ABSTRACT
Thermoanaerobacterium saccharolyticum and Clostridium thermocellum are anaerobic thermophilic bacteria being investigated for their ability to produce biofuels from plant biomass. The bifunctional alcohol and aldehyde dehydrogenase gene, adhE, is present in these bacteria and has been known to be important for ethanol formation in other anaerobic alcohol producers. This study explores the inactivation of the adhE gene in C. thermocellum and T. saccharolyticum. Deletion of adhE reduced ethanol production by >95% in both T. saccharolyticum and C. thermocellum, confirming that adhE is necessary for ethanol formation in both organisms. In both adhE deletion strains, fermentation products shifted from ethanol to lactate production and resulted in lower cell density and longer time to reach maximal cell density. In T. saccharolyticum, the adhE deletion strain lost >85% of alcohol dehydrogenase (ADH) activity. Aldehyde dehydrogenase (ALDH) activity did not appear to be affected, although ALDH activity was low in cell extracts. Adding ubiquinone-0 to the ALDH assay increased activity in the T. saccharolyticum parent strain but did not increase activity in the adhE deletion strain, suggesting that ALDH activity was inhibited. In C. thermocellum, the adhE deletion strain lost >90% of ALDH and ADH activity in cell extracts. The C. thermocellum adhE deletion strain contained a point mutation in the lactate dehydrogenase gene, which appears to deregulate its activation by fructose 1,6-bisphosphate, leading to constitutive activation of lactate dehydrogenase.
IMPORTANCE Thermoanaerobacterium saccharolyticum and Clostridium thermocellum are bacteria that have been investigated for their ability to produce biofuels from plant biomass. They have been engineered to produce higher yields of ethanol, yet questions remain about the enzymes responsible for ethanol formation in these bacteria. The genomes of these bacteria encode multiple predicted aldehyde and alcohol dehydrogenases which could be responsible for alcohol formation. This study explores the inactivation of adhE, a gene encoding a bifunctional alcohol and aldehyde dehydrogenase. Deletion of adhE reduced ethanol production by >95% in both T. saccharolyticum and C. thermocellum, confirming that adhE is necessary for ethanol formation in both organisms. In strains without adhE, we note changes in biochemical activity, product formation, and growth.
INTRODUCTION
Anaerobic bacteria are being investigated for their ability to produce biofuels from biomass. In particular, Thermoanaerobacterium saccharolyticum and Clostridium thermocellum are of interest because of their ability to break down components of lignocellulosic biomass and produce alcohols (1). These thermophilic anaerobes normally produce a mixture of organic acids and ethanol. They have been engineered for increased ethanol yield (2–4) and can produce higher alcohols, such as n-butanol (5), butanediol, and butanol (6), but gaps remain in understanding their metabolism. In particular, there is uncertainty in the genes responsible for ethanol production. The T. saccharolyticum and C. thermocellum genomes contain multiple annotated genes for aldehyde dehydrogenase (ALDH) and alcohol dehydrogenase (ADH) activity which could be responsible for alcohol formation, including adhE. AdhE is a bifunctional enzyme composed of an aldehyde dehydrogenase at the N-terminal domain and an iron-dependent alcohol dehydrogenase at the C-terminal domain, connected by a small linker sequence between the domains. Thus, AdhE can catalyze the two terminal steps in ethanol formation: the reduction of acetyl coenzyme A (acetyl-CoA) to acetaldehyde (i.e., ALDH) and reduction of acetaldehyde to ethanol (i.e., ADH), with two reduced nicotinamide cofactors (i.e., NADH and NADPH) as electron donors.
The adhE gene is thought to be important for alcohol formation, and its function has been studied in a number of other organisms. The adhE gene Escherichia coli (Ec_adhE) is essential for anaerobic growth (7). In Clostridium acetobutylicum, two different adhE genes are expressed during ethanologenesis (Ca_adhE) and solventogenesis (Ca_adhE2) (8). Deletion of adhE in Thermoanaerobacter mathranii (Tm_adhE) resulted in the loss of ethanol formation and an increase of lactate and acetate formation. Biochemical assays of the T. mathranii Tm_adhE deletion strain demonstrated that it was responsible primarily for ALDH activity, as ADH activity was largely unchanged in cell extracts (9). Similarly, deletion of adhE in Thermoanaerobacterium thermosaccharolyticum (Tt_adhE) resulted in the loss of ethanol, butanol, and acetate formation and increased lactate formation (10). Studies in Thermoanaerobacter pseudethanolicus 39E have suggested that AdhB is the critical enzyme responsible for ethanol formation rather than AdhE (11, 12). Another study suggests a more complicated relationship between AdhB and AdhE in this organism, where AdhB is responsible for ethanol formation in early growth and AdhE is responsible for ethanol formation later in growth as ethanol concentrations increase (13). The true role of T. pseudethanolicus adhE (Tp_adhE) in this organism, however, has not yet been confirmed by deletion. A metagenomic study of fermentative bacteria suggests that while adhE in the genome is indicative of the ability to form ethanol (14), it is not required, as Caldanaerobacter subterraneus subsp. tengcongensis makes ethanol without an annotated adhE (14, 15). Transcriptomic and proteomic studies in T. saccharolyticum and C. thermocellum have shown adhE to be highly expressed, although other putative adh genes were detected as well (16–19). AdhE also has been shown to play a role in ethanol tolerance (20).
To clarify the role of adhE in ethanol formation in T. saccharolyticum and C. thermocellum, we deleted adhE in these bacteria and characterized changes in biochemical activity, growth, and fermentation product distribution.
MATERIALS AND METHODS
Biochemical and molecular techniques.
All chemicals were obtained from Fisher Scientific (Pittsburgh, PA) and Sigma-Aldrich (St. Louis, MO) unless otherwise stated. Primers and strains can be found in Table 1. The adhE gene in T. saccharolyticum (Ts_adhE; Tsac_0416) was deleted by allelic replacement using standard techniques in parent strain JW/SL-YS485 (21, 22), and the resulting strain was named LL1076 (also known as strain M3223). For this deletion, the genome region from coordinates 447543 to 449414 was replaced with the resistance cassette from plasmid pMU424 (21). Genome coordinates are based on numbering from RefSeq NC_017992.1 (http://www.ncbi.nlm.nih.gov/refseq/). Transformation and deletion of adhE in C. thermocellum (Ct_adhE; Clo1313_1798) was accomplished with plasmid pJLO19 using previously described methods in strain M1354, a Δhpt strain allowing genetic manipulations (2, 23–25). The resulting strain was named LL1111. Ct_adhE was reinserted in strain LL1111 using plasmid pSH016, restoring Ct_adhE to the wild-type locus and resulting in strain LL1160. Genetic modification was confirmed by PCR, Sanger sequencing, and whole-genome resequencing by the Department of Energy Joint Genome Institute. Genome resequencing data are available from the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/Traces/sra).
TABLE 1.
