Abstract
Purpose
Most men with elevated serum prostate-specific antigen (PSA) levels and negative biopsies require repeat biopsy because of the lack of a sensitive and specific prostate cancer (CaP) detection test. This study evaluated the diagnostic potential of a duplex assay for CaP by quantifying transcript levels of α-methylacyl-CoA racemase (AMACR) and prostate-cancer antigen 3 (PCA3) in urine sediments following prostatic message.
Materials and Methods
Urine sediments from 92 patients, 43 with 49 without CaP were collected after digital rectal examination. Transcript levels of AMACR, PCA3, and PSA in total RNA isolated from these samples were determined by absolute qRT-PCR. AMACR and PCA3 scores were obtained by normalizing the transcript level to that of PSA for each sample and multiplying by 100.
Results
AMACR (p=0.006) and PCA3 (p=0.014) scores, but not serum PSA (p=0.306), discriminated specimens from patients with and without CaP, Receiver-operating-characteristic analysis established the diagnostic cutoff scores for the AMACR and PCA3 tests at 10.7 and 19.9, respectively. As determined from these cutoff scores, the AMACR test has 70% (95% CI, 56-83%) sensitivity and 71% (95% CI, 59-84%) specificity, whereas the PCA3 test has 72% (95% CI, 59-85%) sensitivity and 59% (95% CI, 45-73%) specificity for CaP detection. The combined use of AMACR and PCA3 scores in a dual-marker test increased sensitivity to 81% (95% CI, 70-93%) and specificity to 84% (95% CI, 73-94%).
Conclusions
Urinary AMACR and PCA3 tests were both superior to serum PSA test for detecting CaP. Their combined use in a dual-marker test further improved sensitivity and accuracy and could be used as a surveillance test after repeat negative prostate biopsies.
Keywords: prostate secretion, prostate-specific antigen, quantitative RT-PCR, urine assay, PCA3, biopsy
The widespread use of the serum prostate-specific antigen (PSA) test has led to the detection of prostate cancer (CaP) at an early stage, resulting in a dramatic decrease in CaP-related mortality.1,2 However, elevated serum PSA is not CaP-specific; this biomarker is frequently elevated under conditions that are not related to the presence of CaP, such as trauma, benign prostatic hyperplasia (BPH), and prostatitis.1,2 Increased serum concentrations of PSA from co-existing BPH also can mask the PSA emanating from small cancer foci; thus risk of CaP can be present at any level of serum PSA. Consequently, men with elevated serum PSA must undergo a biopsy to confirm or exclude the presence of CaP. Annually, more than 1 million men with elevated serum PSA underwent prostate biopsy, yet, among them, only one of four would be diagnosed with CaP. Thus, diagnostic assays with higher specificity need to be developed to reduce unnecessary biopsies. Such assays may also be used as surveillance tests, in lieu of biopsies, to follow patients with elevated serum PSA and repeat negative biopsies.
Several genes, including AMACR3,4 and PCA35,6, were found to be significantly over-expressed in CaP and therefore have been investigated as potential diagnostic markers for CaP. PCA3 is a non-coding messenger RNA expressed only in the prostate epithelium and the kidney.5,6 It has been tested as a urine marker for CaP detection in several large-scale studies, and results are promising, with a sensitivity of 58-82% and a specificity of 56-76%.7-9 AMACR encodes an enzyme that regulates peroxisomal beta-oxidation of phytol-derived, branched-chain fatty acids, first reported in 2000 to be overexpressed in CaP.10 Subsequent immunostaining of AMACR in prostate tissues revealed high sensitivity (83-90%) and a remarkable specificity of ∼100% for CaP detection in needle prostate biopsies.3,4 For this reason, immunostaining for AMACR alone or in combination with p63 and/or the high-molecular-weight cytokeratin 34βE12 is currently a standard adjuvant tool to help in the diagnosis of CaP in needle biopsies with ambiguous lesions.4,11 However, the detection of AMACR protein or transcripts in urine or urine sediments as a diagnostic marker for CaP is still limited to only three pilot studies12-14, including our own13, each of which involved fewer than 30 patients. Although results from these studies are encouraging, investigations involving larger cohorts of patients are needed to establish the diagnostic values of such assays. Furthermore, CaP is a highly heterogeneous cancer with variable gene-expression profiles; therefore, a combination of multiple biomarkers would be expected to identify cases that screening for a single marker failed to detect. In this study, two of the more promising biomarkers, AMACR and PCA3, were evaluated simultaneously for the first time using absolute qRT-PCR protocols to determine their individual and combined efficacies in the detection of CaP in 92 patients, 43 with and 49 without CaP.
