Abstract
Single-cell genomics and single-cell transcriptomics have recently emerged as powerful tools to study the biology of single cells at a genome-wide scale. However, a major challenge is to quantify both genomic DNA and mRNA from the same cell, which would allow direct comparison of genomic variation and transcriptome heterogeneity. Here we describe a method that allows the sequencing of genomic DNA and mRNA from the same cell without physical separation of the nucleic acids prior to amplification. We show that such an integrated strategy achieves efficiency similar to methods that sequence either genomic DNA or mRNA from single cells. We use this method to correlate DNA copy number variation to transcriptome variability among individual cells. Finally, we show that genes that display more cell-to-cell variability in transcript numbers are generally associated with reduced copy number loci and vice-versa, implying that copy number variations could potentially drive variability in gene expression between single cells.
One of the central questions in biology is to understand how genotype influences phenotype. Over the past decade, advances in microarrays and, more recently, next-generation sequencing have started to provide the first glimpses of this correlation at the genome-wide level1-4. However, these studies make measurements starting from a large population of cells or complex tissues, thus providing only an average measurement over the entire population. This obscures direct quantification of how genetic variability may impact the transcriptome at the single cell level. Furthermore, since cell populations exposed to the same environment can also exhibit dramatic cell-to-cell variability in gene expression5, our ability to understand the correlation between single-cell genotype and gene expression will require direct measurements of the transcriptome and the genome of the same cell. Recently, single-cell genome sequencing6-11 and single-cell transcriptome sequencing12-21 have emerged as promising tools for quantifying genetic and expression variability between individual cells22,23. However, as these single-cell technologies are limited to quantifying either the transcriptome or the genome, it is currently impossible to explore the relation between genetic and expression variability in single cells. Here, we developed a method to simultaneously quantify both the genome and transcriptome of the same cell.
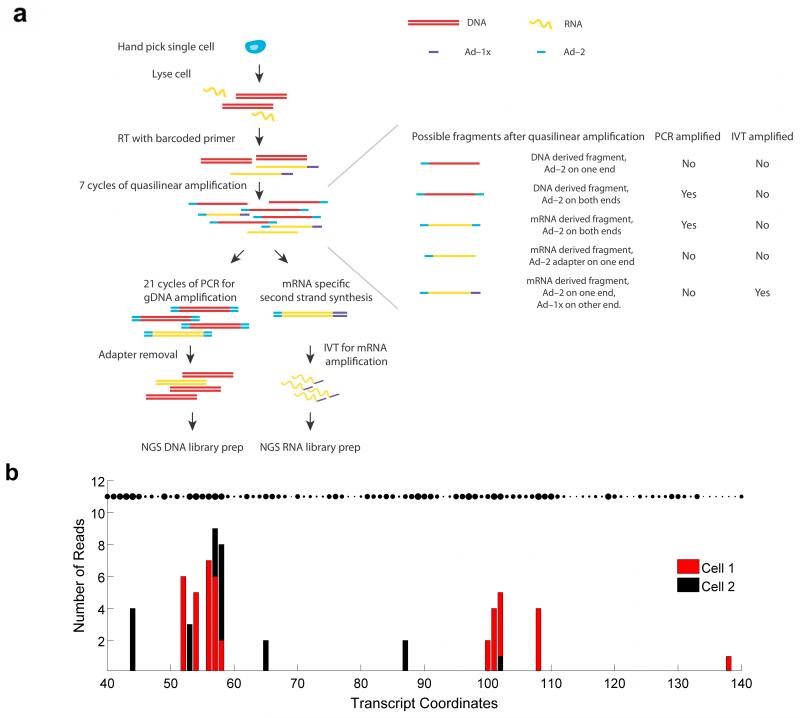
To successfully amplify small quantities of genomic DNA (gDNA) and messenger RNA (mRNA) from single cells in a way that reduces handling, transfer and separation steps, we devised a method (gDNA-mRNA Sequencing, or DR-Seq) that does not involve physical separation of the nucleic acids prior to amplification, thereby minimizing losses and chances of contamination. First, hand-picked single cells are lysed and reverse transcribed using a poly-A primer (called Adapter-1x or Ad-1x) including cell-specific barcodes, a 5′ Illumina adapter and a T7 promoter overhang to convert mRNA to single stranded cDNA (ss cDNA)13 (Fig. 1a). The gDNA and single stranded cDNA are then subjected to quasilinear whole genome amplification, as previously described, using an adapter with a defined 27 nucleotide sequence at the 5′ end followed by 8 random nucleotides (Ad-2)7 (Fig. 1a). After 7 rounds of amplification, the gDNA and cDNA are copied to generate a variety of different short amplicon (0.5–2.5 kb) species, with a majority of amplicons containing adapter Ad-2 at both ends and a small fraction of cDNA derived amplicons containing Ad-2 at one end and Ad-1x at the other (Fig. 1a).
