The Arabidopsis 14-3-3 RCI1A protein plays a critical role in freezing tolerance, partially through an ethylene-dependent signaling pathway. RCI1A interacts with different ACS isoforms to regulate the levels of ethylene that are necessary to promote accurate cold-induced gene expression and freezing tolerance under both control and low-temperature conditions.
Abstract
In plants, the expression of 14-3-3 genes reacts to various adverse environmental conditions, including cold, high salt, and drought. Although these results suggest that 14-3-3 proteins have the potential to regulate plant responses to abiotic stresses, their role in such responses remains poorly understood. Previously, we showed that the RARE COLD INDUCIBLE 1A (RCI1A) gene encodes the 14-3-3 psi isoform. Here, we present genetic and molecular evidence implicating RCI1A in the response to low temperature. Our results demonstrate that RCI1A functions as a negative regulator of constitutive freezing tolerance and cold acclimation in Arabidopsis thaliana by controlling cold-induced gene expression. Interestingly, this control is partially performed through an ethylene (ET)-dependent pathway involving physical interaction with different ACC SYNTHASE (ACS) isoforms and a decreased ACS stability. We show that, consequently, RCI1A restrains ET biosynthesis, contributing to establish adequate levels of this hormone in Arabidopsis under both standard and low-temperature conditions. We further show that these levels are required to promote proper cold-induced gene expression and freezing tolerance before and after cold acclimation. All these data indicate that RCI1A connects the low-temperature response with ET biosynthesis to modulate constitutive freezing tolerance and cold acclimation in Arabidopsis.
INTRODUCTION
For plants, as sessile organisms, extreme temperatures are among the environmental factors that most constrain their geographical distribution and development. During evolution, plants have acquired different adaptive responses to survive extreme temperatures. One of these adaptive responses is cold acclimation, by which many plants from temperate regions increase their constitutive freezing tolerance after exposure to low, nonfreezing temperatures (Levitt, 1980). The complex process of cold acclimation involves many physiological and biochemical changes, such as alterations in lipid composition and the accumulation of sugars and other osmolytes. It is tightly regulated at the transcriptional level, although recent results reveal that it is also under posttranscriptional, translational, and posttranslational control (Viswanathan and Zhu, 2002; Barrero-Gil and Salinas, 2013). Elucidating the signaling pathways that mediate cold acclimation and identifying their components is key to understanding the molecular mechanisms underlying this adaptive response.
In Arabidopsis thaliana, cold acclimation is established through an intricate network of signaling pathways. The best characterized of them is mediated by a family of three AP2-type transcription factors named C-REPEAT/DRE BINDING FACTORs (CBF1 to CBF3) (Medina et al., 2011). These factors are estimated to control the induction of around 12% of the Arabidopsis cold-responsive genes (Fowler and Thomashow, 2002). The expression of each CBF, in turn, is specifically induced by low temperature in a fast and transient manner (Gilmour et al., 1998; Liu et al., 1998; Medina et al., 1999), and their individual overexpression in Arabidopsis results in the constitutive expression of the target genes and increased freezing tolerance (Gilmour et al., 2000; Fowler and Thomashow, 2002). As expected considering their importance in cold acclimation, the expression of CBFs has been shown to be remarkably regulated (Medina et al., 2011). Another relevant signaling pathway involved in cold acclimation is mediated by abscisic acid (ABA). This hormone accumulates in response to low temperature (Lang et al., 1994), and exogenous ABA application increases plant freezing tolerance (Gilmour and Thomashow, 1991). Furthermore, mutants affected in ABA biosynthesis and perception show altered tolerance to freezing temperatures before and after cold acclimation (Llorente et al., 2000). The emerging picture is that, in addition to ABA, other hormones are also involved in cold acclimation. For example, ethylene (ET) accumulates in Arabidopsis, bean (Phaseolus vulgaris), rhododendron (Rhododendron spp), winter rape (Brassica napus), tomato (Solanum lycopersicum), and winter rye (Secale cereale) after exposure to low temperature (Field, 1981; Kacperska and Kubacka-Zgbalska, 1985; Harber and Fuchigami, 1989; Ciardi et al., 1997; Yu et al., 2001; Wang et al., 2012). Moreover, the cold-induced accumulation of ET in winter rye promotes the synthesis of antifreezing proteins and the consequent increase in freezing tolerance (Yu et al., 2001). In addition, exogenous application of ethephon, an ET releaser, enhances rhododendron freezing tolerance (Harber and Fuchigami, 1989), and overexpression in the sense or antisense orientation of TERF2/ERF2, a tomato ET-inducible gene encoding a transcription factor, can increase or decrease freezing tolerance in tomato and tobacco (Nicotiana tabacum), respectively (Zhang and Huang, 2010). In contrast with these data, Shi and colleagues (2012) recently proposed that ET acts as a negative regulator of freezing tolerance and cold acclimation in Arabidopsis, which indicates that the role of this hormone in the low-temperature response still needs further investigation.
14-3-3s are a family of highly conserved regulatory proteins from eukaryotes that have been implicated in protein–protein interactions and mediating signal transduction pathways. They form homodimers and heterodimers in the native state and commonly bind to target proteins that contain specific phosphorylated motifs (Paul et al., 2012). The binding of the 14-3-3s may have different mechanistic consequences on their targets, such as conformational changes, alterations in subcellular localization, variations in intrinsic activity or stability, and changes in affinity to other proteins (Paul et al., 2012). Moreover, different 14-3-3 family members within any organism may carry out isoform-specific functions (Paul et al., 2012). Proteome-wide approaches have revealed that the plant 14-3-3 interactome is comparable in size and functional complexity to its animal counterpart (Oecking and Jaspert, 2009), indicating that plant 14-3-3s may affect many cellular processes. In this sense, it has become increasingly apparent that they mediate the signal transduction associated with plant responses to hormones as well as biotic and abiotic stresses (Denison et al., 2011). Active roles of 14-3-3 proteins in plant hormone signaling have been reported for gibberellic acid, ABA, and brassinosteroids (Gampala et al., 2007; Schoonheim et al., 2007; Nakata et al., 2009). Recently, 14-3-3 proteins were found to interact in vivo with a subset of proteins linked to ET biosynthesis, including several ACC SYNTHASE (ACS) isoforms, regulating their activity (Chang et al., 2009; Huang et al., 2013; Yoon and Kieber, 2013), which strongly suggests that they also play a role in the ET response. Regarding the implication of 14-3-3 proteins in mediating stress signaling, a number of studies have demonstrated that they have an important function in plant responses to biotic stresses (Gökirmak et al., 2010). However, although the expression of 14-3-3 genes is induced by several abiotic stresses, including cold (Jarillo et al., 1994), high salt (Xu and Shi, 2006), and drought (Porcel et al., 2006), as well as phosphorous and iron deficiencies (Wang et al., 2002; Cao et al., 2007), evidence for 14-3-3–mediated abiotic stress responses is still very limited (Yan et al., 2004; Campo et al., 2012; Yang et al., 2013; Zhou et al., 2014).
The Arabidopsis genome contains 13 14-3-3 genes (phi, chi, omega, psi, pi, upsilon, lambda, nu, kappa, mu, epsilon, omicron, and iota) for which transcripts have been detected (Paul et al., 2012). We previously reported that the expression of one of these genes, RARE COLD INDUCIBLE 1A (RCI1A; 14-3-3 psi) is induced by low temperature (Jarillo et al., 1994; Abarca et al., 1999; Roberts et al., 2002). However, RCI1A expression is not induced by ABA, NaCl, or drought stress, suggesting a specific function for this protein in the Arabidopsis response to cold (Jarillo et al., 1994). Here, we examine the role of RCI1A in low-temperature signaling, finding that RCI1A negatively regulates constitutive freezing tolerance and cold acclimation by controlling cold-induced gene expression. This control is accomplished, in part, through an ET-dependent pathway involving physical interaction with different ACS isoforms and a reduced ACS stability. Thus, our work indicates that ET levels promote suitable cold-induced gene expression and freezing tolerance before and after cold acclimation.
RESULTS
Cold Induction of RCI1A Is Regulated at the Transcriptional Level through a CBF- and ABA-Independent Pathway
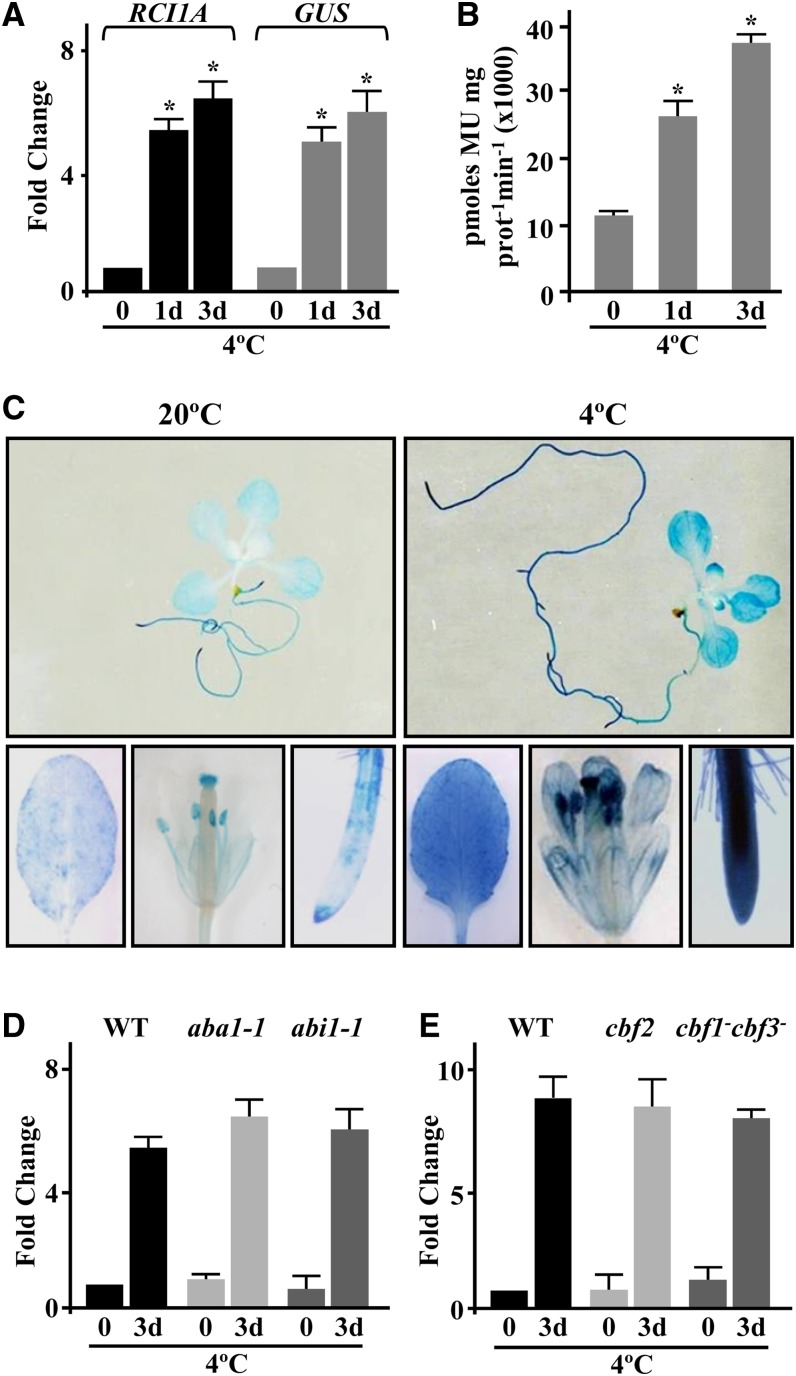
As mentioned above, we previously described RCI1A as an Arabidopsis cold-inducible gene encoding the 14-3-3 psi isoform (Jarillo et al., 1994; Abarca et al., 1999; Roberts et al., 2002). To further understand the molecular mechanisms underlying the cold response of RCI1A, we first generated Arabidopsis transgenic plants containing a fusion between an RCI1A promoter fragment (RCI1APRO; −1978 to +3) and the uidA (β‑glucuronidase [GUS]) reporter gene (RCI1APRO-GUS). Four independent transgenic lines expressing a single copy of RCI1APRO-GUS in homozygosis were analyzed. In all cases, the levels of GUS mRNA and activity increased significantly in response to low temperature, mirroring the expression pattern of endogenous RCI1A (Figures 1A and 1B). These results indicated that the selected promoter region contained all cis-acting elements required for RCI1A cold responsiveness and that the induction of RCI1A expression was positively regulated by low temperature at the transcriptional level. Transgenic plants expressing the RCI1APRO-GUS fusion were then used to follow the expression of RCI1A in different tissues of Arabidopsis in response to cold. Under control conditions, a nearly constitutive weak GUS activity was observed (Figure 1C). When the plants were exposed to 4°C, however, the GUS activity increased substantially (Figure 1C). Consistent with the expression pattern of RCI1A (Jarillo et al., 1994), RCI1APRO-GUS transgenic plants subjected to other related abiotic stresses, such as high salt and drought, or ABA treatment did not exhibit any change in GUS activity (Supplemental Figure 1). We concluded that RCI1A has a ubiquitous expression pattern and is specifically induced in response to low temperature.
