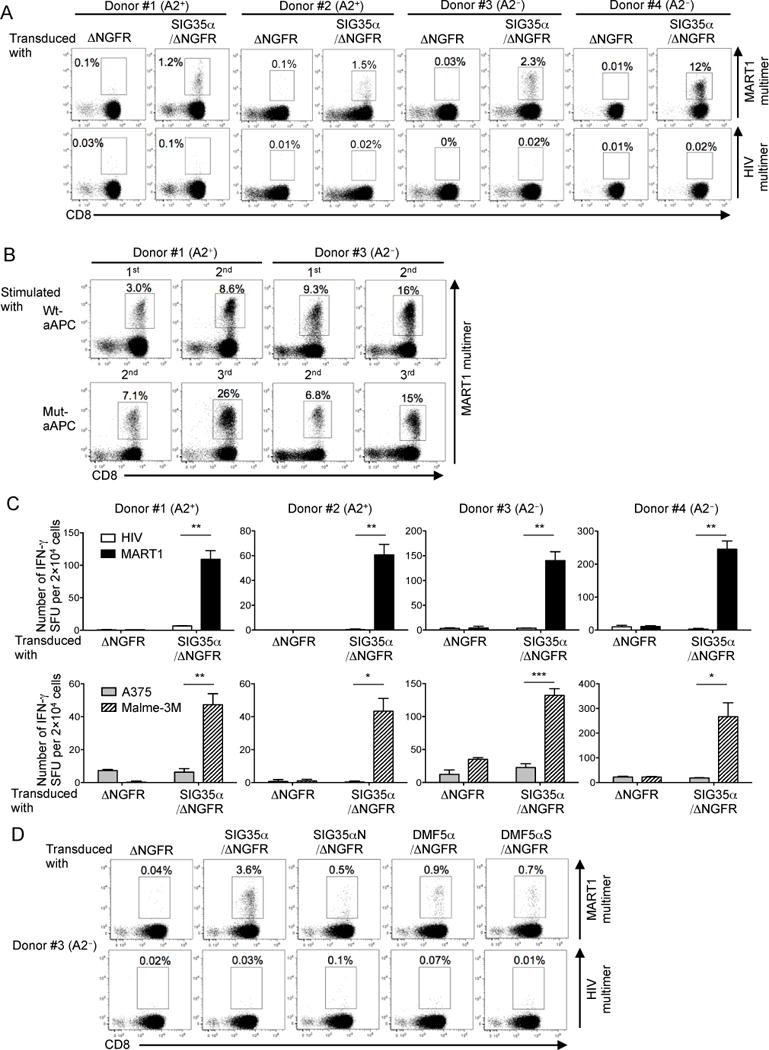
(A), Both HLA-A2+ and A2− peripheral T cells become A2/MART1-reactive upon transduction of chain-centric SIG35α. Peripheral CD8+ T cells freshly isolated from two HLA-A2+ donors (#1 and #2) and two A2− donors (#3 and #4) were retrovirally transduced with ΔNGFR or SIG35α/ΔNGFR and stained with A2/MART1 multimer or A2/HIV multimer in conjunction with anti-CD8 mAb and anti-NGFR mAb. Data shown are gated on ΔNGFR+ cells. Data of donors #1 and #3 are representative of three independent experiments and data of donors #2 and #4 are representative of two independent experiments. (B), SIG35α-transduced A2/MART1 CD8+ T cells expand in an A2/MART1-specific manner. A2+ and A2− CD8+ T cells transduced with SIG35α/ΔNGFR were stimulated with wt-aAPC or mut-aAPC pulsed with wild-type A2/MART1 peptide once a week. Between stimulations, the T cells were supplemented with IL-2 (10 IU/ml) and IL-15 (10 ng/ml) every 3 days. Data depict A2/MART1 multimer staining performed following the first and second stimulations using wt-aAPC and the second and third stimulations utilizing mut-aAPC. Data shown are gated on ΔNGFR+ cells. Representative multimer-staining data from one of two HLA-A2+ donors and one of two A2− donors are shown. (C), Peripheral T cells transduced with SIG35α are highly avid for A2/MART1 recognition. CD8+ T cells following stimulation with wt-aAPC pulsed with wild-type A2/MART1 peptide were used as responder cells in IFN-γ ELISPOT analysis. T2 cells pulsed with 10 μg/ml A2/HIV control peptide or wild-type A2/MART1 peptide were used as stimulator cells (top). The A2+ MART1− melanoma line, A375, and the A2+ MART1+ melanoma line, Malme-3M, were used as stimulator cells (bottom). Data shown are representative of two independent experiments. All experiments were carried out in triplicate and error bars depict SD. *P < 0.05, **P < 0.01, ***P < 0.001. (D), Peripheral CD8+ T cells isolated from an A2− donor #3 were transduced with ΔNGFR, SIG35α/ΔNGFR, SIG35αN/ΔNGFR, DMF5α/ΔNGFR, or DMF5αS/ΔNGFR and stained by A2/MART1 multimer or A2/HIV multimer in conjunction with anti-CD8 mAb and anti-NGFR mAb as described in Fig. 1. Data shown are gated on ΔNGFR+ cells.
Using primary T cells, we also confirmed the significance of the Ser residue located at the V-J junction of SIG35α for its chain centricity shown in Fig. 1. When the SIG35αN mutant was transduced into primary T cells, positivity for A2/MART1 multimer staining drastically decreased compared to when SIG35α was transduced (Fig. 2). The DMF5αS mutant could not significantly upregulate the A2/MART1 multimer positivity over parental DMF5α, again suggesting that a Ser residue is insufficient for the observed α chain centricity of SIG35α and only critical in the context of surrounding CDR3α sequences. As shown in Fig. 2D, although DMF5α-transduced CD8+ T cells were also stained by A2/MART1 multimer, the percentage of A2/MART1 multimer+ T cells was substantially lower compared to SIG35α-transduced T cells. We further compared the A2/MART1 multimer positivity of the T cells transduced with SIG35α or DMF5α in 3 other donors prior to and following antigen-specific expansion (Supplementary Fig. 1B). The percentage of A2/MART1 multimer+ T cells in DMF5α-transduced T cells was consistently lower compared to SIG35α transduced T cells, suggesting that, compared to DMF5α, SIG35α requires less contribution from TCRβ counter-chains to recognize A2/MART1.
SIG35α predominantly pairs with TRBV27 TCRβ chains to recognize A2/MART1
SIG35α expressed in A2/MART1+ T cells paired with various Vβ subfamilies in both A2-positive and negative donors (Fig. 3A). The percentage of the overall transduced T cells expressing each Vβ subfamily is shown in Supplementary Fig. 2. Intriguingly, SIG35α predominantly paired with TRBV27 TCRβ chains to recognize A2/MART1 in all 4 donors tested, which was often observed with A2/MART1-specific T cells isolated from the periphery or tumor sites (23, 24, 31, 49–52). When SIG35α-transduced T cells were costained with anti-TRBV27 mAb and A2/MART1 multimer, large fractions of up to 75% of peripheral TRBV27+ cells were double positive for SIG35α and A2/MART1 multimer (Fig. 3B). These results demonstrate that A2/MART1 specific TCRs can be generated by pairing SIG35α with a large portion of the unrelated TRBV27 TCRβ chain repertoire. Furthermore, they also suggest that the TRBV27 CDR1 and 2β but not CDR3β region primarily regulate the A2/MART1 specificity of SIG35α containing TCRs.
Fig. 3. SIG35α predominantly pairs with TRBV27 TCRβ chains to recognize A2/MART1.
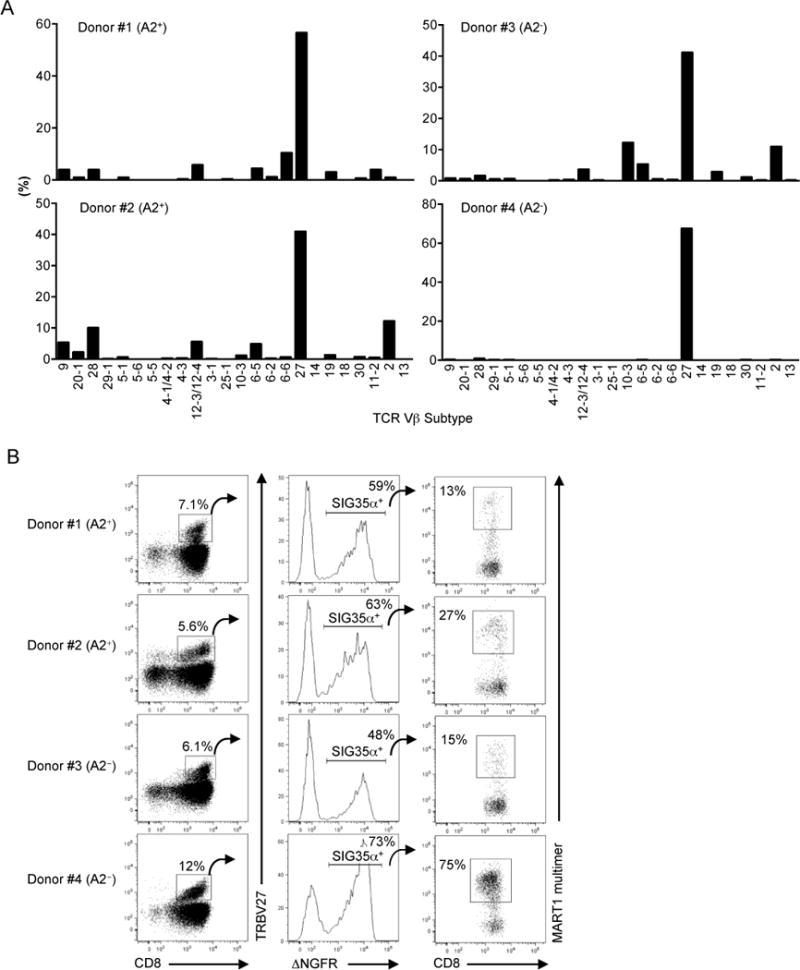
(A), SIG35α/ΔNGFR-transduced peripheral CD8+ T cells from two HLA-A2+ and two A2− donors were stained with A2/MART1 multimer, mAbs for TCR Vβ subtypes, and anti-CD8 mAb. The percentage of A2/MART1 multimer+ CD8+ T cells expressing each subtype is shown. Data shown are gated on ΔNGFR+ cells. (B), A significant proportion of TRBV27 TCRβ chains in the periphery can recognize A2/MART1 when paired with SIG35α. The percentage of A2/MART1 multimer+ cells in CD8+ TRBV27+ T cells transduced with SIG35α/ΔNGFR gene is shown.