Figure 1.
Schematic of DR-Seq for sequencing gDNA and mRNA from the same cell. (a) gDNA and mRNA/cDNA are shown in red and green, respectively. Following single-cell lysis and RT using adapter Ad-1x (purple), gDNA and single stranded cDNA are amplified by Ad-2 (blue) using a quasilinear amplification strategy. The majority of the short amplicons contain Ad-2 at both ends and cDNA-derived amplicons contain Ad-2 at one end and Ad-1x at the other end. The sample is then split into two halves and processed separately to amplify and sequence gDNA or cDNA. (b) Distribution of reads within 100 nucleotides of the gene Dppa5a for two single cells (red and black) as a function of the random priming location by adapter Ad-2. The unique length-based identifiers found in the two cells can be used to count the original number of cDNA molecules within each cell and minimize amplification biases. The figure shows that distinct positions are randomly primed within each cell with high affinity binding sites being preferentially primed. The size of the dots indicate the binding propensity of each location. For most genes such as Dppa5a, the number of theoretical binding sites far exceed the number of length-based identifiers detected, thereby enabling length-based identifiers to accurately estimate the original number of cDNA molecules.
Next, the sample is split into two tubes to further amplify gDNA and cDNA (Fig. 1a). The tube used to sequence gDNA is amplified using PCR. Following sonication, adapter Ad-2 removal, and cell-specific indexed Illumina library preparation, this half is used to sequence gDNA. The tube used to sequence cDNA is converted to double-stranded cDNA and amplified using in vitro transcription such that the amplified RNA (aRNA) is uniquely produced from cDNA but not gDNA (Fig. 1a). 3′ Illumina adapters are then ligated to the aRNA followed by reverse transcription and PCR, allowing quantification of mRNA.
We first applied DR-Seq to a mouse embryonic stem cell line (E14) to validate how the method compares to existing single-cell gDNA or mRNA sequencing techniques. We performed DR-Seq on E14 cells and sequenced the mRNA from 13 single cells together with the gDNA from 3 of these 13 cells. Recently, a few single-cell transcriptomics methods have employed random sequence-based barcodes to identify unique mRNA molecules, thereby significantly reducing PCR and other amplification biases17,20,21. Because cDNA molecules in DR-Seq are randomly primed by Ad-2 during quasilinear amplification, we used the genomic position of such priming events to minimize amplification biases and achieve resolution close to identifying unique mRNA molecules (Supplementary Fig. 1). Because all amplification products that are generated downstream (during quasilinear amplification, in vitro transcription and PCR) from the first randomly primed cDNA derived amplicon retain the same genomic priming location, amplification derived duplicates could be removed to identify unique cDNA molecules. The genomic priming location of the first randomly primed cDNA derived amplicon was called its length-based identifier (Supplementary Fig. 1). For example, although hundreds of reads were detected for the Dppa5a gene, only 34 and 27 unique length-based identifiers were detected in the two single cells shown. A zoomed-in view of 100 nucleotides within this transcript shows that only a few distinct positions are randomly primed in the two cells, with several reads at each genomic coordinate (Fig. 1b). Thus, unique length-based identifiers could potentially reduce amplification biases and technical noise to enable quantification of the original number of cDNA molecules. To demonstrate that length-based identifiers could be used to achieve resolution close to identifying unique transcripts in single cells, we showed that the original cDNA molecules were primed only once on average during the quasilinear amplification steps, thereby enabling length-based identifiers to uniquely tag each original cDNA molecule (Supplementary Fig. 2 and Supplementary Note). Next, we identified the theoretical number of unique binding sites (and therefore length-based identifiers) available for adapter Ad-2 for each gene in the transcriptome to ensure that the original cDNA molecules from each gene could be counted accurately without reaching saturation (Supplementary Fig. 3 and Supplementary Note). For a majority of the genes, we found between 50 to 250 theoretical binding sites, similar to the resolution of 4-bp random barcodes recently used as unique molecule identifiers (UMI) to quantify single-cell transcriptomes20,21 (Supplementary Fig. 4 and Supplementary Note). Finally, we showed that for a majority of expressed genes (>95%), the number of detected length-based identifiers were much smaller than the theoretical number of binding sites, thereby implying that the length-based identifiers do not undercount the number of original cDNA molecules (Supplementary Fig. 5 and Supplementary Note). Together these results show that length-based identifiers in DR-Seq can be used to minimize amplification biases and accurately estimate the underlying distribution of original cDNA molecules.
