Abstract
During spermiogenesis, the postmeiotic phase of mammalian spermatogenesis, transcription is progressively repressed as nuclei of haploid spermatids are compacted through a dramatic chromatin reorganization involving hyperacetylation and replacement of most histones with protamines. Although BRDT functions in transcription and histone removal in spermatids, it is unknown whether other BET family proteins play a role. Immunofluorescence of spermatogenic cells revealed BRD4 in a ring around the nuclei of spermatids containing hyperacetylated histones. The ring lies directly adjacent to the acroplaxome, the cytoskeletal base of the acrosome, previously linked to chromatin reorganization. The BRD4 ring does not form in acrosomal mutant mice. Chromatin immunoprecipitation followed by sequencing in spermatids revealed enrichment of BRD4 and acetylated histones at the promoters of active genes. BRD4 and BRDT show distinct and synergistic binding patterns, with a pronounced enrichment of BRD4 at spermatogenesis-specific genes. Direct association of BRD4 with acetylated H4 decreases in late spermatids as acetylated histones are removed from the condensing nucleus in a wave following the progressing acrosome. These data provide evidence of a prominent transcriptional role for BRD4 and suggest a possible removal mechanism for chromatin components from the genome via the progressing acrosome as transcription is repressed and chromatin is compacted during spermiogenesis.
INTRODUCTION
Mammalian spermatogenesis has emerged as a focus of epigenetic study, as this conserved process requires vast changes in transcription and chromatin organization (1). Spermatogenesis, or the formation of the mature male gamete, takes place in the seminiferous tubules of the testes and begins with the stem-like spermatogonia. Diploid spermatogonia can differentiate into spermatocytes, which enter meiosis to produce four genetically unique haploid round spermatids (see Fig. 1A). During the postmeiotic process of spermiogenesis, a spermatid differentiates into a motile spermatozoon by shedding most of its cytoplasm, forming a flagellum, and compacting the nucleus.
FIG 1.
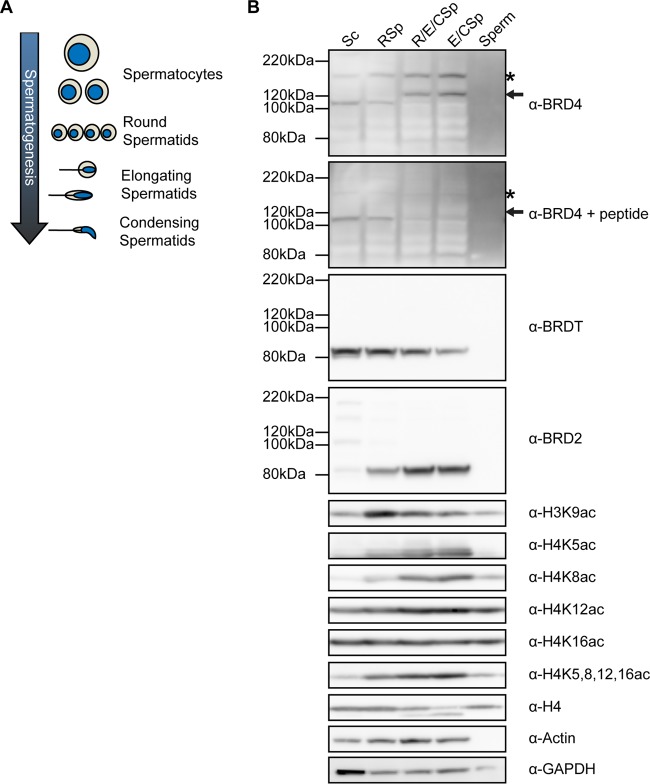
BRD4 is expressed in meiotic cells and spermatids but not in mature sperm. (A) Schematic of the progression of spermatogenesis beginning with meiotic cells (spermatocytes) and progressing through spermiogenesis from round to elongating to condensing spermatids. Changes in cell (tan) and nucleus (blue) size and shape are shown for reference in later figures. (B) Western blot analysis of whole-cell extracts from spermatocytes (Sc lane), round spermatids (RSp lane), elongating/condensing spermatids (E/CSp lane), a mixture of round, elongating, and condensing spermatids (R/E/CSp lane) obtained by STA-PUT velocity sedimentation, and mature sperm (Sperm lane). The asterisks mark the full-length BRD4 isoform, and the arrows indicate a novel shorter BRD4 peptide in spermatids. Peptide competition of BRD4 antibody shows the specificity of the BRD4 antibody.
In mice, nuclear morphology changes dramatically during spermiogenesis: the nucleus is initially round (in round spermatids), then elongates (in elongating spermatids), and finally condenses into a small, hook-like shape (in condensing/condensed spermatids) (see Fig. 1A). This process is necessary for the formation of fertile sperm and involves chromatin compaction and consequent vast transcriptional repression (1). Nuclear compaction is accomplished via nearly complete replacement of canonical histones, some with testis-specific histone variants, but most histones are initially replaced with transition proteins and then with protamines (2). Although several groups have shown that the small percentage of histones that remain associated with the genome in mature sperm are specifically posttranslationally modified and enriched at developmentally important loci (3, 4) and gene regulatory sequences (5), recent studies have provided contrasting evidence that in mouse sperm, histone retention occurs preferentially in large, gene-poor genomic regions (6–8). The mechanism by which almost all histones are removed and degraded has yet to be elucidated, but several important factors in this process have been discovered (9).
Chromatin reorganization during spermiogenesis begins concurrently with acrosome formation and histone hyperacetylation. The acrosome is a cap-like, membrane-bound organelle derived from the Golgi apparatus that covers the apical part of the mature sperm nucleus. This organelle contains digestive enzymes that are released upon contact with the egg to facilitate fertilization. Acrosome biogenesis begins after meiosis is complete and is accomplished via fusion of fragments of the Golgi apparatus at the acroplaxome or the cytoskeletal base of the forming acrosome (10). The acroplaxome consists of actin and keratin and anchors the acrosome to the adjacent nuclear membrane of the spermatid. Recently, several studies have linked acrosome biogenesis to the dramatic chromatin reorganization that takes place during spermiogenesis. Mouse mutants with aberrant acrosome formation produce abnormal, round-headed sperm that show defective nuclear compaction (11–15). Moreover, histone removal in human spermatids takes place adjacent to the acroplaxome as the acrosome progressively caps the nucleus (16). These studies suggest that acrosome biogenesis plays a role in sperm head shaping and nuclear compaction, but the mechanisms by which this may happen are unknown.
After meiosis is complete and acrosome formation has begun, histones become hyperacetylated in the spermatid nucleus (17–19). Histone hyperacetylation is believed to facilitate histone removal either through direct loosening of the chromatin or via binding of bromodomain-containing proteins such as PA200, the activator of the “spermatoproteasome,” and BRDT, the testis-specific BET (bromo- and extraterminal domain) family protein (20–23). Like all BET family members, BRDT contains two bromodomains at its N terminus and an extraterminal domain at its C terminus. Recently, BRDT has been shown to play a dual role during spermatogenesis (24). First, BRDT plays a transcriptional role, binding to acetylated histones and P-TEFb at the promoters of meiotic and postmeiotic genes that are aberrantly repressed in its absence (25). In fact, Brdt knockout or treatment of male mice with JQ1—a small-molecule inhibitor of BET family proteins—results in meiotic arrest and a significant decrease in fertility (25, 26). Second, BRDT may play a structural role in chromatin dynamics during spermiogenesis. Mice expressing BRDT lacking the first bromodomain show defects in fertility caused by abnormal nuclear compaction and chromatin organization during spermiogenesis (20, 27). However, it is unclear whether the defects during spermiogenesis are again due to transcriptional deregulation or rather to decreased binding of BRDT to hyperacetylated histones.
It is also unclear whether other members of the BET family are integral to the process of spermatogenesis, as Brd2 and Brd4 null mouse mutants show embryonic lethality (28, 29). However, these genes are expressed during spermatogenesis at the mRNA and protein levels (30). Interestingly, Brd4 heterozygous null mice show defects in spermatogenesis, although this phenotype has not been well characterized (29). BRD4 has been shown to bind to the acetylated tails of histones H3 and H4 and is generally associated with active gene transcription (31). However, BRD4 also plays nontranscriptional roles such as tethering the human papillomavirus genome to host chromatin during mitosis (32).
In this study, we investigated BRD4 during spermiogenesis. We showed that BRD4 is found in postmeiotic cells and investigated a novel BRD4 ring structure in spermatid nuclei that is closely associated with the acrosome. Moreover, we show by chromatin immunoprecipitation followed by massively parallel DNA sequencing (ChIP-seq) that BRD4 has an unanticipated prominent association with genes expressed in postmeiotic cells. Taken together, our results suggest an interesting mechanism for nuclear protein removal by linking BRD4 and transcription shutdown to acrosome formation during spermiogenesis.
MATERIALS AND METHODS
Antibodies and reagents.
The antibodies and reagents used for immunofluorescence (IF) assays were BRD2 (Abcam ab111078), BRD4 (33), BRDT (Abcam ab5157), H3K9Ac (Active Motif AM39137), H4K5,8,12,16ac (Millipore 05-1355), JQ1-PEG2-biotin (34), lamin B1 (Abcam ab16048 and Santa Cruz sc-373918), phalloidin-488 (Invitrogen A12379), and PNA-488 (Invitrogen L21409). The antibodies and reagents used for Western blot assays were β-actin (Cell Signaling 4970), BRD2 (Life Span Biosciences [amino acids 524 to 573] IHC-plus LS-B923), BRD4 (33), BRDT (Abcam ab5157), cyclin T1 (Santa Cruz H245 sc-10750), glyceraldehyde 3-phosphate dehydrogenase (Fitzgerald 10R-G109a), H3K9Ac (Active Motif AM39137), H4 (Abcam ab10158), H4K5ac (Millipore 07-327), H4K8Ac (Millipore 07-328), H4K12Ac (Millipore 07-595), H4K16Ac (Active Motif AM39167), and H4K5,8,12,16ac (Millipore 05-1355). The antibodies and reagents used for immunoprecipitation (IP) or ChIP were H3 (Abcam ab1791), H3K9ac (Active Motif AM39137), H3K9me3 (Abcam ab8898), H4K5ac (Millipore 07-327), H4K8ac (Millipore 07-328), H4K12ac (Millipore 07-595), H4K16ac (Active Motif AM39167), H4K5,8,12,16ac (Millipore 06-866), and BRD4 (Bethyl A301-985A50).
Mouse models.
All experiments requiring the use of laboratory mice were executed in compliance with all relevant guidelines, regulations, and regulatory agencies. Mouse experiments were conducted under the guidance and approval of the University of Pennsylvania Institutional Animal Care and Use Committee (IACUC). Male 129S6/SvEvTac mice (Taconic Biosciences, Inc., Germantown, NY) and mice with the Hrb gene disrupted (Hrb−/− mice) (11) were maintained and humanely euthanized according to the guidelines of the University of Pennsylvania IACUC.
Mouse spermatogenic cell fractionation.
Spermatogenic cell fractionation was performed by sedimentation of cells prepared from adult mouse testes through a bovine serum albumin (BSA) gradient as previously described (35). Each fractionation experiment used approximately 22 testes. Fractions were analyzed for purity on the basis of cell and nuclear morphology (via 4′,6-diamidino-2-phenylindole [DAPI] staining) and pooled. Mature spermatozoa were obtained from epididymides of adult mice, and contaminating cell types were eliminated by incubation in somatic cell lysis buffer (0.1% SDS, 0.5% Triton X-100 in diethylpyrocarbonate H2O) on ice for 20 min.