Figure 1.
The Accumulation of RCI1A Transcripts Is Transcriptionally Regulated by Low Temperature Independently of CBFs and ABA.
(A) Expression levels of RCI1A and GUS genes, as determined by qPCR, in plants containing the RCI1APRO-GUS fusion exposed for 0, 1, and 3 d to 4°C. Analyses were performed in triplicate with three independent RNA samples from 2-week-old plants. Error bars indicate sd. Asterisks indicate significant differences (P < 0.05) from samples exposed for 0 d to 4°C.
(B) GUS activity in 2-week-old RCI1APRO-GUS seedlings exposed for 0, 1, and 3 d to 4°C. Data are expressed as means of three independent experiments with 10 plants each. Error bars indicate sd. Asterisks indicate significant differences (P < 0.05) from samples exposed for 0 d to 4°C. MU, methylumbelliferone.
(C) Histochemical analysis of GUS activity in whole 2-week-old seedlings and leaves, flowers, and roots from 8-week-old RCI1APRO-GUS plants grown under control conditions (20°C) or exposed for 3 d to 4°C (4°C).
(D) Expression levels of RCI1A, as determined by qPCR, in Landsberg erecta wild-type plants and ABA-deficient (aba-1) and ABA-insensitive (abi1-1) mutants exposed for 0 and 3 d to 4°C. Analyses were performed as described in (A).
(E) Expression levels of RCI1A, as determined by qPCR, in Col-0 wild-type and CBF-deficient (cbf2 and cbf1−cbf3−) plants exposed for 0 and 3 d to 4°C. Analyses were performed as described in (A).
In Arabidopsis, cold-induced gene expression is mainly controlled through two independent signaling pathways, one mediated by the CBF transcription factors and the other by ABA (Xue-Xuan et al., 2010). To determine whether the cold induction of RCI1A was mediated by one of these pathways, we analyzed the accumulation of the corresponding transcripts in response to low temperature in ABA-deficient (aba1-1) or ABA-insensitive (abi1-1) Arabidopsis mutants, in a CBF2 Arabidopsis mutant (cbf2), and in an Arabidopsis transgenic line with highly reduced CBF1 and CBF3 expression (cbf1−cbf3−) (Novillo et al., 2004, 2007). As shown in Figures 1D and 1E, RCI1A displayed a similar expression pattern in the wild type and in aba and cbf plants exposed to 4°C for 3 d, denoting that the cold induction of RCI1A is mediated through a CBF- and ABA-independent pathway.
RCI1A Localizes to the Cytoplasm and Nucleus of Arabidopsis Cells and Accumulates in Response to Low Temperature
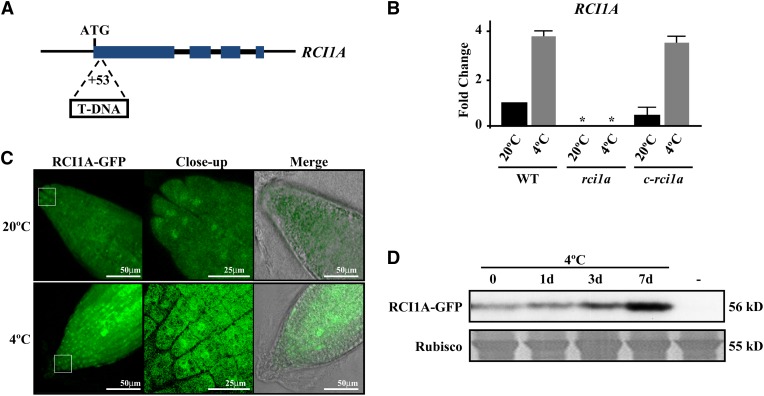
To investigate the function of RCI1A, a transgenic Arabidopsis line containing a single T-DNA inserted 53 bp downstream of the start codon of RCI1A was identified (Figure 2A). The T-DNA insertion completely disrupted RCI1A expression, including its response to 4°C, indicating that this new RCI1A allele (rci1a) was null or highly hypomorphic (Figure 2B). Morphologically, rci1a mutants showed a reduced size compared with wild-type plants throughout Arabidopsis development (Supplemental Figures 2A to 2C). Interestingly, microscopic analysis revealed that rci1a cells were significantly smaller than wild-type ones (Supplemental Figures 2D and 2E), which should be at the origin of the reduced mutant size. rci1a plants transformed with a genomic RCI1A-green fluorescent protein (RCI1A-GFP) fusion driven by the RCI1APRO (c-rci1a) accumulated wild-type levels of RCI1A transcripts under both control and cold conditions (Figure 2B) and recovered the wild-type phenotype (Supplemental Figures 2A to 2E), confirming that the morphological and cellular phenotypes displayed by rci1a mutants were due to the absence of RCI1A expression. In addition, these results demonstrated that the RCI1A-GFP fusion protein was functionally equivalent to RCI1A.
Figure 2.
RCI1A Accumulates in the Nucleus and Cytoplasm of Arabidopsis Cells in Response to Low Temperature.
(A) Schematic representation of the RCI1A gene. Large and small boxes represent exons and introns, respectively. ATG indicates the start codon, and the T-DNA insertion site is shown. The diagram is not drawn to scale.
(B) Expression levels of RCI1A, as determined by qPCR, in Col-0 wild-type plants, rci1a mutants, and rci1a mutants containing the RCI1A-GFP fusion (c-rci1a) grown under control conditions (20°C) or exposed for 3 d to 4°C (4°C). Analyses were performed in triplicate with three independent RNA samples from 2-week-old plants. Error bars indicate sd. Asterisks indicate significant differences (P < 0.05) from wild-type plants.
(C) Confocal laser scanning micrographs of the RCI1A-GFP fusion protein in root tips from 6-d-old c-rci1a seedlings grown under control conditions (20°C) or exposed for 3 d to 4°C (4°C).
(D) Levels of RCI1A-GFP fusion protein (56 kD) in 6-d-old c-rci1a seedlings exposed for 0, 1, 3, and 7 d to 4°C. A lane with Col-0 plants (−) was added as a negative control. The large subunit of Rubisco (55 kD) was used as a loading control.
It has been reported that Arabidopsis 14-3-3s may have different subcellular localizations, which could account for their functional specificity (Paul et al., 2012). Therefore, we decided to determine the subcellular localization of RCI1A under control and cold conditions by examining the distribution of green fluorescence in c-rci1a plants. Confocal microscopy analysis revealed that, at 20°C, GFP activity was evenly distributed in the cytoplasm and nucleus of Arabidopsis root cells (Figure 2C). When c-rci1a plants were treated for 3 d at 4°C, this distribution was maintained, although a considerable increase in GFP activity was detected in both subcellular compartments (Figure 2C). This increase suggested that, concomitant with the accumulation of RCI1A transcripts, the levels of RCI1A protein also accumulated in response to low temperature. To confirm these results, we assessed the amount of RCI1A-GFP fusion protein in c-rci1a plants grown at 20°C or exposed to 4°C for different periods of time. Immunoblot experiments using an anti-GFP monoclonal antibody confirmed that, in fact, RCI1A-GFP was notably more abundant in cold-treated plants (Figure 2D). From these observations, we concluded that RCI1A subcellularly localizes to the cytoplasm and nucleus of Arabidopsis cells, that this localization is not altered under cold conditions, and that the levels of RCI1A increase in response to low temperature.
RCI1A Negatively Regulates Constitutive Freezing Tolerance and Cold Acclimation in Arabidopsis
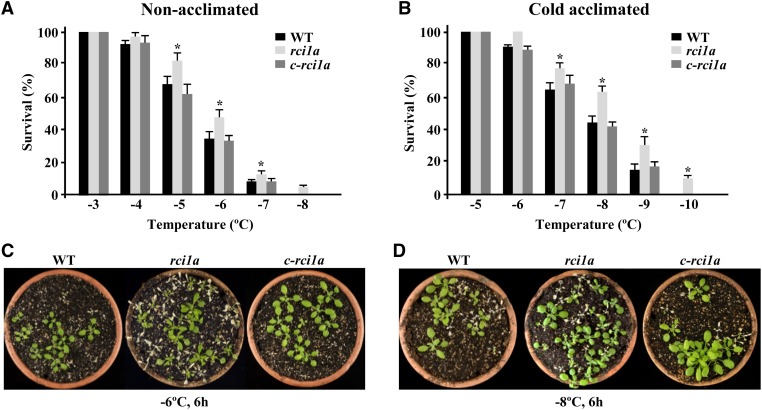
The induction of RCI1A expression and the accumulation of RCI1A under cold conditions suggested that this protein could have a role in Arabidopsis tolerance to freezing temperatures. To investigate this hypothesis, the freezing tolerance of rci1a mutants was analyzed before and after cold acclimation for 7 d at 4°C. Interestingly, rci1a plants in both cases exhibited a freezing tolerance significantly higher than wild-type plants. Their LT50 (temperature that causes 50% lethality) values were −6.3 and −5.4°C, respectively, before cold acclimation (Figures 3A and 3C) and −8.5 and −7.5°C, respectively, after cold acclimation (Figures 3B and 3D). c-rci1a plants showed wild-type capacity to constitutively tolerate freezing and cold acclimate (Figures 3A to 3D), confirming that the tolerant phenotypes displayed by the rci1a mutant were a direct consequence of the absence of RCI1A expression. All these data demonstrated that RCI1A acts as a negative regulator of the constitutive freezing tolerance and cold acclimation response in Arabidopsis.
Figure 3.
RCI1A Negatively Regulates Arabidopsis Constitutive Freezing Tolerance and Cold Acclimation.
(A) and (B) Freezing tolerance of 2-week-old Col-0 wild-type, rci1a, and c-rci1a plants exposed for 6 h to the indicated freezing temperatures before (A) and after (B) being acclimated for 7 d at 4°C. In all cases, freezing tolerance was estimated as the percentage of plants surviving each specific temperature after 7 d of recovery under control conditions. Data are expressed as means of three independent experiments with 50 plants each. Error bars indicate sd. Asterisks indicate significant differences (P < 0.05) from wild-type plants.
(C) and (D) Freezing tolerance of representative nonacclimated (C) and cold-acclimated (D) plants 7 d after being exposed to −6 and −8°C for 6 h, respectively.
[See online article for color version of this figure.]
The tolerance of rci1a mutants to other abiotic stresses related to cold, such as high salt and drought, was also tested. In contrast with freezing tolerance, no differences could be detected between rci1a and wild-type plants when analyzed for their tolerance to these stress conditions (Supplemental Figure 3), suggesting a specific role for RCI1A in Arabidopsis tolerance to freezing temperatures.