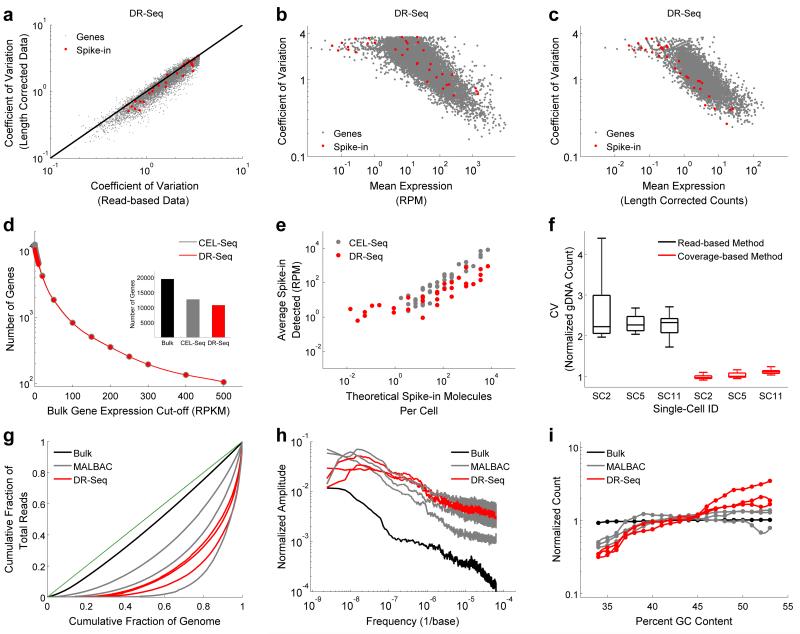
To demonstrate that length-based identifiers reduce technical noise by minimizing amplification biases and perform similar to the recently described random sequence-based UMIs17,20,21, we compared cell-to-cell variability in the expression of endogenous genes before and after correction using length-based identifiers. We found that the coefficient of variation (CV) in expression reduced for a majority of genes (80%) in DR-Seq after correcting the expression using length-based identifiers, similar to the reduction observed in CEL-Seq after correcting the expression using UMIs (Fig. 2a and Supplementary Fig. 6). This suggests that length-based identifiers in DR-Seq reduce technical noise, thereby allowing us to quantify the underlying biological variability in gene expression between single cells. Further, as single cells all contain the same amount of ERCC (External RNA Controls Consortium) spike-in molecules, any cell-to-cell variability detected in these molecules represent technical noise entirely and would therefore be expected to display the lowest CV when compared to endogenous genes with similar mean expression levels. We found that the spike-in molecules typically showed the lowest CV for the entire range of mean expressions only after correcting the read-based DR-Seq data with the length-based identifiers (Fig. 2b,c). We found a similar trend in CEL-Seq after correcting the read-based data with UMIs (Supplementary Fig. 7). As a consequence of this reduction in technical noise, we found that length-based identifiers in DR-Seq and UMIs in CEL-Seq improved cell-to-cell pairwise Pearson correlations in the expression of endogenous genes (Supplementary Fig. 8). Taken together, this data strongly suggests that length-based identifiers in DR-Seq substantially reduce technical noise, thereby allowing us to accurately count the number of original cDNA molecules and capture the underlying biological variability between single cells.
Figure 2.
Development of a computational techniques to reduce technical noise in single-cell DR-Seq sequencing data and comparison of DR-Seq to existing single-cell gDNA or mRNA sequencing methods in the mouse embryonic stem cell line E14. (a) Comparison of the coefficient of variation showed that cell-to-cell variability in the expression of genes reduced after correcting the raw read-based data using length-based identifiers, implying reduction in technical noise in the single-cell transcriptome data of DR-Seq (also see Supplementary Fig. 6). (b) Coefficient of variation versus mean expression of genes for the read-based data. Because each cell contains the same number of spike-in molecules, they are expected to display the lowest noise for a given mean level of expression. The data shows that read-based data contains significant amount of technical noise that obscures biological variability between single cells. (c) After correcting the DR-Seq data using length-based identifiers, spike-in molecules typically display the least noise over the entire range of mean expressions (also see Supplementary Fig. 7). Endogenous genes and spike-in molecules are indicated using gray and red dots, respectively. (d) Comparison of mRNA sequencing results between DR-Seq and CEL-Seq showed that both methods show similar performance in detecting genes above different expression thresholds obtained from bulk mRNA sequencing data. (Inset) Overall, both methods detect similar number of genes (also see Supplementary Fig. 10). (e) Detection of ERCC spike-in molecules in both methods increased monotonically with the expected number of molecules per cell. The figure shows spike-ins that were found in at least 2 single cells. (f) Box plot comparing bin-to-bin variability in gDNA read counts using two different methods for 3 single cells amplified by DR-Seq. The coverage-based method displays approximately two-fold reduction in technical noise compared to the read-based method. The box plots show the coefficient of variation of read distribution over all the autosomes in the mouse genome. (g) Lorenz plots were used to compare single cell gDNA sequencing results between DR-Seq and MALBAC. Lorenz curves were used to assess the uniformity of genome coverage by plotting the cumulative increase in read depth verses the cumulative fraction of genome covered, ordered by increasing coverage. The green line indicates the theoretical limit with reads distributed uniformly across the whole genome. Based on the Lorenz plots, bulk sequencing achieves read distribution close to the theoretical limit. The 6 single cells processed with either DR-Seq or MALBAC display similar distribution of reads across the genome. (h) Power spectrum of read distribution over different genomic length scales are shown for bulk sequencing and single cells processed by DR-Seq and MALBAC. The power spectrum reveals biases in read depth distribution over different ranges of genomic length scales. Bulk sequencing shows the least bias in read distribution with both DR-Seq and MALBAC performing similarly. (i) Read distribution for regions of the genome with different GC content shows that both methods deviate from the expected normalized count of 1 for regions with high and low GC content. This GC bias is corrected prior to estimating copy numbers in single cells.
