Abstract
This review focuses on the interrelationship between ageing and autophagy. There is a striking similarity between the signalling aspects of these two processes. Both ageing and autophagy involve several of the signalling components such as insulin/IGF-1, AMPK, Ras-cAMP-PKA, Sch9 and mTOR. Ageing and ageing-mediated defective autophagy involve accumulation of lipofuscin. Components of anti-ageing and autophagy include SirTs and FoxOs. Nutritional deprivation or calorie restriction as well as several nutriceuticals including resveratrol, spermidine, curcumin and piperine can enhance autophagy and increase lifespan. Such striking similarities indicate that lifespan is strongly dependent on autophagy.
Keywords: autophagy, aging, longevity, resveratrol, AMPK, SirT, FoxO
Introduction
The existence of a Fountain of youth, a legendary spring that can restore the youth of anyone who drinks its water was believed by a large population in the 16th Century influenced by the Spanish explorer Juan Ponce de León y Figueroa [1]. Such a fountain was mentioned also in the writing of Herodotus some thousands of years ago. In the modern era, even though most may not believe in the existence of such a fountain, it is certain that everyone dreams of living longer. Life expectancy has increased tremendously in recent years—in many developed countries including the USA it is close to 80 years, whereas in underdeveloped countries such as Zambia and Angola life expectancy is below 40 years.
The primary parameter responsible for increased life expectancy is certainly the lifestyle [2, 3]. Stress is likely to be the single most important factor that can reduce life expectancy significantly. Although ageing is certainly a genetically and epigenetically regulated process, modification of the lifestyle is likely to be the best way to increase life expectancy. An existing proof of the concept are the Mormons, a specific group of the World population who live longer than average. Among the many reasons why Mormons live longer is possibly their lifestyle forbidding smoking and allowing regular fasting [2, 3] on the first Sunday of each month [4]. In the later study, 59% of the Mormon fasters were diagnosed with heart disease compared to 67% of the non-fasters (after age, weight and other health conditions were taken into account); the non-Mormon fasters showed same benefits as the Mormon fasters.
During day-to-day activities, accumulation of toxins or damaged particles appearing as misfolded or cross-linked aggregated macromolecules retard the biological and physiological cell functions, acting as sinks for important signalling components of the body [5]. Fasting and calorie restriction (CR) have been proven to stimulate anti-ageing processes and prolonging lifespan by activating detoxifying reactions. Ageing, on the other hand, promotes progressive accumulation of damaged macromolecules and organelles [6, 7].
The question arising is how can fasting perform such a critical task of promoting longevity? Autophagy-an intracellular mechanism for degrading long-lived proteins—probably serves as a boost mechanism for cellular clean-up, preventing accumulation of toxic components and promoting longevity [8]. In fact, suppressing autophagy accelerates accumulation of protein aggregates leading to triggering cell death signals [9]. Interestingly, the ageing process involves a co-ordinated program involving a large number of signalling cascades include but not limited to insulin/insulin-like growth factor (IGF)-1, mammalian target of rapamycin (mTOR), JUN NH2-terminal kinase (JNK) and transforming growth factor (TGF) signalling pathways [10–13]. At the same time, the ageing process deactivates several crucial survival protein cascades such as phosphatidylinositol 3-kinase (PI3K)- acutely transforming retrovirus AKT8 in rodent T cell lymphoma (Akt), B cell CLL/Lymphoma-2 (Bcl-2) and/or Sirtuin-Forkhead box (SirT-FoxO) signalling pathways. [14–16]. Autophagy reverses these detrimental signalling processes the same way fasting does [17]. This would tend to suggest that fasting induces autophagy, which in turn would promote anti-ageing signalling. In fact, it is quite true. This review intends to show that anti-ageing effects through lifestyle modification by fasting or chemicals/drugs potentiation of autophagy can clean up the toxic components and induce expression of pro-survival/anti-ageing proteins.
Signalling cascades regulating the ageing process
The large plethora of signalling pathways intersecting at the three major degradative pathways (lysosomal, autophagosomal and proteasomal) is ultimately what decides the balance between ageing and longevity (Fig. 1). Although the death due to ageing for human beings is difficult to correlate because many old people die from other factors such as cardiovascular disease, cancer or diabetes independent of ageing, lifespan and ageing are easy to correlate for lower eukaryotic species. For example, the lifespan of Caenorhabditis elegans can be extended simply by blocking pro-ageing genes [18]. Compared to wild-type C. elegans, mutant species lacking insulin/IGF-1 receptor (Daf-2) live twice their age [19]. Insulin receptor in turn activates class I PI3K-Akt signalling pathway, leading to the inhibition of FoxO-–putative player in the human ageing process (Figs. 1 and 2) [20, 21]). This would tend to suggest that inactivation of PI3K/Akt and activation of FoxOs would extend the lifespan of C. elegans[22, 23]. A mitogen-activated kinase member JNK also modulates FoxO and plays a role in the ageing process [24]. The cytokines also appear to be involved as TGF-β signalling seems to be instrumental for autophagy [25]. In addition, mTOR signalling and AMPK (AMP-activated protein kinase) signalling also appear to be involved in the ageing process [26, 27].
Fig 1.