Strains and primers used in this study
| Strain or primer | Description or sequence | Reference, source, or purpose |
|---|---|---|
| Strains | ||
| T. saccharolyticum | ||
| JW/SL-YS485 | Wild type | Gift from J. Wiegel |
| LL1076 | JW/SL-YS485 ΔadhE::pta ack kanR | This work |
| C. thermocellum | ||
| M1354 | DSM 1313 Δhpt | 2 |
| LL1111 | M1354 ΔadhE ldh(R157L) | This work |
| LL1160 | LL1111 adhE+ ldh(R157L) | This work |
| Primers | ||
| T.sac.adhE.ext.F | CCCTCCCCTTGTACCTTTGTGTCC | P1 primer for amplifying external region of T. saccharolyticum adhE |
| T.sac.adhE.ext.R | GAAAACTTTGGCATCGGCGGC | P1 primer for amplifying external region of T. saccharolyticum adhE |
| T.sac.seq.int.adhE.F | GAGCAAAGCTGCGCCATGAG | P2 primer for amplifying internal region of T. saccharolyticum adhE |
| T.sac.seq.int.adhE.R | CGATGAATAGCGCTTTTTTGC | P2 primer for amplifying internal region of T. saccharolyticum adhE |
| adhE.ext.F | GTGTATTGACTTTGATTGTATTAAACGG | P3 primer for amplifying external region of C. thermocellum adhE |
| adhE.ext.R | GGTGTTTACCGTATGGCAGCACGAAG | P3 primer for amplifying external region of C. thermocellum adhE |
| adhE.int.F | TTGACGATGCCCTTGATAAAGC | P4 primer for amplifying internal region of C. thermocellum adhE |
| adhE.int.R1 | CCTGTTTTTTCATCAGTAATAACCGC | P4 primer for amplifying internal region of C. thermocellum adhE |
Media and growth conditions.
Strains were grown anaerobically at 55°C for all experiments at an initial pH of 6.3 for T. saccharolyticum and pH 7.4 for C. thermocellum. Strains grown for transformation, biochemical characterization, and growth curves were cultured in a modified DSMZ M122 medium containing 5 g/liter cellobiose (2). For analysis of fermentation products, strains were grown in serum bottles (Fisher Scientific) on 50 ml of defined MTC medium for 72 h, as previously described (26), with the following modifications for T. saccharolyticum strains: urea was replaced with ammonium chloride, and thiamine hydrochloride was supplemented at a final concentration of 4 mg/liter. For growth measurements, strains were grown in 200 μl of medium in a 96-well plate. Growth was measured by monitoring absorbance at 600 nm every 5 min for 72 h in a Powerwave XS plate reader as previously described (24). Data for fermentation products and growth rate are averages from biological triplicate experiments.
Analytical techniques.
Fermentation products in the liquid fraction were measured using a Waters (Milford, MA) high-pressure liquid chromatograph (HPLC) with an HPX-87H column with a UV and refractive index detector as previously described (26). H2 was measured using a model 310 SRI Instruments gas chromatograph (Torrence, CA) with a HayeSep D packed column using a thermal conductivity detector and nitrogen carrier gas. Pellet carbon and nitrogen were measured with a Shimadzu TOC-V CPH elemental analyzer with TNM-1 and ASI-V modules (Shimadzu Corp., Columbia, MD) as previously described (26). Mass spectrometry analysis was performed with an HPLC pump (u3000; Dionex, Sunnvale, CA) coupled to an LTQ XL Orbitrap (Thermo Scientific, Waltham, MA) as previously described (28).
Biochemical assays.
Cells for biochemical assays were grown to an optical density at 600 nm (OD600) of 0.3 and harvested by centrifugation at 3,000 × g for 30 min at 4°C. Harvested cells were manipulated in a Coy (Ann Arbor, MI) anaerobic chamber, placed in serum bottles, and stored anaerobically at −80°C. Cell extract was made by incubating cells in a lysis buffer with 1× BugBuster reagent (EMD Millipore, Billerica, MA), 100 mM phosphate buffer (pH 7.0), 5 μM FeSO4, 0.1 mM dithiothreitol (DTT), Ready-Lyse lysozyme (Epicentre Biotechnologies, Madison, WI), and DNase I (Thermo Scientific). Unlysed cells and debris were separated from cell extract by centrifugation for 5 min at 12,000 × g. Protein content was measured using Bio-Rad protein dye reagent with bovine serum albumin (Thermo Scientific) as a standard. Typical protein concentrations of the cell extract ranged from 2 to 10 mg/ml.
All biochemical assays were performed at 55°C in a Coy anaerobic chamber with an 85% N2, 10% CO2, and 5% H2 atmosphere maintained under the anoxic conditions using a palladium catalyst. Alcohol, aldehyde, and lactate dehydrogenase activity were measured based on previously described methods after the oxidation of NAD(P)H at 340 nm (ε = 6,220 M−1 cm−1) (20, 27). In all cases, the final assay volume was 0.8 ml. For the ADH (acetaldehyde reduction) reactions, the anaerobic reaction mixture contained 100 mM Tris-HCl buffer (pH 7.0), 5 μM FeSO4, 0.25 mM NAD(P)H, 18 mM acetaldehyde, 1 mM DTT, and cell extract as indicated. For the ALDH (acetyl-CoA reduction) reactions, the anaerobic reaction mixture contained 100 mM Tris-HCl buffer (pH 7.0), 5 μM FeSO4, 0.25 mM NAD(P)H, 1.25 mM acetyl-CoA, 1 mM DTT, and cell extract. A decrease in absorbance at 340 nm caused by NAD(P)H oxidation was monitored by an Agilent Technologies (Santa Clara, CA) 8453 UV-visible spectrophotometer with Peltier controlled heating set at 55°C. All of the ALDH activity measurements mentioned in this study refer to the reaction in the acetaldehyde-producing direction. In reactions where 2,3-dimethoxy-5-methyl-p-benzoquinone (ubiquinone-0) or dimethyl sulfoxide (DMSO) was added, ubiquinone-0 was added to a final concentration of 2 mM, and the same volume of 100% DMSO was added in controls. The reaction conditions for lactate dehydrogenase (LDH) activity were 200 mM Tris-HCl (pH 7.3), 0.22 mM NADH, 10 mM sodium pyruvate, and 1 mM fructose 1,6-bisphosphate.
Nucleotide sequence accession numbers.
SRA accession numbers for adhE deletion strains of T. saccharolyticum and C. thermocellum are SRX744220 and SRX744221, respectively. GenBank accession numbers for plasmids pJLO19 and pSH016 are KP636798 and KP245915, respectively.
RESULTS
Knockouts of adhE in T. saccharolyticum and C. thermocellum.
