Abstract
Several approaches for the generation of peptide radical cations using ion/ion reactions coupled with either collision induced dissociation (CID) or ultraviolet photo dissociation (UVPD) are described here. Ion/ion reactions are used to generate electrostatic or covalent complexes comprised of a peptide and a radical reagent. The radical site of the reagent can be generated multiple ways. Reagents containing a carbon-iodine (C-I) bond are subjected to UVPD with 266 nm photons, which selectively cleaves the C-I bond homolytically. Alternatively, reagents containing azo functionalities are collisionally activated to yield radical sites on either side of the azo group. Both of these methods generate an initial radical site on the reagent, which then abstracts a hydrogen from the peptide while the peptide and reagent are held together by either electrostatic interactions or a covalent linkage. These methods are demonstrated via ion/ion reactions between the model peptide RARARAA (doubly protonated) and various distonic anionic radical reagents. The radical site abstracts a hydrogen atom from the peptide while the charge site abstracts a proton. The net result is the conversion of a doubly protonated peptide to a peptide radical cation. The peptide radical cations have been fragmented via CID and the resulting product ion mass spectra are compared to the control CID spectrum of the singly protonated, even-electron species. This work is then extended to bradykinin, a more broadly studied peptide, for comparison with other radical peptide generation methods. The work presented here provides novel methods for generating peptide radical cations in the gas phase through ion/ion reaction complexes that do not require modification of the peptide in solution or generation of non-covalent complexes in the electrospray process.
Keywords: Peptide radical cation, ion/ion reaction, ion trap, electrospray ionization
Introduction
Odd-electron molecules play significant roles across many areas of chemistry ranging from small organic compound synthesis to biological macromolecule function.[1] Since the advent of electron ionization (EI), mass spectrometry has proven to be a convenient platform for the study of radical species that are often short-lived or difficult to observe in the solution phase.[2,3] EI, however, is less conducive to the generation of intact biomolecule-ions than the newer soft ionization techniques, such as electrospray ionization (ESI) and matrix-assisted laser desorption ionization (MALDI). The latter techniques generally yield even-electron analytes in various states of protonation or deprotonation. For this reason, peptide ion chemistry studies have been largely devoted to even-electron species. However, it is widely appreciated that structural information that can be derived from tandem mass spectrometry experiments is strongly dependent upon precursor ion-type.[4] For this reason, efforts have been directed to generate odd-electron peptide species of various types,[5] including radical cations[6,7,8]. For example, Julian et al. have extensively demonstrated the homolytic cleavage of C-I bonds upon irradiation with 266 nm photons as a method of producing radical species of peptides, proteins and lipids. This is achieved using covalent modifications such as the iodination of tyrosine[9,10,11] and the attachment of iodobenzene to primary amines of a peptide,[12,13] or using non-covalent interactions between 18-crown-6 ether and primary amines.[12,13,14] These methods result in the generation of gas-phase analyte cations with a radical site. Chu, Siu, Laskin, Tureček, O'Hair and others have used collision-induced dissociation (CID) of transition metal-ligand-peptide complexes generated by ESI to yield [M]+• peptide ions.[15,16,17,18,19,20,21] Collisional activation can be used to generate radicals in other systems as well. Azo and nitroso[22,23,24] functionalities, for example, have been covalently attached to analytes in the solution phase prior to mass spectrometry experiments. Once ionized, these groups undergo homolytic cleavages upon CID, leaving radical sites on the R groups they were bound to after loss of N2 and NO, respectively.
Radical-directed dissociation (RDD) mechanisms underlie many of the structurally diagnostic fragmentation reactions used to identify molecules via EI mass spectrometry.[25] Collisional activation of radical species often yields RDD that provides information complementary to that generated by CID of protonated even-electron ions. While CID of even-electron peptide ions is generally described using the mobile proton model,[26] RDD occurs through radical site migration wherein a series of hydrogen abstraction processes can occur before bond cleavage.[27] Peptide cation radicals tend to exhibit side chain losses via RDD processes, for example. These losses, along with a- and x-type fragments observed in RDD,[28] differ from the fragmentation patterns seen in conventional CID of protonated peptides, where b- and y-type ions are more frequently observed.[29]
Radical ion chemistry has also been used in gas-phase cross-linking experiments to elucidate structural information of various biomolecules. This is achieved via recombination of two radicals within an analyte to link two sites together,[30] or by leveraging the CID-facile azo functionality to break a cross-link, generating a radical electron on the two sites that were originally cross-linked.[24] In both instances, additional CID steps can be used to elucidate the cross-linked sites. Additionally, radical mechanisms have been observed to facilitate disulfide bond cleavage, allowing alternate fragmentation pathways to be observed.[31,32] RDD also has been shown to differentiate isomers such as leucine and isoleucine within peptides.[33] RDD yields complementary information when applied to lipids, providing information about degrees of saturation and locations of double bonds within the fatty acid chains of various lipid types,[34,35,36] whereas conventional CID experiments yield very little structural information.[37,38]
This work is motivated by the possibility for peptide radical cation generation from multiply-protonated peptides within the mass spectrometer via ion/ion reactions. This would provide a novel alternative approach for peptide radical cation generation that requires no alteration of the sample solution or modification of the peptide itself prior to ionization. Ion/ion reactions have been demonstrated to enable a wide range of analyte transformations from one ion-type to another.[39,40] Gas-phase ion/ion reactions of peptide ions proceed either via the rapid transfer of a small charged particle (i.e., proton or electron) or via the formation of a long-lived electrostatic complex comprised of an anion and cation.[41] Using pulsed dual nano electrospray ionization (nESI)[42] for the separate generation of analyte and reagent ions followed by reaction in a multi-pole collision cell (e.g., an electrodynamic ion trap that can store both cations and anions simultaneously) in a tandem mass spectrometer provides a high degree of control over the process of generating peptide radical cations. The identities of the reactants are well-defined via mass-selection using the first stage of the tandem mass spectrometer. The concentrations of the reactants are easily adjusted via the ion accumulation times used for each polarity and the reactions take place in tens-to-hundreds of milliseconds. The gas-phase approach obviates the optimization of solution conditions otherwise required for condensed-phase approaches and allows for ready comparison with the analyte ions generated directly from solution.
In this work we describe several gas-phase ion/ion reaction approaches for generating radical peptide cations using azo-containing reagents as well as reagents that contain C-I bonds. In both of these cases the radical site can be introduced to the peptide through covalent modification using well-studied N-hydroxysuccinimide (NHS)[43,44] gas-phase reactions, or simply through electrostatic interactions of the negatively charged reagent with the positively charged peptide. Using NHS chemistry ultimately yields a covalently modified peptide that contains a radical site, while complexes bound electrostatically ultimately lead to unmodified peptide radical cations. Collisional activation of either of these products results in RDD reactions.
