Significance
Cytochrome c oxidase is the terminal electron acceptor in mitochondria and aerobic bacteria, where O2 reduction is linked to proton pumping across the membrane. Understanding the mechanism by which the enzyme pumps protons requires identification of the so-called proton-loading site, which is the controlling element that assures unidirectionality in the proton flux. The position of this site has been predicted on the basis of theoretical calculations but has not been identified in experimental studies. Here we have used sophisticated biophysical techniques to investigate intraprotein electron and proton transfer in the thermophilic bacterium Thermus thermophilus. The data made it possible to identify the destination of the pumped proton and to unravel the sequence of reactions that leads to proton translocation.
Keywords: cytochrome c oxidase, membrane protein, respiration, cytochrome aa3, electron transfer
Abstract
The ba3-type cytochrome c oxidase from Thermus thermophilus is a membrane-bound protein complex that couples electron transfer to O2 to proton translocation across the membrane. To elucidate the mechanism of the redox-driven proton pumping, we investigated the kinetics of electron and proton transfer in a structural variant of the ba3 oxidase where a putative “pump site” was modified by replacement of Asp372 by Ile. In this structural variant, proton pumping was uncoupled from internal electron transfer and O2 reduction. The results from our studies show that proton uptake to the pump site (time constant ∼65 μs in the wild-type cytochrome c oxidase) was impaired in the Asp372Ile variant. Furthermore, a reaction step that in the wild-type cytochrome c oxidase is linked to simultaneous proton uptake and release with a time constant of ∼1.2 ms was slowed to ∼8.4 ms, and in Asp372Ile was only associated with proton uptake to the catalytic site. These data identify reaction steps that are associated with protonation and deprotonation of the pump site, and point to the area around Asp372 as the location of this site in the ba3 cytochrome c oxidase.
The heme-copper oxygen reductases are membrane-bound proteins in which the reduction of O2 to H2O drives proton pumping, from the negative (n) to the positive (p) side, across the membrane. The free energy from the O2-reduction reaction, conserved in the proton gradient, is used, for example, for transmembrane transport and ATP synthesis. A major fraction of the oxidases known to date can be classified as members of one of three families denoted by the letters A, B, and C (1–3). The A family includes the mitochondrial cytochrome c oxidase (CytcO) as well as the well-studied aa3-type CytcO from Rhodobacter sphaeroides. These enzymes harbor four redox-active metal sites: CuA, the primary electron (e−) acceptor from water-soluble cytochrome c, as well as the intermediate electron acceptor, heme a, and the catalytic site. The latter consists of heme a3 and CuB in close proximity (for a review of the structure and function of oxidases, see refs. 3–10). The A-family bacterial oxidases harbor two functional proton pathways leading from the n-side surface toward the catalytic site. The K pathway is used for transfer of substrate protons from the n-side solution to the catalytic site during reduction of the catalytic site, whereas the D pathway is used for transfer of the remaining substrate protons and all pumped protons after binding of O2 at the catalytic site [the K pathway is not used after O2 binding (11)].
The most-studied member of the B family is the ba3 CytcO from Thermus thermophilus, in which the intermediate electron acceptor is heme b instead of heme a (12–14) (Fig. 1A). The ba3 CytcO uses only one proton pathway for transfer of all protons (15). This pathway overlaps in space with the K pathway of the A-family oxidases, and therefore is referred to as the K-pathway analog. Whereas the A-type oxidases studied to date typically pump ∼1 H+ per e− transferred to O2, the B-type oxidases display a lower stoichiometry of ∼0.5 H+ per e− (16–18).
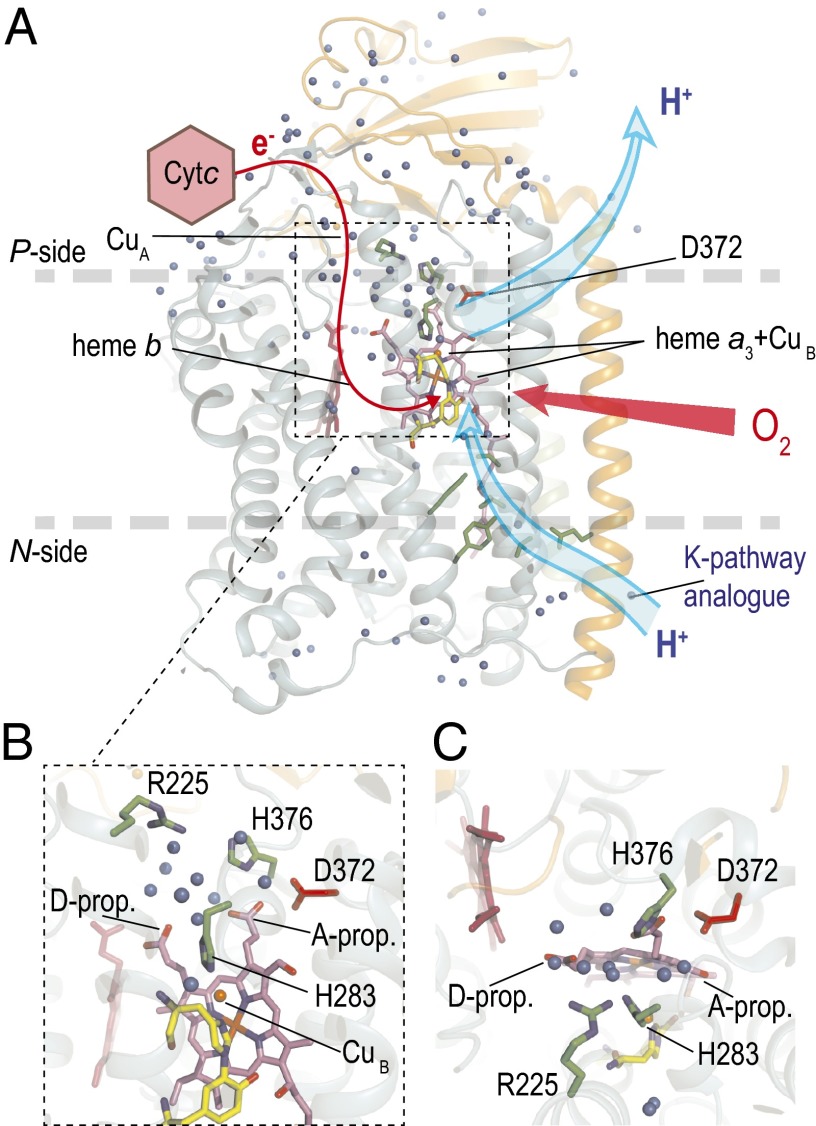
Fig. 1.