RCI1A Negatively Regulates Cold-Induced Gene Expression under Control Conditions and in Response to Low Temperature
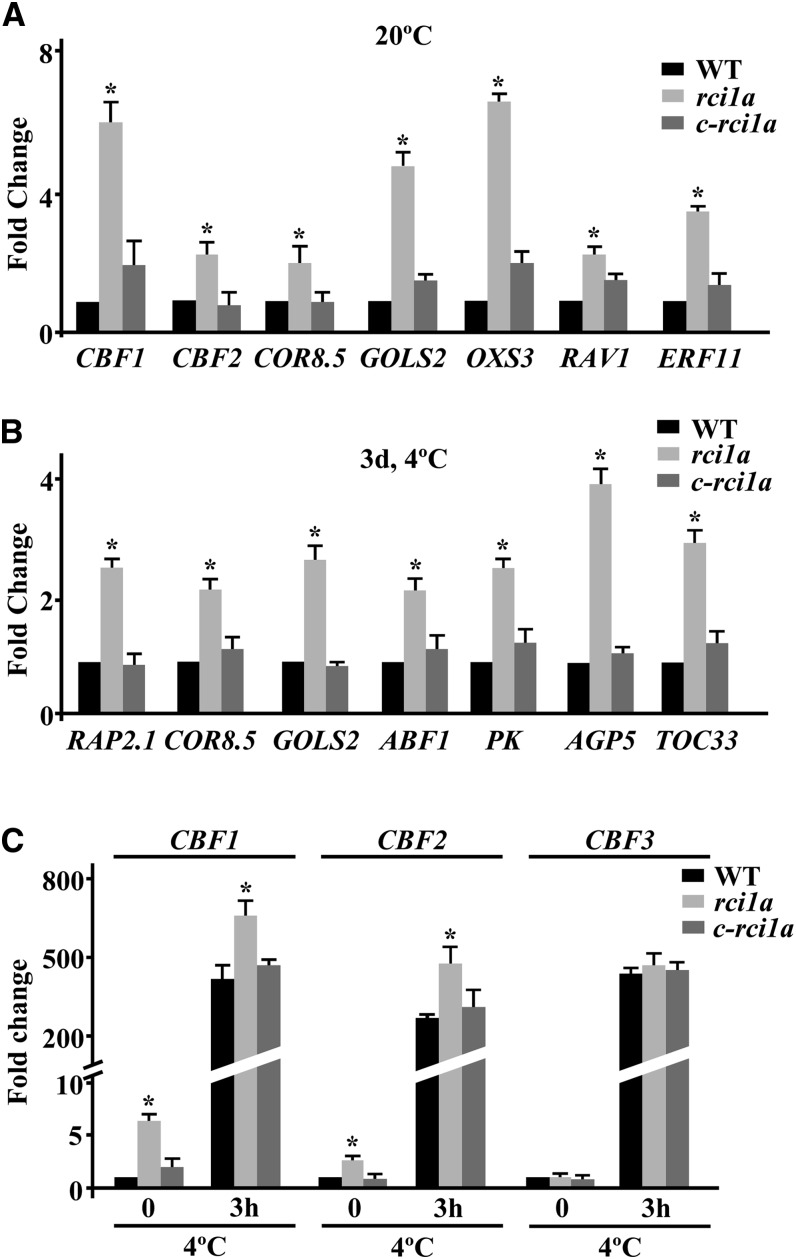
It is well documented that both constitutive freezing tolerance and cold acclimation capacity in Arabidopsis are mainly controlled by changes in gene expression (Medina et al., 2011). Since RCI1A negatively regulates the ability of Arabidopsis to constitutively tolerate freezing and cold acclimate, we assessed the possibility that RCI1A was involved in controlling cold-induced gene expression. Transcript profiling of rci1a plants grown under control conditions allowed us to identify 176 genes whose expression levels were increased at least 2-fold (false discovery rate [FDR] < 0.05) compared with the wild type (Supplemental Data Set 1). Remarkably, 41.4% of these genes (i.e., 73) had been reported to be induced (≥2-fold) in response to low temperature (Kilian et al., 2007) (Supplemental Data Set 2), and some of them, including those encoding the transcription factors CBF1 and CBF2 and their targets COLD REGULATED8.5 (COR8.5) and GALACTINOL SYNTHASE (GOLS2), were reported to have a role in Arabidopsis freezing tolerance (Kasuga et al., 1999; Fowler and Thomashow, 2002; Novillo et al., 2004, 2007). Quantitative PCR (qPCR) experiments confirmed that the expression of these and other cold-inducible genes was significantly higher in rci1a than in wild-type plants, validating the microarray results (Figure 4A). Only 38 genes, apart from RCI1A, showed reduced expression levels (at least 2-fold; FDR < 0.05) in rci1a mutant plants, none of them having been involved in freezing tolerance (Supplemental Data Set 3).
Figure 4.
RCI1A Negatively Regulates Cold-Induced Gene Expression under Both Control and Low-Temperature Conditions.
Expression levels of different cold-inducible genes are shown in 2-week-old Col-0 wild-type, rci1a, and c-rci1a plants grown under control conditions (20°C) (CBF1, CBF2, COR8.5, GOLS2, OXS3, RAV1, and ERF11 genes) (A), exposed for 3 d to 4°C (RAP2.1, COR8.5, GOLS2, ABF1, PK, AGP5, and TOC33 genes) (B), or exposed to 4°C for 0 and 3 h (CBF1, CBF2, and CBF3 genes) (C). In all cases, analyses were performed in triplicate by qPCR with three independent RNA samples. Error bars indicate sd. Asterisks indicate significant differences (P < 0.05) from wild-type plants.
When comparing the global mRNA profiles from rci1a and wild-type plants exposed for 3 d to 4°C, transcripts corresponding to 328 genes were found to be higher by more than 2-fold (FDR < 0.05) in rci1a (Supplemental Data Set 4). In this case, 21.3% out of the 328 genes (i.e., 70) had been described to be cold-induced (≥2-fold) (Kilian et al., 2007) (Supplemental Data Set 5). The increased induction of some of these genes, such as RELATED TO AP2 1 (RAP2.1), COR8.5, GOLS2, ABSCISIC ACID RESPONSIVE ELEMENT BINDING FACTOR1 (ABF1), PYRUVATE KINASE (PK), ARABINOGALACTAN PROTEIN5 (AGP5), and TRANSLOCON AT THE OUTER ENVELOPE MEMBRANE OF CHLOROPLASTS33 (TOC33), in rci1a was verified by qPCR (Figure 4B). The fact that RAP2.1, COR8.5, GOLS2, and ABF1 were CBF targets (Fowler and Thomashow, 2002; Sharabi-Schwager et al., 2010) prompted us to examine whether the induction of the CBF genes themselves was also enhanced in cold-treated rci1a plants. qPCR experiments revealed that, in fact, the induction of CBF1 and CBF2 was significantly higher in rci1a than in wild-type plants exposed for 3 h to 4°C (Figure 4C). The induction of CBF3, however, was similar in mutant and wild-type plants (Figure 4C). Compared with the wild type, only 37 genes, in addition to RCI1A, displayed lower expression levels (at least 2-fold; FDR < 0.05) in the rci1a mutant in response to low temperature, and none of them had been related to freezing tolerance (Supplemental Data Set 6).
c-rci1a plants exhibited wild-type expression patterns for all validated genes (Figures 4A to 4C), demonstrating that the elevated cold-induced gene expression detected in rci1a mutants when growing at 20°C or exposed to 4°C was caused by the absence of RCI1A. Together, the results described above indicated that RCI1A negatively regulates cold-induced gene expression both under control conditions and in response to low temperature, which would account for the increased constitutive freezing tolerance and cold acclimation capacity of rci1a mutant plants.
RCI1A Controls ET Levels in Arabidopsis by Interacting with ACS Isoforms and Reducing ACS Stability
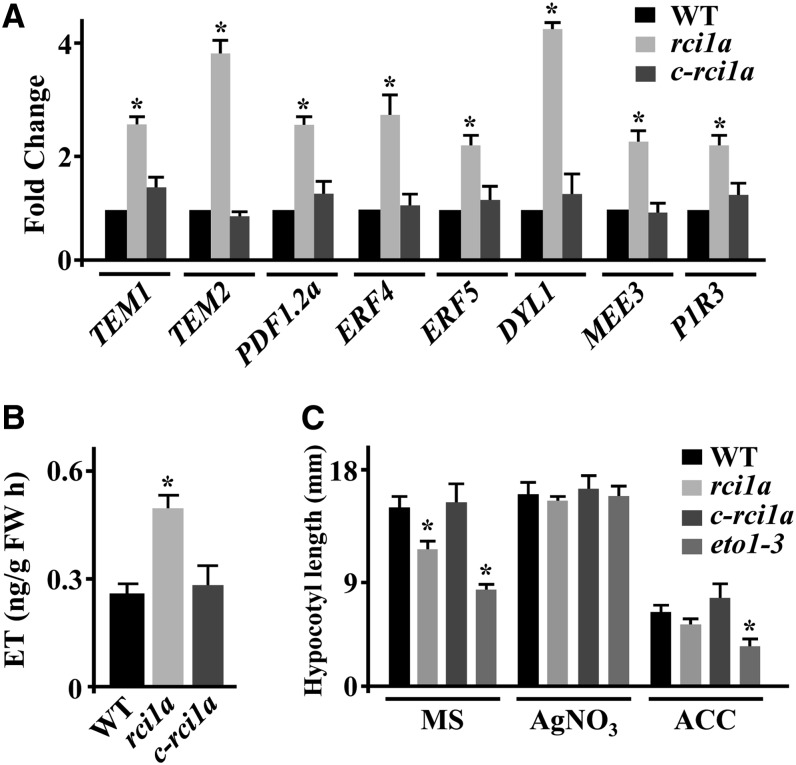
Intriguingly, the transcriptomic analysis revealed that around 18% (i.e., 31) of the genes whose expression under control conditions was higher in rci1a than in wild-type plants had been reported to be upregulated (≥2-fold) by ET (Alonso et al., 2003; Goda et al., 2008) (Supplemental Data Set 7). These data were validated by confirming the enhanced expression of several ET-induced genes, including TEMPRANILLO1 (TEM1), TEM2, PLANT DEFENSIN1.2 (PDF1.2a), ETHYLENE RESPONSE FACTOR4 (ERF4), and ERF5, in rci1a (Figure 5A). These findings suggested that the rci1a mutant could have increased content of ET. Transcript levels corresponding to the genes DORMANCY-ASSOCIATED PROTEIN-LIKE1 (DYL1), MATERNAL EFFECT EMBRYO ARREST3 (MEE3), and P1R3, whose expression is known to be ET-dependent but cold-independent (Kilian et al., 2007; Goda et al., 2008), were also elevated in rci1a compared with the wild type (Figure 5A), further indicating that mutants had higher content of ET than wild-type plants. As expected, c-rci1a plants displayed wild-type expression patterns for all genes analyzed (Figure 5A). ET quantification provided definitive evidence that, in fact, rci1a plants grown at 20°C had higher ET content than the wild type (Figure 5B). Consistent with these results, rci1a seedlings growing in darkness exhibited a significant decrease in hypocotyl elongation compared with the wild-type, resembling that of ethylene overproducer1-3 (eto1-3) mutant seedlings (Kieber et al., 1993) (Figure 5C; Supplemental Figure 4). In the presence of AgNO3, an ET receptor inhibitor (Cancel and Larsen, 2002), the hypocotyl size of rci1a reverted to that of the wild type, as in the case of eto1-3 (Figure 5C; Supplemental Figure 4). In the presence of the ET precursor 1-aminocyclopropane-1-carboxylic acid (ACC), however, rci1a seedlings showed a reduction in hypocotyl size similar to those of wild-type and eto1-3 seedlings (Figure 5C; Supplemental Figure 4), confirming that their short stature when growing in the dark was due to increased ET levels. The analysis of dark-grown c-rci1a seedlings corroborated that the ET-related mutant phenotypes exhibited by rci1a were a direct consequence of the absence of RCI1A (Figures 5B and 5C; Supplemental Figure 4). All these data demonstrated that RCI1A negatively modulates the content of ET in Arabidopsis.
Figure 5.
RCI1A Negatively Regulates ET Levels in Arabidopsis.
(A) Expression levels of TEM1, TEM2, PDF1.2a, ERF4, ERF5, DYL1, MEE3, and P1R3 genes, as determined by qPCR, in 2-week-old Col-0 wild-type, rci1a, and c-rci1a plants grown under control conditions. Analyses were performed in triplicate with three independent RNA samples. Error bars indicate sd. Asterisks indicate significant differences (P < 0.05) from wild-type plants.
(B) Levels of ET, as determined by gas chromatography, in 3-week-old Col-0 wild-type, rci1a, and c-rci1a plants grown under control conditions. Data are expressed as means of three independent experiments with five plants each. Error bars indicate sd. Asterisks indicate significant differences (P < 0.05) from wild-type plants. FW, fresh weight.
(C) Hypocotyl size of 4-d-old Col-0 wild-type, rci1a, c-rci1a, and eto1-3 etiolated seedlings germinated on vertical plates containing MS medium, MS medium supplemented with 10 µM AgNO3, and MS medium supplemented with 0.1 mM ACC. Data are expressed as means of three independent experiments with 10 seedlings each. Error bars indicate sd. Asterisks indicate significant differences (P < 0.05) from wild-type plants.
