Abstract
Sporadic inclusion body myositis (sIBM) has clinical, pathologic and pathomechanistic overlap with some inherited muscle and neurodegenerative disorders. In this study, DNA from 79 patients with sIBM was collected and the sequencing of 38 genes associated with hereditary inclusion body myopathy (IBM), myofibrillar myopathy, Emery–Dreifuss muscular dystrophy, distal myopathy, amyotrophic lateral sclerosis and dementia along with C9orf72 hexanucleotide repeat analysis was performed. No C9orf72 repeat expansions were identified, however; 27 rare (minor allele frequency <1%) missense coding variants in several other genes were identified. One patient carried a p.R95C missense mutation in VCP and another carried a previously reported p.I27V missense mutation in VCP. Mutations in VCP cause IBM associated with Paget’s disease of the bone (PDB) and fronto-temporal dementia (IBMPFD). Neither patient had a family history of weakness or manifested other symptoms reported with VCP mutations such as PDB or dementia. In vitro analysis of these VCP variants found that they both disrupted autophagy similar to other pathogenic mutations. Although no clear genetic etiology has been implicated in sIBM pathogenesis, our study suggests that genetic evaluation in sIBM may be clinically meaningful and lend insight into its pathomechanism.
Keywords: Inclusion body myositis, VCP, Hereditary inclusion body myopathy, Myofibrillar myopathy, Amyotrophic lateral sclerosis
1. Introduction
Sporadic inclusion body myositis (sIBM) is an idiopathic and untreatable myopathy that typically begins in patients over the age of 50 [1]. Patients have a characteristic pattern of involvement with both proximal and distal muscle weakness and a predilection for the knee extensors and wrist and finger flexors. Disease progression leads to significant morbidity with wheelchair confinement often within 10 years of onset [2,3]. The pathogenic mechanism of sIBM is currently unknown. Muscle from patients with sIBM has several myopathologic features that aid in distinguishing sIBM from other inflammatory and inherited muscle disorders. These include endomysial T-cell infiltrates that surround healthy appearing muscle fibers [1]. Vacuoles, classically described as “rimmed,” are present in scattered nonnecrotic fibers [1]. Sarcoplasmic inclusions that are immunoreactive for TARDNA binding protein-43 (TDP-43), p62/SQSTM1 and SMI-31 are also characteristic features [1].
While no hereditary muscle disease consistently has all of these features, some hereditary muscle diseases have a subset of similar pathologic features to sIBM on muscle biopsy and are termed hereditary inclusion body myopathies (hIBM) [1]. Whether genetic variants in hIBM associated proteins are associated with sIBM is not known. Moreover, whether proteins that accumulate in sIBM muscle tissue contribute to muscle pathogenesis is unclear. For example, dementia associated proteins such as β-amyloid and hyperphosphorylated tau have been proposed to accumulate in sIBM tissue implicating amyloid precursor protein processing and microtubule associated protein tau in sIBM pathogenesis [4,5]. Similarly, the identification of TDP-43 and p62/SQSTM1 as specific markers for sIBM pathology has supported the hypothesis that genes mutated in familial ALS may be associated with sIBM pathogenesis [6,7]. Further evidence for this comes from a family of diseases in which mutations in single proteins such as VCP, hnRNPA1, hnRNPA2B1 and matrin-3 can lead to variably penetrant phenotypes that include hIBM, ALS and fronto-temporal dementia [8–10]. Finally, a large group of protein aggregate and vacuolar myopathies can have “rimmed vacuoles” similar to those seen in sIBM patient muscle. The largest component of this group is myofibrillar myopathies and some distal myopathies [11,12].
Some studies have performed targeted genetic mutation analysis in small cohorts of sIBM patients [13–16]. However, disease causing mutations in these genes have not been consistently identified in sIBM patient cohorts. Other studies have focused on risk alleles within populations of sIBM patients such as apolipoprotein E genotypes or HLA subtypes as means to correlate MHC gene alleles with sIBM risk, severity and prognosis [17–19]. Nonetheless, studies systematically evaluating the genetic etiology of sIBM, similar to those performed for other neuromuscular disorders, are lacking [20].
We utilized a targeted next generation sequencing approach to evaluate 38 genes in 79 patients with sIBM. These 38 genes included a wide array of putative candidates and were chosen based upon their association with several muscle diseases including vacuolar, myofibrillar, Emery–Dreifuss and inclusion body myopathies. We also sequenced four genes, including GNE (mutations in GNE are associated with HIBM2, now more appropriately called GNE myopathy), that are essential for sialic acid biosynthesis [21]. Finally, we elected to sequence genes associated with ALS and dementia and evaluate C9orf72 repeat expansions in sIBM patients.
2. Materials and methods
2.1. Study subjects
DNA was collected from 79 patients with a clinical diagnosis of sIBM. 41 patients were identified within the neuromuscular clinic at Washington University and the diagnosis of sIBM was made by that patient’s physician. An additional 38 patients were identified at a Patient Conference and were personally examined by a Washington University Neuromuscular Physician (CCW, MHB, GL, or AP) who found their history and physical exam to be consistent with sIBM. Any patient with a family history of weakness, lack of quadriceps weakness, symptoms beginning before 40 years of age or with upper motor neuron signs were excluded from further analysis. Indeed four patients (two pairs of siblings) were identified at the patient conference and were not further included in our study. One set of siblings was found to be compound heterozygous for previously reported pathogenic GNE variants (NP_001121699.1; p.V727M; p.R42W) [22]. The study population was consistent with previous reports of sIBM patients with 58.2% being male and 41.8% female [2]. The average age at the time of DNA collection was 67 ± 9.4 years. All participants provided informed consent for clinico-genetic studies approved by the Washington University institutional review boards.