Structure of the ba3 CytcO. The electron donor to the ba3 CytcO is cytochrome c552. (A) Electrons are transferred first to CuA and then consecutively to heme b and the catalytic site composed of heme a3 and CuB (red line). Protons are taken up through the K-pathway analog from the negative side of the membrane to the catalytic site as well as to the PLS. The location of residue Asp372, discussed in this work, is shown. (B and C) Close-up views of the protein segment around Asp372 where the PLS may be located. The blue spheres indicate water molecules.
Proton pumping against an electrochemical gradient across the membrane requires a protonatable site with alternating access to the two sides of the membrane. This site, often referred to as the “proton-loading site” (PLS), would initially become protonated specifically from the n side (but not the p side) and then release its proton to the p side (but not the n side) (19–26). The identity of the PLS of the heme-copper oxidases is not known. Nevertheless, assuming that this site is common to all members of the heme-copper oxidase superfamily, there is a limited number of candidates, such as, for example, propionates A or/and D of heme a3, possibly including surrounding water molecules (15, 22, 23, 27–30) (Fig. 1). For the ba3 CytcO, theoretical and experimental data, together with structural analyses, suggest that the ring A propionate of heme a3, including nearby sites, may act as the PLS (12, 15, 27, 29, 31, 32). Results from a more recent study showed that structural perturbations near the heme a3 propionate A resulted in uncoupling of proton pumping from O2 reduction. One particularly interesting case is the replacement of Asp372 by Ile, which yielded 50% active CytcO in which proton pumping was uncoupled from O2 reduction (27). Furthermore, a detailed analysis of electrostatic interactions within a cluster of amino acid residues around the heme a3 propionates of several oxidases (although this study did not include the ba3 CytcO) suggests that a cluster of residues, together with the heme a3 propionic acids, may collectively bind protons (33). This cluster includes residues equivalent to Asp372 in the ba3 CytcO (Fig. 1).
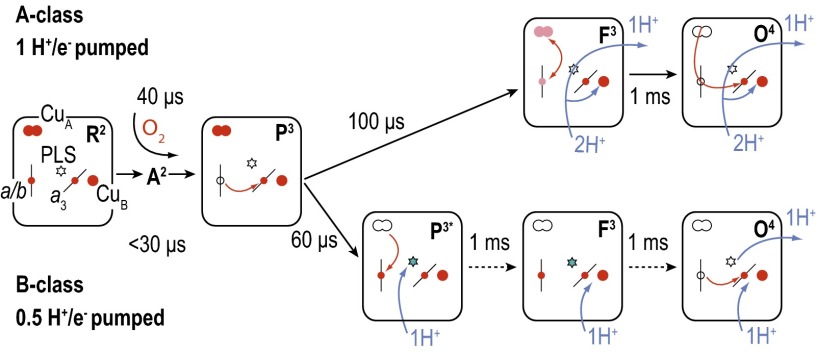
The reaction of the four-electron reduced ba3 CytcO with O2 has been studied using time-resolved spectroscopy after flash photolysis of the blocking CO ligand from heme a3. The sequence of events observed with the ba3 CytcO differs slightly from that observed with the aa3 oxidases (16, 34–37) (Fig. 2). In both the aa3 (here data with the R. sphaeroides aa3-type CytcO are discussed) and ba3 oxidases at neutral pH, O2 binds initially to the reduced heme a3 (state R2; the superscript denotes the number of electrons at the catalytic site) with a time constant of ∼8 μs (at 1 mM O2), forming a state that is denoted A2. After binding of O2, an electron is transferred from heme b to the catalytic site with a time constant of ∼40 μs or ∼15 μs in the aa3 and ba3 CytcOs, respectively, forming state P3 (or PR). In both the aa3 and ba3 CytcO, in the next step there is a net proton uptake from solution with time constants of ∼90 μs and ∼60 μs, respectively. Furthermore, the electron at CuA equilibrates with heme b/heme a over the same timescale, which in the ba3 CytcO results in almost full (re)reduction of heme b. However, there are also significant differences between the two oxidases. Whereas with the aa3 CytcO one proton is transferred to the catalytic site, forming the F3 state, and one proton is simultaneously pumped across the membrane, in the ba3 CytcO there is only proton transfer to a site located at a distance from the catalytic site, suggested to be the PLS. The catalytic site remains in the P3 (PR) state (denoted P3* in Fig. 2) in the ba3 oxidase over this timescale. With the ba3 CytcO, in the next step a proton is transferred from solution to the catalytic site to form state F3 with a time constant of ∼0.8 ms. This reaction approximately overlaps in time with transfer of the fourth electron, from the CuA–heme b equilibrium to the catalytic site, and formation of the oxidized CytcO (state O4). In other words, with the ba3 CytcO, the F3 state is not populated at neutral pH because the P3 → F3 and F3 → O4 reactions display about the same rates [however, at higher pH (>8), the F3 → O4 reaction is slower than formation of F3, which allows observation of both processes, separated in time].
Fig. 2.
Comparison of the reaction schemes with the A- and B-type oxidases. The reaction steps observed after mixing the CytcOs with O2 are shown. The small circles indicate the redox-active metal sites (reduced when red) and the stars indicate the PLS (protonated in blue). The state P3* with the ba3 CytcO is equivalent to P3 in wild-type CytcO, but with a protonated PLS.
The data from the present study show that the initial proton uptake (τ ≅ 65 μs), previously interpreted to be associated with protonation of the PLS, was impaired in the Asp372Ile variant, which is the first variant to our knowledge displaying this behavior. Because the Asp372 residue is located “above” the catalytic site (Fig. 1) in a protein segment that has been implied to be involved in gating of the pumped protons, the results from this study indicate a possible location of the PLS.
