Abstract
BACKGROUND
Although correlation between cytosine-adenine-guanine (CAG) repeat length and age of Huntington disease (HD) onset is well known, improved prediction of onset would be advantageous for clinical trial design and prognostic counseling. We compared genetic, demographic, motor, cognitive, psychiatric, functional and imaging measures for tracking progression and predicting conversion to manifest HD.
METHODS
N=1078 research participants with the gene mutation for HD, but without a rating of 4 on the Diagnostic Confidence Level (DCL) following administration of the 15-item motor assessment of the Unified Huntington’s Disease Rating Scale. Participants were from 33 world wide sites and followed for up to 12 years (mean=5, SD=3·3) over the period 2001–2013. A subset of 225 participants prospectively converted to manifest HD according to the DCL (“meets the operational definition of the unequivocal presence of an otherwise unexplained extrapyramidal movement disorder in a subject at risk for HD” with ≥99% confidence). Joint modeling of longitudinal and survival data was used to examine the extent to which baseline and change of 40 variables analyzed separately was predictive of CAG-adjusted age at motor diagnosis.
FINDINGS
Cross-sectional and longitudinal clinical and imaging measures were significant predictors of motor diagnosis beyond CAG repeat length and age. The strongest predictors in the top three phenotypic domains were total motor score (motor), putamen volume (imaging), and Stroop word test (cognitive). A one standard deviation (SD) difference in total motor score increased the risk of a motor diagnosis by 3·1 times (95% CI=[2·3,4·2]), one SD loss in putamen volume increased risk by 3·3 times ([2·4,4·7]) and one SD cognitive decline increased risk by 2·3 ([1·9,2·9]).
INTERPRETATION
Prediction of HD diagnosis can be considerably improved beyond that obtained by CAG repeat length and age alone. Such knowledge about potential predictors of manifest HD should inform discussions about revisions to guidelines for diagnosis, and prognosis, and counselling, and might be useful in guiding selection of participants and outcome measures for clinical trials.
FUNDING
National Institutes of Health, National Institute of Neurological Disorders and Stroke, and CHDI Foundation, Inc.
INTRODUCTION
Huntington disease (HD) is an autosomal dominant neurodegenerative disease caused by expansion of the trinucleotide cytosine-adenine-guanine (CAG) in the first exon of the Huntingtin gene. There is a well-known relationship between the length of the CAG mutation and the age at disease onset1 although there is also substantial individual variation. Over the past decade, Neurobiological Predictors of Huntington’s Disease (PREDICT-HD; NS040068) and other studies2–11 documented disease-related changes of clinical features and biomarkers in persons with the CAG expansion, but not yet diagnosable with HD.12,13 Useful clinical and biological markers should be predictive of landmark events, such as clinical motor diagnosis. In this study we compare genetic, demographic, motor, cognitive, psychiatric, functional, and imaging measures for predicting conversion to manifest HD in the largest study of gene mutation premanifest participants, culminating in 225 prospectively diagnosed HD patients. Improved predictability of HD diagnosis could advance research design, experimental trials, and clinical care through improved prognosis and earlier intervention.
METHODS
Participants
Participants were 1078 HD gene-expanded (CAG>35) individuals from the PREDICT-HD study who had less than the highest rating on the Diagnostic Confidence Level (DCL) (DCL< 4) of the Unified Huntington’s Disease Rating Scale (UHDRS) at the beginning of the study (see Table 1). Data were collected from 2001 to 2013, and all participants had prior and independent genetic testing for HD. Exclusion criteria included presence of other central nervous system disease, injury, or developmental disorder, or evidence of an unstable medical or psychiatric illness. All participants provided informed written consent (with full study approval by 33 site institutional review boards) and were treated consistent with ethical standards. Mean years in the study was five (SD=3·3) with a range from one to twelve. There were 959 participants (89%) who had two or more waves (years) of data, and 118 who had only one time point (11%) (see web extra material for additional details). A subset of 225 HD gene-expanded participants received a motor diagnosis during the study, according to the DCL (“meets the operational definition of the unequivocal presence of an otherwise unexplained extrapyramidal movement disorder in a subject at risk for HD” with ≥99% confidence). The DCL is administered by a movement disorder specialist after conducting the 15-item standardized motor assessment. PREDICT-HD also had N = 305 non-gene-expanded controls who were used only for an ancillary analysis reported in the web extra material. All abnormalities in clinical and imaging data were forwarded to clinical investigators at the relevant site for additional review and discussion. When the data were found to suggest abnormalities in function or brain imaging, follow-up clinical investigations were encouraged. Findings were reviewed by the Executive Committee, who made decisions regarding use of the data in the study: if a control participant was found to have a previously undetected neurological diagnosis, the participant and all of his or her data were excluded.
Table 1.
Descriptive statistics for non-converters, converters, and the combined sample (mean (SD) or counts (%)).
| Non-Converters | Converters | Combined | |
|---|---|---|---|
| N | 853 | 225 | 1078 |
| CAGa | 42·21 (2·58) | 43·57 (2·85) | 42·49 (2·69) |
| Agea | 38·92 (10·24) | 43·03 (10·31) | 39·78 (10·39) |
| CAPa | 334·89 (82·74) | 436·59 (81·82) | 356·12 (92·30) |
| Duration | 4·28 (3·31) | 6·66 (2·48) | 4·78 (3·30) |
| Educationa | 14·56 (2·62) | 14·08 (2·50) | 14·46 (2·60) |
| Female N | 540 (63%) | 147 (65%) | 687 (64%) |
Measured at study entry. Age and education are expressed in years. Duration=years in the study.
CAG=cytosine-adenine-guanine expansion length. CAP=(Age at Baseline) × (CAG 33·66).
Measures
The web extra material details the 40 longitudinal measures, which were selected based on their sensitivity to the detection and progression of disease.12 Motor variables were total motor score (TMS) from the UHDRS and the chorea, bradykinesia, oculomotor, dystonia and rigidity subdomains from the 15-item standardized motor assessment. Cognitive variables included the Stroop Color and Word Test (three measures), the Symbol Digit Modalities Test, University of Pennsylvania Smell Identification Test, emotion recognition, speeded tapping, time production or paced tapping, and the Trail Making Test. Psychiatric measures included four subscales of the Symptom Checklist 90 (SCL90), the Beck Depression Inventory (BDI), and three subscales of the Frontal Systems Behavioral Scale. Imaging measures included intracranial-corrected volumes (ICV) for putamen, accumbens, caudate, hippocampus, thalamus, globuspallidus, cerebral spinal fluid, lobar white and gray matter. Functional outcome variables included the total functional capacity (TFC) and functional activity scale from the UHDRS, the World Health Organization Disability Assessment Schedule, and the Everyday Cognition Rating Scale (ECOG). Motor diagnosis was defined as a rating of “4” on the Diagnostic Confidence Level (DCL) of the UHDRS.
Statistical analyses
The primary goal was to examine the ability of each variable to predict time to motor diagnosis (first occurrence) over and above CAG repeat length, age, and their interaction. Time to diagnosis and longitudinal change were simultaneously modeled using joint modeling for survival and longitudinal data14,15 (see web extra material for details). The intent was to model progression over the entire lifespan by using the time metric of age adjusted for CAG expansion.