2.2. Molecular genetics
Agilent’s SureDesign website was used to target the exons of 38 genes. Indexed genomic DNA (gDNA) libraries were prepared according to HaloPlex manufacturer’s instructions. 250 ng of gDNA was digested in 8 parallel reactions, then hybridized with biotin-labeled probes designed to recognize and circularize targeted regions. Circularized segments of gDNA were captured using streptavidin-coupled magnetic beads and amplified for sequencing. Samples were pooled in 2 groups and each sequenced by 100 bp paired end reads on a single lane of a HiSeq2000 (Illumina, San Diego, CA). Reads were aligned to the human reference genome (Hg19) with NovoAlign (Novocraft Technologies, Selangor, Malaysia). Variants were called with SAMtools and annotated with SeattleSeq. Coverage across genomic intervals was calculated using BEDTools. Segregation and validation of mutations was assessed with standard polymerase chain reaction (PCR)-based sequencing using Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) for primer design, an Applied Biosystems 3730 DNA Sequencer (Life Technologies, Carlsbad, CA) for sequencing and LaserGene SeqMan Pro version 8.0.2 (DNAStar, Madison, WI) for tracing analysis.
2.3. C9orf72 expansion identification
gDNA samples were screened for the C9orf72 hexanucleotide expansions using repeat-primed polymerase chain reaction (PCR) primers and methods as previously published [23].
2.4. In vitro ATPase assay
Purified recombinant VCP-WT and VCP variants were assayed for intrinsic ATPase activity as previously described [24].
2.5. Tissue culture and immunoblotting
The GFP-fused VCP expression plasmid is previously described [25]. The mutations VCP-R155H, VCP-R95C and VCP-I27V were introduced using site-directed mutagenesis kit (Stratagene, La Jolla, CA). Forty-eight hours post-transfection into U20S cells, cells were harvested and lysates were separated via SDS-PAGE, transferred to nitrocellulose and immunoblotted using the following antibodies [anti-p97/VCP (Fitzgerald, Acton, MA), anti-p62 (Proteintech, Chicago, IL), anti-GAPDH (Sigma-Aldrich), and anti-LC3 (Sigma-Aldrich)] as previously described [24].
3. Results
Our targeted capture and sequencing of 38 candidate genes in 79 subjects with sIBM yielded an average of 320 Mbp (range 140–1000) per subject, with 95 ± 1% of targeted bases covered at ≥10x (92 ± 3% at ≥25x). Variants were filtered for quality and depth, then for those predicted to disrupt coding sequence resulting in 29 ± 7 high-quality coding variants per subject. We hypothesized that variants influencing disease would be rare and focused our analysis on variants that i) had been previously reported as pathogenic, ii) were rare in population control databases (i.e. a global MAF <1%), or iii) were novel. Twenty-seven different heterozygous missense variants met these criteria and were validated by Sanger sequencing (Table 1). No individuals were homozygous or compound heterozygous for any variant or variants within the same gene. Twenty-eight subjects (35% of the cohort) carried one of the identified variants and three subjects carried two variants. Eight of twenty-seven (30%) of these variants were completely absent from large population databases (1000 Genomes project, the Exome Sequencing Project, dbSNP).
Table 1.
Summary of rare <1% minor allele frequency (MAF) missense variants identified in 79 sIBM patients.
| Disease Category | Gene | NCBI Reference Sequence | Protein | Variant | Minor Allele Frequency All/EA/AA |
|---|---|---|---|---|---|
| Hereditary IBM | |||||
| GNE | NM_001190388.1 | UDP-GlcNAc 2-epimerase/N-acetylmannosamine kinase | R30Ha | 0.9029/0.028/2.9 | |
| VCP | NM_007126.3 | Valosin containing protein | R95C | 0/0/0 | |
| I27V | 0.0923/0.035/0.204 | ||||
| HNRNPA2B1 | NM_002137.3 | Heterogeneous nuclear ribonucleoprotein A2/B1 | Y335F | 0/0/0 | |
| HNRNPA1 | NM_031157.2 | Heterogeneous nuclear ribonucleoprotein A1 | |||
| PABPN1 | NM_001199839.1 | Poly(A) binding protein, nuclear 1 | |||
| MYH2 | NM_001100112.1 | Myosin heavy chain-2 | E1681K | 0.0077/0.0116/0 | |
| S1043A | 0.1307/0.186/0.0227 | ||||
| DNAJB6 | NM_058246.3 | DnaJ homolog, subfamily B, member 6 | |||
| Vacuolar myopathy | |||||
| LAMP2 | NM_013995.2 | Lysosomal-associated membrane protein-2 | V391Ib,c | 0.4449/0.654/0.0782 | |
| VMA21 | NM_001017980.3 | VMA21 vacuolar H+-ATPase homolog | |||
| MATR3 | NM_001194954.1 | Matrin-3 | |||
| SIL1 | NM_001037633.1 | Sil1 homolog | |||
| TRIM32 | NM_001099679.1 | Tripartite motif containing 32 | |||
| Myofibrillar myopathy | |||||
| DES | NM_001927.3 | Desmin | |||
