Abstract
Nitrogen and phosphorus are among the most widely used fertilizers worldwide. Nitrate (NO3−) and phosphate (PO43−) are also signaling molecules whose respective transduction pathways are being intensively studied. However, plants are continuously challenged with combined nutritional deficiencies, yet very little is known about how these signaling pathways are integrated. Here we report the identification of a highly NO3−-inducible NRT1.1-controlled GARP transcription factor, HRS1, document its genome-wide transcriptional targets, and validate its cis-regulatory-elements. We demonstrate that this transcription factor and a close homolog repress primary root growth in response to P deficiency conditions, but only when NO3− is present. This system defines a molecular logic gate integrating P and N signals. We propose that NO3− and P signaling converge via double transcriptional and post-transcriptional control of the same protein, HRS1
Keywords: GARP, Nitrate, Transcription Factor, Phosphate, Root Growth, Signal Interaction
Introduction
As sessile organisms, plants have evolved a myriad of adaptive mechanisms to cope with nutritional limitations in their environment. In particular, plants adapt their root development differently according to nutritional cues 1. Nitrate (NO3−) and phosphate (PO43−), two major phyto-macronutrients, are also well known signaling molecules shaping root development through partly defined molecular pathways 2, 3, 4, 5, 6. For the inorganic phosphate (Pi) response, prd 7, lpr1, lpr2 8, pdr2 9, 10, phr1 11, 12 and siz1 13 mutations have been found to affect primary root growth. Concerning NO3−, its effect is more related to lateral root growth, with the implication of several molecular actors (thoroughly reviewed in 4, 14), and has been shown to counteract the effect of glutamate on the primary root growth 15. Despite this knowledge, a mechanism by which a plant integrates the presence or absence of combinations of such key nutritional molecules is still largely unknown. Recently the NLA (Nitrogen Limitation Adaptation) and PHO2 proteins (two ubiquitin ligases) were found to control PO43− transporter trafficking (PHT1 family) which results in a nitrogen-dependent PO43− accumulation in leaves 16, 17, 18, 19. Despite these first hints into the molecular connections between P and N nutrition, nothing has so far been revealed concerning the mechanisms by which NO3− affects PHO2 or NLA activities.
The root tip is known to be at the forefront of Pi sensing 8. It is also the territory of expression of the recently identified nitrate sensor NRT1.1 20, 21. Thus it represents the perfect place for such nutritional signalling interactions. The present work reports such missing mechanism. Here we show that HRS1 and HHO1 are two early NO3−-regulated transcription factors. We document genome-wide HRS1 direct targets and demonstrate that hrs1;hho1 double mutant is involved in the primary root growth repression in response to a combination of N and P signals. We report a potential mechanism to explain this phenotype as HRS1 is under a dual transcriptional and post-transcriptional control. Finally, an in planta genome-wide investigation provides potential signaling pathways under HRS1 and HHO1 influence.
Results
Identification of two NO3− regulated transcription factors
A set of early nitrate-regulated gene clusters have been previously identified in a genome-wide investigation 22. Among these, At1g13300 was one of the most rapidly, and most strongly up-regulated transcription factors (with a significant response recorded within 6 min after treatment). The early and dramatic induction of At1g13300 and several genes involved in nitrate uptake and assimilation have been confirmed in an independent set of experiments (Fig. 1). In roots, NO3− provision triggers the induction of the 2 sentinel gene transcripts (NIR and NRT2.1) after 10 and 30 minutes after treatment, respectively. This validates the conditions of the N-treatment (Fig. 1a). As predicted by previous experiments, At1g13300 accumulation was rapidly and strongly up-regulated (10-fold within 10 min) compared to the KCl mock treatment (Fig. 1a). These observations are consistent with previous reports corresponding to genome-wide or specific RT-qPCR investigations 6, 23, 24, 25.
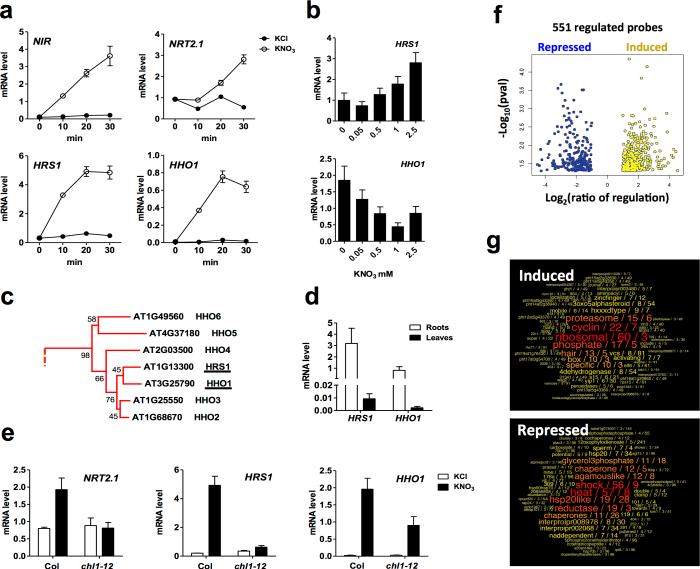
Figure 1. HRS1 and HHO1 are two Arabidopsis transcription factors highly induced by NO3− downstream the nitrate sensor activity. Identification of HRS1 direct targets.
(a) Dynamic nitrate response of mRNA for NIR, NRT2.1 (NO3− responsive sentinels), HRS1 (At1g13300) and HHO1 (At3g25790) in roots of fourteen-day old seedlings treated with 1mM KNO3 or 1mM KCl (as mock treatment), values are means ±SEM (n=4). (b) Steady state level of HRS1 and HHO1 mRNA in roots of plants grown for 14 days on solid media containing 0.5mM KH2PO4 and different KNO 3 concentrations, values are means ±SEM (n=4). (c) A subfamily of the G2-like transcription factors phylogenetic tree built by ClustalW alignment and Maximum Likelihood method, values are bootstraps based on 500 replicates (see Supplementary Fig. 1). (d) HRS1 and HHO1 mRNA levels in roots and leaves of WT plants grown for 14 days on basal MS media containing 0.5mM KH2PO4 and 2.5mM KNO3, values are means ±SEM (n=3). (e) HRS1 and HHO1 nitrate induction is affected in roots of chl1-12 mutant mutated in NRT1.1. WT and chl1-12 fourteen-day old seedlings were treated for 30 minutes with 1mM KNO3 or 1mM KCl (as mock treatment). All transcript levels were quantified by RT-qPCR and normalized to two housekeeping genes (ACT and CLA), values are means ±SEM (n=4). (f) Volcano plot of HRS1 direct regulated targets identified by TARGET (Transient Assay Reporting Genome-wide Effects of Transcription factors) procedure 37. (g) results showing overrepresented terms in the list of the HRS1 direct up and down-regulated genes.