Results
As outlined above, CO binds reversibly to the reduced CytcO catalytic site and the association kinetics reflects the local structure and ligand-induced structural changes. Fourier transform infrared (FTIR) data from earlier studies of the ba3 CytcO indicated that ligand binding to heme a3 is linked to structural or protonation changes around Asp372 (31, 32). Therefore, here we compared the kinetics of CO recombination in the reduced wild-type and Asp372Ile variant of ba3 CytcO from T. thermophilus (Fig. 3). With the wild-type ba3 CytcO, the increase in absorbance induced by the laser flash at t = 0 is associated with dissociation of the CO ligand. The main component of the following absorbance decrease is associated with recombination of CO with heme a3. As seen in Fig. 3, the CO-recombination rate constant was ∼5 s−1 (τ = 200 ms), which is similar to that observed earlier in an infrared study [∼8 s−1 (38, 39); but see also ref. 40]. With the wild-type CytcO, we also observed a small, rapid component (∼5% of the total absorbance decrease) with a rate constant of ∼5,000 s−1 (τ = 200 μs), which may be associated with release of CO from CuB into solution (36) (see Discussion). With the Asp372Ile mutant CytcO, the rate of the rapid component decreased slightly to ∼4,000 s−1 (τ = 250 μs), but its amplitude increased significantly to ∼35% of the total absorbance change. With the Asp372Ile CytcO, the CO recombination to heme a3 displayed two components with rate constants of ∼120 s−1 (25%) and 5 s−1 (40%), where the latter is the same as that observed with the wild-type CytcO.
Fig. 3.
Absorbance changes after light-induced dissociation of CO from reduced ba3 CytcO. The increase in absorbance at 445 nm after the laser flash at t = 0 is associated with dissociation of the CO ligand. The rapid decrease in absorbance is associated with release of CO from CuB, whereas the slower decrease in absorbance is associated with recombination of the CO ligand with heme a3. Experimental conditions: ∼6 µM CytcO in 100 mM Hepes (pH 7.4), 0.05% DDM, 0.1 mM EDTA; 200 µM dithionite (added to the sample after removal of O2).
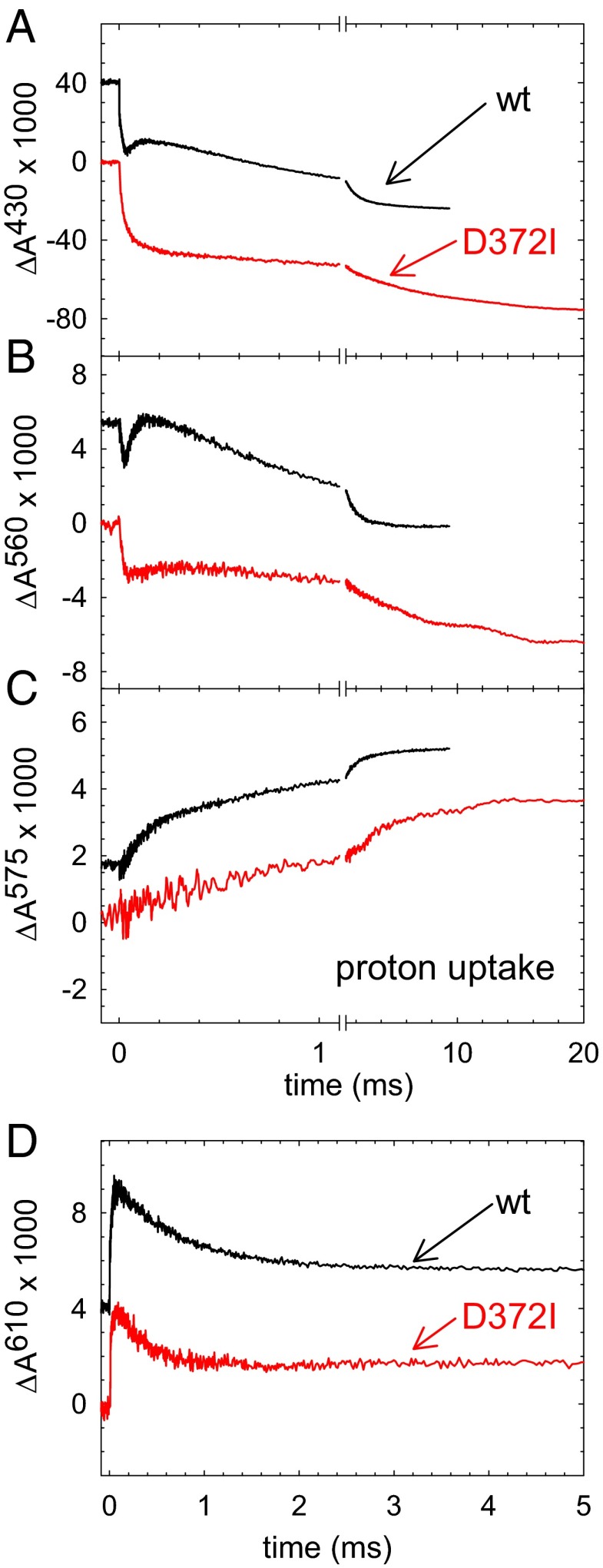
A solution of the fully reduced ba3 CytcO with CO bound at heme a3 was mixed with an oxygen-saturated solution in a stopped-flow apparatus (pH 7.5). The CO ligand was dissociated by a short laser flash ∼30 ms after mixing, which allowed O2 to bind to the catalytic site. Fig. 4 shows absorbance changes at wavelengths characteristic of redox changes of the metal sites as well as changes in the ligation state at the catalytic site (time constants are summarized in Table 1). In Fig. 4A, the unresolved decrease in absorbance at 430 nm is associated with dissociation of the CO ligand and binding of O2. With both the wild-type and Asp372Ile variant of CytcO, this process was followed in time by an increase in absorbance with a time constant of ∼15 μs associated with electron transfer from heme b to the catalytic site. A decrease in absorbance with the same time constant is also seen at 560 nm (Fig. 4B), reflecting the oxidation of heme b. With the wild-type CytcO, this decrease in absorbance at 430 nm was followed in time by an increase in absorbance with a time constant of 65 μs associated with electron transfer from CuA to heme b. With the Asp372Ile variant, this increase in absorbance was not observed at 430 nm (Fig. 4A) and it was very small at 560 nm (Fig. 4B), which indicates that the electron transfer from CuA to heme b was impaired. The final decrease in absorbance occurred with time constants of ∼1.2 ms and 8.4 ms with the wild-type and Asp372Ile variant of CytcO, respectively. Thus, electron transfer to the catalytic site linked to oxidation of the Asp372Ile variant of CytcO was significantly slower with Asp372Ile than with the wild-type CytcO. However, it should be noted that whereas with the wild-type CytcO the electron transfer occurs from heme b, in the Asp372Ile variant the electron is transferred from CuA. At 610 nm (Fig. 4D), the increase in absorbance with a time constant of ∼15 μs is associated with electron transfer from heme b to the catalytic site forming state P3 with both the wild-type and the Asp372Ile variant of CytcO. The following decrease in absorbance is presumably associated decay of the peroxy (P3 or P3*) state concomitant with formation of the ferryl (F3) state, which in the wild-type CytcO occurs with a time constant of ∼0.8 ms. This decrease in absorbance was about a factor of 2 faster with the Asp372Ile mutant (τ ≅ 0.4 ms; Fig. 4D) than with the wild-type CytcO (τ ≅ 0.8 ms).
