Abstract
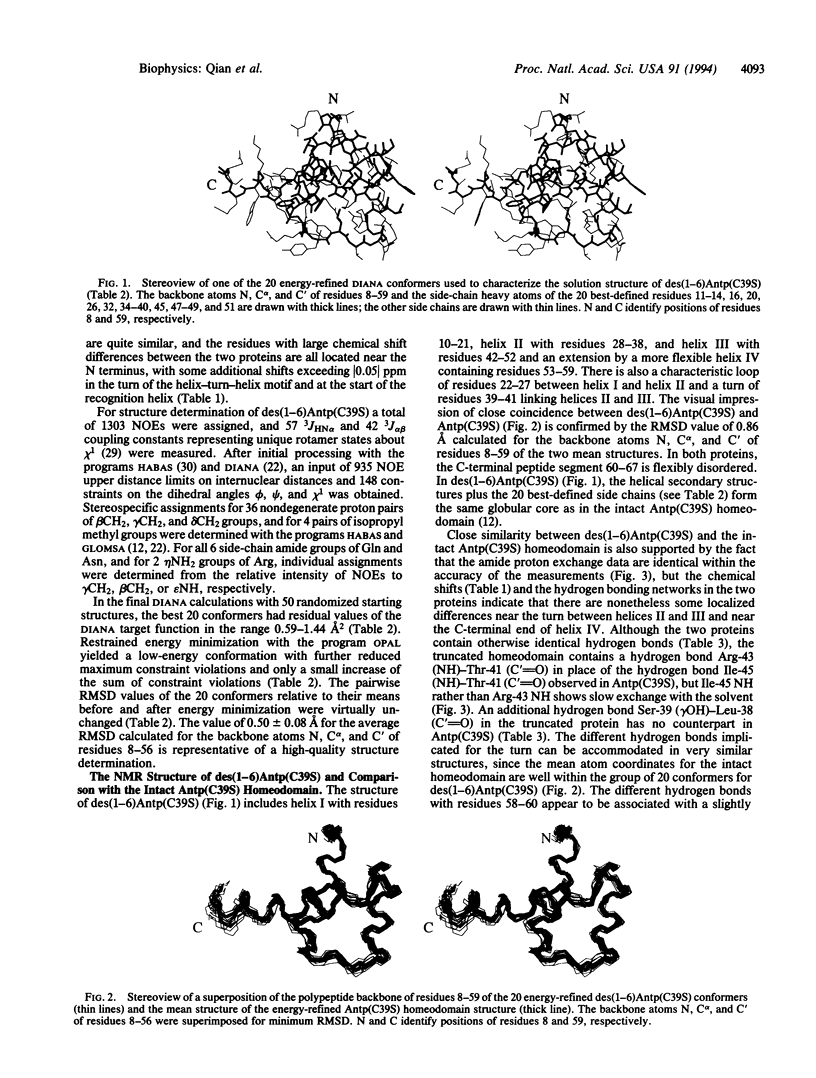
The nuclear magnetic resonance (NMR) solution structure of an N-terminally truncated mutant Antennapedia homeodomain, des(1-6)Antp(C39S), has been determined from 935 nuclear Overhauser effect upper distance constraints and 148 dihedral angle constraints by using the programs DIANA and OPAL. Twenty conformers representing the solution structure of des(1-6)Antp(C39S) have an average root-mean-square distance relative to the mean coordinates of 0.56 A for the backbone atoms of residues 8-59. Comparison with the intact Antp(C39S) homeodomain shows that the two proteins have identical molecular architectures. The removal of the N-terminal residues 1-6, which are flexibly disordered in the intact homeodomain, causes only strictly localized structure variations and does not noticeably affect the adjoining helix I from residues 10-21. The DNA-binding constant of des(1-6)Antp(C39S) is approximately 10-fold reduced relative to the intact Antp(C39S) homeodomain, which can now be attributed to the absence of the previously reported contacts of the N-terminal polypeptide segment of the intact Antp(C39S) homeodomain with the minor groove of the DNA duplex.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Affolter M., Percival-Smith A., Müller M., Billeter M., Qian Y. Q., Otting G., Wüthrich K., Gehring W. J. Similarities between the homeodomain and the Hin recombinase DNA-binding domain. Cell. 1991 Mar 8;64(5):879–880. doi: 10.1016/0092-8674(91)90311-l. [DOI] [PubMed] [Google Scholar]
- Affolter M., Percival-Smith A., Müller M., Leupin W., Gehring W. J. DNA binding properties of the purified Antennapedia homeodomain. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4093–4097. doi: 10.1073/pnas.87.11.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billeter M., Braun W., Wüthrich K. Sequential resonance assignments in protein 1H nuclear magnetic resonance spectra. Computation of sterically allowed proton-proton distances and statistical analysis of proton-proton distances in single crystal protein conformations. J Mol Biol. 1982 Mar 5;155(3):321–346. doi: 10.1016/0022-2836(82)90008-0. [DOI] [PubMed] [Google Scholar]
- Billeter M., Qian Y. Q., Otting G., Müller M., Gehring W., Wüthrich K. Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex. J Mol Biol. 1993 Dec 20;234(4):1084–1093. doi: 10.1006/jmbi.1993.1661. [DOI] [PubMed] [Google Scholar]
- Billeter M., Qian Y., Otting G., Müller M., Gehring W. J., Wüthrich K. Determination of the three-dimensional structure of the Antennapedia homeodomain from Drosophila in solution by 1H nuclear magnetic resonance spectroscopy. J Mol Biol. 1990 Jul 5;214(1):183–197. doi: 10.1016/0022-2836(90)90155-f. [DOI] [PubMed] [Google Scholar]
- Eccles C., Güntert P., Billeter M., Wüthrich K. Efficient analysis of protein 2D NMR spectra using the software package EASY. J Biomol NMR. 1991 Jul;1(2):111–130. doi: 10.1007/BF01877224. [DOI] [PubMed] [Google Scholar]
- Furukubo-Tokunaga K., Flister S., Gehring W. J. Functional specificity of the Antennapedia homeodomain. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6360–6364. doi: 10.1073/pnas.90.13.6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güntert P., Braun W., Wüthrich K. Efficient computation of three-dimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. J Mol Biol. 1991 Feb 5;217(3):517–530. doi: 10.1016/0022-2836(91)90754-t. [DOI] [PubMed] [Google Scholar]
- Güntert P., Qian Y. Q., Otting G., Müller M., Gehring W., Wüthrich K. Structure determination of the Antp (C39----S) homeodomain from nuclear magnetic resonance data in solution using a novel strategy for the structure calculation with the programs DIANA, CALIBA, HABAS and GLOMSA. J Mol Biol. 1991 Feb 5;217(3):531–540. doi: 10.1016/0022-2836(91)90755-u. [DOI] [PubMed] [Google Scholar]