The survival model was a Cox regression model and the longitudinal model was a linear mixed effects regression (LMER) model. The time metric for both was age adjusted for genetic burden (CAG expansion), known as the CAG-Age Product (CAP), CAP = age × (CAG − 33 · 66).16 CAP reflects the cumulative exposure to the effects of mutant huntingtin, and is similar to other CAG and age based measures.17–19 CAP at motor diagnosis or censoring was used for the observation time in the Cox model, and CAP was the longitudinal time metric for the LMER model. It is emphasized that CAP as specified in this analysis is time-varying and represents age adjusted for CAG expansion. Because of the variability in age at study entry, the annual measurements span virtually the entire adult age range, which allows inferences about motor diagnosis risk over the HD lifespan. The natural CAP intercept (baseline) is CAP=0, denoting birth. Predictive power is meaningless at birth because the clinical variables are not measured. We chose the baseline cross-section of CAP=290 as the intercept because this is the value at which motor signs begin to appear in the PREDICT-HD cohort.6 At this baseline the predictors might have sufficient variability to correlate with later motor diagnosis. CAP=290 corresponds to the rounded ages of 40, 35, and 28 for individuals with the CAG sample quartile values of 41, 42, and 44, respectively.
Each outcome was standardized and cubic splines based on five knots (1, 25, 50, 75, 99 percentiles) were used in the LMER portion to model nonlinear change.12,13,20,21 Two joint models were fit for each variable: a reduced model that provided information about a marker’s baseline prediction of the hazard for motor diagnosis (at CAP=290), and a full model that incorporated change of the marker in the prediction. A statistically significant γ estimate meant a variable accounted for variability in the timing of diagnosis over and above CAG expansion and age. The covariates in all models were gender and education. For cognitive variables, depression (BDI) was added to account for mood changes. For imaging variables, field strength was added because some sites updated their scanners during the study. The Hazard Ratio (HR) was computed as exp (γ) and served as the primary effect size (HR-1 was used when the γ estimate was negative). A significant HR indicates a variable that adds to prediction beyond that of CAG and age (the latter variables being indexed by CAP).
A subsequent analysis was performed to characterize the risk of motor diagnosis over the lifespan. Individual fitted values from the LMER spline model were used to obtain baseline values at CAP=290. The baseline information was used in a separate (not joint) Cox model to predict time to diagnosis along with the covariates. The cumulative hazard was estimated based on the fitted models for various baseline predictor values.
An ancillary analysis examined the natural history of key variables from the premanifest phase through diagnosis. All converters were used for this analysis. The time metric was years to diagnosis and cubic spline curves were used with LMER models to allow for nonlinear trends over time.
Role of the funding source
The study sponsors had no role in study design, in the collection, analysis, and interpretation of the data, in the writing of the report or in the decision to submit the paper for publication. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
RESULTS
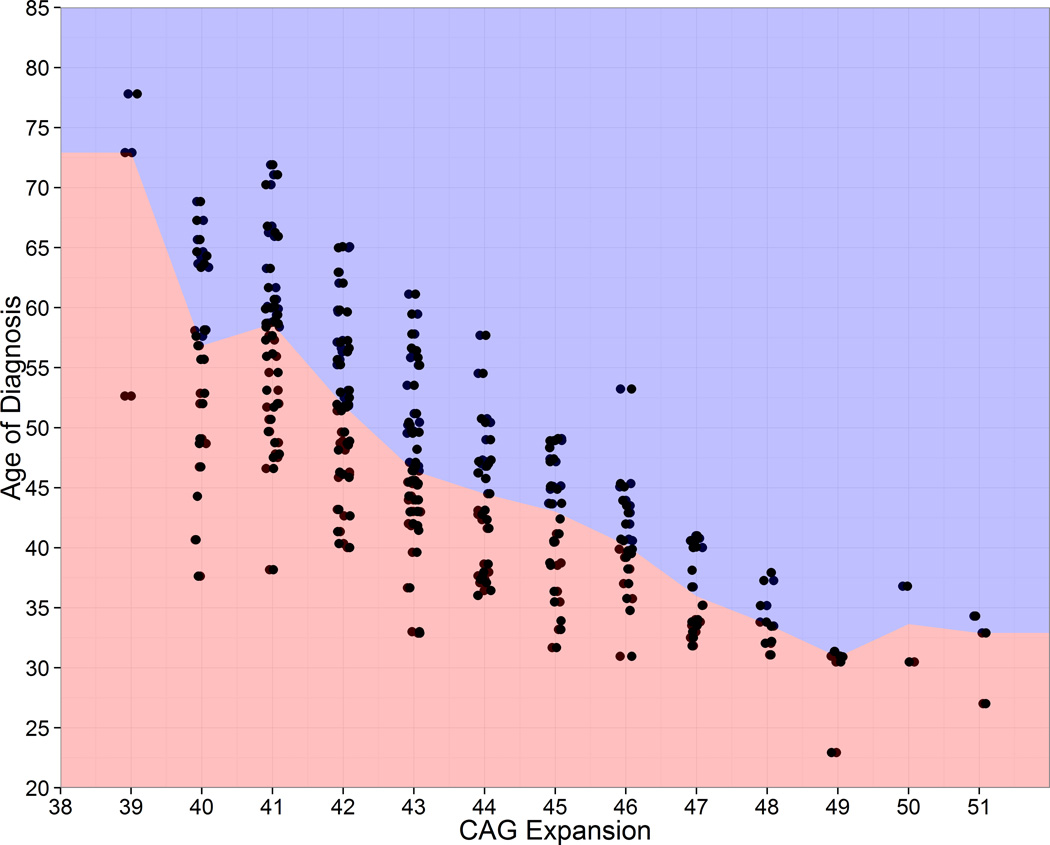
Figure 1 shows age at diagnosis as a function of CAG expansion for the converters who obtained a motor diagnosis during the study. The squared correlation between CAG repeat length and age at HD diagnosis was r2=0·53. For every CAG stratum the figure shows variation for age of diagnosis. Considering CAG=40, the range of diagnosis age is 31 years, and the difference between the first and third quartile is 15 years. Table 1 shows descriptive statistics for converters, non-converters, and the combined sample at study entry. Additional information is listed in the web extra material. Mean CAP at motor diagnosis was 447, which for the sample CAG quartiles of 41, 42, and 44, represents the rounded ages of 61, 54, and 43, respectively.
Figure 1. Observed age of diagnosis by CAG expansion for N=225 prospective converters. Observations were slightly jittered horizontally to reveal overlapping cases.
Figure 1 shows age of diagnosis as a function of CAG expansion for the N=225 people who obtained a motor diagnosis during the 12-year PREDICT-HD study (i.e., the converters). None of the individuals in Figure 1 were diagnosed at study entry and all have repeated measures, some with up to 12 years of observations (mean=5 years). The color change is at the median age for each CAG. As seen in the figure, the age at diagnosis can vary widely for individuals with the same CAG expansion. Considering CAG=40, the range is 31 years, and the difference between the first and third quartile is 15 years. The variability is indicative of the need to include variables in addition to CAG expansion with the goal of improving the accuracy of predicting the time of diagnosis. The main goal of this study is to improve predictive accuracy by identifying variables that are correlated with diagnosis, over and above CAG expansion. CAG=cytosine-adenine-guanine.
Table 2 shows the joint modeling results (each variable was tested separately). The longitudinal variables are grouped by domain and sorted within domain based on the absolute Z-value of γ from the full model. N/A indicates the full model could not be estimated due to a lack of variability in the individual rate of change over time.
Table 2.
