Abstract
A classic T-cell phenotype in Systemic lupus erythematosus (SLE) is the downregulation and replacement of the CD3ζ chain that alters TCR signaling. However, genetic associations with SLE in the human CD247 locus that encodes CD3ζ are not well established and require replication in independent cohorts. Our aim was therefore to examine, localize and validate CD247-SLE association in a large multi-ethnic population. We typed 44 contiguous CD247 SNPs in 8 922 SLE patients and 8 077 controls from four ethnically distinct populations. The strongest associations were found in the Asian population (11 SNPs in intron 1, 4.99×10−4<P<4.15×10−2), where we further identified a five-marker haplotype (rs12141731-rs2949655-rs16859085-rs12144621-rs858554; G-G-A-G-A; Phap=2.12×10−5) that exceeded the most associated single SNP rs858554 (MAFControls=13%; P=4.99×10−4, OR=1.32) in significance. Imputation and subsequent association analysis showed evidence of association (P<0.05) at 27 additional SNPs within intron 1. Cross-ethnic meta-analysis, assuming an additive genetic model adjusted for population proportions, showed 5 SNPs with significant P-values (1.40×10−3<P<3.97×10−2), with one (rs704848) remaining significant after Bonferroni correction (Pmeta=2.66×10−2). Our study independently confirms and extends the association of SLE with CD247, which is shared by various autoimmune disorders and supports a common T cell-mediated mechanism.
INTRODUCTION
Systemic lupus erythematosus (SLE; OMIM 152700) is a chronic and potentially fatal autoimmune disorder characterized by the production of autoantibodies that cause widespread tissue damage. T-cells from patients with SLE have a number of phenotypic and functional abnormalities (1,2). Some of the strongest confirmed genetic associations with SLE obviously affect T-cells, including HLA-DR, which still exceeds all other associations in significance, as well as PTPN22, a TCR signal modifier (3), and PTTG1 affecting miR146a (4) that appears particularly relevant for regulatory T-cells (5). One of the most characteristic aberrations, likely influential in altering intracellular signaling and subsequent aberrant responses of T-cells, is the specific downregulation of the CD3ζ component of the T-cell receptor complex (6,7), CD247. In SLE T-cells, this molecule is specifically replaced by the Fc receptor γ chain that is coupled with a different intracellular signaling pathway (8). In addition to this demonstrated functional relevance, association of genetic polymorphisms within CD247 with SLE has been discovered. Two reports have provided evidence for such an association, identifying two 3' UTR SNPs in strong linkage disequilibrium and showing association with differential CD3ζ expression (9) as well as with SLE (10) in a European population. More recently, several SNPs within CD247 (particularly in intron 1) were also found associated to SLE in Asian populations (11). Because the epidemiology of SLE has demonstrated that the prevalence of disease differs substantially across ethnic groups, it is logical that there exists significant genetic heterogeneity in the causes of SLE across populations (12,13). This has been supported by the differential findings obtained in genome-wide association studies (GWAS) performed in different populations (14-19), with novel loci such as RASGRP3 and WDFY4 found to be associated with SLE in Asian, but not European populations. In this study, in order to further test the association of CD247 gene with SLE in different populations, we typed 44 SNPs in a large multi-ethnic sample with total 17 003 individuals.
RESULTS
Association study and imputation analysis in the Asian population
The strongest associations were found in the Asian population (11 SNPs in intron 1, 4.99×10−4<P<4.15×10−2) (SNPs 14, 17, 24, 26, 27, 28, 30, 31, 32, 35, 36 as identified in Table 1; also see Figure 1). The most associated rs858554 (SNP 31, MAFControls=13%) reached a significance of P=4.99×10−4 (OR[95%CI]=1.32[1.13-1.55]) and a corresponding P=1.50×10−2 after Bonferroni correction for multiple testing.
Table 1.
Results of SNP association in CD247 gene at lq24.2 with SLE in the Asian population
| Genotype numbers | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP ID | SNP reference | Position (Mb) | Gene region | Alleles (minor/major) | MAF | Genotypes | SLE cases (n = 1265) | Controls (n = 1260) | P adj | OR[95% CI] | P cond | OR[95% CI] |
| 3 | rs1917534 | 165663896 | Downstream | A/G | 0.34 | AA/AG/GG | 161/549/527 | 142/539/553 | 0.2003 | 1.08[0.96-1.21] | 0.4342 | 1.05[0.93-1.18] |
| 4 | rs870875 | 165666359 | Downstream | C/A | 0.45 | CC/CA/AA | 257/600/389 | 259/616/367 | 0.5805 | 0.97[0.87-1.08] | 0.7392 | 0.98[0.88-1.10] |