- Güntert P., Wüthrich K. Improved efficiency of protein structure calculations from NMR data using the program DIANA with redundant dihedral angle constraints. J Biomol NMR. 1991 Nov;1(4):447–456. doi: 10.1007/BF02192866. [DOI] [PubMed] [Google Scholar]
- Kissinger C. R., Liu B. S., Martin-Blanco E., Kornberg T. B., Pabo C. O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990 Nov 2;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Marion D., Wüthrich K. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem Biophys Res Commun. 1983 Jun 29;113(3):967–974. doi: 10.1016/0006-291x(83)91093-8. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Krumlauf R. Homeobox genes and axial patterning. Cell. 1992 Jan 24;68(2):283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- Müller M., Affolter M., Leupin W., Otting G., Wüthrich K., Gehring W. J. Isolation and sequence-specific DNA binding of the Antennapedia homeodomain. EMBO J. 1988 Dec 20;7(13):4299–4304. doi: 10.1002/j.1460-2075.1988.tb03328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagayama K., Wüthrich K. Structural interpretation of vicinal proton-proton coupling constants 3JH alpha H beta in the basic pancreatic trypsin inhibitor measured by two-dimensional J-resolved NMR spectroscopy. Eur J Biochem. 1981 Apr;115(3):653–657. doi: 10.1111/j.1432-1033.1981.tb06252.x. [DOI] [PubMed] [Google Scholar]
- Otting G., Qian Y. Q., Billeter M., Müller M., Affolter M., Gehring W. J., Wüthrich K. Protein--DNA contacts in the structure of a homeodomain--DNA complex determined by nuclear magnetic resonance spectroscopy in solution. EMBO J. 1990 Oct;9(10):3085–3092. doi: 10.1002/j.1460-2075.1990.tb07505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- Percival-Smith A., Müller M., Affolter M., Gehring W. J. The interaction with DNA of wild-type and mutant fushi tarazu homeodomains. EMBO J. 1990 Dec;9(12):3967–3974. doi: 10.1002/j.1460-2075.1990.tb07617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y. Q., Billeter M., Otting G., Müller M., Gehring W. J., Wüthrich K. The structure of the Antennapedia homeodomain determined by NMR spectroscopy in solution: comparison with prokaryotic repressors. Cell. 1989 Nov 3;59(3):573–580. doi: 10.1016/0092-8674(89)90040-8. [DOI] [PubMed] [Google Scholar]
- Qian Y. Q., Otting G., Billeter M., Müller M., Gehring W., Wüthrich K. Nuclear magnetic resonance spectroscopy of a DNA complex with the uniformly 13C-labeled Antennapedia homeodomain and structure determination of the DNA-bound homeodomain. J Mol Biol. 1993 Dec 20;234(4):1070–1083. doi: 10.1006/jmbi.1993.1660. [DOI] [PubMed] [Google Scholar]
- Qian Y. Q., Otting G., Furukubo-Tokunaga K., Affolter M., Gehring W. J., Wüthrich K. NMR structure determination reveals that the homeodomain is connected through a flexible linker to the main body in the Drosophila Antennapedia protein. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10738–10742. doi: 10.1073/pnas.89.22.10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Wagner G., Wüthrich K. Sequential resonance assignments in protein 1H nuclear magnetic resonance spectra. Basic pancreatic trypsin inhibitor. J Mol Biol. 1982 Mar 5;155(3):347–366. doi: 10.1016/0022-2836(82)90009-2. [DOI] [PubMed] [Google Scholar]
- Widmer H., Billeter M., Wüthrich K. Three-dimensional structure of the neurotoxin ATX Ia from Anemonia sulcata in aqueous solution determined by nuclear magnetic resonance spectroscopy. Proteins. 1989;6(4):357–371. doi: 10.1002/prot.340060403. [DOI] [PubMed] [Google Scholar]
- Wolberger C., Vershon A. K., Liu B., Johnson A. D., Pabo C. O. Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991 Nov 1;67(3):517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- Zeng W., Andrew D. J., Mathies L. D., Horner M. A., Scott M. P. Ectopic expression and function of the Antp and Scr homeotic genes: the N terminus of the homeodomain is critical to functional specificity. Development. 1993 Jun;118(2):339–352. doi: 10.1242/dev.118.2.339. [DOI] [PubMed] [Google Scholar]