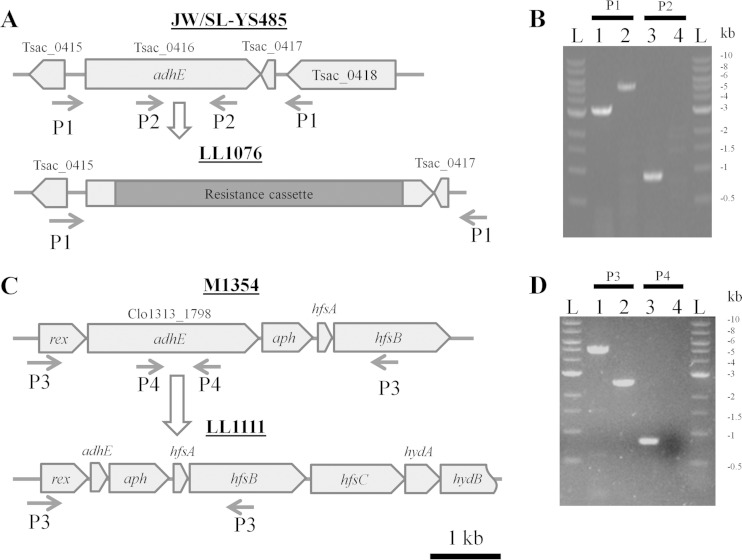
To delete Ts_adhE from T. saccharolyticum strain JW/SL-YS485, we used allelic replacement to insert the resistance cassette from plasmid pMU424 (21) into Ts_adhE and performed selection with kanamycin to generate strain LL1076. Strain LL1076 has a 1.9-kb deletion in Ts_adhE with an insertion of the 4-kb resistance cassette construct in its place, resulting in an increase in size of 2.1 kb at the Ts_adhE locus in the mutant strain, which was confirmed by PCR. In C. thermocellum, we transformed strain M1354 with plasmid pJLO19 to generate strain LL1111. Strain LL1111 contains a 2.3-kb internal deletion in Ct_adhE. Representative deletion strategies and agarose gels of PCRs targeting Ct_adhE regions were run, showing successful genetic modifications of adhE in strains LL1076 and LL1111 (Fig. 1). To confirm no significant polar effects as a result of the Ct_adhE deletion, cell extract samples of LL1111 and M1354 were analyzed by mass spectrometry. Spectral counts of Ct_adhE (Clo1313_1798) showed a >95% reduction, confirming the deletion. The spectral counts of proteins encoded by Clo1313_1799 and Clo1313_1795 to Clo1313_1797 were within 15% between the two strains, suggesting that the in-frame deletion did not have significant polar effects (see Fig. S1 in the supplemental material). Complete genome resequencing of C. thermocellum strain LL1111 revealed a G470T point mutation in the ldh gene, resulting in an amino acid change at position 157 of arginine to leucine. The R157L mutation of ldh was named ldh(R157L) for simple notation. To determine whether the ldh(R157L) mutation had an effect independent of the Ct_adhE deletion, Ct_adhE was reintroduced into LL1111 at the Ct_adhE locus with plasmid pSH016, generating strain LL1160. Strain LL1160 has the wild-type Ct_adhE locus with the ldh(R157L) mutation.
FIG 1.
Deletion of adhE and PCR confirmation in T. saccharolyticum and C. thermocellum. Deletion of T. saccharolyticum adhE with P1 (external) and P2 (internal) confirmation primer pairs. (A) A resistance cassette is transformed into parent strain JW/SL-YS485, deleting adhE and creating strain LL1076. (B) T. saccharolyticum strain JW/SL-YS485 in lane 1 shows the expected 3.2-kb fragment with P1 external primers, and lane 3 shows the 900-bp internal P2 fragment of adhE. LL1076 in lane 2 shows a larger 5.3-kb band due to the replacement of 1.9 kb with the 4-kb disruption construct, and lane 4 shows the loss of the internal P2 band of adhE. (C) Deletion of C. thermocellum adhE with P3 external and P4 internal confirmation primer pairs. M1354 was transformed with plasmid pJLO19 and used to delete adhE to make strain LL1111. C. thermocellum strain M1354 in lane 1 shows the expected P3 external product size of 4.9 kb, and lane 3 shows the 800-bp P4 internal fragment. (D) LL1111 in lane 2 shows a 2.3-kb loss in adhE with a P3 product size of 2.6 kb, and lane 4 shows no P4 fragment. The DNA marker is the 1-kb ladder from New England BioLabs and is marked with an L in panels B and D. Primer sequences can be found in Table 1.
Enzyme assay for aldehyde and alcohol dehydrogenase activity.
To explore biochemical changes caused by inactivation of the adhE gene, we assayed for aldehyde (ALDH) and alcohol dehydrogenase (ADH) activity in cell extracts of adhE deletion strains and their parent strains in both T. saccharolyticum and C. thermocellum (Table 2). We measured low ALDH activity in T. saccharolyticum parent strain JW/SL-YS485 cell extract, similar to previous results (3), which was essentially unchanged in the Ts_adhE deletion strain LL1076. NADH- and NADPH-linked ADH activity was detected in parent strain JW/SL-YS485 cell extracts. In the Ts_adhE deletion strain LL1076, NADH-linked ADH activity decreased to basal levels, while NADPH-linked ADH activity decreased by approximately two-thirds from 0.68 to 0.25 U/mg of cell extract protein. It was surprising to see so little ALDH activity in T. saccharolyticum JW/SL-YS485 cell extracts, which was unchanged in the Ts_adhE deletion strain LL1076. Ethanol is a major fermentation product of T. saccharolyticum, and the AdhE protein should have ALDH activity. We wondered whether there were inhibitors present that prevented us from detecting ALDH activity. Gupta and colleagues reported that detection of high levels of ALDH activity required addition of ubiquinone-0 to their enzyme assay reaction (29). With the addition of ubiquinone-0 to our ALDH enzyme assays, we found that the parent strain JW/SL-YS485 had 5-fold higher ALDH activity (Table 3). The source of inhibition is speculative and will be addressed in Discussion.
TABLE 2.
Specific activities of cell extracts from T. saccharolyticum and C. thermocellum
| Reaction | Sp acta (μmol · min−1 · mg−1 protein) of: |
|||
|---|---|---|---|---|
|
T. saccharolyticum |
C. thermocellum |
|||
| JW/SL-YS485 (parent) | LL1076 (ΔadhE) | M1354 (parent) | LL1111 [ΔadhE ldh(R157L)] | |
| ALDH-NADH | 0.09 (0.01) | 0.09 (0.01) | 2.18 (0.32) | 0.04 (0.03) |
| ALDH-NADPH | 0.05 (0.01) | 0.09 (0.01) | 0.16 (0.04) | 0.10 (0.02) |
| ADH-NADH | 1.52 (0.04) | 0.01 (0.01) | 7.68 (0.13) | 0.03 (0.03) |
| ADH-NADPH | 0.68 (0.03) | 0.25 (0.01) | 0.05 (0.12) | 0.07 (0.03) |
Standard deviations are in parentheses; n = 3. Limit of detection, <0.01.
TABLE 3.
Aldehyde dehydrogenase activity of T. saccharolyticum cell extract with ubiquinone-0
| Substrate | Sp acta (μmol · min−1 · mg−1 protein) of: |
|
|---|---|---|
| JW/SL-YS485 (parent) | LL1076 (ΔadhE) | |
| DMSO only | 0.09 (0.01) | 0.08 (0.03) |
| DMSO plus ubiquinone-0 | 0.43 (0.02) | >0.01 (0.02) |
Standard deviations are in parentheses; n = 3. Limit of detection, <0.01.
