Significance
Bipolar disorder (BD) is a common, severe, and recurrent psychiatric disorder with no known cure and substantial morbidity and mortality. Heritable causes contribute up to 80% of the lifetime risk for BD. Common genetic variation explains ∼25% of this heritable risk. Rare genetic variants may explain additional risk. We identified contributions of rare variants to BD by sequencing the genomes of 200 individuals from 41 families with BD. The two main findings of this study were as follows: rare risk variants for BD were enriched in genes and pathways that regulate diverse aspects of neuronal excitability; and most of these risk variants were noncoding with predicted regulatory functions. These results highlight specific hypotheses for future research and potential therapeutic targets.
Keywords: bipolar disorder, family genomics, regulatory variants, GABAA receptor, voltage-gated calcium channel
Abstract
We sequenced the genomes of 200 individuals from 41 families multiply affected with bipolar disorder (BD) to identify contributions of rare variants to genetic risk. We initially focused on 3,087 candidate genes with known synaptic functions or prior evidence from genome-wide association studies. BD pedigrees had an increased burden of rare variants in genes encoding neuronal ion channels, including subunits of GABAA receptors and voltage-gated calcium channels. Four uncommon coding and regulatory variants also showed significant association, including a missense variant in GABRA6. Targeted sequencing of 26 of these candidate genes in an additional 3,014 cases and 1,717 controls confirmed rare variant associations in ANK3, CACNA1B, CACNA1C, CACNA1D, CACNG2, CAMK2A, and NGF. Variants in promoters and 5′ and 3′ UTRs contributed more strongly than coding variants to risk for BD, both in pedigrees and in the case-control cohort. The genes and pathways identified in this study regulate diverse aspects of neuronal excitability. We conclude that rare variants in neuronal excitability genes contribute to risk for BD.
Bipolar disorder (BD) is a psychiatric disorder characterized by episodes of mania and depression (1). Episodes of mania are marked by elevated or alternatively irritable mood, grandiosity, racing thoughts, rapid speech, diminished need for sleep, and risk-taking behavior. Depression includes sadness, low energy and motivation, decreased ability to experience pleasure, insomnia, and appetite changes. Psychosis with hallucinations and delusions can occur in either state. BD affects 1–2% of the US population, and if untreated, up to 15% of patients die from suicide. Twin and family studies suggest that heritable causes explain 60–80% of lifetime risk for BD (2), with an approximate eightfold relative risk in the siblings of BD probands (3). Several common genetic markers have shown significant and replicable associations in genome-wide association studies (GWASs), including a region near a voltage-gated calcium channel, CACNA1C, and another near a synaptic scaffolding gene, ANK3 (4). Additive effects of common loci detectable on commercially available genomic arrays may be used to predict about 25% of the risk for BD (5), but typically the function and exact location of the causative variants linked to these loci is unknown.
Rare variants may explain additional risk for BD. It is possible that one or a few rare variants of large effect dramatically increase disease risk, resulting in an inheritance model resembling monogenic inheritance in a given family. However, four exome-sequencing and whole-genome sequencing (WGS) studies of BD pedigrees have detected few, if any, plausible variants of large effect (6–9). An alternative oligogenic model posits that different combinations of several uncommon or rare variants of moderate effect cluster in affected individuals and collectively cause disease.
To test the hypothesis that rare variants contribute to risk for BD, we sequenced the genomes of 200 individuals from 41 multiply affected BD pedigrees of European ancestry. We sequenced 2–17 individuals per pedigree (SI Appendix, Table S1 and Figs. S1–S4). We selected clusters of closely related affected individuals, affected individuals separated by multiple meioses, or pairs of individuals discordant for BD. These three strategies are designed to maximize power to detect both monogenic and oligogenic effects. We analyzed these genomes from BD pedigrees together with the genomes of 254 individuals in control pedigrees of European descent.
Results
The genetic complexity and heterogeneity of BD limits power to identify causal genes in a genome-wide analysis (10). To increase power, we focused on 3,087 candidate genes and 325 gene sets (pathways) selected based on two hypotheses about the genetic basis of BD. First, we hypothesized that rare variants contributing to genetic risk for BD are enriched in genes with intrinsic neuronal functions. To test this hypothesis, we analyzed 2,511 candidate genes from proteomics studies of the synapse and curated databases of neuronal genes (SI Appendix). Second, we hypothesized that rare risk variants are enriched at loci with prior evidence from GWASs. To test this hypothesis, we analyzed 669 genes that were located within 100 kb of loci with suggestive associations to BD (P < 1e-4) in a recent GWAS with 7,481 BD cases and 9,250 controls (11) or which had an empirical P < 0.05 in a meta-analysis of this same sample together with four additional cohorts (12). Ninety-three genes overlapped between those with synaptic functions and those with prior evidence from GWAS, for a total of 3,087 candidate genes. For pathway-level analyses, we focused on 325 Gene Ontology, Kyoto Encyclopedia of Genes and Genomes, and BioCarta pathways that were statistically enriched [false discovery rate (FDR) < 0.05] among the 3,087 candidate genes.