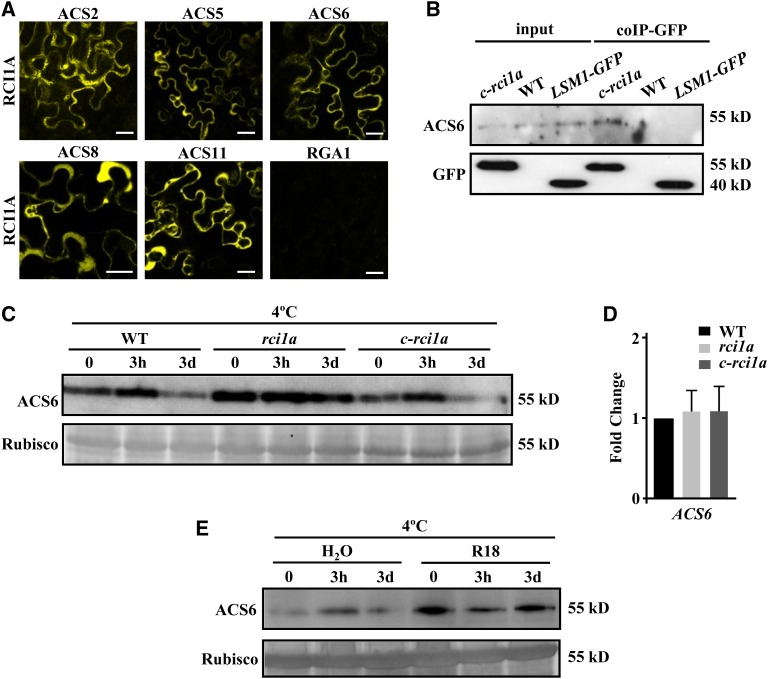
Some 14-3-3 proteins have been reported to regulate ET biosynthesis, and consequently ET levels, by interacting with ACS isoforms and controlling their stability (Chang et al., 2009; Huang et al., 2013; Yoon and Kieber, 2013). We decided to examine the possibility that RCI1A could also control ET content by interacting with ACS proteins. The interactions between RCI1A and ACS2, ACS5, ACS6, ACS8, and ACS11, which represent the three types of ACS isoforms that have been described in Arabidopsis (Tsuchisaka et al., 2009), were examined by means of bimolecular fluorescence complementation (BiFC) (Hu et al., 2002; Walter et al., 2004) in Nicotiana benthamiana leaves. Reconstitution of yellow fluorescent protein (YFP) was observed, in terms of yellow fluorescence, in a high number of N. benthamiana epidermal cells cotransformed with nYFP-RCI1A and all the cYFP-ACS fusions (Figure 6A; Supplemental Figure 5), demonstrating that RCI1A interacts in vivo with components of the three ACS types. Consistent with the cytoplasmic localization described for Arabidopsis ACSs (Yang and Hoffman, 1984), RCI1A–ACS interactions were essentially detected in the cytoplasm of the cells (Figure 6A; Supplemental Figure 5). These interactions were specific, as demonstrated by the fact that we could not observe an interaction between RCI1A and the REPRESSOR OF GA1-3 1 (RGA1) DELLA protein (Figure 6A; Supplemental Figure 5), and not affected by low temperature (Supplemental Figure 5). The in vivo interaction between RCI1A and ACS proteins was confirmed using a coimmunoprecipitation assay with c-rci1a extracts and antibodies against GFP and ACS6, the most abundant ACS isoform in Arabidopsis (Tsuchisaka et al., 2009) (Figure 6B). Then, we analyzed whether this interaction could influence the stability and, therefore, the abundance of ACS proteins. The results showed that, indeed, rci1a mutants grown at 20°C exhibited increased levels of ACS6 compared with wild-type plants (Figure 6C). ACS6 expression, however, was similar in wild-type and mutant plants (Figure 6D). Furthermore, wild-type plants treated with the R18 peptide, which disrupts functional 14-3-3 interactions (Wang et al., 1999), displayed higher levels of ACS6 than untreated plants (Figure 6E). Collectively, all these results strongly suggested that RCI1A negatively regulates the levels of ET in Arabidopsis plants growing under control conditions by directly interacting with ACS isoforms, which would lead to a reduced ACS stability.
Figure 6.
RCI1A Interacts with ACS Isoforms Reducing ACS Stability.
(A) Visualization of in vivo interactions between Arabidopsis RCI1A and ACS2, ACS5, ACS6, ACS8, ACS11, and RGA1 proteins by BiFC assays. Interactions of nYFP-RCI1A/cYFP-ACSs and nYFP-RCI1A/cYFP-RGA1 protein pairs were tested by Agrobacterium-mediated transformation in leaves of N. benthamiana plants grown at 20°C. Bars = 20 μm.
(B) Coimmunoprecipitation of RCI1A and ACS6 proteins. Protein extracts from 2-week-old c-rci1a plants were immunoprecipitated with GFP-Trap beads, and the immunoprecipitated proteins were subsequently analyzed by immunoblotting using anti-GFP or anti-ACS6 antibody. Protein extracts from Col-0 wild-type plants and Col-0 plants containing one copy of an LSM1-GFP fusion were used as negative controls.
(C) Levels of ACS6 protein (55 kD) in 2-week-old Col-0 wild-type, rci1a, and c-rci1a plants exposed for 0 and 3 h and 3 d to 4°C. The large subunit of Rubisco (55 kD) was used as a loading control.
(D) Expression levels of ACS6, as determined by qPCR, in 2-week-old Col-0 wild-type, rci1a, and c-rci1a plants grown under control conditions. Analyses were performed in triplicate with three independent RNA samples. Error bars indicate sd.
(E) Levels of ACS6 protein (55 kD) in 2-week-old Col-0 plants exposed for 0 and 3 h and 3 d to 4°C in the presence of 0 (H2O) or 10 μg/mL R18 peptide (R18). The large subunit of Rubisco (55 kD) was used as a loading control.
[See online article for color version of this figure.]
RCI1A Control of ET Levels Is Required for Correct Cold-Induced Gene Expression under Control Conditions and Accurate Constitutive Freezing Tolerance in Arabidopsis
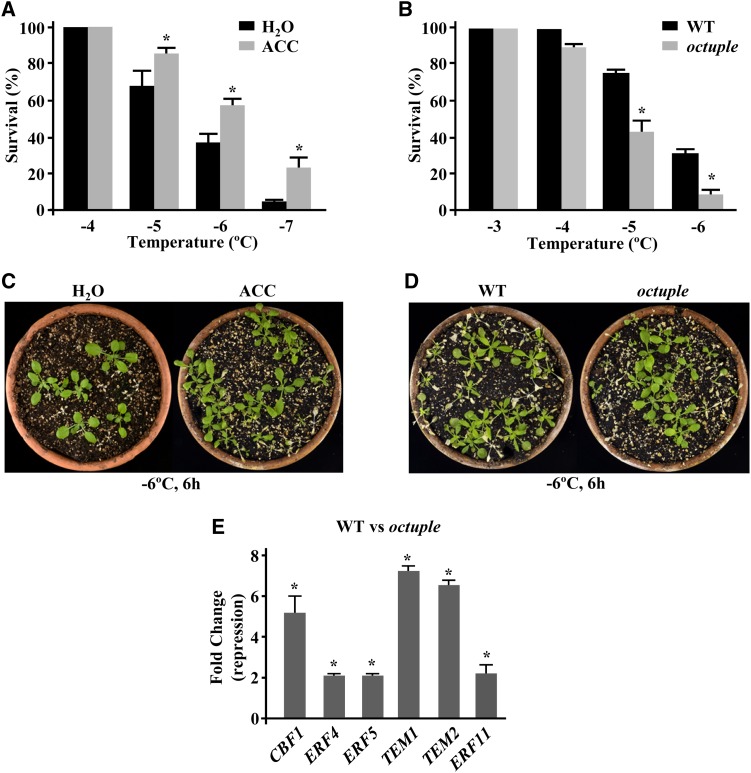
It has been reported that exogenous application of ACC increases freezing tolerance in tomato and tobacco (Zhang and Huang, 2010). Conversely, the exogenous application of CoCl2, an inhibitor of ET biosynthesis, reduces the tolerance of these species to temperatures below 0°C (Zhang and Huang, 2010), suggesting a positive role for ET in regulating plant freezing tolerance. According to this, the elevated levels of ET detected in the rci1a mutant (Figure 5B) might account for its increased constitutive capacity to tolerate freezing (Figures 3A and 3C). This hypothesis was first explored by analyzing the effect of ACC on Arabidopsis intrinsic freezing tolerance. The results revealed that, as in tomato and tobacco, exogenous application of ACC significantly augmented the constitutive freezing tolerance of Arabidopsis (Figures 7A and 7C). The LT50 values obtained were −5.3 and −6.2°C for control and ACC-treated plants, respectively (Figures 7A and 7C). Then, we determined the constitutive freezing tolerance of an Arabidopsis octuple ACS mutant that contains extremely low levels of ET (Tsuchisaka et al., 2009). Consistent with our hypothesis, octuple plants exhibited a significantly lower freezing tolerance than wild-type plants, the corresponding LT50 values being −4.8 and −5.5°C, respectively (Figures 7B and 7D). These findings provided genetic evidence that ET positively regulates Arabidopsis intrinsic freezing tolerance and, therefore, that the high levels of ET in rci1a would account for its increased constitutive freezing tolerance.
Figure 7.
ET Positively Regulates Freezing Tolerance and Cold-Induced Gene Expression in Arabidopsis under Control Conditions.
(A) and (B) Freezing tolerance of 2-week-old nonacclimated plants exposed for 6 h to the indicated freezing temperatures. Tolerance is shown in Col-0 plants treated with water (H2O) or 1 mM ACC (ACC) for 24 h before being exposed to freezing temperatures (A) and in Col-0 wild-type and octuple ACS plants (B). In all cases, freezing tolerance was estimated as the percentage of plants surviving each specific temperature after 7 d of recovery under control conditions. Data are expressed as means of three independent experiments with 50 plants each. Error bars indicate sd. Asterisks indicate significant differences (P < 0.05) from water-treated (A) and wild-type (B) plants.
(C) and (D) Freezing tolerance of representative Col-0 plants treated with water (H2O) or 1 mM ACC (ACC) (C) and Col-0 wild-type and octuple ACS plants (D) 7 d after being exposed to −6°C for 6 h.
(E) Expression levels of CBF1, ERF4, ERF5, TEM1, TEM2, and ERF11 genes, as determined by qPCR, in 2-week-old Col-0 wild-type and octuple ACS plants grown under control conditions. Analyses were performed in triplicate with three independent RNA samples. Error bars indicate sd. Asterisks indicate significant differences (P < 0.05) from wild-type plants.
[See online article for color version of this figure.]
The question arising from the results described above is how ET positively regulates the constitutive capacity of Arabidopsis to tolerate freezing. Interestingly, ∼39% (i.e., 225) of the genes whose expression was described to be upregulated (≥2-fold) in ACC-treated Arabidopsis plants (Alonso et al., 2003; Goda et al., 2008) are also upregulated (≥2-fold) by cold (Kilian et al., 2007) (Supplemental Data Set 8). In addition, a detailed analysis of the microarray data available from the octuple mutant (Tsuchisaka et al., 2009) revealed that around 48% (i.e., 42) of the downregulated (≥2-fold) genes had been reported to be induced (≥2-fold) in response to low temperature (Kilian et al., 2007) (Supplemental Data Set 9). These observations suggested that ET mediates the constitutive expression of many cold-inducible genes and, therefore, could function in Arabidopsis intrinsic freezing tolerance by maintaining appropriate levels of constitutive cold-induced gene expression. Supporting this assumption, 34% (i.e., 25) of the cold-inducible genes whose expression was increased (≥2-fold) in rci1a mutant plants under control conditions had been described to be induced (≥2-fold) by ET as well (Alonso et al., 2003; Goda et al., 2008) (Supplemental Data Set 7). Moreover, the expression of some of these genes, including those encoding the transcription factors CBF1, ERF4, ERF5, TEM1, TEM2, and ERF11, was downregulated (≥2-fold) in octuple plants (Figure 7E). Considering that RCI1A prevents ET biosynthesis (see above), all these data indicated that this 14-3-3 isoform negatively regulates cold-induced gene expression under control conditions and, consequently, Arabidopsis constitutive freezing tolerance, in part, by controlling the endogenous levels of ET.
RCI1A-Regulated Accumulation of ET in Response to Low Temperature Mediates Cold-Induced Gene Expression and the Precise Development of Cold Acclimation in Arabidopsis
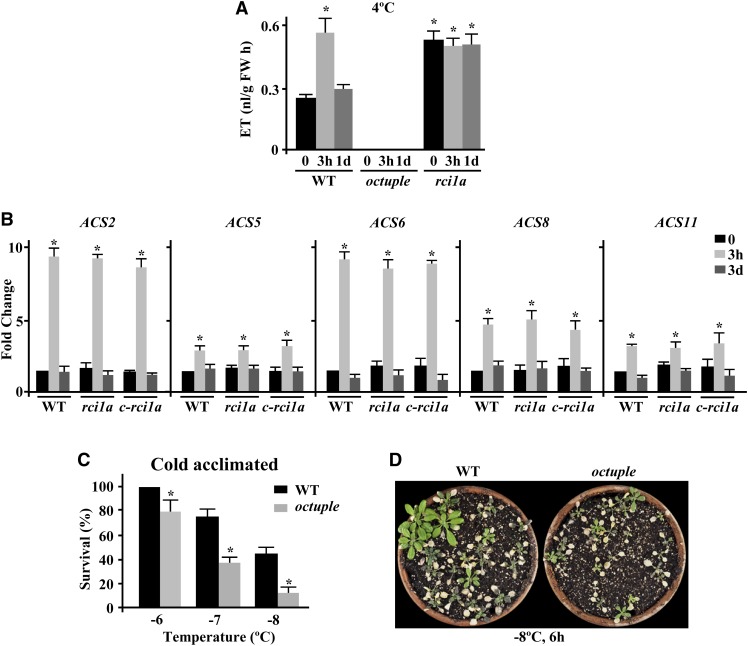
Once we established that ET positively regulates the intrinsic freezing tolerance of Arabidopsis, we considered whether it could also play a positive role in the regulation of cold acclimation in this species, as reported for winter rape, winter rye, and rhododendron (Kacperska and Kubacka-Zgbalska, 1985; Harber and Fuchigami, 1989; Yu et al., 2001). First, we determined ET accumulation in response to low temperature under our growing conditions. In agreement with Wang et al. (2012), when Arabidopsis wild-type plants were exposed to 4°C, a rapid increase of ET was detected (Figure 8A). It is worth noting that the levels of ET attained were similar to those observed in rci1a mutants under control conditions (Figures 5B and 8A). Subsequently, coinciding with the induction of RCI1A (Figure 1A), ET content decreased (Figure 8A). This pattern of ET accumulation in response to low temperature paralleled those of ACS6 protein (Figure 6C) and ACS2, ACS5, ACS6, ACS8, and ACS11 transcripts (Figure 8B). Next, we examined the capacity to cold acclimate of octuple mutants, which as expected did not exhibit an increase of ET when exposed to 4°C (Figure 8A). The results showed that cold-acclimated wild-type plants displayed an LT50 value of −7.8°C, while the LT50 value of cold-acclimated octuple mutants was −6.6°C (Figures 8C and 8D). We concluded that, as in the case of constitutive freezing tolerance, ET also functions as a positive regulator of cold acclimation in Arabidopsis. As anticipated considering that the stability of ACS protein is negatively regulated by RCI1A (see above), the rci1a mutant exposed to 4°C had levels of ACS6 and ET as under control conditions (Figures 6C and 8A), and wild-type plants treated with the R18 peptide in the presence of low temperature recapitulated the accumulation patterns of ACS6 in the rci1a mutant (Figure 6E). These data indicated that the increased capacity of the rci1a mutant to cold acclimate (Figures 3B and 3D) is not due to a higher ET content during cold acclimation and that RCI1A has a critical role in the accumulation of ET that occurs in Arabidopsis in response to low temperature. This role, however, is not accomplished by controlling the cold-induced accumulation of ACS transcripts, since it is identical in cold-treated rci1a and wild-type plants (Figure 8B).