For C. thermocellum, the parent strain M1354 had high NADH-linked alcohol and aldehyde dehydrogenase activity, similar to those in previous reports on ADH and ALDH activity in C. thermocellum (20, 30). Both of these activities were largely lost (<3% of original NADH-linked activity) in the Ct_adhE deletion strain LL1111. Low levels of NADPH-linked alcohol and aldehyde dehydrogenase activities were detected in the parent strain M1354, and these activities were unchanged in the Ct_adhE deletion strain LL1111.
Growth and fermentation products.
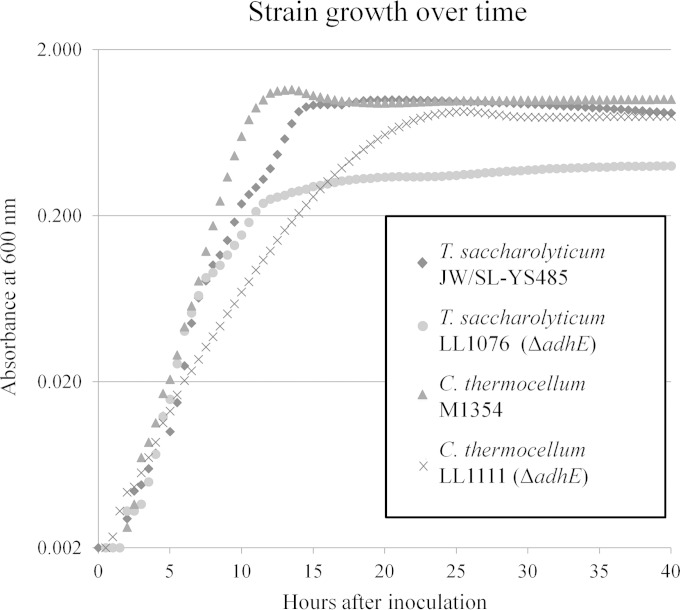
We next compared the growth and product distribution in batch cellobiose fermentations. First, growth was monitored by measuring the change in absorbance every 5 min at 600 nm in a 96-well plate in 200 μl of medium in triplicate for 72 h (Fig. 2). T. saccharolyticum parent strain JW/SL-YS485 reached a maximum OD600 of 1.0 at 19 h. The Ts_adhE deletion strain LL1076 had a similar growth profile for the first 10 h but subsequently exhibited dramatically slower growth, reaching a maximum absorbance of 0.4 after 36 h. The C. thermocellum parent strain M1354 grew to a maximum OD600 of 1.1 in 12 h, while the Ct_adhE deletion strain LL1111 reached a maximum OD600 of 0.8 after 25 h. No significant change in OD600 in any strain was seen after 36 h.
FIG 2.
Growth of C. thermocellum and T. saccharolyticum ΔadhE and parent strains over time. Strains were grown in 200 μl of medium in a 96-well plate reader at 55°C for 72 h. Growth was monitored by taking OD600 readings every 5 min. Data shown are the averages from three replicates at 30-min intervals for the first 40 h. Significant growth was not seen after 36 h.
The slower growth and lower maximum absorbance for the adhE deletion strains suggested that the loss of this enzyme has strong effects on metabolism. To further explore metabolic consequences of the loss of adhE, we grew the strains for 72 h on 0.72 mmol cellobiose (5 g/liter in a 50-ml working volume) in defined medium in closed bottles (Table 4). In both T. saccharolyticum and C. thermocellum, loss of adhE resulted in a >95% reduction of ethanol formation. T. saccharolyticum Ts_adhE deletion strain LL1076 consumed only about 60% of the supplied cellobiose after 72 h. From the 0.44 mmol of cellobiose consumed, the major products were lactate, H2, and acetate (in descending order). LL1076 pellet C and N, measures of cell mass (31), were approximately a third of the values seen for JW/SL-YS485. The loss of pellet C and N is complementary to the growth data from the 96-well plate assay, which showed a significantly lower maximum OD600. The C. thermocellum adhE deletion strain LL1111 consumed all of the supplied cellobiose, with the major products being H2, lactate, and acetate. Strain LL1160, derived from LL1111, with Ct_adhE restored to the wild-type genotype and with the ldh(R157L) genotype, restores ∼75% of the ethanol yield of M1354 (0.77 mmol versus 1.01 mmol). This strain also had increased lactate formation over the parent strain M1354 (0.28 versus 0.02). The increased lactate formation of strain LL1160 over M1354 indicates that the ldh(R157L) genotype has an effect on LDH activity.
TABLE 4.
Fermentation products from T. saccharolyticum and C. thermocellum from 0.72 mmol cellobiose after 72 h of growth
| Product or parameter | Value fora: |
||||
|---|---|---|---|---|---|
|
T. saccharolyticum |
C. thermocellum |
||||
| JW/SL-YS485 (parent) | LL1076 (ΔadhE) | M1354 (parent) | LL1111 [ΔadhE ldh(R157L)] | LL1160 [adhE+ ldh(R157L)] | |
| Cellobiose (mmol) | 0.01 (0.02) | 0.30 (0.08) | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) |
| Lactate (mmol) | 0.17 (0.07) | 0.96 (0.32) | 0.02 (0.00) | 1.12 (0.02) | 0.28 (0.01) |
| Ethanol (mmol) | 1.29 (0.04) | 0.05 (0.01) | 1.01 (0.07) | 0.04 (0.04) | 0.77 (0.01) |
| Acetate (mmol) | 0.82 (0.07) | 0.36 (0.08) | 0.81 (0.04) | 0.73 (0.02) | 0.73 (0.02) |
| Formate (mmol) | 0.03 (0.02) | 0.01 (0.02) | 0.23 (0.07) | 0.05 (0.00) | 0.24 (0.01) |
| H2 (mmol) | 1.94 (0.14) | 0.91 (0.19) | 1.67 (0.03) | 1.96 (0.19) | 1.50 (0.07) |
| Pellet C (mmol) | 0.93 (0.04) | 0.33 (0.08) | 1.25 (0.04) | 1.19 (0.09) | 1.20 (0.04) |
| Pellet N (mmol) | 0.24 (0.04) | 0.08 (0.03) | 0.36 (0.02) | 0.31 (0.06) | 0.29 (0.02) |
| Calculated CDWb (mg) | 25.89 | 9.19 | 34.80 | 33.13 | 33.41 |
| O/R indexc | 0.9 | 1 | 0.8 | 0.8 | 0.8 |
| Final pH | 4.8 | 4.7 | 6.9 | 6.8 | 7.1 |
Standard deviation in are parentheses; n = 3.
Cell dry weight, calculated from pellet C and models from Holwerda et al. (31).
O/R, oxidation/reduction.
LDH activity in C. thermocellum.
To see whether the R157L mutation in ldh(R157L) affected LDH activity, we tested for changes in activity in cell extracts (Table 5). Cell extracts of the parent strain M1354 had LDH activity that was activated by fructose 1,6-bisphosphate (F1,6BP), which is consistent with previous reports (27, 32). In contrast, cell extracts of LL1111 had LDH activity that was unaffected by the addition of F1,6BP. The maximum LDH activity in the LL1111 strain was twice the maximum measured in M1354. The LDH activity of cell extract from LL1160, containing only the ldh(R157L) mutation, also was unaffected by the addition of F1,6BP and had LDH activity similar to that of M1354 with added F1,6BP. This shows that the R157L mutation in Ldh is responsible for the altered regulation seen in the tested cell extracts. Possibilities for altered Ldh regulation are explored in Discussion.
