Abstract
NEDD9 is an established marker of invasive and metastatic cancers. NEDD9 downregulation has been shown to dramatically reduce cell invasion and metastasis in multiple tumors. The mechanisms by which NEDD9 regulates invasion are largely unknown. In the current study, we have found that NEDD9 is required for MMP14 enzymatic recovery/recycling through the late endosomes to enable disengagement of tissue inhibitor of matrix metalloproteinase 2 (TIMP2) and tumor invasion. Depletion of NEDD9 decreases targeting of the MMP14/TIMP2 complex to late endosomes and increases trafficking of MMP14 from early/sorting endosomes back to the surface in a small GTPase Arf6-dependent manner. NEDD9 directly binds to Arf6-GAP, ARAP3, and Arf6 effector GGA3 thereby facilitating the Arf6 inactivation required for MMP14/TIMP2 targeting to late endosomes. Re-expression of NEDD9 or a decrease in Arf6 activity is sufficient to restore MMP14 activity and the invasive properties of tumor cells. Importantly, NEDD9 inhibition by Vivo-Morpholinos, an antisense therapy, decreases primary tumor growth and metastasis in xenograft models of breast cancer. Collectively, our findings uncover a novel mechanism to control tumor cells dissemination through NEDD9/Arf6-dependent regulation of MMP14/TIMP2 trafficking, and validates NEDD9 as a clinically relevant therapeutic target to treat metastatic cancer.
Introduction
NEDD9 is pro-metastatic gene known to be upregulated in different metastatic cancers (1-3). It is a cytoplasmic docking protein required for mesenchymal migration and invasion driven by extracellular matrix (ECM) proteolysis (3-5). Genetic ablation of NEDD9 expression in an MMTV-HER2-driven transgenic model of mammary tumorigenesis led to a 90% decrease in the incidence of cancer (6). As we and others have previously reported, NEDD9 is upregulated during progression to invasive carcinoma (7-9), while depletion of NEDD9 reduces the number of circulating tumor cells by 80% (10). NEDD9 is targeted to invasive protrusions and co-localizes with MMP14. We also have shown that NEDD9 does not affect the expression or secretion of MMPs, but dramatically decreases their activity (10). The clinically relevant strategies to decrease NEDD9 expression in established tumors in vivo to abrogate cancer progression were not yet tested, although the potential implications would be highly beneficial.
Activation of soluble MMPs is triggered by membrane bound MMPs such as MMP14. Activity of MMP14 on the cell surface is regulated by multiple mechanisms, including binding of tissue specific inhibitor of MMP (TIMP2) and endocytosis. It was shown that endocytosis of MMP14 is stimulated upon binding of TIMP2 (11) and almost 80% of MMP14 is recycled back to the cell surface (12), but the fate of TIMP2 post internalization is controversial (11, 13). Upon internalization, MMP14 is localized first to early endosomes and later gets recycled through either early endosomes and the trans-Golgi network (14) or through late endosomes (15, 16). It is currently unknown whether TIMP2 follows the similar pattern of recycling and degradation. We have previously reported that depletion of NEDD9 leads to a decrease in MMP14 activity and an accumulation of the MMP14/TIMP2 complex on cell surface. The molecular mechanisms and clinical significance of this phenomenon are unknown.
One of the central signaling hubs regulating endocytic recycling is associated with small GTPase ADP-ribosylation factor-6 (Arf6). Arf6 activity is critical for recycling from early/sorting Rab4/5 positive endosomes (17). Activity of Arf6 is tightly regulated by a number of GEFs and GAPs to support continuous cycling of Arf6 between GDP and GTP bound forms (18-20). In this study, we identify NEDD9 as a scaffolding component of the ARAP3-Arf6-GGA3 complex, which is required to decrease the levels of active Arf6 and allow for trafficking of MMP14/TIMP2 complex to late endosomes. This knowledge is crucial for understanding the mechanisms of recovery of MMP14 enzymatic activity and for development of new strategies for MMPs inhibition and eradication of metastases. This study outlines mechanisms for NEDD9-driven invasion, as well as provides new avenues for the development of alternative strategies to ablate tumor progression based on the manipulation of NEDD9 and ARAP3.
Results
NEDD9 depletion leads to re-distribution of MMP14 to the cell surface
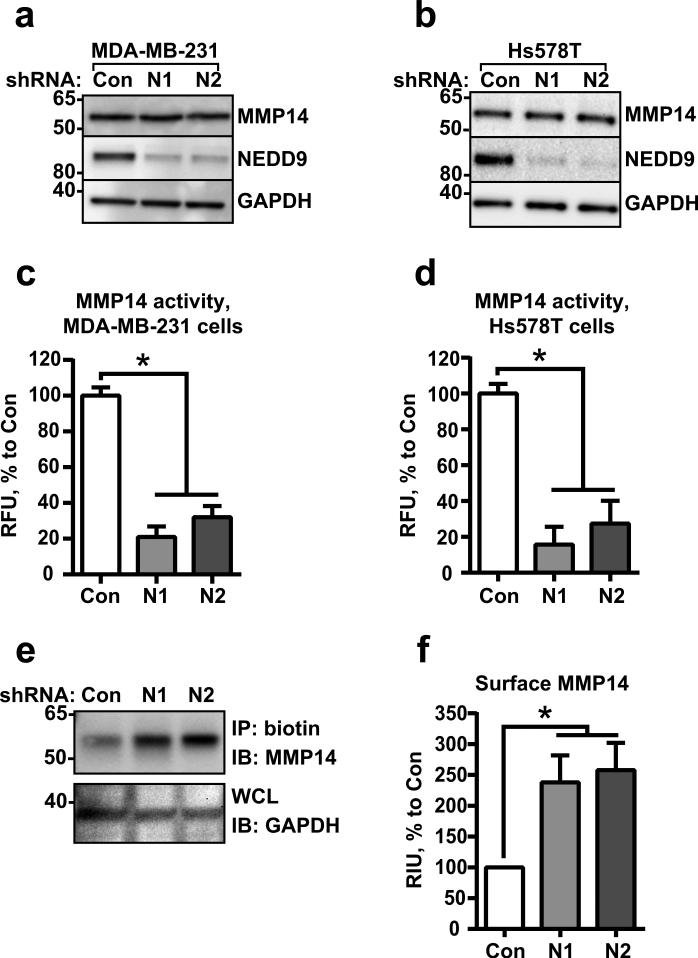
The depletion of NEDD9 in invasive mesenchymal breast tumor cell lines (Fig.S1a) or the genetic ablation of NEDD9 in mouse embryonic fibroblasts (MEFs) from NEDD9 knockout mice leads to a drastic decrease in the activity of soluble and transmembrane MMPs, particularly MMP14 (8, 10). The total amount of MMP14 in the analyzed cells did not change (Fig.1a-b), but the activity, measured by the degradation of MMP14-specific fluorogenic substrate (21), was drastically decreased (Fig.1c-d). Note that the antibodies used for detection of MMP14 are specific and capable of recognizing both pro- and active forms of the enzyme (Fig.S1b-c), thus the initial activation of the pro-enzyme by furin was not affected by NEDD9 depletion. Nevertheless, based on a surface biotinylation study, the plasma membrane (PM) levels of MMP14 had increased two-fold in the shNEDD9 cells (Fig.1e-f). The increase in MMP14 on the cell surface is often due to the deficiency in internalization, which is necessary for proper enzyme function (22, 23).
Figure 1. NEDD9 depletion leads to redistribution of MMP14 to the cell surface.
(a-b) WB analysis of WCL from MDA-MB-231 (a) and Hs578T (b) cells expressing siRNAs against NEDD9 (N1, N2) or non-targeting control siRNA (Con) with anti-NEDD9 (2G9), -MMP14, -GAPDH antibodies. (c-d) MMP14-specific fluorogenic substrate degradation assay in cells as in (a-b). Fluorescence was measured at 328/400 nm excitation/emission. Data plotted as % of RFU to shControl ±S.E.M; n=3; one-way ANOVA with Dunnett's post-hoc analysis for MDA-MB-231: p<0.0001 siCon/shN1 or /N2; Hs578T: p<0.0001 siCon/siN1 or /N2. (e) Western blot analysis of streptavidin agarose precipitated biotinylated MMP14 (IP: biotin) from shCon-, shNedd9 (N1, N2) MDA-MB-231 cells with anti-MMP14 and -GAPDH (loading control). (f) Quantification of WB as in (e), n=3; one-way ANOVA with Dunnett's post-hoc analysis p=0.043, 0.023 shCon/shN1 or/N2.