Western blot analysis.
Cells were resuspended in buffer (20 mM Tris [pH 7.5], 1 mM MgCl2, 1 mM CaCl2, 137 mM NaCl, 10% glycerol, 1% NP-40, EDTA-free Complete protease inhibitor [Roche], 300 nM trichostatin A [Sigma T1952]), and rotated at 4°C for 1 h in the presence of 12.5 U/ml Benzonase (Novagen 70746). Protein content was measured with Bradford dye, and cell lysate containing 25 μg of protein or sperm lysate containing 5 μg of protein was added to each well of a 4 to 12% 1-mm Bis-Tris NuPAGE protein gel. Protein was transferred to a polyvinylidene difluoride membrane and blocked for 1 h in 5% BSA–Tris-buffered saline–Tween 20 (BSA-TBST). Membranes were incubated with primary antibody (in 1% BSA-TBST) for 1 h, washed, incubated with a horseradish peroxidase-conjugated secondary antibody (in 1% BSA-TBST) for 30 min, washed, and detected by enhanced chemiluminescence. For peptide competition, the primary antibody was incubated with 5 μg/ml immunizing peptide for 1 h at room temperature before application to the membrane.
IF assay.
IF assay was performed with 8-μm cryosections of testes from 10-week-old mice or cell suspensions obtained by the spermatogenic cell separation method described above (35). Testes were dissected and immediately placed in 4% paraformaldehyde (PFA; in phosphate-buffered saline [PBS]) or snap-frozen in liquid nitrogen. The PFA-fixed testes were incubated in 15% and then 30% sucrose (in PBS) and embedded in Tissue-Tek O.C.T. compound. Fixed or frozen tissue was cryosectioned onto Superfrost Plus slides (Fisher). Cell suspensions in PBS were allowed to adhere to Superfrost Plus slides for 1 h at 33°C. Snap-frozen sections and cell suspensions were fixed for 15 min in 4% PFA at room temperature. Sections or cells were washed three times for 5 min each time in PBS and incubated with 125 mM glycine (in PBS) for 1 min at room temperature. Sections or cells were washed three times for 5 min each time in PBS and then permeabilized with 0.1% Triton X-100 for 2 min at 4°C. Sections or cells were washed three times for 5 min each time in PBS and then blocked with 3% donkey serum in PBS for 1 h at 37°C. Sections or cells were incubated with primary antibody or 40 μM JQ1-biotin (in 3% donkey serum in PBS) for 1 h at 37°C and washed three times for 5 min each time in PBS. Sections or cells were incubated with 5 μg/ml (in 3% donkey serum in PBS) Alexa Fluor-conjugated secondary antibody (Invitrogen) for 30 min at 37°C and then washed three times for 5 min each time in PBS. Sections or cells were then incubated with DAPI (Invitrogen; 5 μg/ml in 3% donkey serum in PBS) and embedded in Prolong Gold antifade reagent (Invitrogen). Sections or cells were imaged with a Leica TCS SP8 confocal microscope, and pseudo-three-dimensional (pseudo-3D) images were created with Velocity 6 software. Minimal changes to IF images (contrast and pseudocoloring) were made with ImageJ.
ChIP-seq.
ChIP-seq for BRD4 and histone posttranslational modifications (PTMs) was carried out as previously described, with minor modifications (36). Cells were cross-linked in 1% formaldehyde in PBS for 10 min at room temperature. The reaction was quenched with 125 mM glycine in PBS for 5 min at room temperature. After cell lysis, lysates were sonicated for 20 min with a Covaris S220 sonicator (5% duty cycle, 140 W of peak incident power, 200 cycles per burst). For each IP, 500 μg of protein (measured by bicinchoninic acid assay) from the cell lysate, 30 μl of protein G Dynabeads (Life Technologies), and 5 to 10 μg of antibody or IgG (Pierce 31235) were used. ChIP libraries for sequencing were prepared with 5 ng of DNA and the NEBNext Ultra DNA library prep kit for Illumina. Size selection was performed with AMPure XP beads (A63881; Beckman Coulter, Inc.). Libraries were sequenced with a NextSeq 500 machine (Illumina) in accordance with the manufacturer's protocol.
ChIP-seq data analysis.
ChIP-seq data generated with the NextSeq 500 machine were demultiplexed with the bcl2fastq utility (02.14.01.07). Data were then aligned with mouse genomic assembly mm9 by using Bowtie 0.12.7 (parameters -m 1 -best).
University of California Santa Cruz (UCSC) Genome Browser tracks.
Visual tracks of ChIP-seq data were generated in the following way. For each sample, the aligned data file for PCR-duplicated reads was filtered (i.e., any set of aligned reads with the same chromosome, start, and stop coordinates was reduced to a single representative). Coverage maps were then created with the BEDTools utility genomeCoverageBed. Resulting bedGraphs were scaled by using the RPKM (number of reads per kilobase per million sequenced reads) coefficient, a measure of the billions of bases sequenced per sample to correct for sequencing efficiency biases. Finally, an input coverage map was subtracted for the BRD4 coverage map and each histone PTM coverage map. The BRDT data from reference 25 were treated similarly.
Genome compartment plot.
The genomic compartment table was generated by first calling peaks for each histone PTM or BRD4 with SICER (peaks were called against the input as the background; window, fragment, and gap size parameters were fixed at 200 bp; and the false-discovery rate was controlled at 0.1%). Previously published peak locations from reference 25 were used for BRDT. Peak locations were overlapped with the BEDTools intersect utility by using RefSeq promoters, exons, and introns in an exclusive way: if a peak overlapped a promoter, it was removed from consideration for overlap exons or introns, and if it overlapped an exon, it was removed from consideration for overlap introns.
Expression versus ChIP enrichment heat map.
RefSeq transcripts were assessed for expression in round spermatids by loading two previously published replicate data sets (GSM95950 and GSM95951 in GEO series GSE4193) into the Partek Genomics Suite software package. Data were background corrected with gcrma, quantile normalized, and median polished along with all of the other data sets in the GSE4193 series. Promoters (1-kb upstream regions) associated with these expression-scored transcripts were assessed for numbers of aligned tags in each indicated ChIP. Tag counts were then normalized to the millions of tags sequenced, as well as the input. Brightness for each track is scaled to the maximum ChIP value in that track; tracks are sorted in the order of least expressed to most expressed genes in GSM95950.
Expression box plots.
The BRDT target genes used were as described in reference 25. BRD4 target genes were those with a SICER-determined BRD4 peak in the promoter where the peak enrichment was in the 90th percentile of all BRD4 peaks. For each of the three gene sets, expression enrichments were determined by using GSM95950 (data processing is described above) and ChIP-seq enrichments were determined by using normalized tag counts at the promoter (data processing is described above). The P values and W statistics (reflecting expression distribution and promoter acetylation distribution differences between genes bound by BRD4, BRDT, both, or neither) (see Table 2) were estimated by using the one-sided Mann-Whitney test in R.
TABLE 2.
| Fig. 6 panel and comparison | Histone PTM | W statistic | P value |
|---|---|---|---|
| Ba | |||
| BRD4 vs all genes | 2,683,292 | <2.2e−16 | |
| BRDT vs all genes | 1,729,944 | <2.2e−16 | |
| Cobound vs all genes | 378,363 | <2.2e−16 | |
| Cobound vs BRD4 | 181,821 | 2.6e−08 | |
| Cobound vs BRDT | 127,587 | 6.8e−11 | |
| Cb | |||
| BRD4 vs BRDT | H3K9ac | 1,403,278 | <2.2e−16 |
| BRD4 vs BRDT | H4K5ac | 1,484,792 | <2.2e−16 |
| BRD4 vs BRDT | H4K8ac | 1,466,692 | <2.2e−16 |
| BRD4 vs BRDT | H4K12ac | 1,468,998 | <2.2e−16 |
| BRD4 vs BRDT | H4K16ac | 1,444,708 | <2.2e−16 |
| BRD4 vs BRDT | H4Kac | 1,479,768 | <2.2e−16 |
| Cobound vs BRD4 | H3K9ac | 276,948.5 | 7.411e−16 |
| Cobound vs BRDT | H3K9ac | 158,058 | <2.2e−16 |
| Cobound vs BRD4 | H4K5ac | 273,017.5 | 2.435e−14 |
| Cobound vs BRDT | H4K5ac | 162,835.5 | <2.2e−16 |
| Cobound vs BRD4 | H4K8ac | 277,942.5 | 2.972e−16 |
| Cobound vs BRDT | H4K8ac | 162,476.5 | <2.2e−16 |
| Cobound vs BRD4 | H4K12ac | 272,689 | 3.232e−14 |
| Cobound vs BRDT | H4K12ac | 161,513.5 | <2.2e−16 |
| Cobound vs BRD4 | H4K16ac | 271,330.5 | 1.027e−13 |
| Cobound vs BRDT | H4K16ac | 158,641.5 | <2.2e−16 |
| Cobound vs BRD4 | H4Kac | 277,562 | 4.223e−16 |
| Cobound vs BRDT | H4Kac | 162,543 | <2.2e−16 |
Comparison of BRD4-bound, BRDT-bound, and cobound gene expression levels.
Comparison of BRD4-bound, BRDT-bound, and cobound gene promoter acetylation levels.
GO pie charts.
Gene ontology (GO) enrichment analysis of BRD4 target genes, BRDT target genes, and cobound genes was done with DAVID (37). GO terms in the “biological process” hierarchy were collapsed to a single representative term if they shared the same gene; if one GO term's associated target genes were a subset of another's, that GO term was dropped in favor of the other. GO terms were further combined if they had more than 15 transcripts in common. GO terms are represented by their overall gene “real estate.” Each gene is given a vote inversely proportional to the number of terms in which it appears. Each term's weight is the sum of the votes of its genes. Finally, pie charts were simplified by visual inspection.
Promoter heat map.
Genes enriched in the GO category “spermatogenesis” (GO:0007283) were associated with RefSeq transcripts with UniProt and DAVID. For each transcript, a vector describing the ChIP-seq enrichment profile around the transcriptional start site (TSS; 2.5 kb upstream and 2.5 kb downstream) was assessed for BRD4 and BRDT (ChIP-seq enrichment data were normalized to the input, length, and millions of reads as described above). Profiles were sorted from the top to the bottom by overall BRD4 enrichment intensity. The maximum green in each plot was determined by the enrichment value at the 90th percentile.
IP and MS.
Antibody-coupled beads were prepared by incubating 30 μl/IP protein G Dynabeads (Life Technologies) with 10 μg of the primary antibody or rabbit IgG (Pierce 31235) in 0.5% BSA-PBS for 6 h at 4°C with rotation. Cells were resuspended in lysis buffer (20 mM Tris [pH 7.5], 1 mM MgCl2, 1 mM CaCl2, 137 mM NaCl, 10% glycerol, 1% NP-40, EDTA-free Complete protease inhibitor [Roche], 10 mM NaB, 300 nM trichostatin A), after which 12.5 U/ml Benzonase was added. Lysates were incubated for 1 h at 4°C with rotation and then cleared by centrifugation at 14,000 rpm for 10 min. The supernatant was removed, and the protein concentration was measured with Bradford dye. After incubation with antibody, beads were washed three times with 1 ml of buffer. One milligram of protein from the lysate was added to the beads and incubated overnight at 4°C with rotation. Beads were washed five times with 1 ml of buffer. Beads were resuspended in 30 μl of sample buffer and incubated for 5 min at 90°C. The eluate was separated from the beads, separated on a 4 to 12% 1-mm Bis-Tris NuPAGE protein gel, and analyzed for enriched histone modifications compared to input samples by mass spectrometry (MS).