Materials and Methods
Patients and urine sample
Patients were recruited from the urological clinic at the University of Cincinnati Medical Center. Patient characteristics and diagnostic information are listed in Table 1. After signing an informed consent statement under a study protocol approved by the Institutional Review Board, the patients were asked to provide a 20 to 50 ml urine specimen, including the initial portion of the urine, following a digital rectal examination. This was followed by ultrasound-guided prostate biopsy. Urine specimens were centrifuged immediately on site. The sediments were homogenized in 1 to 2 ml of Trizol reagents (Invitrogen, Carlsbad, CA), transported to the research laboratory, and stored at -80°C.
Table 1. Characteristics of patients.
| Prostate cancer | Non-prostate cancer* | |
|---|---|---|
| No. patients | 43 | 49 |
| Age (mean ± SEM) | 69.5 ± 10.5 | 61.7 ± 7.0 |
| Ethnicity: | ||
| White | 42 (97.7%) | 48 (98%) |
| Black | 1 (2.3%) | 1 (2%) |
| Serum PSA (ng/ml) | ||
| < 2.5 | 5 (11.6%) | 10 (20.4%) |
| 2.5 - 10 | 34 (79.1%) | 38 (77.6%) |
| <4.0 | 10 (23.3%) | 22 (44.9%) |
| 4.0 - 10 | 29 (67.4%) | 26 (53.1%) |
| >10 | 4 (7.0%) | 1 (2.0%) |
| Gleason score | ||
| 5-6 | 35 (83.7%) | |
| 7-8 | 7 (16.3%) |
Non-CaP status was determined in patients on a single 10-core biopsy
Note: Percents of patients in each category are shown in parentheses
RNA isolation and quantitative RT-PCR
The RNA isolation and reverse transcription to cDNA were performed as previously described.15 Glycogen (40 μg in 1 ml TRizol reagent) was added as a carrier for enhancing isolation recovery. RT-PCR primers are listed in Table 2 or described previously.15 Transcripts were measured in sediments using absolute quantitative protocols tailored and optimized for each transcript. Cloned plasmid DNA(s) containing each sequence to be amplified were constructed and used to generate a standard curve for quantification of each biomarker gene transcript. The copy number was determined according to the published formula in the instruction manual of Applied Biosystems (Foster City, CA). Assays were conducted with 2× TaqMan Universal PCR Master Mix in an ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). A pooled cDNA sample from multiple LNCaP cell cultures served as the internal control for amplification efficiency. GAPDH transcripts were used to normalize for the amount of cDNA, and PSA transcripts were used for normalization of the number of prostate epithelial cells in the various prostate sediments. All reactions were run in triplicate along with serial dilutions of the corresponding plasmid DNA for the corresponding biomarker to generate a standard curve for absolute quantification of a gene transcript.15 AMACR and PCA3 scores were obtained by normalizing the absolute number of AMACR transcripts or PCA3 transcripts to PSA transcripts and then multiplied by 100, respectively.
Table 2. Oligonucleotide primer sequences.
| Gene Name | GenBank Access # | Primer Name | Sequence (5′ – 3′) | Nucleotide Position |
|---|---|---|---|---|
| AMACR | AF047020 | IAx1-F0 | CGCGGTGTCATGGAGAAACT | 332-351 |
| IAx2-R4 | CTTCCTGACTGGCCAAATCC | 432-413 | ||
| PCA3 | AF103907 | DD3-F5b | CAATGGCAGGGGTGAGAAATAAGA | 275-298 |
| DD3-R3 | CTCCCAGGGATCTCTGTGCTT | 479–459 | ||
| PSA | NM001648 | PSAx1-F0 | GTGACGTGGATTGGTGCTGCA | 75 –95 |
| PSAx1-R1 | CTGGGGGTGCACCAGAACA | 215-197 |
Statistical analysis
The variance of each diagnostic method was stabilized by logarithmic transformation of the AMACR and the PCA3 scores. The association of AMACR score, PCA3 score, or serum PSA levels with CaP diagnosis was analyzed by univariate logistic regression. Receiver-operator-characteristic (ROC) analyses were performed to select optimal cutoff points and to evaluate the performance of each score (sensitivity= TP/(TP+FP); specificity= TN/(TN+FN); positive predictive value (PPV) = TP/number positives (including cancer and cancer-free), and negative predictive value (NPV) = TN/number negatives (including cancer and cancer-free)). For the joint distribution of AMACR and PCA3, sensitivity and specificity were based on the definitions that a ‘positive’ (‘negative’) score is that either AMACR or PCA3 or both are positive (negative). Joint sensitivity and specificity were calculated and compared to each score separately. All analyses and plotting were performed by statistical software SAS 9.1.3 (SAS Institute) and R€ (http://www.r-project.org).