TABLE 5.
Lactate dehydrogenase activity of C. thermocellum cell extract
| Condition | Sp acta (umol · min−1 · mg−1 protein) of strain: |
||
|---|---|---|---|
| M1354 (parent) | LL1111 [ΔadhE ldh(R157L)] | LL1160 [adhE+ ldh(R157L)] | |
| With F16BP | 0.65 (0.02) | 1.44 (0.09) | 0.63 (0.12) |
| Without F16BP | 0.11 (0.03) | 1.27 (0.01) | 0.58 (0.12) |
Standard deviations are in parentheses; n = 3. Limit of detection, 0.01.
DISCUSSION
This study shows that the adhE gene is necessary for ethanol formation in both T. saccharolyticum and C. thermocellum. Although other genes are annotated as aldehyde and alcohol dehydrogenases, substantial ethanol production is not observed in the absence of adhE in both of these organisms.
In T. saccharolyticum, we found that ubiquinone-0 relieves inhibition of ALDH that is present in the cell extract. Gupta and colleagues showed that in E. coli, the inhibition of ALDH activity was due to intermediates of ubiquinone synthesis (29). Ubiquinone synthesis is not believed to occur in anaerobic bacteria (33), although menaquinone synthesis is noted to occur in several related thermophilic anaerobic bacteria (34). Menaquinones are responsible for thiosulfate reduction in Salmonella (35), and T. saccharolyticum can reduce thiosulfate to elemental sulfur (36), perhaps via a similar mechanism. Menaquinone synthesis has yet to be proven in T. saccharolyticum, and whether or not these intermediates interfere with ALDH activity has yet to be proven. The biochemical results nevertheless indicate that the low ALDH value previously observed was due to inhibition, and that AdhE in T. saccharolyticum has both ALDH and ADH activity. With the loss of Ts_adhE, there was no significant ALDH or NADH-specific ADH activity remaining in cell extracts. However, even with the loss of Ts_adhE in strain LL1076, there was significant NADPH-linked ADH activity remaining. A recent proteomic study of T. saccharolyticum detected several candidate alcohol dehydrogenase genes for this activity, Tsac_2087, Tsac_1049, and Tsac_0285, with Tsac_2087 being detected among the top 100 peptides in relative abundance (19). Tsac_2087 is predicted to encode a putative AdhA, which shares high identity (88%) to the AdhA from Thermoanaerobacter ethanolicus JW200. This AdhA has been shown to be NADPH dependent (37) and could be the source of the remaining NADPH-linked ADH activity. The function of AdhA in T. saccharolyticum is unknown.
In T. saccharolyticum, deletion of Ts_adhE resulted in markedly worse growth in terms of both maximum culture density and time to reach that density. Interestingly, the reduction in growth rate appears more pronounced after an OD600 of 0.2. Similarly, the Tt_adhE deletion in T. thermosaccharolyticum showed a growth phenotype comparable to that of the parent strain at an OD600 of <0.3. It also had a slower growth and a final OD600 of less than half of that of its parent strain (10). Despite the similarities in the growth curves of T. saccharolyticum strain LL1076 and T. thermosaccharolyticum with Tt_adhE deleted, there were differences, as the T. thermosaccharolyticum strain exhibited complete cellobiose consumption and a complete loss of acetate formation. In the T. saccharolyticum Ts_adhE deletion strain LL1076, the distribution of fermentation products changed dramatically, most notably with lactate becoming the major end product. Lactate formation in T. saccharolyticum is known to be linked to NADH (40), and perhaps increased lactate formation and incomplete cellobiose consumption reflects an inability of strain LL1076 to oxidize NADH through ethanol formation via AdhE. The inability to consume available cellobiose in T. saccharolyticum was previously noted in several hydrogenase mutants (41) and in general may reflect an inability of strains to maintain redox balance.
In C. thermocellum, biochemical evidence suggests that Ct_adhE is responsible for the majority of both ALDH and ADH activity in cell extracts and that other annotated alcohol/aldehyde dehydrogenases do not play a significant role in ethanol formation. Loss of Ct_adhE in C. thermocellum strain LL1111 caused a shift from ethanol to lactate production. In C. thermocellum, increases in lactate formation have been reported before, particularly in an ethanol-tolerant strain that had a mutated Ct_adhE gene (20) and in a phosphotransacetylase (pta) deletion strain that could not produce acetate (2). Lactate formation is catalyzed mainly by lactate dehydrogenase in C. thermocellum (2), which is strongly activated by F1,6BP (27, 30, 32). The R157L mutation we found in C. thermocellum Ldh corresponds to a conserved residue in several well-characterized lactate dehydrogenases (Fig. 3). Structural studies of the Geobacillus stearothermophilus Ldh show this arginine residue interacts with the F1,6BP phosphate groups through arginine's positively charged guanidinium group, suggesting that the interaction was disrupted in the mutant Ldh protein (42). Mutation of arginine to glutamine in G. stearothermophilus Ldh affected the allosteric regulation of LDH by F1,6BP and stabilized the homotetrameric form, which lowered the Km for pyruvate (43). Based on the observed accumulation of lactate, loss of F1,6BP activation of Ldh in enzyme assays, mutation of the conserved arginine, and structural data linking the arginine residue to F1,6BP, we believe the observed mutation in the ldh of strain LL1111 altered the F1,6BP allosteric regulation in a similar manner. Despite the increased lactate formation in strain LL1160 compared to strain M1354 due to the deregulated Ldh, strain LL1160 had an ethanol yield ∼75% of that of strain M1354. This suggests that the primary cause of the increase in lactate production in LL1111 was due to the loss of Ct_adhE and not ldh(R157L). Although we cannot rule out synergistic effects between ldh(R157L) and the loss of Ct_adhE for increased lactate formation, it is worth noting that the T. saccharolyticum Ts_adhE deletion strain LL1076 produced almost as much lactate as strain LL1111 (0.96 versus 1.12) despite consuming 40% less cellobiose and lacking the ldh(R157L) genotype.
FIG 3.
Mutation of Arg157 in lactate dehydrogenase corresponds to the conserved residue shown to interact with F1,6BP. Alignment of amino acid sequences from C. thermocellum parent strain M1354, ΔadhE strain LL1111, and strain LL1160 and selected lactate dehydrogenases with published structures is shown. Dots represent residues that are identical to those in the top sequence. The boxed residues correspond to a conserved arginine, which is mutated to leucine in strains LL1111 and LL1160.