| 5 | rs1052230 | 165666706 | 3' UTR | C/G | 0.20 | CC/CG/GG | 63/399/783 | 45/401/796 | 0.2575 | 1.08[0.94-1.24] | 0.1747 | 1.10[0.96-1.26] |
| 6 | rs16859030 | 165667879 | Intron 7 | A/G | 0.24 | AA/AG/GG | 77/446/721 | 74/440/726 | 0.7608 | 1.02[0.90-1.16] | 0.8690 | 1.01[0.89-1.15] |
| 7 | rs6668182 | 165668608 | Intron 7 | A/G | 0.23 | AA/AG/GG | 73/421/716 | 73/407/713 | 0.8349 | 1.01[0.89-1.16] | 0.9301 | 1.01[0.88-1.15] |
| 8 | rs953808 | 165670469 | Intron 5 | G/C | 0.20 | GG/GC/CC | 60/398/786 | 41/401/797 | 0.2582 | 1.08[0.94-1.25] | 0.1710 | 1.10[0.96-1.27] |
| 9 | rs1723023 | 165671757 | Intron 4 | A/G | 0.11 | AA/AG/GG | 9/238/954 | 18/245/939 | 0.2060 | 0.89[0.74-1.07] | 0.0861 | 0.85[0.71-1.02] |
| 10 | rs2995082 | 165672870 | Intron 4 | G/A | 0.35 | GG/GA/AA | 144/560/518 | 157/539/526 | 0.8065 | 0.99[0.88-1.11] | 0.5434 | 0.96[0.86-1.09] |
| 11 | rs1404567 | 165675499 | Intron 2 | G/A | 0.29 | GG/GA/AA | 108/487/649 | 100/512/626 | 0.6690 | 0.97[0.86-1.10] | 0.9901 | 1.00[0.88-1.13] |
| 12 | rs1554669 | 165682416 | Intron 1 | G/A | 0.15 | GG/GA/AA | 29/312/905 | 28/297/917 | 0.4660 | 1.06[0.91-1.24] | 0.8027 | 1.02[0.87-1.20] |
| 14 | rs7523907 | 165693872 | Intron 1 | G/A | 0.08 | GG/GA/AA | 11/166/1068 | 13/203/1026 | 3.00E-02 | 0.80[0.66-0.98] | 3.98E-02 | 0.81[0.66-0.99] |
| 15 | rs1723015 | 165699519 | Intron 1 | A/G | 0.48 | AA/AG/GG | 310/580/322 | 279/599/341 | 0.1485 | 1.09[0.97-1.21] | 0.0791 | 1.11[0.99-1.24] |
| 17 | rs2995091 | 165700902 | Intron 1 | G/A | 0.08 | GG/GA/AA | 11/158/1077 | 13/200/1029 | 1.50E-02 | 0.78[0.64-0.95] | 1.99E-02 | 0.79[0.64-0.96] |
| 18 | rs12132416 | 165703507 | Intron 1 | A/G | 0.16 | AA/AG/GG | 38/346/861 | 24/342/876 | 0.2498 | 1.09[0.94-1.27] | 0.1783 | 1.11[0.95-1.29] |
| 19 | rs12036775 | 165703672 | Intron 1 | A/G | 0.29 | AA/AG/GG | 100/514/630 | 111/518/613 | 0.4529 | 0.95[0.84-1.08] | 0.7899 | 0.98[0.87-1.11] |
| 20 | rs7523351 | 165703888 | Intron 1 | G/C | 0.26 | GG/GC/CC | 90/498/658 | 86/455/700 | 0.1479 | 1.10[0.97-1.25] | 0.3888 | 1.06[0.93-1.20] |
| 21 | rs2949659 | 165706163 | Intron 1 | G/A | 0.32 | GG/GA/AA | 135/535/574 | 131/528/583 | 0.6102 | 1.03[0.92-1.16] | 0.3874 | 1.05[0.94-1.19] |
| 24 | rs1214611 | 165715729 | Intron 1 | A/G | 0.43 | AA/AG/GG | 258/592/395 | 199/610/433 | 7.81E-03 | 1.17[1.04-1.30] | 1.25E-02 | 1.16[1.03-1.29] |
| 26 | rs12737372 | 165717740 | Intron 1 | A/G | 0.45 | AA/AG/GG | 240/595/405 | 268/623/345 | 1.96E-02 | 0.88[0.78-0.98] | 0.2695 | 0.93[0.83-1.05] |
| 27 | rs12141731 | 165718065 | Intron 1 | A/G | 0.45 | AA/AG/GG | 244/596/405 | 273/629/340 | 1.25E-02 | 0.87[0.78-0.97] | 0.2091 | 0.93[0.82-1.04] |
| 28 | rs2949655 | 165718475 | Intron 1 | A/G | 0.46 | AA/AG/GG | 245/594/381 | 275/626/319 | 1.28E-02 | 0.87[0.77-0.97] | 0.2082 | 0.92[0.82-1.05] |
| 29 | rs16859085 | 165719959 | Intron 1 | G/A | 0.09 | GG/GA/AA | 5/193/1048 | 10/206/1025 | 0.2380 | 0.89[0.73-1.08] | 3.22E-02 | 0.80[0.65-0.98] |
| 30 | rs12144621 | 165720283 | Intron 1 | G/C | 0.50 | GG/GC/CC | 341/613/292 | 278/627/337 | 3.91E-03 | 1.18[1.05-1.32] | 0.1279 | 1.10[0.97-1.25] |
| 31 | rs858554 | 165721539 | Intron 1 | A/G | 0.13 | AA/AG/GG | 48/312/882 | 19/277/943 | 4.99E-04 | 1.32[1.13-1.55] | NA | NA |
| 32 | rs863455 | 165724449 | Intron 1 | G/A | 0.36 | GG/GA/AA | 173/580/493 | 136/578/528 | 4.15E-02 | 1.13[1.01-1.27] | 0.3327 | 1.06[0.94-1.20] |
| 35 | rs858545 | 165728016 | Intron 1 | A/C | 0.31 | AA/AC/CC | 122/567/556 | 97/533/612 | 1.41E-02 | 1.17[1.03-1.32] | 4.20E-02 | 1.14[1.01-1.29] |
| 36 | rs704848 | 165728498 | Intron 1 | G/C | 0.35 | GG/GC/CC | 166/593/485 | 132/572/536 | 1.36E-02 | 1.16[1.03-1.31] | 0.1191 | 1.10[0.98-1.25] |
| 37 | rs704852 | 165730541 | Intron 1 | C/A | 0.12 | CC/CA/AA | 21/250/974 | 17/260/965 | 0.7842 | 0.98[0.82-1.16] | 0.5492 | 0.95[0.80-1.13] |
| 38 | rs10918706 | 165732746 | Intron 1 | A/G | 0.32 | AA/AG/GG | 142/533/562 | 129/523/584 | 0.3030 | 1.06[0.95-1.20] | 0.4216 | 1.05[0.93-1.18] |
| 39 | rs858543 | 165733923 | Intron 1 | G/A | 0.49 | GG/GA/AA | 314/620/310 | 295/590/356 | 0.0797 | 1.10[0.99-1.23] | 0.0881 | 1.10[0.99-1.23] |
Abbreviations: Mb, Megabases; MAF, minor allele frequency (in controls); OR, odds ratio; 95% CI, 95% confidence interval. The presented genetic association P-values are under the additive model and adjusted for the first three principal components and gender (Padj). The P-values from the association analysis conditioned on the most associated SNP, rs858554 (Pcond), are also indicated. Significant P-values (<0.05) are highlighted in bold.
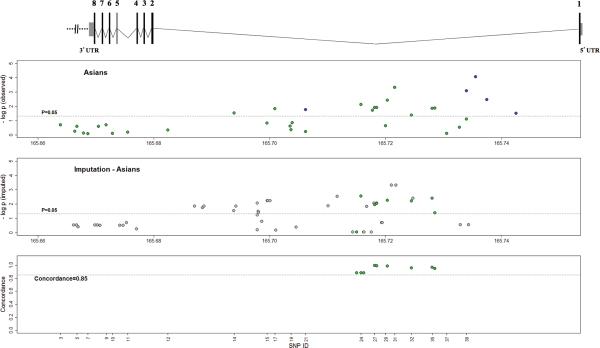
Figure 1.