| CRYAB | NM_001885.1 | αB-crystallin | |||
| BAG3 | NM_004281.3 | BCL2-associated athanogene | H83Q | 0.346/0/1.02 | |
| G414K | 0.0231/0.035/0 | ||||
| T144A | 0/0/0 | ||||
| FHL1 | NM_001159699.1 | Four and a half LIM domains 1 | |||
| FLNC | NM_001127487.1 | Filamin-C | R575W | 0/0/0 | |
| D693A | 0.46/0.536/0.122 | ||||
| R1241C | 0.6692/0.95/0.115 | ||||
| R2331H | 0.2/0.238/0.124 | ||||
| E309K | 0/0/0 | ||||
| R526Q | 0.141/0.153/0.116 | ||||
| LDB3 | NM_001080114.1 | Zasp/LIM domain binding 3 | V118Md | 0.5155/0.698/0.16 | |
| A222T | 0.0384/0.047/0.023 | ||||
| Q414K | 0.0154/0.023/0 | ||||
| MYOT | NM_006790.2 | Myotilin | |||
| Emery–Dreifuss muscular dystrophy | |||||
| EMD | NM_000117.2 | Emerin | |||
| LMNA | NM_001257374.1 | Lamin A/C | G526Ra | 0.0923/0/0.272 | |
| N171S | 0/0/0 | ||||
| Amyotrophic lateral sclerosis | |||||
| C9orf72 | NM_001256054.1 | C9orf72f | |||
| CHMP2b | NM_001244644.1 | Charged multivesicular body protein 2B | |||
| FUS | NM_001170634.1 | FUS | |||
| TARDP | NM_007375.3 | TDP-43 | |||
| TAF15 | NM_003487.3 | TAF15 | |||
| SQSTM1 | NM_001142298.1 | Sequestosome-1 | |||
| UBQLN2 | NM_013444.3 | Ubiquilin 2 | |||
| EWSR1 | NM_001163285.1 | EWSR1 | G464S | 0.9688/1.3023/0.317 | |
| OPTN | NM_001008211.1 | Optineurin | |||
| Dementia | |||||
| GRN | NM_002087.2 | Progranulin | |||
| MAPT | NM_001123066.3 | Microtubule-associated protein tau | V224Ge | 0.3078/0.42/0.091 | |
| R544L | 0/0/0 | ||||
| APP | NM_000484.3 | Amyloid Precursor Protein | T276S | 0/0/0 | |
| R328W | 0/0/0 | ||||
| PSEN1 | NM_000021.3 | Presenilin-1 | |||
| PSEN2 | NM_000447.2 | Presenilin-2 | |||
| Sialic acid biosynthesis | |||||
| CMAS | NM_018686.4 | Cytidine monophosphate N-acetylneuraminic acid synthetase |
|||
| NANP | NM_152667.2 | N-acetylneuraminic acid phosphatase | |||
| NANS | NM_018946.3 | N-acetylneuraminic acid synthase | G82R | 0.0077/0/.0227 | |
African American patient.
Identified in 2 female patients.
Variant in the LAMP2b isoform.
Identified in 3 patients.
Identified in 2 patients.
Coding sequence and expanded repeat analysis.
A single rare missense variant in LDB3 (c.352G > A; p.V118M) recurred in 3/79 (3.8%) unrelated patients in our cohort. Although this frequency is considerably higher than the MAF of 0.5155%, the difference did not meet statistical significance (p = 0.11). Two other patients also carried single rare variants in LDB3 (c.664G > A; p.A222T and c.1240C > A; p.Q414K) with MAF of 0.0384% and 0.0154% respectively. Rare variants were identified in other genes associated with myofibrillar myopathies: FLNC (six variants, two of which were novel) and BAG3 (three variants, one which was novel). One novel missense variant (c.1004C > A; p.Y335F) was identified in HNRNPA2B1, a gene recently shown to cause IBMPFD2 (OMIM 600124) [9]. This variant is not present in population databases and is predicted to affect the nuclear import sequence of hnRNPA2/B1 [26]. No sIBM patients had evidence of C9orf72 GGGGCC repeat expansions as determined by repeat primed PCR.
Seven previously reported putative disease associated variants were identified in five genes: FHL1, DES, MYOT, SQSTM1 and VCP (Table 2). In the case of FHL1, DES, MYOT and SQSTM1 the MAF for each variant was >1% in control populations, suggesting that they are not disease causing. In contrast, two VCP variants (one novel and one previously reported) were identified (c.283G > A; p.R95C and c.79A > G; p.I27V) and were either absent (p.R95C) or very rare (MAF 0.0923%) (p.I27V) in population databases [27–29]. The clinical and pathologic features for the two subjects with VCP mutations are shown in Table 3 and described in the supplemental material. Notably, the subject with the VCP p.R95C mutation fulfilled diagnostic criteria for probable IBM (ENMC 2013 criteria) [38] and possible IBM (Griggs criteria) [39]. He had no family history of weakness, including among his 5 adult children. His father died at 75 years old with a diagnosis of lupus and his mother passed away at age of 96 with type 2 diabetes. One sibling died at 90 years old after 10 years of Alzheimer Disease and before DNA could be collected. The subject’s living siblings (age 75 and 79 years old without symptoms and age 94 years with dementia) agreed to undergo genetic evaluation but were negative for the p.R95C mutation. The subject carrying VCP p.I27V fulfilled ENMC 2013 criteria for clinico-pathologically defined IBM and Griggs criteria for definite IBM. There was no history of weakness, dementia or PDB in his parents, sibling or two children. No further genetic analysis was performed on this family.