These length corrected mRNA sequencing results obtained from DR-Seq were then compared to results obtained from sequencing 33 single E14 cells using CEL-Seq (Supplementary Fig. 9) 13,21. To avoid sampling bias in comparing DR-Seq to CEL-Seq, we chose the 13 cells with the highest read counts out of 33 that were sequenced using CEL-Seq. Despite these stringent criteria, DR-Seq detected approximately similar number of genes as CEL-Seq, with 9,735 genes common between the two methods (Fig 2d (Inset) and Supplementary Fig. 10). Similarly, the number of genes detected above different expression thresholds was very similar between the two methods (Fig. 2d). In addition, gene expression correlations for either method compared to bulk sequencing were similar (Supplementary Table 1). Finally, analysis of synthetic ERCC spike-in RNA molecules showed that 66 different spike-in species from a total of 92 were detected using DR-Seq compared to 51 species detected using CEL-Seq24 (Fig. 2e and Supplementary Fig. 11). This increased sensitivity in detecting low abundance spike-ins is most likely due to the exponential amplification of cDNA-derived amplicons for the remainder of the quasilinear amplification steps in DR-Seq. For the higher range of concentrations, the number of spike-in molecules detected correlated well with the expected number of molecules (Fig. 2e). Further, because we detected linear correlation over 3 orders of magnitude between the detected and theoretical number of spike-in molecules for both CEL-Seq and DR-Seq, this suggested that both methods display similar dynamic range in detecting transcripts (Fig. 2e). Similarly, expression of endogenous genes also spanned over 3 orders of magnitude in both CEL-Seq and DR-Seq, implying that both methods have similar sensitivities in amplifying reverse transcribed cDNA molecules (Fig. 2d). Taken together, analysis of the two methods across these different metrics suggests that DR-Seq performs similar to CEL-Seq and the additional steps involved in amplifying gDNA do not adversely affect the mRNA sequencing results (Supplementary Fig. 12 and Supplementary Note).
To analyze gDNA sequencing results in DR-Seq, reads from the gDNA fraction were mapped to the genome after masking out the coding sequences. This is because the fraction that is used to sequence gDNA also contains reads that originate from cDNA molecules within coding sequences (Fig. 1a). By masking the coding sequences within the genome, such ambiguous reads that might arise from either gDNA or cDNA within coding regions are discarded computationally leaving only reads that arise from gDNA (Supplementary Fig. 13). Because the coding regions make up a very small portion of the genome, such a strategy does not influence copy number calling over large genomic regions (See Online Methods for details). Further, gDNA reads in DR-Seq were distributed into unequal bins to account for the masking of the genome (Supplementary Fig. 14 and Online Methods). Further, to reduce amplification biases introduced during the quasilinear amplification steps, we developed a computational technique to reduce bin-to-bin technical noise in gDNA read counts. During quasilinear amplification, the first amplicons that are generated from the gDNA template do not loop out of the reaction pool and remain templates for the remaining cycles. Thus, differences in the cycle in which gDNA regions are first amplified can introduce bin-to-bin variability and pileup of reads for certain regions of the genome. To correct for this amplification bias, we developed a coverage-based model that allowed us to more accurately count the original amplicons that are generated from the gDNA template, rather than the amplicons that are repeatedly amplified from the quasilinear amplification generated products. Since the coverage-based method is not influenced by amplification duplicates, it reduced technical noise in estimating gDNA counts over the entire genome (Supplementary Note). For the E14 cell line, we found that bin-to-bin technical variability for all the autosomes reduced 2-fold in our coverage-based method compared to the conventional read-based method (Fig. 2f). Further, additional analyses quantifying bin-to-bin technical variability and correlations between single cells revealed that the coverage-based method reduced amplification biases and technical noise, thereby improving copy number calling in cancer genomes (Supplementary Figs. 15,16,17,18, Supplementary Table 2 and Supplementary Note).