We considered variants with minor allele frequencies (MAF) < 5% that were annotated to coding or regulatory functions. We defined gene-disrupting variants as those predicted to render one or more protein isoforms nonfunctional, including frameshift insertions and deletions (indels), stop-gain and stop-loss single nucleotide variants (SNVs), and splice-site variants. We also analyzed protein-coding variants that are predicted to alter just one or a few amino acids, including nonframeshift indels and missense SNVs. Regulatory variants were defined as noncoding variants that may be predicted to impact gene expression, including those that disrupt a transcription factor binding site in a gene’s promoter or putative enhancer sequences, as well as variants located in 5′ or 3′ UTRs (SI Appendix).
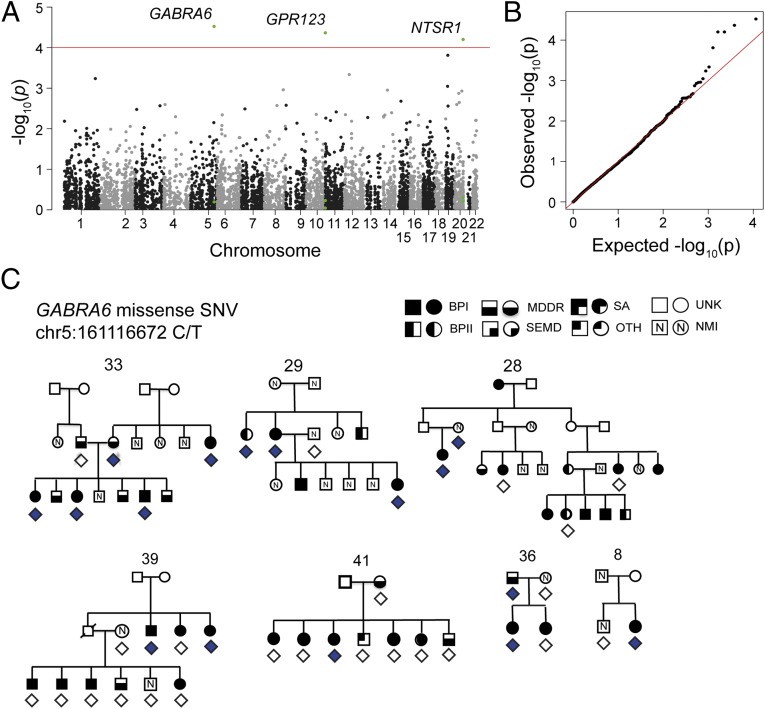
Linear mixed modeling (13) of 5,730 biallelic SNVs with 1–5% allele frequencies and predicted coding or regulatory functions revealed associations between BD and four of these SNVs at an FDR (q) of <10% (Fig. 1 A and B and SI Appendix, Table S2). The top associated SNV was a missense SNV in GABRA6 (rs3811993; T187M; P = 3.0e-5; q = 8.9e-2; Fig. 1C). GABRA6 encodes the α-6 subunit of the GABAA receptor. GABAA receptors are pentameric chloride channels activated by GABA, the major inhibitory neurotransmitter in the CNS. The rs3811993 SNV in GABRA6 results in a threonine to methionine substitution at position 187 of the protein. Combined annotation-dependent depletion (14) (CADD v1.1) analysis predicted that this variant is among the top 0.1% most deleterious SNVs genome-wide (raw score = 3.04; scaled score = 21.8). Sanger sequencing confirmed 100% of the genotype calls from WGS. Targeted sequencing of this variant in an additional 3,014 cases and 1,717 controls from the Genetic Association Information Network (GAIN) (15) and Translational Genomics Research Institute (TGEN) (16) collections identified the rs3811993 variant in an additional seven BD cases and in only a single control.
Fig. 1.
Associations of BD with 5,730 uncommon coding and noncoding SNVs in 3,087 candidate genes. (A) –log10(P values) for the association of individual SNVs with BD, plotted by chromosomal position. P values were calculated using linear mixed models implemented with EMMAX (13). SNVs with significant associations (FDR < 10%) are indicated. (B) Uniform, quantile-quantile plot for the observed vs. null-expectation distribution of P values. Positive deviation from the null indicates the presence of true signal and effective accounting for family and population structure. (C) A missense SNV in the GABA receptor gene GABRA6 was associated with risk for BD (P = 3.0e-5). This SNV was identified in a total of 13 BD cases, 2 major depression cases, and 1 unaffected individual across seven pedigrees. Diamonds below an individual indicate WGS (filled, contains variant; unfilled, does not contain variant). BPI, bipolar disorder, type I; BPII, bipolar disorder, type II; MDDR, major depressive disorder, recurrent; NMI, no mental illness; OTH, other axis I or axis II diagnosis; SA, schizoaffective disorder; SEMD, single episode of major depression; UNK, unknown phenotype (not ascertained).