TRBV27 TCRβ chains that can recognize A2/MART1 when paired with SIG35α are highly heterogeneous and unique
In order to assess the CDR3β heterogeneity of TRBV27 TCRβ genes which paired with SIG35α for A2/MART1 reactivity, TRBV27 TCRβ genes were molecularly cloned from SIG35α+ A2/MART1 multimer positive cells from one A2+ and one A2− donor. Wt- and mut-aAPCs were used to expand SIG35α-transduced T cells with a broad range of avidity. Sequence analysis of the CDR3β region revealed that cloned TRBV27 TCRβ genes were highly heterogeneous (Fig. 4, Table II). We isolated a total of 139 and 38 independent clonotypic TCRβ chains from wt- and mut-aAPCs, respectively, with highly diverse CDR3β sequences and amino acid lengths. No clonotypic TCRβ gene was shared between the two donors (Table II). Only three TCRβ gene clones were shared between A2/MART1 T cells obtained after stimulation with wt- and mut-aAPCs in the A2− donor. No clonotypic TCRβ gene was shared by the T cells expanded by wt- and mut-aAPCs in the A2+ donor. Furthermore, except for Jβ1–3 and 1–6, all Jβ subfamilies were utilized (Fig. 4). These results demonstrate that SIG35α can pair with a highly diverse repertoire of TRBV27 TCRβ chains to constitute a TCR specific for A2/MART1. This confirms that the TRBV27 CDR3β region does not play a significant role in determining the A2/MART1 specificity of SIG35α.
Fig. 4. TRBV27 TCRβ chains which recognize A2/MART1 when paired with SIG35α are highly heterogeneous and unique.
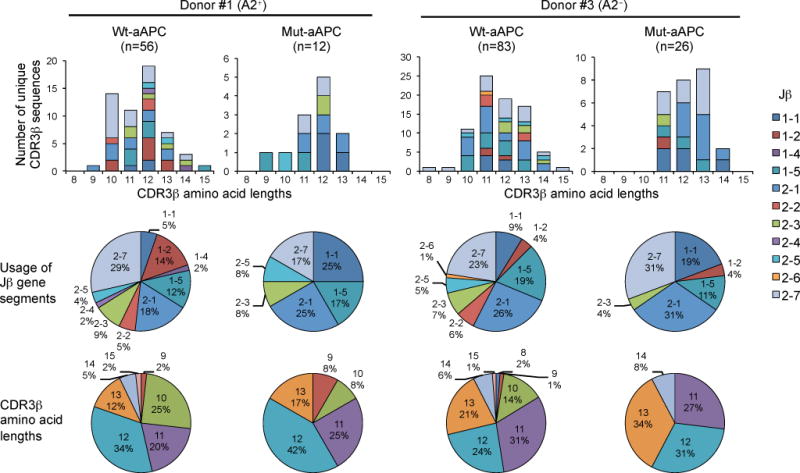
SIG35α/ΔNGFR-transduced CD8+ T cells from the HLA-A2+ donor #1 and the A2− donor #3 were stimulated with wt-aAPC or mut-aAPC pulsed with wild-type A2/MART1 peptide. A2/MART1 multimer+ CD8+ T cells were collected by fluorescence activated cell sorting (>99% purity) and their TRBV27 CDR3β regions were amplified by PCR and sequenced after cloning. The number of unique CDR3β sequences (top), the relative usage of Jβ gene segments (middle), and the CDR3β amino acid lengths (bottom) are depicted separately for the A2+ donor #1 (left) and A2− donor #3 (right). Data were analyzed by the aAPC used for stimulations, wt-aAPC vs. mut-aAPC, in each donor.
Table II.
Sequencing results of TCR TRBV27 chains isolated from A2/MART1 multimer+ CD8+ T cells
| Wt-aAPC stimulation | Mut-aAPC stimulation | Number of shared clonotypes | |||
|---|---|---|---|---|---|
|
|
|||||
| Donor | Number of unique clonotypes | Number of isolates sequenced | Number of unique clonotypes | Number of isolates sequenced | |
| #1 (A2+) | 56 | 190 | 12 | 19 | 0 |
| #3 (A2−) | 83 | 122 | 26 | 89 | 3 |
| Total | 139 | 312 | 38 | 108 | 3 |
The avidity range of A2/MART1 TCRs consisting of SIG35α is very broad and further enhanced by the presence of CD8
To study the avidity range of SIG35α+ A2/MART1 T cells, we randomly selected 5 and 6 clonotypic TRBV27 TCRβ chain genes cloned from SIG35α+ A2/MART1 T cells stimulated by wt- and mut-aAPCs, respectively. These 11 clonotypic TRBV27 TCRβ genes were individually reconstituted along with SIG35α on TCR−/− Jurkat 76 T cells in the presence or absence of CD8αβ (Fig. 5A, Supplementary Fig. 3). All 12 transfectants including the one expressing DMF5 demonstrated comparable surface CD3 expression, suggesting the equivalent expression level of transduced TCR genes (Fig. 5A, top). Except for those expressing Cl. 413 and 523, all transfectants were stained by A2/MART1 multimer in the absence of CD8αβ coreceptor expression, suggesting high structural avidity. When coexpressed with CD8αβ, these two clones became positive for the multimer albeit at a lower level (Fig. 5A, bottom). Coexpression of CD8αβ molecules also enhanced the A2/MART1 multimer staining of other transfectants with higher structural avidity.
Fig. 5. The structural and functional avidity range of A2/MART1 TCRs consisting of SIG35α is very broad and further enhanced by the presence of CD8.
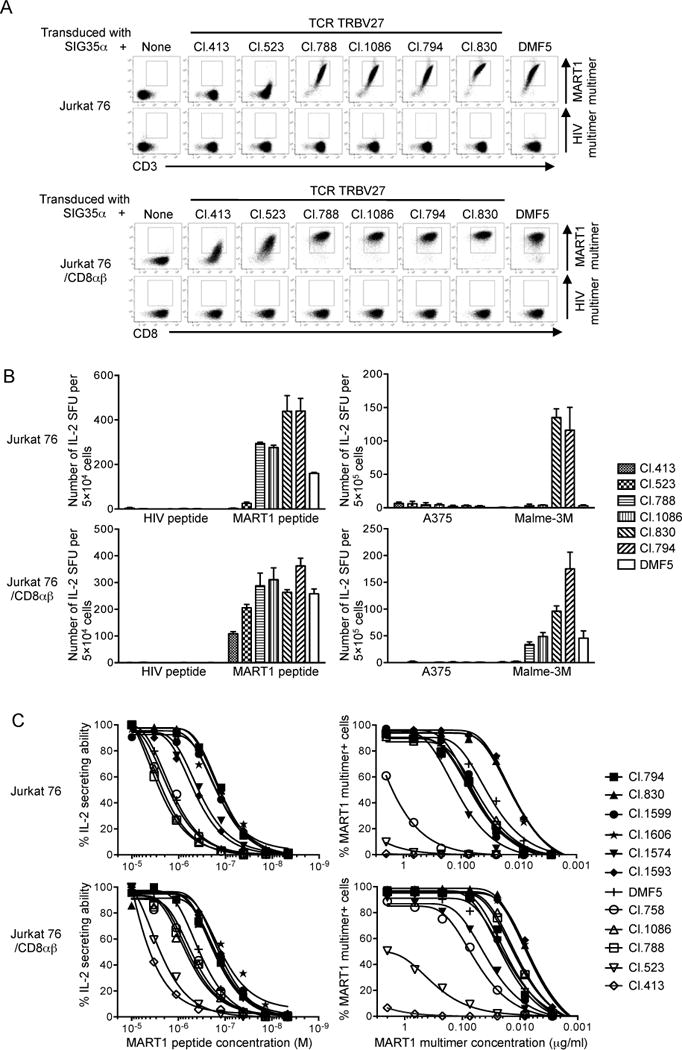
Jurkat 76 cells, which lack the expression of CD8αβ and endogenous TCRs, were retrovirally transduced with CD8αβ to produce Jurkat 76/CD8αβ. Jurkat 76 or Jurkat 76/CD8αβ cells were individually transduced with eleven distinct TRBV27 TCRβ chains along with SIG35α or with DMF5αβ chains. (A), A2/MART1 TCRs reconstituted on Jurkat 76 or Jurkat 76/CD8αβ cells were differentially stained by A2/MART1 multimer. All Jurkat 76 or Jurkat 76/CD8αβ transfectants were stained with 2 μg/ml A2/MART1 or A2/HIV multimer along with anti-CD3 mAb (top) or anti-CD8 mAb (bottom). Data for multimer staining of seven representative Jurkat 76 or Jurkat 76/CD8αβ transfectants are shown. Data for multimer staining of the remaining 5 transfectants are shown in Supplementary Fig. 3. (B), Reconstituted A2/MART1 TCRs are highly avid for A2/MART1 recognition. IL-2 ELISPOT assays were performed using seven representative Jurkat 76 or Jurkat 76/CD8αβ transfectants as responder cells. T2 cells pulsed with 10 μg/ml wild-type A2/MART1 or A2/HIV control peptide were used as stimulator cells (left). The A2+ MART1− melanoma line, A375, and the A2+ MART1+ melanoma line, Malme-3M were used as stimulator cells (right). All experiments were conducted in triplicate and error bars show SD. Data shown are representative of two independent experiments. (C), Reconstituted A2/MART1 TCRs possess a broad range of functional and structural avidities. Functional avidities of Jurkat 76 or Jurkat 76/CD8αβ cells expressing 11 different A2/MART1 TCRβ chains paired with SIG35α and DMF5 are depicted as % IL-2 secreting abilities determined by IL-2 ELISPOT assays using T2 cells pulsed with graded concentrations of wild-type A2/MART1 peptide as stimulator cells (left). Structural avidities of the same transfectants are shown as multimer staining percentages determined by staining with graded concentrations of A2/MART1 multimer (right). Data shown are representative of two independent experiments.