Depletion of NEDD9 does not affect the internalization of MMP14
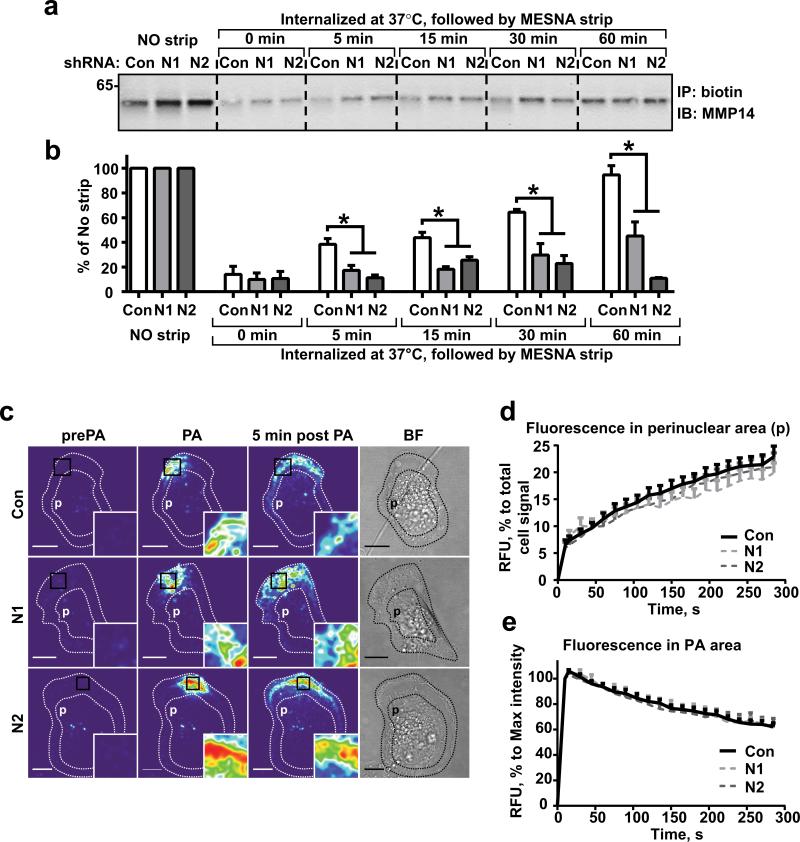
To determine the impact of NEDD9 expression on MMP14 internalization, we performed time course studies on the uptake of biotinylated surface molecules. Initially, the total amount of biotinylated MMP14 on the cell surface was increased in the shNEDD9 cells (Fig.2a, no strip). Internalization was initiated by shifting the temperature to 37°C, causing the fraction of biotinylated MMP14 to be transported inside the cells (IC-MMP14). We found that the shNEDD9 cells contain two-fold less of IC-MMP14 than the shControl when normalized to the original amount of biotinylated PM-MMP14 (no strip) (Fig.2a-b). The amount of IC-MMP14 is the sum of internalization proficiency, intracellular degradation rates, and recycling capabilities of the cells. Since proteasome and lysosome inhibitors were used during the studies, the decrease in biotinylated IC-MMP14 could not be attributed to the increased degradation of the protein post-internalization. However, the decrease could potentially be due to either the decreased internalization of MMP14 or the plasma membrane (PM) in general, or the increased recycling of the internalized surface molecules.
Figure 2. NEDD9 depletion does not affect internalization of MMP14.
(a) Western blot (WB) analysis of immunoprecipitated (IP) biotinylated MMP14 from shCon-, shNEDD9 (N1, N2)-MDA-MB-231cells: no strip-0°C (lines 1-3), the rest incubated at 37°C for indicated times, striped with MESNA, lysed, IPed with streptavidin agarose and probed with anti-MMP14 antibody. (b) Quantification of WBs as in (a), three independent experiments, graphs are mean percent (%) of relative intensity units (RIU) to no strip conditions (100%) ±S.E.M; one-way ANOVA with Dunnett's post-hoc analysis *p= 0.0054, 0.0049, 0.0098, 0.0028 for 5, 15, 30 and 60 min respectively. (c) Representative confocal images of shCon-, shNEDD9 (N1, N2)-MDA-MB-231cells expressing PA-mCherry-MMP14 before photo-activation (pre-PA), after- (PA), and 5 min after PA in a defined area. Scale bar, 10μm; inserts are the enlarged areas of PA; BF-bright field image. (d) Quantification of relative fluorescence intensity units (RFU) of mCherry-MMP14 in cells as in (c) in perinuclear area (p); graph is mean RFU % of total RFU/cell ±S.E.M; 10 cells/per treatment (Con, N1, N2); F test performed for fitted lines, p is non-significant (ns) for shCon/shN1 or /N2. (e) Quantification of relative fluorescence intensity (RFU) of mCherry-MMP14 in cells as in (c) in PA area (black rectangle); graph is mean RFU (%) of max RFU/cell (100%) ±S.E.M; 10 cells/per treatment (Con, N1, N2), F test performed for fitted lines, p is (ns).
Next, we determined the rates of bulk PM internalization using a CellMask-PM stain and live cell imaging. We found that the decay in fluorescence intensity of CellMask at the PM and perimembrane (2-4μm beneath the PM) area in shNEDD9 cells was similar to that of the control (Fig.S1d-e; MoviesS1-S3), suggesting that NEDD9 does not affect bulk PM internalization.
To visualize the trafficking of MMP14, we exogenously expressed previously characterized photoactivatable fluorescent MMP14 (PAmCherry-MMP14 (24)). We imaged the early stages of internalization and translocation of PM-anchored and perimembrane pools of MMP14 after photo-activation in the defined areas (Fig.2c, MoviesS4-S6). The fluorescence intensity of mCherry-MMP14 in the entire cell and perinuclear (near the nucleus) area (p) was measured. The increase in the fluorescence intensity of mCherry-MMP14 in the perinuclear area (Fig.2d) and the decrease in the original photoactivated area (Fig.2e) was nearly identical in both the shControl and shNEDD9 cells, suggesting a similar uptake of the MMP14-positive membrane/vesicles. We conclude that NEDD9 deficiency does not alter the internalization of MMP14.
NEDD9 deficiency leads to an increase in the recycling of MMP14
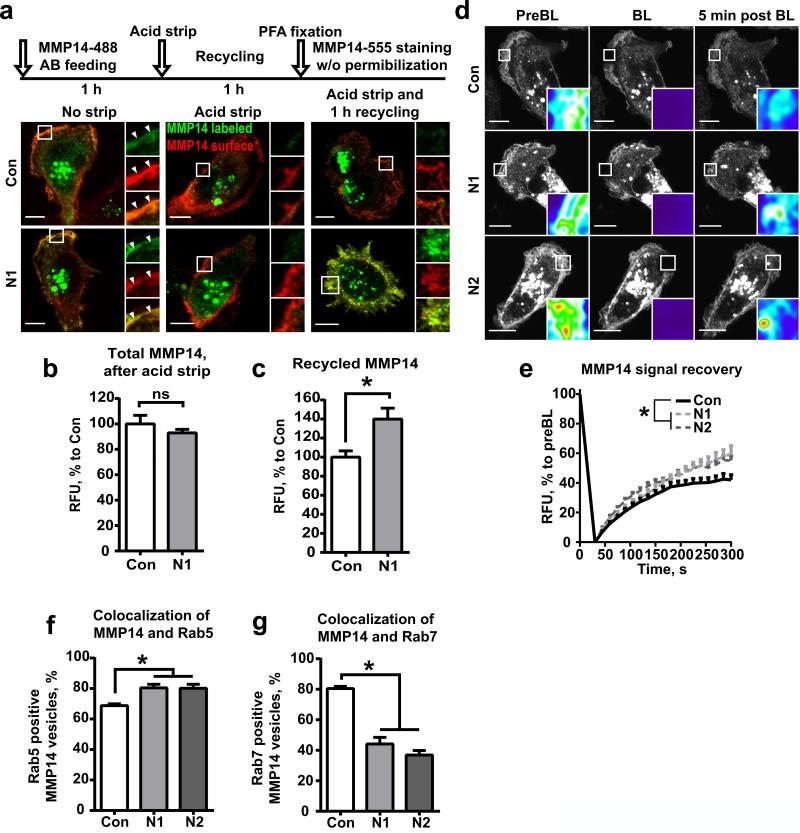
To assess the recycling of MMP14, we used fluorescently labeled anti-MMP14 antibodies in an antibody feeding assay (14). The shControl and shNEDD9 cells were pre-treated with antibodies for 1 hour to allow for MMP14 labeling and internalization of the Ab/MMP14 complex, followed by acidic wash (25) and an additional hour of recycling. Cells were then stained with different anti-MMP14 antibodies to differentiate between the recycled Ab/MMP14 complex and the newly synthesized MMP14. Interestingly, the amount of recycled MMP14, as indicated by co-staining with both anti-MMP14 antibodies, was increased up to 40% in the shNEDD9 cells (Fig.3a-c).
Figure 3. NEDD9 deficiency leads to increase in recycling of MMP14.
(a) Top: Schematic of fluorescently labeled anti-MMP14 (Alexa488) antibody feeding assay. Bottom: Representative 3D reconstructed confocal images of siControl (Con) and siNEDD9 (N1) MDA-MB-231 cells pre-treated with antibodies for 1 hour (left panel-No strip); antibodies were stripped (middle panel-Acid strip), followed by 1 hour of incubation for Ab/MMP14 complex recycling and stained with different anti-MMP14 (Alexa555-right panel) antibody, without permeabilization. Scale bar is 10μm; insets are enlarged areas indicated in the merged image. (b-c) Quantification of RFU of MMP14 total signal (b, after acid strip) and recycled MMP14, as indicated by co-stain with MMP14-488 and MMP14-555 1h after acid strip (c); graphs are mean RFU (%) to shCon ±S.E.M; n=3 (three independent experiments), 10 cells/treatment; two-tailed t-test for total MMP14: p is ns; Recycled MMP14: *p= 0.017 shCon/shN1. (d) Representative images of FRAP analysis of the perimembrane area of siControl (Con) or siNEDD9 (N1, N2) cells expressing mCherry-MMP14 before photobleaching (PreBL), after- (BL), and 5 min post BL; Scale bar is 10μm; insets are enlarged area of BL. (e) Quantification of relative fluorescence intensity (RFU) of MMP14 signal in cells as in (d) at indicated time points in seconds; graphs are mean RFU (%) to original preBL ±S.E.M; 10 cells/treatment; F test performed for fitted lines, p is <0.0001 for shCon/shN1 or /shN2. (f, g) Quantification of colocalization of Rab5 (f) and MMP14 or Rab7 (g) and MMP14 positive vesicles as % of total MMP14 vesicles; graphs are mean % values ±S.E.M; three independent experiments, 10 cells/treatment; one-way ANOVA with Dunnett's post-hoc analysis Rab5: *p=0.0154, 0.0087 for Con/N1 or/N2; Rab7:*p<0.0001for Con/N1 or /N2.
Similarly, FRAP analysis of cells expressing mCherry-MMP14 revealed that shNEDD9 cells recover fluorescent PM-MMP14 to a higher extent, with a 1.3-1.7 fold increase in the mobile fraction and a comparable increase in t1/2 (Fig.3d-e; MoviesS7-S9). Hence, the decrease in NEDD9 expression leads to an increase in recycling of internalized MMP14.