Other significant SNVs were a noncoding SNV in the 5′UTR of GPR123 (rs147825070; P = 4.3e-5; q = 8.9e-2), a brain-specific, orphan G protein-coupled receptor (17), and two SNVs in the 3′ UTR of NTSR1 (P = 6.3e-5; q = 8.9e-2). NTSR1 encodes the receptor for the neuropeptide neurotensin. A murine ntsr1 knockout showed alternations in sleep and affective behaviors (18, 19).
To evaluate the effects of variants too rare to be detected in a single variant association test, we identified functional variants that cosegregated with disease in each BD pedigree and used the empirical distribution of segregating variants in control pedigrees to characterize genes and pathways with an increased burden of rare variants. No individual gene had a significant burden of segregating rare variants in BD pedigrees after correcting for multiple tests, although several genes had suggestive trends (SI Appendix, Table S3). We therefore focused on associations involving pathways and gene sets.
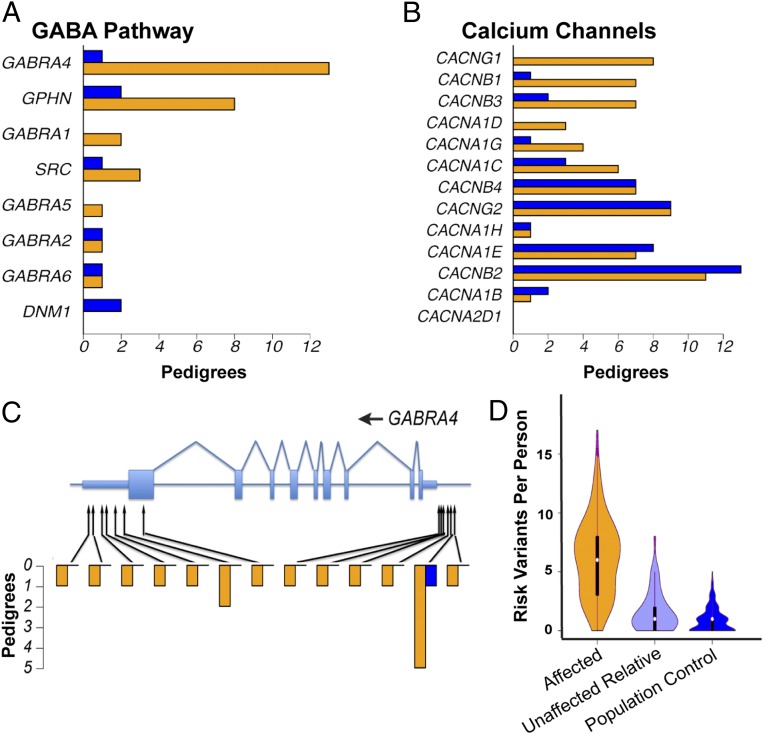
BD pedigrees had an increased burden of rare or uncommon (MAF < 0.05) variants in GABA pathway genes (P = 1.5e-4; q = 0.04; Fig. 2A). Genes encoding GTPases (P = 6.8e-3), calcium/calmodulin-dependent protein (CaM) kinases (P = 1.3e-2), and voltage-gated calcium channels (P = 3.6e-2; Fig. 2B) also had suggestive P values (SI Appendix, Table S4). Several genes from each of these sets contained candidate variants in multiple BD pedigrees but in few control pedigrees. For instance, the GABAA receptor subunits GABRA4 (Fig. 2C) and GABRA1 each contained distinct segregating variants in multiple BD pedigrees, but equivalent variants were 4.3-fold less common in control pedigrees.
Fig. 2.
Pathways enriched for rare variants in BD pedigrees. (A) Number of BD (orange) and control (blue) pedigrees in which a fully or nearly fully segregating coding or regulatory variant was found in a gene assigned to the GABA pathway by BioCarta. The 34 control pedigrees used in this analysis were of European ancestry and were matched for similar size and structure to the BD pedigrees. (B) Number of BD (orange) and control (blue) pedigrees in which a fully or nearly fully segregating coding or regulatory variant was found in a gene assigned to the Gene Ontology term “Voltage-Gated Calcium Channel Complex.” (C) Fully and nearly fully segregating coding and regulatory SNVs and indels in the GABA receptor gene GABRA4. Protein-coding regions and UTRs are shown, respectively, by wide and narrow light blue rectangles. Heights of orange and blue bars indicate the number of BD and control pedigrees in which each variant was fully or nearly fully segregating. (D) Number of risk variants identified in each BD case (orange), unaffected individual in a BD pedigree (light blue), or population control (dark blue). For this analysis, we defined as risk variants all SNVs with P < 0.001 and odds ratios > 1 by mixed model analysis (14 SNVs), as well as SNVs with mixed-model P < 0.05, odds ratios > 1, and an annotation to one of the following pathways with an increased burden of rare variants in BD pedigrees (SI Appendix, Table S4): BioCarta GABA pathway (9 SNVs), voltage-gated calcium channel complex (38 SNVs), CaM kinases (10 SNVs), GTPases (88 SNVs), and glycolysis/tricarboxylic acid cycle (13 SNVs). The widths of polygons are proportional to the number of individuals with each variant count.