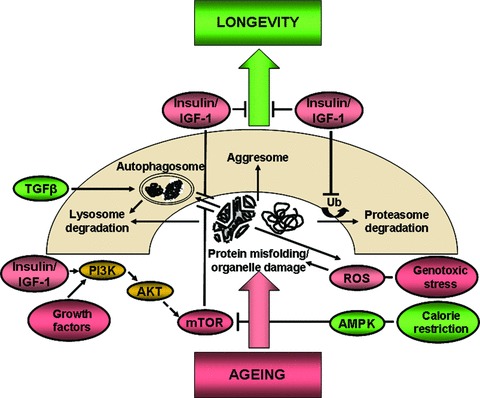
Signalling cascades regulating the ageing process: pathways intersecting along the way from ageing to longevity. AKT, acutely transforming retrovirus AKT8 in rodent T cell lymphoma; AMPK, AMP-activated protein kinase; Bcl-2, B cell CLL/Lymphoma-2; IGF, insulin-like growth factor; mTOR, mammalian target of rapamycin; PI3K, phosphatidylinositol 3-kinase; ROS, reactive oxygen species; TGF, transforming growth factor; Ub, ubiquitin.
Fig 2.
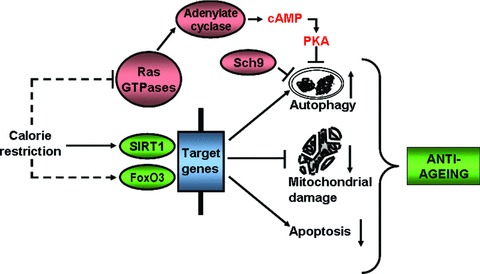
Calorie restriction induced signalling and autophagy interplay in the ageing process. SIRT1, sirtuin 1; FoxO, forkhead box; Sch9, Saccharomyces cerevisiae kinase 9; PKA, protein kinase A.
Ageing and autophagy
Ageing is a process in which the organisms gradually loose their ability to adapt to the changing environments, become more vulnerable to stress and accumulate damaged products due to overall decrease in the protein degradation time. A growing body of evidence indicates that ageing is regulated by autophagic mechanisms. Autophagy also known as autophagocytosis, is a catabolic process involving the degradation of a cell’s own components through the lysosomal machinery. It is an evolutionarily conserved intracellular degradative system that operates to maintain cellular homeostasis. In addition, it is responsible for elimination of damaged organelles and intracellular pathogens as well as aberrant or aggregated proteins that cannot be removed via the proteasomal pathway. Autophagy can promote both cell death and cell survival depending on the specific circumstances. Low levels of autophagy in general is considered to be pro-survival up to a certain threshold at which higher level of autophagy cause cellular stress and therefore death described as type II programmed cell death [28, 29]. Based on the selective uptake of cargo destined for degradation, three different forms of autophagy are known to date:
1. Macroautophagy in which portions of the cytosol and complete organelles are engulfed by double-membrane structures known as autophagosomes (early autophagic vacuoles), which readily fuse with the lysosome to form single membrane structures autophagolysosomes (late autophagic vacuoles) [28]. By this time, luminal content is degraded and resulting elements are returned into the cytosol to undergo metabolic processes. According to the ability of the autophagosomes to degrade different organelle ‘waste’ material, the intracellular engulfment has been referred as pexophagy (in case of peroxisomes [30]), xenophagy (for intracellular bacteria and viruses [31]) or mitophagy (for mitochondria [32]). Very likely, the future will describe other subtypes of autophagy, possibly ERophagy (for endoplasmic reticulum), ribophagy (for ribosomes) or any other cell organellephagy.
During ageing, age-associated increase in reactive oxygen species (ROS) release and consequent mitochondrial DNA (mtDNA) damage occurs (Fig. 1) [33]. The mitochondria quality control system has been tightly associated with autophagy to segregate damaged, potentially harmful ROSs inside the cell [34]. Hypoxia has been associated with modulation of various cellular processes including autophagy [35] and as a cause of mitochondrial dysfunction and tissue damage [36]—the brain, heart [37, 38] and kidney [39] are all known to be affected in this process. In the brain, for example, autophagy has been implicated as the crucial regulator of the ageing process [40]. Oxidative stress in the irreplaceable post-mitotic neurons can cause epigenetic or specific gene promoters silencing in response to unrepaired DNA damage, therefore evading apoptosis [41]. From worms and flies to mice and human beings, induction of autophagy has been associated with prolonged lifespan and decreased neurodegeneration due to decreased accumulation of ubiquitylated proteins [42, 43]. Attenuation of the mTOR signalling pathway has been show to extend lifespan in yeast, worms and flies [44]. Rapamycin, an inhibitor of the mTOR pathway extends the lifespan of treated mice by reducing the toxic-aggregate formation in Huntington’s disease model mice [45].
Autophagy in the heart has dual role: under normal conditions, autophagy has a housekeeping role in the turnover of cytoplasmic constituents; failure to do so, may result in defective autophagic degradation (deficiency of lysosomal-associated membrane protein-2 (LAMP-2) causes cardiomyopathy) [46]; cardiomyocytes expressing polyglutamine exhibit increased autophagosomal content eventually causing heart failure [47]; missense mutation in the aB-crystallin (CryAB) gene increases insoluble CryAB-associated aggregate formation by reducing autophagy causing severe form of desmin-related cardiomyopathy [48]. Autophagy also removes damaged organelles in the heart after cellular distress (hypoxia/ reoxygenation), working against the mitochondrial protein Bnip3 (Bcl-2/adenovirus E1B 19 kD interacting protein) death signalling or apoptosis [49].
Regardless of the organ system involved, CR in general attenuates mitochondrial dysfunction [50] and affects cell adaptation to hypoxia [51]. CR prolongs the lifespan through a silent information regulator 1 (SirT1) by enhancing autophagy [36].
In the kidney, CR attenuated mitochondrial oxidative damage caused by hypoxia via enhancement of SirT1 activity. Hypoxia-induced expression of Bnip3 is positively regulated by the hypoxia-inducible factor 1α (Hif1α) and forkhead box O3 (FoxO3) being essential for enhancement of autophagy (Fig. 2).