Results
Total RNAs were extracted from urine sediments collected from 106 men. Quantitative RT-PCR for the housekeeping gene GAPDH and the prostate-specific gene PSA was performed to assess sufficiency of prostate epithelial cell RNA in specimens. Since our previous study13 showed the AMACR assay has the sensitivity of detecting a single CaP cells, any specimens with less than one copy of PSA transcript per qPCR reaction were considered to have insufficient prostate epithelial cells to yield meaning data, and were excluded from further study. Fourteen specimens or 13% of collected specimens were excluded. A total of 92 usable samples, 43 from patients with CaP and 49 from patients without CaP based on a very recent biopsy were used for analysis in this study.
Univariate logistic regression analysis was first used to evaluate the association between the AMACR score or the PCA3 score with the diagnosis of CaP. Both urinary AMACR scores (coefficient 0.419, p=0.006) and PCA3 scores (coefficient 0.305, p=0.014) significantly discriminated patients with CaP from those with a negative biopsy. In contrast, the serum PSA test (coefficient 0.24, p=0.306) showed no statistically significant differentiation between specimens with CaP and samples without CaP. The median AMACR score was 12.7 in specimens from men with CaP and 5.5 in those from men with a recent negative biopsy. The median PCA3 score was 88.5 in men with a positive biopsy and 15.2 in non-CaP samples.
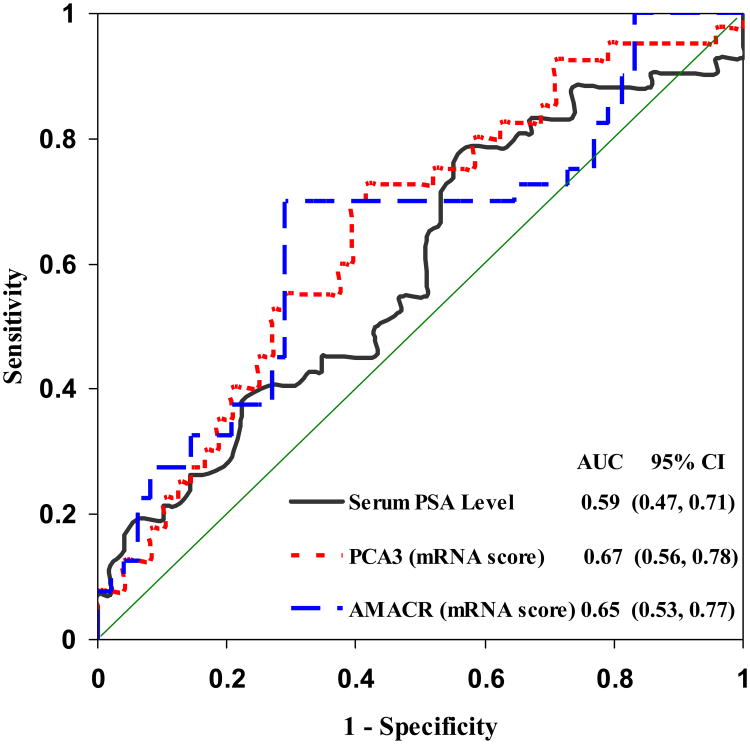
ROC analysis was then used to determine the diagnostic cutoff points for each test to best discriminate between CaP and non-CaP samples (Figure 1).16 The CaP diagnostic cutoff point for the AMACR test was 10.7 and that for the PCA3 test was 19.9. The area(s) under the curve (AUC) was 0.67 (95%CI, 0.56-0.78) (p<0.01) for the PCA3 and 0.65 (95% CI, 0.53-0.77) (p<0.01) for the AMACR test, values that were similar and significantly better than those of the serum PSA test (cutoff value = 4 ng/ml; AUC= 0.59, 95%CI, 0.47-0.71; p>0.05) for CaP detection. Samples with scores higher than the cutoff were classified as “cancer” and those lower than the cutoff, as “non-cancer” by the diagnostic marker in this study (see Supplement 1). Of the 43 specimens from patients with positive biopsies, 30 had AMACR scores ≥10.7 and were correctly diagnosed as “cancer,” thereby yielding a sensitivity of 70% (95% CI, 56-83%); 31 had a PCA3 score ≥19.9, yielding a sensitivity of 72% (95% CI, 59-85%, see Table 3). Of the 49 specimens from patients with a negative biopsy, the AMACR score correctly classified 35 cases in the “non-cancer” category (71% specificity, 95% CI, 59-84%) and the PCA3 score correctly placed 29 samples in