In-gel histone proteins were derivatized twice with a mixture of propionic anhydride and 100 mM ammonium bicarbonate (1:1) for 15 min under vigorous vortexing and then digested with 12.5 ng/μl of trypsin at room temperature overnight. Resultant histone peptides were extracted from the gel, repropionylated twice, and then desalted with C18-based homemade stage tips before MS analysis. Desalted peptides were separated by reverse-phase nanospray liquid chromatography (LC) with the Thermo Scientific Easy-nLC 1000 system and an in-house-packed C18 resin column (15-cm length, 3-μm particle size). Buffer A was water with 0.1% formic acid. Buffer B was acetonitrile with 0.1% formic acid. Histone peptides were eluted by gradients of 2 to 30% buffer B for 35 min and 30 to 98% buffer B for 20 min and then washed with 98% buffer B for 15 min at a flow rate of 200 nl/min. MS was performed with a Thermo Scientific Orbitrap Velos Pro hybrid ion trap-Orbitrap mass spectrometer. Each cycle included one full MS scan (m/z 290 to 1,400, resolution of 60,000, automatic gain control [AGC] target value of 1 × 106), followed by seven data-dependent MS2 scans of the most intense peptide ions using collision-induced dissociation (normalized collision energy of 35%, isolation width of 3 m/z, AGC target value of 1 × 104). In the section between 23 and 45 min, MS2 scans targeting precursor ions with m/z 528.30, 570.84, 768.95, 761.94, and 754.93 (isolation width of 1 m/z) were performed to determine acetylation sites on histone peptides with multiple lysines. Dynamic exclusion of 25 s was used to prevent repeated analysis of the same components. Ions with a charge state of one or more than four and a rejection list of common contaminant ions were excluded from the analysis. Histone peptides were identified on the basis of retention times and tandem MS (MS/MS). Abundance of histone peptides was quantified by integrating the area under each peak in the MS chromatogram with Thermo Scientific Xcalibur Qual Browser. The LC-MS/MS data sets were also analyzed with in-house-developed software as previously described (38).
Accession number.
All data sets in this study can be found at GEO accession number GSE56526.
RESULTS
BRD4 is expressed in meiotic cells and spermatids but not in mature sperm.
Although a transcriptional role for BRDT has been demonstrated during meiosis and spermiogenesis, it is unclear if other BET family proteins play a role in spermatogenesis (25). As mentioned above, Brd4 null heterozygous male mice show spermatogenic defects, suggesting that BRD4 may play a role in spermatogenesis (28, 29). To investigate the expression pattern of BRD4 over the course of spermatogenesis, we analyzed protein levels in different cell types obtained by STA-PUT velocity sedimentation from mature mouse testes (35). By this method, we collected four cell populations (Fig. 1A), (i) a mixture of meiotic cells (spermatocytes [Sc]), (ii) early postmeiotic spermatids (round spermatids [RSp]), (iii) later postmeiotic spermatids (elongating and condensing spermatids [E/CSp]), and (iv) a mixture of early and later spermatids (R/E/CSp). Mature sperm were isolated from the cauda epididymis of wild-type mice.
In addition to microscopic verification of purity via cellular and nuclear morphology (see reference 35 for our methods), we used Western blot analysis of lysates from these cells to confirm relative purity. Although H4 protein is depleted in late spermatids (E/CSp), H4K5ac, H4K8ac, H4K12ac, and H4K16ac, but not H3K9ac, were relatively enriched in these elongating and condensing spermatids (Fig. 1B). This analysis also revealed the presence of the long isoform of BRD4 protein (Fig. 1B, asterisk) in meiotic cells (Sc lane), round spermatids (RSp lane), and elongating/condensing spermatids (E/CSp lane) but not in mature sperm (Sperm lane). A second smaller BRD4 isoform or degradation product was also detected in later spermatids (Fig. 1B, upper panel, arrow). The specificity of the BRD4 antibody for the long and shorter forms was confirmed by peptide competition (Fig. 1B, second panel). The canonical BRD4 short isoform (∼723 amino acids) was not detected in any spermatogenic cells in this analysis. In addition to BRD4, BRDT and BRD2 are expressed over the course of spermatogenesis. While the BRDT expression level is highest in spermatocytes, BRD2 and BRD4 protein levels increase over the course of spermiogenesis.
BRD4 forms a ring around the spermatid nucleus as histones become hyperacetylated.
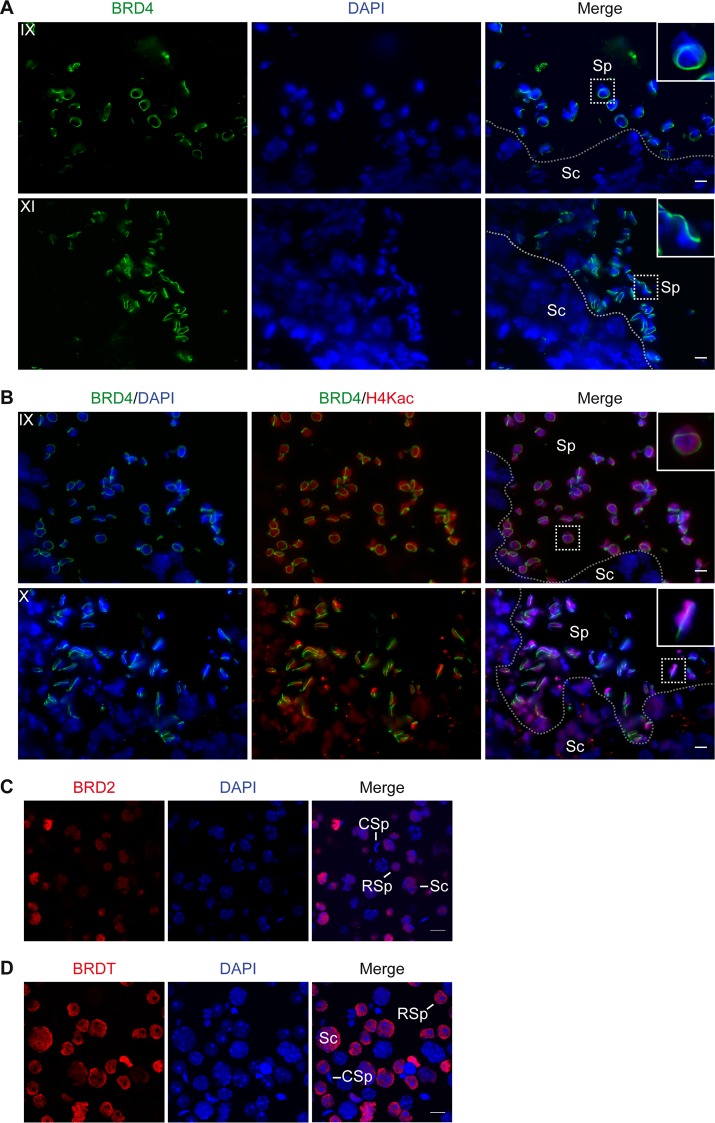
To gain insight into a possible function for BRD4 during spermatogenesis, we determined its subcellular localization by performing indirect IF assays of tissue sections from the testes of adult wild-type mice. Using intact tissue allows the identification of specific steps of spermatogenesis within the seminiferous tubules (39). Interestingly, we detected BRD4 in a distinct, complete ring around the nucleus beginning in approximately stage 7 to 8 spermatids (Sp) (Fig. 2A). The BRD4 ring is not present in spermatogonia or spermatocytes (Sc) (Fig. 2A) and appears concurrently with the postmeiotic hyperacetylation of histones in the nucleus, not seen in spermatocytes (compare Sp to Sc in Fig. 2B). We confirmed that the apparently distinct “ring” is not associated with the entire nuclear periphery by using pseudo-3D images created with z-stacked individual confocal images of early and late elongating spermatid nuclei (see Movies S1A and S1B in the supplemental material, respectively). We found that the BRD4 structure closely changes shape along with the condensing nucleus of the spermatid, from round to oblong, always at the periphery of the DAPI-stained nucleus (Fig. 2A and B; see Movies S1A and S1B in the supplemental material). This ring structure is specific to BRD4, as IF analysis of a mixed population of spermatogenic cells showed BRD2 and BRDT in a diffuse nuclear staining pattern in spermatocytes and round spermatids (Fig. 2C and D).
FIG 2.
BRD4 forms a ring around the spermatid nucleus as histones become hyperacetylated. (A) Indirect IF assay of cryosectioned mouse testis tissue shows that BRD4 (green) forms a ring around the early (top) to late (bottom) elongating spermatid nucleus (DAPI-stained DNA in blue). The ring is absent from all nonspermatid cell types, such as spermatocytes (Sc). (B) Indirect IF assay of cryosectioned mouse testis tissue shows that the BRD4 ring (green) forms at the onset of histone H4 hyperacetylation (red) in the nuclei (DAPI-stained DNA in blue) of early (top) to late (bottom) elongating spermatids. The stage of spermatogenesis is shown in the upper left corner of each panel. Separation of spermatocytes (Sc) and spermatids (Sp) within the seminiferous tubule is indicated with a dotted line. The inset shows a 3× magnification of the spermatids outlined with a dotted square. (C and D) Indirect IF assay of a mixed population of spermatogenic cells shows that BRD2 and BRDT (red) are diffusely localized in the nuclei (DAPI-stained DNA in blue) of spermatocytes (Sc) and round spermatids (RSp) but not condensing spermatids (CSp). Scale bars, 10 μm.
BRD4 forms a ring within the nuclear envelope at the base of the acrosome.
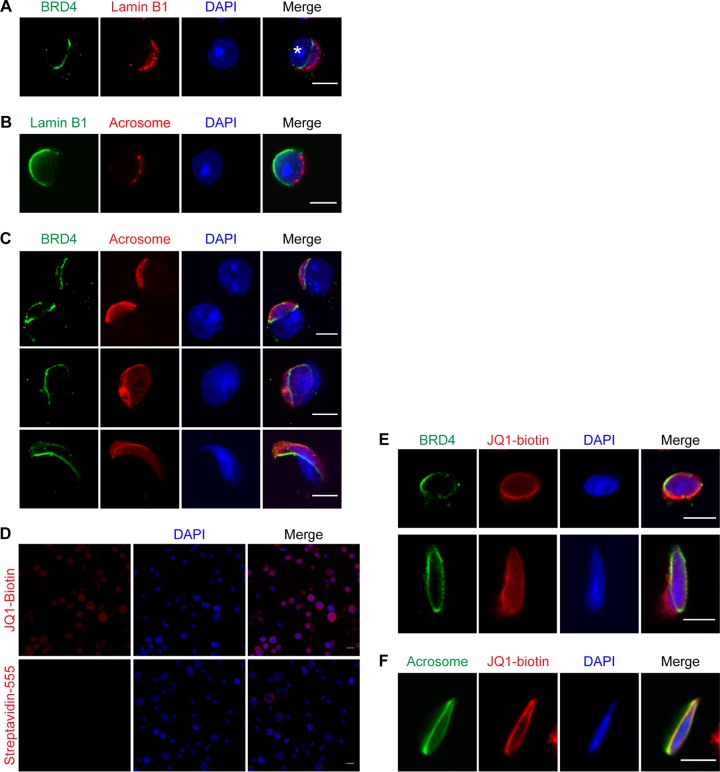
To further investigate the subcellular location of BRD4, we performed an indirect IF assay for BRD4 and lamin B1, a key component of the nuclear membrane. Codetection of these proteins revealed that BRD4 is located in an important transitional region of the nuclear membrane in spermatids (Fig. 3A). More specifically, lamin B1 and nuclear pores become polarized to the posterior end of the spermatid nuclear membrane, clearly distinct from the anterior end, which becomes closely covered by the acrosome (Fig. 3B; acrosome location indicated by an asterisk in Fig. 3A) (40–42). The acrosome is partly anchored to the nuclear envelope by a cytoskeletal plate called the acroplaxome, which forms a ring-like structure very similar to the BRD4 ring in the region where the lamin B1-associated nuclear envelope meets the acrosome-associated nuclear envelope (see Fig. 7D) (10). Therefore, we hypothesized that BRD4 may be associated with the acroplaxome.