During the ageing, the efficiency of autophagic degradation reduces resulting in an accumulation of intracellular waste products [52]. Interestingly, the same signalling pathways appear to regulate both ageing and autophagy [53].
2. Microautophagy is a process where engulfment occurs directly by the lysosomal membrane [54]. Although microautphagy similar to macroautophagy is responsible for the disposal of long-lived proteins and organelles, it does not exhibit adaptation to nutritional deprivation. Its distinction from macroautophagy comes from the fact that here small portions of the cytosol are internalized through small lysosomal invaginations for continuous protein degradation even under normal conditions. Studies of this type of autophagy are still limited to yeast, but implications for importance of this process in antigen presentation and the major histocompatibility complex (MHC) do exist [55, 56].
3. Chaperone-mediated autophagy (CMA) is the third type of autophagic processes in which cytosolic proteins containing specific lysosome-targeting motif are recognized by a complex of chaperone protein such as heat shock cognate protein (hsc) 70 and delivered to the lysosomal membranes. After binding to LAMP2A, the protein becomes unfolded followed by transportation into the lysosomes for degradation [57]. Besides the great interplay among the three types of autophagy, CMA in contrast, specifically targets single soluble cytosolic proteins for translocation across the lysosomal membrane and degradation [58, 59]. This specificity is based upon recognition of an amino acid motif on the protein destined for degradation by a cytosolic chaperone complex [60]. Nevertheless, the role of CMA in the turnover of long-lived proteins and intracellular quality control, as well as it role in different diseases has been similar to other types of autophagy.
Almost all proteins targeted for CMA degradation contain a fingerprint pentapeptide KFERQ recognized by the CMA cytosolic chaperone complex, but the same proteins can also contain targeting signals for other proteolytic systems, the ultimate decision for degradation being made upon current cellular conditions or reasons for degradation. A definite ‘ticket’ for degradation of a substrate via CMA is recognition by the hsc70 chaperone/co-chaperone complex [61].
LAMP2A, a single-span lysosomal membrane protein serves to bind and translocate the protein destined for lysosomal degradation. Upon binding, substrate unfolding takes place and lysosomal lumen associated hsc70 protein (lys-hsc70) together with a lysosomal hsp90 protein somehow translocate the cytosolic protein for degradation, which takes only min. [62].
Under normal conditions, CMA has housekeeping and stress-response function, but it can also be activated by accumulation of misfolded or oxidized proteins or starvation itself [63]. Interestingly, under nutritional deprivation LAMP2A receptor levels increase not due to increased expression of this protein, but due to decreased rate of degradation; supply from the pooled LAMP2A on the luminal side of the lysosome regulate the activity of the CMA during starvation [64]. Several pathologic conditions associate with CMA: lysosomal storage diseases (galactosialidosis, mucolypidosis, Danon’s disease), nephropathies (diabetic, acidotic and chronic kidney disease), neurodegenerative disorders (Alzheimer’s, Parkinson’s and Huntington’s disease), oncogenic and immunological diseases [65]. During ageing, the CMA activity also declines mainly as a consequence of decreased levels of LAMP2A and altered turnover of this protein on the lysosomal membrane [66]. A proof of concept was recently found in an inducible exogenous copy of LAMP2A (Tet-off-LAMP2A mouse) where activation of the transgene in the liver of old mice restored the CMA levels close to those of young mice. Overall, comparing macroautophagy to CMA, one would say that the first one takes place early during nutritionally deprived conditions with the second one being actually activated later, when the macroautophagic activity starts to decrease [67].
Lipofuscin and the mitochondrial-lysosomal axis theory of aging
Accumulated evidence suggests that during the ageing process, the ability of autophagy to digest oxidatively damaged macromolecles and organelles is significantly diminished resulting in an accumulation of long-lived post-mitotic cells such as cardiomyocytes and neurons as well as lipofuscin (a non-degradable intralysosomal polymer) and aberrant proteins forming aggresomes (Fig. 1) [68]. Mitochondria become defective and interaction between the defective mitochondria with lipofuscin-loaded lysosomes promotes cellular ageing process [69]. Lipofuscin, an ageing pigment, is considered a creditable marker for the ageing of cells. Lipofuscin tends to accumulate even at an early age, but rapidly progresses with the advancement of ageing process suggesting the inability of autophagy to handle the garbage disposal capacity. The mitochondrial-lysosomal axis theory of ageing [70] predicts that lysosomal dysfunction associated with ageing process is the hallmark for lipofuscin accumulation. Thus, decline of autophagy during ageing appears to be the cause for lipofuscin accumulation. In fact, both the in vivo and in vitro function of autophagy is reduced with the advancement of age. Interestingly, such reduction in age-dependent autophagy is reversed with fasting and/or CR [53, 66]. In the next section, we will discuss a number of pharmacological manipulations that can induce autophagy when it is needed.
Signalling cascades regulating autophagy
Before discussing pharmacological manipulation to enhance autophagy, it is necessary to understand the signalling processes for autophagy regulation (Fig. 3). Interestingly, there is striking similarities in the signalling pathways between autophagy and ageing (Fig. 4).
Fig 3.