Table 2.
Reported disease associated variants.
| Gene | NCBI Reference Sequence | Protein | Variant | Minor Allele Frequency All/EA/AA | African American | Clinical syndrome previously described |
|---|---|---|---|---|---|---|
| VCP | NM_007126.3 | Valosin containing protein | R95C | 0/0/0 | N | IBMPFD [27] |
| VCP | NM_007126.3 | Valosin containing protein | I27V | 0.092/0.035/0.204 | N | IBMPFD [28]; Parkinsons Disease [29] |
| SQSTM1 | NM_001142298.1 | Sequestosome-1 | E274D | 1.868/2.535/0.567 | N | ALS [30] |
| DES | NM_001927.3 | Desmin | V459I | 1.2/0.012/3.56 | Y | Cardiomyopathy [31] |
| DES | NM_001927.3 | Desmin | A213V | 1.053/1.372/0.431 | N | Cardiomyopathy [31,32]; Myopathy [33] |
| MYOT | NM_006790.2 | Myotilin | K74Q | 1.484/0.0/4.38 | Y | Myofibrillar myopathy [34,35] |
| FHL1 | NM_001159699.1 | Four and half LIM domains protein 1 | D275N | 1.306/1.858/0.339 | Y | Cardiomyopathy [36]; Myopathy [37] |
Table 3.
Clinical summaries of VCP mutation carrying patients.
| VCP mutation | R95C | I27V |
|---|---|---|
| Clinical and Laboratory Features | ||
| Duration >12 months | Yes | Yes |
| Age at onset >45 | Yes | Yes |
| Sporadic | Yesb | Yes |
| Slow progression | Yes | Yes |
| Knee extension weakness ≥ hip flexion weakness | Yes | Yes |
| Finger flexion weakness > shoulder abduction weakness | No | Yes |
| Wrist flexion weakness > wrist extension weakness | No | Yes |
| Finger flexion weakness | Yes | Yes |
| Quadriceps ≤ MRC 4 | Yes | Yes |
| Proximal and distal weakness of arms and legs | Yes | Yes |
| CK no greater than 15xULN | Yes (551 u/L) | Yes (914 u/L; nl 30–200 u/L) |
| EMG consistent | Yes | Yes |
| Presence of Anti-NT5C1A | Noa | N/A |
| Pathologic Features | ||
| Endomysial inflammatory infiltrate | Yes | Yes |
| Invasion of nonnecrotic fibers | Yes | Yes |
| Rimmed vacuoles | Yes | Yes |
| Protein accumulation | Yes | N/A |
| 15–18 nM tubulofilaments | N/A | N/A |
| Intracellular amyloid deposits | No | Yes |
| Up-regulation of MHCI | Noa | N/A |
| Diagnostic Criterion | ||
| ENMC 2013 Diagnosis | Probable IBM | Clinico-pathologically defined IBM |
| Griggs Criteria Diagnosis | Possible IBM | Definite IBM |
Testing performed after identification of VCP mutation.
Two siblings with late onset dementia.
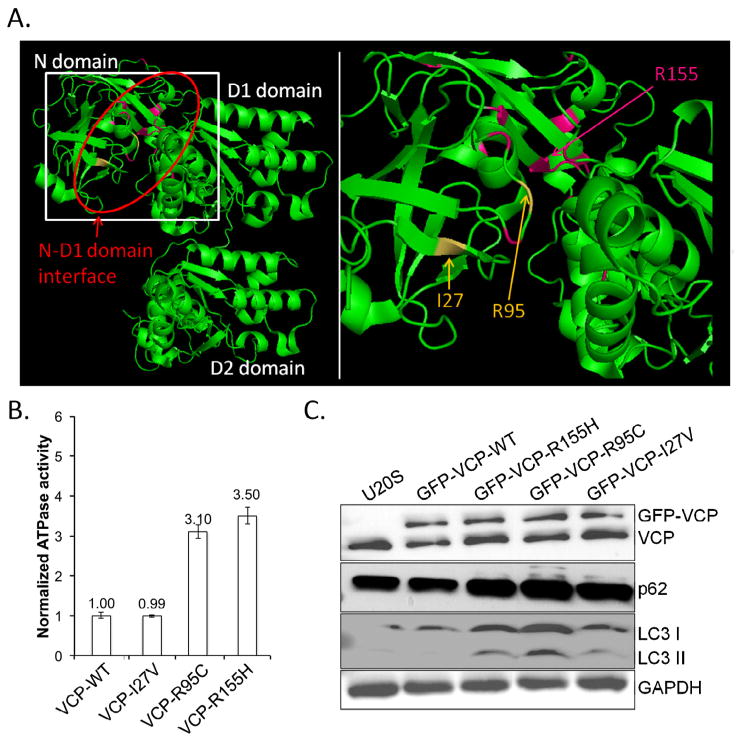
The R95C and I27V variants are within the N-domain of VCP where > 50% of all pathogenic VCP variants exist [40]. In addition, when these mutated residues are identified in the crystal structure of VCP, they reside at the N-D1 domain interface adjacent to all other reported pathogenic mutations (Fig. 1A). Many, but not all, previously reported pathogenic VCP mutations have enhanced basal ATPase activity [24]. To test this, we measured the basal rate of ATP hydrolysis in the presence of recombinant wild-type VCP (VCP-WT), VCP-I27V, VCP-R95C and the most common pathogenic VCP mutation VCP-R155H. The VCP-R95C variant had a 3-fold increase ATPase activity similar to VCP-R155H (Fig. 1B). In contrast, the VCP-I27V variant did not have an increase in basal ATPase activity (Fig. 1B).