Joint modeling results. Within each domain, variables are ranked by absolute Z-value for the association parameter (γ) of the full model.
| Cross-Sectional Prediction (Reduced Model) |
Longitudinal Prediction (Full Model) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variable | Domain | N | N* | Event | γ(SE) | Z-value(p) | HR or HR−1 (95% CI) |
γ(SE) | Z-value(p) | HR or HR−1 (95% CI) |
| TMS | Motor | 1073 | 3661 | 225 | 0·98(0·16) | 6·20(<·0001) | 2·65(1·95,3·61) | 1·12(0·16) | 7·23(<·0001) | 3·07(2·26,4·16) |
| Chorea | Motor | 1073 | 3661 | 225 | 1·09(0·17) | 6·53(<·0001) | 2·99(2·15,4·15) | 1·16(0·16) | 7·08(<·0001) | 3·19(2·31,4·40) |
| Brady | Motor | 1073 | 3661 | 225 | 0·76(0·15) | 5·05(<·0001) | 2·14(1·59,2·87) | 0·89(0·13) | 6·80(<·0001) | 2·44(1·89,3·16) |
| Ocular | Motor | 1073 | 3661 | 225 | 0·73(0·19) | 3·94(<·0001) | 2·07(1·44,2·98) | 0·82(0·20) | 4·07(<·0001) | 2·28(1·53,3·39) |
| Rigidity | Motor | 1073 | 3661 | 225 | 0·29(0·18) | 1·61(0·1073) | 1·33(0·94,1·88) | 0·36(0·17) | 2·17(0·0298) | 1·44(1·04,2·00) |
| Dystonia | Motor | 1073 | 3661 | 225 | 0·66(0·25) | 2·57(0·0101) | 1·93(1·17,3·17) | N/A | N/A | N/A |
| Putamen | Imaging | 980 | 1774 | 147 | −1·11(0·15) | −7·36(<·0001) | 3·03(2·25,4·06) | −1·20(0·17) | −6·95(<·0001) | 3·32(2·37,4·65) |
| Hippocampus | Imaging | 980 | 1774 | 147 | −0·58(0·11) | −5·29(<·0001) | 1·78(1·44,2·21) | −0·64(0·11) | −6·02(<·0001) | 1·90(1·54,2·35) |
| Lobar Gray | Imaging | 967 | 1703 | 146 | −0·59(0·14) | −4·38(<·0001) | 1·81(1·39,2·36) | −0·75(0·13) | −5·95(<·0001) | 2·11(1·65,2·69) |
| CSF | Imaging | 985 | 1785 | 148 | 0·38(0·08) | 4·87(<·0001) | 1·47(1·26,1·71) | 0·48(0·09) | 5·50(<·0001) | 1·62(1·36,1·92) |
| Accumbens | Imaging | 980 | 1774 | 147 | −0·87(0·19) | −4·48(<·0001) | 2·39(1·63,3·49) | −1·00(0·18) | −5·48(<·0001) | 2·71(1·90,3·88) |
| Globus | Imaging | 980 | 1774 | 147 | −1·11(0·26) | −4·29(<·0001) | 3·04(1·83,5·04) | −1·29(0·25) | −5·19(<·0001) | 3·64(2·23,5·92) |
| Caudate | Imaging | 980 | 1774 | 147 | −0·73(0·18) | −4·11(<·0001) | 2·07(1·46,2·92) | −0·85(0·18) | −4·84(<·0001) | 2·34(1·66,3·29) |
| Thalamus | Imaging | 980 | 1774 | 147 | −0·34(0·14) | −2·49(0·0129) | 1·40(1·07,1·84) | −0·35(0·14) | −2·60(0·0093) | 1·42(1·09,1·85) |
| Lobar White | Imaging | 952 | 1659 | 143 | −0·09(0·08) | −1·08(0·2795) | 1·09(0·93,1·29) | −0·12(0·07) | −1·67(0·0946) | 1·13(0·98,1·31) |
| Stroop-Wo | Cognitive | 979 | 2879 | 178 | −0·75(0·11) | −7·13(<·0001) | 2·12(1·72,2·61) | −0·84(0·11) | −7·79(<·0001) | 2·32(1·88,2·87) |
| Smell-ID | Cognitive | 962 | 2139 | 159 | −0·45(0·08) | −5·59(<·0001) | 1·57(1·34,1·83) | −0·54(0·08) | −6·87(<·0001) | 1·72(1·47,2·00) |
| SDMT | Cognitive | 979 | 2876 | 178 | −0·68(0·12) | −5·84(<·0001) | 1·97(1·57,2·48) | −0·72(0·12) | −6·20(<·0001) | 2·05(1·63,2·57) |
| Stroop-Co | Cognitive | 979 | 2877 | 178 | −0·73(0·15) | −4·98(<·0001) | 2·09(1·56,2·78) | −0·81(0·13) | −6·15(<·0001) | 2·25(1·74,2·91) |
| Stroop-In | Cognitive | 979 | 2869 | 178 | −0·76(0·15) | −4·89(<·0001) | 2·13(1·57,2·89) | −0·77(0·13) | −5·88(<·0001) | 2·17(1·68,2·81) |
| Timing | Cognitive | 759 | 1391 | 104 | 0·44(0·12) | 3·58(0·0003) | 1·55(1·22,1·97) | 0·59(0·11) | 5·43(<·0001) | 1·81(1·46,2·23) |
| Sp-Tapping | Cognitive | 764 | 1392 | 104 | 0·38(0·10) | 3·67(0·0002) | 1·47(1·20,1·80) | 0·46(0·09) | 4·86(<·0001) | 1·58(1·32,1·91) |
| EmoRec | Cognitive | 765 | 1406 | 103 | −0·42(0·14) | −2·99(0·0028) | 1·52(1·15,1·99) | −0·52(0·14) | −3·73(0·0002) | 1·68(1·28,2·21) |
| TMT-A | Cognitive | 974 | 2217 | 167 | 0·18(0·07) | 2·50(0·0124) | 1·20(1·04,1·38) | 0·21(0·08) | 2·55(0·0109) | 1·24(1·05,1·45) |
| TMT-B | Cognitive | 970 | 2197 | 165 | 0·16(0·09) | 1·89(0·0592) | 1·18(0·99,1·40) | N/A | N/A | N/A |
| F-Exc-C | Psychiatric | 1002 | 3071 | 191 | 0·53(0·09) | 5·77(<·0001) | 1·69(1·42,2·03) | 0·62(0·09) | 6·64(<·0001) | 1·86(1·55,2·23) |
| S-OC-C | Psychiatric | 1009 | 3120 | 195 | 0·56(0·10) | 5·34(<·0001) | 1·75(1·42,2·15) | 0·64(0·11) | 5·93(<·0001) | 1·90(1·54,2·35) |
| F-Apa-C | Psychiatric | 1002 | 3071 | 191 | 0·37(0·08) | 4·53(<·0001) | 1·45(1·23,1·70) | 0·46(0·08) | 5·37(<·0001) | 1·58(1·34,1·86) |
| S-GSI-C | Psychiatric | 988 | 2943 | 184 | 0·46(0·09) | 4·89(<·0001) | 1·58(1·32,1·90) | 0·51(0·10) | 5·34(<·0001) | 1·67(1·38,2·02) |
| F-Dis-C | Psychiatric | 1002 | 3071 | 191 | 0·36(0·09) | 3·84(0·0001) | 1·43(1·19,1·72) | 0·42(0·09) | 4·56(<·0001) | 1·52(1·27,1·81) |
| S-Hos-C | Psychiatric | 1009 | 3120 | 195 | 0·35(0·11) | 3·35(0·0008) | 1·42(1·16,1·75) | 0·35(0·10) | 3·38(0·0007) | 1·42(1·16,1·75) |
| S-Dep-C | Psychiatric | 987 | 2942 | 184 | 0·45(0·12) | 3·75(0·0002) | 1·57(1·24,1·99) | N/A | N/A | N/A |
| S-Anx-C | Psychiatric | 988 | 2943 | 184 | 0·39(0·10) | 3·94(<·0001) | 1·48(1·22,1·80) | N/A | N/A | N/A |
| BDI | Psychiatric | 816 | 2297 | 137 | 0·25(0·10) | 2·43(0·0149) | 1·28(1·05,1·56) | N/A | N/A | N/A |
| TFC | Functional | 1071 | 3626 | 225 | −0·53(0·10) | −5·15(<·0001) | 1·70(1·39,2·08) | −0·61(0·10) | −6·34(<·0001) | 1·84(1·52,2·22) |