FIG 3.
BRD4 forms a ring within the nuclear envelope at the base of the acrosome. (A) Indirect IF assay of BRD4 (green), lamin B1 (red), and DAPI-stained DNA (blue) in a round spermatid. The asterisk indicates the location of the acrosome. (B) Indirect IF assay of lamin B1 (green), the acrosome (detected with PNA [red]), and DAPI-stained DNA (blue) in a round spermatid. (C) Indirect IF assay of BRD4 (green), acrosome (detected with PNA in red), and DAPI-stained DNA (blue) in round (top), early elongating (middle), and condensing (bottom) spermatids. (D) Indirect IF assay of fluorophore-conjugated streptavidin (red) with (top) or without (bottom) JQ1-biotin and DAPI-stained DNA (blue) in a mixed population of spermatogenic cells. (E) Indirect IF assay of BRD4 (green), JQ1-biotin (red), and DAPI-stained DNA (blue) in late round (top) and elongating (bottom) spermatids. (F) Indirect IF assay of the acrosome (detected with PNA [green]), JQ1-biotin (red), and DAPI-stained DNA (blue) in a condensing spermatid. Scale bars, 5 (A to C, E, and F) and 10 (D) μm.
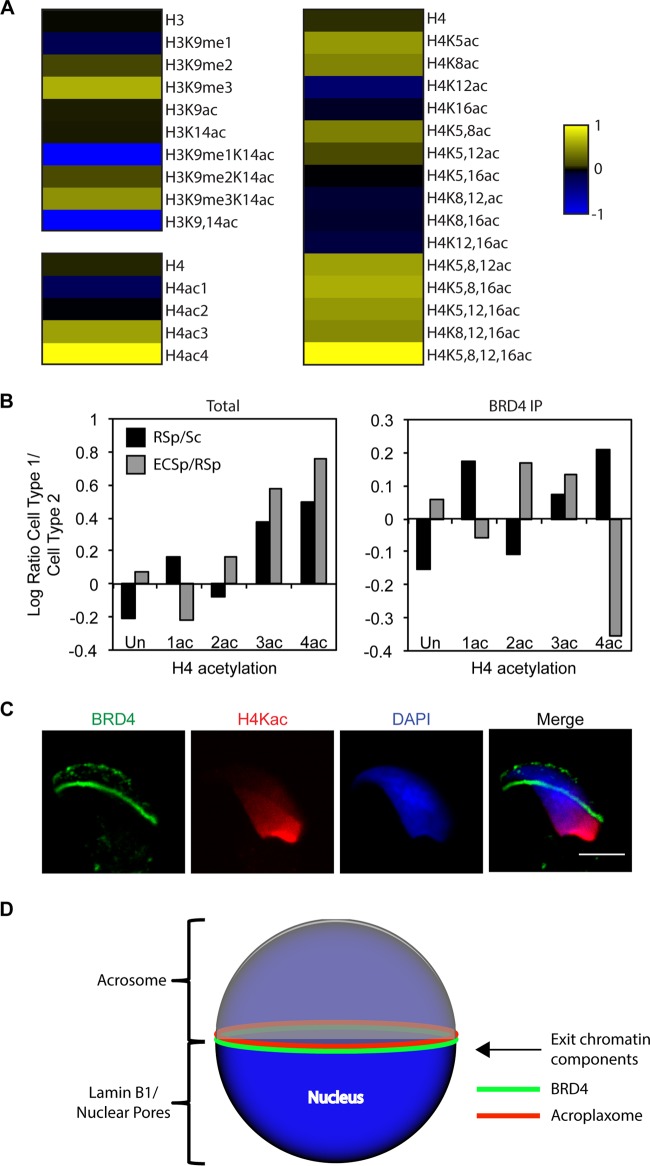
FIG 7.
BRD4 association with polyacetylated histone H4 diminishes in late spermatids as acetylated histones are removed from the condensing nucleus. (A) MS analysis of BRD4-immunoprecipitated histones from round spermatids. Heat map representation of enrichment of acetylation or methylation of histone H3 (amino acids 9 to 17, KSTGGKAPR) or H4 peptide (amino acids 4 to 17, GKGGKGLGKGGAKR) in BRD4-immunoprecipitated chromatin normalized to the total chromatin (log transformed). The raw values used to generate the heat maps are in Table 3. (B) Comparison of degrees of H4 peptide acetylation (amino acids 4 to 17, GKGGKGLGKGGAKR) in total and BRD4-immunoprecipitated (IP) chromatin over the course of spermatogenesis. Bars indicate changes in H4 acetylation from spermatocytes (Sc) to round spermatids (RSp) and from round spermatids to elongating/condensing spermatids (ECSp), respectively, via log-transformed ratio of percentages of acetylated H4 peptide. The raw values used to generate the graphs are in Table 4. (C) Indirect IF assay of BRD4 (green), H4K5,8,12,16ac (red), and DAPI-stained DNA (blue) in a condensing spermatid. Scale bar, 5 μm. (D) Model showing the locations of different cellular features associated with the spermatid nucleus. BRD4 forms a ring structure between the nucleus and the acroplaxome in a key region of the nuclear membrane where the lamin B1- and nuclear-pore-enriched posterior portion meets the acrosome-associated, nuclear-pore-depleted anterior portion.
To determine whether the BRD4 ring is linked to the acrosome/acroplaxome, we performed an IF assay of spermatogenic cells, probing with fluorophore-conjugated peanut agglutinin (PNA) to detect the acrosome or with phalloidin to detect actin in the acroplaxome (Fig. 3C; see Movie S1C in the supplemental material). Indeed, the BRD4 ring appears directly at the base of the acrosome during capping in late round spermatids (top) and persists in elongating (middle) and condensing spermatids (bottom) (Fig. 3C). Using a confocal microscope to create a pseudo-3D image, we discovered that the BRD4 ring lies just adjacent to the actin ring of the acroplaxome but closer to the DAPI-stained nucleus (see Movie S1C in the supplemental material).
To provide additional evidence of the acrosome-associated BRD4 ring, we incubated spermatogenic cells with JQ1-biotin, followed by fluorophore-conjugated streptavidin. JQ1 is a small molecule that binds to the bromodomains of all BET family proteins, which are expressed in the nuclei of multiple spermatogenic cell types (Fig. 2C and D) (30, 34, 43). Thus, JQ1-biotin shows a diffuse nuclear staining pattern in most spermatogenic cell types (Fig. 3D, top panel); however, this staining is not a random artifact of streptavidin binding (Fig. 3D, bottom panel). Importantly, in addition to a diffuse nuclear staining, JQ1-biotin is enriched in a ring structure that overlaps the BRD4 ring in spermatids (late round [top] and elongating [bottom] spermatids in Fig. 3E). Moreover, this JQ1-biotin enrichment is found at the base of the acrosome, providing additional evidence of the existence of the BRD4 ring (Fig. 3F).
Because the BRD4 ring and the acroplaxome are remarkably similar in shape and location, we hypothesized that acrosome formation is needed for BRD4 ring formation. To test this hypothesis, we analyzed Hrb (also known as Agfg1) null mice, which produce infertile sperm that lack acrosomes and have round, poorly compacted nuclei (11). In wild-type male mice, the acrosome is formed by the fusion at the acroplaxome of proacrosomic vesicles derived from the Golgi apparatus (Fig. 4A, top) (44). In Hrb−/− male mice, the proacrosomic vesicles begin to form in round spermatids but are unable to fuse properly to form a mature acrosome (Fig. 4A, bottom). Hrb−/− mice also show defects in nuclear elongation and compaction later in condensing spermatids when the acrosome is completely absent (Fig. 4A, bottom right).
FIG 4.
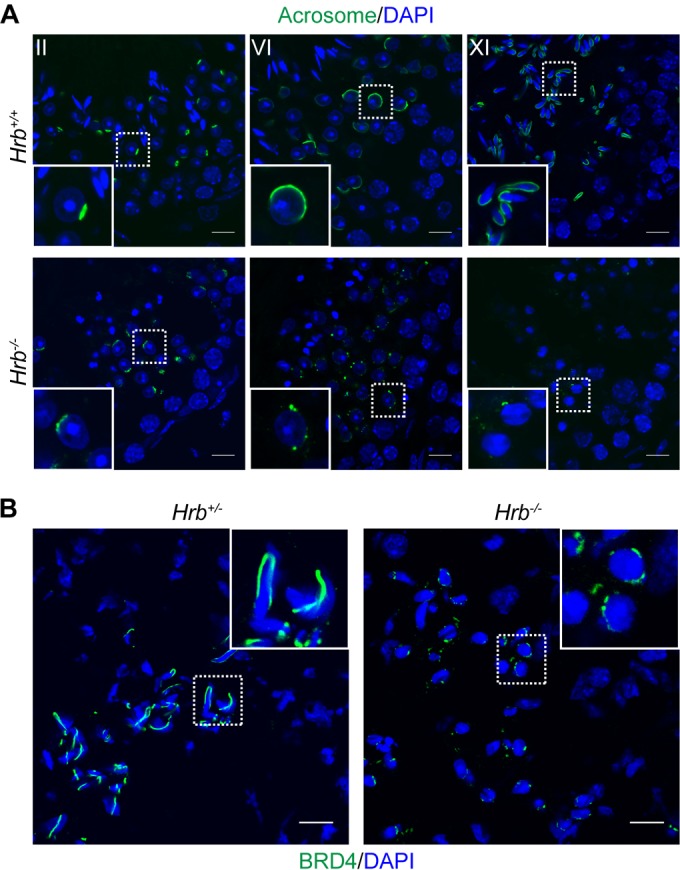
BRD4 does not form a ring in Hrb−/− acrosomal mutant mice. (A) Indirect IF assay of the acrosome (detected with PNA in green) and DAPI-stained DNA (blue) in testis tissue sections from wild-type and Hrb−/− mice. The stage of spermatogenesis is shown in the upper left corner of each panel. (B) Indirect IF assay of BRD4 (green) and DAPI-stained DNA (blue) in elongating spermatids of cryosectioned testis tissues from Hrb+/− (left) and Hrb−/− (right) mice. The inset shows a 3× magnification of the spermatids outlined with a dotted square. Scale bars, 10 μm.