the “non-cancer” category (59% specificity, 95% CI, 45-73%). The AMACR test had a PPV of 68% (95% CI, 54-82%) and a NPV of 73% (95% CI, 60-85%), and the PCA3 test had a PPV of 61% (95% CI, 47-74%) and a NPV of 71% (95% CI, 57-85%, see Table 3). In contrast, the serum PSA test had a specificity of 45% (95% CI, 31-59%) overall and of only 29% (95% CI, 16-41%) for men 60–69 years old in this cohort (see Supplement 2).
Figure 1.
Receiver-operating characteristic curves determined from univariate logistic regressions of each biomarker separately. The urinary PCA3 and AMACR scores and the serum PSA value were transformed as logarithmic scale for creating ROCs. The optimum cutoff points derived from the curve are 10.7 for the urinary AMACR score and 19.9 for urinary PCA3 score; AUC values are listed inside the figure. AMACR scores (dashed line), PCA3 scores (dotted line), and serum PSA values (successive line).
Table 3. Optimum values of sensitivity, specificity, positive predictive value, and negative predictive value from the ROC curves*.
| Diagnostic Method | Cutoff point | Sensitivity % (95% CI) | Specificity % (95% CI) | Positive predictive value (95% CI) | Negative predictive value (95% CI) |
|---|---|---|---|---|---|
| PSA (serum) | 4.0 | 77 (64-89) | 45 (31-59) | 55 (42-68) | 66 (49-82) |
| PCA3 (mRNA) | 19.9 | 72 (59-85) | 59 (45-73) | 61 (47-74) | 71 (57-85) |
| AMACR (mRNA) | 10.7 | 70 (56-83) | 71 (59-84) | 68 (54-82) | 73 (60-85) |
| Duplex assay** | 81 (70-93) | 84 (73-94) |
Each pair of optimum values were the point on the ROC which has the minimum distance to (0,1) or 1-Specificity=0 and Sensitivity=1.
Duplex AMACR-PCA3 assay classified a specimen as “cancer” if either the AMACR or PCA3 score was above their respective cutoff values and as “non-cancer” when both scores were below these values (see TABLE 4).
Next we examined whether the use of both AMACR and PCA3 scores would improve the accuracy/performance of such a dual-marker assay in CaP detection (Table 4). An additional five cancer cases were correctly diagnosed when specimens that had either a PCA3 score or an AMACR score above their respective diagnostic cutoff values were used (Table 4). Thus, the combined use of the two diagnostic markers with the “or” positive criteria identified 35/43 cancer patients correctly for a sensitivity of 81% (95% CI, 70-93%) and increased the specificity to 84% (95% CI, 73-94%). When both tests were negative, 23/49=47% (95% CI, 36-61%) non-cancer patients were correctly classified and 8/43 =19% (95% CI, 7-30%) patients with cancer were incorrectly classified. Interestingly, 8/49=16% (95% CI, 6-27%) patients with negative biopsies showed both AMACR and PCA3 scores above their respective cutoffs.