Experimental
Materials
Direct Red 81 (DR81), 2-iodobenzoic acid, and potassium 2-iodo-5-methylbenzenesulfonate (MIBS), and bradykinin, were purchased from Sigma-Aldrich (St. Louis, MO, USA). 4,4’-Azo-bis(4-cyanovaleric acid) was purchased from Thermo Fisher Scientific, Waltham, MA, USA). The model peptide RARARAA was synthesized by NeoBioSci (Cambridge, MA, USA). Methanol was purchased from Macron Chemicals (part of Avantor, Center Valley, PA, USA). All samples subjected to nESI in this work were comprised of 0.1 mg/mL concentrations of reagent in 1:1 water:methanol.
Mass Spectrometry
Radical generation with collisional activation
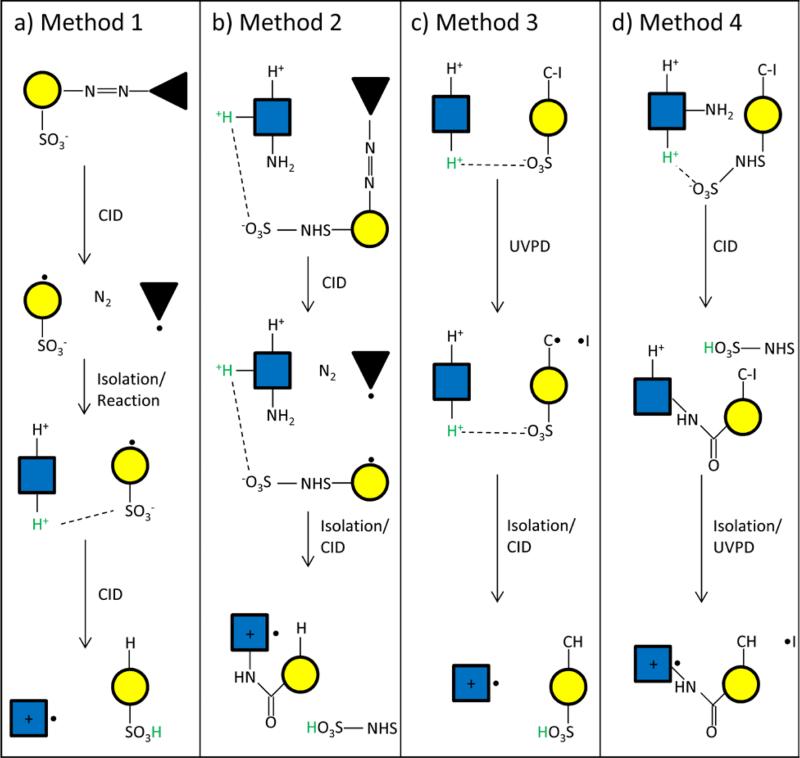
Experiments generating radical peptide cations with collisional activation (Methods 1 and 2 in Figure 1 (a and b)) were performed using a modified QTRAP 4000 hybrid triple quadrupole/linear ion trap (AB SCIEX, Concord, ON, Canada).[45] In Method 1 (Figure 1a), the azo reagent is injected into q2 under beam-type CID conditions to initiate the the azo cleavage, leaving a radical site on a fragment anion that contains a sulfonate. After isolation of this fragment through RF-only isolation, the doubly protonated peptide is injected into q2, undergoing Q1 apex isolation in transit. The reaction cell is operated under mutual storage conditions, allowing the reagent fragment and peptide to form an electrostatic complex. Reaction products are then transferred to Q3, the complex is isolated and collisionally activated to yield the nominal peptide radical cation after hydrogen abstraction from the peptide to the radical site on the anion and proton transfer from the peptide to the sulfonate group of the reagent. The result is a radical cation. No presumption is made regarding the locations of the charge and radical sites in the peptide in Figure 1. In Method 2, the doubly protonated peptide is injected into q2, undergoing isolation in transit as in the previous method. An N-hydroxysulfosuccinimide (sulfo-NHS) ester with an azo containing group (see Figure 1b cartoon) is injected in the same manner and an ion/ion reaction ensues, generating an electrostatic complex that is transferred to Q3 and isolated. Activation of this complex cleaves the azo functionality, leaving the sulfo-NHS ester with an R group, depicted as a yellow circle in Figure 1, containing a radical. Activation of this product results in the loss of sulfo-NHS and the formation of an amide bond between a primary amine or guanidino group on the peptide and the remaining R• group. It is depicted in Figure 1 that a hydrogen atom from the peptide is transferred to the R• group leaving the excess charge and radical site in the peptide. This is consistent with the observed RDD when the covalently modified peptide radical cation is subjected to CID (see below). Product ions in both methods are ultimately mass analyzed via mass-selective axial ejection.[46]
Figure 1.
A simplified outline of the radical generation methods demonstrated in this work. (a) Collisional activation of azo functionalities in electrostatic complexes, (b) collisional activation of azo functionalities combined with covalent modification via NHS reactivity, (c) UVPD of C-I bonds in an electrostatic complex, (d) UVPD of C-I bonds in an R group that has been covalently bound to a peptide via NHS chemistry. Yellow circles indicate the reagent on which the radical is initially generated while the blue squares represent the peptide that eventually undergoes RDD.
Radical generation with UVPD
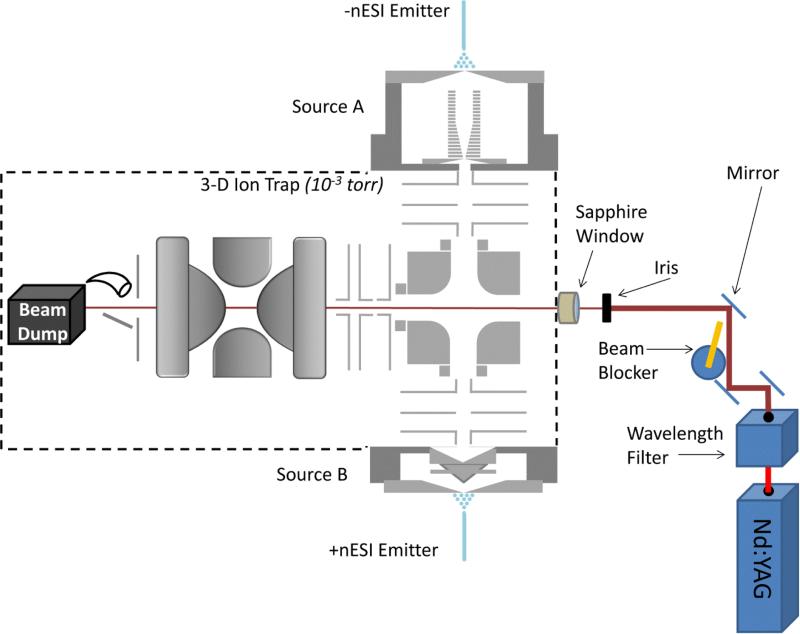
Experiments involving UVPD (Methods 3 and 4 in Figure 1(c and d)) were performed on a homebuilt 3D quadrupole ion trap (QIT) mass spectrometer modified from a previous description.[47] The instrument has been interfaced with a Nd:YAG laser (Continuum, San Jose, CA, USA) that emits 1064 nm photons passing through two second harmonic generators to produce 266 nm pulses at 20 Hz. As Figure 2 depicts, the beam path is controlled by a beam blocker that is triggered to pass photons during UVPD experiments. The beam is then directed through a quartz window into the vacuum housing and ultimately passes through the 3D ion trap along its z-axis where it overlaps with the trapped ion cloud.