| FAS | Functional | 827 | 2326 | 137 | −0·36(0·11) | −3·39(0·0007) | 1·43(1·16,1·76) | −0·40(0·10) | −3·90(<·0001) | 1·49(1·22,1·83) |
| ECOG-C | Functional | 602 | 911 | 101 | 0·37(0·13) | 2·76(0·0058) | 1·44(1·11,1·87) | 0·43(0·13) | 3·22(0·0013) | 1·54(1·18,2·00) |
| WHODAS-C | Functional | 529 | 736 | 67 | 0·48(0·21) | 2·25(0·0245) | 1·61(1·06,2·44) | N/A | N/A | N/A |
| ECOG-P | Functional | 678 | 1093 | 120 | 0·45(0·09) | 5·12(<·0001) | 1·57(1·32,1·86) | N/A | N/A | N/A |
| WHODAS-P | Functional | 581 | 850 | 63 | 0·42(0·14) | 3·11(0·0019) | 1·53(1·17,1·99) | N/A | N/A | N/A |
N=number of subjects. N*=total number of observations summed over subjects and time. Event=number of conversions. HR=hazard ratio (HR−1= inverse HR). TMS=total motor score. Brady=bradykinesia. CSF=cerebrospinal fluid. Hippo=hippocampus. Stroop-Wo=Stroop Color and Word Test – word condition. Stroop-In=Stroop Color and Word Test – interference condition. Stroop-Co=Stroop Color and Word Test – color condition. SDMT=Symbol Digit Modalities Test. Smell-ID=University of Pennsylvania Smell Identification Test (UPSIT). EmoRec=emotion recognition test. Sp-Tapping=speeded tapping. TMT-A=Trail Making Test, Part A. TMT-B=Trail Making Test, Part B. S-OC-C=Symptom Checklist 90. F-Exc-C=Frontal Systems Behavioral Scale – executive subscale – companion rating. S-GSI-C=Symptom Checklist 90 – Global Severity Index – companion rating. F-Apa-C=Frontal Systems Behavioral Scale – apathy subscale – companion rating. F-Dis-C=Frontal Systems Behavioral Rating Scale – disinhibition subscale – companion rating. S-Hos-C=Symptom Checklist 90 – hostility subscale – companion rating. S-Dep-C=Symptom Checklist 90 – depression subscale – companion rating. S-Anx-C=Symptom Checklist 90 – anxiety subscale – companion rating. BDI=Beck Depression Inventory. TFC=total functional capacity. FAS=functional activity scale. ECog-C=Everyday Cognition Rating Scale – Companion Rating. WHODAS-C=World Health Organization Disability Assessment Schedule – companion rating. ECog-P=Everyday Cognition Rating Scale – Participant Rating. WHODAS-P=World Health Organization Disability Assessment Schedule – participant rating. Psych=Psychiatric. All measures in the Imaging Domain were corrected for intracranial volume. N/A=not available because full model could not be estimated.
The column of reduced model γ estimates shows that the baseline information was a significant predictor of the hazard for 37 of 40 variables. A comparison shows that the full model γ estimates were larger in absolute value for every variable that had a full model estimated. Thus, prediction of motor diagnosis based on baseline and longitudinal change information was stronger than using only the former.
Regarding the full model results, the largest motor domain effect size was for TMS (Z=7·23, p<0·0001). The TMSHR indicates a one SD difference increased the risk of diagnosis by 3·07 times (95% CI=[2·26,4·16]) For the imaging domain, putamen volume was the strongest predictor (Z=−6·95, p<0·0001), and a one SD difference increased the risk of diagnosis by 3·32 times ([2·37,4·65]). The strongest cognitive variable was Stroop word (Z=−7·79, p < 0·0001), which increased the risk of diagnosis by 2·32 ([1·88,2·87]). The best psychiatric variable was executive functioning (Z=6·64, p<0·0001) with an increased risk of diagnosis of 1·86 ([1·55,2·23]). TFC was the strongest functional variable (Z=−6·34, p<0·0001), with an increased risk of diagnosis of 1·84 ([1·52,2·22]).
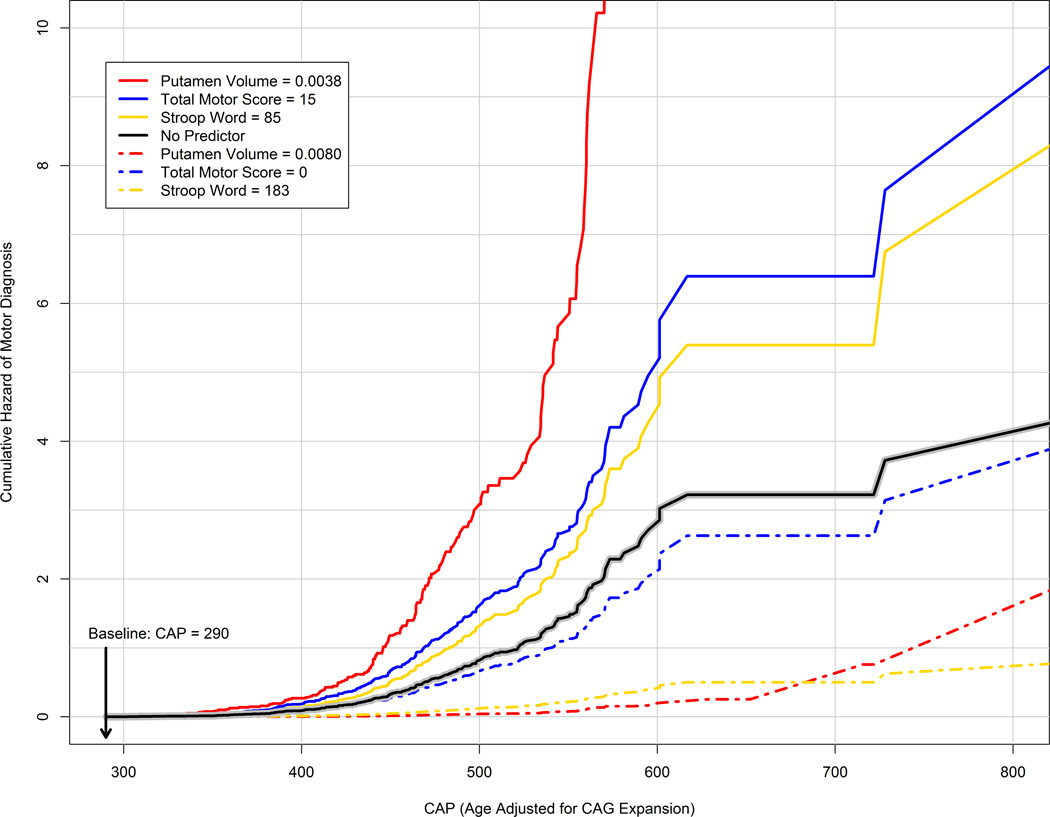
Figure 2 illustrates the Table 2 results for putamen volume, total motor score, and Stroop word test. Each variable was the strongest predictor within the three strongest domains (functional and psychiatric variables, though significant, had weaker prediction in terms of the estimated HR; see Table 2). The figure shows the cumulative hazard (accumulated risk rate) as a function of CAP for the three variables and a model with no predictor (solid black line). The no-predictor model represents the cumulative hazard associated with only CAG expansion and age (both variables are indexed by CAP). As indicated in the figure, the baseline was set to CAP=290, and the predictor curves were generated for values of the variables representing no deterioration (dash-dot lines) and advanced deterioration (solid lines). The no deterioration values (total motor score=0, putamen volume (ICV-corrected)=0·008, Stroop word=183) were the most extreme values in the sample (minimum for total motor score, and maximum for the others). The advanced values (total motor score=15, putamen volume (ICV-corrected)=0·0038, Stroop word=85) were the medians for DCL=3 of the sample. The figure shows when there is advanced deterioration at baseline, the cumulative hazard for the predictors accelerates at a rate faster than when the covariates are ignored. Conversely, when there is no deterioration at baseline, the cumulative hazard for the predictors accelerates at a slower rate than when the covariates are ignored.