Figure 8.
ET Positively Regulates the Capacity of Arabidopsis to Cold Acclimate.
(A) Levels of ET, as determined by gas chromatography, in 3-week-old Col-0 wild-type, octuple ACS, and rci1a plants exposed for 0 and 3 h and 1 d to 4°C. Data are expressed as means of three independent experiments with five plants each. Error bars indicate sd. Asterisks indicate significant differences (P < 0.05) from wild-type plants grown under control conditions. FW, fresh weight.
(B) Expression levels of ACS2, ACS5, ACS6, ACS8, and ACS11 genes, as determined by qPCR, in 2-week-old Col-0 wild-type, rci1a, and c-rci1a plants exposed for 0 and 3 h and 3 d to 4°C. Analyses were performed in triplicate with three independent RNA samples. Error bars indicate sd. Asterisks indicate significant differences (P < 0.05) from wild-type plants grown under control conditions.
(C) Freezing tolerance of 2-week-old Col-0 wild-type and octuple ACS plants exposed for 6 h to the indicated freezing temperatures after being acclimated for 7 d at 4°C. Freezing tolerance was estimated as the percentage of plants surviving each specific temperature after 7 d of recovery under control conditions. Data are expressed as means of three independent experiments with 50 plants each. Error bars indicate sd. Asterisks indicate significant differences (P < 0.05) from wild-type plants.
(D) Freezing tolerance of representative cold-acclimated Col-0 wild-type and octuple ACS plants 7 d after being exposed to −8°C for 6 h.
[See online article for color version of this figure.]
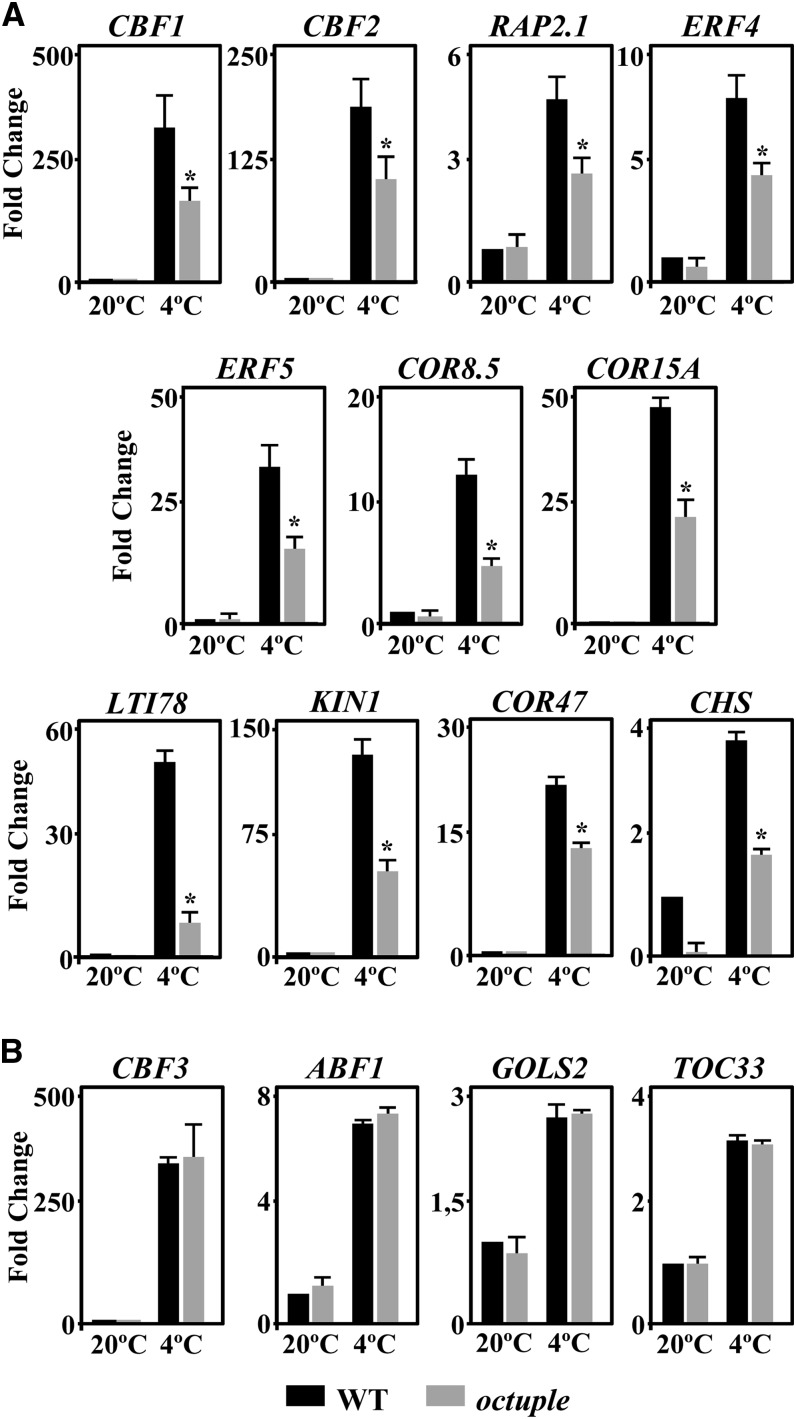
To determine whether ET also functioned in Arabidopsis cold acclimation by mediating the induction of gene expression that takes place during this adaptive response, we examined the expression levels of several cold-inducible genes that were regulated (i.e., CBF1, CBF2, RAP2.1, ERF4, ERF5, COR8.5, ABF1, GOLS2, and TOC3) or not (i.e., CBF3, COR15A, LOW-TEMPERATURE-INDUCED78 [LTI78], KIN1, COR47, and CHALCONE SYNTHASE [CHS]) by RCI1A (see above and Supplemental Table 1) in octuple mutant plants exposed to 4°C. Compared with the wild type, the induction of most of the genes analyzed was significantly lower in the mutant (Figure 9A), indicating that it was ET-dependent and, therefore, that ET was required for the accurate induction of gene expression during cold acclimation. Only the induction of CBF3, ABF1, GOLS2, and TOC33 was not affected in octuple mutant plants (Figure 9B). Collectively, all these findings provided evidence that RCI1A controls the adequate increase of ET that takes place in response to low temperature and that this increase mediates part of the cold-induced gene expression that is required for the correct development of the cold acclimation response in Arabidopsis.
Figure 9.
ET Positively Regulates Cold-Induced Gene Expression in Response to Low Temperature.
Expression levels are shown for different cold-inducible genes in 2-week-old Col-0 wild-type and octuple ACS plants grown under control conditions (20°C) or exposed to 4°C (4°C). In (A), the expression of CBF1, CBF2, RAP2.1, ERF4, ERF5, COR8.5, and COR15A was analyzed after 3 h at 4°C, and that of LTI78, KIN1, COR47, and CHS was analyzed after 1 d at 4°C. In (B), CBF3 and ABF1 expression was examined after 3 h at 4°C, while the expression of GOLS2 and TOC33 was tested after 1 d at 4°C. In all cases, analyses were performed in triplicate by qPCR with three independent RNA samples. Error bars indicate sd. Asterisks indicate significant differences (P < 0.05) from wild-type plants exposed to 4°C.
DISCUSSION
To survive subzero temperatures, plants have evolved sophisticated mechanisms involving altered physiological and biochemical processes. Uncovering the molecular basis governing those mechanisms is of relevance to understand how plants grow and reproduce under adverse environmental conditions and may help in the development of crops with improved freezing tolerance. Here, we present genetic and molecular evidence that RCI1A, the Arabidopsis 14-3-3 psi isoform, plays a critical role in freezing tolerance before and after cold acclimation by ensuring appropriate cold-induced gene expression. This function is accomplished, partially, through an ET-dependent signaling pathway involving direct interaction with different ACS isoforms and a decreased ACS stability. RCI1A negatively modulates ET biosynthesis and contributes to determine the precise levels of ET that are necessary in Arabidopsis to promote accurate cold-induced gene expression and freezing tolerance under both standard and low-temperature conditions.
Histochemical determination of GUS activity in transgenic plants containing an RCI1APRO-GUS fusion revealed that, as described for other Arabidopsis 14-3-3 genes (Paul et al., 2012), RCI1A is ubiquitously expressed under control conditions. Moreover, GUS assays showed that this expression is specifically induced by low temperature, which is in agreement with our previous report (Jarillo et al., 1994). As expected from these results, the levels of RCI1A protein also increase after cold treatment, mirroring those of RCI1A transcripts. RCI1A is uniformly distributed in cytoplasm and nucleus under both control and cold conditions. Our data indicate that the cold induction of RCI1A is regulated at the transcriptional level and that the cis-acting element(s) implicated are contained within its proximal promoter region (<2 kb). This region does not contain any described low-temperature-responsive element (Catala et al., 2012); accordingly, the levels of RCI1A transcripts in CBF-deficient mutants and wild-type plants exposed to 4°C are identical. Furthermore, these levels are also the same in ABA-deficient and ABA-insensitive mutants, suggesting that the induction of RCI1A expression in response to low temperature is mediated by a novel cold-responsive cis-acting element through a CBF- and ABA-independent pathway.
Our genetic and molecular analyses substantiate that RCI1A functions in Arabidopsis constitutive freezing tolerance and cold acclimation by negatively regulating cold-induced gene expression through several pathways. In fact, Arabidopsis mutants deficient in RCI1A show a constitutive freezing tolerance and a capacity to cold acclimate significantly higher than those of wild-type plants, both of them correlating with an enhanced cold-induced gene expression. At 20°C, the expression of 73 cold-inducible genes is higher (≥2-fold) in rci1a than in the wild type. After 3 d of exposure to 4°C, 70 cold-inducible genes are more induced (≥2-fold) in rci1a than in wild-type plants. The elevated levels of transcripts corresponding to these cold-inducible genes in the rci1a mutant should account for its increased constitutive freezing tolerance and cold acclimation capacity. When comparing the 73 and 70 genes, there are 10 in common, including those encoding the transcription factors CBF1 and CBF2, which indicates that RCI1A negatively regulates cold-induced gene expression under standard conditions and in response to low temperature via shared and specific signaling pathways. Interestingly, the expression of some RCI1A-regulated cold-inducible genes is mediated by ET, indicating that, in addition, RCI1A control of cold-induced gene expression is performed through ET-dependent and ET-independent pathways.