Figure 2.
A diagram of the modified QIT that has been interfaced with a Nd:YAG laser.
In Method 3, the doubly protonated peptide is injected into the ion trap and isolated. Injection of the anion containing a C-I bond initiates the ion/ion reaction to form a reaction complex. After a defined reaction time (50-500 ms), the reaction complex is isolated and subjected to UVPD, causing the C-I bond to undergo homolytic cleavage. The remaining electrostatic complex, which has lost the neutral I•, is isolated and collisionally activated resulting in proton transfer from the peptide to the sulfonate group and hydrogen abstraction by the radical site of the reagent to yield a peptide radical cation. Method 4 starts off in a similar fashion to Method 3. The main difference is that along with a C-I bond, the anionic reagent contains a sulfo-NHS ester. Once the ion/ion reaction occurs and the complex is formed, collisional activation drives the NHS chemistry, binding an R group containing a C-I bond to the peptide. UVPD of this product homolytically cleaves the C-I bond, leaving the singly charged peptide attached to a radical-containing R group. Isolations in Methods 3 and 4 are carried out by using resonance or boundary ejection. All products in Methods 3 and 4 are mass analyzed using resonance ejection during an RF amplitude scan with a supplementary AC voltage applied at an optimized amplitude and frequency to yield the appropriate mass extension.[48] All UVPD experiments are carried out by introducing ~5 pulses of 266 nm photons into the QIT along the z-axis through the endcap electrodes. It should be noted that generation of the radical reagent via UVPD prior to electrostatic complex formation was attempted. However, relatively rapid hydrogen abstraction from adventitious species in the ion trap limited the lifetime of the radical anions thereby limiting the utility of this approach.
Results and Discussion
Collisional activation of azo functionalities (Methods 1 and 2)
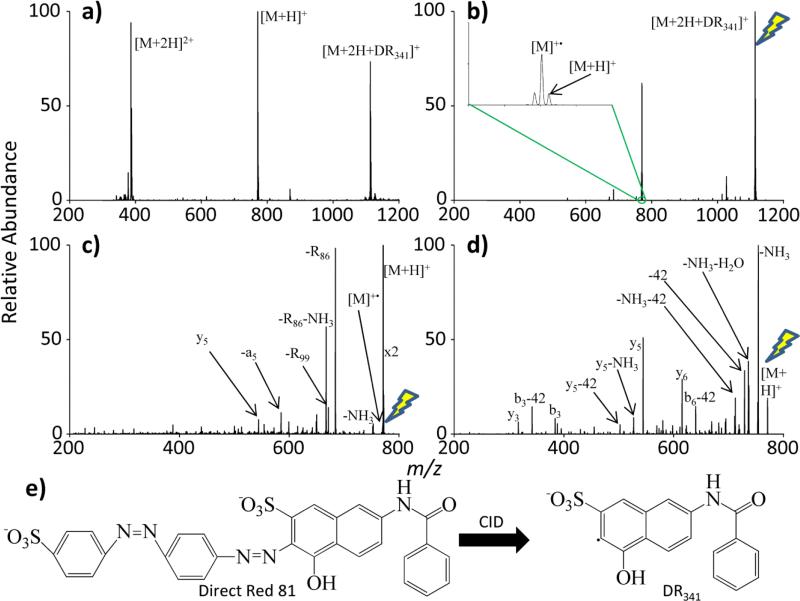
Direct Red 81 is a commercially available dye that contains two azo functionalities which, upon collisional activation, can undergo homolytic cleavage on both sides of the azo group, resulting in a loss of N2 and generating a radical site on both R groups that the azo functionality was bridging together. Experiments using Direct Red 81 to generate radical peptides (Method 1 in Figure 1(a)) followed a general process outlined in Figure 3. Direct Red 81 is deprotonated via negative mode nESI and subjected to beam-type CID during injection into the q2 reaction cell of the modified QTRAP 4000, cleaving the azo groups within Direct Red 81. One of the products of this cleavage is a radical anion with a mass-to-charge (m/z) ratio of 341 (later referred to as DR341). This product is isolated within q2. The doubly protonated peptide RARARAA is then generated via positive mode nESI and isolated in transit to q2. The peptide and the radical reagent are held under mutual storage conditions in the reaction cell (q2). Two major ion/ion reaction pathways are observed in Figure 3(a). Proton transfer yields the [M+H]+ ion, where M=RARARAA, and electrostatic complex formation to yield the [M+2H+ DR341]+ ion.
Figure 3.
The general process of generating radical peptides using collisional activation and electrostatic complexes. (a) The ion/ion reaction between doubly protonated RARARAA (denoted as M) and DR341, (b) collisional activation of the isolated reaction complex, (c) and CID of the resulting radical peptide. (d) The CID spectrum of [RARARAA+H]+ is provided for comparison. (e) The fragmentation of Direct Red 81 into DR341 is provided. The lightning bolt image indicates the precursor ion for each CID experiment. The “x2” in (c) implies that the [M+H]+ peak is twice the abundance displayed in the spectrum.
When the electrostatic complex is subjected to collisional activation, proton transfer to yield the [M+H]+ ion is noted as a minor channel in the inset of Figure 3(b). This ion was kept during isolation to avoid reducing the abundance of radical RARARAA. The major pathway involves hydrogen abstraction from the peptide to the reagent radical site and proton transfer to the sulfonate of the reagent resulting in the radical cation, denoted as [M]+• in Figure 3(b). We emphasize here that no a priori assumption is made regarding the locations of the charge and radical sites and that a radical cation generated via EI may well have a different partitioning of charge and radical sites. In any case, some hydrogen scrambling is expected to occur upon collisional activation. The ion trap CID spectrum of the radical cation is shown in Figure 3(c). This CID experiment yields fragmentation commonly observed in RDD. Most notably, fragmentation of the RARARAA radical cation yields a product that goes through a nominal loss of 86, indicating the radical cleavage of an arginine side chain (indicated by R86 in Figure 3(c))[28] as well as the a5 fragment of RARARAA. These cleavages are not observed in the fragmentation spectrum of the even-electron protonated peptide, as seen in Figure 3(d).