The At1g13300 gene encodes a myb-related transcription factor belonging to the GARP (GOLDEN2, ARR-B, Psr1) family; a homolog to NIGT1 identified in rice 26. The GOLDEN2-like subgroup is composed of 40 protein sequences (AGRIS, http://arabidopsis.med.ohio-state.edu/). It encompasses proteins involved in various processes ranging from the control of chloroplast and leaf development (GLKs 27, KANADIs 28) to nutritional reprogramming in plants (PHR1 and PHL1 29). A phylogenetic tree has been built out of the 40 G2-like protein sequences (Supplementary Fig. 1, see Methods). It shows that At1g13300 belongs to a small group of 7 proteins sharing high sequence similarity to each other, but quite well separated from the rest of the tree (Fig. 1c and Supplementary Fig. 1). This sub-group within the larger GARP family has already been reported on its own 30. We thus kept the proposed nomenclature and extended the relationships between the other members of the GARP family. At1g13300 is named HRS1 (Hypersensitive to low Pi-elicited primary Root Shortening 1) 30. HRS1 is a paralog with its closest neighbor, At3g25790, which has been named HHO1 (HRS1 Homolog 1) 30. It is noteworthy that these two close homologs, HRS1 and HHO1, have recently been identified among the 2,594 gene pairs that were defined in an effort to isolate redundant duplicated genes 31. Interestingly, HHO1 showed a similar pattern of expression to HRS1 with a strong up-regulation (> 50-fold) within the first 20 min of nitrate treatment (Fig. 1a), both being mainly expressed in roots (Fig. 1d). Furthermore, HRS1 and HHO1 mRNA levels in roots are dependent on nitrate concentration in the media. WT seedlings constantly grown for 14 days on media containing increasing NO3− concentrations presented a gradual transcriptional response of HRS1 and HHO1 (Fig. 1b). Interestingly, despite the fact that both genes are up-regulated by NO3− provision, the increasing KNO3 concentration has opposite effects on their steady state expression levels: HRS1 is positively regulated and HHO1 is negatively affected (Fig. 1b). In conclusion, HRS1 and HHO1 are two root-specific, NO3−-controlled transcription factors (found to be responsive in the NR-null mutant in previous genome wide-investigations 32). A meta-analysis 6 also demonstrated that HRS1 and HHO1 are under the control of NRT1.1 protein activity. Indeed, their nitrate induction is strongly and robustly affected in chl1 mutants (defective in the NRT1.1 gene) in 2 independent transcriptomic datasets issued from 2 independent laboratories 25, 33. We also performed analysis in our conditions and indeed recorded defects in nitrate responsiveness in an independent chl1 deletion allele of NRT1.1 (chl1-12; Fig. 1e). In the chl1-12 mutant, the 24-fold induction (after 30 min of NO3− treatment) of HRS1 expression is totally abolished and the HHO1 strong induction (~ 100-fold) is reduced to a third. Finally HRS1 and HHO1 are also under the influence of NLP genes (NIN-like transcription factors) implicated in N-signaling 34 and are found to be bound (assayed by ChIP-seq) by NLP7 35.
Collectively these results demonstrate that the TF paralogs, HRS1 and HHO1 are each strongly NO3− controlled (e.g. are among the most, if not the most, robustly NO3− regulated genes in many datasets 36) and positioned downstream of the early regulators NRT1.1/CHL1 and NLP7-6 activity.
Identification of HRS1 direct targets
At the outset of this study nothing was known about the molecular mechanism downstream of At1g13300 (HRS1). We thus decided to investigate the genome-wide effect of this transcription factor using a transient assay system for TF perturbation that can uncover direct targets only 37. Indeed, we have shown for other TFs (ABI3, bZIP1) that using this system to uncover the direct regulated targets of a transcription factor can give very important clues about its functional role in planta 37, 38. We thus speculated that we could retrieve the functional in planta activity of At1g13300 by studying the set of its direct targets.
To search for such genes directly regulated by At1g13300, we used the TARGET approach described by Bargmann et al. 37. Briefly, protoplasts are isolated from roots of 10-day old Arabidopsis seedling and transformed with the plasmid pBeaconRFP_GR-HRS1, expressing a translational fusion (N-ter) between HRS1 and the rat glucocorticoid receptor (GR) under the control of the pCaMV35S promoter. Protoplasts are treated with i) dexamethasone (DEX; triggers GR-TF fusion entrance in the nucleus) and ii) cycloheximide (CHX; translation inhibitor that prevents activation of indirect targets) 37. Because of the possibility that HRS1 acts as part of a NO3−-regulated protein complex (as it is itself NO3− regulated, see above), we kept NO3− present during the whole TARGET procedure. After FACS (Fluorescent Activated Cell Sorting) selection (based on RFP signal provided by an independent cassette in the plasmid) of transformed protoplasts, total RNA was isolated from GR-HRS1 protoplasts and used for the transcriptomic analysis. The statistical analysis (see Methods) of transcriptome results identified 551 gene probes whose expression was affected by HRS1 nuclear import (e.g. upon DEX treatment nucleus entrance of the GR-HRS1 complex). The corresponding genes were classified as up- and down-regulated HRS1 direct target genes (Fig.1f, Supplementary Data 1).
To search for the functions globally affected by HRS1 activity, we performed two kinds of analysis. First, we determined over-represented GO (Gene Ontology) categories using the Virtual Plant and agriGO platforms 39, 40. Second, we developed our own algorithm that search for over-represented terms in a set of TAIR v10 gene description (see Methods). Both approaches yielded very similar conclusions: At1g13300 (HRS1) collectively induces genes related to “phosphate” and “cell division” (including the terms meristematic activity, cyclin, ribosomal proteins). On the other hand, in list of HRS1 down-regulated genes, the most frequent terms were “heat” “shock” and the most represented functions were linked to response to stresses (Fig. 1g, Supplementary Data 1 and Supplementary Fig. 6). This genome-wide investigation of the At1g13300 HRS1 transcription factor direct targets brought us to be interested in the interaction between NO3− signaling (that controls transcriptional activation of this gene) and P nutrition that seems to be one of the functions under its influence 30. Taken together with the fact that HRS1 belongs to the same TF family as the very well characterized PHR1 gene 11, 12, known to be central in the control of P starvation response, we decided to employ a reverse genetic approach to understand the role of HRS1 in the control of NO3− and Pi signaling interactions.
HRS1 and HHO1 repress primary root growth
The hrs1-1 mutant was obtained from ABRC seeds stock center, and the TDNA insertion and the absence of HRS1 full-length transcripts were confirmed (Supplementary Fig. 2). First, we tested the growth of the hrs1-1 mutant on different media containing +P (0.5mM KH2PO4) or –P combined with different NO3− concentrations (0mM, 0.05mM, 0.5mM, 1mM, 2.5mM). The results didn't lead to any reliable/robust phenotype (Supplementary Fig. 3a), as previously reported 30. We thus hypothesized that this could be due to functional redundancy of the two NO3− induced close relatives HRS1 and HHO1 (Fig. 1c). We thus tested the double mutant hrs1-1;hho1-1 (see molecular characterization in Supplementary Fig. 2) on the same P/N varying conditions. As previously shown 8, the primary root growth of WT plants was impaired on -P conditions. Interestingly however, primary root growth of the double hrs1;hho1 mutant was strongly insensitive to the -P conditions (Fig. 2a,b). This insensitivity is also manifested on transfer experiments (Supplementary Fig. 3b,c). The fact that the hsr1;hho1 double mutant is resistant to -P conditions complements previous observations reporting that the over-expression of HRS1 indeed confers hypersensitivity to low P conditions 30. More importantly, NO3− is required for the manifestation of the hrs1;hho1 double mutant phenotype. Indeed, the phenotype of the hrs1;hho1 double mutant is lost in plants grown on -N/-P conditions. More precisely, the presence of at least 0.05mM NO3− in the media was necessary for the appearance of the root phenotype in response to Pi depleted conditions (Fig. 2c). These results suggest that the HRS1 and HHO1 proteins act as repressors of primary root development specifically when PO43−is absent and NO3− is present in the media. It is thus tempting to propose that the well-known primary root growth repression in response to low P conditions is an active process that is influenced by i) NO3− and ii) the NO3−-regulated transcription factors HRS1 and HHO1.
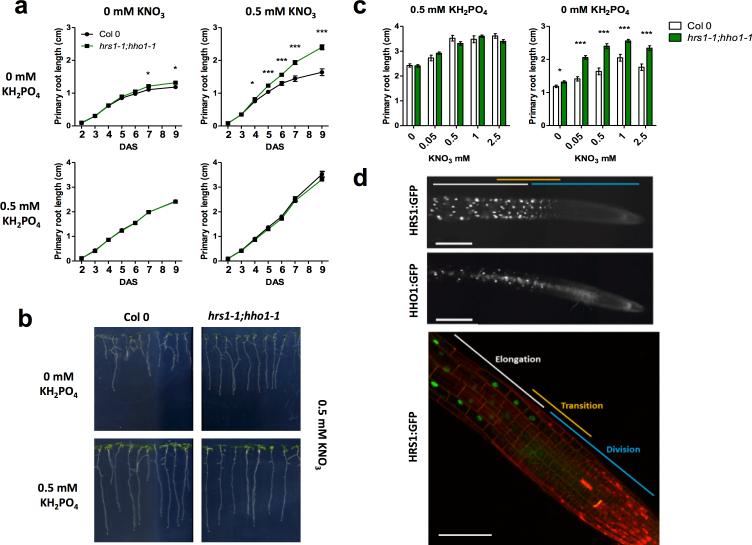
Figure 2. HRS1 and HHO1 are two nuclear factors expressed in meristematic elongating cells repressing root growth in response to combination of P and N availability.