Fig. 4.
Absorbance changes associated with reaction of the Asp372Ile variant and wild-type ba3 CytcO with O2. (A) At 430 nm the absorbance changes are mainly associated with redox reactions at heme b (the initial rapid decrease at t = 0 is associated with CO dissociation). The decrease in absorbance in the range 0–50 μs (τ ≅ 15 μs) is associated with oxidation of heme b. With the wild-type CytcO, the increase in absorbance is associated with electron transfer from CuA to heme b (τ ≅ 65 μs), whereas the final decrease is associated with oxidation of the CytcO (τ ≅ 1.2 ms). With the Asp372Ile variant the increase in absorbance is not seen (cf. CuA → heme b does not take place) and the electron is transferred from CuA to the catalytic site in the last step of the reaction (τ ≅ 8.4 ms). (B) Also at 560 nm, heme b primarily contributes to the absorbance changes. (C) Absorbance changes at 575 nm of the pH dye phenol red. An increase in absorbance corresponds to proton uptake from solution. (D) At 610 nm the increase in absorbance is associated with formation of the P3 (PR) state (τ ≅ 15 μs), whereas the decrease in absorbance is associated with decay of P3 and formation of state F3. Experimental conditions: ∼1 µM CytcO in 100 mM Hepes (pH 7.4), 0.05% DDM, except in C, where the buffer was replaced with 150 mM KCl and 50 μM phenol red was added.
Table 1.
Time constants associated with reaction of the reduced wild-type and Asp372Ile and O2 at pH 7.5
| Reaction | Wild type | Asp372Ile |
| Heme b → catalytic site (A2 → P3) | 15 μs | 15 μs |
| CuA → heme b | 65 μs | Not observed |
| P3 → F3 | 0.8 ms | 0.4 ms |
| Heme b → catalytic site (F3 → O4) | 1.2 ms | — |
| CuA → catalytic site (F3 → O4) | — | 8.4 ms |
| Proton uptake (2 main components) | 65 μs (43%), 0.8 ms (57%) | 65 μs* (9%), 0.5 ms (43%), 8.4 ms (48%) |
Each experiment was repeated 2–15 times with three different samples. The SE in the time constants was <20%.
A component with a fixed time constant of 65 μs was included to make the fit comparable to that with the wild-type CytcO.
With the wild-type ba3, CytcO proton uptake occurs with time constants of ∼65 μs and ∼0.8 ms (Fig. 4C), that is, with the same time constants as the electron transfer from CuA to heme b and formation of the F3 state, respectively (35–37). However, with the Asp372Ile variant of the CytcO, the fast (∼65 μs) proton-uptake component was absent. Instead, two slower components were observed with time constants of ∼0.4 ms and ∼8.4 ms, that is, overlapping in time with the P3 → F3 and F3 → O4 reactions, respectively (Fig. 4C).
Fig. 5A shows the pH dependence of the absorbance decrease at 610 nm in the range of pH 6–10. This absorbance change reflects the formation of F3, which was a factor of ∼2 faster with the Asp372Ile variant than with the wild-type CytcO (see also above) and essentially independent of pH. Fig. 5B shows the pH dependence of the final oxidation of the CytcO (decrease in absorbance at 560 nm). As seen in Fig. 5B, this reaction displayed a relatively strong pH dependence for the wild-type CytcO (36), whereas with the Asp372Ile variant the reaction rate was essentially pH-independent. At pH ∼10, the rates with the wild-type and Asp372Ile variant of CytcO were about the same (∼120 s−1).
Fig. 5.
pH dependence of the F3 and O4 formation rates. The rate constants were obtained from absorbance changes at 610 nm (A; decrease associated with decay of P3) and 560 nm (B; decrease associated with oxidation of the CytcO).
Discussion
Results from earlier studies using infrared spectroscopy indicated a link between ligand binding and changes in structure and/or protonation around Asp372 (31, 32). The CO-photolysis data in Fig. 3 support these conclusions because they show that changes in the structure at Asp372 (the Asp372Ile replacement) result in alteration of the CO-rebinding dynamics. In ba3 variants where residues of the K-pathway analog were altered, no such change in the CO-recombination kinetics was observed, reinforcing the suggestion that Asp372 is linked to the catalytic site. After dissociation from heme a3, CO binds transiently to CuB, after which the ligand is released into solution, presumably triggered by a structural relaxation of the catalytic site (41). With the bovine heart mitochondria CytcOs (42, 43) and R. sphaeroides (44), this event gives rise to a small absorbance decrease at 445 nm with a time constant of 1–2 μs. The CO ligand binds to CuB also in the ba3 oxidase (39, 40), and the release is presumably reflected in the absorbance decrease at 445 nm after CO dissociation with a time constant of 200–250 μs (36) (Fig. 3). This event is slower than observed with the mitochondrial and R. sphaeroides CytcOs, but faster than the corresponding absorbance changes in the infrared region with the ba3 CytcO [τ ≅ 30 ms (40)]. As seen in Fig. 3, the relative amplitude of the 200–250 μs component was significantly increased with the Asp372Ile variant, which indicates that the replacement of Asp372 alters the structural relaxation at the catalytic site that is linked to CO release from CuB.
With the wild-type ba3 CytcO, the first proton, taken up with a time constant of ∼65 μs, is presumably transferred to the PLS (35, 37). A second proton is taken up more slowly, with a time constant of ∼0.8 ms, which leads to the formation of state F3 at the catalytic site (34, 35, 37). The first proton uptake was not observed with the Asp372Ile ba3 CytcO (Figs. 4C and 6). Because this structural variant does not pump protons (27) and in the wild-type CytcO the 65-μs proton is presumably transferred to the PLS (35, 37), the absence of this proton uptake supports the earlier proposals (see Introduction) that the PLS is located in a protein segment involving the Asp372 residue. The electron transfer from CuA to heme b, which in the wild-type ba3 CytcO is synchronized with proton uptake to the PLS (37), was not observed with the Asp372Ile variant (Figs. 4 and 6). This result is consistent with the earlier observation with the aa3 CytcO (45) that in the wild-type CytcO the CuA–heme b electron transfer is induced by protonation of the PLS. In the next step of the reaction, formation of the F3 state, linked to proton uptake to the catalytic site, was a factor of 2 faster with the Asp372Ile mutant than with the wild-type CytcO (∼0.4 ms and ∼0.8 ms, respectively). This difference may be due to a larger negative potential at the catalytic site in state P3 (i.e., after electron transfer from heme b to the catalytic site) in the mutant CytcO because of the absence of a proton at the PLS.