The differences in response to the loss of adhE in T. saccharolyticum and C. thermocellum may be indicative of larger differences in metabolism. Previous comparative deletions of ldh and pta in T. saccharolyticum and C. thermocellum have resulted in different fermentation phenotypes with regard to ethanol yield. Deletions of ldh and pta increased ethanol yields in T. saccharolyticum (3, 21) but not C. thermocellum (2, 27). The genes surrounding adhE in T. saccharolyticum and C. thermocellum suggest major differences between the two strains. Ct_adhE in C. thermocellum appears to be part of a larger operon, including several genes relevant for electron metabolism. Brown and colleagues identified Cthe_0422-0431 (corresponding to Clo1313_1790-1799 in DSM 1313) as a potential 10-gene operon (20). This set of genes is predicted to encode rex, a redox regulator which responds to NADH levels, hfsB (also called hydS), a putative sensory/regulatory hydrogenase, and hydABC, a predicted bifurcating hydrogenase (Fig. 1C). A similar organization recently was identified in a study of Ruminococcus albus hydrogenases, where rex, Ra_adhE, and hydS were in a transcriptional unit with a putative ferredoxin-dependent hydrogenase, and this locus is believed to play an important part in the sensory and regulatory mechanism of electron metabolism in R. albus (44). In C. thermocellum, this gene cluster may play a similar role.
On the other hand, T. saccharolyticum Ts_adhE appears to be in its own operon, with the genes surrounding Ts_adhE predicted to encode proteins with no predicted role in electron metabolism (Fig. 1A). T. thermosaccharolyticum seems to contain a similar genomic locus, with Ts_adhE-flanking genes Tsac_0415 and Tsac_0418 sharing 90% and 78% identity, respectively, with the genes surrounding T. thermosaccharolyticum Tt_adhE, which may explain the similar growth phenotypes in response to the loss of adhE.
Studies have suggested that C. thermocellum generates H2 primarily though bifurcating hydrogenases that use NADH and reduced ferredoxin as electron donors, which generate 2 H2 molecules per NADH and reduced ferredoxin molecule (45, 46). This would allow C. thermocellum to be less reliant on ethanol formation to oxidize NADH generated by glycolysis. Indeed, when C. thermocellum was cocultured with H2-utilizing microbes like methanogens (47) and acetogens (48), ethanol formation dropped by as much as ∼90% and ∼80%, respectively, indicating that under certain conditions, ethanol formation is unimportant for metabolism. R. albus appears to have a similar phenotype: when grown in monoculture, R. albus formed ethanol as a major fermentation product, but when cocultured with the H2-utilizing Wolinella succinogenes, ethanol formation was not observed (49). This is believed to be dependent on bifurcating hydrogenase activity in addition to the regulation of genes in the aforementioned transcriptional unit (44). Bifurcating hydrogenase activity has not yet been shown in C. thermocellum, and the contribution of bifurcating hydrogenase versus other hydrogenases to overall H2 formation has not been clearly established. The fermentation data give some evidence in this regard. It was previously predicted that loss of adhE in C. thermocellum would increase H2 formation (14), and we did see an increase in H2 formation in strain LL1111 versus M1354. This is despite a marked increase in lactate formation, which does not generate reduced ferredoxin for H2 formation.
In contrast, T. saccharolyticum H2 formation is linked with the hfs cluster, which encodes a hydrogenase believed to use only reduced ferredoxin as the electron donor (41). Fermentation of sugars results in NADH formation, which is unable to be oxidized by the ferredoxin-dependent Hfs hydrogenase or AdhE bifunctional alcohol/aldehyde dehydrogenase in the T. saccharolyticum Ts_adhE deletion strain LL1076. Another option for oxidizing NADH is a putative NfnAB (Tsac_2085-6), which is an electron-bifurcating enzyme that couples the oxidation of NADH and reduced ferredoxin to the reduction of 2 NADP+ (50). We noticed that significant NADPH-linked ADH activity remained despite the loss of Ts_adhE, possibly due to AdhA. Interestingly, Tsac_2087, encoding AdhA, is directly upstream of nfnAB. This locus may bear particular importance in ethanol formation. In a high-ethanol-yielding T. saccharolyticum strain, there was increased NADPH-linked ADH and ferredoxin:NADP oxidoreductase activity (3). These activities could be catalyzed by the enzymes encoded by Tsac_2085-7 and provide an explanation for the seemingly coordinated increase in NADPH-linked activity seen in this high-ethanol-yielding strain. Our biochemical data from cell extracts suggests that ALDH activity still relies on AdhE. Ethanol formation could be more important in T. saccharolyticum to maintaining redox balance, as there are fewer metabolic options outside ethanol formation for oxidizing NAD(P)H. The enzymes involved in electron metabolism, especially those involved in ferredoxin reoxidation like NfnAB and hydrogenases, have been poorly studied so far. Understanding their roles in metabolism and product formation will be important for engineering anaerobic microbes for biofuel production.
In conclusion, we have shown that adhE is essential for ethanol production in both C. thermocellum and T. saccharolyticum. In addition, we solved the mystery of low ALDH activity in T. saccharolyticum by adding ubiquinone-0 to the enzyme assay mixture. Finally, we found an interesting mutation in ldh, which affects its regulation by F1,6BP.
Supplementary Material
ACKNOWLEDGMENTS
We thank Johannes P. van Dijken for useful discussions. We thank the Mascoma Corporation for their gift of the T. saccharolyticum adhE deletion strain LL1076 (also known as strain M3223). We thank Richard J. Giannone for proteomic analysis.
The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under contract no. DE-AC02-05CH11231. The BioEnergy Science Center is a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science.
This paper was been authored by Dartmouth College under subcontract no. 4000115284 and contract no. DE-AC05-00OR22725 with the U.S. Department of Energy.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02450-14.