NEDD9 is involved in the regulation of MMP14 trafficking to late endosomes
To understand the route of MMP14 recycling, we assessed the distribution of MMP14 in early and late endosomes. The corresponding compartments were identified by immunofluorescent staining with anti-Rab5 and -Rab7 antibodies. In the shControl cells, about 60% of MMP14 containing vesicles co-localized with Rab5 and about 80% with Rab7. Surprisingly, NEDD9 depletion resulted in a two-fold decrease of MMP14 in the Rab7-vesicles, and an increase of MMP14 residing in the Rab5-vesicles (Fig.3f-g; Fig.S1f-g).
A decrease in MMP14 in late endosomes could arise from an increased degradation of MMP14 or a deficiency of MMP14 sorting/trafficking to late endosomes/lysosomes. To assess the rates of protein degradation, the cells were treated with cycloheximide, an inhibitor of de novo protein synthesis, and the protein levels of MMP14 were measured during a time course study. Surprisingly, the amount of MMP14 protein in shNEDD9 cells was 20% higher than in the control (Fig.S2a-b), suggesting that a decrease in NEDD9 leads to a decrease in the degradation of MMP14. Thus, NEDD9 is most likely to be involved in MMP14 sorting/trafficking to late endosomes.
Note that NEDD9 depletion did not affect the tested membrane compartments (Rab7-late endosomes, Rab11-recycling endosomes, Lamp1-lysosomes) as determined by the immunofluorescent staining (Fig.S2c-d). The trafficking of other proteins to late endosomes/lysosomes such as EGF (Fig.S2e) was also not affected. There were no changes in the acidification of the endosomes upon NEDD9 depletion (Fig.S2f-g), supporting the notion of a specific action by NEDD9 on MMP14 trafficking.
NEDD9 deficiency leads to the enlargement of early/recycling/sorting compartment
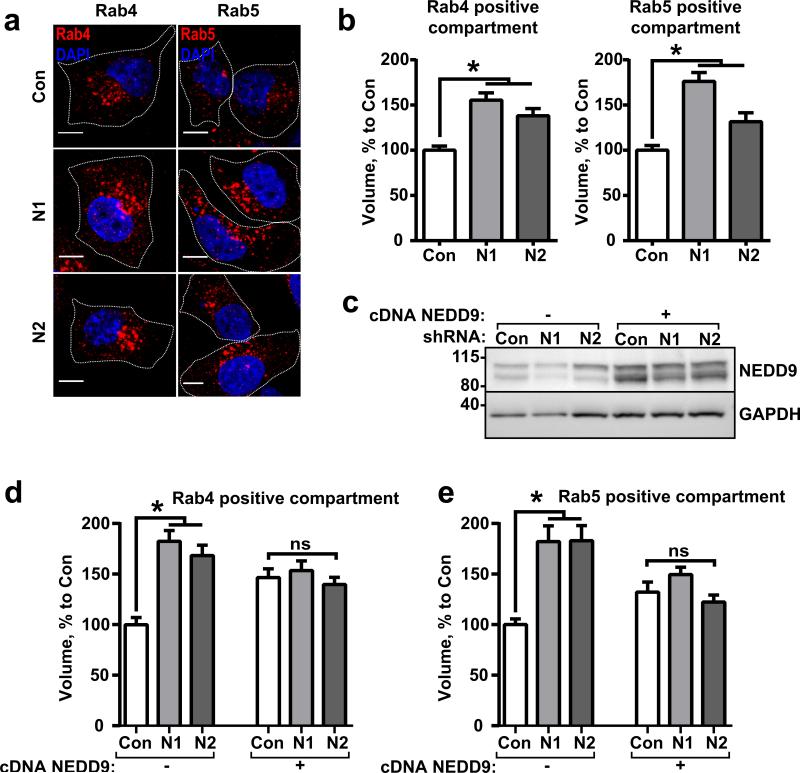
Interestingly, NEDD9 deficiency leads to a two-fold increase in the volume of the early/recycling/sorting compartment (ERS) based on a 3D reconstruction of the confocal images (Fig.4a-b). It is difficult to assess whether the increase was driven by fewer larger vesicles or by an increase in the total number of Rab4/Rab5-positive vesicles due to the tubular shape of the ERS and the many merging points between the differently shaped vesicles. A western blot analysis of the whole cell lysates revealed no change in the Rab-GTPases expression in the shNEDD9 cells (Fig.S3a-d). Nevertheless, re-expression of wild type NEDD9 was sufficient to rescue the phenotype and reduce the volume of the ERS to the control level (Fig.4c-e).
Figure 4. NEDD9 deficiency leads to enlargement of the early/recycling/sorting (ERS) compartment.
(a) Representative images of shControl (Con) and shNEDD9 (N1, N2) MDA-MB-231 cells stained with anti-Rab4 or anti-Rab5 antibodies, DNA (DAPI); Scale bar is 10μm. (b) Quantification of volume of Rab4 and Rab5 positive vesicles based on 3D reconstruction of confocal images; mean volume (%) to control (Con, assigned as 100%) ±S.E.M; three independent experiments, 100 cells/treatment; one-way ANOVA with Dunnett's post-hoc analysis Rab4: *p<0.0001, =0.0002 for Con/N1 or/N2; Rab5:*p<0.0001, =0.0165 for Con/N1 or /N2. (c) Representative image of WB analysis of shControl (Con) and shNEDD9 (N1, N2) MDA-MB-231 cells without (−) or with (+) re-expression of wild type NEDD9 with anti-NEDD9 (2G9) antibodies; GAPDH used as loading control. (d-e) Quantification of volume of Rab4 (d) and Rab5 (e) positive vesicles in cells as in (c) with and without NEDD9 rescue; mean volume as % to control (Con, assigned as 100%) ±S.E.M; three independent experiments, 100 cells/treatment; one-way ANOVA with Dunnett's post-hoc analysis Rab4: *p<0.0001, for Con/N1 or/N2; ns for Con/N1+NEDD9/or N2+NEDD9; Rab5:*p<0.0001 for Con/N1 or /N2; ns for Con/N1+NEDD9/or N2+NEDD9.
Importantly, in the absence of NEDD9, MMP14 gets recycled from the ERS back to the PM (Fig.2-3). This could potentially be associated with the dysregulation of adenosine diphosphateribosylation factor 6 (Arf6)-GTPase (17, 26, 27), which was previously shown to be involved in the recycling of multiple receptors including c-Met (28).
NEDD9 is required to control the levels of active Arf6 GTPase
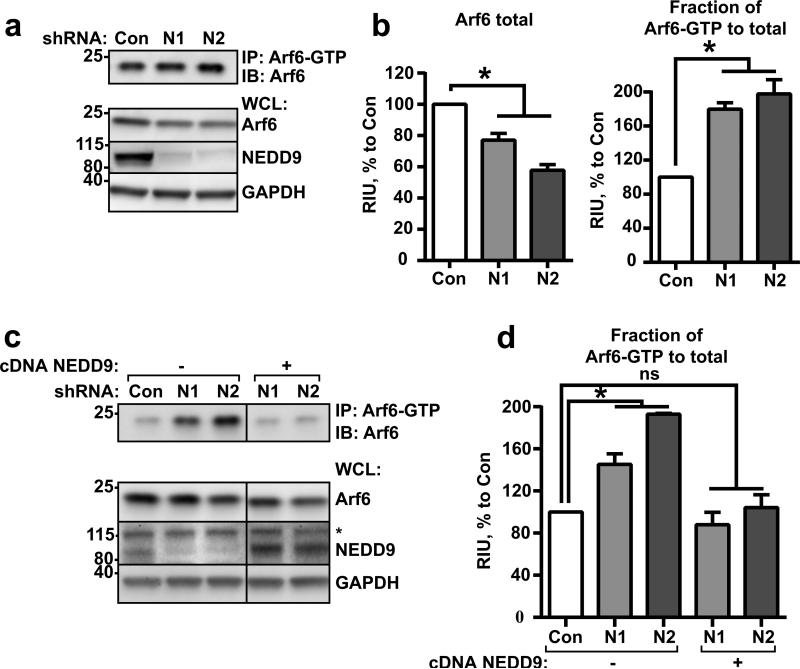
NEDD9 is a scaffolding protein, and through binding to GEFs like DOCK180 and AND-34, it controls the activation of Rap1 (29) and Rac1 (30). Following this trend, we tested the activity of Arf6-GTPase upon NEDD9 manipulation. Surprisingly, overexpression of NEDD9 led to a decrease in Arf6 activity (Fig.S4a), while the knockdown of NEDD9 resulted in approximately a two-fold increase in Arf6 (Fig.5a-b) activity. We did not detect any changes in Arf1 activity upon NEDD9 depletion (Fig.S4b-c), suggesting a specific action by NEDD9 on Arf6. The activation of Arf6 was also indirectly confirmed by an increase in the activity/phosphorylation of the downstream Arf6 target PIP5 alpha kinase (Fig.S4d), and by an increase in membrane invaginations, as previously reported in cells with upregulation of endogenous Arf6 (Fig.S4e) (31). We also noticed a moderate decrease in the total amount of the Arf6 protein in cells with constitutive knockdown of NEDD9 (Fig.5a-b). Interestingly, upon transient siRNA depletion, no changes in the total Arf6 level were observed (Fig.S4f), suggesting the existence of a compensatory mechanism for the excessive amount of active Arf6 or reflecting the destabilization of Arf6 in the absence of NEDD9. Re-expression of exogenous NEDD9 in shNEDD9 cells decreased the amount of active Arf6 (Fig.5c-d). Collectively, our findings suggest that NEDD9 is required to keep activity of Arf6-GTPase balanced, thus providing fine tuning of GTP/GDP bound Arf6 cycling and allowing for cargo sorting either for recycling or degradation.
Figure 5. NEDD9 regulates inactivation of Arf6 GTPase.