Results of association tests with SLE for observed and imputed single-nucleotide polymorphisms (SNPs) in the CD247 gene. The Scaled diagram of the CD247 gene structure is represented above the plots: exons are represented by black boxes and marked with its corresponding number; 5’UTR and 3’UTR are represented by grey boxes; introns are represented by black lines between exons. The top plot shows the negative logarithms of the P-values for genotypic association (under the additive model and adjusted for the first three principal components and gender) for the polymorphisms successfully genotyped by us in the Asian population (green dots), and the significant SNPs from GWAS data (19) (dark blue dots; personal communication from authors, May 2012). The second plot displays the negative logarithms of the P-values for 51 SNPs in chromosome 1 imputed with high quality (SNPs with a minor allele frequency, MAF≥0.05, and SNP INFO≥0.80, grey dots), including SNPs that were previously genotyped (green dots). The bottom graph displays the rate of concordance of observed and imputed genotypes. Broken horizontal lines in top and second plots indicate a significance level of P=0.05. In all plots, the SNPs that had been initially genotyped are represented with green dots.
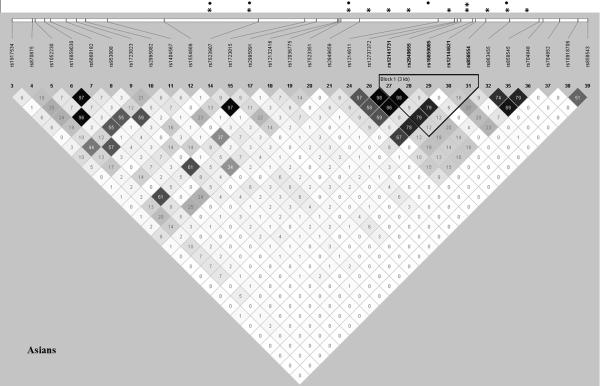
Several of the 11 significant SNPs were in very strong LD (r2>0.75): 14 and 17; 26, 27, 28 and 30; 32, 35 and 36. SNP 24 had moderate to strong LD with SNPs 26, 27, 28 and 30 (0.57≥r2≥0.67). The most significant, SNP 31, however showed weak LD with all other SNPs in our dataset (r2<0.25) (Figure 2). Four SNPs (SNPs 14, 17, 24, 35) remained nominally associated with SLE after conditional logistic regression analysis based on rs858554 (SNP 31), and one newly gained significance: rs16859085 (SNP 29) (Table 1, Figure 2). This suggests the existence of multiple genetic variants within CD247 implicated in SLE.
Figure 2.
Linkage disequilibrium plot for the 30 genotyped single-nucleotide polymorphisms (SNPs) in CD247 in the Asian population. This plot was obtained using the genotyping data from our study with Haploview 4.2 using the pairwise R-square color scheme in a grey scale. The position of the most significantly associated haplotype is indicated. *Significant P-value under the additive model and adjusted for the first three principal components and gender (Padj<0.05); **Significant P-value overpassing Bonferroni correction (Padj<0.0017); ●Significant P-value from the association analysis conditioned on the most significantly associated SNP, rs858554 (Pcond<0.05).
Haplotypic association analysis in the Asian population identified a five-marker haplotype containing five SNPs in intron 1 (rs12141731-rs2949655-rs16859085-rs12144621-rs858554; G-G-A-G-A; identified in Figure 2) showing robust association with SLE (Phap=2.12×10−5).
Even though we investigated 42 SNPs in CD247, a proportion of the genetic variation in the region was not assessed because of the size of the gene (Figure 1). To evaluate the potential association of unobserved polymorphisms in this gene in the Asian population, we imputed SNPs in chromosome 1 using data from HapMap as well as the genotypes observed at the 30 fully genotyped markers. In the CD247 gene, we obtained imputed genotypes meeting minimum quality standards (MAF in controls > 0.05 and SNP INFO > 0.8) for 51 SNPs, including 9 of the genotyped SNPs (Figure 1; identified with the SNP ID in Supplementary Table S1). Previously genotyped SNPs were imputed using the observed genotypes at the other SNPs, and a concordance rate >85% between imputed and observed genotypes was obtained (Figure 1).
From the 51 imputed SNPs, 27 (including 7 of the genotyped SNPs) were associated with SLE susceptibility (P≤0.05) (Supplementary Table S1, Figure 1), the most significant of which were rs858557, rs858556 and rs858553 (all with: P=4.82×10−4, SNP INFO=1.04). All these polymorphisms are located in intron 1 close to our most strongly associated typed SNP rs858554.
Our most significant findings are consistent with those from a previous report in Asian populations (11) that resulted from the examination of GWAS data (19). In these studies, 14 SNPs in the CD247 gene locus (including both upstream and downstream regions of the gene) were found to be significantly associated with SLE, five of which were inside the CD247 gene (personal communication from authors of (19), May 2012), all located in intron 1 (as indicated by the dark blue dots in Figure 1). In our study, the 11 significant SNPs were also all located in intron 1 (although in different variants; as indicated by the green dots in Figure 1).
The plot of pairwise LD of the genotyped SNPs in our Asian samples (Figure 2) showed very similar LD patterns to the plot of CHB HapMap samples (Supplementary Figure S1), supporting the use of this reference dataset to check linkage between the significant SNPs in our Asian cohort and those in CD247 from the GWAS data (19). We can see that the significant GWAS SNPs and our SNPs (black squares and asterisks, respectively, in Supplementary Figure S1) are physically close but in different LD blocks. Namely, the most significant SNPs in both studies, our rs858554 (SNP 31) and the GWAS rs704853, are in two different blocks located in intron 1. Furthermore, all the significant GWAS SNPs are in weak LD (r2<0.25) among themselves and with our associated SNPs (Supplementary Figure S1). Taken together, the results of both studies complement each other, pointing to the existence of different variants in the same gene region that are not in strong LD and were observed independently, which strengthens the general result. Imputation did not return results for the top significant variants in Li et al. (11) and GWAS (19).
Non-Asian populations multiethnic association study, and meta-analysis
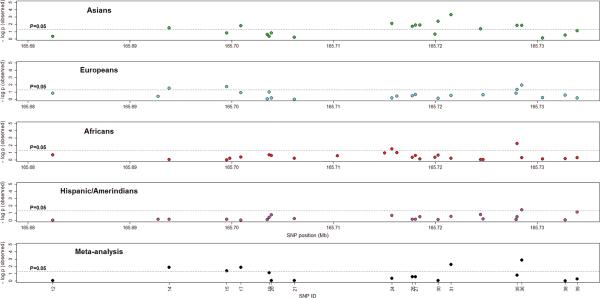
Five SNPS were significantly associated with SLE in the European ancestry samples (1.12×10−2<P<4.51×10−2) including four SNPs within intron 1 (SNPs 14, 15, 35 and 36) and one downstream of CD247 (SNP 1). In the other ethnicities, 3 SNPs were associated in African ancestry (SNPs 6, 24 and 35, 5.92×10−3<P<2.95×10−2), and 1 SNP in the Hispanic/Amerindian (SNP 36 P=3.39×10−2) populations (Figure 3, Supplementary Table S2). None of these SNPs, however, remained significant upon Bonferroni correction for multiple testing. Nevertheless, several of these significant SNPs were common to the associated SNPs in the Asian cohort, namely SNPs 14, 35 and 36 in the European ancestry, SNPs 24 and 35 in the African ancestry, and SNP 36 in the Hispanic/Amerindian ancestry (Figure 3).