Focusing on a more restricted set of functional variants—rare (MAF < 0.01) gene-disrupting SNVs and indels—provided additional support for an association of BD with ion channel genes (SI Appendix, Table S5). We identified 437 genes that contained a segregating, rare, disrupting variant in any BD pedigree. These genes were enriched for nicotinic acetylcholine channels (three genes; P = 4.4e-4; q = 7.3e-2; SI Appendix, Table S5). The broader set of metal-ion transmembrane transporters also had a suggestive P value (11 genes; P = 3.1e-3), with rare, disrupting mutations in BD pedigrees in CACNA1G, CCS, CHRNA6, CHRNB4, CHRND, KCNQ5, KCNS1, MS4A2, SLC39A2, SLC40A1, and TRPM1 (SI Appendix, Table S5).
We obtained convergent evidence for several of these pathways by evaluating their expression patterns in the brain. Of the 27 pathways that had an increased burden of rare variants in BD pedigrees (SI Appendix, Tables S4 and S5), 8 pathways were enriched for genes that were differentially expressed in postmortem dorsolateral prefrontal cortex from BD cases vs. controls (20) (SI Appendix, Table S6). The pathways enriched for differentially expressed genes included the GABA pathway, the voltage-gated calcium channel complex, CaM kinases, and the sonic hedgehog (SHH) pathway.
Combining results from variant- and pathway-level models suggested that most affected individuals inherited multiple risk variants and that most of these variants were located in noncoding, presumptively regulatory regions. This analysis included 164 uncommon risk variants with evidence from both mixed modeling and pathway analysis. Genomes from affected individuals contained on average 6 of 164 risk variants compared with 1 of 164 in unaffected relatives and population controls (Fig. 2D). Genomes of affected individuals typically contained risk-associated SNVs from multiple enriched pathways (SI Appendix, Figs. S9 and S10). One hundred forty-five of these 164 risk variants (88%) were located in promoters and 5′ and 3′ UTRs, whereas only 19 were nonsynonymous coding variants. For example, in GABRA4, 12 of the 13 variants that cosegregated with BD in one or more pedigrees were located in either the 3′ UTR or promoter (Fig. 2C). Counts of risk variants in individuals could be influenced by false-positive and false-negative effects, and it is possible that large effect variants not considered in our analyses (e.g., copy number variants) reside in some of these pedigrees. Nevertheless, the available evidence suggests that genetic risk in most of these pedigrees involves oligogenic combinations of rare variants, most often located in promoters and 5′ and 3′ UTRs, perhaps in combination with common variants.
We obtained independent evidence for the roles of 26 genes by sequencing their coding and regulatory regions in 3,014 European-American BD cases and 1,717 controls from the GAIN (15) and TGEN (16) collections. Of these 26 genes, 20 were identified in this study (10 voltage-gated calcium channel subunits, 5 GABAA receptor subunits, and 5 CaM-kinase signaling genes), and 6 genes were implicated by published GWASs and candidate gene studies (ANK3, TENM4, NTRK2, NTRK1, NGF, and BDNF; SI Appendix, Table S7). Samples were sequenced in pools containing DNA from up to 37 individuals. This strategy yielded accurate allele frequency estimates, even for rare variants (SI Appendix, Table S8 and Figs. S7–S11).
We found relatively little evidence that protein-coding variants in these 26 candidate genes are associated with BD. No gene had a significant sequence kernel association test (SKAT) (21) or C-alpha (22) P value in models that considered the combined effects of protein-coding variants with both risk and protective variants (Table 1 and SI Appendix, Table S9). A single gene, ANK3, reached statistical significance in a gene burden model that assumes only risk variants and not protective variants are present in a gene (P = 1.8e-3, q = 2.2e-2).
Table 1.
Genes with rare variant associations with BD in 3,014 European-American cases and 1,717 controls
| Gene | Coding | Regulatory | ||||
| No. of SNVs | P value | q value | No. of SNVs | P value | q value | |
| CACNA1C | 6 | 3.8e-2 | 0.22 | 238 | <1.0e-5 | 2.6e-4 |
| CACNA1B | 1 | NA | NA | 50 | 4.3e-4 | 4.1e-3 |
| CACNG2 | 0 | NA | NA | 43 | 4.7e-4 | 4.1e-3 |
| CAMK2A | 2 | 0.22 | 0.48 | 58 | 1.4e-3 | 8.6e-3 |
| CACNA1D | 19 | 0.43 | 0.74 | 111 | 1.6e-3 | 8.6e-3 |
| NGF | 2 | 0.74 | 0.88 | 17 | 8.4e-3 | 3.6e-2 |
Targeted sequencing was performed for genes encoding 10 calcium channels, 5 GABA receptors, and 5 calmodulin-dependent protein kinases with evidence for association with BD from whole-genome sequencing and for 6 candidate genes with evidence from GWAS. We evaluated associations between BD and combinations of rare risk variants and protective variants in each gene using SKAT, and we report empirical P values from 100,000 bootstrap permutations. Coding variants and regulatory variants were tested separately. Results are shown for six genes with significant rare variant associations to BD (q < 0.05). Results for all 26 genes are provided in SI Appendix Tables S9 and S10. NA, not applicable.