Figure 2. Cumulative hazard (accumulated risk rate) of motor diagnosis by CAG-Age Product (CAP) for various baseline predictor values.
CAP is age corrected for CAG expansion and is the time metric of the horizontal axis. The solid black line denotes the cumulative hazard for a model with no predictor representing prediction based only on CAG and age (as summarized by CAP). The dash-dot lines are risk profiles for predictors representing no deterioration at baseline (e.g., Total Motor Score=0), whereas the colored solid lines are risk profiles for predictors representing advanced deterioration at baseline (e.g., Total Motor Score = 15). Putamen volume is ICV-corrected. As a reference, CAP=290 corresponds to age 31 and CAP=600 corresponds to age 64 for a person with CAG=43 (the sample mean). CAP=Age × (CAG − 33·66).CAG=cytosine-adenine-guanine.
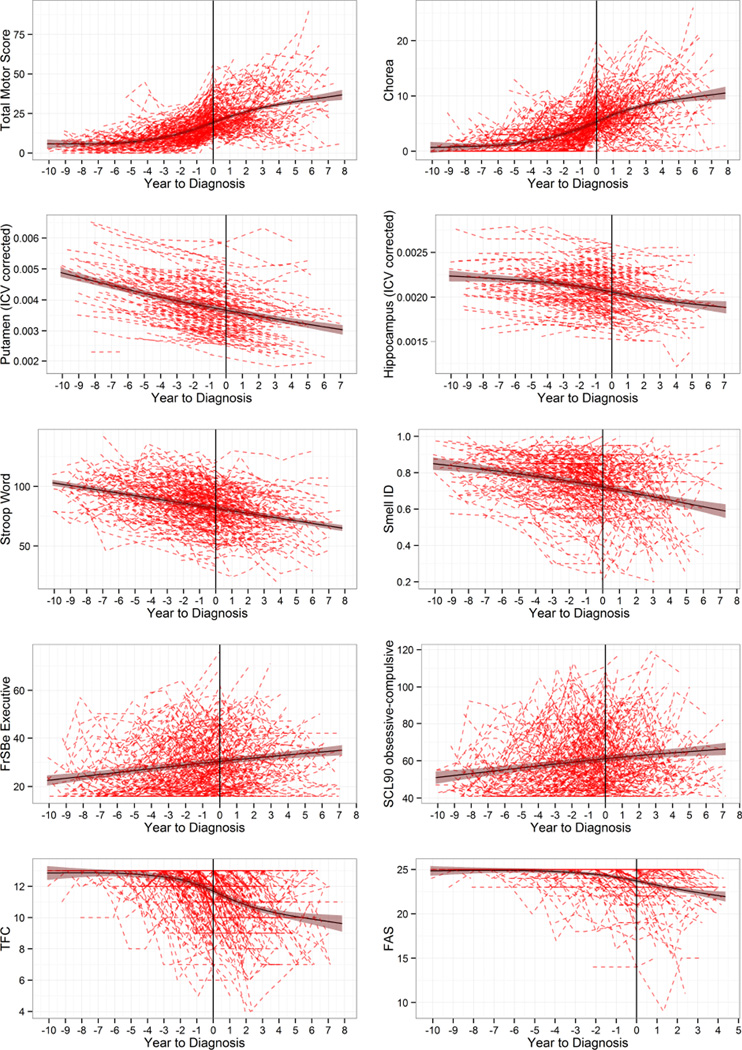
Figure 3 shows individual empirical curves (dashed lines) and fitted spline curves (solid lines) with 95% CIs for the converters. The vertical line in each panel denotes year of diagnosis (set to year=0). The two strongest predictors in each domain (based on absolute Z-values) from Table 2 were selected for graphing. The web extra material lists descriptive information on all the variables for the converters at study entry and time of diagnosis.
Figure 3. Trajectories of motor, imaging, cognitive, functional, and psychiatric variables for N=225 converters.
The top-two strongest predictors in each domain were selected (see Table 2).Dashed lines are individual empirical data, and solid lines are cubic spline curves (shading shows 95% confidence interval). ICV=Intracranial-corrected volumes. Smell ID=Smell Identification (University of Pennsylvania Smell Identification Test). FrSBe=Frontal Systems Behavior Scale. SCL90=Symptom Checklist 90. TFC=total functional capacity. FAS=functional activity scale.
DISCUSSION
The current findings show several clinical and biological measures that can improve the prediction of HD diagnosis above and beyond that obtained by CAG repeat length and age alone. The strongest predictors (in terms of absolute Z-values in Table 2) were in the domains of motor (total motor score was the strongest predictor), imaging (putamen volume), and cognitive (Stroop word test). Psychiatric and functional variables were significant but relatively weak predictors in the sense of having estimated hazard ratios less than two. Findings suggest models of HD onset prediction can be considerably improved using straightforward clinical assessment. Volumetric MRI measures provide additional powerful predictors.
Figure 2 summarizes the statistical results in illustrating the importance of using putamen volume, total motor score, or Stroop word test in estimating the risk of future motor diagnosis in addition to CAG and age (and their interaction; see CAP definition above). The no-predictor curve represents the risk that is associated with CAP alone. Because CAP is age adjusted for CAG, the cumulative risk increases as CAP elapses because the likelihood of motor diagnosis increases as people age. As a result, the no-predictor curve represents the accumulated risk rate that one would predict based only on CAG and age (and their interaction). On the other hand, when a predictor from Table 2 is considered, the risk profile is modified based on the predictor value at baseline. The modification can result in a very different risk profile compared to using CAP alone. Figure 2 indicates for people with advanced deterioration at baseline, the risk of motor diagnosis is greater when a predictor is used, whereas for people with no or little deterioration, the risk is less. Thus, information from a clinical predictor is informative for future motor diagnosis risk assessment over and above CAG and age. Further research is critical to detect additional genetic, environmental, and biological contributions that may lead to new avenues for intervention.
A follow-up analysis not reported showed that using the composite of the three variables in Figure 2 did not improve upon using the variables alone. A complication of Figure 2 is that the maximum Stroop word value may not represent the absence of deterioration, but rather an advanced education level and/or advanced level of intelligence. The converse also holds for low Stroop word scores. However, the relation of Figure 3 still holds; superior or inferior Stroop word test performance at baseline alters the future risk assessment (similar comments regarding putamen volume and TMS also apply).
As expected, the best motor predictor of HD diagnosis was TMS, since diagnosis is based on motor findings. This underscores the value of the motor examination, even in the premanifest period. This result is consistent with previous findings of subtle motor abnormalities years before diagnosis, which may accelerate just prior to diagnosis.6,22 Subdomains for chorea, bradykinesia, and oculomotor abnormalities were also predictive.
The strongest cognitive variable was the Stroop word test, a timed reading task. Previously, we documented 19 cognitive tasks that showed significant longitudinal change prior to motor diagnosis.12 The present results suggest that performance on just one of the most robust of these tests can significantly improve diagnostic prediction. The most robust cognitive measures require just a few minutes to administer and can be used in a variety of settings, making them valuable for research design and clinical practice.