Figure 3.
Results of association tests with SLE and meta-analysis in the four cohorts in our study, specifically in intron 1 of the CD247 gene. The plots show the negative logarithm of the P-value of genotypic association (under the additive model and adjusted for the first three principal components and gender) for the observed polymorphisms genotyped in the: Asians (first plot; 30 SNPs; green dots); Europeans (second plot; 31 SNPs; blue dots); Africans (third plot; 33 SNPs; red dots); and Hispanic/Amerindians (fourth plot; 30 SNPs; pink dots). The bottom plot shows the negative logarithm of the P-value for the meta-analysis (under the additive model and adjusted for the first three principal components and gender). Only SNPs with association results in the four study populations were tested (19 SNPs; black dots).
The significant haplotype identified in the Asian population was not associated in these three populations although the LD structures were similar (Supplementary Figure S2).
Cross-ethnic meta-analysis of the four populations, assuming an additive genetic model and adjusted for population proportions, showed 5 SNPs with significant P-values (1.40×10−3<P<3.97×10−2), all located in intron 1 of CD247 (Table 2, Figure 3). One marker was still significant after Bonferroni correction for multiple testing: rs704848 (SNP 36) with Pmeta=2.66×10−2.
Table 2.
Results of meta-analysis testing in the CD247 gene with SLE
| Asian | European | African | Hispanic / Amerindian | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP ID | SNP reference | Position (Mb) | Gene region | Allelesa | P-value | OR[95% CI] | Allelesa | P-value | OR[95% CI] | Allelesa | P-value | OR[95% CI] | Allelesa | P-value | OR[95% CI] | Pmeta |
| 5 | rs1052230 | 165666706 | 3′ UTR | C/G | 0.2575 | 1.08[0.94-1.24] | C/G | 0.9290 | 1.00[0.90-1.10] | C/G | 0.4898 | 1.05[0.92-1.20] | C/G | 0.8993 | 1.01[0.83-1.23] | 0.4442 |
| 9 | rs1723023 | 165671757 | Intron 4 | A/G | 0.2060 | 0.89[0.74-1.07] | A/G | 0.5936 | 0.98[0.92-1.05] | A/G | 0.0924 | 0.86[0.73-1.03] | A/G | 0.7990 | 0.98[0.85-1.13] | 0.0771 |
| 11 | rs1404567 | 165675499 | Intron 2 | G/A | 0.6690 | 0.97[0.86-1.10] | G/A | 0.2269 | 0.94[0.85-1.04] | G/A | 0.1472 | 1.14[0.95-1.37] | G/A | 0.8441 | 0.98[0.80-1.20] | 0.7039 |
| 12 | rs1554669 | 165682416 | Intron 1 | G/A | 0.4660 | 1.06[0.91-1.24] | G/A | 0.1464 | 0.90[0.79-1.04] | G/A | 0.1958 | 1.08[0.96-1.21] | G/A | 0.9670 | 1.00[0.82-1.21] | 0.9154 |
| 14 | rs7523907 | 165693872 | Intron 1 | G/A | 3.00E-02 | 0.80[0.66-0.98] | G/A | 2.85E-02 | 0.93[0.86-0.99] | G/A | 0.8992 | 1.01[0.91-1.11] | G/A | 0.6858 | 0.97[0.85-1.11] | 1.40E-02 |
| 15 | rs1723015 | 165699519 | Intron 1 | A/G | 0.1485 | 1.09[0.97-1.21] | G/A | 1.83E-02 | 0.92[0.86-0.99] | G/A | 0.9620 | 1.00[0.91-1.10] | G/A | 0.6715 | 1.03[0.91-1.17] | 3.97E-02 |
| 17 | rs2995091 | 165700902 | Intron 1 | G/A | 1.50E-02 | 0.78[0.64-0.95] | G/A | 0.1160 | 0.95[0.88-1.01] | G/A | 0.4225 | 0.93[0.79-1.10] | G/A | 0.9499 | 1.00[0.86-1.15] | 1.42E-02 |
| 19 | rs12036775 | 165703672 | Intron 1 | A/G | 0.4529 | 0.95[0.84-1.08] | A/G | 0.0931 | 1.06[0.99-1.14] | A/G | 0.1951 | 1.07[0.97-1.19] | A/G | 0.3792 | 1.06[0.93-1.20] | 0.0707 |
| 20 | rs7523351 | 165703888 | Intron 1 | G/C | 0.1479 | 1.10[0.97-1.25] | G/C | 0.6238 | 1.03[0.92-1.15] | G/C | 0.2454 | 0.93[0.83-1.05] | G/C | 0.1771 | 0.87[0.70-1.07] | 0.8799 |
| 21 | rs2949659 | 165706163 | Intron 1 | G/A | 0.6102 | 1.03[0.92-1.16] | G/A | 0.9564 | 1.00[0.88-1.13] | G/A | 0.6354 | 1.03[0.92-1.16] | G/A | 0.5844 | 0.95[0.81-1.13] | 0.8530 |
| 24 | rs1214611 | 165715729 | Intron 1 | A/G | 7.81E-03 | 1.17[1.04-1.30] | A/G | 0.6487 | 0.98[0.92-1.05] | G/A | 2.95E-02 | 1.11[1.01-1.23] | A/G | 0.2017 | 0.92[0.81-1.05] | 0.4358 |
| 26 | rs12737372 | 165717740 | Intron 1 | A/G | 1.96E-02 | 0.88[0.78-0.98] | A/G | 0.3261 | 0.96[0.89-1.04] | A/G | 0.4384 | 1.08[0.90-1.29] | A/G | 0.6820 | 1.03[0.89-1.20] | 0.2823 |
| 27 | rs12141731 | 165718065 | Intron 1 | A/G | 1.25E-02 | 0.87[0.78-0.97] | A/G | 0.2225 | 0.96[0.89-1.03] | A/G | 0.2380 | 1.10[0.94-1.29] | A/G | 0.6833 | 1.03[0.90-1.18] | 0.2664 |
| 30 | rs12144621 | 165720283 | Intron 1 | G/C | 3.91E-03 | 1.18[1.05-1.32] | C/G | 0.6756 | 1.02[0.94-1.10] | C/G | 0.2346 | 1.08[0.95-1.23] | C/G | 0.7843 | 1.02[0.88-1.18] | 0.8532 |
| 31 | rs858554 | 165721539 | Intron 1 | A/G | 4.99E-04 | 1.32[1.13-1.55] | A/G | 0.2795 | 1.04[0.97-1.11] | A/G | 0.6303 | 1.03[0.93-1.13] | G/A | 0.2774 | 0.93[0.82-1.06] | 5.80E-03 |
| 35 | rs858545 | 165728016 | Intron 1 | A/C | 1.41E-02 | 1.17[1.03-1.32] | A/C | 4.51E-02 | 1.07[1.00-1.15] | C/A | 5.92E-03 | 1.15[1.04-1.27] | A/C | 0.3180 | 1.07[0.94-1.22] | 0.1583 |
| 36 | rs704848* | 165728498 | Intron 1 | G/C | 1.36E-02 | 1.16[1.03-1.31] | C/G | 1.12E-02 | 0.92[0.85-0.98] | C/G | 0.5043 | 1.04[0.93-1.16] | C/G | 3.39E-02 | 0.87[0.76-0.99] | 1.40E-03 |