(a) Representative image of WB analysis with anti-Arf6. Top: pull-down of Arf6-GTP from shControl (Con) and shNEDD9 (N1, N2) MDA-MB-231 cells; Bottom: whole cell lysate (WCL) used for pull-down, WB with anti-Arf6, -NEDD9 (2G9), -GAPDH antibodies. (b) Quantification of relative intensity units (RIU) of WBs from (a), Arf6-total in WCL and Arf6-GTP in pull-down); three independent experiments; graphs are mean RIU as % to control (Con, assigned as 100%) ±S.E.M. one-way ANOVA with Dunnett's post-hoc analysis Arf6 total: *p=0.0013, <0.0001 for Con/N1 or/N2; Arf6-GTP: *p=0.0009, 0.0002 for Con/N1 or/N2. (c) Top: pull-down of Arf6-GTP from shControl (Con), shNEDD9 (N1, N2) or shNEDD9 (N1, N2) re-expressing wild type NEDD9 (+) MDA-MB-231 cells. WB with anti-Arf6 antibody; Bottom: WCL used for pull-down, WB with anti-Arf6, -NEDD9, -GAPDH antibodies. * indicate non-specific band of p130Cas. (d) Quantification of RIU as described in (b) of Arf6-GTP in three independent experiments; graphs are mean RIU as % to control (Con, assigned as 100%) ±S.E.M. one-way ANOVA with Dunnett's post-hoc analysis Arf6-GTP: *p=0.0226, 0.0005 for Con/N1 or/N2; ns for Con/N1+NEDD9 or N2+NEDD9.
NEDD9 binds to Arf6-GAP, ARAP3, and the effector GGA3
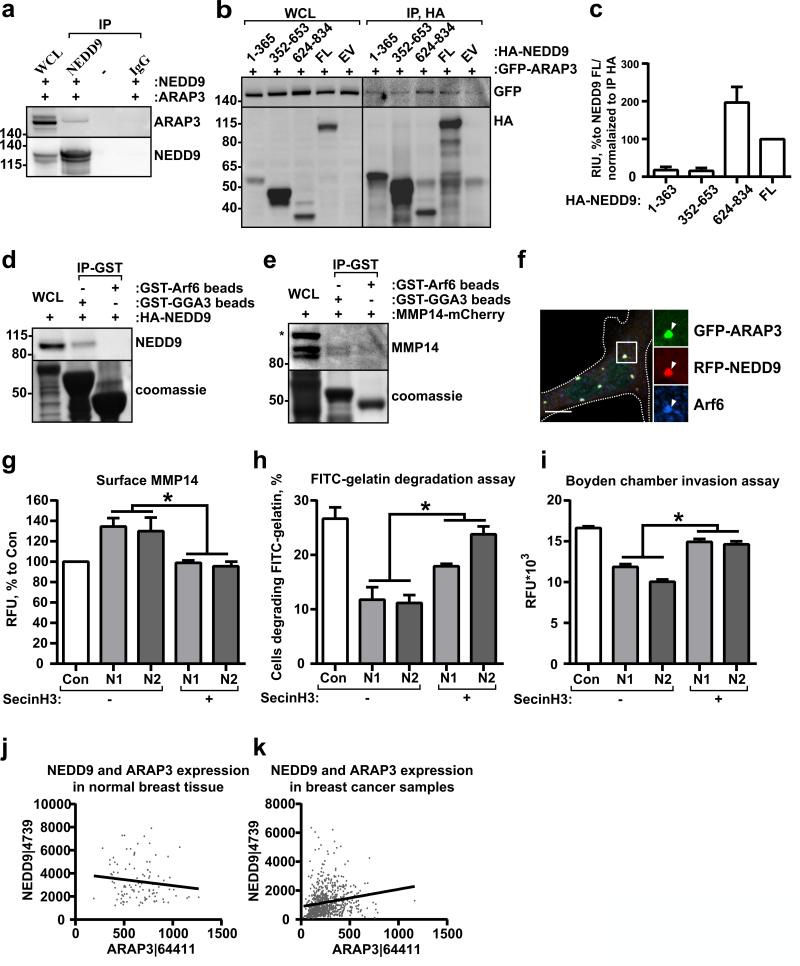
To delineate molecular mechanisms of the NEDD9-driven decrease in Arf6 activity, we screened multiple Arf6-GTPase Activating Proteins (GAPs) (Fig.6a, Fig.S5a). We found that NEDD9 specifically binds to ARAP3 (Fig.6a). The minimum binding fragment resides in the C-terminal domain of NEDD9 (626-834aa) (Fig.6b-c). Interestingly, the Arf6-GTP downstream effector, adaptor protein GGA3, was also present in the complex with NEDD9 (Fig.6d), suggesting that the NEDD9/ARAP3 complex is targeted to the active Arf6/GGA3 complex to promote GTP hydrolysis. We also found MMP14 in the complex with GGA3 (Fig.6e), indicating the involvement of Arf6-GGA3 in MMP14 trafficking.
Figure 6. NEDD9 binds to Arf6-GAP, ARAP3, and effector GGA3.
(a) Representative images of IP with anti-NEDD9 or non-specific IgG from HEK293T cells cotransfected with GFP-ARAP3 and RFP-NEDD9 plasmids; WB with anti-ARAP3 and -NEDD9 antibodies. (b) Delineation of NEDD9 domains involved in interaction: IP with anti-HA from HEK293T cells co-transfected with GFP-ARAP3 and HA-NEDD9 truncation mutants (amino acids as indicated), full length cDNA (FL) or empty vector (EV) plasmids; WB with anti-GFP and -HA antibodies. (c) Quantification of WB as in (b) from in three independent experiments; graphs are mean values as % to FL (assigned as 100%) ±S.E.M. (d) GST pull-down (IP-GST) from HEK293T cells transfected with HA-NEDD9 using GST-GGA3 or GST-Arf6 proteins; WB with anti-NEDD9 antibodies. (e) GST pull-down (IP-GST) from HEK293T cells transfected with mCherry-MMP14 using GST-GGA3 or GST-Arf6 proteins; WB with anti-MMP14 antibodies. (f) Immunofluorescent analysis of MDA-MB-231 cells co-transfected with RFPNEDD9 (red) and GFP-ARAP3 (green) and stained for Arf6 (blue). Scale bar is 10μm. Insets on the right are enlarged areas indicated in the merged image. (g) FACS analysis of MMP14 surface staining in shControl (Con) or shNEDD9 (N1, N2) MDA-MB-231 cells treated with vehicle (-) or SecinH3 (+); three independent experiments; graphs are mean values of RFU % to Con (assigned as 100%) ±S.E.M; one-way ANOVA with Dunnett's post-hoc analysis; Vehicle treated: *p<0.05 (shCon/shN1 or N2); SecinH3 treated: *p<0.0099, 0.0211 (shCon/shN1 or N2). (h) Quantification of matrix degradation in FITC-gelatin degradation assay of cells, as in (g), 100 cells/cell line/experiment in 3 independent experiments. Results presented as % of cells degrading matrix, ±S.E.M; one-way ANOVA with Tukey's post-hoc analysis N1 or N2: *p=0.024, 0.002 (Vehicle/SecinH3). (i) Boyden chamber matrigel invasion assay, cells as in (gh). Data plotted as RFU; one-way ANOVA with Tukey's post-hoc analysis N1 or N2: *p<0.0001 (Vehicle/SecinH3). (j-k) Spearman correlation analysis of NEDD9 and ARAP3 mRNA expression in normal (j) breast tissue or breast cancer biopsies (k) based on TCGA data base; n=105 (for Normal breast), n=791 (for Cancer breast), r=-0.1161, p-ns for normal, r=0.3035, p<0.0001 for tumor.
To define subcellular localization of these proteins, we expressed fluorescently labeled NEDD9 and ARAP3 in tumor cells. Most of the NEDD9 and ARAP3 proteins co-localized at the Arf6-positive vesicles (Fig.6f), suggesting that the membrane associated Arf6-GTP/GGA3 complex recruits NEDD9 and ARAP3 thereby promoting Arf6 inactivation and complex dissociation.
Decrease in Arf6 activity is sufficient to restore PM-MMP14 levels and its activity
To determine the impact of Arf6 activity on MMP14 trafficking, we assessed the level of PM-MMP14 upon partial inhibition of Arf6 by an antagonist of the cytohesin family GEFs, SecinH3, which was previously characterized for Arf6 (32). Treatment of the shNEDD9 cells with SecinH3 or re-expression of wtNEDD9 (10) rescued the phenotype leading to a decrease in the amount of PM-MMP14 (Fig.6g). Moreover, application of SecinH3 restored the degradation capacity and the invasion deficiency (Fig.6h-i) of shNEDD9 tumor cells, indirectly indicating an increase in the activity of MMP14 and soluble MMPs. A publicly available TCGA dataset analysis of ARAP3 and NEDD9 mRNA expression revealed a significant correlation in expression of these proteins in breast cancer biopsies, but not in normal tissue (Fig.6j-k), implying that cooperation between ARAP3 and NEDD9 could be critical for tumor progression (33).