Fig. 1.
A) Crystallographic rendering of a single VCP monomer with the N, D1 and D2 domains indicated. Red oval emphasizes the N-D1 domain interface where all known disease mutations reside. Disease mutated residues are in pink (R155 is the most commonly mutated residue) and the I27 and R95 residues mutated in this report are in yellow. Box denotes region of enlargement. B) Recombinant human wild-type VCP (VCP-WT), or disease mutant associated VCPs (VCP-I27V, VCP-R95C and VCP-R155H) were purified from E. coli and the basal ATP hydrolysis rate assayed. The ATPase activity of VCP-WT was arbitrarily designated as 1. C) Representative immunoblot from three independent experiments for VCP, LC3, p62/SQSTM1 and actin of U20S cells transiently expressing VCP-WT-GFP, VCP-R155H-GFP, VCP-R95C-GFP or VCP-I27V-GFP for 2 days. Note the selective increase in both p62/SQSTM1 and LC3II in disease mutant expressing cells.
Previous studies have found that autophagosome maturation is disrupted by pathogenic VCP mutations as demonstrated by the accumulation of the autophagic substrate p62 and the autophagosome protein LC3II [41]. To assess the pathogenic nature of the variants identified in sIBM patients, we transiently transfected VCP-WT, VCP-R155H, VCP-R95C and VCP-I27V fused to a C-terminal green fluorescent protein tag into U20S cells and evaluated the levels of VCP, p62, LC3 and GAPDH via immunoblot. As previously described, VCP-R155H expression results in an increase in p62 and LC3II levels [41]. Similarly, VCP-R95C and VCP-I27V expression caused an increase in p62 and LC3II consistent with the previously defined disruption in autophagosome maturation by VCP disease mutations [41].
4. Discussion
We performed targeted sequencing of 38 “high probability” genes associated with clinical syndromes having phenotypic and pathogenic similarities to sIBM, in 78 patients with sIBM; making it the largest genetic study performed to date for sIBM. Our goal was to assess whether rare missense coding variants in these genes known to be associated with hereditary IBM, myopathies with IBM-like pathology, ALS or dementia are present in sIBM patients and may begin to explain the sporadic nature of this enigmatic disease. This is of course a significant limitation of our study and biases our analysis to a subset of genes. Moreover we presuppose that rare coding variants will be we pathogenic. While this strategy has been used for other “sporadic” neuromuscular disorders, it seems clear that whole exome sequencing of these patients will reveal other relevant risk alleles [20].
Two patients meeting diagnostic criteria for sIBM carried pathogenic mutations in VCP. The p.R95C variant in VCP had been previously mentioned as causing disease [27] and occurs at an amino acid residue found mutated in other IBMPFD pedigrees (p.R95G and p.R95H) [10,42]. Our subject had no clinical or lab evidence of PDB (serum alkaline phosphatase was normal (65 u/L; nl 38–126 u/L) and plain film X-rays of hips, pelvis and spine were normal) or dementia. In addition, he had no family history of weakness but did have two siblings with late onset dementia. The original review of his left deltoid muscle biopsy revealed chronic myopathic changes, several small angular fibers with rimmed vacuoles, increased endomysial cellularity, and one region of focal invasion (Fig. S1). Subsequent to the genetic identification of his p.R95C variant, we re-examined the muscle biopsy and performed MHCI immunostaining which showed no upregulation. Additionally, banked serum was examined for anti-NT5C1A autoantibodies, suggested to be specific for sIBM, but they were not found [43,44]. Re-evaluation of the clinical phenotype by chart review showed that at one point in his clinical assessments, wrist and finger extensor weakness was greater than that of the wrist and finger flexors. Importantly, although his phenotype was distinctive from classical sIBM, he still fulfilled the 2013 ENMC proposed criteria for probable IBM. The absence of MHCI immunostaining, the absence of anti-NT5C1A autoantibodies, and atypical phenotypic features in patients with vacuolar myopathies may push one toward identifying a genetic etiology.
The VCP p.I27V variant has been reported in three patients with neurologic diseases consistent with IBMPFD, but also in control subjects [28,29]. Since being reported in IBMPFD, the variant has been found in population screening, with a MAF of 0.09% in the Exome Sequencing Project. This is considerably more prevalent than sIBM. This patient had a typical sIBM pattern of weakness with quadriceps and wrist and finger flexor weakness. Moreover, his biopsy was consistent with sIBM. However retrospective immune marker analysis was not able to be performed. Notably this patient did not have any reported family history for weakness, dementia or PDB. Unfortunately, due to the small family structure and late onset of his phenotype, we were unable to perform segregation analysis. It has been suggested that the p.I27V VCP variant could either be a pathologic mutation with incomplete penetrance or a risk allele for neurologic disease since it has been reported in unaffected control patients [29]. Structural modeling, biochemical and in vitro assays were performed on both the VCP I27V and R95C mutations and were consistent with them being pathogenic (Fig. 1A–C). Interestingly, in contrast to the R95C mutation, the I27V variant behaved similar to VCP-WT with regard to intrinsic ATPase activity. An elevation in ATPase activity has been reported for many but not all pathogenic VCP mutations [24,45]. In particular, we recently identified an E185K missense variant in VCP that segregated in a large family with late onset CMT2 [24]. Similar to the VCP-I27V variant, VCP-E185K disrupted autophagic function in cell culture but did not have an intrinsic elevation in ATPase function [24]. Perhaps the normal ATPase activity portends a milder VCP disease phenotype as in the case of CMT2 and the VCP-E185K variant or incomplete penetrance and increased risk in patients carrying the VCP-I27V mutation.