| 38 | rs10918706 | 165732746 | Intron 1 | A/G | 0.3030 | 1.06[0.95-1.20] | A/G | 0.2756 | 0.96[0.89-1.03] | A/G | 0.6949 | 1.03[0.89-1.20] | A/G | 0.8676 | 1.01[0.86-1.20] | 0.9309 |
| 39 | rs858543 | 165733923 | Intron 1 | G/A | 0.0797 | 1.10[0.99-1.23] | A/G | 0.6240 | 1.02[0.94-1.10] | A/G | 0.5157 | 1.04[0.93-1.16] | A/G | 0.0677 | 1.15[0.99-1.32] | 0.5225 |
Abbreviations: Mb, Megabases; Pmeta, meta-analysis P-values; Pmetacorr, Bonferroni corrected meta-analysis P-values; OR, odds ratio; 95% CI, 95% confidence interval.
minor/major allele.
rs704848 marker is still significant after Bonferroni correction for multiple testing (Pmeta=2.66×10−2).
The meta-analysis P-values are under the additive model and adjusted for the first three principal components and gender. Only SNPs with association results in the four study populations were tested. Significant P-values (<0.05) are highlighted in bold.
DISCUSSION
In this multiethnic association study, we independently validated and extended the previous association of CD247 genetic variants with SLE, primarily in the Asian population.
Two studies have previously found an association of the 3’UTR of this gene with reduced expression of CD3ζ (9) and SLE (10). In contrast, our discoveries highlight genetic association in Asians in the 5’ region (intron1) of CD247. This is consistent with recent studies performed in Asian populations (11). Considering the ethnic heterogeneities in the epidemiology of SLE (12,13), these observations suggest a particular association of CD247 genetic variants in Asian populations. Although pointing to heterogeneity in the genetic association of CD247 with SLE, most importantly, these results further support and highlight the implication of this gene in SLE.
The CD247 gene spans 88 kb and has been mapped to chromosome 1q24.2. The first intron spans about 78 kb, followed by seven other exons of the gene. The 11 significant SNPs in the Asian population and 78% of the significant SNPs in the other three populations tested lie in intron 1, suggesting a possible role in the regulation of CD247 expression (11). This region is further highlighted by the imputation analysis (27 imputed SNPs reached significance) and haplotypic association (Phap=2.12×10−5) in the Asians, and by an overall significant meta-analysis of all four populations.
Gorman et al. (9) found two SNPs (in high LD), rs1052230 and rs1052231 in 3’UTR of the gene being associated with CD247 expression levels in both SLE patients and healthy controls. However, only weak association with disease risk was found for haplotypes in the 3’UTR region of the gene. In addition, Warchoł et al. (10) found that rs1052231 conferred increased risk of incidence of SLE. In our study, SNP rs1052230 did not show significant disease association (P=0.2575), and imputation on rs1052231 was neither significant (P=0.2950). These discrepancies from our results suggest an implicit genetic heterogeneity in the different populations while principally providing further evidence of the involvement of CD247 in SLE susceptibility.
Interestingly, other studies on autoimmune diseases also reported their main findings in intron 1 of CD247 (20-25), supporting a common mechanism behind the involvement of this gene in the etiology of these autoimmune disorders. A recent GWAS on systemic sclerosis (SSc), an autoimmune disease that shares some autoantibody and clinical features with SLE, identified CD247 as a major susceptibility gene (rs2056626, located in intron 1, P=3.39×10−9) (20). This association with SSc was replicated in two other cohorts (21,22). In our study, rs2056626 was not genotyped but was found to be significantly associated when imputed (P=1.42×10−2). Furthermore, this SNP is in strong LD (r2=0.75) with rs7523907 (SNP 14) (using HapMap data; release 23), which had P=3.00×10−2 in our study. A meta-analysis of GWAS in celiac disease and rheumatoid arthritis identified several non-HLA shared loci, among which the SNP rs864537 in intron 1 of CD247 (Pcombined=2.20×10−11) (23). In our study, rs864537 was not genotyped (or imputed) and it is not in LD with any of our SNPs (using HapMap data; release 23). Several GWAS also showed suggestive association of CD247 with Crohn's disease (summarized in Wang et al., (24)), with the relevant SNPs being rs704853, rs12061855, rs1799704, rs2988276 and rs870875 (P=1.80×10−3<P<2.40×10−2). The SNP rs870875 was tested in our study but with no association, and rs2988276 had a borderline association using imputed data (P=3.98×10−2). None of these SNPs is in high LD with any of our variants (using HapMap data; release 23). Recently, a novel association with CD247 (rs1773560, in intron 1) was also identified for juvenile idiopathic arthritis (P=2.57×10−7) (25). This SNP showed an imputed association in our study (P=1.83×10−2) and is in strong LD (r2=0.71) with rs7523907 (SNP 14, significant in our SLE study) and with rs2056626 (r2=0.94) (found associated with SSc) (using HapMap data; release 23).