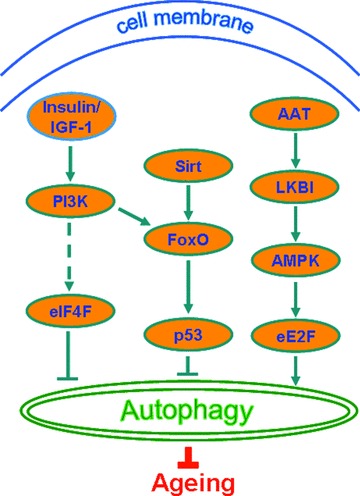
Signalling cascades regulating autophagy. AAT, area at risk; AMPK, AMP-activated protein kinase; FoxO, forkhead box; IGF, insulin-like growth factor; LKBI, tumour suppressor kinase; PI3K, phosphatidylinositol 3-kinase; SirT, Sirtuin; eE2F, eukaryotic elongation factor 2F; eIF4F, eukaryotic translation initiation factor 4F.
Fig 4.
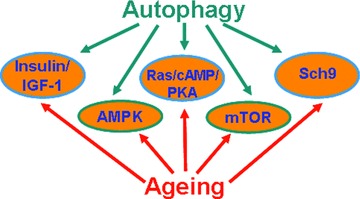
Similarity in the signalling pathways of autophagy and ageing. AMPK, AMP-activated protein kinase; IGF, insulin-like growth factor; mTOR, mammalian target of rapamycin; PKA, protein kinase A; Sch9, Saccharomyces cerevisiae kinase 9.
1. mTOR signalling. In yeast, there are two functionally distinct mTOR complexes, one (TORC1) is rapamycin sensitive and responds to nutrition and the other (TORC2) is rapamycin insensitive and does not respond to nutrition [71]. If mTOR signalling is inhibited, physiological characteristics of starvation appear including induction of autophagy [72]. In yeast cells, autophagy is controlled by TORC1 through autophagy-related genes (Atg) 1, Atg13 and Atg17, all being involved in the formation of the autophagosomes and inactivation of TORC1 by rapamycin alters the phosphorylation of Atg13 thereby inducing autophagy [73]. Reduction of mTOR signalling by RNAi or knocking down of mTOR, let-363/CeTOR or the regulatory associated protein of mTOR (RAPTOR) can extend the lifespan in C. elegans[74]. Insulin/IGF-1 signalling negatively regulates autophagy through mTOR as described later [75].
2. Insulin/IGF-1. Similar to ageing, insulin/IGF-1 pathway is critically involved in the autophagy process. It has been known that low insulin levels can induce autophagy (such as during fasting) whereas high insulin concentration suppresses autophagy [76]. Although the precise relationship of insulin/IGF-1 between ageing and longevity is not clear, autophagy appears to be involved in the regulation of ageing through this signalling pathway. This hypothesis is supported by the observation that knockdown of ATG7 and ATG12 partially inhibits the lifespan extension of the insulin-like receptor Daf-2 mutants and causes significant shortening of the lifespan of wild-type worms [77]. Increased longevity of Daf-2 mutants is partially inhibited by the mutants of Atg6 (also known as Beclin-1), Atg-18 or Atg 8 (ubiquitin-like protein also known as LC3) [78]. These reports suggest that Insulin/IGF-1 signalling pathway is involved in the autophagy regulation of ageing. This concept receives further support from the observations that mutations of Daf-2, Age-1 (encoding the catalytic subunits of PI3K) and Pdk-1 [encoding PDK-1 (pyruvate dehydrogenase kinase)] extend lifespan [79].
3. Ras-cAMP-PKA pathway. This pathway plays a crucial role in the stress response and proliferation as well as longevity (Fig. 2) [80]. In a recent study, three autophagy-related proteins have been identified, Atg1, Atg13 and Atg 18, as protein kinase A (PKA) substrates in Saccharomyces cerevisiae, which act at the early stages of autophagy indicating that Ras/PKA inhibits early in the process [81]. In response to nutrient-rich conditions, two Ras GTPases, Ras1 and Ras2 activate adenylate cyclase to produce cAMP, which after binding results in the dissociation of PKA regulatory subunit Bcy1 leading to the activation of PKA catalytic subunits Tpk1, Tpk2 and Tpk3 [82]. A recent study has indicated that Ras/PKA regulates the lifespan based on the observation that type 5 adenylyl cyclase knockout mice live 30% longer compared to corresponding wild-type littermates [83].
4. Sch9 pathway. Sch9, the yeast PKB homologue, is a negative regulator of autophagy as its hyperactivation reduces autophagy when mTOR is inactivated by rapamycin whereas simultaneous inactivation of Sch9 and PKA without suppression of TORC1 induces autophagy, suggesting that they control autophagy in parallel (Fig. 2) [84]. It appears that longevity phenotype of PKA and Sch9 signalling are linked to each other with autophagy activation as Msn2/4 and Rim15 are involved in PKA- and Sch9-mediated regulation of autophagy [84]. PKA, Sch9 and mTOR are likely to regulate the biological processes in concert because they integrate nutrient signalling and cellular growth [85].
5. AMPK pathway. In response to metabolic stress, AMPK activates cellular processes to restore intracellular energy, and thus functions like an energy sensor [86]. In a recent study, AMPK was found to activate the lifespan of C. elegans and promote autophagy in human cells [87]. Another related study showed that myocardial ischaemia stimulated autophagy through an AMPK-dependent mechanism [88]. Ischaemic areas at risk (AAT) are capable of inducing the tumour suppressor kinase LKBI, which acting through the AMPK pathway and eukaryotic elongation factor 2F (eE2F) can activate autophagy (Fig. 3).
6. FoxO/p53 pathway. The PI3K pathway working through Akt/PKB as well as the Sirtuins have been found to activate FoxO (Fig. 3). Up-regulation of FOXO induces autophagy as shown in Drosophila, C. elegans and mouse muscle fibres [89]. Crosstalking with p53, FoxO and sirtuins can regulate longevity and enhance autophagy [90].