After making these improvements to reduce technical noise, gDNA sequencing results from DR-Seq were compared to results obtained from sequencing 3 single E14 cells using MALBAC7, with all single cells sequenced at 0.6-2.5x sequencing depth. To identify sequencing biases and differences in coverage, we used Lorenz plots to compare cumulative read depth versus cumulative fraction of the genome covered, ordered by increasing coverage. The diagonal represents the theoretical limit with reads uniformly distributed over the entire genome (Fig. 2g). Bulk gDNA sequencing, without the need for any whole genome amplification, achieves a read distribution close to the theoretical limit. Both DR-Seq and MALBAC, relying on quasilinear amplification using random primers to amplify the genome, display greater coverage biases than bulk sequencing but perform similar to each other (Fig. 2g). Furthermore, to assess systematic biases and drifts in read distribution along the length of the genome, power spectra showed that both DR-Seq and MALBAC showed more bias over large genomic scales (i.e., low frequencies), with both methods performing similarly across the entire range of genomic scales (Fig. 2h). Finally, analysis of GC sequencing bias showed that regions of the genome with high and low GC content deviated from the expected normalized counts25 (Fig. 2i). Both DR-Seq and MALBAC showed similar trends in GC bias, with DR-Seq displaying modestly higher bias, possibly due to the extra round of quasilinear amplification performed in DR-Seq compared to MALBAC. However, as the GC bias is easily corrected for, this bias does not influence the final gDNA analysis (Supplementary Fig. 18). Taken together, these metrics suggest that combined gDNA and mRNA sequencing from the same cell using DR-Seq performs similarly to existing methods for sequencing either the genome or transcriptome of single cells (Fig. 2).
We next applied DR-Seq to a breast cancer cell line SK-BR-3 to understand how copy number variations in single cancer cells influence gene expression programs. We applied DR-Seq to 21 single SK-BR-3 cells and sequenced mRNA from all these cells and the gDNA from 7 out of these 21 single cells. We detected 12,205 genes and as with the E14 dataset, average expression of genes from these single cells showed similar correlation to bulk mRNA sequencing (Pearson r = 0.66, Spearman r = 0.69) (Supplementary Fig. 19a,b and Supplementary Table 1). Similarly, detection of spike-ins correlated well with the expected numbers of molecules (Supplementary Fig. 19c,d). gDNA from the 7 single cells was sequenced at 0.6-1.6x depth (Supplementary Table 3). Sequencing coverage and GC bias were similar to that observed in single E14 cells (Supplementary Fig. 20a-c).
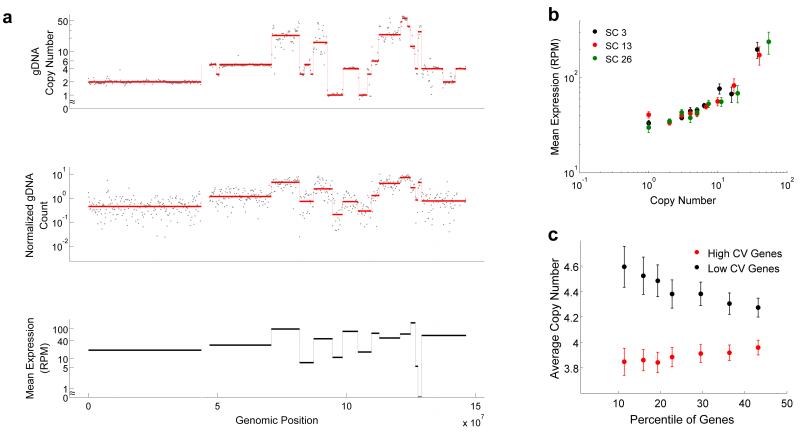
After correcting for GC bias, the circular binary segmentation (CBS) algorithm was used to detect breakpoints26. Figures 3a shows that raw data and breakpoint detection for Chr 8 from one cell correlated well with copy number changes detected in bulk sequencing. Similarly, breakpoint detection over the entire genome for all the single cells correlated well with the bulk sequencing results (Supplementary Fig. 21). The median read counts for each of the segments were used to estimate copy numbers in single cells (Supplementary Figs. 18a,21, Supplementary Table 2 and Supplementary Note). We also developed a model to estimate confidence intervals for the copy numbers that are called by our algorithm (Supplementary Fig. 22 and Supplementary Note). Further, the mean copy numbers over all the single cells correlated well with the bulk sequencing copy numbers over the entire genome (Supplementary Figs. 17 and 18b,c). Finally, we also detected significant cell-to-cell variability in copy numbers over certain regions of the genome (Supplementary Figs. 17 and 23). We performed DNA Fluorescence In Situ Hybridization (FISH) over 4 genomic loci that span a large spectrum of copy numbers and found that the mean copy numbers detected by DR-Seq and DNA FISH were in good agreement (Supplementary Fig. 24)27. Notably, we also found that the distribution of copy numbers for these 4 loci in single cells amplified by DR-Seq were not statistically different from distributions obtained by DNA FISH (p > 0.01 and Supplementary Table 4). These results showed that DR-Seq has the sensitivity to capture heterogeneity in copy numbers across single cells.