Table 4. Concordance analysis of AMACR versus PCA3 in prostate urine specimens.
| A. CaP urine specimen (n=43) | |||
|---|---|---|---|
|
| |||
| AMACR + | AMACR - | Sum | |
| PCA3+ | 26 | 5 | 31 |
| PCA3− | 4 | 8 | 12 |
| Sum | 30 | 13 | 43 |
| B. Non-CaP urine specimen (n=49) | |||
|---|---|---|---|
| AMACR - | AMACR + | Sum | |
| PCA3 - | 23 | 6 | 29 |
| PCA3 + | 12 | 8 | 20 |
| Sum | 35 | 14 | 49 |
| C. Summary of performance of different tests in all urine specimens | ||||
|---|---|---|---|---|
|
| ||||
| CaP, % (case/total) | Non-CaP, % (case/total) | |||
| AMACR + | 70 | (30/43) | 29 | (14/49) |
| PCA + | 72 | (31/43) | 41 | (20/49) |
| AMACR + or PCA3 + | 81 | (35/43) | 53 | (26/49) |
| AMACR - or PCA3 - | 40 | (17/43) | 84 | (41/49) |
| AMACR + and PCA3 + | 60 | (26/43) | 16 | (8/49) |
| AMACR - and PCA3 - | 19 | (8/43) | 47 | (23/49) |
Discussion
AMACR expression in CaP tissues has been investigated extensively in immunohistochemistry studies as a single marker or in an AMACR-p63-HWMK triple-marker test to improve the diagnosis of CaP in needle biopsies.3,4 However, since AMACR is localized primarily in the mitochondria or peroxisomes its development as a blood biomarker for CaP detection is thus unlikely. In contrast, the acini of the exocrine prostate communicates directly into the urethra, thus allowing AMACR or its transcripts to be detected from sloughed-off prostate epithelial cells in urine voids. In a feasibility study, western blot analysis detected AMACR in urine specimens of all 12 CaP patients (100% sensitivity) and in only 5 of the 12 non-CaP patients (42% specificity) after a biopsy.12 Two other studies focused on qRT-PCR-quantification of urinary AMACR transcript levels as diagnostic endpoints; ours reported a sensitivity of 70% (detected 7 of 10 CaP cases) and a specificity of 100% (9 CaP-free cases) when AMACR transcripts were measured in urine sediments after a digital rectal examination, but before a biopsy, was conducted.13,14 Zehentner et al used similar study design to reveal elevated AMACR transcript levels in 5 of 7 patients with T1-T2 CaP.14 However, in a large cohort of 234, Laxman et al found that the urinary AMACR transcript, as a single marker, did not have sufficient diagnostic power for CaP.17 Instead, they reported that the multiplexed quantification of GOLPH2, SPINK1, and PCA3 transcript expression and TMPRSS2:ERG status yielded an assay with a sensitivity of 66% and a specificity of 76%, thus showing promise for CaP detection. Taken together, these reports prompted us to re-evaluate the diagnostic potential of urinary AMACR transcript alone or in combination with the widely tested transcript of PCA3 in a duplex assay for CaP detection in a cohort of 92 patients. Today, published results for urinary PCA3 tests for CaP diagnosis give specificities from 58-82% and 56-76% respectively.7-9
In this study, both urinary AMACR score or PCA3 score used alone to diagnose CaP in 92 specimens (43 CaP, 48 non-CaP) exhibited good to excellent performance, with the AMACR test demonstrating a sensitivity of 70% and a specificity of 71% and the PCA3 test having a sensitivity of 72% and a specificity of 59% for CaP detection. Clearly, both assays outperformed serum PSA value as a CaP diagnostic marker with regard to their higher specificity. More significant, we demonstrated that the use of urinary AMACR and PCA3 scores in combination enhanced both the sensitivity and the specificity by 10% for both parameters to 81% and 84%, respectively. In such a combination test, the possibility that non-CaP patients have both an AMACR score and a PCA3 score above their respective cutoff point values is 16% whereas the possibility that CaP patients have negative results in both the AMACR and PCA3 assays is only 19%. Thus, if our results are validated independently, we envision that patients with both an AMACR score and a PCA3 score above cutoff point values would be strongly recommended for further evaluation, such as a prostate biopsy, while patients with both an AMACR and a PCA3 score below their respective cutoff point values would probably not need an immediate biopsy. The latter group could be followed with serum PSA tests in conjunction with the duplex AMACR-PCA3 urinary assay. This approach may have particular appeal for elderly patients or those with other health conditions who are not suited for frequent biopsies.