Pharmacological manipulations to induce autophagy
There are several ways by which autophagy can be induced. The most common and well known technique is fasting. In this section, some of these techniques will be briefly discussed.
1. Fasting/CR. In most species, fasting and CR have been associated with an enhancement of lifespan by simultaneously inducing autophagy. Fasting or CR reduces plasma insulin levels and indeed exposure of Daf-2 mutant worms to CR extends lifespan, suggesting CR and insulin/IGF-1 signalling function in parallel in regulating ageing [91]. A recent study indicates that Sch9 and mTOR pathways mediate the lifespan extension of CR [92].
2. Resveratrol. Morselli et al. have shown that transgenic expression of SirT1 induces autophagy in human cells in vitro and in C. elegans in vivo[93]. Caloric restriction and resveratrol promotes longevity through the SirT1-dependent induction of autophagy [94] and improved mitochondrial function [95]. The knockdown or knockout of SirT1 prevented the induction of autophagy by resveratrol and by nutrient deprivation in human cells as well as by dietary restriction in C. elegans. Conversely, SirT1 was not required for the induction of autophagy by rapamycin or p53 inhibition, neither in human cells nor in C. elegans. The knockdown of pharmacological inhibition of SirT1 enhanced the vulnerability of human cells to metabolic stress, unless they were stimulated to undergo autophagy by treatment with rapamycin or p53 inhibition. Along the similar lines, resveratrol and dietary restriction only prolonged the lifespan of autophagy-proficient nematodes, whereas these beneficial effects of longevity were abolished by the knockdown of the essential autophagic modulator Beclin-1. This study indicates that autophagy is universally required for the lifespan-prolonging effects of caloric restriction and pharmacological SirT1 activators. Recently, the resveratrol’s ability to directly activate SirT1 and its anti-ageing effect have been questioned [96–98].
3. Spermidine. Spermidine, a precursor of spermine, is a ubiquitous polycation, which is synthetized from putrescine. These polyamines including putrescine, spermidine and spermine participate in diverse biological processes. Exogenous supply of spermidine prolongs the lifespan of several organisms including yeast, nematodes and flies simultaneously reducing age-related oxidative protein damage in mice, indicating this compound may fulfill the definition of anti-ageing drug. Spermidine induces autophagy in cultured yeast and mammalian cells, as well as in nematodes and flies and genetic inactivation of essential autophagy genes abolishes the lifespan prolonging effect of spermidine in yeast, nematodes and flies [99, 100].
4. Other chemical promoters of autophagy. Although not proven beyond any doubt, several nutriceuticals including curcumin [101], piperine [102], trehalose [103] and lithium [104] have been found to induce autophagy. Whether such autophagy is linked with lifespan extension is not yet known [103].
Future perspectives
There is overwhelming evidence that cellular mechanisms and signalling pathways regulating ageing are related to autophagy. An increase in autophagy extends life span via CR and insulin signalling. Key factors that regulate longevity and enhance autophagy such as p53, FOXOs and sirtuins, as well as protein translation factors such as eIF4E and eE2F are among the putative targets in the fight against ageing. Nutriceuticals including resveratrol, spermidine, curcumin and piperine all seem to work in a complex way towards enhancement of autophagy. Future targeting of players involved in the autophagic pathway holds promise in anti-ageing or lifespan extension research.
Conflict of interest
None of the authors have any conflict of interest.
References
- 1.Garry DJ, Masino AM, Naseem RH, et al. Ponce de Leon’s Fountain: stem cells and the regenerating heart. Am J Med Sci. 2005;329:190–201. doi: 10.1097/00000441-200504000-00005. [DOI] [PubMed] [Google Scholar]
- 2.Breslow L, Enstrom JE. Persistence of health habits and their relationship to mortality. Prev Med. 1980;9:469–83. doi: 10.1016/0091-7435(80)90042-0. [DOI] [PubMed] [Google Scholar]
- 3.Enstrom JE, Breslow L. Lifestyle and reduced mortality among active California Mormons, 1980–2004. Prev Med. 2008;46:133–6. doi: 10.1016/j.ypmed.2007.07.030. [DOI] [PubMed] [Google Scholar]
- 4.Horne BD, May HT, Anderson JL, et al. Usefulness of routine periodic fasting to lower risk of coronary artery disease in patients undergoing coronary angiography. Am J Cardiol. 2008;102:814–9. doi: 10.1016/j.amjcard.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willis MS, Townley-Tilson WH, Kang EY, et al. Sent to destroy: the ubiquitin proteasome system regulates cell signaling and protein quality control in cardiovascular development and disease. Circ Res. 106:463–78. doi: 10.1161/CIRCRESAHA.109.208801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirkwood TB. A systematic look at an old problem. Nature. 2008;451:644–7. doi: 10.1038/451644a. [DOI] [PubMed] [Google Scholar]
- 7.Hekimi S, Guarente L. Science. Vol. 299. (New York, NY): 2003. Genetics and the specificity of the aging process; pp. 1351–4. [DOI] [PubMed] [Google Scholar]
- 8.Salminen A, Kaarniranta K. Regulation of the aging process by autophagy. Trends Molec Med. 2009;15:217–24. doi: 10.1016/j.molmed.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Gurusamy N, Das DK. Is autophagy a double-edged sword for the heart. Acta Physiol Hungarica. 2009;96:267–76. doi: 10.1556/APhysiol.96.2009.3.2. [DOI] [PubMed] [Google Scholar]