TIMP2/ MMP14 complex targets to Rab7 endosomes
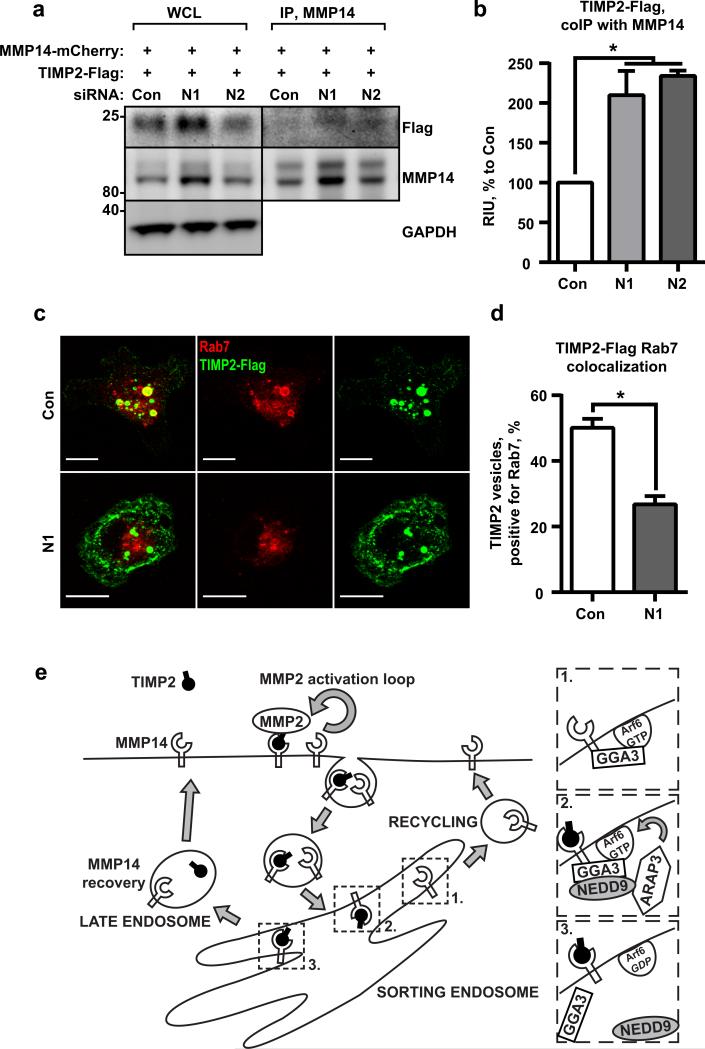
Previously, our group reported an accumulation of an endogenous MMP inhibitor, TIMP2, on the surface of the NEDD9 deficient cells as a potential mechanism for MMP14 inactivation upon depletion of NEDD9 (10). To test if PM-MMP14 forms a complex with TIMP2 and visualize it, we expressed mCherry-MMP14 and Flag-tagged-TIMP2 in shControl and shNEDD9 cells. Increased amounts of the TIMP2/MMP14 complex were co-immunoprecipitated from shNEDD9 cells (Fig.7a-b), confirming our previous findings and suggesting a potential decrease in TIMP2/MMP14 dissociation under NEDD9 deficient conditions. Note that no changes in the expression or secretion of TIMP2 were detected upon the manipulation of NEDD9 (10). To identify the cause of the increase in the TIMP2/MMP14 complex, we determined the fate of the internalized TIMP2. The cells expressing mCherry MMP14 and TIMP2-Flag were fed with anti-Flag antibodies to visualize the TIMP2 internalization. The shNEDD9 cells showed a two-fold decrease in targeting TIMP2 to late endosomes based on co-localization with Rab7-positive vesicles (Fig.7c-d). We also noted a robust increase in surface TIMP2 upon NEDD9 depletion. Overall, these data suggest that the TIMP2/MMP14 complexes are dissociating/degrading less efficiently in NEDD9 deficient cells due to the impaired targeting to the late endosomes, where TIMP2 gets released from MMP14 in a pH dependent-manner (13), (34). However, further studies are required to delineate the fate of TIMP2 targeted to late endosomes and the potential for TIMP2 recycling (11, 13).
Figure 7. TIMP2/ MMP14 complex targets to Rab7 endosomes.
(a)WB analysis of WCL and IP with anti-MMP14 antibodies from siControl (Con) or siNEDD9 (N1, N2) MDA-MB-231 cells transfected with mCherry-MMP14 and Flag-tag fusion TIMP2; WB with anti-Flag and -MMP14 antibodies. (b) Quantification of WB as in (a) from three independent experiments; graphs are mean RIU values as % to siCon (assigned as 100%) ±S.E.M; one-way ANOVA with Dunnett's post-hoc analysis *p=0.0368 or 0.0216 for siCon/siN1 or N2; (c) Representative 3D reconstructed confocal images of cells as in (a) stained with anti-Rab7 (red) and anti-Flag antibodies (green); Scale bar is 10μm. (d) Quantification of colocalization of Rab7 and TIMP2 positive vesicles, mean % of TIMP2/Rab7 positive vesicles ±S.E.M; three independent experiments, 10 cells/treatment; two-tailed t-test:*p= 0.0003 for Con/N1. (e) Schematic representation of the model depicting action of NEDD9 on Afr6 through recruitment of ARAP3 to Arf6/GGA3/MMP14 complex.
Collectively, our findings suggest a model (Fig.7e) in which NEDD9 scaffolds the interaction between ARAP3 and the Arf6-GTP/GGA3/MMP14 complex at the Arf6-positive ERS compartment to facilitate GTP hydrolysis, Arf6 inactivation, and dissociation from the ERS along with GGA3. This would allow vesicle maturation and trafficking to late endosomes for MMP14 enzyme recovery.
Downregulation of NEDD9 via siNEDD9-Vivo-Morpholino treatment decreases tumor progression in a xenograft model of breast cancer
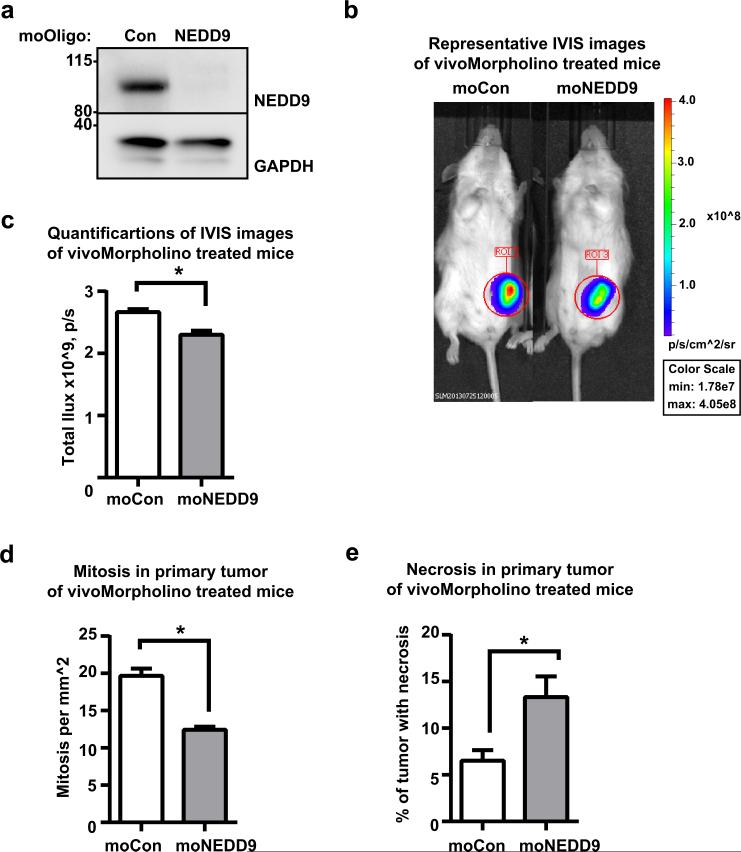
NEDD9 overexpression is a feature of invasive human cancers with increased metalloproteinase activity. Unfortunately, the anti-MMP cancer therapy did not yield the expected results in the clinic, hence targeting alternative processes that control activity of MMPs might prove to be a viable anti-metastasis strategy. Therefore, we wanted to determine whether NEDD9 knockdown via treatment with antisense oligonucleotides targeting NEDD9 that have been modified for in vivo applications (moNEDD9-Vivo-Morpholinos) (Fig.8a-b) affects tumor growth and metastasis in a mouse xenograft model using MDA-MB-231LN cells. We found a significant decrease in the size of primary tumors and metastases in lungs of mice treated with moNEDD9 compared to moControl after only 1 week of treatment (Fig.8c-e). The primary tumors exhibited a striking decrease in viability and proliferation based on the mitotic index and the percentage of necrotic tissue evaluated by pathology. These results indicate that NEDD9 depletion in established highly metastatic tumors decreases growth and further dissemination of breast cancer cells. Importantly, these data provide the first clinical evidence of the potent anti-tumorigenic activity of NEDD9 targeted siRNAs in vivo and evaluates NEDD9 as a novel therapeutic target.
Figure 8. Downregulation of NEDD9 expression via siNEDD9-Vivo-Morpholino treatment decreases tumor progression in xenograft model of breast cancer.
(a) WB analysis of WCL with anti-NEDD9 antibodies of moControl (moCon) or moNEDD9 MDA-MB-231 treated cells. (b) Representative images of bioluminescence (BLI) of mice orthotopically injected with MDA-MB-231LN cells after 1 weeks of treatment. (c) Quantification of the primary tumor growth based on BLI, n=4/group, mean total photon flux +/−S.E.M, t-test p=0.0266 after 1 weeks of treatment; (d) Quantification of mitotic cells in primary tumor +/−S.E.M after 1 weeks of treatment, n=4 in each group, t-test, p<0.0001; (e) Quantification of percentage of necrotic area in primary tumor +/−S.E.M after 1 weeks of treatment, n=4 in each group, p=0.0081.
Discussion
Recent studies have established NEDD9 as an essential factor for invasive (1, 2) and metastatic behavior in tumors (3-5). The results of this study establish a novel role for NEDD9 as a key regulator of Arf6-dependent trafficking of MMP14, highlighting the basic mechanism utilized by NEDD9 to support cancer cell invasion and metastasis. The cancer cell lines (35) used in this study utilize the mesenchymal type of invasion (36), however we are not excluding the possibility of similar effects of NEDD9 on collective cell migration, where the leading cells (or recruited fibroblasts) often utilize MMP-dependent invasion (37). This mechanism might be involved in regulation of other known NEDD9 functions including focal adhesion disassembly (38) and integrin signaling (39).
Expression and activity of MMP14 is critical for normal morphogenesis and tumor progression (40-42). While nearly 80% of the internalized MMP14 is recycled, little is known about the regulation of this process (12, 22). Several studies reported recycling of MMP14 from early and late endosomes (14, 15, 24), but the key molecules defining the route of recycling for MMP14 are still unknown. Even less is known about the fate of TIMP2, an MMP14 ligand involved in MMP2 activation (11-13). As we previously indicated, NEDD9 depletion leads to a decrease in activation of MMP2 and MMP9, thus affecting the ability of cancer cells to invade (43). The TIMP2 is internalized with MMP14 (12), and is either degraded (11) or recycled (13). Interestingly, the documented pH required for TIMP2 dissociation (13) is reached only in the late endosomes (34). Thus, recycling of the MMP14/TIMP2 complex through late endosomes might be required for reactivation of the TIMP2 bound enzyme.