To determine if acrosome development is required for BRD4 ring formation, we performed an indirect IF assay of cryosectioned testis tissue from adult Hrb+/− (which have normal spermatogenesis) and Hrb−/− mice (11). Strikingly, the BRD4 ring does not form properly in Hrb−/− mice (Fig. 4B, right side). A small amount of BRD4 can be seen around spermatid nuclei in Hrb−/− mice, but the conspicuous ring structure never develops and exists only in small fragments, if at all, in late-stage spermatids (Fig. 4B, right side). Because the acroplaxome forms partially, although aberrantly, in Hrb−/− acrosomal mutant mice, it is possible that the BRD4 ring is able to form partially because of an association with certain components of the acroplaxome that may be able to assemble in Hrb−/− mice despite the lack of a functional acrosome (45).
BRD4 is enriched at the promoters of active genes in spermatids.
It is possible that BRD4 is present diffusely throughout the nucleus in round spermatids, undetectable by IF assay until it is present in a higher concentration in the ring structure in very late round spermatids. To determine if BRD4 interacts with the genome in spermatids, we analyzed the genome-wide enrichment of BRD4 by ChIP-seq. We analyzed round spermatids to characterize BRD4 binding in the context of the distinct postmeiotic gene expression program, as a previous study did for BRDT (25). We also determined the genome-wide localization of various other histone PTMs to assess whether BRD4 shows a binding preference for any of these PTMs (all of which were normalized to the input). All of the ChIP-seq data alignment information is in Table 1.
TABLE 1.
ChIP-seq data alignment information
| ChIP | No. of reads |
% aligned | % uniquea | % genome coverage | ||
|---|---|---|---|---|---|---|
| Total | Aligned | Unique | ||||
| Input | 54,990,464 | 35,623,672 | 32,940,684 | 64.78 | 92.47 | 90.64 |
| H3 | 71,828,757 | 37,798,741 | 30,151,960 | 52.62 | 79.77 | 82.96 |
| H3K9me3 | 84,339,867 | 39,799,707 | 27,591,619 | 47.19 | 69.33 | 75.92 |
| H3K9ac | 67,856,707 | 48,595,679 | 34,042,406 | 71.62 | 70.05 | 93.67 |
| H4K5ac | 66,596,815 | 46,239,262 | 34,065,138 | 69.43 | 73.67 | 93.73 |
| H4K8ac | 79,795,471 | 48,838,021 | 29,541,448 | 61.20 | 60.49 | 81.28 |
| H4K12ac | 71,252,246 | 46,341,541 | 33,485,169 | 65.04 | 72.26 | 92.14 |
| H4K16ac | 77,287,145 | 49,326,598 | 28,973,938 | 63.82 | 58.74 | 79.72 |
| H4ac | 70,049,830 | 48,662,721 | 32,799,658 | 69.47 | 67.40 | 90.25 |
| BRD4 | 84,730,635 | 47,671,985 | 24,864,246 | 56.26 | 52.16 | 68.41 |
Total aligned reads compared to uniquely mapped reads for each ChIP-seq sample.
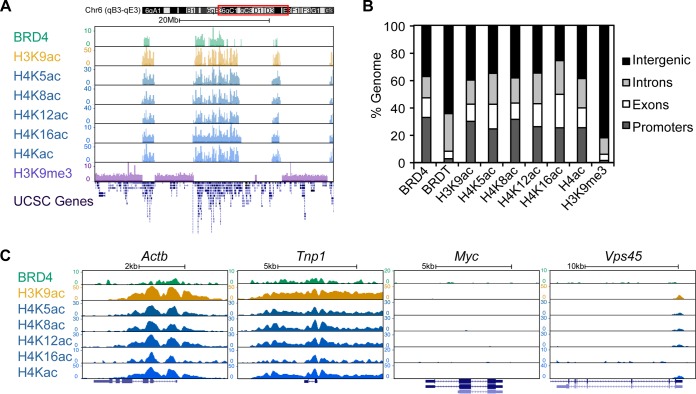
Upon initial examination of the ChIP-seq data, it is evident that BRD4 and H3 and H4 acetylation (H3/H4ac) are enriched in genic regions of the genome, as opposed to an established heterochromatin PTM, H3K9me3, which is enriched in large intergenic regions (Fig. 5A). Indeed, bioinformatic analysis revealed that the majority of the BRD4, H3ac, and H4ac peaks are located within promoters (1 kbp [kb] upstream of the TSS) or genes (introns or exons), unlike H3K9me3, which is located primarily in intergenic regions (Fig. 5B). Upon closer examination, BRD4 is enriched at the TSS of active housekeeping genes such as Actb and active spermatogenesis-specific genes such as Tnp1 (Fig. 5C). Conversely, BRD4 is not found at repressed housekeeping genes such as Myc (Fig. 5C). BRD4 is not bound to all active genes, even though some, such as Vps45, may be enriched for H3 or H4 acetylation (Fig. 5C).
FIG 5.
BRD4 is enriched at the promoters of active genes in round spermatids. (A) ChIP-seq of BRD4 and various histone PTMs reveals that BRD4 and histone H3 and H4 acetylation are associated with gene-rich regions of the genome while H3K9me3 is enriched in gene-poor intergenic regions. UCSC-defined genes are shown at the bottom. Genomic location is indicated at the top. The y axis is ChIP enrichment (normalized to the input). The x axis is DNA sequence. (B) Percentages of defined ChIP-seq peaks of BRD4, BRDT, and various histone PTMs in promoter (1 kb upstream of the TSS), gene (intron versus exon), and intergenic regions in round spermatids. (C) ChIP-seq of BRD4 and various histone PTMs at housekeeping (Actb) and spermatogenesis-specific (Tnp1) transcriptionally active genes and an inactive gene, Myc. BRD4 is not present at all gene promoters that are enriched for H3/H4 acetylation (Vps45). UCSC-defined genes are shown at the bottom in dark blue. The y axis is ChIP enrichment (normalized to the input). The x axis is DNA sequence.
In addition, we did not observe significant differences in enrichment patterns in acetylation at different H3 and H4 residues. In general, H3K9ac, H4K5ac, H4K8ac, H4K12ac, and H4K16ac appear to be enriched surprisingly similarly at the TSSs of active genes (Fig. 5A and C). Although BRD4 is not present at the TSSs of all active genes, heat map analysis revealed that levels of BRD4, H3K9ac, and H4ac at gene promoters (green in Fig. 6A) show a strong correlation with the transcriptional activity levels of those genes in round spermatids (red in Fig. 6A) (transcription data from references 46 and 47).
FIG 6.
BRD4 and BRDT bind to different subsets of transcriptionally active genes in round spermatids. (A) Heat map representation of ChIP-seq enrichment at promoters of all genes (green) compared to the levels of transcription (Txn) of these genes (red) in round spermatids. Brightness indicates higher levels of enrichment or transcription. (B) Box-and-whisker plot of transcription levels of genes that are bound by BRD4, BRDT, or both. Shown are the overall transcription levels of spermatogenesis-specific genes (Sperm.) and all genes, as a reference. (C) Box-and-whisker plot of enrichment levels of different histone PTMs at the gene promoters (1 kb upstream of the TSS) bound by BRD4, BRDT, or both (presence or absence of binding is indicated by a plus or minus sign, respectively, at the bottom). (D) Pie chart representations of GO terms of genes enriched for BRD4 (left), BRDT (middle), or both (right) in round spermatids. (E) ChIP-seq of BRD4, BRDT, and various histone PTMs at promoters of genes present in the topmost represented GO term category from the corresponding pie chart in panel D. UCSC-defined genes are shown at the bottom in blue. The y axis is ChIP enrichment (normalized to the input). The x axis is DNA sequence. (F) Heat map representation of BRD4 and BRDT enrichment around the TSS (±2.5 kb) of spermatogenesis-specific genes in round spermatids. Brightness indicates higher levels of enrichment.
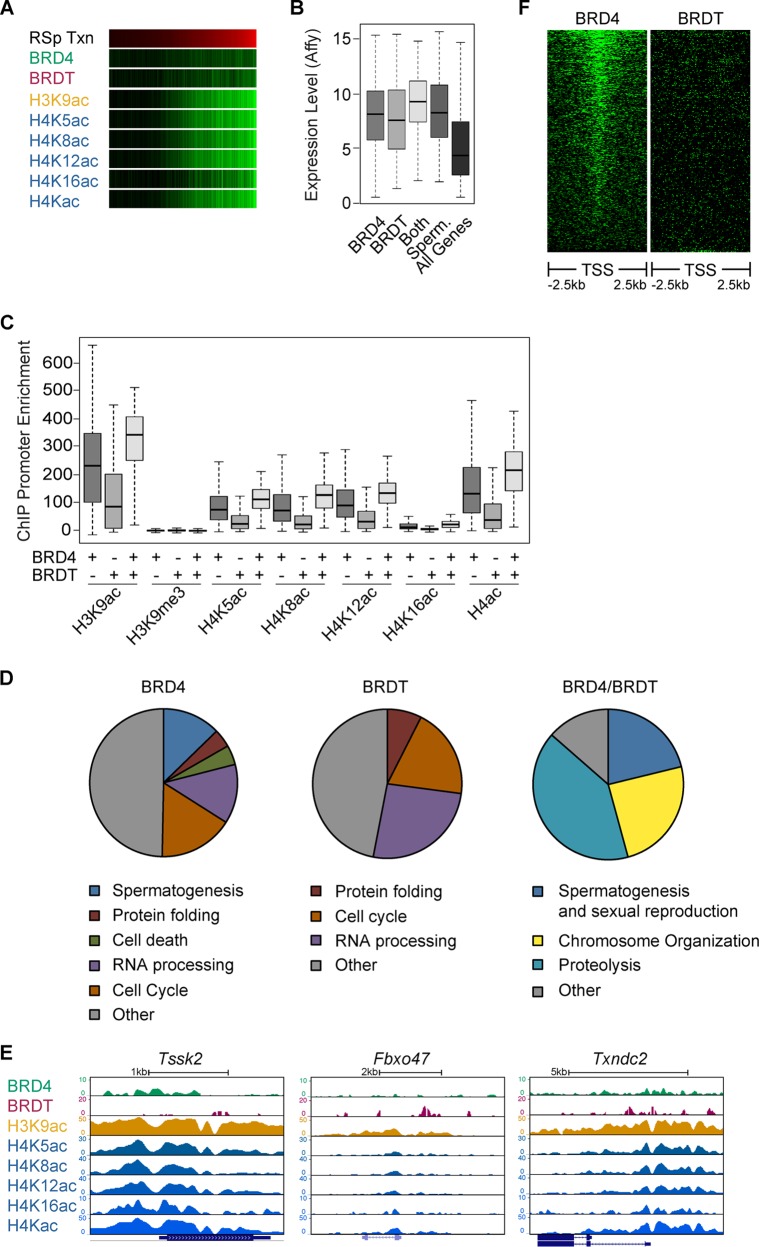
We compared our BRD4 ChIP-seq data in round spermatids to previously published BRDT ChIP-seq data obtained with the same cell type (25). Although the previous study showed that BRDT binds to the TSSs of active genes in round spermatids, we found the majority of the BRDT peaks (∼64%) to be present in intergenic regions of the genome and only 3% of the peaks within gene promoters (Fig. 5B). However, BRDT enrichment at gene promoters does correlate with the transcriptional activity of those genes (Fig. 6A). These data suggest that while most BRDT is found in intergenic regions, the small percentage located at gene promoters correlates well with transcriptional activity.