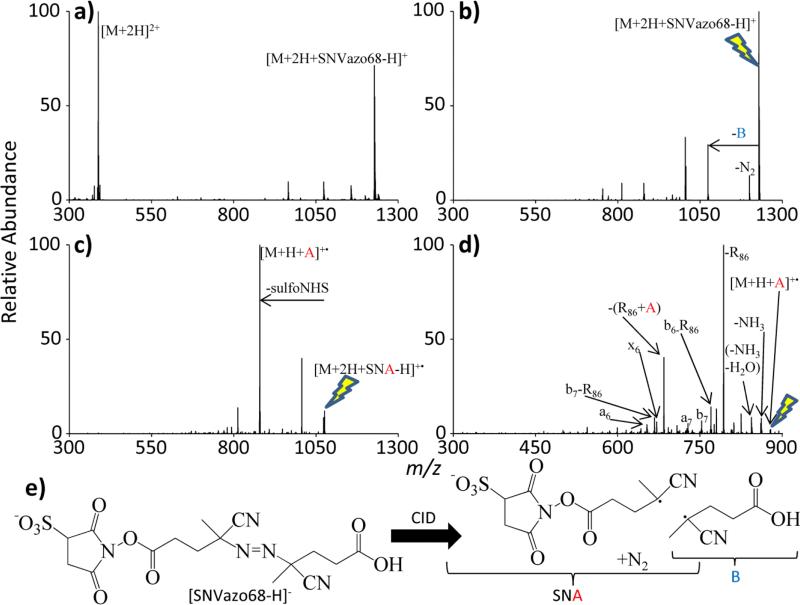
A radical site was also introduced into the peptide via covalent modification of the peptide RARARAA (Method 2 in Figure 1(b)). In order to carry this out, 4,4’-azo-bis(4-cyanovaleric acid) (also known as Vazo 68) was converted into a N-hydroxysulfosuccinimide (sulfo-NHS) reagent using previously published methods.[43,44] The gas-phase CID-based ion/ion reaction method for generating peptides with covalently bound radical sites is demonstrated in Figure 4.
Figure 4.
A demonstration of the process of covalently binding a radical site to a peptide through ion/ion reactions. (a) Ion/ion reaction of the doubly protonated peptide RARARAA (denoted as M) and [SNVazo68-H]−, (b) CID of the resulting electrostatic complex, (c) CID of the remaining electrostatic complex to drive the sulfo-NHS chemistry, and (d) fragmentation of the covalently modified radical peptide. (e) The fragmentation of SNVazo68 into SNA and B is provided. The lightning bolt image indicates the precursor ion for each CID experiment.
In Figure 4(a), doubly protonated RARARAA and the modified 4,4’-azo-bis(4-cyanovaleric acid) anionic reagent (referred to as SNVazo68) undergo an ion/ion reaction to form a singly charged electrostatic complex. Upon CID of this complex (Figure 4(b)), the R group on the side of the azo group that does not contain a sulfonate anion (designated as fragment B, whereas the side containing a sulfonate is denoted as fragment A) is lost after the nitrogen loss that is characteristic of azo functionalities. Once the B loss product is isolated ([M+H+SNA]+•), it is again subjected to CID to drive the N-hydroxysulfosuccinimide chemistry as previously described[43,44]. In Figure 4(c), the loss of sulfo-NHS indicates an amide bond formation between the reagent and either a primary amine on the peptide[38] or an unprotonated arginine side-chain.[49] Fragmentation of this product ([M+H+A]+• in Figure 4(d)) yields the characteristic R86 loss from an unmodified arginine side chain and the loss of the analogous fragment from the side chain of a modified arginine, indicated as –(R86+ A) in Figure 4(d). The control CID spectrum of [RARARAA+H]+, given in Figure 3(d), shows a quite different fragmentation pattern.
UVPD of C-I bonds (Methods 3 and 4)
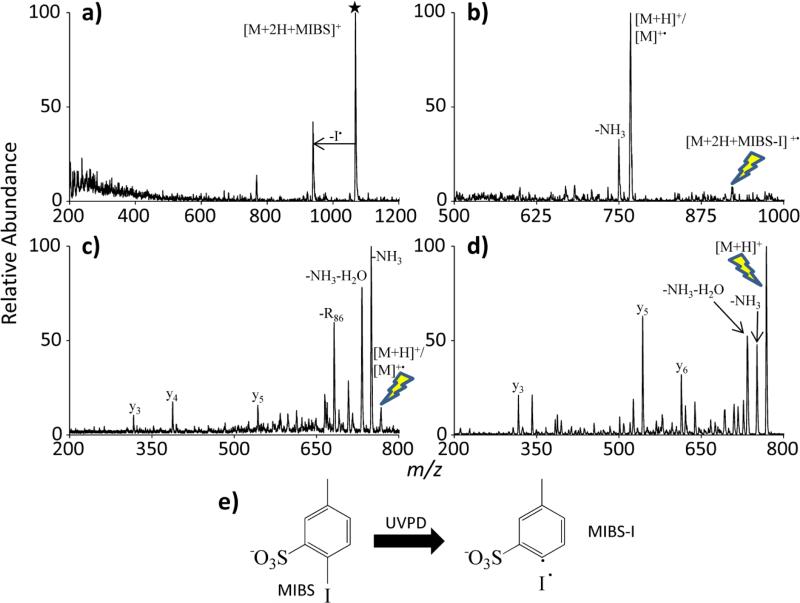
While the azo cleavage pathway is facile in collisional activation experiments, it is not the sole pathway. Methods 1 and 2 are convenient as many ion traps are CID capable, making azo bond cleavage a more widely available option. However, UVPD experiments can provide additional selectivity. Provided there are no other chromophores that absorb at 266 nm, C-I bond cleavage is exclusive during UVPD. As previously mentioned, radical peptides in this study were generated by using electrostatic interactions between a reagent with a C-I bond and a peptide or by covalently binding a C-I bond to a peptide and subjecting the product to UVPD. 2-iodo-5-methylbenzenesulfonate (MIBS) (see structure given in Figure 5) was found to be a suitable anionic reagent for hydrogen abstraction via UVPD of C-I bonds to generate the initial radical site. A typical experiment in which an ion/ion reaction results in the generation of an electrostatic complex that is then subjected to UVPD (Method 3 in Figure 1(c)) is demonstrated in Figure 5.