(a) Primary root growth measurement over time of Col and hrs1-1;hho1-1 seedlings grown on P/N combinations. In +P/-N conditions the WT and mutant slopes overlap. (b) 9 day-old seedlings grown on 0.5 mM of NO3− and PO43− under different conditions (0-0.5 mM added). (c) Effect of different nitrate concentrations in the media on the primary root growth of hrs1-1;hho1-1 9 day-old seedlings. Values represent the means ± SEM (n=30). Asterisks indicate significant differences from WT plants (* P<0.05; ** P<0.01; *** P<0.001; Student's t test). (d) Epifluorescent and confocal imaging of pHRS1:HRS1:GFP and pHHO1:HHO1:GFP expressing plants (scale bars correspond to 200 μm).
HRS1 and HHO1 are expressed in elongating root cell nuclei
In order to further investigate the in planta roles of the HRS1 and HHO1 proteins, we constructed native promoter-gene-GFP lines for these two transcription factors. Epi-fluorescence and confocal imaging of transgenic pHRS1:HRS1:GFP and pHHO1:HHO1:GFP plants indicate that the two proteins are expressed and localized in the nucleus of epidermal and cortex cells in elongating roots (Fig. 2d and Supplementary Fig. 4). HRS1 and HHO1 expression is found in the transition domain of the root apical meristem and in the elongation zone (Fig. 2d). The expression of HRS1 was also visible in root hair cells (Supplementary Fig. 4). The localization of these two proteins supports the idea that HRS1 and HHO1 are nuclear-localized transcription factors involved in the control of the root elongation.
HRS1 binds to two different cis regulatory elements
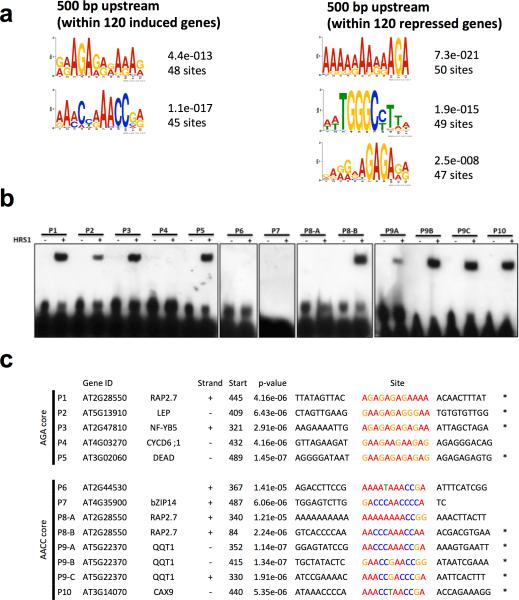
In order to identify cis-regulatory elements (CRE) directly targeted by HRS1 protein, we searched for conserved motifs in the promoters of the most strongly up- and down-regulated target genes. Thus, we examined the 500bp promoter sequences upstream the transcription start site for the 120 top induced and repressed genes (based on fold regulation upon DEX treatment) from the TARGET transcriptome analysis (presented Fig. 1f). Five strongly significant motifs were retrieved using the MEME algorithm 41. The two cis-motifs, which were overrepresented in the promoters of genes up-regulated by HRS1, display the consensus sequences AGANNNAAA and AAACNNAACC. By contrast, the HRS1 down-regulated genes share three over represented motifs: AANNAGA, TGGGC and GAGA (Fig. 3a). Among these last three cis-motifs, the TGGGC(C/T) motif was already known as the core binding sequence of the cis-regulatory element for TCP (Teosinte branched1, Cycloidea, PCF) transcription factors 42. Inspired by the consensus sequences above, we defined 5 motifs (M1 to M5) (Supplementary Fig. 5c and Table 1) that were tested in vitro for direct binding by HRS1 in Electophoretic Mobility Shift Assays (EMSA). The HRS1 recombinant protein produced a shift for motifs #1, 2, 3 and 5. The shifts were completely reverted by the competition of a 200-fold molar excess of the same unlabeled probe (Supplementary Fig. 5d). No band shift was detected for motif #4, which corresponds to the TCP binding site. This means that TCP transcription factors may be partners of HRS1 mediating its repressive transcriptional activity. Motifs #1, #2 and #3 in the HRS1 induced genes share the core sequence AGA and the motif #5 the core AACC (already known as part of the GLK1/2 binding site 27). This is in accordance with the recent finding that a particular transcription factor can bind to distinct cis-elements 43. The specificity of HRS1 binding to the selected DNA probes was confirmed by EMSA analysis, using unlabeled probes mutated on the above described core sequences (AGA and AACC). Ten-, 25-, and 50-fold molar excess of unlabeled mutant probes could not compete with HRS1 binding to motifs #1, #3 and #5, thus confirming that HRS1 binding is maintained by AGA and AACC core sequences (Supplementary Fig. 5e).
Figure 3. Identification of Cis-Regulatory Elements and binding of HRS1 to the promoters of “meristem related” genes.
(a) Weight matrix representation of the motifs retrieved by the MEME algorithm analysis from the 500bp sequences upstream the transcription start sites of the top 120 direct up and down-regulated HRS1 target genes. (b) EMSA analysis on 30bp promoter fragment from HRS1 directly regulated genes (from TARGET analysis); biotin labeled DNA probes (20 fmol), HRS1-GST protein (50 ng). (c) List of the promoter fragments sequences used for EMSA analysis in a. For each promoter the gene ID, the common name and the strand position is indicated.
In order to obtain functional explanations of the root developmental phenotype described Fig. 2a-c, we tested if HRS1 was also able to bind native CRE elements found in promoters of genes categorized as “meristem related genes”. For that, we used 30bp promoter fragments as EMSA probes. HRS1 produced a significant shift for 9 of 13 promoter fragments tested (Fig. 3b,c). Promoters have been selected as they are; i) induced by HRS1 in the TARGET system (Fig. 1f); and ii) they are classified by GO ontology as being involved in meristematic activity. To summarize, HRS1 is able to bind the promoter sequences of ERF/AP2 transcription factors - RAP2-7 (RELATED TO AP2.7; AT2G28550), LEP (LEAFY PETIOLE; AT5G13910), the NF-YB5 (Nuclear Factor Y, subunit B5; AT2G47810) transcription factor, the helicase DEAD (AT3G02060), the DNA binding protein QQT1 (QUATRE QUART 1; AT5G22370) involved in embryo development, and the cation calcium exchanger CAX9 (CATION CALCIUM EXCHANGER 9; AT3G14070).
These results show that HRS1 possesses two distinct target cis-regulatory elements found in HRS1 activated genes belonging to developmental processes pathways.