Because both BRD4 and BRDT were found to bind to the promoters of active genes in round spermatids, we sought to investigate possible differences between the roles of these BET family proteins. We defined a list of genes bound by BRD4 on the basis of the presence of peaks 1 kb upstream of the TSS. We then intersected this list with a list of approximately 1,544 genes previously shown to be bound by BRDT in round spermatids (25). Thus, we generated three categories of genes: BRD4 bound (approximately 2,093 genes), BRDT bound (approximately 1,347 genes), and cobound (approximately 197 genes). Genes bound only by BRD4 showed an average expression level slightly higher than that of genes bound only by BRDT (Fig. 6B). However, the average expression levels of BRD4- or BRDT-bound genes were approximately 2-fold higher than the average expression level of all genes and similar to the average expression level of spermatogenesis-specific genes (P < 2.2e−16 for BRD4 and BRDT bound in Fig. 6B). Interestingly, genes that were cobound by BRD4 and BRDT show the highest average transcription level (P < 2.2e−16 in Fig. 6B). These same trends can be seen in the levels of H3 and H4 acetylation at genes bound by BRD4, BRDT, or both (Fig. 6C). Histone H3/H4 acetylation levels are higher at the promoters of genes that are bound only by BRD4 but are highest at the promoters of genes that are cobound by BRD4 and BRDT (see Table 2 for P values in Fig. 6B and C). As a control, H3K9me3 enrichment is extremely low at the promoters of genes bound by BRD4 and BRDT.
Next, we performed a GO analysis of BRD4-bound, BRDT-bound, or cobound genes to determine if BRD4 and BRDT could possibly regulate different categories of genes. Categories of housekeeping genes such as “RNA processing” or “protein folding” can be found in genes bound by BRD4 or BRDT only (Fig. 6D and E, left two panels). However, spermatogenesis-specific genes are enriched in the set of genes bound by BRD4 only or cobound by BRD4 and BRDT (Fig. 6D and E, left and right panels). Indeed, heat map analysis of BRD4 and BRDT enrichment at the TSSs of all spermatogenesis-specific genes reveals a strong positioning of BRD4 (Fig. 6F). These data suggest that BRD4 and BRDT both play a role in the activation of transcription in postmeiotic spermatids but that BRD4 may play a particularly strong role in the activation of spermatogenesis-specific genes.
The necessity of BRD4 in transcriptional activation during spermiogenesis is difficult to determine in the current absence of a conditional-knockout mouse or a reliable spermatogenic cell culture system. Knockdowns in the germ line are extremely difficult to generate, and staged cell populations are not easily obtainable or manipulated in cell culture. Moreover, treatment of male mice with JQ1 results in a meiotic arrest before spermiogenesis (26). Thus, we are currently unable to directly test the function of BRD4 during spermiogenesis.
BRD4 association with polyacetylated histone H4 diminishes in late spermatids as acetylated histones are removed from the condensing nucleus.
Although our ChIP-seq data provide correlative evidence of BRD4 binding to both acetylated histone H3 and H4, they do not demonstrate direct binding to these histone PTMs. To investigate the composition of BRD4-associated chromatin, we performed IP of BRD4 in round spermatids (Fig. 7A). Total cell lysate and BRD4-immunoprecipitated proteins were separated by SDS-PAGE and analyzed by MS. We quantified the fraction of peptides bearing different combinations of acetyl and methyl PTMs for histones H3 and H4 in total chromatin and BRD4-immunoprecipitated chromatin (Table 3). When the values of immunoprecipitated peptides were normalized to the values of total chromatin in round spermatids, we noticed approximately 4- and 10-fold tri- and tetra-acetylated H4 peptide enrichment, respectively (log-transformed ratios in Fig. 7A, bottom left panel; Table 3 contains the raw values). Specifically, various combinations of tri- and tetra-acetylated H4 peptides (H4K5ac, H4K8ac, H4K12ac, and H4K16ac) were highly enriched in immunoprecipitated chromatin, but all of these most highly enriched combinations include H4K5ac or H4K8ac (Fig. 7A, right side, and Table 3). In general, H3K9ac is not enriched in BRD4-immunoprecipitated chromatin and H3K14ac is only slightly enriched when combined with different degrees of H3K9 methylation (Fig. 7A, upper left). The reason for the relative absence of H3 acetylation and higher H3K9me3 is not clear.
TABLE 3.
MS analysisa of BRD4-immunoprecipitated histones from round spermatids
| Peptide and modification | Avg IP/total | SD |
|---|---|---|
| H3 9-17 | ||
| None | 1.02 | 0.18 |
| H3K9me1 | 0.65 | 0.28 |
| H3K9me2 | 1.26 | 0.73 |
| H3K9me3 | 4.24 | 1.51 |
| H3K9ac | 1.11 | 1.58 |
| H3K14ac | 1.09 | 0.08 |
| H3K9me1K14ac | 0.09 | 0.13 |
| H3K9me2K14ac | 1.35 | 1.09 |
| H3K9me3K14ac | 3.04 | 2.97 |
| H3K9,14ac | 0.00 | 0.00 |
| H4 4-17 | ||
| None | 1.14 | 0.20 |
| H4Kac1 | 0.61 | 0.12 |
| H4Kac2 | 0.98 | 0.08 |
| H4Kac3 | 3.72 | 1.04 |
| H4Kac4 | 10.07 | 5.17 |
| None | 1.17 | 0.21 |
| H4K5ac | 3.31 | 2.85 |
| H4K8ac | 2.67 | 4.89 |
| H4K12ac | 0.48 | 0.51 |
| H4K16ac | 0.87 | 0.42 |
| H4K5,8ac | 2.54 | 0.96 |
| H4K5,12ac | 1.36 | 0.87 |
| H4K5,16ac | 0.99 | 0.33 |
| H4K8,12ac | 0.83 | 0.35 |
| H4K8,16ac | 0.85 | 0.24 |
| H4K12,16ac | 0.80 | 0.13 |
| H4K5,8,12ac | 3.73 | 2.69 |
| H4K5,8,16ac | 4.18 | 1.51 |
| H4K5,12,16ac | 3.30 | 2.68 |
| H4K8,12,16ac | 2.82 | 1.54 |
| H4K5,8,12,16ac | 10.93 | 6.10 |
Quantification of the degrees of acetylation or methylation of histone H3 (amino acids 9 to 17, KSTGGKAPR) or H4 (amino acids 4 to 17, GKGGKGLGKGGAKR) peptide. Shown are average ratios of the percentage of H3 or H4 peptide in BRD4-immunoprecipitated chromatin to the total chromatin. Standard deviations were calculated for two and four biological replicates for H3 and H4, respectively.
We then quantified the change in total and BRD4-immunoprecipitated H4 acetylation over the course of spermatogenesis (raw values are in Table 4). We generated ratios of H4 acetylation in round spermatids versus spermatocytes and compared these to the ratio of H4 acetylation in elongating/condensing spermatids versus round spermatids. First, tri- and tetra-acetylated H4 peptide levels increased significantly in total chromatin over the course of spermiogenesis (from spermatocytes to round spermatids to elongating/condensing spermatids) (Fig. 7B, left graph). Although levels of BRD4-immunopreceipitated mono-, tri-, and tetra-acetylated H4 increased from spermatocytes to round spermatids, the levels of tetra-acetylated H4 decreased significantly from round spermatids to elongating/condensing spermatids (Fig. 7B, right graph). Interestingly, the immunofluorescent signal of hyperacetylated histones was depleted from the nuclear region underlying the acrosome (see Movie S1D in the supplemental material), as was shown in human spermatids (16), and adjacent to the BRD4 ring (Fig. 7C). Taken together, these data suggest a localization of BRD4 from the genome to the ring structure, followed by removal of hyperacetylated histones from the genome as the acrosome caps the nucleus (model in Fig. 7D).
TABLE 4.
MS analysisa of BRD4-immunoprecipitated histone H4 during spermatogenesis
| Cell type and modification | Total avg % | Total % SD | BRD4 IP avg % | BRD4 IP % SD |
|---|---|---|---|---|
| Sc | ||||
| None | 48.75 | 0.89 | 53.01 | 18.06 |
| 1ac | 39.41 | 1.37 | 27.42 | 2.80 |
| 2ac | 10.45 | 2.52 | 9.88 | 8.31 |
| 3ac | 0.94 | 0.15 | 3.60 | 2.42 |
| 4ac | 0.45 | 0.10 | 6.09 | 4.52 |
| RSp | ||||
| None | 30.39 | 0.46 | 37.40 | 4.57 |
| 1ac | 57.27 | 1.28 | 40.78 | 2.51 |
| 2ac | 8.67 | 1.13 | 7.70 | 0.49 |
| 3ac | 2.26 | 0.01 | 4.27 | 1.52 |
| 4ac | 1.42 | 0.32 | 9.85 | 3.08 |
| E/CSp | ||||
| None | 35.91 | 0.72 | 42.74 | 7.08 |
| 1ac | 34.90 | 3.29 | 35.69 | 3.77 |
| 2ac | 12.51 | 0.63 | 11.43 | 1.50 |
| 3ac | 8.55 | 0.76 | 5.78 | 1.22 |
| 4ac | 8.12 | 1.18 | 4.35 | 0.59 |
Quantification of the degrees of acetylation (% total peptide) of histone H4 peptide (amino acids 4 to 17, GKGGKGLGKGGAKR) in total and BRD4-immunoprecipitated chromatin from spermatocytes (Sc), round spermatids (RSp), and elongating/condensing spermatids (E/CSp). Standard deviations were calculated for two biological replicates.
DISCUSSION
Mammalian spermatogenesis results in a specialized sperm cell with a highly compacted nucleus. A hallmark of this nuclear compaction is the removal of almost all histones from the genome, with a small percentage retained at developmentally important loci (3, 4) and repetitive DNA sequences (7, 8). While the mechanism of mass histone removal and degradation remains unclear, it is believed that histone hyperacetylation and thus bromodomain-containing proteins, especially BRDT, are integral to this process (2, 20, 21, 25, 27). Surprisingly little is known about the involvement of the other BET family members—BRD2, BRD3, and BRD4—during spermiogenesis. A previous study used immunohistochemical analysis of testis tissue to show BRD4 expression specifically in spermatogonia (30). In this study, however, we found that BRD4 is expressed during meiotic and postmeiotic phases of mouse spermatogenesis by several approaches. First, we detected BRD4 gene expression in meiotic and postmeiotic cells by Western blotting and reverse transcription-quantitative PCR (Fig. 1B and data not shown). Second, we immunoprecipitated BRD4 from meiotic and postmeiotic cells and detected its association with expressed genes and with acetylated histones (48) (Fig. 5 to 7). Thus, BRD4 protein is present during the postmeiotic phase of spermatogenesis.
Our characterization of BRD4 in spermatids provides evidence of an interesting mechanism by which transcription is attenuated by the progressive removal of BRD4 itself and acetylated histones via the acrosome. It is possible that this is a general mechanism for the removal of transcriptionally relevant proteins. We initially observed BRD4 in a novel ring-like structure that is closely associated, both spatially and functionally, with the acrosome/acroplaxome. However, our unprecedented combination of IP/MS and ChIP-seq analysis of BRD4 with endogenous antibodies demonstrated the in vivo binding of BRD4 throughout the chromatin of postmeiotic cells. Importantly, we performed these analyses with round spermatids, just before the BRD4 ring first appears and an increase in histone H4 acetylation can be detected by Western blot assay, IF assay, and MS (Fig. 1B, 2A and B, and 7B). Also, in this cell type, we were able to capture the chromatin state before the initiation of histone replacement, vast compaction, and transcriptional shutdown.