Figure 5.
A step by step demonstration of radical generation via UVPD of an electrostatic complex: (a) UVPD of the electrostatic complex between RARARAA (denoted as M) and MIBS, (b) CID of the resulting iodine loss product and (c) CID of the radical peptide. (d) CID of the even electron [M+H]+ is provided for comparison. (e) The C-I bond cleavage via UVPD of MIBS is provided. The lightning bolt image indicates the precursor ion for each CID experiment. The star symbol indicates the precursor ion for the UVPD experiment.
A doubly protonated peptide (RARARAA) is reacted with a singly deprotonated iodine-containing reagent (MIBS). The resulting electrostatic complex is subjected to UVPD using ~5 pulses of 266 nm photons, cleaving the C-I bond homolytically. In Figure 5(a) the elevated noise level present in the mass range of 200-400 m/z presumably arises from ionization of background species from electrons sputtered by scattered laser light. The ions generated in this way are eliminated in subsequent ion isolation steps. The photoproduct generated via iodine loss in Figure 5a has a radical site originating at the location of the C-I bond. Collisional activation of this product causes the complex to follow one of two pathways similar to those previously mentioned using electrostatic complexes and azo functionalities: proton transfer with no hydrogen transfer, or proton transfer with hydrogen abstraction from the peptide. The latter reaction leads to the radical cation whereas the former reaction yields the protonated molecule. The homebuilt 3D QIT in which the UVPD experiments are carried out does not have the resolving power to differentiate the protonated peptide from the radical cation, nor can it isolate a single m/z peak. Hence, the end product peak in Figure 5(b) is denoted as [M+H]+/[M]+•. Collisional dissociation of this product (Figure 5(c)) displays fragments commonly observed in RDD, most notably a nominal 86 loss, indicating the radical cleavage of an arginine side chain (compare with the CID spectrum of Figure 3(c) obtained using the QTRAP instrument). This fragment is not observed in the collisional activation of [RARARAA+H]+ (Figure 5(d)) (compare with the analogous spectrum generated using the QTRAP instrument in Figure 3(d)).
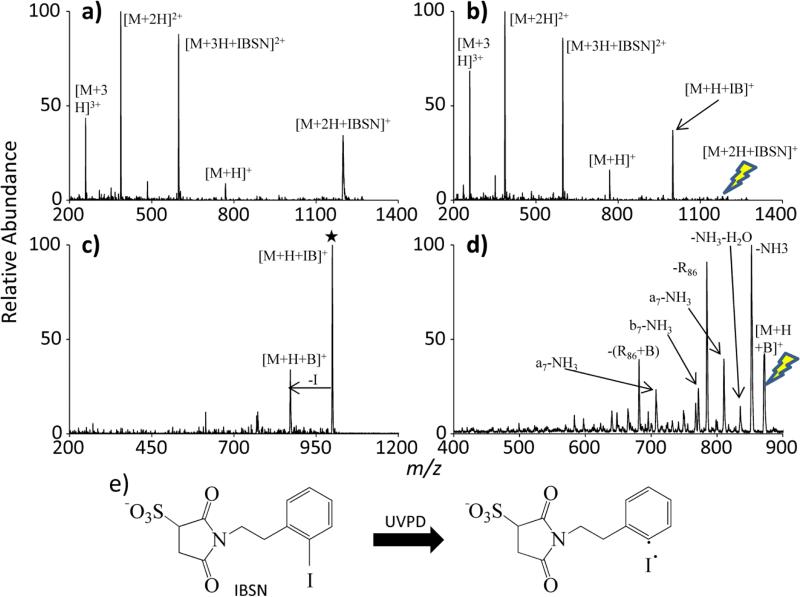
As with the collision-based methods for generating radicals, UVPD-cleavable sites can also be covalently bound to the peptide to ultimately yield RDD. This is once again achieved using sulfo-NHS chemistry. 2-iodobenzoic acid is converted to a sulfo-NHS reagent using the same method mentioned above for modification of 4,4’-azo-bis(4-cyanovaleric acid), yielding what is referred to as iodobenzene sulfo-NHS (IBSN). Figure 6 demonstrates the process of covalently attaching iodobenzene to RARARAA in the gas phase via an ion/ion reaction, followed by UVPD to create a radical cation that ultimately undergoes CID to generate RDD fragments. The ion/ion reaction between doubly protonated RARARAA and singly deprotonated IBSN yields an electrostatic complex as seen in Figure 6(a). This complex is then collisionally activated (without prior isolation) to initiate the sulfo-NHS chemistry (Figure 6(b)), which generates a peptide with a covalently attached iodobenzene moiety. With iodobenzene attached to a primary amine or guanidino group on the peptide, the product is subjected to UVPD (~5 pulses of 266 nm photons), homolytically cleaving the C-I bond. The resulting radical modified peptide exhibits typical RDD fragments when collisionally activated. The main signature again is the nominal 86 loss from an arginine side chain, both modified (-R86) and unmodified (-R86+B where B is the radical benzene moiety).
Figure 6.
The process of covalently binding a radical site to peptide. (a) Ion/ion reaction between doubly protonated RARARAA (denoted as M) and IBSN; (b) CID of the reaction complex; (c) UVPD of the modified peptide; (d) fragmentation of the modified radical peptide. (e) The C-I bond cleavage via UVPD of IBSN is provided. The lightning bolt image indicates the precursor ion of CID experiments. The star symbol indicates the precursor ion in UVPD experiments.