Fig. 6.
Summary of results. A comparison of the reaction steps linked to proton uptake observed with the wild-type and Asp372Ile ba3 CytcOs.
The F3 → O4 reaction with the A-type oxidases is linked to proton uptake to the catalytic site and proton pumping (46, 47). Also with the ba3 oxidase, this reaction step is linked to proton pumping (16). However, with the wild-type ba3 CytcO, the F3 → O4 reaction is not associated with any net proton uptake, presumably because the proton stored at the PLS is released with the same time constant as proton uptake to the catalytic site (which has to be taken up to form the oxidized state, O4) (35–37). With the Asp372Ile variant, on the other hand, the O4 state was associated with a net proton uptake from solution (Fig. 4C). This observation is consistent with the absence of the 65-μs proton uptake after formation of P3. If the PLS does not become protonated, the F3 → O4 reaction is not associated with proton release from the PLS and only the net proton uptake to the catalytic site during formation of O4 is observed (Fig. 6).
Another difference between the Asp372Ile variant and the wild-type CytcO is that formation of the oxidized state was a factor of ∼7 slower in the variant. The F3 → O4 reaction involves a coupled electron and proton transfer to the catalytic site, where the overall rate, kFO, is determined by the fraction with reduced heme a3 in state F3, (in the electron equilibrium involving CuA, heme b, and the catalytic site), multiplied by the proton-transfer rate to the catalytic site, kH (45, 48, 49): . Consequently, even if the P3 → F3 reaction rate is unchanged or faster (cf. value of kH), the F3 → O4 rate may be slowed if is diminished, as was observed earlier with a structural variant of the R. sphaeroides aa3 oxidase (45). With the wild-type ba3 CytcO, upon formation of state F3, heme b is essentially fully (re)reduced such that during F3 → O4 the “fourth” electron is transferred directly from heme b. In the Asp372Ile structural variant, on the other hand, in state F3 the electron equilibrium is shifted away from heme b [only a small fraction of heme b is (re)reduced; Fig. 4 A and B], which indicates that the heme b apparent midpoint potential in the transiently formed F3 state is lower than in the wild-type CytcO. This difference is attributed to the unprotonated PLS in state F3. Because amino acid residue 372 is located even closer to heme a3/CuB than to heme b, a similar decrease in the apparent midpoint potential is expected for the catalytic site in the F3 state. As a result, during the F3 → O4 reaction, the fraction with reduced catalytic site, (see above), would be smaller with the Asp372Ile variant than with the wild-type CytcO, which would result in a slower F3 → O4 reaction even though proton transfer to the catalytic site (cf. the P3 → F3 reaction) is accelerated by a factor of 2.
Another reason for the slowed F3 → O4 reaction with the Asp372Ile variant may be an effect on changes in structure around the K pathway and the catalytic site. Such changes in structure are required for gating the unidirectional flow of protons and must also involve the PLS, and are likely to modulate the proton-transfer rate through the K pathway. Consequently, it is likely that a structural modification near or at the PLS would alter the proton-transfer rate through the K pathway.
With the wild-type ba3 CytcO, the F3 → O4 reaction rate is pH-dependent and drops by about a factor of 10 from pH 6 to 10. In contrast, with the Asp372Ile oxidase, this rate was essentially pH-independent (Fig. 5). With the wild-type ba3 CytcO, the reactions that occur before P3 → F3, that is, proton uptake to the PLS (τ ≅ 65 μs) and proton uptake to form state F3 (τ ≅ 0.8 ms), display essentially pH-independent kinetics [a decrease by a factor of ∼2 over 5 pH units (37)]. The unique feature of the F3 → O4 reaction with the wild-type CytcO is that it is linked to the release of a pumped proton. Consequently, the pH dependence of the F3 → O4 reaction could be associated with deprotonation of the PLS or changes in structure that are linked to this reaction. The decrease in the F3 → O4 rate with increasing pH would then reflect the degree of protonation of the PLS. Based on our data, we speculate that with the Asp372Ile variant the PLS is always deprotonated and therefore the F3 → O4 rate is approximately the same as that for the fully deprotonated PLS with the wild-type CytcO (i.e., at high pH).
Earlier FTIR spectroscopy data suggested that a possible acceptor for pumped protons is a cluster involving Asp372, a water molecule, and the ring A propionate of heme a3 (31). The same protein segment was suggested on the basis of theoretical calculations, which indicated that His376, which is also hydrogen-bonded to the ring A propionate, may accept protons from the n side of the membrane (29). Moreover, proton pumping may also involve a water molecule bridging the heme a3 D and A propionates (15, 27, 50), which, together with the CuB ligand His283, was implied to be the PLS. These conclusions were also supported by data from studies of the structural variants His376Asn and Asp372Ile (investigated here), which display a significant O2-reduction activity that is uncoupled from proton pumping (27). However, the results from the same study also showed that the uncoupling is specific to certain replacements and not to the replacement position. For example, upon replacement of His376 by Phe or Asp372 by Val, proton pumping was maintained. Seemingly inconclusive results were also obtained with structural variants at the position equivalent to the ba3 CytcO Asp372 from other organisms. With the R. sphaeroides aa3 CytcO, the effect of replacement of Asp407 by Ala, Asn, or Cys was studied and the data suggested that the residue has no role in proton pumping (51). Similar results were obtained with the bo3 quinol oxidase (52). With the Paracoccus denitrificans CytcO, the Asp399Leu replacement displayed an activity of 7% of that of the wild-type CytcO and no proton pumping. However, with the Asp399Asn replacement the activity was ∼60%, and no effect on proton pumping was observed (53). Collectively, these results, together with those discussed above for the ba3 CytcO, indicate that structural changes around propionate A of heme a3 can result in uncoupling of proton pumping. However, on the basis of these studies, no unique structural elements could be identified as the PLS of the heme-copper oxidases. Furthermore, not all of these modified residues are conserved among the heme-copper oxidases. These observations support the proposal that a cluster consisting of a number of amino acid residues, water molecules, and propionates A and D of heme a3 together act as a PLS (15, 27, 50). In a recent study, continuum electrostatics simulations with different aa3 CytcO crystal structures were used to show that the PLS is not a single site but rather includes a large number of sites, which interact with the heme a3 propionates (33). The pKas and changes in these values determine the protonation state of the PLS. If the pKas of the propionates are sufficiently low, the PLS does not become protonated and the CytcO would reduce O2 to water, but without linking this reaction to proton pumping (i.e., proton pumping is uncoupled from O2 reduction) (33). The contribution of the different residues of the PLS to its net protonation state depends on the composition of the PLS and therefore varies between CytcOs from different organisms. In other words, the position of the PLS would be the same in all oxidases, but its composition would be different and be fine-tuned for each structure. Consequently, the effects of mutations are likely to be different.