HRS1 is post-transcriptionally controlled by P provision
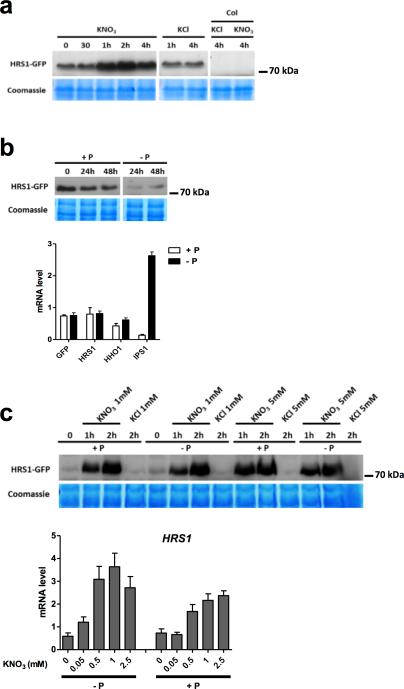
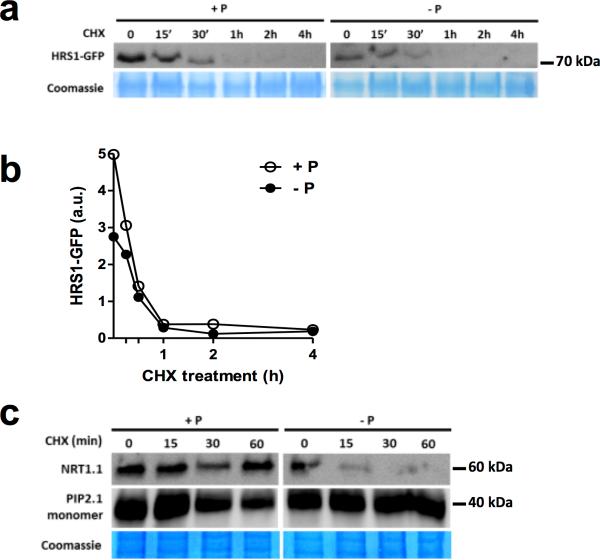
Since HRS1 is clearly strongly transcriptionally activated by the NO3− signaling pathway (Fig. 1a), and it is involved in the convergence of Pi and NO3− signals (Fig. 2), we investigated how Pi and NO3− might affect the levels of HRS1 protein. As expected for such a NO3−-induced transcript (Fig. 1), the HRS1 protein strongly accumulates in response to NO3− treatment (Fig. 4a). Interestingly, in whole roots, HRS1:GFP protein accumulation was reduced after 24 and 48 hours of Pi deprivation (Fig. 4b). In the same conditions (48h Pi starvation), HRS1 mRNA levels are not changed, even if the plant felt the Pi deprivation (as reported by IPS1, Fig. 4b). These findings suggest a possible post-transcriptional regulation of HRS1 by Pi provision. Interestingly this interaction of NO3− transcriptional induction of HRS1, together with its post-transcriptional regulation in response to –P, could constitute a mechanism that entangles both nutritional signals. To confirm this hypothesis, the NO3− transcriptional regulation has to be conserved independent of the preceding P treatment. In order to demonstrate this, we verified that the transcriptional regulation of HRS1 by NO3− is maintained in P varying conditions (Fig. 4c). Furthermore, we observed that the accumulation of the HRS1 protein accumulation is not affected by P provision in short term experiments. This demonstrates that NO3− and -P signals, act early (within minutes) and late (within days), at the transcriptional and post-transcriptional levels, respectively (Fig. 4). To delve further into one potential mechanism of the P effect on HRS1 activity, we analyzed HRS1-GFP protein half-life in P varying conditions (Fig. 5a,b). We recorded that –P treatment shifts the HRS1 half-life from ~20 to 30 min. Interestingly, this mechanism is not restricted to HRS1, since another nitrate-regulated protein is under this kind of regulation (Fig. 5c). Indeed, we have found that the nitrate transporter/sensor NRT1.1 is strongly destabilized upon –P conditions (as compared to another membrane protein PIP2.1, Fig. 5c). This demonstrates that P deficiency conditions may broadly affect nitrate-regulated proteins to convey a layer of the NO3− and PO43− signal interaction.
Figure 4. P and NO3− provisions influence HRS1 protein accumulation, but the P signal does not affect its mRNA accumulation and NO3− response.
(a) HRS1-GFP protein accumulation is induced in response to 1mM NO3− treatment. (b) HRS1-GFP protein accumulation is affected by P provision; 48h P starvation (reported by IPS1 sentinel regulation) does not affect HRS1 (endogenous gene), HHO1, and HRS1:GFP mRNA accumulation. (c) Rapid NO3− transcriptional activation of HRS1 and subsequent protein accumulation is maintained regardless P provision. Different NO3− levels induce: i) HRS1-GFP protein accumulation in roots of 48h P starved plants and ii) HRS1 transcript accumulation in plant roots grown (14 days) on P varying media. Values are means ±SEM (n=3).
Figure 5. P provision affects HRS1 and NRT1.1 protein stability.
(a) CHX (100μM) treatment affects HRS1-GFP accumulation differentially in +P and -P conditions. (b) Quantification of immunoblot signal. (c) P starvation accelerates the protein degradation rate of the nitrate transporter/sensor NRT1.1, but not the aquaporin PIP2.1 (used as non nitrate-inducible control).
Genome-wide effect of HRS1 and HHO1 on root gene expression
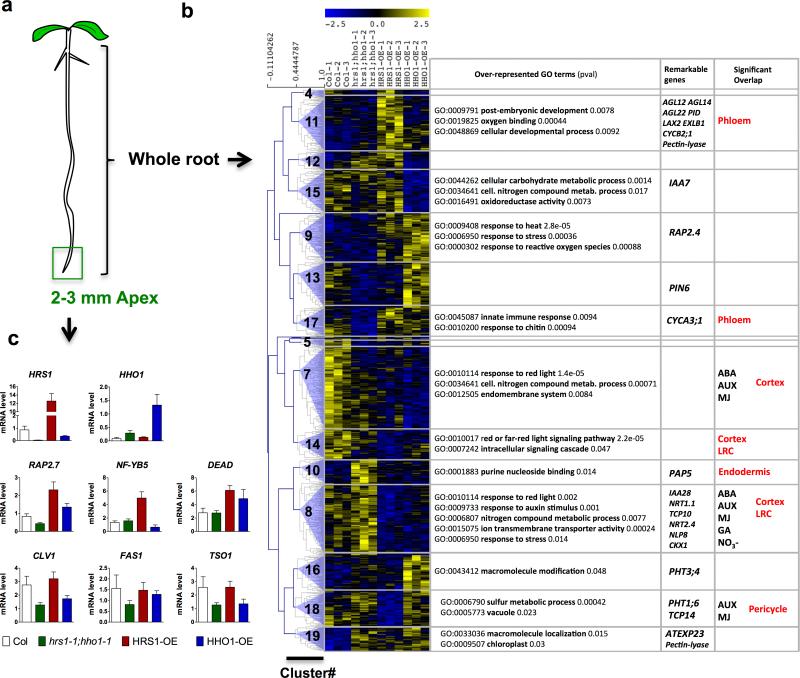
In order to better document the in planta effects from the modification of the HRS1/HHO1 transcription factors, plantlets (Col, hrs1;hho1, HRS1-OE, HHO1-OE) were grown in -P/+NO3− conditions for 6 days in three independent experiments. Whole roots (Fig. 6a) were harvested to measure the effect of these HRS1/HHO1 genetic modifications using last generation Affymetrix™ chips (Arabidopsis gene1.1ST array, Supplementary Data 2). Analysis was performed as previously described in references 44, 45 (for details see Methods). Analysis of variance followed by a post hoc Tukey test yielded 1,125 HRS1/HHO1 regulated probes corresponding to 969 unambiguous genes. A clustering analysis helped to understand the dominant mode of regulation triggered by the mutation or the over-expression of HRS1 and HHO1 (Fig. 6b). Among these 969 HRS1/HHO1 regulated genes, 22 genes have been identified to be direct HRS1 targets based on the TARGET approach (11 up and 11 down-regulated, Supplementary Data 1). This relatively low number is however higher than one would expect by chance (Monte Carlo test pval<0.05). Among these HRS1 direct targets is NRT1.1 which is found in cluster #8. Since, we have shown that NRT1.1 is a key regulator of HRS1 primary NO3− induction (Fig.1e), this demonstrates a potential feedback mechanism of HRS1 on its own regulator, as is often found in regulatory networks. The 969 HRS1/HHO1 regulated genes have been subjected to GO enrichment analysis through the agriGO web site 40 followed by a REVIGO analysis in order to reduce and summarize the number of GO terms detected 46. Interestingly, this analysis demonstrates that “phosphorus metabolism”, “hormone metabolism”, “starch metabolism”, “developmental process”, “cellular nitrogen compound metabolism”, “post embryonic development”, “response to red light”, “response to abiotic stimulus” are [among others (Supplementary Fig. 6)] enriched in this list. This demonstrates that i) HRS1/HHO1 indeed affect the processes that have been predicted by the cell-based TARGET system approach (Fig. 1g and Supplementary Fig. 6), and ii) that HRS1/HHO1 may have other functions, as the one described herein, some of which that was predicted by other studies (ref 47, 48 and discussed below).