In addition to VCP variants, we identified another five subjects with previously reported disease associated variants in DES, MYOT, FHL1 and SQSTM1. All of these variants have MAFs >1.0% making them unlikely to be disease causing. However, it was recently suggested that the p.A213V desmin variant, while not being a disease-causing variant, could be disease-associated or a risk factor for cardiomyopathy [32]. Whether this may similarly be true for sIBM or other muscle disorders is unclear and warrants further assessment in larger cohorts of patients.
We also noted a potential excess of rare variants in LDB3 and other myofibrillar myopathy related proteins (FLNC and BAG3). Although our study was not powered to perform either single variant or rare-variant burden associations, this finding warrants further analysis of this gene in larger groups of patients. Interestingly, this finding is in line with a recent study evaluating genetic variants in candidate genes in 21 Japanese sIBM patients; where they also identified a patient with a novel LDB3 variant [13].
It is well established that some hereditary muscle diseases can mimic that of sIBM, and even meet varied clinical criteria [46]. Therefore it is tempting to dismiss that a patient with a clinical diagnosis of sIBM and later found to have a genetic etiology was “misdiagnosed.” This is a limitation of many genetic studies. Indeed similar clinically reported DNA collections (e.g. NINDS Human Genetics DNA and Cell Line Repository at Coriell) in which the investigating researcher may not have immediate access to all clinical information are an expeditious means of performing large scale human genetic studies [29,47]. Regardless, since 38 of the 79 patients in our study were identified at a patient support conference and only a history and physical exam was utilized to corroborate a diagnosis of sIBM, we were not able to classify all of our patients into ENMC 2013 or Griggs criteria. However it is reassuring that the patient identified with the VCP I27V variant, who was identified at a patient conference, fulfilled ENMC 2013 and Griggs criteria for clinic-pathologically defined and definite IBM when all clinical material was later evaluated.
The future of research and clinical treatment for acquired muscle diseases such as sIBM is rapidly evolving and will dramatically change with the advent of inexpensive and comprehensive genetic testing. Moreover, clinical trials and therapeutic interventions for sIBM are currently in development. The identification of potential genetic risk factors, genetic modifiers or genetic elements associated with treatment response for sIBM will be of equal value to that of properly genetically diagnosing sIBM patients. Our study offers an initial glimpse into the genetic variation seen in clinically reported sIBM patients. This will be the group of patients seeking clinical care and enrolling in clinical trials. Indeed the patient with the VCP I27V mutation was enrolled in a sIBM clinical trial further emphasizing that probing the genetics of sIBM is critical for patient care and research development. It may not be surprising that rare missense genetic variants will be identified in sIBM patients. Whether these variant, including the ones identified in this manuscript are solely causal or risk alleles for sIBM remains to be established. Future studies in larger populations of patients and utilizing whole exome or whole genome sequencing will be necessary to define whether these rare genetic variants are indeed genetic modifiers of sIBM.
Supplementary Material
Acknowledgments
We thank Amir Dori, Taha Bali, Cindy Ly, Arun Varadacharry, Paul Cooper, and Leo Wang for help with patient assessment. Supported by a research agreement from Ultragenyx Pharmaceuticals, Novato CA to CCW. Other funding included NIH AG031867 (CCW), AG042095 (CCW), NS055980 (RHB), NS069669 (RHB), and NS075094 (MBH). The Muscular Dystrophy Association (CCW), the Myositis Association (CCW), and the Hope Center for Neurological Disorders (CCW and MBH). RHB holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund. Dr. Weihl had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Appendix: Supplementary material
Supplementary data to this article can be found online at doi:10.1016/j.nmd.2014.12.009.