BRD4 has traditionally been associated with euchromatin, active transcription, or mitotic bookmarking (38, 49–54). Our ChIP-seq data suggest that BRD4 plays a similar role in transcriptional activation in round spermatids. BRD4, H3K9ac, and H4K5ac, H4K8ac, H4K12ac, and H4K16ac are present at the TSSs of active genes in round spermatids, and their enrichment correlates with transcription levels (Fig. 5 and 6A). While BRD4 is known to bind to polyacetylated histone H4 in vitro, our study is the first to confirm this preference for acetylated H4 (especially polyacetylated H4 modified at K5 and K8) over H3 by in vivo IP/MS with an antibody against endogenous BRD4 (Fig. 7A) (38, 55, 56).
When we compared our ChIP-seq data to those from a recent study of BRDT in the same cell type (25), we found that BRD4 and BRDT enrichment at gene promoters correlates with the transcriptional activity of those genes (Fig. 6A). BRD4 is especially enriched at spermatogenesis-specific genes and shows a very robust positioning around their TSSs, suggesting that this BET family protein may play a strong role in their activation (Fig. 6F). Interestingly, genes that were cobound by BRD4 and BRDT showed higher average transcription and histone acetylation levels than genes bound only by BRD4 or BRDT (Fig. 6B and C). It is unclear how BRD4, BRDT, or both would be recruited to specific subsets of genes, but these data suggest that synergistic binding of these two BET family proteins could lead to higher histone acetylation and expression levels via increased recruitment of the P-TEFb complex, as has been shown for both BRD4 and BRDT (25, 53, 54).
Although BRDT binds to a considerable number of gene promoters, we found that the majority of BRDT peaks occur in intergenic regions of the genome (Fig. 5B). Interestingly, a recent study demonstrated that nucleosomes that are retained in the mature mouse sperm genome show 10-fold overrepresentation in promoter regions (57). Because BRDT has also been implicated in the histone-to-protamine transition via removal of acetylated histones, we propose that BRDT may show a binding preference for regions where histones are largely evicted from the mature sperm genome, perhaps even as early as the round spermatid stage (20, 23, 25). In the future, it will be interesting to investigate the genome-wide binding patterns of other BET family proteins over the course of spermatogenesis.
Because transcription is largely deactivated as the genome is highly compacted during spermiogenesis, it is reasonable to postulate that BRD4 and other transcriptional coactivators must be removed from the genome during this process. Indeed, Western blot analysis showed that BRDT, BRD2, and BRD4 are largely absent from mature sperm (Fig. 1B). It is unclear, however, how histones and so many other chromatin components may be removed and/or degraded during this nuclear condensation. In approximately spermatid stage 7 to 8, when histone hyperacetylation and nuclear elongating/compaction begin, the BRD4 ring appears just adjacent to the acroplaxome (Fig. 1B, 2B, and 3C; see Movies S1A to S1C in the supplemental material). At the same stage of spermiogenesis, we observed by Western blot assay a BRD4 peptide with a slightly lower molecular weight than the full-length BRD4 isoform (Fig. 1B). It is possible that the BRD4 ring contains a spermatid-specific isoform that binds to acetylated histones or other acetylated chromatin-associated proteins in order to provide a tethering force (via the acroplaxome) for chromatin compaction and reorganization. Indeed, a growing body of evidence implicates BRD4 in structural roles such as tethering, insulating, and maintaining higher-order chromatin structure (58–61). Alternatively, because BRD4 is undetectable in mature sperm by IF and Western blot assays, this smaller BRD4 peptide may simply be a degradation product that is created during the removal of BRD4 from the genome via the acroplaxome-associated ring.
Interestingly, the BRD4 ring does not form in acrosomal mutant mice, which show nuclear compaction and fertility defects that are highly similar to human globozoospermia, a condition in which the acrosome is malformed or absent, the sperm head is round, and chromatin compaction is abnormal (Fig. 4) (11–15). Chromatin compaction during spermiogenesis may be incomplete in these mutant mice because of abnormal retention of chromatin-associated proteins and histones. Future ChIP-seq studies with acrosomal mutant mice will elucidate potential defects in the chromatin signature characteristic of spermiogenesis.
Our observations strongly support an increasing body of evidence that acrosome formation plays a key role in nuclear compaction and chromatin remodeling during spermiogenesis. Our mouse IF assay data (Fig. 7C; see Movie S1D in the supplemental material) and those of a recent study of human spermatids show that acetylated histones in the nucleus are depleted first in the region directly adjacent to the acrosome, where initial DNA compaction occurs (16). Moreover, various chromatin-associated proteins known to be involved in chromatin compaction, such as H1T2, are found in the nucleus adjacent to the acrosome (62). Finally, it has been suggested that the acroplaxome is able to provide contractile force to the compacting nucleus (10). It is unclear whether histones are degraded within the nucleus or shuttled out of the nucleus first during spermiogenesis; however, recent studies have shown that histone removal, while delayed, still occurs in mice lacking PA200, the acetylated histone-binding activator of the spermatogenesis-specific proteasome (21).
As histone removal, followed by degradation, appears to be the most-supported model, the acroplaxome is in an ideal location to facilitate this potential shuttling (22). It has been shown that the nuclear membrane underlying the expanding acrosomal cap is devoid of any nuclear pores and may be impenetrable to exiting nuclear components (42). If the acrosome is coupled to the extensive removal of histones and other chromatin components from the genome, the force-providing acroplaxome could act to facilitate shuttling at the base of the acrosome, where nuclear pores and lamins B1 and B3 are still present. Future studies, such as MS analysis, will provide critical insight into potential binding partners or PTMs of BRD4 that may localize this protein to the acroplaxome in spermatids. Regardless, this striking BRD4 localization suggests an interesting link between a chromatin component known to bind to acetylated histones and extranuclear spermatogenic structures.
Our combined approaches of immunofluorescence, biochemistry, MS, and ChIP-seq suggest that BRD4 may play a fundamental role in the transcription of spermatogenesis-specific genes and then in the transition from the transcriptionally active genome of early postmeiotic spermatids to the highly compact, transcriptionally silent genome of mature sperm. Importantly, our characterization of BRD4 contributes to the growing body of evidence that dramatic chromatin events taking place in the nucleus during spermiogenesis may be directly affected by extranuclear changes in cell structure and composition. Further investigation should reveal other proteins that are also involved in this process and elucidate the mechanism by which histones and other chromatin components are removed from the genome in an apparently acrosome-dependent manner. Finally, it would be interesting to further investigate how BET family proteins function during spermiogenesis first to activate transcription and then to repress it by potentially removing acetylated histones from the spermatid genome. The future study of various steps in this intricate process of transcriptional repression and nuclear compaction will lead to a better understanding of chromatin dynamics during spermatogenesis, epigenetic signatures in sperm, and mammalian fertility.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Berger lab, especially Parisha Shah, for all their support and advice. We thank Jérôme Govin for his guidance. We thank Saadi Khochbin for his advice and sharing his BRDT ChIP-seq data. We thank Jan Van Deursen for sharing his Hrb−/− mice. We thank James Bradner for sharing JQ1-biotin. We thank Andrea Stout of the Cell and Developmental Microscopy Core for her help with IF imaging. We thank Joseph Grubb and Jonathan Schug of the University of Pennsylvania Functional Genomics Core for their help with ChIP-seq.
Support to J.M.B. was from the T32 Genetics Training Grant at the University of Pennsylvania (GM008216). Support to S.L.B. was from NIH grants GM055360 and U54-HD068157. B.A.G. acknowledges funding from NIH grant GM110174 and Innovator grant DP2OD007447 from the Office of the Director. R.G.M. was supported by NIH grants R01HD048837 and U54HD068157.
We have no competing financial interests.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01328-14.
REFERENCES
- 1.Rajender S, Avery K, Agarwal A. 2011. Epigenetics, spermatogenesis and male infertility. Mutat Res 727:62–71. doi: 10.1016/j.mrrev.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Govin J, Caron C, Lestrat C, Rousseaux S, Khochbin S. 2004. The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur J Biochem 271:3459–3469. doi: 10.1111/j.1432-1033.2004.04266.x. [DOI] [PubMed] [Google Scholar]
- 3.Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schubeler D, Stadler MB, Peters AH. 2010. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol 17:679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- 4.Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. 2009. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, Platts AE, Saida M, Steger K, Tedder P, Miller D. 2009. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res 19:1338–1349. doi: 10.1101/gr.094953.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carone BR, Hung JH, Hainer SJ, Chou MT, Carone DM, Weng Z, Fazzio TG, Rando OJ. 2014. High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Dev Cell 30:11–22. doi: 10.1016/j.devcel.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer-Ficca ML, Lonchar JD, Ihara M, Bader JJ, Meyer RG. 2013. Alteration of poly(ADP-ribose) metabolism affects murine sperm nuclear architecture by impairing pericentric heterochromatin condensation. Chromosoma 122:319–335. doi: 10.1007/s00412-013-0416-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samans B, Yang Y, Krebs S, Sarode GV, Blum H, Reichenbach M, Wolf E, Steger K, Dansranjavin T, Schagdarsurengin U. 2014. Uniformity of nucleosome preservation pattern in mammalian sperm and its connection to repetitive DNA elements. Dev Cell 30:23–35. doi: 10.1016/j.devcel.2014.05.023. [DOI] [PubMed] [Google Scholar]
- 9.Gaucher J, Reynoird N, Montellier E, Boussouar F, Rousseaux S, Khochbin S. 2010. From meiosis to postmeiotic events: the secrets of histone disappearance. FEBS J 277:599–604. doi: 10.1111/j.1742-4658.2009.07504.x. [DOI] [PubMed] [Google Scholar]
- 10.Kierszenbaum AL, Tres LL. 2004. The acrosome-acroplaxome-manchette complex and the shaping of the spermatid head. Arch Histol Cytol 67:271–284. doi: 10.1679/aohc.67.271. [DOI] [PubMed] [Google Scholar]
- 11.Kang-Decker N, Mantchev GT, Juneja SC, McNiven MA, van Deursen JM. 2001. Lack of acrosome formation in Hrb-deficient mice. Science 294:1531–1533. doi: 10.1126/science.1063665. [DOI] [PubMed] [Google Scholar]
- 12.Fujihara Y, Satouh Y, Inoue N, Isotani A, Ikawa M, Okabe M. 2012. SPACA1-deficient male mice are infertile with abnormally shaped sperm heads reminiscent of globozoospermia. Development 139:3583–3589. doi: 10.1242/dev.081778. [DOI] [PubMed] [Google Scholar]