T cells are considered to be central to the pathogenesis of SLE because aberrations in their functionality are very likely strongly contributing to the altered immune responses and overproduction of pathogenic autoantibodies (26). CD247 encodes the T-cell receptor zeta chain (CD3ζ), a component of the T-cell receptor (TCR)-CD3 complex (27). TCRζ is a pivotal component of the TCR signaling machinery and vital for T cell activation. A defective expression of the CD3ζ-chain has been associated with autoimmune diseases including SLE (6, 7, 28) and rheumatoid arthritis (29,30), but also other conditions such as tumors and chronic infection (31). It is one established reason for various functional alterations in T cells in these conditions that TCR signaling through CD3ζ is replaced by FcRγ (8) and its associated Syk pathway that enhances calcium and cytoskeletal reactions (32). This mechanism could be responsible for the shared association of several autoimmune diseases with CD247. Another effect that seems particularly relevant for SLE is that CD3ζ signaling reportedly augments IL-2 production (7), indicating that its loss likely contributes to the defective IL-2 production that characterizes T cells in SLE (33). Potential mechanisms as per how autoimmunity-associated genetic variants exert their effects may include differences in expression, splicing and posttranslational processing, but their relevance is still not clear (34). Our findings confirm the relevance of these effects for SLE pathogenesis and highlight that the development of SLE is influenced by mechanisms shared with other autoimmune diseases, which involve a role of the TCR signaling pathway that should be further characterized. This is a part of several GWAS-identified risk loci shared between SLE and other autoimmune disorders pointing to common immunological mechanisms (35). In this study, we provide a replication establishing CD247 as a genetic risk factor for SLE, which generates new implications for the pathogenesis of the disease and might lead to new therapeutic targets for disease management.
PATIENTS AND METHODS
Study design
The genotype data used in this study were generated as a part of a joint effort of more than 40 investigators from around the world. These investigators contributed samples, funding, and hypotheses on a combined array containing ~35,000 SNPs (Figure S1 from Lessard et al. (36)). The Oklahoma Medical Research Foundation (OMRF) served as the coordinating center, ran the arrays, and sent the data to a central facility for quality control at Wake Forest Medical Center. These data were then distributed back to the investigators, who requested the SNPs, for final analysis of their own respective hypotheses.
Patient and control samples
A total of 17 003 samples (8 922 SLE patients and 8 077 healthy controls; 4 with unknown disease status) from four main populations with Asian, Hispanic/Amerindian, European and African ancestry were initially enrolled in this multiethnic study. Details regarding the characteristics of the study participants in each dataset were previously described (37). The samples were assembled at the Oklahoma Medical Research Foundation (OMRF) after collection in multiple institutions around the world, following ethics committee approval and informed consent in accordance with the Declaration of Helsinki. Patients were classified with SLE based upon using the American College of Rheumatology criteria (38).
Genotyping
A total of 44 SNPs in the CD247 region and 347 ancestral-informative markers (AIMs) were genotyped using the Illumina iSelect technology (Illumina, San Diego, CA, USA). Extensive quality control was performed following stringent criteria to select the SNPs to be used in the analysis, namely well-defined cluster scatter plots, >90% call rates across the entire study and in this specific set of SNPs, deviations from Hardy-Weinberg equilibrium with P > 0.01 in controls and P > 0.0001 in cases (using the PLINK (39) Hardy-Weinberg analysis), total proportion missing <5%, and P > 0.05 for differential missingness between cases and controls. Only SNPs with MAF > 5% in both case and controls groups were analysed for association in each population.
Samples with <90% call rate, excess heterozygosity, as well as first-degree relatives, duplicates and individuals with self-reported vs. genetically determined gender inconsistencies were excluded from the analysis as previously described (37).
EIGENSTRAT (40) was used to identify population substructure within the samples based on AIMs. The AIMs were selected to distinguish four continental ancestral populations: Africans, Europeans, American Indians, and East Asians (41,42). Principal components from EIGENSTRAT outputs were used to identify genetic outliers from each population cluster (as described in (37)). After quality control a total of 1 452 samples were excluded. The final meta-dataset used in the analysis consisted of 15 551 subjects (8 214 SLE cases and 7 337 controls): 2 488 Asian, 2 247 Hispanic/Amerindian, 7 248 European and 3 568 African. Characteristics of the study participants in each dataset are described in Table 3.
Table 3.
Demographic characteristics of the four populations (after quality control)
| Population Ancestry | Samples after QC | Cases | Age of onset (mean ±SD) | Controls | Male | Female |
|---|---|---|---|---|---|---|
| Asian | 2 488 | 1 246 | 26.4 ± 0.3 | 1 242 | 245 | 2 243 |
| European | 7 248 | 3 842 | 33.6 ± 0.3 | 3 406 | 1 452 | 5 796 |
| African | 3 568 | 1 669 | 34.0 ± 0.3 | 1 899 | 713 | 2 855 |
| Hispanic / Amerindians | 2 247 | 1 457 | 29.5 ± 0.4 | 790 | 199 | 2 048 |
| Total | 15 551 | 8 214 | 7 337 | 2 609 | 12 942 |
Abbreviations: QC, quality control.
Populations: African ancestry includes 274 Gullah and 3 294 other African Americans; Hispanic/Amerindian ancestry includes 1 252 Hispanics and 995 Native Americans. Information for Age of onset was available for most of the cases in each population.
Two SNPs (rs1214603 and rs10918694) were excluded due to genotyping failure. Of the 42 SNPs with genotyping results, three further were excluded in all the four populations (rs2995087, rs1214604 and rs704855), nine more in the Asians, nine more in the Hispanic/Amerindians, eight more in the Europeans and six more in the African ancestry (African American/Gullah) samples (Table 1, Supplementary Table S2) due to quality control issues previously described (37). A final set of 39 SNPs were successfully genotyped in at least one population (SNP ID 1–39; listed in Table 1 and Supplementary Table S2): 30 in the Asian population; 30 in the Hispanic/Amerindians; 31 in the Europeans; and 33 in the Africans.
Statistical Analysis
Multiple logistic regression (PLINK (39); additive genetic model) was used to test for SLE association. Analysis was adjusted for the first three principal components calculated from AIMs, and gender. Conditional analyses based on the most strongly associated SNP (rs858554) (results expressed as conditional P [Pcond] values) were performed with logistic regression using PLINK (39), (additive genetic model; adjusted for the first three principal components and gender). Results were considered significant below the conventional level of P<0.05. Correction for multiple testing was performed using the conservative Bonferroni method.
Haplotypic association was tested using PLINK (39) sliding window analysis. Linkage disequilibrium (LD) plots for each cohort were created using Haploview 4.2 (43). We also used the HapMap CHB (Han Chinese from Beijing, China, n=84) reference dataset (downloaded from the International HapMap Project website; HapMap3, release 2; chr1:165663570..165742500) to construct the LD plot of the reference Asian population and check linkage between the significant SNPs in our Asian cohort and those in CD247 from the GWAS data that we had access to (19).
Meta-analysis of the 19 SNPs with association data for the four populations were calculated using Stouffer's Ztrend method implemented in METAP (44), weighted by sample size and taking into account effect directions.