By contrast, we identified significant associations between risk for BD and rare, noncoding promoter and enhancer variants that were predicted to impact transcription factor binding sites (23) in 6 of the 26 candidate genes (SKAT; q < 0.05; Table 1 and SI Appendix, Table S10): CACNA1B, CACNA1C, CACNA1D, CACNG2, CAMK2A, and NGF. In addition, CACNG1, GABRA4, TENM4, and GABRA1 had nominally significant P < 0.05. Similar results were obtained with C-alpha and with alternative strategies for data normalization (SI Appendix, Table S10). No gene reached statistical significance in a gene burden model assuming only risk variants are present (SI Appendix, Table S10), so the genes with significant SKAT P values are likely to contain a combination of risk, protective, and neutral variants.
Discussion
There are two main findings in this study. First, rare risk variants for BD were enriched in genes and pathways that regulate neuronal excitability, with the strongest signal in two groups of ion channels: GABAA receptors and voltage-gated calcium channels. Second, most of the risk variants that we discovered are noncoding variants with predicted regulatory effects.
Rare variants in ion channel genes—especially subunits of GABAA receptors and voltage-gated calcium channels—were associated with risk for BD both in families and in the case-control cohort. GABAA receptors are widely distributed in the brain and are the primary channels through which GABA inhibits the excitability of neurons in the central nervous system (24, 25). Abnormalities in GABA function have been postulated for many years in bipolar disorder and are supported by the observation of decreased CSF GABA in bipolar patients (26), as well as alterations in GABA subunit ratios in the postmortem brain (27). Positive allosteric modulators of GABAA receptors (benzodiazepines) are used adjunctively in the treatment of mania, a key aspect of BD. Mechanistic studies of the risk variants identified in this study could lead to personalized therapies focused on specific GABAA receptor subunits.
Voltage-gated calcium channels in the extracellular membrane of postsynaptic neurons are activated by membrane depolarization. Channel activation causes an influx of Ca2+ ions into the cell, leading to increased neuronal excitability, changes in gene expression, and other physiological changes. Voltage-gated calcium channel variants have been among the most consistent signals from psychiatric GWASs (5). This is consistent with preexisting data regarding the importance of calcium signaling in BD. Although early studies of the calcium channel blocker verapamil suggested efficacy, more recent studies of calcium channel blockers have been mixed, suggesting that a subset of patients may respond (28). Lymphocytes of patients with bipolar disorder have also been shown to have an elevated calcium response to stimulation (29). Rare risk variants in these voltage-gated calcium channel genes have been discovered in schizophrenia and autism (10, 30). An important goal of future studies will be to elucidate functional consequences of the risk variants in these channels and their impact on each psychiatric disorder.
It is likely that additional classes of genes contribute to risk for BD. We found an elevated burden of risk variants in nicotinic acetylcholine receptors and CaM kinases. Others have reported variants in potassium channels and other gene sets (7, 12). As larger sample sizes become available, it will be possible to conduct unbiased genome-wide analyses. All of the associations reported in this study should be replicated with larger sample sizes.
The symptoms of BD are derived from biological activities at multiple scales, including the activities of networks of neurons. Neuronal networks and the individuals harboring these networks can have stable attractor states. These attractor states may be stable despite instability of any one neuron or shifts in the architecture of the network driving these responses. BD is characterized by episodic symptoms, so genetic architecture of BD is likely to promote episodic rather than permanent changes in state.
Genetic variation contributes to architecture shifts both by altering coding for the physical elements of the network and by regulatory rewiring. These changes could result in neuropsychiatric disease either by causing a network to persistently reach a perturbed attractor state or to episodically move between weaker attractors, such as a manic state or a depressed state. A mutation that would be predicted to have a weak effect by current variant-annotation algorithms—including regulatory and missense variation at nonconserved positions—might be sufficient particularly in combination with other weak variants to shift the architecture of the network enough to cause BD by weakening barriers between attractors. This weakening may include destabilization of the normal euthymic state by increasing neuronal excitability. Quite possibly, stronger perturbations to the very same genes that cause BD would cause other disorders, such as schizophrenia, by perturbing the network to a larger extent. Our observations are consistent with this model, at least for many families with segregating BD. In these families, many variants of weak effects together in an individual increase risk for BD.