In this work, we demonstrate that, as described in other species (Field, 1981; Kacperska and Kubacka-Zgbalska, 1985; Harber and Fuchigami, 1989; Ciardi et al., 1997; Yu et al., 2001), low temperature provokes a transient increase of ET in Arabidopsis that is preceded by the accumulation of cold-inducible ACS transcripts and, consequently, of ACS protein. More importantly, we also show that an Arabidopsis octuple ACS mutant containing very low amounts of ET (Tsuchisaka et al., 2009) exhibits a decreased capacity to constitutively tolerate freezing temperatures and to cold acclimate, which provides direct genetic evidence that ET acts as a positive regulator of Arabidopsis constitutive freezing tolerance and cold acclimation. In addition, the expression levels of many cold-inducible genes, including both RCI1A-regulated and RCI1A-nonregulated ones, is severely compromised in the octuple mutant when growing under standard conditions or upon exposure to low temperature, indicating that ET is involved in Arabidopsis constitutive freezing tolerance and cold acclimation by mediating cold-induced gene expression. A positive role for ET in regulating the constitutive freezing tolerance of tomato and tobacco was also reported by Zhang and Huang (2010). Similarly, ET was also described to increase the capacity of rhododendron plants to cold acclimate (Harber and Fuchigami, 1989). Moreover, it has been demonstrated that ET biosynthesis is required for the accumulation of antifreeze proteins in winter rye during cold acclimation (Yu et al., 2001), and ET has been proposed to protect mitochondrial activity in Arabidopsis during cold acclimation (Wang et al., 2012). In contrast with our findings and all previous reports positively associating ET with plant responses to low temperatures (Field, 1981; Kacperska and Kubacka-Zgbalska, 1985; Harber and Fuchigami, 1989; Ciardi et al., 1997; Yu et al., 2001; Wang et al., 2012), Shi et al. (2012) described that ET production is reduced by cold treatment and that ET negatively regulates freezing tolerance in Arabidopsis. These different results could be due, in all likelihood, to the different conditions employed in these studies to grow plants. While we and the other groups used plants grown on soil, all experiments performed by Shi and collaborators (2012) were conducted with seedlings grown on Murashige and Skoog (MS) medium in Petri dishes, which implies that, among other differences, they were exposed to a high relative humidity. Intriguingly, it has been reported that the environmental relative humidity has a decisive influence on the ET levels of cold-treated plants. Indeed, the production of ET in response to low temperature is inhibited under conditions of high relative humidity (Guye et al., 1987; Janowiak and Dürffling, 1995). Consistent with this interpretation, we found that, contrary to the results obtained with plants grown on soil, rci1a mutant seedlings grown in Petri dishes on MS medium displayed impaired capacity to tolerate freezing and to cold acclimate (Supplemental Figure 6). Therefore, growing conditions must be seriously considered when studying ET regulation of the plant response to low temperature. Moreover, it was recently shown that soil-grown eto1-15 mutants exhibit a salt-tolerant phenotype that is not observed when grown in Petri dishes (Jiang et al., 2013), further supporting the critical role that growing conditions play in ET-dependent Arabidopsis responses to abiotic stresses.
The data obtained from this study also uncover an important function for RCI1A in controlling Arabidopsis levels of ET under both standard and low-temperature conditions. In plants growing at 20°C, the absence of RCI1A leads to an increase in ET content similar to that induced by cold in wild-type Arabidopsis (∼2-fold), revealing that it negatively modulates the intrinsic levels of ET and that such an increase should substantially contribute to the constitutive freezing tolerance observed in the rci1a mutant. Furthermore, we show that the role of RCI1A in ET homeostasis is performed through direct interaction with ACS proteins, which leads to reduced ACS stability. RCI1A physically interacts with multiple ACS isoforms, including ACS2, ACS5, ACS6, ACS8, and ACS11, and rci1a mutants contain an increased content of ACS6, comparable to that detected in cold-treated wild-type plants, without having affected ACS gene expression. As already mentioned, in response to low temperature, ACS protein and, consequently, ET accumulate in wild-type plants. The levels of ACS6 protein and ET increase rapidly and subsequently decrease, coinciding with the later induction and accumulation of RCI1A, which is consistent with the proposed function for RCI1A as a negative regulator of ACS stability. Moreover, as expected, in rci1a mutants exposed to 4°C, ACS6 and ET levels remain as under control conditions. ACS2, ACS5, ACS6, ACS8, and ACS11 transcripts also accumulate in response to low temperature in wild-type plants, but this accumulation is not affected in rci1a mutants. In agreement also with a role for RCI1A in destabilizing ACS protein, wild-type plants treated with the R18 peptide recapitulate the accumulation patterns of ACS6 in rci1a mutants under both standard and cold conditions. All these findings indicate that RCI1A regulates the content of ET in Arabidopsis not at the transcriptional level but posttranslationally, by reducing the stability of ACS protein. The role of 14-3-3 proteins in regulating ET levels has already been documented in potato (Solanum tuberosum), where the antisense expression of an endogenous 14-3-3 gene induces ET accumulation (Szopa, 2002). More recently, it has been reported that the Arabidopsis 14-3-3 proteins positively regulate ET biosynthesis by interacting with and stabilizing ACS isoforms (Yoon and Kieber, 2013). Although apparently conflicting at first sight, our data and these results may be compatible. In fact, available evidence indicates that there are functional specificities in the 14-3-3 isoforms (Fu et al., 2000; Börnke, 2005; Paul et al., 2012; Pallucca et al., 2014), and Yoon and Kieber (2013) used the 14-3-3 omega isoform in their experiments instead of RCI1A. Furthermore, it has also been reported that one 14-3-3 isoform may have different functions depending on the binding sites it recognizes in a given client protein (Ganguly et al., 2005). It is perfectly conceivable, therefore, that Arabidopsis 14-3-3 isoforms omega and RCI1A have different roles in regulating ACS stability.
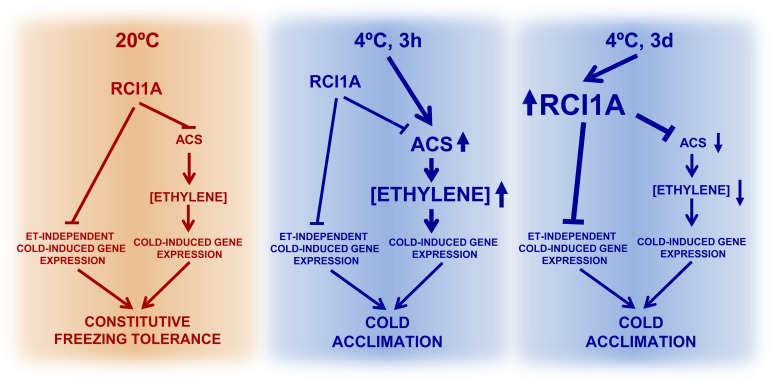
Based on the results described here, a hypothetical model for RCI1A function in freezing tolerance and cold acclimation is presented in Figure 10. Under standard conditions, steady state levels of RCI1A would negatively control the expression of a number of cold-inducible genes to ensure the appropriate levels of their corresponding transcripts and, consequently, the adequate Arabidopsis constitutive freezing tolerance. The expression of several of these genes is mediated by ET and would be controlled by RCI1A by maintaining the correct levels of ACS protein and, therefore, of ET. The expression of the rest of the genes would be controlled by RCI1A via ET-independent pathways. When plants are exposed to low temperature, the expression of multiple ACS genes is rapidly induced and the levels of ACS and ET increase, overcoming the control of RCI1A. This rise in ET would mediate the accurate induction of many cold-regulated genes that are required for the full development of the cold acclimation response in Arabidopsis. Later, subsequent to the induction of ACS genes, low temperature induces the expression of RCI1A, which results in an increase of RCI1A levels. The increase of RCI1A would contribute to restore the basal levels of ACS protein and, consequently, of ET, which would ensure the suitable cold induction patterns of the ET-mediated cold-regulated genes. In addition, the increased levels of RCI1A would control the cold induction patterns of other genes whose response to low temperature is not mediated by ET. Although further studies are needed to reveal the complete repertoire of mechanisms through which RCI1A mediates cold signaling, our data provide new insights to advance our understanding of the molecular basis of plant tolerance to freezing temperatures.
Figure 10.
Proposed Model for RCI1A Function in the Arabidopsis Response to Low Temperature.
RCI1A plays a critical role in Arabidopsis freezing tolerance, before and during cold acclimation, by controlling cold-induced gene expression through ET-dependent and ET-independent pathways. Lines with arrowheads denote positive regulation, and lines with blunt heads represent negative regulation.
METHODS
Plant Materials
Arabidopsis thaliana Columbia-0 (Col-0) and Landsberg erecta ecotypes, and mutants aba1-1, abi1-1, eto1-3, and octuple ACS, were obtained from the Nottingham Arabidopsis Stock Centre. The Arabidopsis cbf2 mutant, as well as the cbf1−cbf3− and LSM1-GFP transgenic lines, were generated previously in our laboratory (Novillo et al., 2004, 2007; Perea-Resa et al., 2012). To obtain the RCI1APRO-GUS fusion, a 1981-bp (−1978 to +3) promoter fragment from RCI1A was amplified with appropriate primers (Supplemental Table 1) and cloned into the pBI101 binary vector (Clontech). The RCI1APRO-RCI1A-GFP (c-rci1a) fusion was obtained by amplifying the RCI1A genomic region, including the RCI1APRO fragment, with appropriate primers (Supplemental Table 1) and cloning the resulting PCR product into the pGWB4 Gateway binary vector (Nakagawa et al., 2007). The RCI1APRO-GUS and c-rci1a fusions were verified by sequencing and introduced in Col-0 and rci1a, respectively, via Agrobacterium tumefaciens C58C1 using the floral dip method (Clough and Bent, 1998). All transgenic lines were genetically determined to have the fusions integrated at a single locus in homozygosis. For BiFC assays, full-length cDNAs corresponding to RCI1A, ACS2, ACS5, ACS6, ACS8, ACS11, and RGA1 genes were amplified with appropriate primers (Supplemental Table 1) to incorporate convenient cloning sequences at their 5′ and 3′ ends. The PCR products generated were then cloned into the pSPYNE-35S and pSPYCE-35S binary vectors (Walter et al., 2004), kindly provided by Jörg Kudla (Westfälische Wilhelms-Universität), using the In-Fusion HD Cloning Kit (Clontech), sequenced, and introduced in Agrobacterium strain C58C1 for subsequent agroinfiltration in Nicotiana benthamiana leaves (see below).
Growth Conditions and Treatments
Plants and seedlings were grown at 20°C under long-day photoperiods (16 h of cool-white fluorescent light, photon flux of 90 μmol m−2 s−1) in pots containing a mixture of organic substrate and vermiculite (3:1, v/v) or in Petri dishes containing MS medium (Murashige and Skoog, 1962) supplemented with 1% Suc and solidified with 0.8% (w/v) agar, respectively. Low-temperature treatments for expression analysis were performed by transferring plants to a growth chamber set to 4°C for different periods of time under a long-day photoperiod (16 h of cool-white fluorescent light, photon flux of 40 μmol m−2 s−1). Subsequently, plants were immediately frozen in liquid N2 and stored at −80°C until use. Etiolated seedlings were obtained by germinating seeds under dark conditions for 4 d at 20°C on vertical plates containing MS medium supplemented with 1% Suc. Treatments with 10 μM AgNO3 and 0.1 mM ACC were performed as described (Cancel and Larsen, 2002). Treatments with the R18 peptide were realized on 2-week-old wild-type seedlings grown in soil under standard conditions transferred to MS liquid medium supplemented with 10 μg/mL R18 peptide (Sigma) and exposed to 4°C for different periods of time. For histochemical analysis of GUS activity, cold treatments were performed on 2-week-old RCI1APRO-GUS seedlings and 8-week-old RCI1APRO-GUS plants grown under standard conditions and subsequently transferred to a growth chamber set to 4°C for an additional 3 d. NaCl and ABA treatments were conducted with 2-week-old transgenic seedlings grown under standard conditions and then transferred to Petri dishes containing MS medium supplemented with 100 mM NaCl and 100 μM ABA, respectively, during an additional 3 d. The dehydration treatment was accomplished by transferring 2-week-old RCI1APRO-GUS transgenic seedlings grown under standard conditions to a filter paper soaked in water and allowing them to dehydrate until they lost 30% of their fresh weight.
In vivo freezing assays were performed on 2-week-old plants as described previously (Catalá et al., 2011). For freezing experiments with ACC-treated plants, 2-week-old plants were sprayed with a 1 mM ACC solution containing 0.1% Tween 20, or with 0.1% Tween 20 for controls, 1 d before being exposed to freezing temperatures. In vitro freezing assays were performed in 2-week-old seedlings as reported by Xin and Browse (1998). For NaCl tolerance analysis, 5-d-old seedlings grown under standard conditions were transferred to new plates with MS medium supplemented with 100 mM NaCl. Tolerance to NaCl was estimated as the number of green leaves and the percentage of remaining fresh weight 2 weeks after the treatment. For drought tolerance evaluation, 2-week-old seedlings grown under standard conditions were transferred to a filter paper soaked in water and allowed to dehydrate for 2 d. Tolerance was measured as the percentage of initial fresh weight that remained after treatment. In all cases, values were statistically analyzed by means of one-way ANOVA and the Dunnett test, taking P < 0.05 as significant, using the GraphPad Prism6 (GraphPad Software) statistical analysis software.
Isolation of T-DNA Insertion Mutants in RCI1A
An rci1a mutant was identified from a collection of 60,000 Arabidopsis T-DNA insertion lines (Alonso et al., 2003) by PCR screening with specific oligonucleotides for the left and right borders of the T-DNA and for RCI1A (Supplemental Table 1). DNA sequencing showed that the T-DNA insertion in the mutant was 53 bp downstream of the start codon of RCI1A.