- 10.Jung CH, Ro SH, Cao J, et al. mTOR regulation of autophagy. FEBS Lett. 584:1287–95. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurmasheva RT, Houghton PJ. IGF-I mediated survival pathways in normal and malignant cells. Biochim Biophys Acta. 2006;1766:1–22. doi: 10.1016/j.bbcan.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Lorin S, Pierron G, Ryan KM, et al. Evidence for the interplay between JNK and p53-DRAM signalling pathways in the regulation of autophagy. Autophagy. 6:153–4. doi: 10.4161/auto.6.1.10537. [DOI] [PubMed] [Google Scholar]
- 13.Bursch W, Karwan A, Mayer M, et al. Cell death and autophagy: cytokines, drugs, and nutritional factors. Toxicology. 2008;254:147–57. doi: 10.1016/j.tox.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 14.Hietakangas V, Cohen SM. Regulation of tissue growth through nutrient sensing. Ann Rev Genet. 2009;43:389–410. doi: 10.1146/annurev-genet-102108-134815. [DOI] [PubMed] [Google Scholar]
- 15.Chipuk JE, Moldoveanu T, Llambi F, et al. The BCL-2 family reunion. Molec. Cell. 37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salminen A, Kaarniranta K. SIRT1: regulation of longevity via autophagy. Cell Signal. 2009;21:1356–60. doi: 10.1016/j.cellsig.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 17.Bergamini E, Cavallini G, Donati A, et al. The role of autophagy in aging: its essential part in the anti-aging mechanism of caloric restriction. Ann NY Acad Sci. 2007;1114:69–78. doi: 10.1196/annals.1396.020. [DOI] [PubMed] [Google Scholar]
- 18.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–60. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Kenyon C, Chang J, Gensch E, et al. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–4. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 20.Ogg S, Paradis S, Gottlieb S, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–9. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 21.Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–9. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- 22.Paradis S, Ailion M, Toker A, et al. A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes Dev. 1999;13:1438–52. doi: 10.1101/gad.13.11.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin K, Dorman JB, Rodan A, et al. Science. Vol. 278. (New York, NY): 1997. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans; pp. 1319–22. [DOI] [PubMed] [Google Scholar]
- 24.Wang MC, Bohmann D, Jasper H. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. 2005;121:115–25. doi: 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 25.Danielpour D, Song K. Cross-talk between IGF-I and TGF-beta signaling pathways. Cytokine Growth Factor Rev. 2006;17:59–74. doi: 10.1016/j.cytogfr.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Oldham S, Hafen E. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 2003;13:79–85. doi: 10.1016/s0962-8924(02)00042-9. [DOI] [PubMed] [Google Scholar]
- 27.Apfeld J, O’Connor G, McDonagh T, et al. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 2004;18:3004–9. doi: 10.1101/gad.1255404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bursch W, Ellinger A, Kienzl H, et al. Active cell death induced by the anti-estrogens tamoxifen and ICI 164 384 in human mammary carcinoma cells (MCF-7) in culture: the role of autophagy. Carcinogenesis. 1996;17:1595–607. doi: 10.1093/carcin/17.8.1595. [DOI] [PubMed] [Google Scholar]
- 29.Petrovski G, Zahuczky G, Katona K, et al. Clearance of dying autophagic cells of different origin by professional and non-professional phagocytes. Cell Death Differ. 2007;14:1117–28. doi: 10.1038/sj.cdd.4402112. [DOI] [PubMed] [Google Scholar]
- 30.Sakai Y, Oku M, Van Der Klei IJ, et al. Pexophagy: autophagic degradation of peroxisomes. Biochim. Biophys. Acta. 2006;1763:1767–75. doi: 10.1016/j.bbamcr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 31.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–62. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 32.Yu L, Strandberg L, Lenardo MJ. The selectivity of autophagy and its role in cell death and survival. Autophagy. 2008;4:567–73. doi: 10.4161/auto.5902. [DOI] [PubMed] [Google Scholar]
- 33.Cutler RG. Human longevity and aging: possible role of reactive oxygen species. Ann NY Acad Sci. 1991;621:1–28. doi: 10.1111/j.1749-6632.1991.tb16965.x. [DOI] [PubMed] [Google Scholar]
- 34.Gottlieb RA, Carreira RS. Autophagy in health and disease: V. Mitophagy as a way of life. Am J Physiol. 2010;299:C203–10. doi: 10.1152/ajpcell.00097.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, Hamdane D, Im SC, et al. Cytochrome b5 inhibits electron transfer from NADPH-cytochrome P450 reductase to ferric cytochrome P450 2B4. J Biol Chem. 2008;283:5217–25. doi: 10.1074/jbc.M709094200. [DOI] [PubMed] [Google Scholar]
- 36.Kume S, Uzu T, Horiike K, et al. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest. 120:1043–55. doi: 10.1172/JCI41376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pepe S. Mitochondrial function in ischaemia and reperfusion of the ageing heart. Clin Exp Pharmacol Physiol. 2000;27:745–50. doi: 10.1046/j.1440-1681.2000.03326.x. [DOI] [PubMed] [Google Scholar]
- 38.Gurusamy N, Das DK. Autophagy, redox signaling, and ventricular remodeling. Antioxidants Redox Signal. 2009;11:1975–88. doi: 10.1089/ars.2009.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wada T, Pippin JW, Nangaku M, et al. Dexamethasone’s prosurvival benefits in podocytes require extracellular signal-regulated kinase phosphorylation. Nephron. 2008;109:e8–19. doi: 10.1159/000131892. [DOI] [PubMed] [Google Scholar]