Except for the one expressing Cl. 413, all Jurkat 76 transfectants tested recognized wild-type A2/MART1 peptide pulsed on target cells in the absence of CD8αβ coreceptor (Fig. 5B, left). Coexpression of CD8αβ enabled the Jurkat 76 cells expressing Cl. 413 to also be reactive (Fig. 5B, left). Jurkat 76 transfectants expressing Cl. 830 and 794 possessed higher functional avidity compared with other transfectants and recognized A2+ MART1+ Malme-3M tumor cells in the absence of CD8αβ (Fig. 5B, right). In our experimental condition, the functional avidity of DMF5-transduced CD8αβ− Jurkat 76 cells was insufficient to recognize A2+ MART1+ Malme-3M tumor cells. However, when CD8αβ molecules were expressed, all transfectants, except for the ones expressing Cl. 413 and 523, were able to recognize A2+ MART1+ Malme-3M tumor cells. The Cl. 413- and 523-expressing transfectants were unable to detect Malme-3M even in the presence of CD8αβ coexpression (Fig. 5B, right). To further demonstrate the specific recognition of A2+ MART1+ tumor cells by the reconstituted A2/MART1 TCRs, we generated A2+ MART1+ A375/MART1, A2+ MART1+ SK-MEL-28/A2, and A2+ MART1low Malme-3M/siMART1 cells (Supplementary Fig. 4A, B). Using these tumor cells as target cells, we demonstrated the A2/MART1-restricted recognition by the Jurkat 76/CD8αβ transfectants individually expressing the eleven distinct clonotypic A2/MART1 TCRs (Table III). Furthermore, we evaluated the cross-reactivity of these TCRs to MART1-related peptides derived from normal human proteins, which were reported by Dutoit et al. (53) (Table IV). The number of MART1-related peptides recognized by the 11 Jurkat 76/CD8αβ TCR transfectants (2.6 ± 1.0, mean ± SD) is not significantly higher compared to the 5 A2/MART1 CTL clones (2.4 ± 1.7, mean ± SD) reported by Dutoit’s group (53).
Table III.
Specific recognition of HLA-A2+ MART1+ tumor cells by Jurkat 76/CD8αβ reconstituted with A2/MART1 TCRs
| TRBV27 chain
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stimulator cells | HLA-A2 | MART1 | Cl.413 | Cl.523 | Cl.788 | Cl.1086 | Cl.758 | Cl.1593 | Cl.1574 | Cl.1599 | Cl.1606 | Cl.830 | Cl.794 | DMF5 |
| A375 | + | − | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| A375/MART1 | + | + | <10 | <10 | 21 (6) | 20 (5) | 45 (7) | 98 (8) | 55 (9) | 170 (3) | 94 (10) | 153 (8) | 197 (10) | 33 (3) |
| Malme-3M/siControl | + | + | <10 | <10 | 18 (5) | 19 (8) | 64 (6) | 73 (15) | 60 (11) | 143 (18) | 76 (10) | 99 (8) | 151 (15) | 39 (5) |
| Malme-3M/siMART1 | + | Low | <10 | <10 | 14 (5) | 14 (6) | 50 (6) | 54 (5) | 45 (9) | 99 (10) | 48 (4) | 75 (9) | 113 (11) | 27 (5) |
| SK-MEL-28 | − | + | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| SK-MEL-28/A2 | + | + | <10 | <10 | 34 (6) | 31 (4) | 67 (9) | 101 (14) | 81 (4) | 128 (14) | 70 (6) | 115 (10) | 147 (10) | 40 (6) |
| SK-MEL-37 | + | − | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| K562 | − | − | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
Tumor cell recognition was assessed using various target cells, which did or did not express HLA-A2 and/or MART1, as stimulator cells in IL-2 ELISPOT assays. A half million Jurkat 76/CD8αβ cells, which were individually transduced with eleven distinct clonotypic TRBV27 TCRβ chains along with SIG35α or with DMF5αβ chains, were used as responder cells. Mean values of SFUs in triplicate samples are shown. SDs of triplicates are shown in parentheses.
Table IV.
Recognition of MART1–related peptides by Jurkat 76/CD8αβ reconstituted with A2/MART1 TCRs
| TRBV27 chain
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Protein | Peptide sequence | Cl.413 | Cl.523 | Cl.788 | Cl.1086 | Cl.758 | Cl.1593 | Cl.1574 | Cl.1606 | Cl.1599 | Cl.830 | Cl.794 | DMF5 |
| 1 | KIAA0935 | RVTDEAGHPV | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| 2 | cMOAT2 | NVADIGLHDV | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| 3 | SLC1A1 | VLTGLAIHSI | <10 | <10 | <10 | <10 | <10 | <10 | 29 | 42 | <10 | <10 | <10 | <10 |
| 4 | P47 | RISDIRLFIV | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| 5 | Prostaglandin transporter | LLAGIGTVPI | 100 | 248 | 153 | 100 | 205 | 272 | 296 | 277 | 241 | 189 | 283 | 174 |
| 6 | ABC transporter MOAT-C | RISDIGLADL | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| 7 | KIAA0735 | LISGIGIGGA | <10 | 33 | <10 | <10 | <10 | 32 | 100 | 111 | 31 | 23 | 36 | <10 |
| 8 | Hypothetical 20 kD protein | RISAIILHPN | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| 9 | Endothelin-1 receptor | RVQGIGIPLV | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| 10 | G-protein coupled receptor RE2 | RITDLGLSPH | <10 | 64 | 35 | <10 | 124 | 43 | 215 | 213 | 168 | 17 | 248 | 80 |
| 11 | IGHG1 | RLSELAIFGV | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| 12 | Monocarboxylate transporter 8 | AVAFIGLHTS | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| 13 | MRP3 | NVADIGFHDV | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| 14 | Wild type MART1 | AAGIGILTV | 20 | 102 | 100 | 163 | 198 | 270 | 235 | 216 | 218 | 112 | 272 | 144 |
Peptide recognition was assessed using T2 cells loaded with 10 μg/ml of the indicated peptide as stimulator cells in IL-2 ELISPOT assays. Thirteen MART1-related peptides were reported by Dutoit et al. (53). Fifty thousand Jurkat 76/CD8αβ cells, which were individually transduced with eleven distinct TRBV27 TCRβ chains along with SIG35α or with DMF5αβ chains, were used as responder cells. Mean values of SFUs in triplicate samples are shown.
We then systemically evaluated and compared structural and functional avidities of all Jurkat 76 transfectants in the absence or presence of CD8αβ. As shown in Fig. 5C, these transfectants demonstrated a wide range of structural and functional avidities which can be generally augmented by the CD8αβ coexpression. Data for structural and functional avidities of all transfectants are summarized in Table V. These results demonstrate that A2/MART1 T cells expressing SIG35α can possess a broad spectrum of avidity (>2 log orders), which is regulated by the CDR3β sequence in the context of CDR1/2β sequence of the TCR counterchains.
Table V.