The utility of brain imaging markers in the detection of HD has been documented in several studies over the past decade.2,23,24 Striatal volume consistently distinguishes gene-mutation positives from negatives and tracks disease progression. The current study demonstrates imaging variables were among the best predictors of diagnosis in premanifest HD, and their preeminence offers biological validity for the models presented. The use of imaging variables may translate to advances in clinical trial design used either in selection criteria or as an outcome measure. Imaging may also be useful in clinical care and education, although broad dissemination of imaging predictors would require standardization of acquisition and analysis protocols for clinical care.
Findings validate and extend results from other studies using smaller samples, shorter follow-up and varying endpoints.2–11 The integration of these findings with the literature provide strong evidence that cognitive, motor, and imaging deficits are evident prior to traditional motor diagnosis and may provide an opportunity for earlier intervention, treatment and support. The predictive utility of the markers suggested by this research can be immediately integrated into clinical trial design and can be used to begin to advance clinical care through refined diagnostic and prognostic guidelines.
There is considerable evidence that the diagnosis of HD is made relatively late in the disease course, after a high proportion of persons already show significant cognitive decline,13,25 psychiatric abnormalities,26–28 motor impairment,6,22 and at a time when over half of their striatal volume is lost.2 Notably, many persons are diagnosed after major changes in functioning have occurred (loss of usual employment or driving) as well as a reduction in basic activities of daily living (requiring financial or care assistance).29–32 It is possible that an earlier diagnosis might be more beneficial to intervention and life planning.33,34 Our analysis concerned the predictive ability of individual markers, and it might be possible to translate these findings into revised diagnostic and prognostic criteria for HD, which could advance research and clinical care.
The trajectories illustrated in Figure 3 suggest some interesting models regarding the course of the disease. Using the largest known sample of prospectively followed converters (N=225), the graphs suggest that many of the clinical markers of disease progression (i.e., cognitive, sensory, and psychiatric variables) progress in a near linear fashion and decline in concert with biological markers of brain imaging abnormalities. Additionally, it suggests motor and functional variables progress in a nonlinear fashion, which is reflected by the fact that motor signs and functional impairment become evident only at certain points of disease progression. There are several possible explanations for the variations in disease progression. One explanation may be that atrophy of each individual brain region proceeds relatively linearly, beginning with the striatum, but as additional brain regions begin to undergo degeneration and dysfunction, their combined effect causes acceleration of the clinical expression of disease. An alternative threshold hypothesis is that at some point, a threshold is surpassed, triggering acceleration of motor and functional deficits. Researchers making the critical choice of outcome measures for clinical trials might benefit from the data provided so study designs can enhance the possibility of documenting therapeutic effects, should they occur. Given the variation in motor and functional changes across the disease course, it is likely that subject selection at varying disease epochs could drastically alter interpretations made about an intervention.
It is important to consider qualifications of these research findings. The baseline for prediction was defined at a point for which it is known the PREDICT-HD sample has the earliest detectible change in motor signs.6 Should other samples suggest the examination of additional cross sections in the premanifest period, the estimates may vary accordingly. It is encouraging to note, however, that the present findings mirror those reported by studies of smaller samples followed for shorter duration.11 Replication with other samples will continue to refine these predictive models. Translation of these models into clinical care may require further research to determine how such information can be integrated into genetic counseling in a safe and productive manner to promote understanding. Advances in diagnosis and prognosis will require clinical consensus as well as a white paper to best communicate standard criteria and clinical care practices. Implementation of new diagnostic and prognostic criteria requires patient-centered clinical outcome research to document best practices for HD families who choose to have greater prognostic information
Additional caveats concern the variability observed in this study. The graphs of converters (Figure 3) indicate a relatively wide range of values associated with motor diagnosis (time=0), especially for TMS (min=2, max=56). As discussed in the methods section, individuals might have had different motor examiners over time, which could inflate the TMS variability. Early in the study, substantial TMS variation was noted and efforts were made to help assure data integrity as outlined in the web extra material. Another source of heterogeneity was introduced by the upgrading of scanners at the sites (1.5T to 3T). Scanner strength was adjusted for, both in the image processing and the statistical analysis (see web extra material). It is noted that despite the substantial observed variability, both the TMS and the imaging variables were among the strongest predictors. Thus, sources of variance such as different raters and scanners did not overcome the predictive power of the variables. Future studies that constrain sources of variance by having the same scanners or the same motor raters might show even larger effect sizes than reported here.
The detection and tracking of early clinical signs and symptoms in HD is critical to choosing outcome measures useful for clinical trials. Treatments with face validity (impacting on symptoms of disability in motor, cognitive, psychiatric and functional domains) can be essential components of clinical trials and often mandated by regulatory agencies. The predictive measures reported here might have value in the selection of specific types of research participants and might help choose outcomes that are associated with a critically meaningful endpoint – that of being diagnosed.
Supplementary Material
Panel: Research in context.
Systematic review
On August 25, 2014, we searched PubMed and Medline for English-language articles with human participants age 19 or older with the following search terms: “Huntington disease”, “longitudinal”, “prospective”, “onset”, and “diagnosis”. Reference lists of found articles were also reviewed. Since no previous PREDICT-HD study examined comprehensive prediction of diagnosis, they were excluded. Seven publications were found using prospective data to predict HD diagnosis based on motor criteria. Sample sizes for participants prospectively diagnosed were 21 to 70 and number of years followed were from 2.5 to 5.
Interpretation
Number of converters prospectively followed in the literature were smaller in size by over 320% and number of years followed was from 2.5 to 5. Four of the seven found studies only considered cognitive predictors, one study only considered dietary predictors, and the remaining two studies examined various comprehensive predictors of prospective diagnosis. Using data from the Huntington Study Group, Langbehn and Paulsen5 found cognitive, motor and self-reported symptoms to be predictive of traditional motor diagnosis. Although traditional motor criteria for diagnosis was not examined in Tabrizi,11 findings suggest cognitive, quantitative motor and imaging predictors of motor onset. The current study is the first to use comprehensive longitudinal assessments to prospectively predict traditional motor diagnosis in HD. Joint modeling of longitudinal change and time to HD diagnosis revealed several significant phenotypic and biologic predictors that might be useful as endpoints in clinical trials and for selecting samples. Findings fill a gap in the literature by identifying predictors that are important for HD diagnosis over and above CAG expansion and age, demonstrating that baseline status and longitudinal change are important in prediction of a relevant outcome in the progression of HD. Illustrations of our statistical analysis reveal cumulative relative risk of diagnosis profiles for persons in extremes of the disease course (far from diagnosis at baseline versus close to diagnosis at baseline). Our results provide insights regarding the nature of HD progression and show that a relatively brief clinical assessment can enhance prediction of motor diagnosis.
ACKNOWLEDGEMENTS
This research is supported by the National Institutes for Health, National Institute of Neurological Disorders and Stroke (5R01NS040068) awarded to JSP, CHDI Foundation, Inc (A3917) awarded to JSP, Cognitive and Functional Brain Changes in Preclinical Huntington’s Disease (HD) (5R01NS054893) awarded to JSP, Enterprise Storage In A Collaborative Neuroimaging Environment (1S10RR023392) awarded to HJJ, BRAINS Morphology and Image Analysis (5R01NS050568), National Alliance for Medical Image Computing (NAMIC), Functional Connectivity in Premanifest Huntington’s Disease (1U01NS082083), and Basal Ganglia Shape Analysis and Circuitry in Huntington ’s Disease (1U01NS082085) awarded to CAR.