In our study, we stumbled upon a paradoxical observation where the depletion of NEDD9 resulted in a decrease in MMP14 activity, concomitant with an increase in the amount of PMMMP14. Our data suggests that the main form of MMP14 present in tested cells is active furin processed MMP14 (Fig.S1c). NEDD9 deficiency does not interfere with furin-dependent activation of MMP14 based on no changes in MMP14 size or internalization (44), but rather leads to a decreased targeting of the TIMP2/MMP14 complex to late endosomes and an increase in recycling from the ERS compartment. This indicates the switch from recycling through late endosomes to the ERS recycling pathway.
NEDD9 is known to activate small GTPase Rac1 (30), cdc42 (45) and Rap1, driving mesenchymal migration and T-cell activation (29). Therefore, we decided to check whether NEDD9 regulates MMP14 recycling through small GTPases. Surprisingly, NEDD9, was involved in inactivation of Arf6-GTPase. Arf6 regulates key aspects of endocytic recycling (46), (47). The activity of Arf6 is closely controlled by a number of GEFs including GEP100 (48) and ARNO (49), and also GAPs such as ACAP1 (50), ASAP1 and ARAP3. We found that NEDD9 interacts with ARAP3 and the Arf6 downstream effector GGA3 (28). The NEDD9 depletion phenotype resembles the previously published aluminum fluoride induced increase in the activity of endogenous Arf6 (27) rather than overexpression of a constitutively active Arf6, Arf6Q67L (31, 51, 52). A two-fold increase in the amount of Arf6-GTP, in the case of NEDD9 depletion, did not result in an accumulation of large actin-positive vesicles, but instead both increased the volume of vesicles in the early endocytic compartment and also Arf6-dependent recycling to the PM, similar to a phenotype previously reported (53). Moreover, in the case of NEDD9 depletion, there is no overexpression of Arf6 protein, but rather a decrease, indicating the existence of a potential feedback loop to reduce the activity of Arf6 via a decrease in protein stability or expression.
The increase in activity of endogenous Arf6 upon NEDD9 depletion was further confirmed by an analysis of its downstream target – phosphatidylinositol-4-phosphate-5-kinase (PIP5K) (26). Activation of PIP5K could explain the increased number of membrane invaginations and ruffling in NEDD9 deficient cells, which was also previously reported in cells with increased Arf6 activity (54). NEDD9 deficiency does not abolish proper sorting of cargoes or maturation of endosomes, as shown by live cell imaging using RFP-Rab5/or Rab7 and Alexa647-EGF. However, the distribution of MMP14 to different endosomal compartments is affected and its recycling is re-routed. The same redistribution was observed for TIMP2. Previously, a similar phenomenon was documented for GGA3, which is required for targeting the c-Met receptor for recycling instead of degradation (28). In our current work, we found MMP14 in a complex with GGA3, suggesting the possibility of similar regulation of MMP14. We proposed a model where NEDD9 controls the switch of MMP14 recycling through the ERS versus late endosomes (Fig.7e). Previously MMP14 was reported to be recycled through a Rab8 dependent mechanism (55). We did not observe any changes in the level of Rab8 (Fig.S3c-d), however we are not excluding the possibility of crosstalk between Arf6 and Rab8 in regulation of MMP14 trafficking, as the Arf6 and Rab8 positive compartments are largely overlapping (56).
Ablation of the MMP14 gene causes severe developmental defects (57), suggesting that the activity of MMP14 cannot be substituted by any other MMP. Attempts at developing selective MMP14 small molecule inhibitors have been so far unsuccessful (58). Hence, binding partners that modulate MMP14 activity may represent an attractive avenue for MMP14 inhibition. Alteration of NEDD9 expression in tumor cells thus provides a novel regulatory mechanism for controlling invasive properties via MMP14 (59). The decrease in tumor progression upon inhibition of NEDD9 by antisense therapy in human breast cancer xenografts supports this notion and might allow for a targeted decrease in MMP activity in the tumor, thus providing a novel therapeutic approach to block the spreading of cancer.
Materials and Methods
Plasmids and cell culture
Cell lines HEK293T (ATCC), Hs578T (ATCC), and MDA-MB-231LN (Caliper LifeScience) were grown based on the manufacturer's recommendations. shRNAs and siRNAs against NEDD9, non-targeting control (ThermoFisher Scientific), and NEDD9-cDNA were prepared as previously described (7). mRFP-Rab5, and -Rab7 were purchased from Addgene (plasmid #14437, #14436) (60). Plasmids mCherry-MMP14 and PAmCherry-MMP14 were a gift from Dr.Chavrier (Institute Curie, France) and Dr.Machesky (The Beatson Institute for Cancer Research, Scotland) (24). Human TIMP2-cDNA (ThermoFisher Scientific) was sub-cloned into pcDNA3.1-3x-Flag plasmid (gift from Dr.Frisch, West Virginia University, USA).
ECM coated Boyden chamber invasion assays
Assays were carried out according to the manufacturer's protocol and as previously described (7).
Fluorescent-gelatin degradation assay
Fluorescence-activated cell sorting
Assay was performed as previously described (10).
Protein expression, western blotting, and immunoprecipitation
For pull down (PD) assays, the recombinant GST-tagged GGA3-VHS-GAT domain was produced in Bl21(DE3) E.coli as previously described (62). Western blotting was done as previously described (62). Primary antibodies against NEDD9 mAb (2G9) (62), PIP5K, phS309-PIP5K, MMP14 (Novus International), Flag (Sigma-Aldrich), Arf6, (Santa-Cruz Biotech), GAPDH (Millipore), GFP, EEA1 (BD Bioscience), GGA3, RFP (ThermoFisher), GFP, LAMP1/2 (NIH Hybridoma Bank), Rab4, Rab5, Rab7, Rab8, Rab11 (Cell Signaling Technology) were used. Bands were digitized and quantified using a digital documentation and image analysis system (Syngene).
Immunofluorescence
Cells were processed as previously described (62). Primary antibodies against Rab4, 5, 7, 9, 11 (Cell Signaling Technology), Lamp1 (NIH Hybridoma Bank), Arf6 (Santa-Cruz Biotech), MMP14 (Novus International), MMP14-488 (R&D Systems), EEA1 (BD Bioscience) were used. Secondary antibodies included AlexaFluor 405, 488, 555, 647 (LifeTechnologies). Images were captured using a standard setting by a confocal microscope LSM510 (Zeiss). All images represent whole cell 3D reconstructed projections with 0.25μm steps.
Cell surface Biotinylation assay
Cells pretreated with 10μM MG132 and 50μM Leupeptin (FisherScientific) were labeled at 4°C with Sulfo-NHS-S-S-biotin (ThermoFisher Scientific) in PBS, followed by incubation at 37°C for internalization of surface molecules. Residual surface biotinylation was removed by incubation with 200mM MESNA (Sigma) for 30 minutes. Cells were homogenized in PTY buffer (62), and biotinylated molecules were pulled down with streptavidin-conjugated agarose (Millipore) and used for western blotting.
Live cell imaging Endocytosis
. Cells were plated on delta-T4 glass bottom dishes (FisherScientific), labeled with EGF-647 or CellMask DeepRed (LifeTechnologies) in the presence of 0.45M sucrose, and imaged by a LSM510 (Zeiss) or NikonLiveScan SFC microscope equipped with perfect focus x-y stage control and z-axis motor (Nikon), Photometrics QuantEM CCD camera (Photometrics), and a 60X PlanFluor, NA1.21 objective. Images were captured every 5 seconds for 30 minutes. NIS (Nikon) and ImageJ (NIH) software were used for data processing and analysis. To compare rates of internalization, small areas corresponding to cell margins were selected and fluorescence intensities throughout the movie were measured.
FRAP analysis
Cells were transfected with mCherry-MMP14 and siRNAs against NEDD9, plated on glass bottom dishes (InVitro Scientific), imaged with LSM510 (Zeiss) microscope both before and after photo-bleaching of the designated area every 15 seconds for 5 minutes. The collected movies were analyzed with ImageJ software (NIH).
Trafficking of PAmCherry-MMP14
Cells were transfected with mCherry-MMP14 and siNEDD9, plated on glass bottom dishes (InVitro Scientific), and imaged 24 hours later with a LSM510 (Zeiss) confocal microscope both before and after photo-activation of the designated area every 15 seconds for 5 minutes. The collected movies were analyzed with ImageJ software (NIH).
MMP14 recycling assay
Cells were transfected with mCherry-MMP14 and siNEDD9. Mouse monoclonal anti-MMP14 antibodies conjugated with AlexaFluor-488 (R&D Systems) were added to the cells for 1 hour at 37°C. Cells were washed with acidic strip solution (25), and the Ab/MMP14 complex was allowed to recycle for 1 hour at 37°C, followed by fixation with 4% PFA/PBS and processed for immunofluorescence (without permeabilization).
MMP14/TIMP2 intracellular distribution
The cells were transfected with mCherry-MMP14 or PAmCherry-MMP14 and TIMP2-Flag. The anti-MMP14-488 or anti-Flag antibodies were added to the cells for 1 hour at 37°C, fixed with 4% PFA/PBS and processed for immunofluorescence.
Transmission electron microscopy
Cells were fixed with 2.5% glutaraldehyde/2% paraformaldehyde, post fixed in 1% osmium tetroxide for 1 hour, stained with uranyl acetate, and processed for conventional ultrathin sectioning. Sections were post stained with uranyl acetate and lead citrate, and imaged with a Zeiss Libra120 transmission electron microscope.
Arf6/Arf1 activity assessment
The assay was performed using an Active Arf6 Pull-Down and Detection Kit (Pierce) or Arf1 Activation Assay Kit (Cell Biolabs) according to the manufacturer's protocols.