Figure 6. Probing the effect of HRS1 and HHO1 mutations and over-expressions in –P deficient root transcriptome.
(a) 6 day old plantlets were grown on –P/+NO3− conditions (b), or on +P/+NO3− media and then transferred 2 days to –P/+NO3− conditions (c). (b) Clustering of the 969 HRS1 regulated genes from whole roots of plantlets grown on –P/+NO3− media. For each cluster, a selection of over-represented GO terms are given, as well as remarkable genes belonging to the cluster. Clusters overlap with NO3− 32; hormonal 62 responsive genes, or root cell-type specific markers 52 have been measured using the GeneSect algorithm 22. Significant overlaps (pval<0.05) are reported in the 3rd column. (c) RT-qPCR analysis on selected HRS1 direct targets on RNA extracted from 2-3 mm sections of the root apex in a transfer experiment Values are means ±SEM (n=3).
In order to obtain further insights into the altered gene responses triggered by altered expression of HRS1/HHO1, a clustering analysis uncovered 19 clusters of mis-regulated genes (Supplementary Data 2). The overall analysis of the 19 clusters demonstrates that even if HRS1 and HHO1 seem to play redundant roles in the control of primary root growth (Fig. 2), they may control at the same time the same set of genes (such as in cluster #9,17,7,14,10,8) or different genes (such as in clusters #4,11,12,15,13,16). AgriGO was then used to determine the molecular coherence of these HRS1/HHO1 regulated gene clusters into potential biomodules (i.e.: co-regulated genes having a particular function 49). Interestingly, in the clusters for which HRS1 and HHO1 have a coherent effect on gene regulation (control the same set of genes), the GO terms are related to “stress responsiveness”, “response to ROS”. Moreover, it seems that HRS1 over-expression is the one that triggers the most regulation on genes responsible for growth control (i.e.: cluster #11). Taken together this could suggest that HRS1 and HHO1 play redundant functions due to their phylogenetic proximity, but might control different set of genes depending on the genomic context. However, since the two TFs converge toward the control of ROS signalling related genes, and that ROS have recently been involved in the control of primary root meristematic activity 50 or cell size 51, it is tempting to propose such mechanism could be downstream of HRS1/HHO1 influence. It is however important to note that UPB1 does not belong to the 969 HRS1/HHO1 regulated genes based on whole root transcriptome studies. These hypotheses will need further experiments to be fully validated.
One hypothesis related to the modularity of the HRS1 vs. HHO1 effect, is that they can affect different genes according to possible tissue context. To document this possibility, the different HRS1/HHO1 controlled gene clusters were overlapped to set of genes defined as markers of different root cell layers 52. Interestingly clusters #11,7,17,14,10,8,18 appear to significantly (pval<0.05- Monte Carlo test named GeneSect22) overlap with cell-type specific markers (3rd column Fig. 6b). Thus, since the transcriptome is performed at the whole root level, this overlap may show that some clustering properties are related to different activities of the two TFs in different cell types. Again, this will need to be fully validated by following further experimental studies.
The most striking cluster of this analysis is #8. Indeed, it gathers genes being more highly expressed in the double hrs1-1;hho1-1 mutant, and repressed by HRS1 or HHO1 over-expression (Fig. 6b). Interestingly this cluster comprises genes known to be responsive to NO3− (including NRT1.1 and NRT2.4, among others) and hormones. Indeed, “response to auxin stimulus” is also an over-represented GO term in this gene cluster. This observation opens perspectives concerning the role of auxin signaling in the control of root developmental mechanisms that could explain the effect of HRS1 and HHO1 in –P conditions.
In order to consolidate the fact that HRS1 binds cis-elements of 6 genes found to be direct targets of HRS1 that are activated in the cell based system (Fig. 1 and Fig. 3), we wondered why these genes are not found in the 969 HRS1 regulated genes detected in whole roots? Since the P effect is very much localized to the root apex, and likely not affected by the whole plant P status (according to transfer experiments Supplementary Fig. 3), we hypothesized that the regulation by P may be restricted to the most apical part of the root. To validate this hypothesis, we performed transfer experiments from +P/+N to -P/+N media. The tip of the primary root (2 to 3 millimeters) was harvested from the WT, hrs1-1;hho1-1 double mutant and over-expressors and RNA used for qPCR experiments. This revealed that RAP2.7, NF-YB5 and the DEAD helicase are indeed regulated by HRS1 over-expression at the root apex (Fig. 6c). Several meristem related genes (CLV1, FAS1, TSO1), defined has direct targets by the cell based TARGET system (Supplementary Data 1) were also measured and displayed mis-regulation in the hrs1;hho1 double mutant, but with no effect of the TF over-expression (Fig. 6c). This demonstrates that these genes may be indeed under the direct influence of HRS1 in plants and are potential candidates controlling meristematic activity together with the previously evoked pathways that include auxin and ROS signaling (refers to clusters #8 and #9,15,11 respectively, Fig. 6b). In order to decipher the different regulatory pathways below the action of the genes known to sensitize primary root to local P deficiency, it will be important to confront whole genome expression reprogramming induced by lpr1/2 8, pdr2 9 and hrs1;hho1 in order to define if these regulatory modules share common signaling pathways. Further reverse genetic studies will also be needed to validate that the potential regulators of meristematic activity such as CLV1, FAS1, TSO1, RAP2.7, or NFYB5 are indeed part of the HRS1/HHO1 mediated response.
Finally, we believe that this transcriptomic analysis provides an important proxy towards understanding the other roles of HRS1 in plants. Indeed, it has been shown to be controlling germination 53, or to confer drought or salt stress tolerance 54. Again the general functions regulated by HRS1 such as “ROS signaling” as well the important overlap with ABA responsive genes are important keys to understand the underlying mechanisms behind the previously reported phenotypes.
Discussion
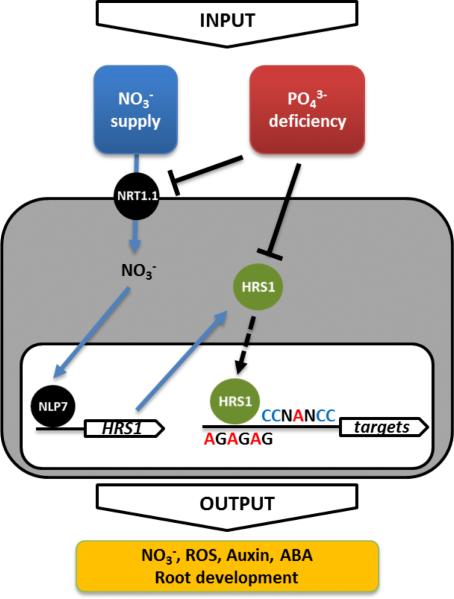
As we take together the above observations, we propose the model described in Fig. 7. HRS1 is under a dual regulation: i) a NO3− induced (NRT1.1/CHL1 NLP7 dependent) transcriptional control, and ii) a Pi regulated post-transcriptional control. In response to these two signals, HRS1, controls primary root growth through possibly: i) its action on two independent cis-regulatory elements contained in promoters of activated genes; ii) its effect on pathways such as ROS or Auxin signaling. These two possibilities are not exclusive. Our observations can, to some extent, be related to the recent work of Kellermeier et al. (2014) 55, which makes a clear demonstration that nutrient signalling interaction is rather a general rule than an exception.
Figure 7. Proposed model: NO3− and P limitation signals are integrated by the control of HRS1 at the transcriptional and post-transcriptional level, respectively.
Upon NO3− treatment, the NRT1.1 and NLP7 regulatory module induces rapid HRS1 transcript and protein accumulation. The prolonged P deficiency condition negatively affects the accumulation of HRS1 and modifies its regulatory activity. Furthermore, the P limitation signal seems to control the NO3− sensor (NRT1.1) protein half-life. This delineates a working model to understand several points of NO3− and PO43− signal interactions via the HRS1 pathway.