REFERENCES
- 1.Olson DG, McBride JE, Shaw AJ, Lynd LR, Joe Shaw A, Lynd LR. 2012. Recent progress in consolidated bioprocessing. Curr Opin Biotechnol 23:396–405. doi: 10.1016/j.copbio.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 2.Argyros DA, Tripathi SA, Barrett TF, Rogers SR, Feinberg LF, Olson DG, Foden JM, Miller BB, Lynd LR, Hogsett DA, Caiazza NC. 2011. High ethanol titers from cellulose by using metabolically engineered thermophilic, anaerobic microbes. Appl Environ Microbiol 77:8288–8294. doi: 10.1128/AEM.00646-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaw AJ, Podkaminer KK, Desai SG, Bardsley JS, Rogers SR, Thorne PG, Hogsett DA, Lynd LR. 2008. Metabolic engineering of a thermophilic bacterium to produce ethanol at high yield. Proc Natl Acad Sci U S A 105:13769–13774. doi: 10.1073/pnas.0801266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng Y, Olson DG, Zhou J, Herring CD, Joe Shaw A, Lynd LR. 2013. Redirecting carbon flux through exogenous pyruvate kinase to achieve high ethanol yields in Clostridium thermocellum. Metab Eng 15:151–158. doi: 10.1016/j.ymben.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Bhandiwad A, Shaw AJ, Guss A, Guseva A, Bahl H, Lynd LR. 2014. Metabolic engineering of Thermoanaerobacterium saccharolyticum for n-butanol production. Metab Eng 21:17–25. doi: 10.1016/j.ymben.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 6.Holwerda EK, Thorne PG, Olson DG, Amador-Noguez D, Engle NL, Tschaplinski TJ, van Dijken JP, Lynd LR. 2014. The exometabolome of Clostridium thermocellum reveals overflow metabolism at high cellulose loading. Biotechnol Biofuels 7:155. doi: 10.1186/s13068-014-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trinh CT, Li J, Blanch HW, Clark DS. 2011. Redesigning Escherichia coli metabolism for anaerobic production of isobutanol. Appl Environ Microbiol 77:4894–4904. doi: 10.1128/AEM.00382-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fontaine L, Meynial-Salles I, Girbal L, Yang X, Croux C, Soucaille P. 2002. Molecular characterization and transcriptional analysis of adhE2, the gene encoding the NADH-dependent aldehyde/alcohol dehydrogenase responsible for butanol production in alcohologenic cultures of Clostridium acetobutylicum ATCC 824. J Bacteriol 184:821–830. doi: 10.1128/JB.184.3.821-830.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao S, Mikkelsen MJ. 2010. Identification and overexpression of a bifunctional aldehyde/alcohol dehydrogenase responsible for ethanol production in Thermoanaerobacter mathranii. J Mol Microbiol Biotechnol 19:123–133. doi: 10.1159/000321498. [DOI] [PubMed] [Google Scholar]
- 10.Bhandiwad A, Guseva A, Lynd L. 2013. Metabolic Engineering of Thermoanaerobacterium thermosaccharolyticum for increased n-butanol production. AiM 3:46–51. doi: 10.4236/aim.2013.31007. [DOI] [Google Scholar]
- 11.Burdette D, Zeikus JG. 1994. Purification of acetaldehyde dehydrogenase and alcohol dehydrogenases from Thermoanaerobacter ethanolicus 39E and characterization of the secondary-alcohol dehydrogenase (2 degrees Adh) as a bifunctional alcohol dehydrogenase–acetyl-CoA reductive thioesterase. Biochem J 302:163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hemme CL, Fields MW, He Q, Deng YY, Lin L, Tu Q, Mouttaki H, Zhou A, Feng X, Zuo Z, Ramsay BD, He Z, Wu L, Van Nostrand J, Xu J, Tang YJ, Wiegel J, Phelps TJ, Zhou J. 2011. Correlation of genomic and physiological traits of Thermoanaerobacter species with biofuel yields. Appl Environ Microbiol 77:7998–8008. doi: 10.1128/AEM.05677-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pei J, Zhou Q, Jiang Y, Le Y, Li H, Shao W, Wiegel J. 2010. Thermoanaerobacter spp. control ethanol pathway via transcriptional regulation and versatility of key enzymes. Metab Eng 12:420–428. doi: 10.1016/j.ymben.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Carere CR, Rydzak T, Verbeke TJ, Cicek N, Levin DB, Sparling R. 2012. Linking genome content to biofuel production yields: a meta-analysis of major catabolic pathways among select H2 and ethanol-producing bacteria. BMC Microbiol 12:295. doi: 10.1186/1471-2180-12-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soboh B, Linder D, Hedderich R. 2004. A multisubunit membrane-bound [NiFe] hydrogenase and an NADH-dependent Fe-only hydrogenase in the fermenting bacterium Thermoanaerobacter tengcongensis. Microbiology 150:2451–2463. doi: 10.1099/mic.0.27159-0. [DOI] [PubMed] [Google Scholar]
- 16.Rydzak T, McQueen PD, Krokhin OV, Spicer V, Ezzati P, Dwivedi RC, Shamshurin D, Levin DB, Wilkins JA, Sparling R. 2012. Proteomic analysis of Clostridium thermocellum core metabolism: relative protein expression profiles and growth phase-dependent changes in protein expression. BMC Microbiol 12:214. doi: 10.1186/1471-2180-12-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riederer A, Takasuka TE, Makino S, Stevenson DM, Bukhman YV, Elsen NL, Fox BG. 2011. Global gene expression patterns in Clostridium thermocellum as determined by microarray analysis of chemostat cultures on cellulose or cellobiose. Appl Environ Microbiol 77:1243–1253. doi: 10.1128/AEM.02008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raman B, McKeown CK, Rodriguez M, Brown SD, Mielenz JR. 2011. Transcriptomic analysis of Clostridium thermocellum ATCC 27405 cellulose fermentation. BMC Microbiol 11:134. doi: 10.1186/1471-2180-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Currie DH, Guss AM, Herring CD, Giannone RJ, Johnson CM, Lankford PK, Brown SD, Hettich RL, Lynd LR. 2014. Profile of secreted hydrolases, associated proteins, and SlpA in Thermoanaerobacterium saccharolyticum during the degradation of hemicellulose. Appl Environ Microbiol 80:5001–5011. doi: 10.1128/AEM.00998-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown SD, Guss AM, Karpinets TV, Parks JM, Smolin N, Yang S, Land ML, Klingeman DM, Bhandiwad A, Rodriguez M, Raman B, Shao X, Mielenz JR, Smith JC, Keller M, Lynd LR. 2011. Mutant alcohol dehydrogenase leads to improved ethanol tolerance in Clostridium thermocellum. Proc Natl Acad Sci U S A 108:13752–13757. doi: 10.1073/pnas.1102444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaw AJ, Covalla SF, Hogsett DA, Herring CD. 2011. Marker removal system for Thermoanaerobacterium saccharolyticum and development of a markerless ethanologen. Appl Environ Microbiol 77:2534–2536. doi: 10.1128/AEM.01731-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desai SG, Guerinot ML, Lynd LR. 2004. Cloning of L-lactate dehydrogenase and elimination of lactic acid production via gene knockout in Thermoanaerobacterium saccharolyticum JW/SL-YS485. Appl Microbiol Biotechnol 65:600–605. doi: 10.1007/s00253-004-1575-9. [DOI] [PubMed] [Google Scholar]
- 23.Olson DG, Lynd LR. 2012. Transformation of Clostridium thermocellum by electroporation. Methods Enzymol 510:317–330. doi: 10.1016/B978-0-12-415931-0.00017-3. [DOI] [PubMed] [Google Scholar]