Correlative analysis of NEDD9 and ARAP3 in breast cancer
TCGA (NIH) data sets of 791 human breast cancer microarrays and 105 normal tissue microarrays were used for analysis with the GraphPad Prism package (GraphPad Software, Inc.) with Spearman correlation analysis.
MMP14 Activity assay
4×104 cells/well were plated on a 96-well plate. MMP14 fluorogenic substrate (Millipore) was added in L15-medium for 4 hours. Fluorescence was measured by plate-reader SynergyH4 (BioTec) at 328/400mn excitation/emission.
Animal Bioluminescence Imaging (BLI) of mammary fat pad xenografts
Nod/SCID/gamma (NSG) immunodeficient mice (Jackson Laboratory) were housed in the West Virginia University Animal Facility under pathogen-free conditions with an approved Institutional Animal Care and Use Committee protocol. The tumor injection and analysis was performed as previously described (8, 11). The moNEDD9- or moControl-VivoMorpholino (GeneTool LLC) treatment was initiated 2 weeks post tumor cell injection. Animals with tumors of similar volume were injected by IP with 12.5 mg/kg of Vivo-Morpholinos dissolved in PBS daily for 1 week. The average lung and primary tumor radiance was measured at each time point. The mammary fat pads and lungs were collected at the end of the study, fixed, and pathologically analyzed for the number of mitotic tumor cells, percentage of necrosis, and number/size of metastases as described (8) by a pathologist (M.B.S).
Statistical Analysis
Statistical comparisons were made using two-tailed student's t test. When more than two groups were analyzed, one-way analysis of variance (ANOVA) was used. P≤0.05 was considered to be significant. Experimental values were reported as the means ± S.E.M. All calculations of statistical significance were performed using the GraphPad Prism package (GraphPad Software, Inc.).
Supplementary Material
Acknowledgements
We thank Mrs. Yoho and Drs. Schaller and Weed for critically reading the manuscript and providing administrative support during manuscript preparation. We also thank the West Virginia University Microscope Imaging Facility, supported by the Mary Babb Randolph Cancer Center and NIH grants P20 RR016440, P30 RR032138/GM103488, and P20 RR016477. The WVU Flow Cytometry Core Facility was supported by NIH grants P30GM103488, P30RR032138, and RCP1101809. We also thank Mr. Farrugia for his assistance in the analysis of the microarray data. This work was supported by grants from the NIH-NCI (CA148671 to ENP), the Susan G. Komen For the Cure Foundation (KG100539 to ENP), and in part by NIH/NCRR (5 P20 RR016440-09).
Footnotes
Disclaimers: This manuscript contains original work only and has not been published or submitted elsewhere. All authors have directly participated in the planning, execution and analysis of this study and approved the submitted version of this manuscript.
Conflicts of interest: The authors declare no conflict of interest.
References
- 1.Kim M, Gans JD, Nogueira C, Wang A, Paik JH, Feng B, et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125(7):1269–81. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Kondo S, Iwata S, Yamada T, Inoue Y, Ichihara H, Kichikawa Y, et al. Impact of the integrin signaling adaptor protein NEDD9 on prognosis and metastatic behavior of human lung cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18(22):6326–38. doi: 10.1158/1078-0432.CCR-11-2162. [DOI] [PubMed] [Google Scholar]
- 3.Speranza MC, Frattini V, Pisati F, Kapetis D, Porrati P, Eoli M, et al. NEDD9, a novel target of miR-145, increases the invasiveness of glioblastoma. Oncotarget. 2012;3(7):723–34. doi: 10.18632/oncotarget.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn J, Sanz-Moreno V, Marshall CJ. The metastasis gene NEDD9 product acts through integrin beta3 and Src to promote mesenchymal motility and inhibit amoeboid motility. Journal of cell science. 2012;125(Pt 7):1814–26. doi: 10.1242/jcs.101444. [DOI] [PubMed] [Google Scholar]
- 5.O'Neill GM, Seo S, Serebriiskii IG, Lessin SR, Golemis EA. A new central scaffold for metastasis: parsing HEF1/Cas-L/NEDD9. Cancer research. 2007;67(19):8975–9. doi: 10.1158/0008-5472.CAN-07-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Little JL, Serzhanova V, Izumchenko E, Egleston BL, Parise E, Klein-Szanto AJ, et al. A requirement for Nedd9 in luminal progenitor cells prior to mammary tumorigenesis in MMTV HER2/ErbB2 mice. Oncogene. 2013 doi: 10.1038/onc.2012.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ice RJ, McLaughlin SL, Livengood RH, Culp MV, Eddy ER, Ivanov AV, et al. NEDD9 depletion destabilizes Aurora A kinase and heightens the efficacy of Aurora A inhibitors: implications for treatment of metastatic solid tumors. Cancer research. 2013;73(10):3168–80. doi: 10.1158/0008-5472.CAN-12-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucas JT, Jr., Salimath BP, Slomiany MG, Rosenzweig SA. Regulation of invasive behavior by vascular endothelial growth factor is HEF1-dependent. Oncogene. 2010;29(31):4449–59. doi: 10.1038/onc.2010.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin Y, Li F, Zheng C, Wang Y, Fang Z, Guo C, et al. NEDD9 promotes lung cancer metastasis through epithelial-mesenchymal transition. International journal of cancer Journal international du cancer. 2013 doi: 10.1002/ijc.28568. [DOI] [PubMed] [Google Scholar]
- 10.McLaughlin SL, Ice RJ, Rajulapati A, Kozyulina PY, Livengood RH, Kozyreva VK, et al. NEDD9 Depletion Leads to MMP14 Inactivation by TIMP2 and Prevents Invasion and Metastasis. Molecular cancer research : MCR. 2013 doi: 10.1158/1541-7786.MCR-13-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maquoi E, Frankenne F, Baramova E, Munaut C, Sounni NE, Remacle A, et al. Membrane type 1 matrix metalloproteinase-associated degradation of tissue inhibitor of metalloproteinase 2 in human tumor cell lines. J Biol Chem. 2000;275(15):11368–78. doi: 10.1074/jbc.275.15.11368. [DOI] [PubMed] [Google Scholar]
- 12.Remacle A, Murphy G, Roghi C. Membrane type I-matrix metalloproteinase (MT1-MMP) is internalised by two different pathways and is recycled to the cell surface. Journal of cell science. 2003;116(Pt 19):3905–16. doi: 10.1242/jcs.00710. [DOI] [PubMed] [Google Scholar]
- 13.Zucker S, Hymowitz M, Conner C, DeClerck Y, Cao J. TIMP-2 is released as an intact molecule following binding to MT1-MMP on the cell surface. Experimental cell research. 2004;293(1):164–74. doi: 10.1016/j.yexcr.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Ma D, Keski-Oja J, Pei D. Co-recycling of MT1-MMP and MT3-MMP through the trans-Golgi network. Identification of DKV582 as a recycling signal. J Biol Chem. 2004;279(10):9331–6. doi: 10.1074/jbc.M312369200. [DOI] [PubMed] [Google Scholar]
- 15.Hakulinen J, Sankkila L, Sugiyama N, Lehti K, Keski-Oja J. Secretion of active membrane type 1 matrix metalloproteinase (MMP-14) into extracellular space in microvesicular exosomes. Journal of cellular biochemistry. 2008;105(5):1211–8. doi: 10.1002/jcb.21923. [DOI] [PubMed] [Google Scholar]
- 16.Williams KC, Coppolino MG. Phosphorylation of membrane type 1-matrix metalloproteinase (MT1-MMP) and its vesicle-associated membrane protein 7 (VAMP7)-dependent trafficking facilitate cell invasion and migration. J Biol Chem. 2011;286(50):43405–16. doi: 10.1074/jbc.M111.297069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nature reviews Molecular cell biology. 2006;7(5):347–58. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 18.Anders N, Jurgens G. Large ARF guanine nucleotide exchange factors in membrane trafficking. Cellular and molecular life sciences : CMLS. 2008;65(21):3433–45. doi: 10.1007/s00018-008-8227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakagami H. The EFA6 family: guanine nucleotide exchange factors for ADP ribosylation factor 6 at neuronal synapses. Tohoku J Exp Med. 2008;214(3):191–8. doi: 10.1620/tjem.214.191. [DOI] [PubMed] [Google Scholar]
- 20.Spang A, Shiba Y, Randazzo PA. Arf GAPs: gatekeepers of vesicle generation. FEBS Lett. 2010;584(12):2646–51. doi: 10.1016/j.febslet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Netzel-Arnett S, Mallya SK, Nagase H, Birkedal-Hansen H, Van Wart HE. Continuously recording fluorescent assays optimized for five human matrix metalloproteinases. Anal Biochem. 1991;195(1):86–92. doi: 10.1016/0003-2697(91)90299-9. [DOI] [PubMed] [Google Scholar]
- 22.Uekita T, Itoh Y, Yana I, Ohno H, Seiki M. Cytoplasmic tail-dependent internalization of membrane-type 1 matrix metalloproteinase is important for its invasion-promoting activity. The Journal of cell biology. 2001;155(7):1345–56. doi: 10.1083/jcb.200108112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anilkumar N, Uekita T, Couchman JR, Nagase H, Seiki M, Itoh Y. Palmitoylation at Cys574 is essential for MT1-MMP to promote cell migration. FASEB J. 2005;19(10):1326–8. doi: 10.1096/fj.04-3651fje. [DOI] [PubMed] [Google Scholar]
- 24.Yu X, Zech T, McDonald L, Gonzalez EG, Li A, Macpherson I, et al. N-WASP coordinates the delivery and F-actin-mediated capture of MT1-MMP at invasive pseudopods. The Journal of cell biology. 2012;199(3):527–44. doi: 10.1083/jcb.201203025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Worthylake R, Opresko LK, Wiley HS. ErbB-2 amplification inhibits down-regulation and induces constitutive activation of both ErbB-2 and epidermal growth factor receptors. J Biol Chem. 1999;274(13):8865–74. doi: 10.1074/jbc.274.13.8865. [DOI] [PubMed] [Google Scholar]