Imputation Analysis
SNPs not directly genotyped in the CD247 region for the Asian population, where we had the strongest associations, were imputed with PLINK (39) using HapMap Phase II and specific reference panels for the Asian population (Release 23; 161 230 SNPs on chromosome 1, 90 JPT+CHB founders). For every imputed SNP, PLINK provides an information content metric INFO, ranging from 0 to 1 (although it can be greater than 1 occasionally). A higher INFO value generally means a better SNP imputation. All imputed SNPs with MAF smaller than 0.05 and with INFO<0.8 were excluded. For genotyped SNPs, PLINK calculates the concordance rate among observed and imputed genotypes (Figure 1).
Supplementary Material
Acknowledgements
We thank the SLE patients and the healthy controls for their collaboration in this study. We also thank the entire OMRF team for organizing this study. We thank all the authors of the “Genome-Wide Association Study in Asian Populations Identifies Variants in ETS1 and WDFY4 Associated with Systemic Lupus Erythematosus” (Yang et al. 2010) paper for sharing the data information on the GWAS results on the CD247 region. We also thank the University of Alabama Birmingham Center for Clinical and Translational Science (CCTS). This study was supported by the: National Institutes of Health grants [UL1RR025741] to R.R.G., Northwestern University Feinberg School of Medicine, [K24AR002138, P602AR30692, P01AR49084] to R.P.K., G.S.A., E.E.B., University of Alabama Birmingham, L.M.P., National Institute of Arthritis and Musculoskeletal and Skin Diseases, R.R.G.), [UL1TR000165 and P01AI083194] to R.P.K., [RO1AR43814] to B.P.T., University of California Los Angeles, [P60AR053308, UL1TR000004] to L.C., University of California San Francisco, [AR43727] to M.A.P., Johns Hopkins University, [R21AI070304] to S.A.C., University of Colorado School of Medicine, [RO1AR057172] to C.O.J., University of Southern California, [UL1RR025014 and R01AR051545-03] to A.M.S., Seattle Children's Research Institute Arthritis Foundation, [UL1RR029882 and P60AR062755] to G.S.G. and D.L.K., Medical University of South Carolina, [P30AR53483, U19AI082714, P30GM103510, U01AI101934] to J.A.J. and J.M.G., [AI063274, AR056360 and AI083194] to P.M.G., Oklahoma Medical Research Foundation, [R37AI024717, P01083194, P01AR049084] to J.H., Cincinnati Children's Hospital Medical Center; the US Departments of Defense [PR094002] to J.H. and Veterans Affairs to J.H.; the Alliance for Lupus Research to L.C., B.P.T.; a Kirkland Scholar Award to L.C.; Korea Healthcare technology R & D project [A121983] funded by the Ministry for Health and Welfare, Republic of Korea to S.C.B.; the Swedish Research Council and Instituto de Salud Carlos III grant [PS09/00129] cofinanced by FEDER funds of the European Union to M.E.A.R.; Fundação para a Ciência e Tecnologia (FCT, Portugal) fellowships [SFRH/BPD/29354/2006] to M.Martins and [SFRH/BPD/34648/2007] to C.F.
Footnotes
Conflits of Interest Statement. None declared.
Supplementary Information accompanies this paper on Genes and Immunity website (http://www.nature.com/gene)
REFERENCES
- 1.Crispín JC, Kyttaris V, Juang YT, Tsokos GC. Systemic lupus erythematosus: new molecular targets (review). Ann Rheum Dis. 2007;66:65–69. doi: 10.1136/ard.2007.078493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crispín JC, Liossis SN, Kis-Toth K, Lieberman LA, Kyttaris VC, Juang YT, et al. Pathogenesis of human systemic lupus erythematosus: recent advances (review). Trends Mol Med. 2010;16:47–57. doi: 10.1016/j.molmed.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guerra SG, Vyse TJ, Cunninghame Graham DS. The genetics of lupus: a functional perspective (review). Arthritis Res Ther. 2012;14:211–222. doi: 10.1186/ar3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Löfgren SE, Frostegård J, Truedsson L, Pons-Estel BA, D'Alfonso Witte ST, Lauwerys BR, et al. Genetic association of miRNA-146a with systemic lupus erythematosus in Europeans through decreased expression of the gene. Genes Immun. 2012;13:268–274. doi: 10.1038/gene.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–929. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liossis SN, Ding XZ, Dennis GJ, Tsokos GC. Altered pattern of TCR/CD3-mediated protein-tyrosyl phosphorylation in T cells from patients with systemic lupus erythematosus. Deficient expression of the T cell receptor zeta chain. J Clin Invest. 1998;101:1448–1457. doi: 10.1172/JCI1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nambiar MP, Fisher CU, Warke VG, Krishnan S, Mitchell JP, Delaney N, et al. Reconstitution of deficient T cell receptor zeta chain restores T cell signaling and augments T cell Receptor/CD3-induced interleukin-2 production in patients with systemic lupus erythematosus. Arthritis Rheum. 2003;48:1948–1955. doi: 10.1002/art.11072. [DOI] [PubMed] [Google Scholar]
- 8.Enyedy EJ, Nambiar MP, Liossis SN, Dennis G, Kammer GM, Tsokos GC. Fc epsilon receptor type I gamma chain replaces the deficient T cell receptor zeta chain in T cells of patients with systemic lupus erythematosus. Arthritis Rheum. 2001;44:1114–1121. doi: 10.1002/1529-0131(200105)44:5<1114::AID-ANR192>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 9.Gorman CL, Russell AI, Zhang Z, Cunninghame Graham D, Cope AP, Vyse TJ. Polymorphisms in the CD3Z gene influence TCRz expression in Systemic Lupus Erythematosus patients and healthy controls. J Immunol. 2008;180:1060–1070. doi: 10.4049/jimmunol.180.2.1060. [DOI] [PubMed] [Google Scholar]
- 10.Warchoł T, Piotrowski P, Lianeri M, Cieślak D, Wudarski M, Hrycaj P, et al. The CD3Z 844 T>A polymorphism within the 3’-UTR of CD3Z confers increased risk of incidence of systemic lupus erythematosus. Tissue Antigens. 2009;74:68–72. doi: 10.1111/j.1399-0039.2009.01264.x. [DOI] [PubMed] [Google Scholar]
- 11.Li R, Yang W, Zhang J, Hirankarn N, Pan HF, Mok CC, et al. Association of CD247 with systemic lupus erythematosus in Asian Populations. Lupus. 2012;21:75–83. doi: 10.1177/0961203311422724. [DOI] [PubMed] [Google Scholar]
- 12.Hopkinson ND, Doherty M, Powell RJ. Clinical features and race-specific incidence/prevalence rates of systemic lupus erythematosus in a geographically complete cohort of patients. Ann Rheum Dis. 1994;53:675–680. doi: 10.1136/ard.53.10.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danchenko N, Satia JA, Anthony MS. Epidemiology of systemic lupus erythematosus: a comparison of worldwide disease burden (review). Lupus. 2006;15:308–318. doi: 10.1191/0961203306lu2305xx. [DOI] [PubMed] [Google Scholar]