One way to accomplish transmembrane proton translocation in CytcO is to couple changes in the alternating proton access of the PLS to the n and p sides, to changes in its collective pKa (54). A transmembrane proton electrochemical gradient of 180 meV is equal to the free energy required to shift the pKa of this group by ∼3 units. Consequently, to accomplish unidirectional proton pumping across the membrane, a PLS should have a ΔpKa (pKa,n − pKa,p) >3 (the upper limit is the available free energy). Structural modifications within the PLS would typically result in changes in the values of pKa,n, pKa,p, and ΔpKa, but changes in the protonation and deprotonation of the PLS would depend on the values of these parameters relative to the pH and the transmembrane proton electrochemical gradient. Consequently, different structural modifications at a specific site or region may render very different effects on the pumping stoichiometry (see above).
We note that in the wild-type ba3 oxidase there is a delay between proton uptake to the PLS (τ ≅ 65 μs) and release from the PLS (τ ≅ 1.2 ms), that is, the PLS is protonated in the time between these events. Thus, absence of the 65-μs component in the Asp372Ile variant indicates that the structural alteration changes the properties (e.g., pKas; see above) of the PLS itself such that it cannot be protonated. Furthermore, it is unlikely that the structural alteration slows proton transfer through the exit pathway (55), because we would then expect to see proton uptake from solution.
In summary, the data from the present study show that the uncoupling of the proton pump is caused by slowed protonation of the PLS. Furthermore, the data point to a general location of the pump site in the CytcO and allowed us to identify a specific reaction step in the sequence of electron- and proton-transfer events that is associated with protonation of the PLS.
Materials and Methods
His-tagged wild-type ba3 CytcO was expressed and purified as described previously (37), and kept at 4 °C in 5 mM Hepes (pH 7.4), 0.05% dodecyl-β-d-maltoside (DDM; Glycon).
For the flow-flash measurements, a sample containing ba3 CytcO (∼5 μM CytcO in 2 mM Hepes, pH 7.4, 0.05% DDM) was transferred to an anaerobic cuvette and air was exchanged for N2 on a vacuum line. The CytcO was reduced upon adding 0.5 μM phenazine methosulfate (PMS) and 2 mM Na ascorbate. After incubation until full reduction of the CytcO, the atmosphere was exchanged for CO. Redox reactions and CO binding were followed spectroscopically (Cary 4000; Agilent Technologies). The reaction of the reduced CytcO with O2 was monitored spectrophotometrically using a locally modified stopped-flow apparatus (Applied Photophysics) as described (56). Briefly, the CytcO solution (2 mM Hepes, pH 7.5, 0.05% DDM) was mixed 1:5 with an oxygen-saturated solution (100 mM buffer, pH 6–10, 0.05% DDM). The reaction was initiated after 30 ms by flash photolysis of the CytcO–CO complex (Nd:YAG laser; Quantel; 10 ns, 532 nm, 200 mJ). Kinetic traces were recorded at specific wavelengths using a digital oscilloscope. The following buffers were used: 2-(N-morpholino)ethanesulfonic acid (MES) (pH 6), Hepes (pH 7–8), 2-(Cyclohexylamino)ethanesulfonic acid (CHES) (pH 9), N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) (pH 10). Proton uptake from solution was monitored as described (57). Briefly, the CytcO solution was run over a PD-10 column (GE Healthcare), where the buffer was exchanged for 150 mM KCl at pH ∼7.4 in 0.05% DDM. The CytcO was then diluted in the same buffer to a concentration of ∼5 μM and placed in a Thunberg cuvette (see above). The CytcO was mixed 1:5 with an unbuffered but pH-adjusted (pH ∼7.4) solution containing 150 mM KCl, 0.05% DDM, and 50 μM phenol red, and the absorbance changes at 575 nm were detected as a function of time.
Acknowledgments
Supported by grants from the Swedish Research Council (to P.B., P.Ä., and C.v.B.) and Grant HL 16101 from the National Institutes of Health (to R.B.G.). C.v.B. was supported by a fellowship from the Swiss National Science Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Pereira MM, Santana M, Teixeira M. A novel scenario for the evolution of haem-copper oxygen reductases. Biochim Biophys Acta. 2001;1505(2-3):185–208. doi: 10.1016/s0005-2728(01)00169-4. [DOI] [PubMed] [Google Scholar]
- 2.Hemp J, Gennis RB. Diversity of the heme-copper superfamily in archaea: Insights from genomics and structural modeling. Results Probl Cell Differ. 2008;45:1–31. doi: 10.1007/400_2007_046. [DOI] [PubMed] [Google Scholar]
- 3.Lee HJ, Reimann J, Huang Y, Ädelroth P. Functional proton transfer pathways in the heme-copper oxidase superfamily. Biochim Biophys Acta. 2012;1817(4):537–544. doi: 10.1016/j.bbabio.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Hosler JP, Ferguson-Miller S, Mills DA. Energy transduction: Proton transfer through the respiratory complexes. Annu Rev Biochem. 2006;75:165–187. doi: 10.1146/annurev.biochem.75.062003.101730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshikawa S, et al. Proton pumping mechanism of bovine heart cytochrome c oxidase. Biochim Biophys Acta. 2006;1757(9-10):1110–1116. doi: 10.1016/j.bbabio.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Brzezinski P, Gennis RB. Cytochrome c oxidase: Exciting progress and remaining mysteries. J Bioenerg Biomembr. 2008;40(5):521–531. doi: 10.1007/s10863-008-9181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brzezinski P, Ädelroth P. Design principles of proton-pumping haem-copper oxidases. Curr Opin Struct Biol. 2006;16(4):465–472. doi: 10.1016/j.sbi.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson-Miller S, Hiser C, Liu J. Gating and regulation of the cytochrome c oxidase proton pump. Biochim Biophys Acta. 2012;1817(4):489–494. doi: 10.1016/j.bbabio.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rich PR, Maréchal A. Functions of the hydrophilic channels in protonmotive cytochrome c oxidase. J R Soc Interface. 2013;10(86):20130183. doi: 10.1098/rsif.2013.0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaila VRI, Verkhovsky MI, Wikström M. Proton-coupled electron transfer in cytochrome oxidase. Chem Rev. 2010;110(12):7062–7081. doi: 10.1021/cr1002003. [DOI] [PubMed] [Google Scholar]