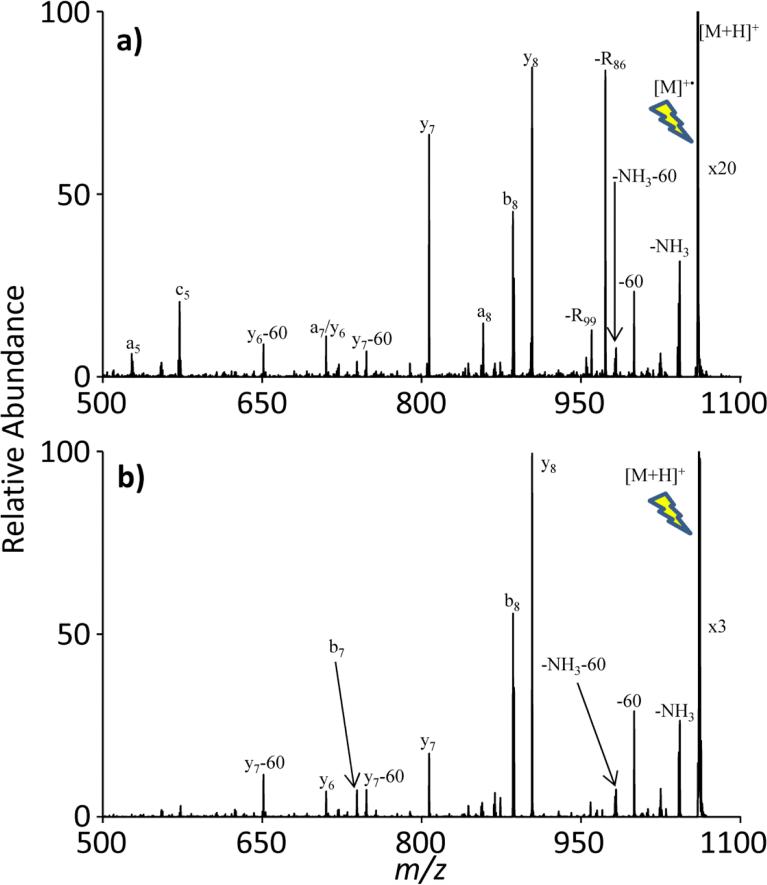
While the peptide RARARAA makes for a good system to test radical generation techniques given the on/off nature of the R86 loss depending upon spin state, the methods for the generation of unmodified radical cations were also demonstrated on bradykinin (RPPGFSPFR), a widely studied polypeptide in mass spectrometry. For example, the bradykinin radical cation was generated using collisional dissociation of azo functionalities in Direct Red 81, as described in Method 1. RDD of radical bradykinin results in the prevalent loss of R86, along with another arginine sidechain cleavage, R99. As previously observed[28], RDD leads to the generation of some a- series ions including the a5 through a8 series in Figure 7(a) while they are absent in the CID spectrum of the singly protonated, even-electron bradykinin (Figure 7(b)). It should be noted that the nominal 60 loss in both spectra is commonly seen in the collisional dissociation of peptides containing a C-terminal arginine.[50]
Figure 7.
A comparison of the CID spectra of radical bradykinin generated through (a) collisional activation of azo functionalities and (b) collisional activation of singly protonated, even-electron bradykinin. The lightning bolt image indicates the precursor ion for each CID experiment.
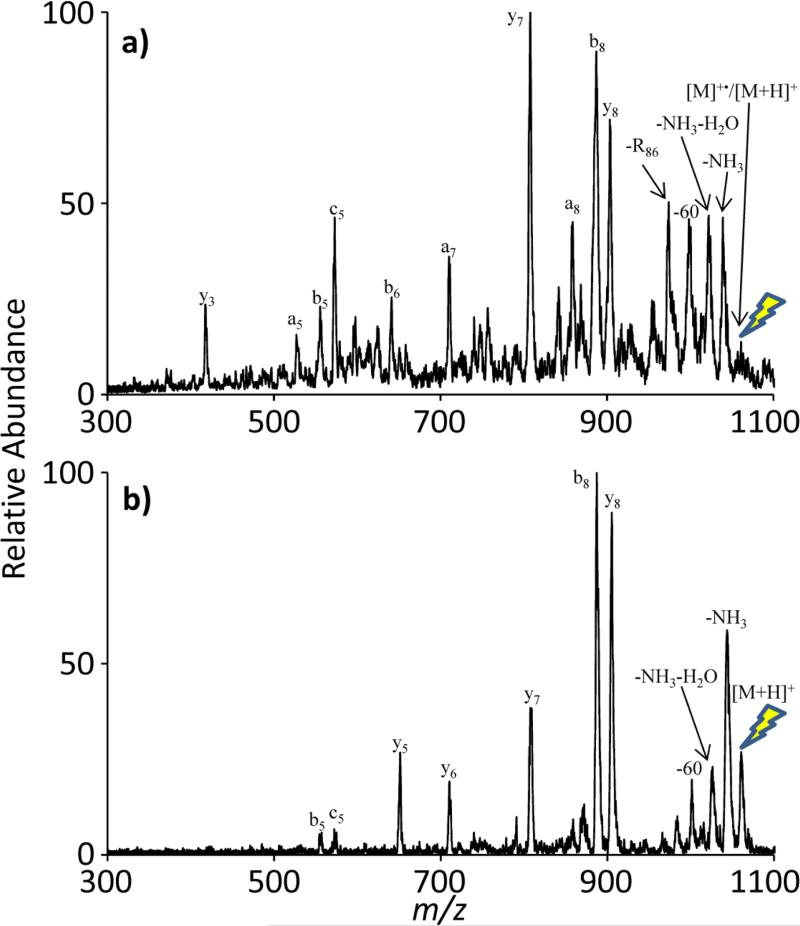
UVPD of the electrostatic complex between doubly protonated bradykinin and singly deprotonated MIBS (Method 3) also generates a bradykinin radical cation. CID of the radical cation produces the fragmentation spectrum seen in Figure 8(a). (Note that the experiments of Figure 7 and 8 were conducted using different instruments, which underlies the difference in resolving powers apparent in the spectra.) The RDD fragments observed in Figure 8(a) clearly distinguish the radical cation from the protonated molecule (see the CID spectrum of Figure 8(b)). As with the comparison of Figure 7, the R86 loss and an ions are unique to the radical cation.
Figure 8.
A comparison of CID experiments of (a) radical bradykinin generated through UVPD and (b) singly protonated, even-electron bradykinin. The lightning bolt image indicates the precursor ion for each CID experiment.
Conclusions
This work demonstrates four gas-phase methods for converting multiply-protonated peptides to radical cations via ion/ion reactions. This can be done using either electrostatic interactions or covalent modifications to bring a radical site into close proximity with a peptide. The radical sites can be generated either collisionally, as demonstrated here with azo compounds, or photolytically, as demonstrated here via UVPD of a C-I bond. The main novelty associated with the methods described here derives from the generation of the electrostatic or covalent interactions in the gas-phase rather than in solution. The relatively rapid gas-phase approaches enable a high degree of control over the identities of the reactants, their concentrations, and the interaction times. They also avoid the generation of complex mixtures in solution and concomitant complications in ionization that may arise from condensed-phase methods for the generation of complexes or covalently modified peptides. The use of oppositely charged ions to generate complexes avoids volatility problems, since both the analyte and reagent ions are generated via ESI, which might arise with ion/molecule strategies. This work extends the range of ion transformation processes that are accessible via ion/ion reactions to include the generation of radical cations so that RDD information can be obtained from ions generated by electrospray.
Acknowledgement
This work was supported by the National Institutes of Health under Grant GM 45372.