Gene Expression Analysis
Total RNA was extracted using the Purezol reagent (Bio-Rad) according to the manufacturer’s protocol. RNA samples were treated with DNase I (Roche) and quantified with a Nanodrop spectrophotometer (Thermo Scientific). For real-time qPCR, cDNAs were prepared with the iScript cDNA synthesis kit (Bio-Rad) and then amplified using the Bio-Rad iQ2 thermal cycler, the SsoFast EvaGreen Supermix (Bio-Rad), and gene-specific primers (Supplemental Table 1). The relative expression values were determined using the At4g24610 gene as a reference (Czechowski et al., 2005). All reactions were realized in triplicate employing three independent RNA samples. In all cases, values were statistically analyzed by means of one-way ANOVA and the Dunnett test, taking P < 0.05 as significant, using the GraphPad Prism6 (GraphPad Software) statistical analysis software.
The Expression Browser tool of The Bio-Analytic Resource for Plant Biology (http://bar.utoronto.ca) (Toufighi et al., 2005) was used to determine the genes from our microarray data that, in addition of being upregulated in the rci1a mutant, were cold- and/or ET-induced. To uncover the cold-induced genes, selected settings were “AtGenExpress-stress series” as data set and “cold stress (Kilian et al., 2007)” as research area. To uncover the ET-induced genes, selected settings were “AtGenExpress-hormone series” as data set and “ACC time course in wild type seedlings (Goda et al., 2008)” as research area. In both cases, all tissue types, growth stages, and time points were considered, output options were set to “Average of replicate treatments relative to average of appropriate control,” and induction was only contemplated when fold change was equal to or higher than 2-fold.
The total number of cold-inducible genes in Arabidopsis (5252) was estimated from the data described by Kilian et al. (2007), considering those genes whose expression was induced at least 2-fold in at least one time point or tissue analyzed. The genes identified by Alonso et al. (2003) as induced (≥2-fold) by ET (244) and those reported by Goda et al. (2008) to be induced (≥2-fold) by ACC treatment in at least one time point analyzed (335) were considered to generate the list of no redundant Arabidopsis ET-inducible genes (545).
Determination of GUS Activity
GUS activity in Arabidopsis transgenic plants and seedlings containing the fusion RCI1APRO-GUS was detected and measured as described (Medina et al., 2001). GUS activity values were statistically analyzed by means of one-way ANOVA and the Dunnett test, taking P < 0.05 as significant, using the GraphPad Prism6 (GraphPad Software) statistical analysis software.
Microscopy
Subcellular localization of the RCI1A-GFP fusion protein in c-rci1a was performed in roots from 6-d-old seedlings. Transient expression of fusion proteins for BiFC assays was analyzed 3 d after agroinfiltration in leaves of 3-week-old N. benthamiana plants grown at 25°C, as reported by English et al. (1997). Microscopy images were collected using a TCS SP2 confocal laser spectral microscope (Leica Microsystems). The excitation lines for imaging GFP and YFP fusions were 488 and 514 nm, respectively. For cell perimeter measurements, individual cells from leaves of 3-week-old Col-0 and rci1a plants were visualized with the confocal microscope and artificially colored using Adobe Photoshop CS3. Cell perimeters were determined using ImageJ software (http://rsbweb.nih.gov/ij/), and measurements ± sd were obtained for n ≥ 25. Values were statistically analyzed by means of one-way ANOVA and the Dunnett test, taking P < 0.05 as significant, using the GraphPad Prism6 (GraphPad Software) statistical analysis software.
Immunoblot Analysis
For protein blot analysis, protein extracts were obtained as described (Catalá et al., 2011) from 6-d-old c-rci1a seedlings exposed to 4°C for different periods (0, 1, 3, and 7 d). Extracts were also obtained from 2-week-old Col-0, rci1a, and c-rci1a plants grown under control conditions or exposed to 4°C for 3 h and 3 d. Protein samples (40 µg) were resolved by electrophoresis on 12% SDS-polyacrylamide gels and electrophoretically transferred to a polyvinylidene difluoride membrane (Bio-Rad), according to the manufacturer’s protocol. Rabbit polyclonal anti-GFP (ab290; Abcam) and goat polyclonal anti-ACS6 (aC-20; Santa Cruz Biotechnology) antibodies were used as primary antibodies, and horseradish peroxidase–conjugated anti-rabbit (sc-2004; Santa Cruz Biotechnology) and anti-goat (sc-2020; Santa Cruz Biotechnology) antibodies were used as secondary antibodies, respectively. Signal detection was performed with the ECL Western detection kit (Amersham Biosciences). The 55-kD Rubisco subunit was used as a loading control.
Coimmunoprecipitation Assay
Two-week-old Col-0, c-rci1a, and transgenic plants containing an LSM1-GFP fusion (Perea-Resa et al., 2012) grown under control conditions were used for this assay. Coimmunoprecipitation was performed as described previously by Yoon and Kieber (2013) with some modifications. Protein extracts were isolated from plants incubated overnight in 50 μM MG132 solution and suspended in coimmunoprecipitation buffer (100 mM sodium phosphate, pH 7.0, 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 1 mM NaVO3, 5 mM NaF, 0.1% Triton X-100, 1× protease inhibitor cocktail [Roche], and 1 mM phenylmethylsulfonyl fluoride). Extracts were then kept on ice for 30 min, and after clarifying, supernatants were incubated overnight with 25 μL of GFP-Trap (ChromoTek). Subsequently, beads were washed three times with wash buffer (100 mM sodium phosphate, pH 7.0, 250 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 1 mM NaVO3, 5 mM NaF, 0.1% Triton X-100, 1× protease inhibitor cocktail [Roche], and 1 mM phenylmethylsulfonyl fluoride) and eluted with boiled 2× SDS buffer followed by immunoblotting with anti-ACS6 (ab290; Abcam) antibody.
Microarray Analysis
Total RNA from 2-week-old Col-0 and rci1a plants grown at 20°C or exposed for an additional 3 d to 4°C was extracted using the RNeasy kit (Qiagen), and three biological replicates were independently hybridized per transcriptomic comparison. RNA amplification and labeling were performed basically as reported (Goda et al., 2008). Hybridization was performed on Agilent Arabidopsis Oligo Microarrays version 3 (catalog number G2519F-015059) in accordance with the manufacturer’s specifications. The statistical significance of the results was evaluated with FIESTA software (http://bioinfogp.cnb.csic.es). Genes with an FDR-corrected P value of <0.05 and a fold change ≥+2-fold or ≤−2-fold were selected for consideration. Data from these microarray experiments have been deposited in the Gene Expression Omnibus database under accession number GSE51859.
ET Quantification
Three-week-old plants grown under standard conditions or exposed to cold were placed into 12-mL glass vials that were maintained open for 1 h at 20 or 4°C, respectively. After sealing the vials with rubber stoppers, they were kept at the same temperature conditions for 3 and 24 h. Then, 1 mL of gas was withdrawn from each vial using a gas-tight syringe and left for 2 min to equilibrate. Gas samples were subsequently injected into a gas chromatograph (Varian 3900) fitted with a Porapak Q column and a flame ionization detector. The detector and injector were operated at 200 and 170°C, respectively, whereas the oven temperature was 50°C. The carrier gas was nitrogen at a flow rate of 40 mL/min. The ET produced (nL·g−1 fresh weight·h−1) was determined by comparing with an ET standard. All measurements were realized in triplicate employing five independent samples. Values were statistically analyzed by means of one-way ANOVA and the Dunnett test, taking P < 0.05 as significant, using the GraphPad Prism6 (GraphPad Software) statistical analysis software.
Accession Numbers
The microarray data were submitted to the Gene Expression Omnibus site (www.ncbi.nlm.nih.gov/geo) under the accession number GSE51859. Sequence data for the genes described in this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: RCI1A (At5g38480), CBF1 (At4g25490), CBF2 (At4g25470), CBF3 (At4g25480), COR8.5 (At2g23120), GOLS2 (At1g56600), RAP2.1 (At1g46768), ABF1 (At1g49720), PK (At2g36580), AGP5 (At1g35230), TOC33 (At1g02280), TEM1 (At1g25560), TEM2 (At1g68840), PDF1.2a (At5g44420), ERF4 (At3g15210), ERF5 (At5g47230), DYL1 (At1g28330), MEE3 (At2g21650), P1R3 (At3g29370), ACS2 (At1g01480), ACS5 (At3g51770), ACS8 (At4g37770), ACS11 (At4g08040), RGA (At2g01570), COR15A (At2g42540), LTI78 (At5g52310), KIN1 (At5g15960), COR47 (At1g20440), and CHS (At5g13930).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. RCI1A Expression Is Not Regulated by NaCl, Dehydration, or ABA Treatments.
Supplemental Figure 2. Phenotypic Characterization of rci1a Mutant.
Supplemental Figure 3. RCI1A Is Not Involved in Arabidopsis Tolerance to Drought and Salt Stress.
Supplemental Figure 4. rci1a Mutants Display an ET Overproducer Phenotype.
Supplemental Figure 5. RCI1A Interacts with Multiple ACS Isoforms.
Supplemental Figure 6. rci1a Mutants Show Decreased Constitutive Freezing Tolerance and Cold Acclimation Capacity When Growing on MS in Petri Dishes.
Supplemental Table 1. Oligonucleotide Sequences of Primers Used in This Study.
Supplemental Data Set 1. Genes Whose Expression Is Upregulated in the rci1a Mutant at 20°C.
Supplemental Data Set 2. Cold-Induced Genes Whose Expression Is Upregulated in the rci1a Mutant at 20°C.
Supplemental Data Set 3. Genes Whose Expression Is Downregulated in the rci1a Mutant at 20°C.
Supplemental Data Set 4. Genes Whose Expression Is Upregulated in the rci1a Mutant after 3 d at 4°C.
Supplemental Data Set 5. Cold-Induced Genes Whose Expression Is Upregulated in the rci1a Mutant after 3 d at 4°C.
Supplemental Data Set 6. Genes Whose Expression Is Downregulated in the rci1a Mutant after 3 d at 4°C.
Supplemental Data Set 7. ET-Induced Genes Whose Expression Is Upregulated in the rci1a Mutant at 20°C.
Supplemental Data Set 8. Response to Cold of ET-Induced Genes.
Supplemental Data Set 9. Response to Cold of Downregulated Genes in the acs octuple Mutant.
Supplementary Material
Acknowledgments
We thank J. Kudla for the pSPYNE-35S and pSPYCE-35S vectors and J.J. Sanchez-Serrano and R. Solano for discussions and comments. This work was supported by the Spanish Secretary of Research, Development, and Innovation (Grants CSD2007-00057, EUI2009-04074, and BIO2010-17545), by grants from the Gordon and Betty Moore Fundation (GMBF3034) and from the National Science Foundation (MCB 1122250) to J.R.E., and by a JAE-DOC contract from the Consejo Superior de Investigaciones Científicas to R.C.
AUTHOR CONTRIBUTIONS
R.C. designed the research, performed research, and analyzed data. R.L.-C. performed research and analyzed data. M.M.C. and T.A. performed research. J.M.A. and J.R.E. generated the rci1a mutant. J.S. designed the research, analyzed data, and wrote the article.