Functional and structural avidities of the A2/MART1 TCRs
| Clone | Donor | aAPC used for stimulation | TRBV | CDR3β | TRBJ | Functional avidity* without CD8 | Functional avidity with CD8 | Structural avidity† without CD8 | Structural avidity with CD8 |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| EC50 (μM) | EC50 (μM) | EC50 (μg/ml) | EC50 (μg/ml) | ||||||
| Cl.794 | #3 (A2−) | Mut-aAPC | 27 | CASSLLGDYGYTF | 1-2 | 0.12 | 0.16 | 0.06 | 0.02 |
| Cl.830 | #3 (A2−) | Mut-aAPC | 27 | CASSLGGAYEQYF | 2-7 | 0.13 | 0.14 | 0.01 | 0.006 |
| Cl.1599 | #1 (A2+) | Mut-aAPC | 27 | CASSFLGAMAEAFF | 1-1 | 0.14 | 0.16 | 0.06 | 0.02 |
| Cl.1606 | #1 (A2+) | Mut-aAPC | 27 | CASSLLGSYEQYF | 2-7 | 0.16 | 0.12 | 0.01 | 0.006 |
| Cl.1574 | #1 (A2+) | Mut-aAPC | 27 | CASSPWERINTEAFF | 1-1 | 0.35 | 0.15 | 0.1 | 0.03 |
| Cl.1593 | #1 (A2+) | Mut-aAPC | 27 | CASGNNQPQHF | 1-5 | 0.44 | 0.14 | 0.01 | 0.006 |
| DMF5‡ | 6-4 | CASSLSFGTEAFF | 1-1 | 1.4 | 0.33 | 0.03 | 0.02 | ||
| Cl.758 | #3 (A2−) | Wt-aAPC | 27 | CASSPRLAGDGELFF | 2-2 | 1.6 | 0.46 | 2.7 | 0.06 |
| Cl.1086 | #3 (A2−) | Wt-aAPC | 27 | CASSLHGPGGYTF | 1-2 | 2.4 | 0.63 | 0.04 | 0.01 |
| Cl.788 | #3 (A2−) | Wt-aAPC | 27 | CASGPSYEQYF | 2-7 | 2.9 | 0.57 | 0.05 | 0.01 |
| Cl.523 | #3 (A2−) | Wt-aAPC | 27 | CASGSYEQYF | 2-7 | n.m | 2.7 | n.m | 0.3 |
| Cl.413 | #1 (A2+) | Wt-aAPC | 27 | CASSVFGGDMGEKLFF | 1-4 | n.m | 10 | n.m | n.m |
n.m, not measurable.
Functional avidity, expressed as EC50 in μM, was defined as the concentration of peptide required to achieve 50% of maximal response.
Structural avidity, expressed as EC50 in μg/ml, was defined as the concentration of A2/MART1 multimer required to achieve half maximal multimer staining.
TCR chain centricity is commonly observed with HLA-restricted antitumor TCRs
We next investigated whether the observed TCR chain centricity is unique to A2/MART1, which is known to have an exceptionally high precursor frequency (21, 22), or ubiquitous to other HLA-restricted tumor-associated antigens. The TCR gene, clone 1G4, is specific for A2/NY-ESO-1 peptide (54, 55). The TCR 1G4α and β chains harbor TRAV21/J6 and TRBV6-5/J2-2, respectively. A TCRα chain 1G4α variant, called clone 1G4LYα, derived from 1G4 carries two amino acid substitutions at the CDR3α region, which demonstrates a higher TCR affinity when paired with 1G4β (56). Peripheral T cells transfected with any of 1G4α, 1G4β, or 1G4LYα showed positivity for A2/NY-ESO-1 multimer staining (Fig. 6A, left). The expanded A2/NY-ESO-1-specific T cells expressing 1G4α were polyclonal but predominantly positive for TRBV6-5 (Fig. 6A, right). The T cells expressing 1G4α hemichain recognized A2+ target cells pulsed with A2/NY-ESO-1 but not A2/HIV control peptide (Fig. 6B, left). Furthermore, they were capable of recognizing unpulsed A2+ NY-ESO-1+ Me275 tumor cells but not A2+ NY-ESO-1− SK-MEL-21 nor A2− NY-ESO-1− SK-MEL-28 tumor cells (Fig. 6B, right). To further confirm the specificity of the T cells expressing 1G4α hemichain, A2-positive SK-MEL-21 and A2-negative SK-MEL-28 were ectopically transduced with full-length NY-ESO-1 to generate A2+ SK-MEL-21/NY-ESO-1 and A2− SK-MEL-28/NY-ESO-1 (Supplementary Fig. 4C). The 1G4α hemichain-transduced T cells recognized A2+ NY-ESO-1+ SK-MEL-21/NY-ESO-1 but not A2− NY-ESO-1+ SK-MEL-28/NY-ESO-1 (Fig. 6B, right). These results strongly suggest that the 1G4α hemichain-transduced T cells possess functional avidity sufficient to recognize naturally processed and presented A2/NY-ESO-1 peptide in a specific manner. Taken all together, these results strongly suggest that the observed chain centricity of HLA-restricted self-antigen-specific TCRs is a prevailing phenomenon, and can be exploited to efficiently isolate highly avid T cells and encoded TCRs specific for any HLA-restricted tumor-associated and pathogen-derived antigen.
Fig. 6. TCR chain centricity is observed with other HLA-restricted antitumor TCRs.
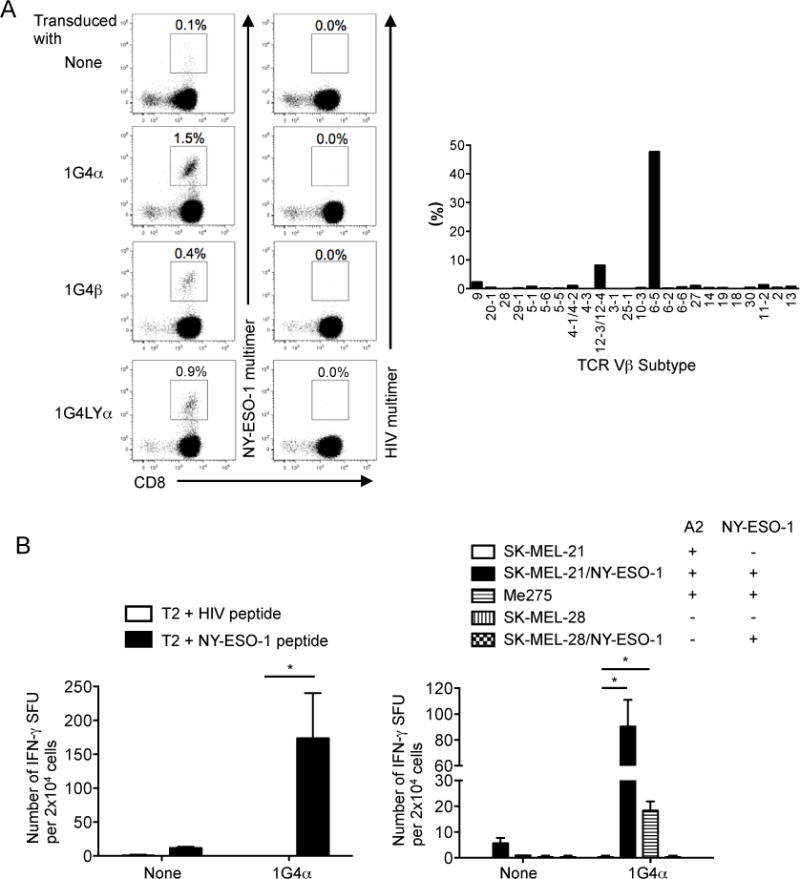
(A), Peripheral CD8+ T cells transduced with 1G4α, 1G4β, or 1G4LYα recognize A2/NY-ESO-1. Peripheral CD8+ T cells transduced with 1G4α, 1G4β, or 1G4LYα were stimulated with IL-21-secreting wt-aAPC pulsed with heteroclitic A2/NY-ESO-1 peptide once a week. Between stimulations, IL-2 (10 IU/ml) and IL-15 (10 ng/ml) were added every 3 days. Data for A2/NY-ESO-1 multimer staining conducted after second stimulation are shown (left). Data are representative of two donors. 1G4α-transduced CD8+ T cells were costained with A2/NY-ESO-1 multimer, mAbs for TCR TRBV subtypes, and anti-CD8 mAb. The percentage of A2/NY-ESO-1 multimer+ CD8+ T cells expressing each subtype after second stimulation is shown (right). (B), Peripheral T cells transduced with 1G4α hemichain are highly avid for A2/NY-ESO-1 recognition. 1G4α-transduced or non-transduced CD8+ T cells were stimulated with mut-aAPC pulsed with heteroclitic NY-ESO-1 peptide and used as responder cells in IFN-γ ELISPOT analysis. T2 cells pulsed with 10 μg/ml A2/HIV control or A2/NY-ESO-1 peptide were used as stimulator cells (left). Various target cells, which did or did not express HLA-A2 and/or NY-ESO-1, were used as stimulator cells (right). Experiments were carried out in triplicate and error bars depict SD. *P < 0.05.
Discussion
We have shown that in the absence of constraints imposed by thymic selection, a single clonotypic TCR hemichain with chain centricity can, in conjunction with a heterogeneous repertoire of TCR counterchains, constitute functional self antigen-specific TCRs with a broad range of affinity. A chain-centric TCR hemichain determines antigen-specificity of T cells, while the paired TCR hemichain lacking chain centricity regulates avidity without perturbing antigen-specificity.
When reconstituted on T cells, about one half of clonotypic TCRs randomly selected from de novo generated A2/MART1 TCR repertoires demonstrated higher avidity compared to DMF5, a naturally occurring A2/MART1 TCR with one of the highest affinities that has been used in TCR gene transfer clinical trials (57). These results demonstrate the following 3 steps may serve as a general strategy to isolate high affinity antigen-specific TCRs by overcoming the hurdles of central and peripheral tolerance: 1) generation of a thymically unselected TCR repertoire by transducing an antigen-specific TCR hemichain regardless of its affinity into human peripheral T cells; 2) enrichment of high avidity T cells by delivering a controlled magnitude of antigen-specific stimulation using our artificial aAPC-based system; and 3) cloning and selection of TCR counterchains.