We observed striking heterogeneity both for variants and for genes. Therefore, altered neuronal network states in BD arise from different sets of risk variants in different individuals, showing that in BD these unstable physiological network fates can be reached by many distinct molecular mechanisms (31). Theoretical and experimental studies in animal models have demonstrated that identical activity in neuronal circuits can arise from many different patterns of ion channel gene expression (32, 33). Similarly, we hypothesize that cellular differences in neuronal excitability underlying susceptibility to mania and depression can arise from distinct combinations of ion channel variants.
Data from this and other studies suggest an important role for the regulation of gene expression in psychiatric disorders. We find a predominance of rare noncoding variants with predicted regulatory functions affecting ion channel genes, both in pedigrees and in the case-control cohort. For several pathways, we identify both genetic regulatory burden and differential expression changes, emphasizing the value of multiple data types for systems biology analyses. Consistent with our hypothesis that BD should be driven largely by variants of mild molecular effect, most of these regulatory variants will have relatively mild effects, at least compared with gene-disrupting variants. Common disease-associated SNPs are also enriched in regulatory regions across several diseases (34, 35), including well-characterized risk variants for psychiatric disorders (36, 37). Increasingly, it is becoming possible to predict the effects of noncoding variants on brain gene expression. These predictions, together with the identification of risk variants from genetic studies such as we present in this report, are the building blocks for constructing gene regulatory networks. A next step for research will be to model and predict risk for BD and potential response to particular therapies with such gene regulatory networks.
Materials and Methods
Whole-Genome Sequencing and Analysis.
The 41 BD pedigrees were drawn from a set of 972 multiply affected pedigrees collected by the National Institute of Mental Health (NIMH) Genetics Initiative and by sites at the University of California, San Diego; the University of California, San Francisco; and the University of Chicago. This sample has been described previously (38). DNA derived from whole blood or from lymphoblastoid cell lines was obtained from the Rutgers University Cell and DNA Repository (Rutgers, NJ) and from the Corriell Institute (Camden, NJ).
Population controls were drawn from a collection of >1,200 genomes in an in-house collection at the Institute for Systems Biology (Seattle, WA). The control pedigrees were originally ascertained on a variety of nonpsychiatric diseases, including a large pedigree segregating a monogenic form of cardiomyopathy (39), 10 pedigrees with Adams-Oliver syndrome (40, 41), 3 pedigrees with Fanconi anemia, and other ongoing studies. Individuals in the control pedigrees are likely to have a rate of BD comparable to that in the broader population (∼1–2%).
Whole-genome sequencing was performed to >40× coverage by Complete Genomics. SNVs, insertions, and deletions were called with the Complete Genomics analysis pipeline version 2.0 or 2.2, relative to the human reference genome GRCh37. We annotated variants to RefSeq gene models with Ingenuity Variant Analysis (Qiagen). We used additional annotations from the Family Genomics Workflow (42), including allele frequency estimates from Kaviar (43). Linear mixed modeling of single variant associations was implemented with EMMAX (13). Associations of genes and pathways with BD were calculated by comparing the frequency of fully and nearly fully segregating variants in BD pedigrees to an empirical distribution in control pedigrees. See SI Appendix for further details.
Targeted Sequencing and Analysis.
For each of 26 selected genes, we targeted all exons from knownGene gene models (University of California Santa Cruz Genome Browser). In addition, we targeted the following noncoding regions with putative regulatory functions: the complete 5′ and 3′ UTRs; the core promoter 1–1,000 bp upstream of each transcriptional start site (TSS); and putative enhancers within 5 kb of each TSS, defined by DNase hypersensitive regions from the Encyclopedia of DNA Elements project (44).
Targeted sequencing was performed on 3,014 BD type 1 cases and 1,717 neurologically cleared controls of European-American ancestry. Samples were primarily from the GAIN and TGEN collections and also included 169 BD cases from a new prospective sample of lithium responsiveness in BD. DNA derived from whole blood or from lymphoblastoid cell lines was obtained from the Rutgers University Cell and DNA Repository. Samples were combined into 149 pools, with each pool containing equimolar quantities of DNA from 16 to 37 individuals. We performed PCR amplification of 3,061 250-bp target regions within the 26 selected genes using TruSeq Custom Amplicons (Illumina). We used 8-bp dual indexing to provide a unique barcode for each pool and performed paired-end 250-bp sequencing on four flowcells of a HiSeq2500 sequencer.
Sequence reads were aligned to the human reference genome (GRCh37) using Burrows-Wheeler Alignment (45), followed by indel realignment with the Genome Analysis Toolkit (46). We used SNVer (47) to call SNVs in each pool and to estimate per-pool allele frequencies. Allele counts of low-frequency variants were normalized to adjust for stochastic variation in read depth. Statistical associations between the rare variants in each gene and BD affection status were assessed with the SKAT (21) and C-alpha (22). We report empirical P values from 100,000 resampling permutations. See SI Appendix for further details.