- 40.Bishop NA, Lu T, Yankner BA. Neural mechanisms of ageing and cognitive decline. Nature. 464:529–35. doi: 10.1038/nature08983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu T, Pan Y, Kao SY, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–91. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 42.Simonsen A, Cumming RC, Brech A, et al. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–84. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 43.Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 44.Schieke SM, Finkel T. Mitochondrial signaling, TOR, and life span. Biol Chem. 2006;387:1357–61. doi: 10.1515/BC.2006.170. [DOI] [PubMed] [Google Scholar]
- 45.Ravikumar B, Rubinsztein DC. Can autophagy protect against neurodegeneration caused by aggregate-prone proteins. Neuroreport. 2004;15:2443–5. doi: 10.1097/00001756-200411150-00001. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka Y, Guhde G, Suter A, et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–6. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 47.Pattison JS, Robbins J. Protein misfolding and cardiac disease: establishing cause and effect. Autophagy. 2008;4:821–3. doi: 10.4161/auto.6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tannous P, Zhu H, Johnstone JL, et al. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proc Natl Acad Sci USA. 2008;105:9745–50. doi: 10.1073/pnas.0706802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamacher-Brady A, Brady NR, Logue SE, et al. Response to myocardial ischemia/ reperfusion injury involves Bnip3 and autophagy. Cell Death Differ. 2007;14:146–57. doi: 10.1038/sj.cdd.4401936. [DOI] [PubMed] [Google Scholar]
- 50.Masoro EJ. Caloric restriction and aging: an update. Exp Gerontol. 2000;35:299–305. doi: 10.1016/s0531-5565(00)00084-x. [DOI] [PubMed] [Google Scholar]
- 51.Kim HJ, Jung KJ, Yu BP, et al. Modulation of redox-sensitive transcription factors by calorie restriction during aging. Mech Ageing Dev. 2002;123:1589–95. doi: 10.1016/s0047-6374(02)00094-5. [DOI] [PubMed] [Google Scholar]
- 52.De Meyer GR, De Keulenaer GW, Martinet W. Role of autophagy in heart failure associated with aging. Heart Failure Rev. 2010;15:423–30. doi: 10.1007/s10741-010-9166-6. [DOI] [PubMed] [Google Scholar]
- 53.Topisirovic I, Sonenberg N. Cell biology. Burn out or fade away. Science. 2010;327:1210–1. doi: 10.1126/science.1187497. [DOI] [PubMed] [Google Scholar]
- 54.Farre JC, Krick R, Subramani S, et al. Turnover of organelles by autophagy in yeast. Curr Opin Cell Biol. 2009;21:522–30. doi: 10.1016/j.ceb.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12:1542–52. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crotzer VL, Blum JS. Cytosol to lysosome transport of intracellular antigens during immune surveillance. Traffic. 2008;9:10–6. doi: 10.1111/j.1600-0854.2007.00664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kettern N, Dreiseidler M, Tawo R, et al. Chaperone-assisted degradation: multiple paths to destruction. Biol Chem. 2010;391:481–9. doi: 10.1515/BC.2010.058. [DOI] [PubMed] [Google Scholar]
- 58.Dice JF. Chaperone-mediated autophagy. Autophagy. 2007;3:295–9. doi: 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- 59.Cuervo AM. Chaperone-mediated autophagy: selectivity pays off. Trends Endocrinol Metab: TEM. 2010;21:142–50. doi: 10.1016/j.tem.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–9. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 61.Agarraberes FA, Dice JF. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J Cell Sci. 2001;114:2491–9. doi: 10.1242/jcs.114.13.2491. [DOI] [PubMed] [Google Scholar]
- 62.Bandyopadhyay U, Cuervo AM. Chaperone-mediated autophagy in aging and neurodegeneration: lessons from alpha-synuclein. Exp Gerontol. 2007;42:120–8. doi: 10.1016/j.exger.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 63.Kiffin R, Christian C, Knecht E, et al. Activation of chaperone-mediated autophagy during oxidative stress. Molec Biol Cell. 2004;15:4829–40. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cuervo AM, Dice JF. Regulation of lamp2a levels in the lysosomal membrane. Traffic. 2000;1:570–83. doi: 10.1034/j.1600-0854.2000.010707.x. [DOI] [PubMed] [Google Scholar]
- 65.Kon M, Cuervo AM. Chaperone-mediated autophagy in health and disease. FEBS Lett. 584:1399–404. doi: 10.1016/j.febslet.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–13. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 67.Cuervo AM, Knecht E, Terlecky SR, et al. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995;269:C1200–8. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- 68.Terman A, Gustafsson B, Brunk UT. Autophagy, organelles and ageing. J Pathol. 2007;211:134–43. doi: 10.1002/path.2094. [DOI] [PubMed] [Google Scholar]
- 69.Terman A, Kurz T, Navratil M, et al. Mitochondrial turnover and aging of long-lived postmitotic cells: the mitochondrial-lysosomal axis theory of aging. Antioxidants Redox Signal. 12:503–35. doi: 10.1089/ars.2009.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brunk UT, Terman A. The mitochondrial-lysosomal axis theory of aging: accumulation of damaged mitochondria as a result of imperfect autophagocytosis. Eur J Biochem/FEBS. 2002;269:1996–2002. doi: 10.1046/j.1432-1033.2002.02869.x. [DOI] [PubMed] [Google Scholar]
- 71.Loewith R, Jacinto E, Wullschleger S, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Molec Cell. 2002;10:457–68. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 72.Rohde J, Heitman J, Cardenas ME. The TOR kinases link nutrient sensing to cell growth. J Biol Chem. 2001;276:9583–6. doi: 10.1074/jbc.R000034200. [DOI] [PubMed] [Google Scholar]