To isolate high affinity TCRs, several different strategies have been developed. Utilizing phage and yeast display systems, many groups screened libraries of TCRs with random amino acid substitutions in any of 6 CDR regions (58–60). Other groups undertook a similar strategy using T cells as host cells for screening (56, 61–63). Computational structure-based methods for high affinity TCR design and engineering have also been reported (64–67). In most of these studies, the libraries screened are comprised of TCRs with fixed lengths of CDR loops in which amino acids were only substituted but not deleted nor inserted. In this regard, our strategy is unique, since it can screen TCRs with various amino acid lengths of CDR3 regions as shown in Fig. 4. It is well known that the mutations in CDR1/2 regions upregulate the overall TCR:pMHC affinity by mainly enhancing the affinity between TCR and MHC but not TCR and pMHC (68, 69). Accordingly, high affinity TCRs with CDR1/2 mutations often lead to the loss of peptide specificity (56, 70). In contrast, our strategy uses native sequences that do not incorporate any mutations to the CDR regions. And yet, there still remains a risk that high affinity TCRs cloned using our strategy carry unwanted off-target toxicities. Any TCR used in TCR gene therapy must still be confirmed to lack unwanted on and off-target toxicities (71–73).
The SIG35α TCR chain can recognize A2/MART1 when paired with TRBV5-1 in addition to TRBV27 (Table I). While comprehensive analysis is awaited, preliminary experiments indeed confirmed that TRBV5-1 TCRβ chains isolated from SIG35α-transduced A2/MART1 multimer+ T cells recognized A2/MART1 when reconstituted with SIG35α (data not shown). However, the majority of endogenous TCRβ chains in SIG35α+ A2/MART1 T cells bore TRBV27 but not TRBV5-1 (Fig. 3A). These results suggest that, in order to recognize A2/MART1, SIG35α requires a lower contribution from TRBV27 compared to TRBV5-1 TCRβ chains. This is underpinned by the fact that the CDR3 region of TRBV27 TCRβ chains that recognized A2/MART1 in association with SIG35α was highly heterogeneous (Fig. 4).
As shown in Fig. 2D and Supplementary Fig. 1B, SIG35α but not SIG35αN, DMF5α, nor DMF5αS demonstrated potent chain centricity when transduced into primary T cells. Importantly, however, peripheral T cells forced to express either of SIG35αN, DMF5α, or DMF5αS also showed substantially higher, albeit low, A2/MART1 multimer positivity (0.5–0.9%) compared to ΔNGFR-transduced control T cells (0.02–0.1%). Furthermore, the transduction of peripheral T cells with A2/NY-ESO-1 hemichains also rendered them positive for specific A2 multimer staining as shown in Fig. 6A. Taken into account that TCRs are intrinsically highly crossreactive and that a single TCR can recognize more than a million different peptides (74), chain centricity is likely to be an inherent and shared attribute of many, if not all, TCRs.
Although the number of donors studied in this study is limited, there were no apparent differences in the heterogeneity of A2/MART1 TCRβ chains cloned from HLA-A2+ and A2− donors (Fig. 4, Table II). Furthermore, avidities of A2/MART1 TCRβ chains isolated from A2+ and A2− donors did not seem to differ when reconstituted with SIG35α on human T cells (Table V). These results suggest that HLA-restricted thymic selection does not affect TCR hemichain repertoires that can constitute functional TCRs in conjunction with a chain-centric TCR counterchain. Also, this raises the possibility that a TCR hemichain without chain centricity can constitute TCRs specific for various HLA-restricted antigens when paired with cognate antigen-specific chain-centric TCR counterchains. It has been recently noted that the overlap in the naïve CD8+ CDR3 sequence repertoires of any two of the individuals is approximately 7,000-fold larger than predicted and seems to be independent of the degree of HLA matching (75). Importantly, these sequencing studies were performed at a population level but not a single cell level, and, therefore, did not consider pairings of clonotypic TCRα and β chains. Our results suggest that pairings of TCRα and β chains can be a critical determinant of TCR repertoire diversity, and that different pairing can obviously make a de novo TCR repertoire and greatly enlarge its size.
Adoptive transfer of TCR gene-modified T cells is a feasible and promising treatment modality of cancer immunotherapy (15, 57, 76). When peripheral T cells are transduced with therapeutic TCRαβ genes, four different TCR chain pairings can be formed, including the therapeutic TCRαβ, the endogenous TCRαβ, and two mispaired TCRαβ dimers comprised of the introduced TCRα or β with the endogenous TCRβ or α chains. These four potential TCRαβ dimers each compete for a fixed amount of endogenous CD3 complexes. Consequently, the density of the therapeutic TCR dimers on cell surface is reduced, leading to the decreased T cell avidity (77). Moreover, the mispaired TCRs may acquire unwanted specificity for unknown antigens, which can evoke harmful autotoxicities (78, 79). To facilitate the matched pairing of the introduced TCR, a number of different approaches have been developed (80). The use of mouse instead of human TCR constant regions (81), the introduction of additional cysteine residues into TCR constant regions (82, 83), the usage of stabilized Vα/Vβ single chain TCRs (84), and a knockdown of endogenous TCRs by zinc-finger nucleases (85) or small-interfering RNAs (39), have been studied in vitro and in vivo. While the transduction of both TCRα and β chains generates two types of mispaired TCRs, the transduction of TCR hemichain alone produces only one TCR mispairing. Accordingly, in theory, transducing a single TCR hemichain alone would reduce in half the issues associated with the transduction of TCR heterodimers. However, it is still mandatory to carefully monitor for possible unwanted harmful autotoxicities caused by the transduction of a TCR hemichain. In addition, it would still be necessary to knock down endogenous TCR hemichain of the same class as the introduced hemichain. It should be noted that our aAPC-based system to expand antigen-specific CD8+ T cells has been successfully translated into the clinic (29). Adoptive transfer of antitumor T cells generated in vitro using the system induced sustained clinical responses in patients with advanced cancer without any in vivo modulation such as cytokine administration or lymphodepletion (20). Clinical trials where patients are infused with antitumor T cells redirected by a chain-centric TCR hemichain and subsequently enriched by the aAPC-based system are warranted.
Supplementary Material
Acknowledgments
We would like to thank Paula Rajkumar for helpful assistance. Jurkat 76 is a generous gift from Dr. Heemskerk, Leiden University Medical Centre. A2/MART1 TCR, clone DMF5, and A2/NY-ESO-1 TCR, clone 1G4, genes have been generously provided by Dr. Rosenberg, National Cancer Institute.
This work was supported by NIH grant R01 CA148673 (NH); Ontario Institute for Cancer Research Clinical Investigator Award IA-039 (NH); The Princess Margaret Cancer Foundation (MOB, NH); Guglietti Fellowship Award (TO); Knudson Postdoctoral Fellowship (KC); Frederick Banting and Charles Best Canada Graduate Scholarship (TG).
Abbreviations in this paper
- aAPC
artificial APC
- pMHC
peptide/MHC
- wt-aAPC
wild-type HLA-A2 aAPC
- mut-aAPC
mutated HLA-A2 aAPC
Footnotes
Author contributions
M.N. and N.H. designed the project. M.N., Y.Y., T.O., S.T., T.G., and K.C. performed the experimental work. M.O.B. provided human samples. M.N., M.O.B., and N.H. analyzed the results and wrote the manuscript.
Competing financial interests
S.T. is an employee of Takara Bio, Inc. This study was partly sponsored by Takara Bio, Inc. The University Health Network has filed a provisional patent application related to this study on which NH, MN, and TO are named as inventors.