Supplementary Material
Acknowledgments
Most importantly, we thank the families who have participated in and contributed to these studies. We also thank the Rutgers University Cell and DNA Repository for transforming cell lines and providing DNA samples. Biomaterials and phenotypic data were obtained for control subjects from the National Institute of Mental Health (NIMH) Schizophrenia Genetics Initiative, which were collected by the “Molecular Genetics of Schizophrenia II” collaboration. Data and biomaterials were collected as part of 11 projects (Study 40) that participated in the NIMH Bipolar Disorder Genetics Initiative (MH59545, MH059534, MH59533, MH59553, MH60068, MH059548, MH59535, MH59567, MH059556, and 1Z01MH002810-01), which was also supported by NIH Grants P50CA89392, from the National Cancer Institute, and 5K02DA021237, from the National Institute of Drug Abuse. This work was supported by the University of Luxembourg–Institute for Systems Biology Strategic Partnership, National Institute of General Medical Sciences Center for Systems Biology Grant P50 GM076547, NIMH Grant R01 MH094483, and the Intramural Research Program of the NIMH (F.J.M., principal investigator). J.A. is a Gordon and Betty Moore Foundation Fellow of the Life Sciences Research Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: Whole-genome sequences have been deposited in the Database of Genotypes and Phenotypes study “Genomic Landscape of Bipolar Disorder,” www.ncbi.nlm.nih.gov/gap (accession no. phs000865).
1A complete list of members of The Bipolar Genome Study is provided in SI Appendix.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424958112/-/DCSupplemental.
References
- 1.Goodwin FK, Jamison KR. Manic-Depressive Illness: Bipolar Disorders and Recurrent Depression. Oxford Univ Press; Oxford: 2007. [Google Scholar]
- 2.Merikangas KR, Low NCP. The epidemiology of mood disorders. Curr Psychiatry Rep. 2004;6(6):411–421. doi: 10.1007/s11920-004-0004-1. [DOI] [PubMed] [Google Scholar]
- 3.Craddock N, Sklar P. Genetics of bipolar disorder. Lancet. 2013;381(9878):1654–1662. doi: 10.1016/S0140-6736(13)60855-7. [DOI] [PubMed] [Google Scholar]
- 4.Ferreira MAR, et al. Wellcome Trust Case Control Consortium Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40(9):1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee SH, et al. Cross-Disorder Group of the Psychiatric Genomics Consortium; International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45(9):984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Georgi B, et al. Genomic view of bipolar disorder revealed by whole genome sequencing in a genetic isolate. PLoS Genet. 2014;10(3):e1004229. doi: 10.1371/journal.pgen.1004229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strauss KA, et al. A population-based study of KCNH7 p.Arg394His and bipolar spectrum disorder. Hum Mol Genet. 2014;23(23):6395–6406. doi: 10.1093/hmg/ddu335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruceanu C, et al. Family-based exome-sequencing approach identifies rare susceptibility variants for lithium-responsive bipolar disorder. Genome. 2013;56(10):634–640. doi: 10.1139/gen-2013-0081. [DOI] [PubMed] [Google Scholar]
- 9.Fiorentino A, et al. Analysis of ANK3 and CACNA1C variants identified in bipolar disorder whole genome sequence data. Bipolar Disord. 2014;16(6):583–591. doi: 10.1111/bdi.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Purcell SM, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506(7487):185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Psychiatric GWAS Consortium Bipolar Disorder Working Group Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43(10):977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nurnberger JI, Jr, et al. Psychiatric Genomics Consortium Bipolar Group Identification of pathways for bipolar disorder: A meta-analysis. JAMA Psychiatry. 2014;71(6):657–664. doi: 10.1001/jamapsychiatry.2014.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang HM, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42(4):348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith EN, et al. Genome-wide association study of bipolar disorder in European American and African American individuals. Mol Psychiatry. 2009;14(8):755–763. doi: 10.1038/mp.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith EN, et al. Genome-wide association of bipolar disorder suggests an enrichment of replicable associations in regions near genes. PLoS Genet. 2011;7(6):e1002134. doi: 10.1371/journal.pgen.1002134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagerström MC, et al. The evolutionary history and tissue mapping of GPR123: Specific CNS expression pattern predominantly in thalamic nuclei and regions containing large pyramidal cells. J Neurochem. 2007;100(4):1129–1142. doi: 10.1111/j.1471-4159.2006.04281.x. [DOI] [PubMed] [Google Scholar]