References
- 1.Dean RT, Fu SL, Stocker R, Davies MJ. Biochemistry and pathology of radical-mediated protein oxidation. Biochem. J. 1997;324:1. doi: 10.1042/bj3240001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammerum S. Distonic radical cations in gaseous and condensed phase. Mass Spectrom. Rev. 1988;42:123. [Google Scholar]
- 3.Stirk KM, Kiminkinen LKM, Kenttäma HI. Ion-molecule reactions of distonic radical cations. Chem. Rev. 1992;92:1649. [Google Scholar]
- 4.McLuckey SA, Mentinova M. Ion/Neutral, Ion/Electron, Ion/Photon, and Ion/Ion Interactions in Tandem Mass Spectrometry: Do we need them all? Are they enough? J. Am. Soc. Mass Spectrom. 2011;22:3. doi: 10.1007/s13361-010-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tureček F, Julian RR. Peptide radicals and cation radicals in the gas phase. Chem. Rev. 2013;113:6691. doi: 10.1021/cr400043s. [DOI] [PubMed] [Google Scholar]
- 6.Tureček F. Copper-Biomolecule Complexes in the Gas Phase. The Ternary Way. Mass Spectrom. Rev. 2007;26:563. doi: 10.1002/mas.20137. [DOI] [PubMed] [Google Scholar]
- 7.Hopkinson AC. Radical Cations of Amino Acids and Peptides: Structures and Stabilities. Mass Spectrom Rev. 2009;28:655. doi: 10.1002/mas.20229. [DOI] [PubMed] [Google Scholar]
- 8.Chu IK, Laskin J. Formation of peptide radical ions through dissociative electron transfer in ternary metal-ligand-peptide complexes. Eur. J. Mass Spectrom. 2011;17:543. doi: 10.1255/ejms.1156. [DOI] [PubMed] [Google Scholar]
- 9.Ly T, Julian RR. Residue-specific radical-directed dissociation of whole proteins in the gas phase. J. Am. Chem. Soc. 2008;130:351. doi: 10.1021/ja076535a. [DOI] [PubMed] [Google Scholar]
- 10.Ly T, Julian RR. Elucidating the tertiary structure of protein ions in vacuo with site specific photoinitiated reactions. J. Am. Chem. Soc. 2010;132:8602–8609. doi: 10.1021/ja910665d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Q, Yin S, Loo JL, Julian RR. Radical directed dissociation for facile identification of iodotyrosine residues using electrospray ionization mass spectrometry. Anal. Chem. 2010;82:3826. doi: 10.1021/ac100256v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ly T, Zhang X, Sun Q, Moore B, Tao Y, Julian RR. Rapid, quantitative, and site specific synthesis of biomolecular radicals from a simple photocaged precursor. Chem. Commun. 2011;47:2835. doi: 10.1039/c0cc03363d. [DOI] [PubMed] [Google Scholar]
- 13.Hamdy OM, Lam S, Julian RR. Identification of inherently antioxidant regions in proteins with radical-directed dissociation mass spectrometry. Anal. Chem. 2014;86:3653. doi: 10.1021/ac500425f. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Julian RR. Exploring radical migration pathways in peptides with positional isomers, deuterium labeling and molecular dynamic simulations. J. Am. Soc. Mass Spectrom. 2013;24:524. doi: 10.1007/s13361-012-0540-6. [DOI] [PubMed] [Google Scholar]
- 15.Laskin J, Yang Z, Lam C, Chu IK. Charge-remote fragmentation of odd-electron peptide ions. Anal. Chem. 2007;79:6607. doi: 10.1021/ac070777b. [DOI] [PubMed] [Google Scholar]
- 16.Vaisar T, Gatlin CL, Tureček F. Oxidation of peptide-copper complexes by alkali metal cations in the gas phase. J. Am. Chem. Soc. 1996;118:5314. [Google Scholar]
- 17.Vaisar T, Gatlin CL, Tureček F. Metal-ligand redox reactions in gas-phase quaternary peptide-metal complexes by electrospray ionization mass spectrometry. Int. J. Mass Spectrom. Ion Processes. 1997;162:77–87. [Google Scholar]
- 18.Hopkinson AC, Siu KWM. Peptide Radical Cations. In: Laskin J, Lifshitz C, editors. Principles of mass spectrometry applied to biomolecules. Wiley-Interscience; Hoboken, NJ: 2006. pp. 301–335. ISBN-13978-0-471-72184-0. [Google Scholar]
- 19.Barlow CK, McFayden WD, O'Hair RAJ. Formation of cationic peptide radicals by gas-phase redox reactions with trivalent chromium, manganese, iron, and cobalt complexes. J. Am. Chem. Soc. 2005;127:6109. doi: 10.1021/ja043088f. [DOI] [PubMed] [Google Scholar]
- 20.Wee S, O'Hair RAJ, McFayden WD. Side-chain radical losses from radical cations allows distinction of leucine and isoleucine residues in the isomeric pepides Gly-XXX Arg. Rapid Commun. Mass Spectrom. 2002;16:884. doi: 10.1002/rcm.658. [DOI] [PubMed] [Google Scholar]
- 21.Hao Q, Song T, Ng DCM, Quan Q, Siu C, Chu IK. Arginine-Facilitated Isomerization: Radical-Induced Dissociation of Aliphatic Radical Cationic Glycylarginyl(iso)leucine Tripeptides. J. Phys. Chem. B. 2012;116:7627. doi: 10.1021/jp301882p. [DOI] [PubMed] [Google Scholar]
- 22.Hao G, Gross SS. Electrospary tandem mass spectrometry analysis of S-and N-nitrosopeptides. facile loss of NO and radical-induced fragmentation. J. Am. Soc. Mass Spectrom. 2006;17:1725. doi: 10.1016/j.jasms.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 23.Hodyss R, Cox HA, Beauchamp JL. Bioconjugates for tunable peptide fragmentation. free radical initiated peptide sequencing (FRIPS). J. Am. Chem. Soc. 2005;127:12436. doi: 10.1021/ja052042z. [DOI] [PubMed] [Google Scholar]
- 24.Falvo F, Fiebig L, Schäfer M. Presentation of a homobifunctional azo-reagent for protein structure analysis by collision-induced dissociative chemical cross-linking: proof -of-principle. Int. J. Mass Spectrom. 2013;354-355:26. [Google Scholar]
- 25.McLafferty FW, Tureček F. Interpretation of Mass Spectra. 4th Ed. University Science Books; Mill Valley, CA: 1993. ISBN 0-935702-25-3. [Google Scholar]