Figure 3.
Applying DR-Seq to the SK-BR-3 cell line to understand how copy number variations affect gene expression in single cells. (a) Top panel shows raw gDNA data (dots) and different copy numbers (red line) identified using the CBS algorithm26 for Chr 8 in bulk sequencing data. The middle panel shows raw data (dots) and median read counts (red line) identified using CBS for one single cell (SC13). Visual comparison of the top and middle panels show that most breakpoints are reliably detected in single cells and patterns of level changes between bulk and single cell gDNA sequencing are well correlated. The median read depths for each segment in single cells and the bulk copy numbers are used to estimate copy number variations in single cells (Supplementary Note). For each median level identified from the single cell gDNA data (middle panel), mean expression of genes within each level was calculated (black lines in lower panel). The lower panel shows that the mean expression of genes within each segment correlates well with the median gDNA levels. (b) Genome-wide quantification of mean expression of genes within different copy number regions shows a monotonic increase in average expression with increase in copy number for 3 single cells (also see Supplementary Fig. 25). (c) For a large range of mean expressions (5-400 RPM), genes exhibiting the highest and lowest noise (quantified as coefficient of variation, or CV) were identified. The x-axis shows the percentage of most noisy and least noisy genes that were considered in the analysis. The data shows that the noisiest genes are associated with low copy number regions and vice versa (also see Supplementary Fig. 27). Error bars represent standard error in estimating the mean obtained by bootstrapping the data.
Next, comparison of copy number variations in Chr 8 to levels of mRNA expression in this single cell showed that the average expression of genes within each segment appeared to be strongly correlated to the copy number of that genomic region (Fig. 3a). To quantify this correlation on a genome-wide scale, we calculated the mean expression of genes within different copy number regions for each of the single cells. We observed a monotonic increase in mean expression with increase in copy number on a genome-wide level across different single cells (Fig. 3b and Supplementary Fig. 25). This increase in expression with copy number provided additional validation that DR-Seq was sensitive enough to simultaneously detect changes in copy numbers and transcript counts from the same cell (Supplementary Fig. 26).
Finally, we investigated if DNA copy number variations within the cancer genome could be an important regulator of gene expression variability. We found that genes that display more cell-to-cell variability in transcript numbers were generally associated with reduced copy number loci and vice-versa, implying that CNVs could potentially drive variability in gene expression between single cells (Fig. 3c, Supplementary Figs. 27 and 28 and Supplementary Note).
In summary, we developed a method that allows combined gDNA and mRNA sequencing from the same cell using a single-pot strategy. Similar integrated strategies might be used in the future to determine the correlation between DNA methylation and transcription, or nucleosome positioning and transcription, in single cells. Additionally, integrated gDNA and mRNA single-cell sequencing might provide enhanced sensitivity to lineage tracing studies in tumors and healthy tissue (Supplementary Table 5).
Online Methods
Tissue culture
E14 cells were cultured in DMEM (Gibco) supplemented with 15% FBS (Gibco), 2mM GlutaMax (Gibco), 0.1 mM MEM nonessential amino acids, 0.1 mM β-mercaptoethanol (Sigma), 1% Pen/Strep (Gibco) and 1000U LIF/ml (ESGRO) on gelatinized petri dishes. SK-BR-3 cells were cultured in McCoy’s 5a Medium Modified (ATCC) with 10% FBS and 1% Pen/Strep. Cells were grown at 37°C and 5% CO2.
Cell picking
Trypsinized single cells were picked using a mouth pipet with a 30 μm glass capillary under a stereomicroscope. Picked cells were deposited in the center of the lid of a 0.2 ml PCR tube and snap frozen in liquid nitrogen.