We thank the PREDICT-HD sites, the study participants, the National Research Roster for Huntington Disease Patients and Families, the Huntington’s Disease Society of America and the Huntington Study Group. This publication was supported by the National Center for Advancing Translational Sciences, and the National Institutes of Health (NIH), through Grant 2 UL1 TR000442-06. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
JSP, CAR and HJW received grants from the National Institutes of Health. JSP and EHA received grants from CHDI Foundation, Inc. JSP has served on an advisory board for Lundbeck, LLC, and has a consulting agreement with ProPhase, LLC.
PREDICT-HD INVESTIGATORS, COORDINATORS, MOTOR RATERS, COGNITIVE RATERS
Isabella De Soriano, Courtney Shadrick, and Amanda Miller (University of Iowa, Iowa City, Iowa, USA);
Edmond Chiu, Joy Preston, Anita Goh, Stephanie Antonopoulos, and Samantha Loi (St. Vincent’s Hospital, The University of Melbourne, Kew, Victoria, Australia);
Phyllis Chua, and Angela Komiti (The University of Melbourne, Royal Melbourne Hospital, Melbourne, Australia);
Lynn Raymond, Joji Decolongon, Mannie Fan, and Allison Coleman (University of British Columbia, Vancouver, British Columbia, Canada);
Christopher A. Ross, Mark Varvaris, Maryjane Ong, and Nadine Yoritomo (Johns Hopkins University, Baltimore, Maryland, USA);
William M. Mallonee and Greg Suter (Hereditary Neurological Disease Centre, Wichita, Kansas, USA);
Ali Samii, Emily P. Freney, and Alma Macaraeg (University of Washington and VA Puget Sound Health Care System, Seattle, Washington, USA);
Randi Jones, Cathy Wood-Siverio, and Stewart A. Factor (Emory University School of Medicine, Atlanta, Georgia, USA);
Roger A. Barker, Sarah Mason, and Natalie Valle Guzman (John van Geest Centre for Brain Repair, Cambridge, UK);
Elizabeth McCusker, Jane Griffith, Clement Loy, Jillian McMillan and David Gunn (Westmead Hospital, Sydney, Australia);
Michael Orth, Sigurd Süβmuth, Katrin Barth, Sonja Trautmann, Daniela Schwenk, and Carolin Eschenbach (University of Ulm, Ulm, Germany);
Kimberly Quaid, Melissa Wesson, and Joanne Wojcieszek (Indiana University School of Medicine, Indianapolis, IN, USA);
Mark Guttman, Alanna Sheinberg, Albie Law, and Irita Karmalkar (Centre for Addiction and Mental Health, University of Toronto, Markham, Ontario, Canada);
Susan Perlman and Brian Clemente (UCLA Medical Center, Los Angeles, California, USA);
Michael D. Geschwind, Sharon Sha, Joseph Winer, and Gabriela Satris (University of California San Francisco, San Francisco, California, USA);
Tom Warner and Maggie Burrows (National Hospital for Neurology and Neurosurgery, London, UK);
Anne Rosser, Kathy Price, and Sarah Hunt (Cardiff University, Cardiff, Wales, UK);
Frederick Marshall, Amy Chesire, Mary Wodarski, and Charlyne Hickey (University of Rochester, Rochester, New York, USA);
Peter Panegyres, Joseph Lee, Maria Tedesco, and Brenton Maxwell (Neurosciences Unit, Graylands, Selby-Lemnos & Special Care Health Services, Perth, Australia);
Joel Perlmutter, Stacey Barton, and Shineeka Smith (Washington University, St. Louis, Missouri, USA);
Zosia Miedzybrodzka, Daniela Rae, Vivien Vaughan, and Mariella D’Alessandro (Clinical Genetics Centre, Aberdeen, Scotland, UK);
David Craufurd, Judith Bek, and Elizabeth Howard (University of Manchester, Manchester, UK);
Pietro Mazzoni, Karen Marder, and Paula Wasserman (Columbia University Medical Center, New York, New York, USA);
Rajeev Kumar, Diane Erickson, Christina Reeves, and Breanna Nickels (Colorado Neurological Institute, Englewood, Colorado, USA);
Vicki Wheelock, Lisa Kjer, Amanda Martin, and Sarah Farias (University of California Davis, Sacramento, California, USA);
Wayne Martin, Oksana Suchowersky, Pamela King, Marguerite Wieler, and Satwinder Sran (University of Alberta, Edmonton, Alberta, Canada);
Anwar Ahmed, Stephen Rao, Christine Reece, Alex Bura, and Lyla Mourany (Cleveland Clinic Foundation, Cleveland, Ohio, USA).
Executive Committee
Jane S. Paulsen, Principal Investigator, Jeffrey D. Long, Hans J. Johnson, Thomas Brashers-Krug, Phil Danzer, Amanda Miller, H. Jeremy Bockholt, and Kelsey Montross (University of Iowa).
Scientific Consultants
Deborah Harrington (University of California, San Diego); Holly Westervelt (Rhode Island Hospital/Alpert Medical School of Brown University); Elizabeth Aylward (Seattle Children’s Research Institute); David J. Moser, Janet Williams, Nancy Downing, Vincent A. Magnotta, Jatin Vaidya, Daniel O’Leary, and Kim Eun Young (University of Iowa); Stephen Rao (Cleveland Clinic).
Core Sections
Biostatistics: Jeffrey D. Long, Ji-In Kim, Spencer Lourens (University of Iowa); Ying Zhang, and Wenjing Lu (University of Indiana).
Ethics: Cheryl Erwin (Texas Tech University Health Sciences Center); Thomas Brashers-Krug and Janet Williams (University of Iowa); Martha Nance (University of Minnesota).
Biomedical Informatics: H. Jeremy Bockholt, Jason Evans, and Roland Zschiegner (University of Iowa).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONTRIBUTORS
JSP, CAR, CJE, HJJ, and EHA contributed to study design. JSP, CJE, JKW, HJJ, EHA, HJB, and RAB participated in data collection. JSP, JDL, HJJ, EHA, YZ, and HJB conducted data analysis. JSP, CAR, DLH, JKW, HJJ, EHA, YZ, and HJB contributed to data interpretation. HJB contributed to data integration. JSP, JDL, CAR, JKW, HJW, and YZ wrote the article. JSP, JDL, DLH, HJW, HJJ, EHA, YZ, HJB, and RAB edited the manuscript for important intellectual content. JDL constructed article figures and tables and JSP conducted a literature search. CJE provided analysis and interpretation for appropriate ethical content and handling of confidential or sensitive information. HJW oversaw data quality control and training of the study cognitive core. JSP provided study supervision and obtained funding for the study written about in the article. All authors had access to the data and gave approval for submission of the manuscript.
DECLARATION OF INTERESTS
JDL, CAR, DLH, CJE, JKW, HJJ, EHA, YZ, HJB, and RAB declare no competing interests.