Our work provides a potential mechanism for understanding how two key plant mineral nutrients (PO43− and NO3−) interact to control root growth, an issue of global importance since, nitrogen and phosphorus are the two most widely used fertilizers worldwide that maintain plant growth and production. For instance, a recent work by Delgado-Baquerizo et al. showed that, at the ecological level, drying lands might impact dramatically nitrogen and phosphorus ratios available for plants, thus jeopardizing food production worldwide 56. The molecular events, mediated by HRS1 and HHO1 in plants, that integrate two mineral-related signaling pathways, is part of the fundamental knowledge needed to tackle such key environmental challenge in a global warming context.
Methods
Plant material
All A. thaliana plants were in the Columbia background. Mutant hrs1-1 (SALK_067074), hho1-1 (SAIL_28_D03) and mutant chl1-12 (Salk_034596) were obtained from ABRC seeds stock center. The hrs1-1;hho1-1 double mutant has been obtained by crossing. Despite the presence of a highly accumulated chimeric RNA in the hho1-1 line (characterized in Supplementary Fig. 2c) the production of a full-length gene product is knocked out (Supplementary Fig. 2b). The promoter-gene-GFP lines for tissue localization were obtained by cloning HRS1 and HHO1 from genomic sequences, bringing respectively the 3kb and 2.5kb upstream promoter region and the gene, into pMDC107 Gateway-compatible vector. Over-expressor lines were obtained by cloning HRS1 and HHO1 coding sequences into pMDC32 Gateway-compatible vector 57, using primers listed in Supplementary Data 1. The constructs were transferred to Agrobacterium tumefaciens strain GV3101 and used for the Arabidopsis transformation by the floral dip method 58. Transgenic plants were selected by antibiotic resistance and T3 homozygote descendants were used for analysis.
Growth conditions and treatments
For NO3− treatment experiment, plants were grown in sterile hydroponic conditions as described in 59. Hydroponic media consisted of MS basal salt medium containing no nitrate and supplemented with 3 mM sucrose, 0.5 mM ammonium succinate, MES buffered at pH 5.7 (0.5 g.l−1). Plants were grown for 15 days in day/night cycles (16/8h; 65 μmol photons m−2.s−1) at 22°C. Plants were transferred to an equivalent fresh nitrogen-free medium for 24h and then treated with nitrogen as 1mM KNO3 or 1mM KCl, as mock-treatment. Roots were sampled at different time points after the treatment and immediately frozen in liquid nitrogen. For P starvation experiments, Col-0 and hrs1-1;hho1-1 seeds were sown in on the surface of solid media consisting of MS basal salt medium nitrogen and phosphorous-free supplemented with KNO3 at different concentrations (0.05mM, 0.5mM, 1mM, 2.5mM), 0.5mM KH2PO4 for P-sufficient condition, 3mM sucrose, MES (0.5 g.l−1) and 0.8% (w/v) agarose. Different volumes of 1mM KCl solution were added to the media to keep the K+ concentration constant among different conditions. Plants were grown vertically for 9 days in day/night cycles (16/8h; 90 μmol photons m−2.s−1) at 22°C.
For P starvation experiments in liquid media, plants were grown for 15 days in sterile hydroponics conditions, in MS media containing 5mM KNO3 and 1mM KH2PO4. Plants were transferred toward an equivalent KH2PO4-free fresh medium (–P) or 1mM KH2PO4 media (+P) for 48h. For half-life experiment CHX was used at 100 μM. All growth and gene expression experiments are supported by at least three independent experiments.
Root growth measurements
Beginning 2 days after seed sowing, two Petri dishes per condition (i.e. at least 30 plants) were scanned every day at 400dpi. Root length was measured using Optimas 6.1 software and statistical differences between genotypes were calculated using Student's t test.
Real-Time qPCR analysis
Total RNA was isolated from Arabidopsis roots and shoots using TriReagent® (Molecular Research center Inc.) and digested with DNAseI (SIGMA-ALDRCH, St Louis, USA). Total RNAs were then reverse transcribed to one-strand cDNA using Thermo™ script RT (Invitrogen) according to the manufacturer's protocol. Gene expression was determined by RT-qPCR (LightCycler® 480; Roche Diagnostics, Basel, Switzerland) using gene-specific primers (listed in Supplementary Data 1) and LightCycler® 480 SYBR Green I Master mix (Roch, IN, USA). Expression levels of tested genes were normalized to expression levels of the ACTIN and CLATHRIN genes.
TF perturbation assays in the TARGET system
The TARGET procedure has been performed as previously described in Bargmann et al. 37. Protoplasts were treated with 35μM cycloheximide (CHX), and 10μM dexamethasone (DEX). The Red Fluorescent Protein was used as marker selection for fluorescent-activated cell sorting (FACS) of successfully transformed protoplasts. 1mM NO3− was maintained during the whole procedure. RNA was extracted and amplified for hybridization with ATH1 Affymetrix™ chips. Data were analyzed with R. Mas5 normalized data were extracted and analyzed through a t-test procedure for DEX response (pval < 0.05, fold regulation > 2X, corresponding to a FDR < 0.1).
Microscopy
Col-0, pHRS1:HRS1:GFP and pHHO1:HHO1:GFP seedlings were grown on half-strength MS media for 8 days. The same liquid media was used for mounting plantlets during image capture. Fluorescence imaging was performed using the Olympus BX61 microscope. Samples were excited at 470nm and emission was collected between 500 and 535 nm. Confocal images were performed with a Zeiss LSM510 Meta laser scanning microscope. Root cell walls were stained with propidium iodine 10 μg.ml−1 (SIGMA) and nucleus with DAPI 1 μg.ml−1 (SIGMA). 3D root reconstructions were obtained from confocal Z series using the Imaris software.
Semantic gene enrichment analysis
In order to identify the most represented gene functions for each gene list, the gene descriptions of targets were retrieved from TAIR web site (http://www.arabidopsis.org/index.jsp) and the list was analyzed using the software developed in [R] (http://www.r-project.org/). Briefly, given the list of 551 HRS1 up-/down- regulated genes, the software counts the occurrence of each term of the description and compares it with the occurrence of the same term in 1000 random lists of the same size. Results are presented as a cloud of words, whose color and size are correlated to their occurrence.
Phylogenetic analysis
The phylogeny reconstruction was inferred by using the Maximum Likelihood method. The 40 sequences coding for G2-like proteins were retrieved from the AGRIS (Arabidopsis Gene Regulatory Information Server; http://arabidopsis.med.ohio-state.edu/) database. The bootstrap values were obtained based on 500 replicates. Evolutionary analysis was conducted in MEGA5 software 60.
Expression and purification of recombinant GST-HRS1 protein
HRS1 CDS was first cloned in pDONR207™, using primers listed in Supplementary Data 1 and then transferred to pDEST15 vector (Invitrogen) by LR reaction following the manufacturer's instructions. The GST-HRS1 fusion protein was expressed in E.coli Rosetta™ 2(DE3)pLysS (Novagen, Darmstadt, Germany) cells after induction with 1mM IPTG for 16h at 22°C. Bacteria were harvested, suspended in PBS buffer containing lysozyme from chicken egg white (SIGMA) and complete protease inhibitor cocktail (Roche) and sonicated. The protein extract was purified on glutathione-sepharose beads (GE Healthcare, Freiburg, Germany), eluted with 10mM reduced-glutathione (SIGMA), 50mM Tris buffer and dialyzed overnight in 150mM NaCl, 50mM HEPES, pH 7.4 buffer.