- 13.Lin YN, Roy A, Yan W, Burns KH, Matzuk MM. 2007. Loss of zona pellucida binding proteins in the acrosomal matrix disrupts acrosome biogenesis and sperm morphogenesis. Mol Cell Biol 27:6794–6805. doi: 10.1128/MCB.01029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao N, Kam C, Shen C, Jin W, Wang J, Lee KM, Jiang L, Xia J. 2009. PICK1 deficiency causes male infertility in mice by disrupting acrosome formation. J Clin Invest 119:802–812. doi: 10.1172/JCI36230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao R, Ito C, Natsume Y, Sugitani Y, Yamanaka H, Kuretake S, Yanagida K, Sato A, Toshimori K, Noda T. 2002. Lack of acrosome formation in mice lacking a Golgi protein, GOPC. Proc Natl Acad Sci U S A 99:11211–11216. doi: 10.1073/pnas.162027899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Vries M, Ramos L, Housein Z, De Boer P. 2012. Chromatin remodelling initiation during human spermiogenesis. Biol Open 1:446–457. doi: 10.1242/bio.2012844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimes SR Jr, Henderson N. 1984. Hyperacetylation of histone H4 in rat testis spermatids. Exp Cell Res 152:91–97. doi: 10.1016/0014-4827(84)90232-5. [DOI] [PubMed] [Google Scholar]
- 18.Meistrich ML, Trostle-Weige PK, Lin R, Bhatnagar YM, Allis CD. 1992. Highly acetylated H4 is associated with histone displacement in rat spermatids. Mol Reprod Dev 31:170–181. doi: 10.1002/mrd.1080310303. [DOI] [PubMed] [Google Scholar]
- 19.Govin J, Escoffier E, Rousseaux S, Kuhn L, Ferro M, Thevenon J, Catena R, Davidson I, Garin J, Khochbin S, Caron C. 2007. Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J Cell Biol 176:283–294. doi: 10.1083/jcb.200604141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shang E, Nickerson HD, Wen D, Wang X, Wolgemuth DJ. 2007. The first bromodomain of Brdt, a testis-specific member of the BET sub-family of double-bromodomain-containing proteins, is essential for male germ cell differentiation. Development 134:3507–3515. doi: 10.1242/dev.004481. [DOI] [PubMed] [Google Scholar]
- 21.Qian MX, Pang Y, Liu CH, Haratake K, Du BY, Ji DY, Wang GF, Zhu QQ, Song W, Yu Y, Zhang XX, Huang HT, Miao S, Chen LB, Zhang ZH, Liang YN, Liu S, Cha H, Yang D, Zhai Y, Komatsu T, Tsuruta F, Li H, Cao C, Li W, Li GH, Cheng Y, Chiba T, Wang L, Goldberg AL, Shen Y, Qiu XB. 2013. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell 153:1012–1024. doi: 10.1016/j.cell.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goudarzi A, Shiota H, Rousseaux S, Khochbin S. 2014. Genome-scale acetylation-dependent histone eviction during spermatogenesis. J Mol Biol 426:3342–3349. doi: 10.1016/j.jmb.2014.02.023. [DOI] [PubMed] [Google Scholar]
- 23.Pivot-Pajot C, Caron C, Govin J, Vion A, Rousseaux S, Khochbin S. 2003. Acetylation-dependent chromatin reorganization by BRDT, a testis-specific bromodomain-containing protein. Mol Cell Biol 23:5354–5365. doi: 10.1128/MCB.23.15.5354-5365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bryant JM, Berger SL. 2012. Low-hanging fruit: targeting Brdt in the testes. EMBO J 31:3788–3789. doi: 10.1038/emboj.2012.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaucher J, Boussouar F, Montellier E, Curtet S, Buchou T, Bertrand S, Hery P, Jounier S, Depaux A, Vitte A-L, Guardiola P, Pernet K, Debernardi A, Lopez F, Holota H, Imbert J, Wolgemuth DJ, Gérard M, Rousseaux S, Khochbin S. 2012. Bromodomain-dependent stage-specific male genome programming by Brdt. EMBO J 31:3809–3820. doi: 10.1038/emboj.2012.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matzuk Martin M, McKeown Michael R, Filippakopoulos P, Li Q, Ma L, Agno Julio E, Lemieux Madeleine E, Picaud S, Yu Richard N, Qi J, Knapp S, Bradner James E. 2012. Small-molecule inhibition of BRDT for male contraception. Cell 150:673–684. doi: 10.1016/j.cell.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berkovits BD, Wolgemuth DJ. 2011. The first bromodomain of the testis-specific double bromodomain protein Brdt is required for chromocenter organization that is modulated by genetic background. Dev Biol 360:358–368. doi: 10.1016/j.ydbio.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shang E, Wang X, Wen D, Greenberg DA, Wolgemuth DJ. 2009. Double bromodomain-containing gene Brd2 is essential for embryonic development in mouse. Dev Dyn 238:908–917. doi: 10.1002/dvdy.21911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Houzelstein D, Bullock SL, Lynch DE, Grigorieva EF, Wilson VA, Beddington RSP. 2002. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol Cell Biol 22:3794–3802. doi: 10.1128/MCB.22.11.3794-3802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shang E, Salazar G, Crowley TE, Wang X, Lopez RA, Wang X, Wolgemuth DJ. 2004. Identification of unique, differentiation stage-specific patterns of expression of the bromodomain-containing genes Brd2, Brd3, Brd4, and Brdt in the mouse testis. Gene Expr Patterns 4:513–519. doi: 10.1016/j.modgep.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Voigt P, Reinberg D. 2011. BRD4 jump-starts transcription after mitotic silencing. Genome Biol 12:133. doi: 10.1186/gb-2011-12-11-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBride AA, Jang MK. 2013. Current understanding of the role of the Brd4 protein in the papillomavirus lifecycle. Viruses 5:1374–1394. doi: 10.3390/v5061374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamonica JM, Deng W, Kadauke S, Campbell AE, Gamsjaeger R, Wang H, Cheng Y, Billin AN, Hardison RC, Mackay JP, Blobel GA. 2011. Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes. Proc Natl Acad Sci U S A 108:E159-168. doi: 10.1073/pnas.1102140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders L, Guenther MG, Qi J, Fan ZP, Marineau JJ, Rahl PB, Lovén J, Sigova AA, Smith WB, Lee TI, Bradner JE, Young RA. 2014. Genome-wide localization of small molecules. Nat Biotechnol 32:92–96. doi: 10.1038/nbt.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bryant JM, Meyer-Ficca ML, Dang VM, Berger SL, Meyer RG. 2013. Separation of spermatogenic cell types using STA-PUT velocity sedimentation. J Vis Exp 80:50648. doi: 10.3791/50648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shah PP, Donahue G, Otte GL, Capell BC, Nelson DM, Cao K, Aggarwala V, Cruickshanks HA, Rai TS, McBryan T, Gregory BD, Adams PD, Berger SL. 2013. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev 27:1787–1799. doi: 10.1101/gad.223834.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 38.LeRoy G, Chepelev I, Dimaggio PA, Blanco MA, Zee BM, Zhao K, Garcia BA. 2012. Proteogenomic characterization and mapping of nucleosomes decoded by Brd and HP1 proteins. Genome Biol 13:R68. doi: 10.1186/gb-2012-13-8-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meistrich ML, Hess RA. 2013. Assessment of spermatogenesis through staging of seminiferous tubules. Methods Mol Biol 927:299–307. doi: 10.1007/978-1-62703-038-0_27. [DOI] [PubMed] [Google Scholar]
- 40.Schütz W, Alsheimer M, Ollinger R, Benavente R. 2005. Nuclear envelope remodeling during mouse spermiogenesis: postmeiotic expression and redistribution of germline lamin B3. Exp Cell Res 307:285–291. doi: 10.1016/j.yexcr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 41.Vester B, Smith A, Krohne G, Benavente R. 1993. Presence of a nuclear lamina in pachytene spermatocytes of the rat. J Cell Sci 104(Pt 2):557–563. [DOI] [PubMed] [Google Scholar]
- 42.Ho HC. 2010. Redistribution of nuclear pores during formation of the redundant nuclear envelope in mouse spermatids. J Anat 216:525–532. doi: 10.1111/j.1469-7580.2009.01204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. 2010. Selective inhibition of BET bromodomains. Nature 468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abou-Haila A, Tulsiani DR. 2000. Mammalian sperm acrosome: formation, contents, and function. Arch Biochem Biophys 379:173–182. doi: 10.1006/abbi.2000.1880. [DOI] [PubMed] [Google Scholar]
- 45.Kierszenbaum AL, Tres LL, Rivkin E, Kang-Decker N, van Deursen JM. 2004. The acroplaxome is the docking site of Golgi-derived myosin Va/Rab27a/b-containing proacrosomal vesicles in wild-type and Hrb mutant mouse spermatids. Biol Reprod 70:1400–1410. doi: 10.1095/biolreprod.103.025346. [DOI] [PubMed] [Google Scholar]
- 46.Mulugeta Achame E, Wassenaar E, Hoogerbrugge JW, Sleddens-Linkels E, Ooms M, Sun ZW, van IWF, Grootegoed JA, Baarends WM. 2010. The ubiquitin-conjugating enzyme HR6B is required for maintenance of X chromosome silencing in mouse spermatocytes and spermatids. BMC Genomics 11:367. doi: 10.1186/1471-2164-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, Lee JT. 2006. Postmeiotic sex chromatin in the male germline of mice. Curr Biol 16:660–667. doi: 10.1016/j.cub.2006.01.066. [DOI] [PubMed] [Google Scholar]
- 48.Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dey A, Nishiyama A, Karpova T, McNally J, Ozato K. 2009. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol Biol Cell 20:4899–4909. doi: 10.1091/mbc.E09-05-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mochizuki K, Nishiyama A, Jang MK, Dey A, Ghosh A, Tamura T, Natsume H, Yao H, Ozato K. 2008. The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. J Biol Chem 283:9040–9048. doi: 10.1074/jbc.M707603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Z, He N, Zhou Q. 2008. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol 28:967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, Taylor MJ, Johns C, Chicas A, Mulloy JC, Kogan SC, Brown P, Valent P, Bradner JE, Lowe SW, Vakoc CR. 2011. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 54.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 55.Liu Y, Wang X, Zhang J, Huang H, Ding B, Wu J, Shi Y. 2008. Structural basis and binding properties of the second bromodomain of Brd4 with acetylated histone tails. Biochemistry 47:6403–6417. doi: 10.1021/bi8001659. [DOI] [PubMed] [Google Scholar]
- 56.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci U S A 100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Erkek S, Hisano M, Liang CY, Gill M, Murr R, Dieker J, Schubeler D, van der Vlag J, Stadler MB, Peters AH. 2013. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat Struct Mol Biol 20:868–875. doi: 10.1038/nsmb.2599. [DOI] [PubMed] [Google Scholar]
- 58.You J, Croyle JL, Nishimura A, Ozato K, Howley PM. 2004. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117:349–360. doi: 10.1016/S0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- 59.Baxter MK, McPhillips MG, Ozato K, McBride AA. 2005. The mitotic chromosome binding activity of the papillomavirus E2 protein correlates with interaction with the cellular chromosomal protein, Brd4. J Virol 79:4806–4818. doi: 10.1128/JVI.79.8.4806-4818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Floyd SR, Pacold ME, Huang Q, Clarke SM, Lam FC, Cannell IG, Bryson BD, Rameseder J, Lee MJ, Blake EJ, Fydrych A, Ho R, Greenberger BA, Chen GC, Maffa A, Del Rosario AM, Root DE, Carpenter AE, Hahn WC, Sabatini DM, Chen CC, White FM, Bradner JE, Yaffe MB. 2013. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature 498:246–250. doi: 10.1038/nature12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang R, Li Q, Helfer CM, Jiao J, You J. 2012. Bromodomain protein Brd4 associated with acetylated chromatin is important for maintenance of higher-order chromatin structure. J Biol Chem 287:10738–10752. doi: 10.1074/jbc.M111.323493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martianov I, Brancorsini S, Catena R, Gansmuller A, Kotaja N, Parvinen M, Sassone-Corsi P, Davidson I. 2005. Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc Natl Acad Sci U S A 102:2808–2813. doi: 10.1073/pnas.0406060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.