- 11.Svahn E, Faxén K, Gennis RB, Brzezinski P. Proton pumping by an inactive structural variant of cytochrome c oxidase. J Inorg Biochem. 2014;140:6–11. doi: 10.1016/j.jinorgbio.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 12.Soulimane T, et al. Structure and mechanism of the aberrant ba(3)-cytochrome c oxidase from Thermus thermophilus. EMBO J. 2000;19(8):1766–1776. doi: 10.1093/emboj/19.8.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tiefenbrunn T, et al. High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment. PLoS ONE. 2011;6(7):e22348. doi: 10.1371/journal.pone.0022348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luna VM, Chen Y, Fee JA, Stout CD. Crystallographic studies of Xe and Kr binding within the large internal cavity of cytochrome ba3 from Thermus thermophilus: Structural analysis and role of oxygen transport channels in the heme-Cu oxidases. Biochemistry. 2008;47(16):4657–4665. doi: 10.1021/bi800045y. [DOI] [PubMed] [Google Scholar]
- 15.Chang HY, Hemp J, Chen Y, Fee JA, Gennis RB. The cytochrome ba3 oxygen reductase from Thermus thermophilus uses a single input channel for proton delivery to the active site and for proton pumping. Proc Natl Acad Sci USA. 2009;106(38):16169–16173. doi: 10.1073/pnas.0905264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siletsky SA, et al. Time-resolved single-turnover of ba3 oxidase from Thermus thermophilus. Biochim Biophys Acta. 2007;1767(12):1383–1392. doi: 10.1016/j.bbabio.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 17.Kannt A, et al. Electrical current generation and proton pumping catalyzed by the ba3-type cytochrome c oxidase from Thermus thermophilus. FEBS Lett. 1998;434(1-2):17–22. doi: 10.1016/s0014-5793(98)00942-9. [DOI] [PubMed] [Google Scholar]
- 18.Han H, et al. Adaptation of aerobic respiration to low O2 environments. Proc Natl Acad Sci USA. 2011;108(34):14109–14114. doi: 10.1073/pnas.1018958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popović DM, Stuchebrukhov AA. Proton pumping mechanism and catalytic cycle of cytochrome c oxidase: Coulomb pump model with kinetic gating. FEBS Lett. 2004;566(1-3):126–130. doi: 10.1016/j.febslet.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 20.Quenneville J, Popović DM, Stuchebrukhov AA. Combined DFT and electrostatics study of the proton pumping mechanism in cytochrome c oxidase. Biochim Biophys Acta. 2006;1757(8):1035–1046. doi: 10.1016/j.bbabio.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 21.Sharpe MA, Ferguson-Miller S. A chemically explicit model for the mechanism of proton pumping in heme-copper oxidases. J Bioenerg Biomembr. 2008;40(5):541–549. doi: 10.1007/s10863-008-9182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blomberg MRA, Siegbahn PEM. The mechanism for proton pumping in cytochrome c oxidase from an electrostatic and quantum chemical perspective. Biochim Biophys Acta. 2012;1817(4):495–505. doi: 10.1016/j.bbabio.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 23.Kaila VRI, Sharma V, Wikström M. The identity of the transient proton loading site of the proton-pumping mechanism of cytochrome c oxidase. Biochim Biophys Acta. 2011;1807(1):80–84. doi: 10.1016/j.bbabio.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 24.Johansson AL, et al. Proton-transport mechanisms in cytochrome c oxidase revealed by studies of kinetic isotope effects. Biochim Biophys Acta. 2011;1807(9):1083–1094. doi: 10.1016/j.bbabio.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chakrabarty S, Namslauer I, Brzezinski P, Warshel A. Exploration of the cytochrome c oxidase pathway puzzle and examination of the origin of elusive mutational effects. Biochim Biophys Acta. 2011;1807(4):413–426. doi: 10.1016/j.bbabio.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsson MH, Warshel A. Monte Carlo simulations of proton pumps: On the working principles of the biological valve that controls proton pumping in cytochrome c oxidase. Proc Natl Acad Sci USA. 2006;103(17):6500–6505. doi: 10.1073/pnas.0510860103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang HY, et al. Exploring the proton pump and exit pathway for pumped protons in cytochrome ba3 from Thermus thermophilus. Proc Natl Acad Sci USA. 2012;109(14):5259–5264. doi: 10.1073/pnas.1107345109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wikström M, Verkhovsky MI. Mechanism and energetics of proton translocation by the respiratory heme-copper oxidases. Biochim Biophys Acta. 2007;1767(10):1200–1214. doi: 10.1016/j.bbabio.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Fee JA, Case DA, Noodleman L. Toward a chemical mechanism of proton pumping by the B-type cytochrome c oxidases: Application of density functional theory to cytochrome ba3 of Thermus thermophilus. J Am Chem Soc. 2008;130(45):15002–15021. doi: 10.1021/ja803112w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goyal P, Lu J, Yang S, Gunner MR, Cui Q. Changing hydration level in an internal cavity modulates the proton affinity of a key glutamate in cytochrome c oxidase. Proc Natl Acad Sci USA. 2013;110(47):18886–18891. doi: 10.1073/pnas.1313908110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koutsoupakis C, Soulimane T, Varotsis C. Probing the Q-proton pathway of ba3-cytochrome c oxidase by time-resolved Fourier transform infrared spectroscopy. Biophys J. 2004;86(4):2438–2444. doi: 10.1016/S0006-3495(04)74300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koutsoupakis C, Soulimane T, Varotsis C. Ligand binding in a docking site of cytochrome c oxidase: A time-resolved step-scan Fourier transform infrared study. J Am Chem Soc. 2003;125(48):14728–14732. doi: 10.1021/ja036107e. [DOI] [PubMed] [Google Scholar]