EMSA
Purified GST-HRS1 recombinant protein was used to determine DNA binding by EMSA. ssDNA oligonucleotides (listed in Supplementary Data 1) were biotin labeled using the Biotin 3’ End DNA Labeling Kit (Thermo Scientific) and complementary pairs were annealed to make dsDNA probes. The binding of recombinant protein (50 ng) to the biotin labeled probes (20 fmol) was carried out in a reaction mixture containing 10mM Tris, 50mM KCl, 1mM DTT, pH7.5, 2.5% glycerol, 5mM MgCl2, 1μg poly (dI-dC) and 0.05% NP-40. After incubation at 22°C for 30min the protein-probe mixture was separated in a 4% polyacrylamide native gel and transferred to a Biodyne B Nylon membrane by capillary action in 20xSSC buffer overnight (Thermo Scientific). After UV-crosslinking (254nm) for 90sec at 120mJ .cm−2, the migration of biotin-labeled probes was detected using streptavidin-horseradish peroxidase conjugates in the Chemioluminescent Nucleic Acid Detection Module (Thermo Scientific) and exposed to X-ray film. As a negative control, we tested that the GST tag has no affinity for HRS1 related CREs (Supplementary Fig. 5).
Protein extraction and Immunoblot analysis
Roots were sampled and frozen in liquid nitrogen. Total proteins were obtained using the Plant Total Protein Extraction Kit (Sigma) and quantified with the PerceTM660nm Protein Assay using a BSA standard curve. Membrane proteins were obtained by disrupting roots in a buffer containing a plant anti-protease cocktail (Sigma-Aldrich) and anti-phosphatases (30 mM glycerophosphate, 5 mM molybdate, and 10 mM NaF). The whole membrane fraction was then isolated by centrifugation (100000g, 4h) on a 55% sucrose cushion. Immunoblot analysis was performed on 40-50 μg of proteins using anti-GFPHRP 1:2500 (Miltenyi Biotec, 130-091-833), anti-NRT1.1 1:5000 (AS12 2611, Agrisera) and anti-PIP2.1 1:5000 61. Coomassie Brilliant Blue staining of blots was used to control protein levels after electro-transfer. Un-cropped versions of the blots are provided in Supplementary Fig. 7 and 8. Band intensity quantification was performed using a chemioluminiescent image analyzer LAS3000 (Fujifilm) and ImageGauge (Fujifilm) software.
Transcriptome analysis
The transcriptome analysis was performed using the GeneChip® Whole Transcript (WT) Expression Array following the manufacturers protocol. Total RNA was isolated from 6-day old roots grown on media containing 2.5mM KNO3 and no PO43− (three independent experiments). RIN were checked by microfluidic analysis in an Agilent 2100 Bioanalyzer. cDNA were prepared from 150ng of total RNA following the WT Plus Reagent Kit protocol, hybridized on Affymetrix® Array Strip and processed on the GeneAtlas® System. Dataset analysis was performed with R. A one-way ANOVA model was applied (4 levels: Col, hrs1-1;hho1-1, HRS1-OE, HHO1-OE) followed by a post hoc Tukey test [R functions aov() and TukeyHSD()]. Any probe having a significant pval<0.05 for the ANOVA or the Tukey test was kept for further analysis. Clustering analysis was performed with MeV software (distance, Pearson correlation).
Supplementary Material
Acknowledgements
We thank, Benoît Lacombe, Frederic Gaymard for comments and discussions; Elodie Jublanc from the Montpellier Rio Imaging (MRI) core facility, for confocal assistance and 3D imaging reconstruction; Tony Sierra for technical assistance during Gene Atlas analysis; Veronique Santoni for the PIP2.1 antibody; Franck Lecocq, Rogatien Picaud, Guy Ruiz, Thierry Dessup, and Hugues Baudot for technical support and plant care. This work was supported by the French Agence Nationale de la Recherche [NitroNet: ANR 11 PDOC 020 01] and Centre National de la Recherche Scientifique [PEPS Bio math Info 2012–2013: SuperRegNet] to G.K. Work on the transcriptional networks was supported NIH R01-GM032877 to G.C. and NIH NRSA-GM095273 to AMC. Results on root architecture responses are supported by NSF MCB-0929338 to GC. Bioinformatics analysis was supported by the VirtualPlant platform (www.virtualplant.org) developed under NSF DBI-0445666 to GC. Work by R.W. and N.C. was supported by the National Science Foundation grants (IOS-1021380 and MCB-0929338).
Footnotes
Author contributions
G.K. designed the project. A.M., A.M-C., E.R., W.S., R.W., S.R. and G.K. performed experiments and analyzed the data. A.M, A.G., N.M.C, S.R., G.M.C and G.K. contributed to the study design during the project course. A.M., and G.K. wrote the paper.
Competing financial interests statement
The authors declare no competing financial interests.
Accession codes
Microarray data associated with this study has been deposited in NCBI GEO database under the accession code GSE64352.
References
- 1.Gruber BD, Giehl RF, Friedel S, von Wiren N. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 2013;163:161–179. doi: 10.1104/pp.113.218453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peret B, Clement M, Nussaume L, Desnos T. Root developmental adaptation to phosphate starvation: better safe than sorry. Trends Plant Sci. 2011;16:442–450. doi: 10.1016/j.tplants.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez JM, Vidal EA, Gutierrez RA. Integration of local and systemic signaling pathways for plant N responses. Curr. Opin. Plant Biol. 2012;15:185–191. doi: 10.1016/j.pbi.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 4.Vidal EA, Gutierrez RA. A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr. Opin. Plant Biol. 2008;11:521–529. doi: 10.1016/j.pbi.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Gojon A, Nacry P, Davidian JC. Root uptake regulation: a central process for NPS homeostasis in plants. Curr. Opin. Plant Biol. 2009;12:328–338. doi: 10.1016/j.pbi.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 6.Krouk G, Crawford NM, Coruzzi GM, Tsay YF. Nitrate signaling: adaptation to fluctuating environments. Curr. Opin. Plant Biol. 2010;13:266–273. doi: 10.1016/j.pbi.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Camacho-Cristobal JJ, Rexach J, Conejero G, Al-Ghazi Y, Nacry P, Doumas P. PRD, an Arabidopsis AINTEGUMENTA-like gene, is involved in root architectural changes in response to phosphate starvation. Planta. 2008;228:511–522. doi: 10.1007/s00425-008-0754-9. [DOI] [PubMed] [Google Scholar]
- 8.Svistoonoff S, et al. Root tip contact with low-phosphate media reprograms plant root architecture. Nature genet. 2007;39:792–796. doi: 10.1038/ng2041. [DOI] [PubMed] [Google Scholar]
- 9.Ticconi CA, et al. ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristems to phosphate availability. Proc. Natl. Acad. Sci. USA. 2009;106:14174–14179. doi: 10.1073/pnas.0901778106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ticconi CA, Delatorre CA, Lahner B, Salt DE, Abel S. Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. Plant J. 2004;37:801–814. doi: 10.1111/j.1365-313x.2004.02005.x. [DOI] [PubMed] [Google Scholar]
- 11.Rubio V, et al. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001;15:2122–2133. doi: 10.1101/gad.204401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bustos R, et al. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010;6:e1001102. doi: 10.1371/journal.pgen.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miura K, et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. U S A. 2005;102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vidal EA, Tamayo KP, Gutierrez RA. Gene networks for nitrogen sensing, signaling, and response in Arabidopsis thaliana. Wiley interdiscip Rev Syst Biol Med. 2010;2:683–693. doi: 10.1002/wsbm.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walch-Liu P, Forde BG. Nitrate signalling mediated by the NRT1.1 nitrate transporter antagonises L-glutamate-induced changes in root architecture. Plant J. 2008;54:820–828. doi: 10.1111/j.1365-313X.2008.03443.x. [DOI] [PubMed] [Google Scholar]