References
- 1.Weihl CC, Pestronk A. Sporadic inclusion body myositis: possible pathogenesis inferred from biomarkers. Curr Opin Neurol. 2010;23:482–8. doi: 10.1097/WCO.0b013e32833d3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox FM, Titulaer MJ, Sont JK, Wintzen AR, Verschuuren JJ, Badrising UA. A 12-year follow-up in sporadic inclusion body myositis: an end stage with major disabilities. Brain. 2011;134:3167–75. doi: 10.1093/brain/awr217. [DOI] [PubMed] [Google Scholar]
- 3.Benveniste O, Guiguet M, Freebody J, et al. Long-term observational study of sporadic inclusion body myositis. Brain. 2011;134:3176–84. doi: 10.1093/brain/awr213. [DOI] [PubMed] [Google Scholar]
- 4.Askanas V, Engel WK, Alvarez RB. Light and electron microscopic localization of beta-amyloid protein in muscle biopsies of patients with inclusion-body myositis. Am J Pathol. 1992;141:31–6. [PMC free article] [PubMed] [Google Scholar]
- 5.Askanas V, Engel WK, Bilak M, Alvarez RB, Selkoe DJ. Twisted tubulofilaments of inclusion body myositis muscle resemble paired helical filaments of Alzheimer brain and contain hyperphosphorylated tau. Am J Pathol. 1994;144:177–87. [PMC free article] [PubMed] [Google Scholar]
- 6.Nogalska A, Terracciano C, D’Agostino C, King Engel W, Askanas V. p62/SQSTM1 is overexpressed and prominently accumulated in inclusions of sporadic inclusion-body myositis muscle fibers, and can help differentiating it from polymyositis and dermatomyositis. Acta Neuropathol. 2009;118:407–13. doi: 10.1007/s00401-009-0564-6. [DOI] [PubMed] [Google Scholar]
- 7.Weihl CC, Temiz P, Miller SE, et al. TDP-43 accumulation in inclusion body myopathy muscle suggests a common pathogenic mechanism with frontotemporal dementia. J Neurol Neurosurg Psychiatry. 2008;79:1186–9. doi: 10.1136/jnnp.2007.131334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson JO, Pioro EP, Boehringer A, et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci. 2014;17:664–6. doi: 10.1038/nn.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim HJ, Kim NC, Wang YD, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–73. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watts GD, Wymer J, Kovach MJ, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–81. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 11.Clemen CS, Herrmann H, Strelkov SV, Schroder R. Desminopathies: pathology and mechanisms. Acta Neuropathol. 2013;125:47–75. doi: 10.1007/s00401-012-1057-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Udd B. Distal myopathies – new genetic entities expand diagnostic challenge. Neuromuscul Disord. 2012;22:5–12. doi: 10.1016/j.nmd.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Cai H, Yabe I, Sato K, et al. Clinical, pathological, and genetic mutation analysis of sporadic inclusion body myositis in Japanese people. J Neurol. 2012;259:1913–22. doi: 10.1007/s00415-012-6439-0. [DOI] [PubMed] [Google Scholar]
- 14.Seelen M, Visser AE, Overste DJ, et al. No mutations in hnRNPA1 and hnRNPA2B1 in Dutch patients with amyotrophic lateral sclerosis, frontotemporal dementia, and inclusion body myopathy. Neurobiol Aging. 2014:35. doi: 10.1016/j.neurobiolaging.2014.01.152. [DOI] [PubMed] [Google Scholar]
- 15.Vasconcelos OM, Raju R, Dalakas MC. GNE mutations in an American family with quadriceps-sparing IBM and lack of mutations in s-IBM. Neurology. 2002;59:1776–9. doi: 10.1212/01.wnl.0000039780.13681.ad. [DOI] [PubMed] [Google Scholar]
- 16.Mezei MM, Mankodi A, Brais B, et al. Minimal expansion of the GCG repeat in the PABP2 gene does not predispose to sporadic inclusion body myositis. Neurology. 1999;52:669–70. doi: 10.1212/wnl.52.3.669. [DOI] [PubMed] [Google Scholar]
- 17.Badrising UA, Schreuder GM, Giphart MJ, et al. Associations with autoimmune disorders and HLA class I and II antigens in inclusion body myositis. Neurology. 2004;63:2396–8. doi: 10.1212/01.wnl.0000148588.15052.4c. [DOI] [PubMed] [Google Scholar]
- 18.Garlepp MJ, Tabarias H, van Bockxmeer FM, Zilko PJ, Laing B, Mastaglia FL. Apolipoprotein E epsilon 4 in inclusion body myositis. Ann Neurol. 1995;38:957–9. doi: 10.1002/ana.410380619. [DOI] [PubMed] [Google Scholar]
- 19.Harrington CR, Anderson JR, Chan KK. Apolipoprotein E type epsilon 4 allele frequency is not increased in patients with sporadic inclusion-body myositis. Neurosci Lett. 1995;183:35–8. doi: 10.1016/0304-3940(94)11108-u. [DOI] [PubMed] [Google Scholar]
- 20.Lattante S, Conte A, Zollino M, et al. Contribution of major amyotrophic lateral sclerosis genes to the etiology of sporadic disease. Neurology. 2012;79:66–72. doi: 10.1212/WNL.0b013e31825dceca. [DOI] [PubMed] [Google Scholar]
- 21.Nishino I, Carrillo-Carrasco N, Argov Z. GNE myopathy: current update and future therapy. J Neurol Neurosurg Psychiatry. 2014 doi: 10.1136/jnnp-2013-307051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huizing M, Rakocevic G, Sparks SE, et al. Hypoglycosylation of alpha-dystroglycan in patients with hereditary IBM due to GNE mutations. Mol Genet Metab. 2004;81:196–202. doi: 10.1016/j.ymgme.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 23.Harms MB, Neumann D, Benitez BA, et al. Parkinson disease is not associated with C9ORF72 repeat expansions. Neurobiol Aging. 2013:34. doi: 10.1016/j.neurobiolaging.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez MA, Feely SM, Speziani F, et al. A novel mutation in VCP causes Charcot-Marie-Tooth Type 2 disease. Brain. 2014 doi: 10.1093/brain/awu224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weihl CC, Dalal S, Pestronk A, Hanson PI. Inclusion body myopathy-associated mutations in p97/VCP impair endoplasmic reticulum-associated degradation. Hum Mol Genet. 2006;15:189–99. doi: 10.1093/hmg/ddi426. [DOI] [PubMed] [Google Scholar]