References
- 1.Turner SJ, Doherty PC, McCluskey J, Rossjohn J. Structural determinants of T-cell receptor bias in immunity. Nature reviews Immunology. 2006;6:883–894. doi: 10.1038/nri1977. [DOI] [PubMed] [Google Scholar]
- 2.Venturi V, Price DA, Douek DC, Davenport MP. The molecular basis for public T-cell responses? Nature reviews Immunology. 2008;8:231–238. doi: 10.1038/nri2260. [DOI] [PubMed] [Google Scholar]
- 3.Miles JJ, Douek DC, Price DA. Bias in the alphabeta T-cell repertoire: implications for disease pathogenesis and vaccination. Immunology and cell biology. 2011;89:375–387. doi: 10.1038/icb.2010.139. [DOI] [PubMed] [Google Scholar]
- 4.Dong L, Li P, Oenema T, McClurkan CL, Koelle DM. Public TCR use by herpes simplex virus-2-specific human CD8 CTLs. Journal of immunology. 2010;184:3063–3071. doi: 10.4049/jimmunol.0903622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennan RM, Petersen J, Neller MA, Miles JJ, Burrows JM, Smith C, McCluskey J, Khanna R, Rossjohn J, Burrows SR. The impact of a large and frequent deletion in the human TCR beta locus on antiviral immunity. Journal of immunology. 2012;188:2742–2748. doi: 10.4049/jimmunol.1102675. [DOI] [PubMed] [Google Scholar]
- 6.Archbold JK, Macdonald WA, Gras S, Ely LK, Miles JJ, Bell MJ, Brennan RM, Beddoe T, Wilce MC, Clements CS, Purcell AW, McCluskey J, Burrows SR, Rossjohn J. Natural micropolymorphism in human leukocyte antigens provides a basis for genetic control of antigen recognition. The Journal of experimental medicine. 2009;206:209–219. doi: 10.1084/jem.20082136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Godfrey DI, Rossjohn J, McCluskey J. The fidelity, occasional promiscuity, and versatility of T cell receptor recognition. Immunity. 2008;28:304–314. doi: 10.1016/j.immuni.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Yokosuka T, Takase K, Suzuki M, Nakagawa Y, Taki S, Takahashi H, Fujisawa T, Arase H, Saito T. Predominant role of T cell receptor (TCR)-alpha chain in forming preimmune TCR repertoire revealed by clonal TCR reconstitution system. The Journal of experimental medicine. 2002;195:991–1001. doi: 10.1084/jem.20010809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouneaud C, Kourilsky P, Bousso P. Impact of negative selection on the T cell repertoire reactive to a self-peptide: a large fraction of T cell clones escapes clonal deletion. Immunity. 2000;13:829–840. doi: 10.1016/s1074-7613(00)00080-7. [DOI] [PubMed] [Google Scholar]
- 10.Kawakami Y, Eliyahu S, Delgado CH, Robbins PF, Rivoltini L, Topalian SL, Miki T, Rosenberg SA. Cloning of the gene coding for a shared human melanoma antigen recognized by autologous T cells infiltrating into tumor. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:3515–3519. doi: 10.1073/pnas.91.9.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coulie PG, Brichard V, Van Pel A, Wolfel T, Schneider J, Traversari C, Mattei S, De Plaen E, Lurquin C, Szikora JP, Renauld JC, Boon T. A new gene coding for a differentiation antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. The Journal of experimental medicine. 1994;180:35–42. doi: 10.1084/jem.180.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yee C, Thompson JA, Byrd D, Riddell SR, Roche P, Celis E, Greenberg PD. Adoptive T cell therapy using antigen-specific CD8+ T cell clones for the treatment of patients with metastatic melanoma: in vivo persistence, migration, and antitumor effect of transferred T cells. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16168–16173. doi: 10.1073/pnas.242600099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber J, Boswell W, Smith J, Hersh E, Snively J, Diaz M, Miles S, Liu X, Obrocea M, Qiu Z, Bot A. Phase 1 trial of intranodal injection of a Melan-A/MART-1 DNA plasmid vaccine in patients with stage IV melanoma. Journal of immunotherapy. 2008;31:215–223. doi: 10.1097/CJI.0b013e3181611420. [DOI] [PubMed] [Google Scholar]
- 14.Pittet MJ, Speiser DE, Lienard D, Valmori D, Guillaume P, Dutoit V, Rimoldi D, Lejeune F, Cerottini JC, Romero P. Expansion and functional maturation of human tumor antigen-specific CD8+ T cells after vaccination with antigenic peptide. Clinical cancer research : an official journal of the American Association for Cancer Research. 2001;7:796s–803s. [PubMed] [Google Scholar]
- 15.Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC, Sherry RM, Royal RE, Topalian SL, Kammula US, Restifo NP, Zheng Z, Nahvi A, de Vries CR, Rogers-Freezer LJ, Mavroukakis SA, Rosenberg SA. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mackensen A, Meidenbauer N, Vogl S, Laumer M, Berger J, Andreesen R. Phase I study of adoptive T-cell therapy using antigen-specific CD8+ T cells for the treatment of patients with metastatic melanoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24:5060–5069. doi: 10.1200/JCO.2006.07.1100. [DOI] [PubMed] [Google Scholar]
- 17.Khammari A, Labarriere N, Vignard V, Nguyen JM, Pandolfino MC, Knol AC, Quereux G, Saiagh S, Brocard A, Jotereau F, Dreno B. Treatment of metastatic melanoma with autologous Melan-A/MART-1-specific cytotoxic T lymphocyte clones. The Journal of investigative dermatology. 2009;129:2835–2842. doi: 10.1038/jid.2009.144. [DOI] [PubMed] [Google Scholar]
- 18.Jager E, Hohn H, Necker A, Forster R, Karbach J, Freitag K, Neukirch C, Castelli C, Salter RD, Knuth A, Maeurer MJ. Peptide-specific CD8+ T-cell evolution in vivo: response to peptide vaccination with Melan-A/MART-1. International journal of cancer Journal international du cancer. 2002;98:376–388. doi: 10.1002/ijc.10165. [DOI] [PubMed] [Google Scholar]
- 19.Cormier JN, Salgaller ML, Prevette T, Barracchini KC, Rivoltini L, Restifo NP, Rosenberg SA, Marincola FM. Enhancement of cellular immunity in melanoma patients immunized with a peptide from MART-1/Melan A. The cancer journal from Scientific American. 1997;3:37–44. [PMC free article] [PubMed] [Google Scholar]
- 20.Butler MO, Friedlander P, Milstein MI, Mooney MM, Metzler G, Murray AP, Tanaka M, Berezovskaya A, Imataki O, Drury L, Brennan L, Flavin M, Neuberg D, Stevenson K, Lawrence D, Hodi FS, Velazquez EF, Jaklitsch MT, Russell SE, Mihm M, Nadler LM, Hirano N. Establishment of antitumor memory in humans using in vitro-educated CD8+ T cells. Science translational medicine. 2011;3:80ra34. doi: 10.1126/scitranslmed.3002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zippelius A, Pittet MJ, Batard P, Rufer N, de Smedt M, Guillaume P, Ellefsen K, Valmori D, Lienard D, Plum J, MacDonald HR, Speiser DE, Cerottini JC, Romero P. Thymic selection generates a large T cell pool recognizing a self-peptide in humans. The Journal of experimental medicine. 2002;195:485–494. doi: 10.1084/jem.20011658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romero P, Valmori D, Pittet MJ, Zippelius A, Rimoldi D, Levy F, Dutoit V, Ayyoub M, Rubio-Godoy V, Michielin O, Guillaume P, Batard P, Luescher IF, Lejeune F, Lienard D, Rufer N, Dietrich PY, Speiser DE, Cerottini JC. Antigenicity and immunogenicity of Melan-A/MART-1 derived peptides as targets for tumor reactive CTL in human melanoma. Immunological reviews. 2002;188:81–96. doi: 10.1034/j.1600-065x.2002.18808.x. [DOI] [PubMed] [Google Scholar]
- 23.Trautmann L, Labarriere N, Jotereau F, Karanikas V, Gervois N, Connerotte T, Coulie P, Bonneville M. Dominant TCR V alpha usage by virus and tumor-reactive T cells with wide affinity ranges for their specific antigens. European journal of immunology. 2002;32:3181–3190. doi: 10.1002/1521-4141(200211)32:11<3181::AID-IMMU3181>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 24.Dietrich PY, Le Gal FA, Dutoit V, Pittet MJ, Trautman L, Zippelius A, Cognet I, Widmer V, Walker PR, Michielin O, Guillaume P, Connerotte T, Jotereau F, Coulie PG, Romero P, Cerottini JC, Bonneville M, Valmori D. Prevalent role of TCR alpha-chain in the selection of the preimmune repertoire specific for a human tumor-associated self-antigen. Journal of immunology. 2003;170:5103–5109. doi: 10.4049/jimmunol.170.10.5103. [DOI] [PubMed] [Google Scholar]
- 25.Cole DK, Yuan F, Rizkallah PJ, Miles JJ, Gostick E, Price DA, Gao GF, Jakobsen BK, Sewell AK. Germ line-governed recognition of a cancer epitope by an immunodominant human T-cell receptor. The Journal of biological chemistry. 2009;284:27281–27289. doi: 10.1074/jbc.M109.022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 27.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nature immunology. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 28.Adams JJ, Narayanan S, Liu B, Birnbaum ME, Kruse AC, Bowerman NA, Chen W, Levin AM, Connolly JM, Zhu C, Kranz DM, Garcia KC. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity. 2011;35:681–693. doi: 10.1016/j.immuni.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butler MO, Hirano N. Human cell-based artificial antigen-presenting cells for cancer immunotherapy. Immunological reviews. 2014;257:191–209. doi: 10.1111/imr.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heemskerk MH, Hoogeboom M, de Paus RA, Kester MG, van der Hoorn MA, Goulmy E, Willemze R, Falkenburg JH. Redirection of antileukemic reactivity of peripheral T lymphocytes using gene transfer of minor histocompatibility antigen HA-2-specific T-cell receptor complexes expressing a conserved alpha joining region. Blood. 2003;102:3530–3540. doi: 10.1182/blood-2003-05-1524. [DOI] [PubMed] [Google Scholar]