- 26.Brown FD, Rozelle AL, Yin HL, Balla T, Donaldson JG. Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. The Journal of cell biology. 2001;154(5):1007–17. doi: 10.1083/jcb.200103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radhakrishna H, Klausner RD, Donaldson JG. Aluminum fluoride stimulates surface protrusions in cells overexpressing the ARF6 GTPase. The Journal of cell biology. 1996;134(4):935–47. doi: 10.1083/jcb.134.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parachoniak CA, Luo Y, Abella JV, Keen JH, Park M. GGA3 functions as a switch to promote Met receptor recycling, essential for sustained ERK and cell migration. Dev Cell. 2011;20(6):751–63. doi: 10.1016/j.devcel.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu JJ, Lavau CP, Pugacheva E, Soderblom EJ, Moseley MA, Pendergast AM. Abl family kinases modulate T cell-mediated inflammation and chemokine-induced migration through the adaptor HEF1 and the GTPase Rap1. Sci Signal. 2012;5(233):ra51. doi: 10.1126/scisignal.2002632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, et al. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135(3):510–23. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 31.Peters PJ, Hsu VW, Ooi CE, Finazzi D, Teal SB, Oorschot V, et al. Overexpression of wild-type and mutant ARF1 and ARF6: distinct perturbations of nonoverlapping membrane compartments. The Journal of cell biology. 1995;128(6):1003–17. doi: 10.1083/jcb.128.6.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hafner M, Schmitz A, Grune I, Srivatsan SG, Paul B, Kolanus W, et al. Inhibition of cytohesins by SecinH3 leads to hepatic insulin resistance. Nature. 2006;444(7121):941–4. doi: 10.1038/nature05415. [DOI] [PubMed] [Google Scholar]
- 33.Radisky ES, Radisky DC. Matrix metalloproteinase-induced epithelial-mesenchymal transition in breast cancer. J Mammary Gland Biol Neoplasia. 2010;15(2):201–12. doi: 10.1007/s10911-010-9177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmid S, Fuchs R, Kielian M, Helenius A, Mellman I. Acidification of endosome subpopulations in wild-type Chinese hamster ovary cells and temperature-sensitive acidification-defective mutants. The Journal of cell biology. 1989;108(4):1291–300. doi: 10.1083/jcb.108.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blick T, Widodo E, Hugo H, Waltham M, Lenburg ME, Neve RM, et al. Epithelial mesenchymal transition traits in human breast cancer cell lines. Clinical & experimental metastasis. 2008;25(6):629–42. doi: 10.1007/s10585-008-9170-6. [DOI] [PubMed] [Google Scholar]
- 36.Friedl P, Locker J, Sahai E, Segall JE. Classifying collective cancer cell invasion. Nature cell biology. 2012;14(8):777–83. doi: 10.1038/ncb2548. [DOI] [PubMed] [Google Scholar]
- 37.Giampieri S, Pinner S, Sahai E. Intravital imaging illuminates transforming growth factor beta signaling switches during metastasis. Cancer research. 2010;70(9):3435–9. doi: 10.1158/0008-5472.CAN-10-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong J, Baquiran JB, Bonakdar N, Lees J, Ching YW, Pugacheva E, et al. NEDD9 stabilizes focal adhesions, increases binding to the extra-cellular matrix and differentially effects 2D versus 3D cell migration. PloS one. 2012;7(4):e35058. doi: 10.1371/journal.pone.0035058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cabodi S, del Pilar Camacho-Leal M, Di Stefano P, Defilippi P. Integrin signalling adaptors: not only figurants in the cancer story. Nat Rev Cancer. 2010;10(12):858–70. doi: 10.1038/nrc2967. [DOI] [PubMed] [Google Scholar]
- 40.Hemmann S, Graf J, Roderfeld M, Roeb E. Expression of MMPs and TIMPs in liver fibrosis - a systematic review with special emphasis on anti-fibrotic strategies. J Hepatol. 2007;46(5):955–75. doi: 10.1016/j.jhep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 41.Oblander SA, Zhou Z, Galvez BG, Starcher B, Shannon JM, Durbeej M, et al. Distinctive functions of membrane type 1 matrix-metalloprotease (MT1-MMP or MMP-14) in lung and submandibular gland development are independent of its role in pro-MMP-2 activation. Dev Biol. 2005;277(1):255–69. doi: 10.1016/j.ydbio.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 42.Polette M, Nawrocki-Raby B, Gilles C, Clavel C, Birembaut P. Tumour invasion and matrix metalloproteinases. Crit Rev Oncol Hematol. 2004;49(3):179–86. doi: 10.1016/j.critrevonc.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 43.McLaughlin SL, Ice RJ, Rajulapati A, Kozyulina PY, Livengood RH, Kozyreva VK, et al. NEDD9 depletion leads to MMP14 inactivation by TIMP2 and prevents invasion and metastasis. Molecular cancer research : MCR. 2014;12(1):69–81. doi: 10.1158/1541-7786.MCR-13-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Remacle AG, Rozanov DV, Fugere M, Day R, Strongin AY. Furin regulates the intracellular activation and the uptake rate of cell surface-associated MT1-MMP. Oncogene. 2006;25(41):5648–55. doi: 10.1038/sj.onc.1209572. [DOI] [PubMed] [Google Scholar]
- 45.Cai D, Felekkis KN, Near RI, O'Neill GM, van Seventer JM, Golemis EA, et al. The GDP exchange factor AND-34 is expressed in B cells, associates with HEF1, and activates Cdc42. J Immunol. 2003;170(2):969–78. doi: 10.4049/jimmunol.170.2.969. [DOI] [PubMed] [Google Scholar]
- 46.Schweitzer JK, Sedgwick AE, D'Souza-Schorey C. ARF6-mediated endocytic recycling impacts cell movement, cell division and lipid homeostasis. Semin Cell Dev Biol. 2011;22(1):39–47. doi: 10.1016/j.semcdb.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paleotti O, Macia E, Luton F, Klein S, Partisani M, Chardin P, et al. The small G-protein Arf6GTP recruits the AP-2 adaptor complex to membranes. J Biol Chem. 2005;280(22):21661–6. doi: 10.1074/jbc.M503099200. [DOI] [PubMed] [Google Scholar]
- 48.Sabe H, Hashimoto S, Morishige M, Ogawa E, Hashimoto A, Nam JM, et al. The EGFR GEP100-Arf6-AMAP1 signaling pathway specific to breast cancer invasion and metastasis. Traffic. 2009;10(8):982–93. doi: 10.1111/j.1600-0854.2009.00917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santy LC, Ravichandran KS, Casanova JE. The DOCK180/Elmo complex couples ARNO-mediated Arf6 activation to the downstream activation of Rac1. Curr Biol. 2005;15(19):1749–54. doi: 10.1016/j.cub.2005.08.052. [DOI] [PubMed] [Google Scholar]
- 50.Fang Z, Miao Y, Ding X, Deng H, Liu S, Wang F, et al. Proteomic identification and functional characterization of a novel ARF6 GTPase-activating protein, ACAP4. Molecular & cellular proteomics : MCP. 2006;5(8):1437–49. doi: 10.1074/mcp.M600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.Aikawa Y, Martin TF. ARF6 regulates a plasma membrane pool of phosphatidylinositol(4,5)bisphosphate required for regulated exocytosis. The Journal of cell biology. 2003;162(4):647–59. doi: 10.1083/jcb.200212142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shteyn E, Pigati L, Folsch H. Arf6 regulates AP-1B-dependent sorting in polarized epithelial cells. The Journal of cell biology. 2011;194(6):873–87. doi: 10.1083/jcb.201106010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Radhakrishna H, Donaldson JG. ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. The Journal of cell biology. 1997;139(1):49–61. doi: 10.1083/jcb.139.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Radhakrishna H, Al-Awar O, Khachikian Z, Donaldson JG. ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangements. Journal of cell science. 1999;112(Pt 6):855–66. doi: 10.1242/jcs.112.6.855. [DOI] [PubMed] [Google Scholar]
- 55.Bravo-Cordero JJ, Marrero-Diaz R, Megias D, Genis L, Garcia-Grande A, Garcia MA, et al. MT1-MMP proinvasive activity is regulated by a novel Rab8-dependent exocytic pathway. The EMBO journal. 2007;26(6):1499–510. doi: 10.1038/sj.emboj.7601606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hattula K, Furuhjelm J, Tikkanen J, Tanhuanpaa K, Laakkonen P, Peranen J. Characterization of the Rab8-specific membrane traffic route linked to protrusion formation. Journal of cell science. 2006;119(Pt 23):4866–77. doi: 10.1242/jcs.03275. [DOI] [PubMed] [Google Scholar]
- 57.Holmbeck K, Bianco P, Caterina J, Yamada S, Kromer M, Kuznetsov SA, et al. MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell. 1999;99(1):81–92. doi: 10.1016/s0092-8674(00)80064-1. [DOI] [PubMed] [Google Scholar]
- 58.Pahwa S, Stawikowski MJ, Fields GB. Monitoring and Inhibiting MT1-MMP during Cancer Initiation and Progression. Cancers (Basel) 2014;6(1):416–35. doi: 10.3390/cancers6010416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strongin AY. Proteolytic and non-proteolytic roles of membrane type-1 matrix metalloproteinase in malignancy. Biochimica et biophysica acta. 2010;1803(1):133–41. doi: 10.1016/j.bbamcr.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vonderheit A, Helenius A. Rab7 associates with early endosomes to mediate sorting and transport of Semliki forest virus to late endosomes. PLoS Biol. 2005;3(7):e233. doi: 10.1371/journal.pbio.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clark ES, Whigham AS, Yarbrough WG, Weaver AM. Cortactin is an essential regulator of matrix metalloproteinase secretion and extracellular matrix degradation in invadopodia. Cancer research. 2007;67(9):4227–35. doi: 10.1158/0008-5472.CAN-06-3928. [DOI] [PubMed] [Google Scholar]
- 62.Pugacheva EN, Golemis EA. The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2 kinases at the centrosome. Nature cell biology. 2005;7(10):937–46. doi: 10.1038/ncb1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.