REFERENCES
- 1.Andrew SE, Goldberg YP, Kremer B, et al. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington's disease. Nat Genet. 1993;4:398–403. doi: 10.1038/ng0893-398. [DOI] [PubMed] [Google Scholar]
- 2.Aylward EH, Liu D, Nopoulos PC, et al. Striatal volume contributes to the prediction of onset of Huntington disease in incident cases. Biol Psychiatry. 2012;71:822–828. doi: 10.1016/j.biopsych.2011.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandt J, Inscore AB, Ward J, et al. Neuropsychological deficits in Huntington's disease gene carriers and correlates of early "conversion". J Neuropsychiatry Clin Neurosci. 2008;20:466–472. doi: 10.1176/appi.neuropsych.20.4.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrington DL, Smith MM, Zhang Y, Carlozzi NE, Paulsen JS PREDICT-HD Investigators of the Huntington Study Group. Cognitive domains that predict time to diagnosis in prodromal Huntington disease. J Neurol Neurosurg Psychiatry. 2012;83:612–619. doi: 10.1136/jnnp-2011-301732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langbehn DR, Paulsen JS Huntington Study Group. Predictors of diagnosis in Huntington disease. Neurology. 2007;68:1710–1717. doi: 10.1212/01.wnl.0000261918.90053.96. [DOI] [PubMed] [Google Scholar]
- 6.Long JD, Paulsen JS, Marder K, et al. Tracking motor impairments in the progression of Huntington's disease. Mov Disord. 2014;29:311–319. doi: 10.1002/mds.25657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marder K, Gu Y, Eberly S, et al. Relationship of Mediterranean diet and caloric intake to phenoconversion in Huntington disease. JAMA Neurol. 2013;70:1382–1388. doi: 10.1001/jamaneurol.2013.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paulsen JS, Zhao H, Stout JC, et al. Clinical markers of early disease in persons near onset of Huntington's disease. Neurology. 2001;57:658–662. doi: 10.1212/wnl.57.4.658. [DOI] [PubMed] [Google Scholar]
- 9.Snowden JS, Craufurd D, Thompson J, Neary D. Psychomotor, executive, and memory function in preclinical Huntington's disease. J Clin Exp Neuropsychol. 2002;24:133–145. doi: 10.1076/jcen.24.2.133.998. [DOI] [PubMed] [Google Scholar]
- 10.Solomon AC, Stout JC, Weaver M, et al. Ten-year rate of longitudinal change in neurocognitive and motor function in prediagnosis Huntington disease. Mov Disord. 2008;23:1830–1836. doi: 10.1002/mds.22097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tabrizi SJ, Scahill RI, Owen G, et al. Predictors of phenotypic progression and disease onset in premanifest and early-stage Huntington's disease in the TRACK-HD study: analysis of 36-month observational data. Lancet Neurol. 2013;12:637–649. doi: 10.1016/S1474-4422(13)70088-7. [DOI] [PubMed] [Google Scholar]
- 12.Paulsen JS, Long JD, Johnson HJ, et al. Clinical and Biomarker Changes in Premanifest Huntington Disease Show Trial Feasibility: A Decade of the PREDICT-HD Study. Front Aging Neurosci. 2014;6:78. doi: 10.3389/fnagi.2014.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paulsen JS, Smith MM, Long JD PREDICT-HD investigators and coordinators of the Huntington Study Group. Cognitive decline in prodromal Huntington Disease: implications for clinical trials. J Neurol Neurosurg Psychiatry. 2013;84:1233–1239. doi: 10.1136/jnnp-2013-305114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diggle PJ, Sousa I, Chetwynd AG. Joint modelling of repeated measurements and time-to-event outcomes: the fourth Armitage lecture. Stat Med. 2008;27:2981–2998. doi: 10.1002/sim.3131. [DOI] [PubMed] [Google Scholar]
- 15.Wulfsohn MS, Tsiatis AA. A joint model for survival and longitudinal data measured with error. Biometrics. 1997;53:330–339. [PubMed] [Google Scholar]
- 16.Zhang Y, Long JD, Mills JA, et al. Indexing disease progression at study entry with individuals at-risk for Huntington disease. American Journal of Medical Genetics Part B-Neuropsychiatric Genetics. 2011;156B:751–763. doi: 10.1002/ajmg.b.31232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langbehn DR, Hayden MR, Paulsen JS PREDICT-HD Investigators. CAG-repeat length and the age of onset in Huntington disease (HD): A review and validation study of statistical approaches. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:397–408. doi: 10.1002/ajmg.b.30992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Penney JB, Jr, Vonsattel JP, MacDonald ME, Gusella JF, Myers RH. CAG repeat number governs the development rate of pathology in Huntington's disease. Ann Neurol. 1997;41:689–692. doi: 10.1002/ana.410410521. [DOI] [PubMed] [Google Scholar]
- 19.Ross CA, Aylward EH, Wild EJ, et al. Huntington disease: natural history, biomarkers and prospects for therapeutics. Nat Rev Neurol. 2014;10:204–216. doi: 10.1038/nrneurol.2014.24. [DOI] [PubMed] [Google Scholar]
- 20.Paulsen JS, Hayden M, Stout JC, et al. Preparing for preventive clinical trials: the Predict-HD study. Arch Neurol. 2006;63:883–890. doi: 10.1001/archneur.63.6.883. [DOI] [PubMed] [Google Scholar]
- 21.Paulsen JS, Langbehn DR, Stout JC, et al. Detection of Huntington's disease decades before diagnosis: the Predict-HD study. J Neurol Neurosurg Psychiatry. 2008;79:874–880. doi: 10.1136/jnnp.2007.128728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biglan KM, Ross CA, Langbehn DR, et al. Motor abnormalities in premanifest persons with Huntington's disease: the PREDICT-HD study. Mov Disord. 2009;24:1763–1772. doi: 10.1002/mds.22601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bohanna I, Georgiou-Karistianis N, Hannan AJ, Egan GF. Magnetic resonance imaging as an approach towards identifying neuropathological biomarkers for Huntington's disease. Brain Res Rev. 2008;58:209–225. doi: 10.1016/j.brainresrev.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Kloppel S, Henley SM, Hobbs NZ, et al. Magnetic resonance imaging of Huntington's disease: preparing for clinical trials. Neuroscience. 2009;164:205–219. doi: 10.1016/j.neuroscience.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duff K, Paulsen J, Mills J, et al. Mild cognitive impairment in prediagnosed Huntington disease. Neurology. 2010;75:500–507. doi: 10.1212/WNL.0b013e3181eccfa2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beglinger LJ, Paulsen JS, Watson DB, et al. Obsessive and compulsive symptoms in prediagnosed Huntington's disease. J Clin Psychiatry. 2008;69:1758–1765. doi: 10.4088/jcp.v69n1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duff K, Paulsen JS, Beglinger LJ, Langbehn DR, Stout JC Predict-HD Investigators of the Huntington Study Group. Psychiatric symptoms in Huntington's disease before diagnosis: the Predict-HD study. Biol Psychiatry. 2007;62:1341–1346. doi: 10.1016/j.biopsych.2006.11.034. [DOI] [PubMed] [Google Scholar]
- 28.Epping EA, Mills JA, Beglinger LJ, et al. Characterization of depression in prodromal Huntington disease in the neurobiological predictors of HD (PREDICT-HD) study. J Psychiatr Res. 2013;47:1423–1431. doi: 10.1016/j.jpsychires.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beglinger LJ, O'Rourke JJ, Wang C, et al. Earliest functional declines in Huntington disease. Psychiatry Res. 2010;178:414–418. doi: 10.1016/j.psychres.2010.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Downing NR, Kim JI, Williams JK, et al. WHODAS 2.0 in prodromal Huntington disease: measures of functioning in neuropsychiatric disease. Eur J Hum Genet. 2013 doi: 10.1038/ejhg.2013.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams J, Downing N, Vaccarino AL, Guttman M, Paulsen JS. Self Reports of Day-to-Day Function in a Small Cohort of People with Prodromal and Early HD. PLoS Curr. 2011;3:RRN1254. doi: 10.1371/currents.RRN1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paulsen JS, Wang C, Duff K, et al. Challenges assessing clinical endpoints in early Huntington disease. Mov Disord. 2010;25:2595–2603. doi: 10.1002/mds.23337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothstein M, Siegal G. Health Information Technology and Physicians’ Duty to Notify Patients of New Medical Developments. Hous J Health L & Policy. 2012;12:93–136. [Google Scholar]
- 34.Williams JK, Erwin C, Juhl A, et al. Personal factors associated with reported benefits of Huntington disease family history or genetic testing. Genetic Testing and Molecular Biomarkers. 2010;14:629–636. doi: 10.1089/gtmb.2010.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.