- 33.Lu J, Gunner MR. Characterizing the proton loading site in cytochrome c oxidase. Proc Natl Acad Sci USA. 2014;111(34):12414–12419. doi: 10.1073/pnas.1407187111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smirnova I, et al. Single mutations that redirect internal proton transfer in the ba3 oxidase from Thermus thermophilus. Biochemistry. 2013;52(40):7022–7030. doi: 10.1021/bi4008726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Ballmoos C, Ädelroth P, Gennis RB, Brzezinski P. Proton transfer in ba(3) cytochrome c oxidase from Thermus thermophilus. Biochim Biophys Acta. 2012;1817(4):650–657. doi: 10.1016/j.bbabio.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 36.von Ballmoos C, Lachmann P, Gennis RB, Ädelroth P, Brzezinski P. Timing of electron and proton transfer in the ba(3) cytochrome c oxidase from Thermus thermophilus. Biochemistry. 2012;51(22):4507–4517. doi: 10.1021/bi300132t. [DOI] [PubMed] [Google Scholar]
- 37.von Ballmoos C, Gennis RB, Ädelroth P, Brzezinski P. Kinetic design of the respiratory oxidases. Proc Natl Acad Sci USA. 2011;108(27):11057–11062. doi: 10.1073/pnas.1104103108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Einarsdóttir O, Killough PM, Fee JA, Woodruff WH. An infrared study of the binding and photodissociation of carbon monoxide in cytochrome ba3 from Thermus thermophilus. J Biol Chem. 1989;264(5):2405–2408. [PubMed] [Google Scholar]
- 39.Woodruff WH. Coordination dynamics of heme-copper oxidases. The ligand shuttle and the control and coupling of electron transfer and proton translocation. J Bioenerg Biomembr. 1993;25(2):177–188. doi: 10.1007/BF00762859. [DOI] [PubMed] [Google Scholar]
- 40.Koutsoupakis K, Stavrakis S, Pinakoulaki E, Soulimane T, Varotsis C. Observation of the equilibrium CuB-CO complex and functional implications of the transient heme a3 propionates in cytochrome ba3-CO from Thermus thermophilus. Fourier transform infrared (FTIR) and time-resolved step-scan FTIR studies. J Biol Chem. 2002;277(36):32860–32866. doi: 10.1074/jbc.M204943200. [DOI] [PubMed] [Google Scholar]
- 41.Pilet E, Jasaitis A, Liebl U, Vos MH. Electron transfer between hemes in mammalian cytochrome c oxidase. Proc Natl Acad Sci USA. 2004;101(46):16198–16203. doi: 10.1073/pnas.0405032101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heitbrink D, Sigurdson H, Bolwien C, Brzezinski P, Heberle J. Transient binding of CO to Cu(B) in cytochrome c oxidase is dynamically linked to structural changes around a carboxyl group: A time-resolved step-scan Fourier transform infrared investigation. Biophys J. 2002;82(1 Pt 1):1–10. doi: 10.1016/S0006-3495(02)75368-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Einarsdóttir O, et al. Photodissociation and recombination of carbonmonoxy cytochrome oxidase: Dynamics from picoseconds to kiloseconds. Biochemistry. 1993;32(45):12013–12024. doi: 10.1021/bi00096a011. [DOI] [PubMed] [Google Scholar]
- 44.Namslauer A, Brändén M, Brzezinski P. The rate of internal heme-heme electron transfer in cytochrome c oxidase. Biochemistry. 2002;41(33):10369–10374. doi: 10.1021/bi025976y. [DOI] [PubMed] [Google Scholar]
- 45.Karpefors M, Ädelroth P, Zhen Y, Ferguson-Miller S, Brzezinski P. Proton uptake controls electron transfer in cytochrome c oxidase. Proc Natl Acad Sci USA. 1998;95(23):13606–13611. doi: 10.1073/pnas.95.23.13606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verkhovsky MI, Morgan JE, Verkhovskaya ML, Wikström M. Translocation of electrical charge during a single turnover of cytochrome-c oxidase. Biochim Biophys Acta. 1997;1318(1-2):6–10. [Google Scholar]
- 47.Faxén K, Gilderson G, Ädelroth P, Brzezinski P. A mechanistic principle for proton pumping by cytochrome c oxidase. Nature. 2005;437(7056):286–289. doi: 10.1038/nature03921. [DOI] [PubMed] [Google Scholar]
- 48.Verkhovsky MI, Morgan JE, Wikström M. Control of electron delivery to the oxygen reduction site of cytochrome c oxidase: A role for protons. Biochemistry. 1995;34(22):7483–7491. doi: 10.1021/bi00022a023. [DOI] [PubMed] [Google Scholar]
- 49.Brändén G, et al. The protonation state of a heme propionate controls electron transfer in cytochrome c oxidase. Biochemistry. 2005;44(31):10466–10474. doi: 10.1021/bi0502745. [DOI] [PubMed] [Google Scholar]
- 50.Daskalakis V, Farantos SC, Guallar V, Varotsis C. Regulation of electron and proton transfer by the protein matrix of cytochrome c oxidase. J Phys Chem B. 2011;115(13):3648–3655. doi: 10.1021/jp1115993. [DOI] [PubMed] [Google Scholar]
- 51.Qian J, et al. Aspartate-407 in Rhodobacter sphaeroides cytochrome c oxidase is not required for proton pumping or manganese binding. Biochemistry. 1997;36(9):2539–2543. doi: 10.1021/bi962721+. [DOI] [PubMed] [Google Scholar]
- 52.Thomas JW, Puustinen A, Alben JO, Gennis RB, Wikström M. Substitution of asparagine for aspartate-135 in subunit I of the cytochrome bo ubiquinol oxidase of Escherichia coli eliminates proton-pumping activity. Biochemistry. 1993;32(40):10923–10928. doi: 10.1021/bi00091a048. [DOI] [PubMed] [Google Scholar]
- 53.Pfitzner U, et al. Tracing the D-pathway in reconstituted site-directed mutants of cytochrome c oxidase from Paracoccus denitrificans. Biochemistry. 2000;39(23):6756–6762. doi: 10.1021/bi992235x. [DOI] [PubMed] [Google Scholar]
- 54.Wikström M, Krab K, Saraste M. Cytochrome c Oxidase. A Synthesis. Academic; London: 1981. [Google Scholar]
- 55.Popović DM, Stuchebrukhov AA. Proton exit channels in bovine cytochrome c oxidase. J Phys Chem B. 2005;109(5):1999–2006. doi: 10.1021/jp0464371. [DOI] [PubMed] [Google Scholar]
- 56.Brändén M, et al. On the role of the K-proton transfer pathway in cytochrome c oxidase. Proc Natl Acad Sci USA. 2001;98(9):5013–5018. doi: 10.1073/pnas.081088398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smirnova I, et al. Functional role of Thr-312 and Thr-315 in the proton-transfer pathway in ba3 cytochrome c oxidase from Thermus thermophilus. Biochemistry. 2010;49(33):7033–7039. doi: 10.1021/bi100749p. [DOI] [PMC free article] [PubMed] [Google Scholar]