- 31.Butler MO, Lee JS, Ansen S, Neuberg D, Hodi FS, Murray AP, Drury L, Berezovskaya A, Mulligan RC, Nadler LM, Hirano N. Long-lived antitumor CD8+ lymphocytes for adoptive therapy generated using an artificial antigen-presenting cell. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:1857–1867. doi: 10.1158/1078-0432.CCR-06-1905. [DOI] [PubMed] [Google Scholar]
- 32.Hirano N, Butler MO, Xia Z, Ansen S, von Bergwelt-Baildon MS, Neuberg D, Freeman GJ, Nadler LM. Engagement of CD83 ligand induces prolonged expansion of CD8+ T cells and preferential enrichment for antigen specificity. Blood. 2006;107:1528–1536. doi: 10.1182/blood-2005-05-2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rimoldi D, Rubio-Godoy V, Dutoit V, Lienard D, Salvi S, Guillaume P, Speiser D, Stockert E, Spagnoli G, Servis C, Cerottini JC, Lejeune F, Romero P, Valmori D. Efficient simultaneous presentation of NY-ESO-1/LAGE-1 primary and nonprimary open reading frame-derived CTL epitopes in melanoma. Journal of immunology. 2000;165:7253–7261. doi: 10.4049/jimmunol.165.12.7253. [DOI] [PubMed] [Google Scholar]
- 34.Chen YT, Stockert E, Jungbluth A, Tsang S, Coplan KA, Scanlan MJ, Old LJ. Serological analysis of Melan-A(MART-1), a melanocyte-specific protein homogeneously expressed in human melanomas. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:5915–5919. doi: 10.1073/pnas.93.12.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang S, Cohen CJ, Peng PD, Zhao Y, Cassard L, Yu Z, Zheng Z, Jones S, Restifo NP, Rosenberg SA, Morgan RA. Development of optimal bicistronic lentiviral vectors facilitates high-level TCR gene expression and robust tumor cell recognition. Gene therapy. 2008;15:1411–1423. doi: 10.1038/gt.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitamura T. New experimental approaches in retrovirus-mediated expression screening. International journal of hematology. 1998;67:351–359. doi: 10.1016/s0925-5710(98)00025-5. [DOI] [PubMed] [Google Scholar]
- 37.Imataki O, Ansen S, Tanaka M, Butler MO, Berezovskaya A, Milstein MI, Kuzushima K, Nadler LM, Hirano N. IL-21 can supplement suboptimal Lck-independent MAPK activation in a STAT-3-dependent manner in human CD8(+) T cells. Journal of immunology. 2012;188:1609–1619. doi: 10.4049/jimmunol.1003446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purbhoo MA, Boulter JM, Price DA, Vuidepot AL, Hourigan CS, Dunbar PR, Olson K, Dawson SJ, Phillips RE, Jakobsen BK, Bell JI, Sewell AK. The human CD8 coreceptor effects cytotoxic T cell activation and antigen sensitivity primarily by mediating complete phosphorylation of the T cell receptor zeta chain. The Journal of biological chemistry. 2001;276:32786–32792. doi: 10.1074/jbc.M102498200. [DOI] [PubMed] [Google Scholar]
- 39.Okamoto S, Mineno J, Ikeda H, Fujiwara H, Yasukawa M, Shiku H, Kato I. Improved expression and reactivity of transduced tumor-specific TCRs in human lymphocytes by specific silencing of endogenous TCR. Cancer research. 2009;69:9003–9011. doi: 10.1158/0008-5472.CAN-09-1450. [DOI] [PubMed] [Google Scholar]
- 40.Ansen S, Butler MO, Berezovskaya A, Murray AP, Stevenson K, Nadler LM, Hirano N. Dissociation of its opposing immunologic effects is critical for the optimization of antitumor CD8+ T-cell responses induced by interleukin 21. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:6125–6136. doi: 10.1158/1078-0432.CCR-08-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirano N, Butler MO, Xia Z, Berezovskaya A, Murray AP, Ansen S, Kojima S, Nadler LM. Identification of an immunogenic CD8+ T-cell epitope derived from gamma-globin, a putative tumor-associated antigen for juvenile myelomonocytic leukemia. Blood. 2006;108:2662–2668. doi: 10.1182/blood-2006-04-017566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirano N, Butler MO, Xia Z, Berezovskaya A, Murray AP, Ansen S, Nadler LM. Efficient presentation of naturally processed HLA class I peptides by artificial antigen-presenting cells for the generation of effective antitumor responses. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12:2967–2975. doi: 10.1158/1078-0432.CCR-05-2791. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka M, Butler MO, Ansen S, Imataki O, Berezovskaya A, Nadler LM, Hirano N. Induction of HLA-DP4-restricted anti-survivin Th1 and Th2 responses using an artificial antigen-presenting cell. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:5392–5401. doi: 10.1158/1078-0432.CCR-10-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butler MO, Ansen S, Tanaka M, Imataki O, Berezovskaya A, Mooney MM, Metzler G, Milstein MI, Nadler LM, Hirano N. A panel of human cell-based artificial APC enables the expansion of long-lived antigen-specific CD4+ T cells restricted by prevalent HLA-DR alleles. International immunology. 2010;22:863–873. doi: 10.1093/intimm/dxq440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Butler MO, Imataki O, Yamashita Y, Tanaka M, Ansen S, Berezovskaya A, Metzler G, Milstein MI, Mooney MM, Murray AP, Mano H, Nadler LM, Hirano N. Ex vivo expansion of human CD8+ T cells using autologous CD4+ T cell help. PloS one. 2012;7:e30229. doi: 10.1371/journal.pone.0030229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li LP, Lampert JC, Chen X, Leitao C, Popovic J, Muller W, Blankenstein T. Transgenic mice with a diverse human T cell antigen receptor repertoire. Nature medicine. 2010;16:1029–1034. doi: 10.1038/nm.2197. [DOI] [PubMed] [Google Scholar]
- 47.Carrasco YR, Ramiro AR, Trigueros C, de Yebenes VG, Garcia-Peydro M, Toribio ML. An endoplasmic reticulum retention function for the cytoplasmic tail of the human pre-T cell receptor (TCR) alpha chain: potential role in the regulation of cell surface pre-TCR expression levels. The Journal of experimental medicine. 2001;193:1045–1058. doi: 10.1084/jem.193.9.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson LA, Heemskerk B, Powell DJ, Jr, Cohen CJ, Morgan RA, Dudley ME, Robbins PF, Rosenberg SA. Gene transfer of tumor-reactive TCR confers both high avidity and tumor reactivity to nonreactive peripheral blood mononuclear cells and tumor-infiltrating lymphocytes. Journal of immunology. 2006;177:6548–6559. doi: 10.4049/jimmunol.177.9.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wieckowski S, Baumgaertner P, Corthesy P, Voelter V, Romero P, Speiser DE, Rufer N. Fine structural variations of alphabetaTCRs selected by vaccination with natural versus altered self-antigen in melanoma patients. Journal of immunology. 2009;183:5397–5406. doi: 10.4049/jimmunol.0901460. [DOI] [PubMed] [Google Scholar]
- 50.Valmori D, Dutoit V, Lienard D, Lejeune F, Speiser D, Rimoldi D, Cerundolo V, Dietrich PY, Cerottini JC, Romero P. Tetramer-guided analysis of TCR beta-chain usage reveals a large repertoire of melan-A-specific CD8+ T cells in melanoma patients. Journal of immunology. 2000;165:533–538. doi: 10.4049/jimmunol.165.1.533. [DOI] [PubMed] [Google Scholar]
- 51.Sensi M, Traversari C, Radrizzani M, Salvi S, Maccalli C, Mortarini R, Rivoltini L, Farina C, Nicolini G, Wolfel T, et al. Cytotoxic T-lymphocyte clones from different patients display limited T-cell-receptor variable-region gene usage in HLA-A2-restricted recognition of the melanoma antigen Melan-A/MART-1. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:5674–5678. doi: 10.1073/pnas.92.12.5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cole DK, Edwards ES, Wynn KK, Clement M, Miles JJ, Ladell K, Ekeruche J, Gostick E, Adams KJ, Skowera A, Peakman M, Wooldridge L, Price DA, Sewell AK. Modification of MHC anchor residues generates heteroclitic peptides that alter TCR binding and T cell recognition. Journal of immunology. 2010;185:2600–2610. doi: 10.4049/jimmunol.1000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dutoit V, Rubio-Godoy V, Pittet MJ, Zippelius A, Dietrich PY, Legal FA, Guillaume P, Romero P, Cerottini JC, Houghten RA, Pinilla C, Valmori D. Degeneracy of antigen recognition as the molecular basis for the high frequency of naive A2/Melan-a peptide multimer(+) CD8(+) T cells in humans. The Journal of experimental medicine. 2002;196:207–216. doi: 10.1084/jem.20020242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen JL, Dunbar PR, Gileadi U, Jager E, Gnjatic S, Nagata Y, Stockert E, Panicali DL, Chen YT, Knuth A, Old LJ, Cerundolo V. Identification of NY-ESO-1 peptide analogues capable of improved stimulation of tumor-reactive CTL. Journal of immunology. 2000;165:948–955. doi: 10.4049/jimmunol.165.2.948. [DOI] [PubMed] [Google Scholar]
- 55.Boulter JM, Glick M, Todorov PT, Baston E, Sami M, Rizkallah P, Jakobsen BK. Stable, soluble T-cell receptor molecules for crystallization and therapeutics. Protein engineering. 2003;16:707–711. doi: 10.1093/protein/gzg087. [DOI] [PubMed] [Google Scholar]
- 56.Robbins PF, Li YF, El-Gamil M, Zhao Y, Wargo JA, Zheng Z, Xu H, Morgan RA, Feldman SA, Johnson LA, Bennett AD, Dunn SM, Mahon TM, Jakobsen BK, Rosenberg SA. Single and dual amino acid substitutions in TCR CDRs can enhance antigen-specific T cell functions. Journal of immunology. 2008;180:6116–6131. doi: 10.4049/jimmunol.180.9.6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson LA, Morgan RA, Dudley ME, Cassard L, Yang JC, Hughes MS, Kammula US, Royal RE, Sherry RM, Wunderlich JR, Lee CC, Restifo NP, Schwarz SL, Cogdill AP, Bishop RJ, Kim H, Brewer CC, Rudy SF, VanWaes C, Davis JL, Mathur A, Ripley RT, Nathan DA, Laurencot CM, Rosenberg SA. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114:535–546. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, Moysey R, Molloy PE, Vuidepot AL, Mahon T, Baston E, Dunn S, Liddy N, Jacob J, Jakobsen BK, Boulter JM. Directed evolution of human T-cell receptors with picomolar affinities by phage display. Nature biotechnology. 2005;23:349–354. doi: 10.1038/nbt1070. [DOI] [PubMed] [Google Scholar]