- 24.Tripathi SA, Olson DG, Argyros DA, Miller BB, Barrett TF, Murphy DM, McCool JD, Warner AK, Rajgarhia VB, Lynd LR, Hogsett DA, Caiazza NC. 2010. Development of pyrF-based genetic system for targeted gene deletion in Clostridium thermocellum and creation of a pta mutant. Appl Environ Microbiol 76:6591–6599. doi: 10.1128/AEM.01484-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guss AM, Olson DG, Caiazza NC, Lynd LR. 2012. Dcm methylation is detrimental to plasmid transformation in Clostridium thermocellum. Biotechnol Biofuels 5:30. doi: 10.1186/1754-6834-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou J, Olson DG, Argyros DA, Deng Y, van Gulik WM, van Dijken JP, Lynd LR. 2013. Atypical glycolysis in Clostridium thermocellum. Appl Environ Microbiol 79:3000–3008. doi: 10.1128/AEM.04037-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van der Veen D, Lo J, Brown SD, Johnson CM, Tschaplinski TJ, Martin M, Engle NL, van den Berg RA, Argyros AD, Caiazza NC, Guss AM, Lynd LR. 2013. Characterization of Clostridium thermocellum strains with disrupted fermentation end-product pathways. J Ind Microbiol Biotechnol 40:725–734. doi: 10.1007/s10295-013-1275-5. [DOI] [PubMed] [Google Scholar]
- 28.Olson DG, Tripathi SA, Giannone RJ, Lo J, Caiazza NC, Hogsett DA, Hettich RL, Guss AM, Dubrovsky G, Lynd LR. 2010. Deletion of the Cel48S cellulase from Clostridium thermocellum. Proc Natl Acad Sci U S A 107:17727–17732. doi: 10.1073/pnas.1003584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta S, Mat-Jan F, Latifi M, Clark DP. 2000. Acetaldehyde dehydrogenase activity of the AdhE protein of Escherichia coli is inhibited by intermediates in ubiquinone synthesis. FEMS Microbiol Lett 182:51–55. doi: 10.1111/j.1574-6968.2000.tb08872.x. [DOI] [PubMed] [Google Scholar]
- 30.Lamed R, Zeikus JG. 1980. Ethanol production by thermophilic bacteria: relationship between fermentation product yields of and catabolic enzyme activities in Clostridium thermocellum and Thermoanaerobium brockii. J Bacteriol 144:569–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holwerda EK, Ellis LD, Lynd LR. 2013. Development and evaluation of methods to infer biosynthesis and substrate consumption in cultures of cellulolytic microorganisms. Biotechnol Bioeng 110:2380–2388. doi: 10.1002/bit.24915. [DOI] [PubMed] [Google Scholar]
- 32.Ozkan M, Yilmaz EI, Lynd LR, Ozcengiz G, Özkan M, Ebru Y, Özcengiz G. 2004. Cloning and expression of the Clostridium thermocellum L-lactate dehydrogenase gene in Escherichia coli and enzyme characterization. Can J Microbiol 851:845–851. doi: 10.1139/w04-071. [DOI] [PubMed] [Google Scholar]
- 33.Collins MD, Jones D. 1981. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol Rev 45:316–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto K, Murakami R, Takamura Y. 1998. Isoprenoid quinone, cellular fatty acid composition and diaminopimelic acid isomers of newly classified thermophilic anaerobic Gram-positive bacteria. FEMS Microbiol Lett 161:351–358. doi: 10.1111/j.1574-6968.1998.tb12968.x. [DOI] [Google Scholar]
- 35.Stoffels L, Krehenbrink M, Berks BC, Unden G. 2012. Thiosulfate reduction in Salmonella enterica is driven by the proton motive force. J Bacteriol 194:475–485. doi: 10.1128/JB.06014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee Y-E, Jain MK, Lee C, Zeikus JG. 1993. Taxonomic distinction of saccharolytic thermophilic anaerobes: description of Thermoanaerobacterium xylanolyticum gen. nov., sp. nov., and Thermoanaerobacterium saccharolyticum gen. nov., sp. nov.; reclassification of Thermoanaerobium brockii, Clostridium. Int J Syst Bacteriol 43:41–51. doi: 10.1099/00207713-43-1-41. [DOI] [Google Scholar]
- 37.Holt PJ, Williams RE, Jordan KN, Lowe CR, Bruce NC. 2000. Cloning, sequencing and expression in Escherichia coli of the primary alcohol dehydrogenase gene from Thermoanaerobacter ethanolicus JW200. FEMS Microbiol Lett 190:57–62. doi: 10.1111/j.1574-6968.2000.tb09262.x. [DOI] [PubMed] [Google Scholar]
- 38.Reference deleted.
- 39.Reference deleted.
- 40.Shaw JA, Jenney FE, Adams MWW, Lynd LR. 2008. End-product pathways in the xylose fermenting bacterium, Thermoanaerobacterium saccharolyticum. Enzyme Microb Technol 42:453–458. doi: 10.1016/j.enzmictec.2008.01.005. [DOI] [Google Scholar]
- 41.Shaw AJ, Hogsett DA, Lynd LR. 2009. Identification of the [FeFe]-hydrogenase responsible for hydrogen generation in Thermoanaerobacterium saccharolyticum and demonstration of increased ethanol yield via hydrogenase knockout. J Bacteriol 191:6457–6464. doi: 10.1128/JB.00497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wigley DB, Gamblin SJ, Turkenburg JP, Dodson EJ, Piontek K, Muirhead H, Holbrook JJ. 1992. Structure of a ternary complex of an allosteric lactate dehydrogenase from Bacillus stearothermophilus at 2.5 A resolution. J Mol Biol 223:317–335. doi: 10.1016/0022-2836(92)90733-Z. [DOI] [PubMed] [Google Scholar]
- 43.Clarke AR, Wigley DB, Barstow DA, Chia WN, Atkinson T, Holbrook JJ. 1987. A single amino acid substitution deregulates a bacterial lactate dehydrogenase and stabilizes its tetrameric structure. Biochim Biophys Acta 913:72–80. doi: 10.1016/0167-4838(87)90234-2. [DOI] [PubMed] [Google Scholar]
- 44.Zheng Y, Kahnt J, Kwon IH, Mackie RI, Thauer RK. 2014. Hydrogen formation and its regulation in Ruminococcus albus: involvement of an electron-bifurcating [FeFe]-hydrogenase, of a non electron-bifurcating [FeFe]-hydrogenase and of a putative hydrogen-sensing [FeFe]-hydrogenase. J Bacteriol 196:3840–3852. doi: 10.1128/JB.02070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schut GJ, Adams MWW. 2009. The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J Bacteriol 191:4451–4457. doi: 10.1128/JB.01582-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rydzak T, Grigoryan M, Cunningham ZJ, Krokhin OV, Ezzati P, Cicek N, Levin DB, Wilkins JA, Sparling R. 2014. Insights into electron flux through manipulation of fermentation conditions and assessment of protein expression profiles in Clostridium thermocellum. Appl Microbiol Biotechnol 98:6497–6510. doi: 10.1007/s00253-014-5798-0. [DOI] [PubMed] [Google Scholar]
- 47.Weimer P, Zeikus J. 1977. Fermentation of cellulose and cellobiose by Clostridium thermocellum in the absence of Methanobacterium thermoautotrophicum. Appl Environ Microbiol 33:289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le Ruyet P, Dubourguier HC, Albagnac G. 1984. Homoacetogenic fermentation of cellulose by a coculture of Clostridium thermocellum and Acetogenium kivui. Appl Environ Microbiol 48:893–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iannotti EL, Kafkewitz D, Wolin MJ, Bryant MP. 1973. Glucose fermentation products of Ruminococcus albus grown in continuous culture with Vibrio succinogenes: changes caused by interspecies transfer of H2. J Bacteriol 114:1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang S, Huang H, Moll J, Thauer RK. 2010. NADP+ reduction with reduced ferredoxin and NADP+ reduction with NADH are coupled via an electron-bifurcating enzyme complex in Clostridium kluyveri. J Bacteriol 192:5115–5123. doi: 10.1128/JB.00612-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.