- 73.Kamada Y, Funakoshi T, Shintani T, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–13. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vellai T, Takacs-Vellai K, Zhang Y, et al. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 75.Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- 76.Mortimore GE, Poso AR, Lardeux BR. Mechanism and regulation of protein degradation in liver. Diabetes/Metab Rev. 1989;5:49–70. doi: 10.1002/dmr.5610050105. [DOI] [PubMed] [Google Scholar]
- 77.Hars ES, Qi H, Ryazanov AG, et al. Autophagy regulates ageing in C. elegans. Autophagy. 2007;3:93–5. doi: 10.4161/auto.3636. [DOI] [PubMed] [Google Scholar]
- 78.Toth ML, Sigmond T, Borsos E, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–8. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- 79.Mihaylova VT, Borland CZ, Manjarrez L, et al. The PTEN tumor suppressor homolog in Caenorhabditis elegans regulates longevity and dauer formation in an insulin receptor-like signaling pathway. Proc Natl Acad Sci USA. 1999;96:7427–32. doi: 10.1073/pnas.96.13.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thevelein JM, Cauwenberg L, Colombo S, et al. Nutrient-induced signal transduction through the protein kinase A pathway and its role in the control of metabolism, stress resistance, and growth in yeast. Enzyme Microb Technol. 2000;26:819–25. doi: 10.1016/s0141-0229(00)00177-0. [DOI] [PubMed] [Google Scholar]
- 81.Budovskaya YV, Stephan JS, Reggiori F, et al. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem. 2004;279:20663–71. doi: 10.1074/jbc.M400272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Portela P, Moreno S. Glucose-dependent activation of protein kinase A activity in Saccharomyces cerevisiae and phosphorylation of its TPK1 catalytic subunit. Cell Signal. 2006;18:1072–86. doi: 10.1016/j.cellsig.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 83.Yan L, Vatner DE, O’Connor JP, et al. Type 5 adenylyl cyclase disruption increases longevity and protects against stress. Cell. 2007;130:247–58. doi: 10.1016/j.cell.2007.05.038. [DOI] [PubMed] [Google Scholar]
- 84.Yorimitsu T, Zaman S, Broach JR, et al. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Molec Biol Cell. 2007;18:4180–9. doi: 10.1091/mbc.E07-05-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pedruzzi I, Dubouloz F, Cameroni E, et al. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Molec Cell. 2003;12:1607–13. doi: 10.1016/s1097-2765(03)00485-4. [DOI] [PubMed] [Google Scholar]
- 86.Hardie DG, Hawley SA. AMP-activated protein kinase: the energy charge hypothesis revisited. Bioessays. 2001;23:1112–9. doi: 10.1002/bies.10009. [DOI] [PubMed] [Google Scholar]
- 87.Liang J, Shao SH, Xu ZX, et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nature Cell Biol. 2007;9:218–24. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- 88.Takagi H, Matsui Y, Hirotani S, et al. AMPK mediates autophagy during myocardial ischemia in vivo. Autophagy. 2007;3:405–7. doi: 10.4161/auto.4281. [DOI] [PubMed] [Google Scholar]
- 89.Hands SL, Proud CG, Wyttenbach A. mTOR’s role in ageing: protein synthesis or autophagy. Aging. 2009;1:586–97. doi: 10.18632/aging.100070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.You H, Mak TW. Crosstalk between p53 and FOXO transcription factors. Cell Cycle. 2005;4:37–8. doi: 10.4161/cc.4.1.1401. [DOI] [PubMed] [Google Scholar]
- 91.Houthoofd K, Johnson TE, Vanfleteren JR. Dietary restriction in the nematode Caenorhabditis elegans. J Gerontol. 2005;60:1125–31. doi: 10.1093/gerona/60.9.1125. [DOI] [PubMed] [Google Scholar]
- 92.Kaeberlein M, Powers RW, 3rd, Steffen KK, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–6. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 93.Morselli E, Maiuri MC, Markaki M, et al. The life span-prolonging effect of sirtuin-1 is mediated by autophagy. Autophagy. 6:186–8. doi: 10.4161/auto.6.1.10817. [DOI] [PubMed] [Google Scholar]
- 94.Gurusamy N, Lekli I, Mukherjee S, et al. Cardioprotection by resveratrol: a novel mechanism via autophagy involving the mTORC2 pathway. Cardiovasc Res. 86:103–12. doi: 10.1093/cvr/cvp384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lagouge M, Argmann C, Gerhart-Hines Z, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–22. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 96.Alcain FJ, Villalba JM. Sirtuin activators. Expert Opin Therap Patents. 2009;19:403–14. doi: 10.1517/13543770902762893. [DOI] [PubMed] [Google Scholar]
- 97.Kaeberlein M, McDonagh T, Heltweg B, et al. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005;280:17038–45. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 98.Pacholec M, Bleasdale JE, Chrunyk B, et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem. 285:8340–51. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Eisenberg T, Knauer H, Schauer A, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–14. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 100.Madeo F, Eisenberg T, Buttner S, et al. Spermidine: a novel autophagy inducer and longevity elixir. Autophagy. 6:160–2. doi: 10.4161/auto.6.1.10600. [DOI] [PubMed] [Google Scholar]
- 101.Jia YL, Li J, Qin ZH, et al. Autophagic and apoptotic mechanisms of curcumin-induced death in K562 cells. J Asian Natural Products Res. 2009;11:918–28. doi: 10.1080/10286020903264077. [DOI] [PubMed] [Google Scholar]
- 102.Curcumin, piperine could play role in preventing breast cancer. Cancer Biol Therapy. 2009;8:i–i. [PubMed] [Google Scholar]
- 103.Sarkar S, Davies JE, Huang Z, et al. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–52. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 104.Sarkar S, Floto RA, Berger Z, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170:1101–11. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]