- 59.Holler PD, Holman PO, Shusta EV, O’Herrin S, Wittrup KD, Kranz DM. In vitro evolution of a T cell receptor with high affinity for peptide/MHC. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5387–5392. doi: 10.1073/pnas.080078297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weber KS, Donermeyer DL, Allen PM, Kranz DM. Class II-restricted T cell receptor engineered in vitro for higher affinity retains peptide specificity and function. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:19033–19038. doi: 10.1073/pnas.0507554102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kessels HW, van Den Boom MD, Spits H, Hooijberg E, Schumacher TN. Changing T cell specificity by retroviral T cell receptor display. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:14578–14583. doi: 10.1073/pnas.97.26.14578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chervin AS, Aggen DH, Raseman JM, Kranz DM. Engineering higher affinity T cell receptors using a T cell display system. Journal of immunological methods. 2008;339:175–184. doi: 10.1016/j.jim.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmid DA, Irving MB, Posevitz V, Hebeisen M, Posevitz-Fejfar A, Sarria JC, Gomez-Eerland R, Thome M, Schumacher TN, Romero P, Speiser DE, Zoete V, Michielin O, Rufer N. Evidence for a TCR affinity threshold delimiting maximal CD8 T cell function. Journal of immunology. 2010;184:4936–4946. doi: 10.4049/jimmunol.1000173. [DOI] [PubMed] [Google Scholar]
- 64.Haidar JN, Pierce B, Yu Y, Tong W, Li M, Weng Z. Structure-based design of a T-cell receptor leads to nearly 100-fold improvement in binding affinity for pepMHC. Proteins. 2009;74:948–960. doi: 10.1002/prot.22203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zoete V, Irving MB, Michielin O. MM-GBSA binding free energy decomposition and T cell receptor engineering. Journal of molecular recognition : JMR. 2010;23:142–152. doi: 10.1002/jmr.1005. [DOI] [PubMed] [Google Scholar]
- 66.Irving M, Zoete V, Hebeisen M, Schmid D, Baumgartner P, Guillaume P, Romero P, Speiser D, Luescher I, Rufer N, Michielin O. Interplay between T cell receptor binding kinetics and the level of cognate peptide presented by major histocompatibility complexes governs CD8+ T cell responsiveness. The Journal of biological chemistry. 2012;287:23068–23078. doi: 10.1074/jbc.M112.357673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pierce BG, Hellman LM, Hossain M, Singh NK, Vander Kooi CW, Weng Z, Baker BM. Computational design of the affinity and specificity of a therapeutic T cell receptor. PLoS computational biology. 2014;10:e1003478. doi: 10.1371/journal.pcbi.1003478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dunn SM, Rizkallah PJ, Baston E, Mahon T, Cameron B, Moysey R, Gao F, Sami M, Boulter J, Li Y, Jakobsen BK. Directed evolution of human T cell receptor CDR2 residues by phage display dramatically enhances affinity for cognate peptide-MHC without increasing apparent cross-reactivity. Protein science : a publication of the Protein Society. 2006;15:710–721. doi: 10.1110/ps.051936406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Madura F, Rizkallah PJ, Miles KM, Holland CJ, Bulek AM, Fuller A, Schauenburg AJ, Miles JJ, Liddy N, Sami M, Li Y, Hossain M, Baker BM, Jakobsen BK, Sewell AK, Cole DK. T-cell receptor specificity maintained by altered thermodynamics. The Journal of biological chemistry. 2013;288:18766–18775. doi: 10.1074/jbc.M113.464560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao Y, Bennett AD, Zheng Z, Wang QJ, Robbins PF, Yu LY, Li Y, Molloy PE, Dunn SM, Jakobsen BK, Rosenberg SA, Morgan RA. High-affinity TCRs generated by phage display provide CD4+ T cells with the ability to recognize and kill tumor cell lines. Journal of immunology. 2007;179:5845–5854. doi: 10.4049/jimmunol.179.9.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cameron BJ, Gerry AB, Dukes J, Harper JV, Kannan V, Bianchi FC, Grand F, Brewer JE, Gupta M, Plesa G, Bossi G, Vuidepot A, Powlesland AS, Legg A, Adams KJ, Bennett AD, Pumphrey NJ, Williams DD, Binder-Scholl G, Kulikovskaya I, Levine BL, Riley JL, Varela-Rohena A, Stadtmauer EA, Rapoport AP, Linette GP, June CH, Hassan NJ, Kalos M, Jakobsen BK. Identification of a Titin-derived HLA-A1-presented peptide as a cross-reactive target for engineered MAGE A3-directed T cells. Science translational medicine. 2013;5:197ra103. doi: 10.1126/scitranslmed.3006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Linette GP, Stadtmauer EA, Maus MV, Rapoport AP, Levine BL, Emery L, Litzky L, Bagg A, Carreno BM, Cimino PJ, Binder-Scholl GK, Smethurst DP, Gerry AB, Pumphrey NJ, Bennett AD, Brewer JE, Dukes J, Harper J, Tayton-Martin HK, Jakobsen BK, Hassan NJ, Kalos M, June CH. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood. 2013;122:863–871. doi: 10.1182/blood-2013-03-490565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morgan RA, Chinnasamy N, Abate-Daga D, Gros A, Robbins PF, Zheng Z, Dudley ME, Feldman SA, Yang JC, Sherry RM, Phan GQ, Hughes MS, Kammula US, Miller AD, Hessman CJ, Stewart AA, Restifo NP, Quezado MM, Alimchandani M, Rosenberg AZ, Nath A, Wang T, Bielekova B, Wuest SC, Akula N, McMahon FJ, Wilde S, Mosetter B, Schendel DJ, Laurencot CM, Rosenberg SA. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. Journal of immunotherapy. 2013;36:133–151. doi: 10.1097/CJI.0b013e3182829903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wooldridge L, Ekeruche-Makinde J, van den Berg HA, Skowera A, Miles JJ, Tan MP, Dolton G, Clement M, Llewellyn-Lacey S, Price DA, Peakman M, Sewell AK. A single autoimmune T cell receptor recognizes more than a million different peptides. The Journal of biological chemistry. 2012;287:1168–1177. doi: 10.1074/jbc.M111.289488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Robins HS, Srivastava SK, Campregher PV, Turtle CJ, Andriesen J, Riddell SR, Carlson CS, Warren EH. Overlap and effective size of the human CD8+ T cell receptor repertoire. Science translational medicine. 2010;2:47ra64. doi: 10.1126/scitranslmed.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Robbins PF, Morgan RA, Feldman SA, Yang JC, Sherry RM, Dudley ME, Wunderlich JR, Nahvi AV, Helman LJ, Mackall CL, Kammula US, Hughes MS, Restifo NP, Raffeld M, Lee CC, Levy CL, Li YF, El-Gamil M, Schwarz SL, Laurencot C, Rosenberg SA. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:917–924. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thomas S, Stauss HJ, Morris EC. Molecular immunology lessons from therapeutic T-cell receptor gene transfer. Immunology. 2010;129:170–177. doi: 10.1111/j.1365-2567.2009.03227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bendle GM, Linnemann C, Hooijkaas AI, Bies L, de Witte MA, Jorritsma A, Kaiser AD, Pouw N, Debets R, Kieback E, Uckert W, Song JY, Haanen JB, Schumacher TN. Lethal graft-versus-host disease in mouse models of T cell receptor gene therapy. Nature medicine. 2010;16:565–570. doi: 10.1038/nm.2128. 561p following 570. [DOI] [PubMed] [Google Scholar]
- 79.van Loenen MM, de Boer R, Amir AL, Hagedoorn RS, Volbeda GL, Willemze R, van Rood JJ, Falkenburg JH, Heemskerk MH. Mixed T cell receptor dimers harbor potentially harmful neoreactivity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10972–10977. doi: 10.1073/pnas.1005802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Govers C, Sebestyen Z, Coccoris M, Willemsen RA, Debets R. T cell receptor gene therapy: strategies for optimizing transgenic TCR pairing. Trends in molecular medicine. 2010;16:77–87. doi: 10.1016/j.molmed.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 81.Cohen CJ, Zhao Y, Zheng Z, Rosenberg SA, Morgan RA. Enhanced antitumor activity of murine-human hybrid T-cell receptor (TCR) in human lymphocytes is associated with improved pairing and TCR/CD3 stability. Cancer research. 2006;66:8878–8886. doi: 10.1158/0008-5472.CAN-06-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cohen CJ, Li YF, El-Gamil M, Robbins PF, Rosenberg SA, Morgan RA. Enhanced antitumor activity of T cells engineered to express T-cell receptors with a second disulfide bond. Cancer research. 2007;67:3898–3903. doi: 10.1158/0008-5472.CAN-06-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kuball J, Dossett ML, Wolfl M, Ho WY, Voss RH, Fowler C, Greenberg PD. Facilitating matched pairing and expression of TCR chains introduced into human T cells. Blood. 2007;109:2331–2338. doi: 10.1182/blood-2006-05-023069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aggen DH, Chervin AS, Schmitt TM, Engels B, Stone JD, Richman SA, Piepenbrink KH, Baker BM, Greenberg PD, Schreiber H, Kranz DM. Single-chain ValphaVbeta T-cell receptors function without mispairing with endogenous TCR chains. Gene therapy. 2012;19:365–374. doi: 10.1038/gt.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Provasi E, Genovese P, Lombardo A, Magnani Z, Liu PQ, Reik A, Chu V, Paschon DE, Zhang L, Kuball J, Camisa B, Bondanza A, Casorati G, Ponzoni M, Ciceri F, Bordignon C, Greenberg PD, Holmes MC, Gregory PD, Naldini L, Bonini C. Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nature medicine. 2012;18:807–815. doi: 10.1038/nm.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