- 14.International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN) Harley JB, Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozyrev SV, Abelson AK, Wojcik J, Zaghlool A, Linga Reddy MV, Sanchez E, et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:211–216. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- 16.Graham RR, Cotsapas C, Davies L, Hackett R, Lessard CJ, Leon JM, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 19.Yang W, Shen N, Ye DQ, Liu Q, Zhang Y, Qian XX, et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 2010;6:e1000841. doi: 10.1371/journal.pgen.1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radstake TR, Gorlova O, Rueda B, Martin JE, Alizadeh BZ, Palomino-Morales R, et al. Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nat Genet. 2010;42:426–429. doi: 10.1038/ng.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorlova O, Martin JE, Rueda B, Koeleman BP, Ying J, Teruel M, et al. Identification of novel genetic markers associated with clinical phenotypes of systemic sclerosis through a genome-wide association strategy. PLoS Genet. 2011;7:e1002178. doi: 10.1371/journal.pgen.1002178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dieudé P, Boileau C, Guedj M, Avouac J, Ruiz B, Hachulla E, et al. Independent replication establishes CD247 gene as a genetic systemic sclerosis susceptibility factor. Ann Rheum Dis. 2011;70(9):1695–1696. doi: 10.1136/ard.2010.147009. [DOI] [PubMed] [Google Scholar]
- 23.Zhernakova A, Stahl EA, Trynka G, Raychaudhuri S, Festen EA, Franke L, et al. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet. 2011;7:e1002004. doi: 10.1371/journal.pgen.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K, Zhang H, Kugathasan S, Annese V, Bradfield JP, Russell RK, et al. Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn Disease. Am J Hum Genet. 2009;84:399–405. doi: 10.1016/j.ajhg.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinks A, Cobb J, Sudman M, Eyre S, Martin P, Flynn E, et al. Investigation of rheumatoid arthritis susceptibility loci in juvenile idiopathic arthritis confirms high degree of overlap. Ann Rheum Dis. 2012;71:1117–1121. doi: 10.1136/annrheumdis-2011-200814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moulton VR, Tsokos GC. Abnormalities of T cell signaling in systemic lupus erythematosus (review). Arthritis Res Ther. 2011;13(2):207. doi: 10.1186/ar3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Call ME, Wucherpfennig KW. Molecular mechanisms for the assembly of the T cell receptor-CD3 complex (review). Mol Immunol. 2004;40:1295–1305. doi: 10.1016/j.molimm.2003.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pang M, Setoyama Y, Tsuzaka K, Yoshimoto K, Amano K, Abe T, et al. Defective expression and tyrosine phosphorylation of the T cell receptor zeta chain in peripheral blood T cells from systemic lupus erythematosus patients. Clin Exp Immunol. 2002;129:160–168. doi: 10.1046/j.1365-2249.2002.01833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berg L, Rönnelid J, Klareskog L, Bucht A. Down-regulation of the T cell receptor CD3 zeta chain in rheumatoid arthritis (RA) and its influence on T cell responsiveness. Clin Exp Immunol. 2000;120:174–182. doi: 10.1046/j.1365-2249.2000.01180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romagnoli P, Strahan D, Pelosi M, Cantagrel A, van Meerwijk JP. A potential role for protein tyrosine kinase p56(lck) in rheumatoid arthritis synovial fluid T lymphocyte hyporesponsiveness. Int Immunol. 2001;13:305–312. doi: 10.1093/intimm/13.3.305. [DOI] [PubMed] [Google Scholar]
- 31.Baniyash M. TCR zeta-chain downregularion: curtailing an excessive inflammatory immune response (review). Nat Rev Immunol. 2004;4:675–687. doi: 10.1038/nri1434. [DOI] [PubMed] [Google Scholar]
- 32.Krishnan S, Juang YT, Chowdhury B, Magilavy A, Fisher CU, Nguyen H, et al. Differential expression and molecular associations of Syk in systemic lupus erythematosus T cells. J Immunol. 2008;181(11):8145–8152. doi: 10.4049/jimmunol.181.11.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tenbrock K, Juang YT, Kyttaris VC, Tsokos GC. Altered signal transduction in SLE T cells (review). Rheumatology. 2007;46(10):1525–1530. doi: 10.1093/rheumatology/kem154. [DOI] [PubMed] [Google Scholar]
- 34.Takeuchi T, Suzuki K, Kondo T, Yoshimoto K, Tsuzaka K. CD3 ζ defects in systemic lupus erythematosus (review). Ann Rheum Dis. 2012;71(Suppl2):i78–81. doi: 10.1136/annrheumdis-2011-200641. [DOI] [PubMed] [Google Scholar]
- 35.Deng Y, Tsao B. Genetic susceptibility to systemic lupus erythematosus in the genomic era (review). Nat Rev Rheumatol. 2010;6:683–692. doi: 10.1038/nrrheum.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lessard CJ, Adrianto I, Ice JA, Wiley GB, Kelly JA, Glenn SB, et al. Identification of IRF8, TMEM39A, and IKZF3-ZPBP2 as susceptibility loci for systemic lupus erythematosus in a large-scale multiracial replication study. Am J Hum Genet. 2012;90(4):648–660. doi: 10.1016/j.ajhg.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lessard CJ, Adrianto I, Kelly JA, Kaufman KM, Grundahl KM, Adler A, et al. Identification of a Systemic Lupus Erythematosus Susceptibility Locus at 11p13 between PDHX and CD44 in a Multiethnic Study. Am J Hum Genet. 2011;88:83–91. doi: 10.1016/j.ajhg.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus (letter). Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 39.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 41.Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, et al. A high-density admixture map for disease gene discovery in African Americans. Am J Hum Genet. 2004;74:1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Halder I, Shriver M, Thomas M, Fernandez JR, Frudakis T. A panel of ancestry informative markers for estimating individual biogeographical ancestry and admixture from four continents: Utility and applications. Hum Mutat. 2008;29:648–658. doi: 10.1002/humu.20695. [DOI] [PubMed] [Google Scholar]
- 43.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 44.Whitlock MC. Combining probability from independent tests: The weighted Z-method is superior to Fisher's approach. J Evol Biol. 2005;18:1368–1373. doi: 10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.