DR-Seq
First strand cDNA synthesis was performed by adding 2 μL of reaction mix containing 0.2 μL first strand buffer (MessageAmp II, Life technologies), 0.4 μL of dNTP mix (MessageAmp II, Life technologies), 0.1 μL Arrayscript (MessageAmp II, Life technologies), 0.1 μL RNAse inhibitor (MessageAmp II, Life technologies), 0.2 μL RT primer with cell specific barcode (Ad-1x)13, 0.2 μL 1:500000 diluted ERCC spike-in mix 1 (Life technologies) and 0.05% IGEPAL in water. The first strand cDNA synthesis and lysis reaction mix together with the spike-in molecules were added directly to the drop in the lid of the tube containing a single cell. Samples were incubated in a PCR machine with lid and block set to 42C for 15 minutes after which the samples were spun down and incubated for another 105 minutes. After first strand synthesis, samples were incubated for 10 minutes at 80°C. Quasilinear amplification buffer containing 6.0 μL ThermoPol buffer (NEB) 1.0 μL 10 mM dNTP mix, 26 μL water and 0.15 μL 50 μM primer mix (Ad-2)7 was added to each sample. Samples were incubated for 3 minutes at 94 °C to denature the DNA. Seven cycles of quasilinear amplification was performed (10°C for 45 seconds, 15°C for 45 seconds, 20°C for 45 seconds, 30°C for 45 seconds, 40°C for 45 seconds, 50°C for 45 seconds, 65°C for 2 minutes, 95°C for 20 seconds, 58°C for 40 seconds and then immediately quench on ice). Prior to each cycle 0.6 μL polymerase mix containing 2U Bst large fragment (NEB) and 0.8U Pyrophage 3173 exo- (Lucigen) was added. Note that the 58°C for 40 seconds step, prior to quenching the reaction on ice, is not performed for the first quasilinear amplification round. After 7 sounds of quasilinear amplification, samples were split in two. One half of the sample was processed for gDNA sequencing, the other half was processed for mRNA sequencing.
For mRNA sequencing, second strand synthesis of the quasilinear amplified cDNA was performed using the P1 primer (5′ - CGATTGAGGCCGGTAATAC - 3′) in a single cycle of PCR (94°C for 20 sec, 51°C for 20 sec, 72°C for 7 min). After this, samples with non-overlapping barcodes were pooled and cleaned up on a cDNA purification column (MessageAmp II, Life technologies), and eluted twice with 9 μL of water at 55°C. Next, the volume of the sample was reduced to 6.4 μL using a SpeedVac®. In vitro transcription (IVT) mix containing 1.6 μL 10x IVT buffer, 1.6 μL ATP, 1.6 μL GTP, 1.6 μL CTP, 1.6 μL UTP and 1.6 μL enzyme mix (MessageAmp II, Life technologies) were added to the samples and incubated at 37°C for 13 hours. After IVT, the aRNA was immediately cleaned up without fragmentation using the aRNA clean up columns (MessageAmp II, Life Technologies) and the aRNA was eluted twice in 12 μL of warm water at 55°C. After clean up, aRNA quality was assessed on a bioanalyzer (Agilent) Eukaryote Total RNA Pico chip. Library preparation was performed as previously described13.
For DNA sequencing, the other half of the quasilinear amplification product was amplified further by PCR. PCR mix containing 1.0 μL 10mM dNTP, 3 μL Thermopol buffer (10X), 0.2 μL 100 μM primer P2 (5′ - GTGAGTGATGGTTGAGGTAGTGTGGAG - 3′) and 1.0 μL Deep VentR (exo-) polymerase (NEB) was added to each sample for a final volume of 68 μL. PCR was performed as follows, 21 cycles of (94°C for 20 seconds, 59°C for 20 seconds, 65°C for 1 minute, 72°C for 2 minutes), and 72°C for 5 minutes at the end. After PCR, the quality of the products were assessed by agarose gel electrophoresis and the samples were cleaned up using a PCR purification column (Qiagen) (Supplementary Fig. 29). Next, to remove adapter Ad-2 from the PCR product prior to preparing Illumina libraries, another PCR was done starting with 80 ng of product from the previous step. PCR mix containing 0.3 μL of 50 μM primer P3 with a 5′ biotinylated end (5′ - GTGAGCTGGAGTTGAGGTAGTGTGGAG - 3′), 5 μL Thermopol buffer (10X), 1 μL 10mM dNTP and 1 μL Deep VentR (exo-) polymerase (NEB) was added to each sample for a final volume of 50 μL. PCR was performed as follows, 94°C for 2 minutes, then 4 cycles of (94°C for 20 seconds, 46°C for 20 seconds, 65°C for 1 minute and 72°C for 2 minutes) and 9 cycles of (94°C for 20 seconds, 59°C for 20 seconds, 65°C for 1 minute and 72°C for 2 minutes). The PCR product was sheared using a sonicator (Biorupter®) on the low power setting with 15 cycles of 1 minute (30 seconds On, 30 seconds Off) with constant cooling at 4°C. The sheared products were then cleaned up using a PCR purification column (Qiagen) and eluted in 50 μL water. The final product distribution was verified on a bioanalyzer (Agilent) High Sensitivity DNA chip to have an average product size of approximately 300 bp. The DNA products were then added to Dynabeads MyOne Streptavidin C1 beads (Life Technologies) in 50 μL 2× BW buffer (10 mM Tris-HCl, 1mM EDTA and 2mM NaCl). After immobilizing the DNA products on the beads for 15 minutes, the biotinylated DNA was separated using a magnetic stand and the supernatant was stored. The biotinylated DNA was digested on the magnetic beads and the beads were washed twice with 50 μL 1× BW buffer. These two washes were then combined with the first supernatant and purified using a PCR purification column (Qiagen). Finally, Illumina libraries were prepared with different index primers for each single cell using the NEBNext Ultra DNA Library Prep Kit for Illumina® (NEB).