Glossary
- ABA
abscisic acid
- ET
ethylene
- LT50
temperature that causes 50% lethality
- FDR
false discovery rate
- qPCR
quantitative PCR
- BiFC
bimolecular fluorescence complementation
- MS
Murashige and Skoog
- Col-0
Columbia-0
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Abarca D., Madueño F., Martínez-Zapater J.M., Salinas J. (1999). Dimerization of Arabidopsis 14-3-3 proteins: Structural requirements within the N-terminal domain and effect of calcium. FEBS Lett. 462: 377–382. [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Barrero-Gil J., Salinas J. (2013). Post-translational regulation of cold acclimation response. Plant Sci. 205-206: 48–54. [DOI] [PubMed] [Google Scholar]
- Börnke F. (2005). The variable C-terminus of 14-3-3 proteins mediates isoform-specific interaction with sucrose-phosphate synthase in the yeast two-hybrid system. J. Plant Physiol. 162: 161–168. [DOI] [PubMed] [Google Scholar]
- Campo S., Peris-Peris C., Montesinos L., Peñas G., Messeguer J., San Segundo B. (2012). Expression of the maize ZmGF14-6 gene in rice confers tolerance to drought stress while enhancing susceptibility to pathogen infection. J. Exp. Bot. 63: 983–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancel J.D., Larsen P.B. (2002). Loss-of-function mutations in the ethylene receptor ETR1 cause enhanced sensitivity and exaggerated response to ethylene in Arabidopsis. Plant Physiol. 129: 1557–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao A., Jain A., Baldwin J.C., Raghothama K.G. (2007). Phosphate differentially regulates 14-3-3 family members and GRF9 plays a role in Pi-starvation induced responses. Planta 226: 1219–1230. [DOI] [PubMed] [Google Scholar]
- Catala, R., Diaz, A., and Salinas, J. (2012). Molecular responses to extreme temperatures. In Plant Biotechnology and Agriculture Prospects for the 21st Century, A. Altman and P.M. Hasegawa, eds (Amsterdam: Elsevier Academic Press), pp. 287–301. [Google Scholar]
- Catalá R., Medina J., Salinas J. (2011). Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 16475–16480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang I.F., Curran A., Woolsey R., Quilici D., Cushman J.C., Mittler R., Harmon A., Harper J.F. (2009). Proteomic profiling of tandem affinity purified 14-3-3 protein complexes in Arabidopsis thaliana. Proteomics 9: 2967–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciardi J.A., Deikman J., Orzolek M.D. (1997). Increased ethylene synthesis enhances chilling tolerance in tomato. Physiol. Plant. 101: 333–340. [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison F.C., Paul A.L., Zupanska A.K., Ferl R.J. (2011). 14-3-3 proteins in plant physiology. Semin. Cell Dev. Biol. 22: 720–727. [DOI] [PubMed] [Google Scholar]
- English J.J., Davenport G.F., Elmayan T., Vaucheret H., Baulcombe D.C. (1997). Requirement of sense transcription for homology-dependent virus resistance and trans-inactivation. Plant J. 12: 597–603. [Google Scholar]
- Field R.J. (1981). The effect of low temperature on ethylene production by leaf tissues of Phaseolus vulgaris L. Ann. Bot. (Lond.) 47: 215–221. [Google Scholar]
- Fowler S., Thomashow M.F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Subramanian R.R., Masters S.C. (2000). 14-3-3 proteins: Structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 40: 617–647. [DOI] [PubMed] [Google Scholar]
- Gampala S.S., et al. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly S., Weller J.L., Ho A., Chemineau P., Malpaux B., Klein D.C. (2005). Melatonin synthesis: 14-3-3-dependent activation and inhibition of arylalkylamine N-acetyltransferase mediated by phosphoserine-205. Proc. Natl. Acad. Sci. USA 102: 1222–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour S.J., Thomashow M.F. (1991). Cold acclimation and cold-regulated gene expression in ABA mutants of Arabidopsis thaliana. Plant Mol. Biol. 17: 1233–1240. [DOI] [PubMed] [Google Scholar]
- Gilmour S.J., Sebolt A.M., Salazar M.P., Everard J.D., Thomashow M.F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124: 1854–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour S.J., Zarka D.G., Stockinger E.J., Salazar M.P., Houghton J.M., Thomashow M.F. (1998). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16: 433–442. [DOI] [PubMed] [Google Scholar]
- Goda H., et al. (2008). The AtGenExpress hormone and chemical treatment data set: Experimental design, data evaluation, model data analysis and data access. Plant J. 55: 526–542. [DOI] [PubMed] [Google Scholar]
- Gökirmak T., Paul A.L., Ferl R.J. (2010). Plant phosphopeptide-binding proteins as signaling mediators. Curr. Opin. Plant Biol. 13: 527–532. [DOI] [PubMed] [Google Scholar]
- Guye M.G., Vigh L., Wilson J.M. (1987). Chilling-induced ethylene production in relation to chill-sensitivity in Phaseolus spp. J. Exp. Bot. 38: 680–690. [Google Scholar]
- Harber, R.M., and Fuchigami, L.H. (1989). Ethylene-Induced Stress Resistance. (Boca Raton, FL: CRC Press). [Google Scholar]
- Hu C.D., Chinenov Y., Kerppola T.K. (2002). Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9: 789–798. [DOI] [PubMed] [Google Scholar]
- Huang S.J., Chang C.L., Wang P.H., Tsai M.C., Hsu P.H., Chang I.F. (2013). A type III ACC synthase, ACS7, is involved in root gravitropism in Arabidopsis thaliana. J. Exp. Bot. 64: 4343–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowiak F., Dürffling K. (1995). Chilling-induced changes in the contents of 1-aminocyclopropane-1-carboxylic acid (ACC) and its N-malonyl conjugate (MACC) in seedlings of two maize inbreds differing in chilling tolerance. J. Plant Physiol. 147: 257–262. [Google Scholar]
- Jarillo J.A., Capel J., Leyva A., Martínez-Zapater J.M., Salinas J. (1994). Two related low-temperature-inducible genes of Arabidopsis encode proteins showing high homology to 14-3-3 proteins, a family of putative kinase regulators. Plant Mol. Biol. 25: 693–704. [DOI] [PubMed] [Google Scholar]
- Jiang C., Belfield E.J., Cao Y., Smith J.A., Harberd N.P. (2013). An Arabidopsis soil-salinity-tolerance mutation confers ethylene-mediated enhancement of sodium/potassium homeostasis. Plant Cell 25: 3535–3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacperska A., Kubacka-Zgbalska M. (1985). Is lipoxygenase involved in the formation of ethylene from ACC? Physiol. Plant. 64: 333–338. [Google Scholar]
- Kasuga M., Liu Q., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17: 287–291. [DOI] [PubMed] [Google Scholar]
- Kieber J.J., Rothenberg M., Roman G., Feldmann K.A., Ecker J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441. [DOI] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D’Angelo C., Bornberg-Bauer E., Kudla J., Harter K. (2007). The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50: 347–363. [DOI] [PubMed] [Google Scholar]
- Lang V., Mantyla E., Welin B., Sundberg B., Palva E.T. (1994). Alterations in water status, endogenous abscisic acid content, and expression of rab18 gene during the development of freezing tolerance in Arabidopsis thaliana. Plant Physiol. 104: 1341–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt, J. (1980). Responses of Plants to Environmental Stress. Chilling, Freezing, and High Temperature Stresses, 2nd ed. (New York: Academic Press). [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente F., Oliveros J.C., Martínez-Zapater J.M., Salinas J. (2000). A freezing-sensitive mutant of Arabidopsis, frs1, is a new aba3 allele. Planta 211: 648–655. [DOI] [PubMed] [Google Scholar]
- Medina J., Bargues M., Terol J., Pérez-Alonso M., Salinas J. (1999). The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 119: 463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina J., Catalá R., Salinas J. (2001). Developmental and stress regulation of RCI2A and RCI2B, two cold-inducible genes of Arabidopsis encoding highly conserved hydrophobic proteins. Plant Physiol. 125: 1655–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina J., Catalá R., Salinas J. (2011). The CBFs: Three Arabidopsis transcription factors to cold acclimate. Plant Sci. 180: 3–11. [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Nakata M., Yuasa T., Takahashi Y., Ishida S. (2009). CDPK1, a calcium-dependent protein kinase, regulates transcriptional activator RSG in response to gibberellins. Plant Signal. Behav. 4: 372–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novillo F., Alonso J.M., Ecker J.R., Salinas J. (2004). CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 3985–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novillo F., Medina J., Salinas J. (2007). Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad. Sci. USA 104: 21002–21007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oecking C., Jaspert N. (2009). Plant 14-3-3 proteins catch up with their mammalian orthologs. Curr. Opin. Plant Biol. 12: 760–765. [DOI] [PubMed] [Google Scholar]
- Pallucca R., Visconti S., Camoni L., Cesareni G., Melino S., Panni S., Torreri P., Aducci P. (2014). Specificity of ε and non-ε isoforms of Arabidopsis 14-3-3 proteins towards the H+-ATPase and other targets. PLoS ONE 9: e90764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A.L., Denison F.C., Schultz E.R., Zupanska A.K., Ferl R.J. (2012). 14-3-3 phosphoprotein interaction networks—Does isoform diversity present functional interaction specification? Front. Plant Sci. 3: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Resa C., Hernández-Verdeja T., López-Cobollo R., del Mar Castellano M., Salinas J. (2012). LSM proteins provide accurate splicing and decay of selected transcripts to ensure normal Arabidopsis development. Plant Cell 24: 4930–4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcel R., Aroca R., Cano C., Bago A., Ruiz-Lozano J.M. (2006). Identification of a gene from the arbuscular mycorrhizal fungus Glomus intraradices encoding for a 14-3-3 protein that is up-regulated by drought stress during the AM symbiosis. Microb. Ecol. 52: 575–582. [DOI] [PubMed] [Google Scholar]
- Roberts M.R., Salinas J., Collinge D.B. (2002). 14-3-3 proteins and the response to abiotic and biotic stress. Plant Mol. Biol. 50: 1031–1039. [DOI] [PubMed] [Google Scholar]
- Schoonheim P.J., Sinnige M.P., Casaretto J.A., Veiga H., Bunney T.D., Quatrano R.S., de Boer A.H. (2007). 14-3-3 adaptor proteins are intermediates in ABA signal transduction during barley seed germination. Plant J. 49: 289–301. [DOI] [PubMed] [Google Scholar]
- Sharabi-Schwager M., Samach A., Porat R. (2010). Overexpression of the CBF2 transcriptional activator in Arabidopsis suppresses the responsiveness of leaf tissue to the stress hormone ethylene. Plant Biol. (Stuttg.) 12: 630–638. [DOI] [PubMed] [Google Scholar]
- Shi Y., Tian S., Hou L., Huang X., Zhang X., Guo H., Yang S. (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24: 2578–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szopa J. (2002). Transgenic 14-3-3 isoforms in plants: The metabolite profiling of repressed 14-3-3 protein synthesis in transgenic potato plants. Biochem. Soc. Trans. 30: 405–410. [DOI] [PubMed] [Google Scholar]
- Toufighi K., Brady S.M., Austin R., Ly E., Provart N.J. (2005). The Botany Array Resource: E-northerns, expression angling, and promoter analyses. Plant J. 43: 153–163. [DOI] [PubMed] [Google Scholar]
- Tsuchisaka A., Yu G., Jin H., Alonso J.M., Ecker J.R., Zhang X., Gao S., Theologis A. (2009). A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics 183: 979–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan C., Zhu J.K. (2002). Molecular genetic analysis of cold-regulated gene transcription. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357: 877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438. [DOI] [PubMed] [Google Scholar]
- Wang B., Yang H., Liu Y.C., Jelinek T., Zhang L., Ruoslahti E., Fu H. (1999). Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry 38: 12499–12504. [DOI] [PubMed] [Google Scholar]
- Wang H., Huang J., Liang X., Bi Y. (2012). Involvement of hydrogen peroxide, calcium, and ethylene in the induction of the alternative pathway in chilling-stressed Arabidopsis callus. Planta 235: 53–67. [DOI] [PubMed] [Google Scholar]
- Wang Y.H., Garvin D.F., Kochian L.V. (2002). Rapid induction of regulatory and transporter genes in response to phosphorus, potassium, and iron deficiencies in tomato roots. Evidence for cross talk and root/rhizosphere-mediated signals. Plant Physiol. 130: 1361–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z., Browse J. (1998). Eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc. Natl. Acad. Sci. USA 95: 7799–7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W.F., Shi W.M. (2006). Expression profiling of the 14-3-3 gene family in response to salt stress and potassium and iron deficiencies in young tomato (Solanum lycopersicum) roots: Analysis by real-time RT-PCR. Ann. Bot. (Lond.) 98: 965–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue-Xuan X., Hong-Bo S., Yuan-Yuan M., Gang X., Jun-Na S., Dong-Gang G., Cheng-Jiang R. (2010). Biotechnological implications from abscisic acid (ABA) roles in cold stress and leaf senescence as an important signal for improving plant sustainable survival under abiotic-stressed conditions. Crit. Rev. Biotechnol. 30: 222–230. [DOI] [PubMed] [Google Scholar]
- Yan J., He C., Wang J., Mao Z., Holaday S.A., Allen R.D., Zhang H. (2004). Overexpression of the Arabidopsis 14-3-3 protein GF14 lambda in cotton leads to a “stay-green” phenotype and improves stress tolerance under moderate drought conditions. Plant Cell Physiol. 45: 1007–1014. [DOI] [PubMed] [Google Scholar]
- Yang J.L., Chen W.W., Chen L.Q., Qin C., Jin C.W., Shi Y.Z., Zheng S.J. (2013). The 14-3-3 protein GENERAL REGULATORY FACTOR11 (GRF11) acts downstream of nitric oxide to regulate iron acquisition in Arabidopsis thaliana. New Phytol. 197: 815–824. [DOI] [PubMed] [Google Scholar]
- Yang S.F., Hoffman N.E. (1984). Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 1984: 155–189. [Google Scholar]
- Yoon G.M., Kieber J.J. (2013). 14-3-3 regulates 1-aminocyclopropane-1-carboxylate synthase protein turnover in Arabidopsis. Plant Cell 25: 1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.M., Griffith M., Wiseman S.B. (2001). Ethylene induces antifreeze activity in winter rye leaves. Plant Physiol. 126: 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Huang R. (2010). Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol. Biol. 73: 241–249. [DOI] [PubMed] [Google Scholar]
- Zhou H., Lin H., Chen S., Becker K., Yang Y., Zhao J., Kudla J., Schumaker K.S., Guo Y. (2014). Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell 26: 1166–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.