- 26.Dongre AR, Jones JL, Somogyi A, Wysocki VH. Influence of peptide composition, gas-phase basicity, and chemical modification on fragmentation efficiency: Evidence for the mobile proton model. J. Am. Chem. Soc. 1996;118:8365. [Google Scholar]
- 27.Moore BN, Ly T, Julian RR. Radical conversion and migration in electron capture dissociation. J. Am. Chem. Soc. 2011;133:6997. doi: 10.1021/ja1096804. [DOI] [PubMed] [Google Scholar]
- 28.Sun QY, Nelson H, Ly T, Stoltz BM, Julian RR. Side chain chemistry mediates backbone fragmentation in hydrogen deficient peptide radicals. J. Proteome Res. 2009;8:958. doi: 10.1021/pr800592t. [DOI] [PubMed] [Google Scholar]
- 29.Wee S, O'Hair RAJ, McFadyen WD. Comparing the gas-phase fragmentation reactions of protonated radical cations of the tripeptides GXR. Int. J. Mass Spectrom. 2004;234:101. [Google Scholar]
- 30.Zhang X, Julian RR. Photoinitiated intramolecular diradical cross-linking of polyproline peptides in the gas phase. Phys. Chem. Chem. Phys. 2012;14:16243. doi: 10.1039/c2cp42242e. [DOI] [PubMed] [Google Scholar]
- 31.Lee M, Lee Y, Kang M, Park H, Seong Y, Sung BJ, Moon B, Oh HB. Disulfide bond cleavage in TEMPO-free radical initiated peptide sequencing mass spectrometry. J. Mass Spectrom. 2011;46:830. doi: 10.1002/jms.1955. [DOI] [PubMed] [Google Scholar]
- 32.Stinson CA, Xia Y. Radical induced disulfide bond cleavage within peptides via ultraviolet irradiation of an electrospray plume. Analyst. 2013;138:2840. doi: 10.1039/c3an00303e. [DOI] [PubMed] [Google Scholar]
- 33.Vaisar T, Gatlin CL, Rao RD, Seymour JL, Türecek F. Sequence information, distinction and quantification of C-terminal leucine and isoleucine in ternary complexes of tripeptides with Cu(II) and 2,2′-bipyridine. J. Mass Spectrom. 2001;36:306. doi: 10.1002/jms.135. [DOI] [PubMed] [Google Scholar]
- 34.Pham HT, Julian RR. Mass shifting and radical delivery with crown ether attachment for separation and analysis of phophatidylethanolamine lipids. Anal Chem. 2014;86:3020. doi: 10.1021/ac403754j. [DOI] [PubMed] [Google Scholar]
- 35.Pham HT, Julian RR. Radical delivery and fragmentation for structural analysis of glycerophospholipids. Int J. Mass Spectrom. 2014;370:58. [Google Scholar]
- 36.Pham HT, Ly T, Trevitt AJ, Mitchell TW, Blanksby SJ. Differentiation of complex lipid isomers by radical-directed dissociation mass spectrometry. Anal Chem. 2012;84:7525. doi: 10.1021/ac301652a. [DOI] [PubMed] [Google Scholar]
- 37.Brügger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc. Natl. Acac. Sci. U.S.A. 1997;94:2339. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han X, Gross RW. Structural determination of picomole amounts of phospholipids via electrospray ionization tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 1995;6:1202. doi: 10.1016/1044-0305(95)00568-4. [DOI] [PubMed] [Google Scholar]
- 39.Pitteri SJ, McLuckey SA. Recent Developments in the Ion/Ion Chemistry of High-Mass Multiply Charged Ions. Mass Spectrom. Rev. 2005;24:931. doi: 10.1002/mas.20048. [DOI] [PubMed] [Google Scholar]
- 40.Prentice BM, McLuckey SA. Gas-Phase Ion/Ion Reactions of Peptides and Proteins: Acid/Base, Redox, and Covalent Chemistries. Chem. Commun. 2013;49:947. doi: 10.1039/c2cc36577d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McLuckey SA. Biomolecule Ion/Ion Reactions. In: Laskin J, Lifshitz C, editors. Principles of Mass Spectrometry Applied to Biomolecules. Wiley-Interscience; Hoboken, NJ: 2006. pp. 519–564. ISBN-13978-0-471-72184-0. [Google Scholar]
- 42.Xia Y, Liang X, McLuckey SA. Pulsed Dual Electrospray Ionization for Ion/Ion Reactions. J. Am. Soc. Mass Spectrom. 2005;16:1750. doi: 10.1016/j.jasms.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 43.Mentinova M, McLuckey SA. Covalent Modification of Gaseous Peptide Ions with N Hydroxysuccinimide Ester Reagent Ions. J. Am. Chem. Soc. 2010;132:18248. doi: 10.1021/ja107286p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mentinova M, Barefoot NZ, McLuckey SA. Solution Versus Gas-Phase Modification of Peptide Cations with NHS-Ester Reagents. J. Am. Soc. Mass Spectrom. 2012;23:282. doi: 10.1007/s13361-011-0291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xia Y, Wu J, McLuckey SA, Londry FA, Hager JW. Mutual Storage Mode Ion/Ion Reactions in a Hybrid Linear Ion Trap. J. Am. Soc. Mass Spectrom. 2004;16:71. doi: 10.1016/j.jasms.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 46.Londry FA, Hager JW. Mass Selective Axial Ion Ejection from a Linear Quadrupole Ion Trap. J. Am. Soc. Mass Spectrom. 2003;14:1130. doi: 10.1016/S1044-0305(03)00446-X. [DOI] [PubMed] [Google Scholar]
- 47.Wells JM, Chrisman PA, McLuckey SA. “Dueling” ESI: Instrumentation to study ion/ion reaction of electrospray-generated cations and anions. J. Am. Soc. Mass Spectrom. 2002;13:614. doi: 10.1016/S1044-0305(01)00364-6. [DOI] [PubMed] [Google Scholar]
- 48.Kaiser RE, Cooks RG, Stafford GC, Syka JEP, Hemberger PH. Operation of a quadrupole ion trap mass spectrometer to achieve high mass/charge ratios. Int J. Mass Spectrom. Ion Processes. 1991;106:79. [Google Scholar]
- 49.McGee WM, Mentinova M, McLuckey SA. Gas-phase conjugation to arginine residues in polypeptide ions via N-hydroxysuccinimide ester-based reagent ions. J. Am. Chem. Soc. 2012;134:11412. doi: 10.1021/ja304778j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deery MJ, Summerfield SG, Buzy A, Jenning KR. A mechanism for the loss of 60 u from peptides containing an arginine residue at the C-terminus. J. Am. Mass Spectrom. 1997;8:253. [Google Scholar]