It has been reported that about 15-25% patients with a negative biopsy will have a positive CaP diagnosis in subsequent biopsies. 18 In the present study, we have identified 8 cases of “non-cancer” based on a single-negative biopsy result (i.e. 16% of the “non-cancer” group). However, based on the fact that these specimens all have both an AMACR score and a PCA3 score above their respective cutoff point values it is likely that they fall into the “false negative” group. With vigilant follow-ups, these cases may turn out to be CaP cases. In this regard, this assay may have utilities as an adjuvant assay to serum PSA for early detection of patients who have a normal biopsy. However, because tumor size and stage, infection, and the vigor of the prostatic massage may cause insufficient number of prostate epithelia cells shed to urine void, which could diminish the sensitivity of the assay. With the advent of new biomarker for CaP, inclusion of additional biomarker could further enhance this test.
Surprisingly, in our current study, the overall specificity for the serum PSA for diagnosis of CaP was close to 45% and significantly higher than values commonly documented in the literature (∼27% using 4 ng/ml as the cutpoint value).1,2 This apparent higher performance could be due to an over-representation of younger patients (<60 year olds) in our cohort. Once the relationships of serum PSA values to CaP were categorized according to age groups (Supplement 1), the apparently better performance of the assay than in the published studies disappeared. Indeed, similar to the literature, the specificity of serum PSA reached only 29% in men between 61 and 69 years old in our cohort.1,2 Our finding and previously reported data indicated that the performance of serum PSA value, as a single diagnostic marker, for CaP detection is poor in men older than 60 years because >50% would have some degree of BPH.1,2 Thus, repeat biopsy in older men with abnormal serum PSA commonly are negative for CaP. These outcomes strongly support the development of a minimally invasive surveillance/auxiliary test for CaP, in lieu of biopsies, to follow older patients who have elevated serum PSA levels but repeat negative biopsies.
Obviously, the future trend for CaP detection and management will include the continued improvement of minimal invasive diagnostic or surveillance assays, such as the one described here, using a combination of specific biomarkers to achieve high performance. A careful selection of biomarkers and independent validations with larger cohorts are crucial in achieving this goal. The use of such an assay as an adjuvant to the serum PSA test is expected to improve routine care in specific elderly populations and reduce health and financial costs of unnecessary prostate biopsies.
Conclusion
We report for the first time the utility of the combination of two of the most extensively studied CaP biomarkers, AMACR and PCA3, for CaP detection from urine specimens. As single-marker assays, the urinary AMACR and PAC3 tests had similar specificities, but the AMACR assay had a higher specificity. However, a duplex assay based on the use of both the AMACR and the PCA scores greatly improved the accuracy/performance of the assay. This duplex assay, if independently validated, may serve as a surveillance/adjuvant assay to the serum PSA test for monitoring elderly patients with repeat negative biopsies.
Supplementary Material
Supplement 1. Quantification of transcripts of AMACR, PCA3 in urinary sediments by qPCR
Supplement 2. Sensitivity and specificity of biomarker assays in different age groups
Acknowledgments
We are grateful to Barbara Bracken for assistance in patient information enlistment and sample collection and Eric Smith and Chensi Ouyang for critical reading of the manuscript. Research is supported in part by an award from the Department of Defense Prostate Cancer Research Program # W81XWH-06-1-0433 (SMH) and a NIH grant ES006096 (SMH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Etzioni R. Statistical issues in the evaluation of screening and early detection modalities. Urol Oncol. 2008;26:308. doi: 10.1016/j.urolonc.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 2.Stamey TA, Caldwell M, McNeal JE, Nolley R, Hemenez M, Downs J. The prostate specific antigen era in the United States is over for prostate cancer: what happened in the last 20 years? J Urol. 2004;172:1297. doi: 10.1097/01.ju.0000139993.51181.5d. [DOI] [PubMed] [Google Scholar]