- 26.Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–60. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimonis VE, Fulchiero E, Vesa J, Watts G. VCP disease associated with myopathy, Paget disease of bone and frontotemporal dementia: review of a unique disorder. Biochim Biophys Acta. 2008;1782:744–8. doi: 10.1016/j.bbadis.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Rohrer JD, Warren JD, Reiman D, et al. A novel exon 2 I27V VCP variant is associated with dissimilar clinical syndromes. J Neurol. 2011;258:1494–6. doi: 10.1007/s00415-011-5966-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Majounie E, Traynor BJ, Chio A, et al. Mutational analysis of the VCP gene in Parkinson’s disease. Neurobiol Aging. 2012:33. doi: 10.1016/j.neurobiolaging.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubino E, Rainero I, Chio A, et al. SQSTM1 mutations in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Neurology. 2012;79:1556–62. doi: 10.1212/WNL.0b013e31826e25df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor MR, Slavov D, Ku L, et al. Prevalence of desmin mutations in dilated cardiomyopathy. Circulation. 2007;115:1244–51. doi: 10.1161/CIRCULATIONAHA.106.646778. [DOI] [PubMed] [Google Scholar]
- 32.Kostareva A, Sjoberg G, Gudkova A, et al. Desmin A213V substitution represents a rare polymorphism but not a mutation and is more prevalent in patients with heart dilation of various origins. Acta Myol. 2011;30:42–5. [PMC free article] [PubMed] [Google Scholar]
- 33.Goudeau B, Rodrigues-Lima F, Fischer D, et al. Variable pathogenic potentials of mutations located in the desmin alpha-helical domain. Hum Mutat. 2006;27:906–13. doi: 10.1002/humu.20351. [DOI] [PubMed] [Google Scholar]
- 34.Bar H, Goudeau B, Walde S, et al. Conspicuous involvement of desmin tail mutations in diverse cardiac and skeletal myopathies. Hum Mutat. 2007;28:374–86. doi: 10.1002/humu.20459. [DOI] [PubMed] [Google Scholar]
- 35.Olive M, Goldfarb LG, Shatunov A, Fischer D, Ferrer I. Myotilinopathy: refining the clinical and myopathological phenotype. Brain. 2005;128:2315–26. doi: 10.1093/brain/awh576. [DOI] [PubMed] [Google Scholar]
- 36.Friedrich FW, Wilding BR, Reischmann S, et al. Evidence for FHL1 as a novel disease gene for isolated hypertrophic cardiomyopathy. Hum Mol Genet. 2012;21:3237–54. doi: 10.1093/hmg/dds157. [DOI] [PubMed] [Google Scholar]
- 37.Schoser B, Goebel HH, Janisch I, et al. Consequences of mutations within the C terminus of the FHL1 gene. Neurology. 2009;73:543–51. doi: 10.1212/WNL.0b013e3181b2a4b3. [DOI] [PubMed] [Google Scholar]
- 38.Rose MR, Group EIW. 188th ENMC International Workshop: inclusion body myositis, 2–4 December 2011, Naarden, The Netherlands. Neuromuscul Disord. 2013;23:1044–55. doi: 10.1016/j.nmd.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 39.Griggs RC, Askanas V, DiMauro S, et al. Inclusion body myositis and myopathies. Ann Neurol. 1995;38:705–13. doi: 10.1002/ana.410380504. [DOI] [PubMed] [Google Scholar]
- 40.Weihl CC, Pestronk A, Kimonis VE. Valosin-containing protein disease: inclusion body myopathy with Paget’s disease of the bone and fronto-temporal dementia. Neuromuscul Disord. 2009;19:308–15. doi: 10.1016/j.nmd.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ju JS, Fuentealba RA, Miller SE, et al. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol. 2009;187:875–88. doi: 10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaleem M, Zhao A, Hamshere M, Myers AJ. Identification of a novel valosin-containing protein polymorphism in late-onset Alzheimer’s disease. Neurodegener Dis. 2007;4:376–81. doi: 10.1159/000105158. [DOI] [PubMed] [Google Scholar]
- 43.Larman HB, Salajegheh M, Nazareno R, et al. Cytosolic 5′-nucleotidase 1A autoimmunity in sporadic inclusion body myositis. Ann Neurol. 2013;73:408–18. doi: 10.1002/ana.23840. [DOI] [PubMed] [Google Scholar]
- 44.Pluk H, van Hoeve BJ, van Dooren SH, et al. Autoantibodies to cytosolic 5′-nucleotidase 1A in inclusion body myositis. Ann Neurol. 2013;73:397–407. doi: 10.1002/ana.23822. [DOI] [PubMed] [Google Scholar]
- 45.Niwa H, Ewens CA, Tsang C, Yeung HO, Zhang X, Freemont PS. The role of the N-domain in the ATPase activity of the mammalian AAA ATPase p97/VCP. J Biol Chem. 2012;287:8561–70. doi: 10.1074/jbc.M111.302778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lloyd TE, Mammen AL, Amato AA, Weiss MD, Needham M, Greenberg SA. Evaluation and construction of diagnostic criteria for inclusion body myositis. Neurology. 2014;83:426–33. doi: 10.1212/WNL.0000000000000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abramzon Y, Johnson JO, Scholz SW, et al. Valosin-containing protein (VCP) mutations in sporadic amyotrophic lateral sclerosis. Neurobiol Aging. 2012:33. doi: 10.1016/j.neurobiolaging.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.