- 16.Kant S, Peng M, Rothstein SJ. Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in arabidopsis. PLoS Genet. 2011;7:e1002021. doi: 10.1371/journal.pgen.1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin WY, Huang TK, Chiou TJ. NITROGEN LIMITATION ADAPTATION, a Target of MicroRNA827, Mediates Degradation of Plasma Membrane-Localized Phosphate Transporters to Maintain Phosphate Homeostasis in Arabidopsis. Plant Cell. 2013;26:454–464. doi: 10.1105/tpc.113.116012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng M, et al. Adaptation of Arabidopsis to nitrogen limitation involves induction of anthocyanin synthesis which is controlled by the NLA gene. Journal Exp. Bot. 2008;59:2933–2944. doi: 10.1093/jxb/ern148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khamis S, Lamaze T, Lemoine Y, Foyer C. Adaptation of the Photosynthetic Apparatus in Maize Leaves as a Result of Nitrogen Limitation : Relationships between Electron Transport and Carbon Assimilation. Plant Physiol. 1990;94:1436–1443. doi: 10.1104/pp.94.3.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krouk G, et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell. 2010;18:927–937. doi: 10.1016/j.devcel.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 21.Ho CH, Lin SH, Hu HC, Tsay YF. CHL1 functions as a nitrate sensor in plants. Cell. 2009;138:1184–1194. doi: 10.1016/j.cell.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Krouk G, Mirowski P, LeCun Y, Shasha DE, Coruzzi GM. Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol. 2010;11:R123. doi: 10.1186/gb-2010-11-12-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheible WR, et al. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 2004;136:2483–2499. doi: 10.1104/pp.104.047019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang R, Gutierrez RA, Hoffman M, Xing X, Chen M, Coruzzi G, Crawford NM. Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 2004;136:2512–2522. doi: 10.1104/pp.104.044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu HC, Wang YY, Tsay YF. AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 2009;57:264–278. doi: 10.1111/j.1365-313X.2008.03685.x. [DOI] [PubMed] [Google Scholar]
- 26.Sawaki N, et al. A nitrate-inducible GARP family gene encodes an auto-repressible transcriptional repressor in rice. Plant Cell Physiol. 2013;54:506–517. doi: 10.1093/pcp/pct007. [DOI] [PubMed] [Google Scholar]
- 27.Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA. GLK Transcription Factors Coordinate Expression of the Photosynthetic Apparatus in Arabidopsis. Plant Cell. 2009;21:1109–1128. doi: 10.1105/tpc.108.065250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS. KANADI regulates organ polarity in Arabidopsis. Nature. 2001;411:706–709. doi: 10.1038/35079629. [DOI] [PubMed] [Google Scholar]
- 29.Bustos R, et al. A Central Regulatory System Largely Controls Transcriptional Activation and Repression Responses to Phosphate Starvation in Arabidopsis. PLoS Genet. 2010;6:e1001102. doi: 10.1371/journal.pgen.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, et al. Overexpressing HRS1 confers hypersensitivity to low phosphate-elicited inhibition of primary root growth in Arabidopsis thaliana. J. Integr. Plant Biol. 2009;51:382–392. doi: 10.1111/j.1744-7909.2009.00819.x. [DOI] [PubMed] [Google Scholar]
- 31.Bolle C, et al. GABI-DUPLO: a collection of double mutants to overcome genetic redundancy in Arabidopsis thaliana. Plant J. 2013;75:157–171. doi: 10.1111/tpj.12197. [DOI] [PubMed] [Google Scholar]
- 32.Wang R, et al. Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 2004;136:2512–2522. doi: 10.1104/pp.104.044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang R, Xing X, Wang Y, Tran A, Crawford NM. A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol. 2009;151:472–478. doi: 10.1104/pp.109.140434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konishi M, Yanagisawa S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nature Commun. 2013;4:1617. doi: 10.1038/ncomms2621. [DOI] [PubMed] [Google Scholar]
- 35.Marchive C, et al. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nature Commun. 2013;4:1713. doi: 10.1038/ncomms2650. [DOI] [PubMed] [Google Scholar]
- 36.Canales J, Moyano TC, Villarroel E, Gutiérrez RA. Systems analysis of transcriptome data provides new hypotheses about Arabidopsis root response to nitrate treatments. Frontiers Plant. Sci. 2014;5 doi: 10.3389/fpls.2014.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bargmann BO, et al. TARGET: a transient transformation system for genome-wide transcription factor target discovery. Mol. Plant. 2013;6:978–980. doi: 10.1093/mp/sst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Para A, et al. Hit-and-run transcriptional control by bZIP1 mediates rapid nutrient signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2014;111:10371–10376. doi: 10.1073/pnas.1404657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katari MS, et al. VirtualPlant: a software platform to support systems biology research. Plant Physiol. 2010;152:500–515. doi: 10.1104/pp.109.147025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38:W64–70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bailey TL, et al. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giraud E, et al. TCP Transcription Factors Link the Regulation of Genes Encoding Mitochondrial Proteins with the Circadian Clock in Arabidopsis thaliana. Plant Cell. 2010;22:3921–3934. doi: 10.1105/tpc.110.074518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franco-Zorrilla JM, Lopez-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA. 2014;111:2367–2372. doi: 10.1073/pnas.1316278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Obertello M, Krouk G, Katari MS, Runko SJ, Coruzzi GM. Modeling the global effect of the basic-leucine zipper transcription factor 1 (bZIP1) on nitrogen and light regulation in Arabidopsis. BMC Sys. Biol. 2010;4:111. doi: 10.1186/1752-0509-4-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krouk G, et al. A systems approach uncovers restrictions for signal interactions regulating genome-wide responses to nutritional cues in Arabidopsis. PLoS Comput. Biol. 2009;5:e1000326. doi: 10.1371/journal.pcbi.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Canales J, Moyano TC, Villarroel E, Gutierrez RA. Systems analysis of transcriptome data provides new hypotheses about Arabidopsis root response to nitrate treatments. Frontiers Plant Sci. 2014;5:22. doi: 10.3389/fpls.2014.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alvarez JM, et al. Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. 2014;80:1–13. doi: 10.1111/tpj.12618. [DOI] [PubMed] [Google Scholar]
- 49.Nero D, Krouk G, Tranchina D, Coruzzi GM. A system biology approach highlights a hormonal enhancer effect on regulation of genes in a nitrate responsive “biomodule”. BMC Sys. Biol. 2009;3:59. doi: 10.1186/1752-0509-3-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsukagoshi H, Busch W, Benfey PN. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell. 2010;143:606–616. doi: 10.1016/j.cell.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 51.Lu D, Wang T, Persson S, Mueller-Roeber B, Schippers JH. Transcriptional control of ROS homeostasis by KUODA1 regulates cell expansion during leaf development. Nature Commun. 2014;5:3767. doi: 10.1038/ncomms4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bargmann BO, et al. A map of cell type-specific auxin responses. Mol. Sys. Biol. 2013;9:688. doi: 10.1038/msb.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu C, et al. HRS1 acts as a negative regulator of abscisic acid signaling to promote timely germination of Arabidopsis seeds. PLoS One. 2012;7:e35764. doi: 10.1371/journal.pone.0035764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mito T, Seki M, Shinozaki K, Ohme-Takagi M, Matsui K. Generation of chimeric repressors that confer salt tolerance in Arabidopsis and rice. Plant Biotechnol. J. 2011;9:736–746. doi: 10.1111/j.1467-7652.2010.00578.x. [DOI] [PubMed] [Google Scholar]
- 55.Kellermeier F, Armengaud P, Seditas TJ, Danku J, Salt DE, Amtmann A. Analysis of the Root System Architecture of Arabidopsis Provides a Quantitative Readout of Crosstalk between Nutritional Signals. Plant Cell. 2014;26:1480–1496. doi: 10.1105/tpc.113.122101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Delgado-Baquerizo M, et al. Decoupling of soil nutrient cycles as a function of aridity in global drylands. Nature. 2013;502:672–676. doi: 10.1038/nature12670. [DOI] [PubMed] [Google Scholar]
- 57.Curtis MD, Grossniklaus U. A Gateway Cloning Vector Set for High-Throughput Functional Analysis of Genes in Planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clough SJ, Bent AF. Floral dip: a simplified method forAgrobacterium-mediated transformation ofArabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 59.Krouk G, Mirowski P, LeCun Y, Shasha D, Coruzzi G. Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol. 2010;11:R123. doi: 10.1186/gb-2010-11-12-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santoni V, Vinh J, Pflieger D, Sommerer N, Maurel C. A proteomic study reveals novel insights into the diversity of aquaporin forms expressed in the plasma membrane of plant roots. Biochem J. 2003;373:289–296. doi: 10.1042/BJ20030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.