- 3.Jiang Z, Woda BA. Diagnostic utility of alpha-methylacyl CoA racemase (P504S) on prostate needle biopsy. Adv Anat Pathol. 2004;11:316. doi: 10.1097/01.pap.0000146924.14246.be. [DOI] [PubMed] [Google Scholar]
- 4.Adley BP, Yang XJ. Application of alpha-methylacyl coenzyme A racemase immunohistochemistry in the diagnosis of prostate cancer: a review. Anal Quant Cytol Histol. 2006;28:1. [PubMed] [Google Scholar]
- 5.Bussemakers MJ, van Bokhoven A, Verhaegh GW, Smit FP, Karthaus HF, Schalken JA, et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59:5975. [PubMed] [Google Scholar]
- 6.de Kok JB, Verhaegh GW, Roelofs RW, Hessels D, Kiemeney LA, Aalders TW, et al. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002;62:2695. [PubMed] [Google Scholar]
- 7.Tinzl M, Marberger M, Horvath S, Chypre C. DD3PCA3 RNA analysis in urine--a new perspective for detecting prostate cancer. Eur Urol. 2004;46:182. doi: 10.1016/j.eururo.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 8.van Gils MP, Hessels D, van Hooij O, Jannink SA, Peelen WP, Hanssen SL, et al. The time-resolved fluorescence-based PCA3 test on urinary sediments after digital rectal examination; a Dutch multicenter validation of the diagnostic performance. Clin Cancer Res. 2007;13:939. doi: 10.1158/1078-0432.CCR-06-2679. [DOI] [PubMed] [Google Scholar]
- 9.Marks LS, Fradet Y, Deras IL, Blasé A, Mathis J, Aubin SM, et al. PCA3 molecular urine assay for prostate cancer in men undergoing repeat biopsy. Urology. 2007;69:532. doi: 10.1016/j.urology.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 10.Xu J, Stolk JA, Zhang X, Silva SJ, Houghton RL, Matsumura M, et al. Identification of differentially expressed genes in human prostate cancer using subtraction and microarray. Cancer Res. 2000;60:1677. [PubMed] [Google Scholar]
- 11.Molinié V, Hervé JM, Lugagne PM, Lebret T, Botto H. Diagnostic utility of a p63/alpha-methyl coenzyme A racemase (p504s) cocktail in ambiguous lesions of the prostate upon needle biopsy. BJU Int. 2006;97:1109. doi: 10.1111/j.1464-410X.2006.06069.x. [DOI] [PubMed] [Google Scholar]
- 12.Rogers CG, Yan G, Zha S, Gonzalgo ML, Isaacs WB, Luo J, et al. Prostate cancer detection on urinalysis for alpha methylacyl coenzyme a racemase protein. J Urol. 2004;172:1501. doi: 10.1097/01.ju.0000137659.53129.14. [DOI] [PubMed] [Google Scholar]
- 13.Zielie PJ, Mobley JA, Ebb RG, Jiang Z, Blute RD, Ho SM. A novel diagnostic test for prostate cancer emerges from the determination of alpha-methylacyl-coenzyme a racemase in prostatic secretions. J Urol. 2004;172:1130. doi: 10.1097/01.ju.0000133560.87118.4d. [DOI] [PubMed] [Google Scholar]
- 14.Zehentner BK, Secrist H, Zhang X, Hayes DC, Ostenson R, Goodman G, et al. Detection of alpha-methylacyl-coenzyme-A racemase transcripts in blood and urine samples of prostate cancer patients. Mol Diagn Ther. 2006;10:397. doi: 10.1007/BF03256217. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Leung YK, Ho SM. AP-2 regulates the transcription of estrogen receptor (ER)-beta by acting through a methylation hotspot of the 0N promoter in prostate cancer cells. Oncogene. 2007;26:7346. doi: 10.1038/sj.onc.1210537. [DOI] [PubMed] [Google Scholar]
- 16.Huang D, Casale GP, Tian J, Wehbi NK, Abrahams NA, Kaleem Z, et al. Quantitative fluorescence imaging analysis for cancer biomarker discovery: application to beta-catenin in archived prostate specimens. Cancer Epidemiol Biomarkers Prev. 2007;16:1371–81. doi: 10.1158/1055-9965.EPI-06-0718. [DOI] [PubMed] [Google Scholar]
- 17.Laxman B, Morris DS, Yu J, Siddiqui J, Cao J, Mehra R, et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008;68:645. doi: 10.1158/0008-5472.CAN-07-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nesrallah L, Nesrallah A, Antunes AA, Leite KR, Srougi M. The role of extended prostate biopsy on prostate cancer detection rate: a study performed on the bench. Int Braz J Urol. 2008;34:563–70. doi: 10.1590/s1677-55382008000500004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement 1. Quantification of transcripts of AMACR, PCA3 in urinary sediments by qPCR
Supplement 2. Sensitivity and specificity of biomarker assays in different age groups