- 18.Cáceda R, Kinkead B, Owens MJ, Nemeroff CB. Virally mediated increased neurotensin 1 receptor in the nucleus accumbens decreases behavioral effects of mesolimbic system activation. J Neurosci. 2005;25(50):11748–11756. doi: 10.1523/JNEUROSCI.4282-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fitzpatrick K, et al. Altered sleep and affect in the neurotensin receptor 1 knockout mouse. Sleep. 2012;35(7):949–956. doi: 10.5665/sleep.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akula N, et al. RNA-sequencing of the brain transcriptome implicates dysregulation of neuroplasticity, circadian rhythms and GTPase binding in bipolar disorder. Mol Psychiatry. 2014;19(11):1179–1185. doi: 10.1038/mp.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee S, et al. NHLBI GO Exome Sequencing Project—ESP Lung Project Team Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91(2):224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neale BM, et al. Testing for an unusual distribution of rare variants. PLoS Genet. 2011;7(3):e1001322. doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyle AP, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Craddock N, et al. Wellcome Trust Case Control Consortium (WTCCC) Strong genetic evidence for a selective influence of GABAA receptors on a component of the bipolar disorder phenotype. Mol Psychiatry. 2010;15(2):146–153. doi: 10.1038/mp.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benes FM, Berretta S. GABAergic interneurons: Implications for understanding schizophrenia and bipolar disorder. Neuropsychopharmacology. 2001;25(1):1–27. doi: 10.1016/S0893-133X(01)00225-1. [DOI] [PubMed] [Google Scholar]
- 26.Mann JJ, et al. Anxiety in major depression and cerebrospinal fluid free gamma-aminobutyric acid. Depress Anxiety. 2014;31(10):814–821. doi: 10.1002/da.22278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fatemi SH, Folsom TD, Rooney RJ, Thuras PD. Expression of GABAA α2-, β1- and ε-receptors are altered significantly in the lateral cerebellum of subjects with schizophrenia, major depression and bipolar disorder. Transl Psychiatr. 2013;3:e303. doi: 10.1038/tp.2013.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levy NA, Janicak PG. Calcium channel antagonists for the treatment of bipolar disorder. Bipolar Disord. 2000;2(2):108–119. doi: 10.1034/j.1399-5618.2000.020204.x. [DOI] [PubMed] [Google Scholar]
- 29.Kato T, et al. Mechanisms of altered Ca2+ signalling in transformed lymphoblastoid cells from patients with bipolar disorder. Int J Neuropsychopharmacol. 2003;6(4):379–389. doi: 10.1017/S1461145703003717. [DOI] [PubMed] [Google Scholar]
- 30.Krumm N, O’Roak BJ, Shendure J, Eichler EE. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 2014;37(2):95–105. doi: 10.1016/j.tins.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang S, Eichler G, Bar-Yam Y, Ingber DE. Cell fates as high-dimensional attractor states of a complex gene regulatory network. Phys Rev Lett. 2005;94(12):128701. doi: 10.1103/PhysRevLett.94.128701. [DOI] [PubMed] [Google Scholar]
- 32.Marder E, Goaillard J-M. Variability, compensation and homeostasis in neuron and network function. Nat Rev Neurosci. 2006;7(7):563–574. doi: 10.1038/nrn1949. [DOI] [PubMed] [Google Scholar]
- 33.Schulz DJ, Goaillard J-M, Marder E. Variable channel expression in identified single and electrically coupled neurons in different animals. Nat Neurosci. 2006;9(3):356–362. doi: 10.1038/nn1639. [DOI] [PubMed] [Google Scholar]
- 34.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kellis M, et al. Defining functional DNA elements in the human genome. Proc Natl Acad Sci USA. 2014;111(17):6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou X, et al. Transcription factor SP4 is a susceptibility gene for bipolar disorder. PLoS ONE. 2009;4(4):e5196. doi: 10.1371/journal.pone.0005196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caspi A, et al. Influence of life stress on depression: Moderation by a polymorphism in the 5-HTT gene. Science. 2003;301(5631):386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 38.Badner JA, et al. Genome-wide linkage analysis of 972 bipolar pedigrees using single-nucleotide polymorphisms. Mol Psychiatry. 2012;17(8):818–826. doi: 10.1038/mp.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu H, et al. A unified test of linkage analysis and rare-variant association for analysis of pedigree sequence data. Nat Biotechnol. 2014;32(7):663–669. doi: 10.1038/nbt.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lehman A, et al. Diffuse angiopathy in Adams-Oliver syndrome associated with truncating DOCK6 mutations. Am J Med Genet A. 2014;164A(10):2656–2662. doi: 10.1002/ajmg.a.36685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stittrich A-B, et al. Mutations in NOTCH1 cause Adams-Oliver syndrome. Am J Hum Genet. 2014;95(3):275–284. doi: 10.1016/j.ajhg.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roach JC, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328(5978):636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glusman G, Caballero J, Mauldin DE, Hood L, Roach JC. Kaviar: An accessible system for testing SNV novelty. Bioinformatics. 2011;27(22):3216–3217. doi: 10.1093/bioinformatics/btr540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei Z, Wang W, Hu P, Lyon GJ, Hakonarson H. SNVer: A statistical tool for variant calling in analysis of pooled or individual next-generation sequencing data. Nucleic Acids Res. 2011;39(19):e132. doi: 10.1093/nar/gkr599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.