Applying DR-Seq to E14 and SK-BR-3 cells, the typical success rate in amplifying single cells was approximately 70% (21/30 for SK-BR-3 and 13/18 for E14 cells).
Libraries were sequenced on an Illumina Hi-seq 2500. cDNA libraries from DR-Seq were sequenced with 100 bp paired-end sequencing and the gDNA and other cDNA libraries (from CEL-Seq or bulk) were sequenced with 50 bp or 100 bp paired-end sequencing.
Bioinformatic Analysis
For bulk mRNA and CEL-Seq libraries, paired end sequencing reads were aligned to the transcriptome using Burrows-Wheeler Aligner (BWA) with default parameters. For single cell mRNA processed using DR-Seq, the Ad-2 adapter sequence was trimmed computationally from the right mate and then aligned to the transcriptome using BWA with default parameters. For the E14 cells, we used the RefSeq gene models based on the mouse genome release mm10. For the SK-BR-3 cells, we used the RefSeq gene models based on the human genome release hg19. For bulk mRNA sequencing both mates of each read were mapped to the transcriptome. For CEL-Seq and DR-Seq, the right mate of each read pair was mapped to the transcriptome and the ERCC spike-ins. The left mate was used to identify the cell from which the transcript came based on the cell-specific barcode. Reads mapping to more than one region were distributed uniformly. For the bulk mRNA sequencing libraries, PCR duplicates were then removed to obtain the dataset used in all the analysis. The left mate of the CEL-Seq libraries also contained a 4-bp random sequence, introduced during reverse transcription, to count unique cDNA molecules, as previously described (Supplementary Fig. 6)21. Length-based identifiers were determined for each read in the single-cell mRNA libraries processed by DR-Seq using the first coordinate of the right mate after trimming off adapter Ad-2 (Fig. 1b). The length-based identifiers were used to minimize amplification biases and achieve resolution close to identifying unique cDNA molecules (Fig. 2a,b,c, Supplementary Figs. 6,7,8 and Supplementary Note).
For bulk gDNA and MALBAC libraries, paired end sequencing reads were aligned to the genome release mm10 for mouse cells (E14) and to the genome release hg19 for human cells (SK-BR-3) using BWA with default parameters. For the single cell gDNA libraries processed by DR-Seq, paired end sequencing reads were aligned to a masked genome mm10 for mouse cells and to a masked genome hg19 for human cells using BWA with default parameters. The masked genomes mm10 and hg19 were created by replacing all the coding sequences within the genome with “N”. This is because the fraction used to sequence gDNA contains sequences that could originate from the cDNA within coding regions (Fig 1a and Supplementary Fig. 13). By masking the coding sequences within the genome, such ambiguous reads that might arise from either gDNA or cDNA are discarded computationally leaving only reads that arise from gDNA. This does not pose a problem for calling copy number variations because the coding region constitutes only approximately 2% of the genome. Therefore, gDNA sequencing results obtained from DR-Seq can be used to quantify copy number variations and single nucleotide variants in the genome (Fig. 3a, Supplementary Fig. 21 and Supplementary Table 5). Next, all PCR duplicates within mapped reads from the bulk, MALBAC or DR-Seq libraries are removed. As the first step towards quantifying the gDNA data, the genome is divided into bins. To account for the masking of the genome in the DR-Seq data, the start and end coordinates of each bin are chosen such that the length of all bins are the same after excluding coding regions within each bin. This variable binning strategy provides a more accurate description of the distribution of reads within each bin as reads that map to coding regions are masked from the analysis (Supplementary Figs. 13 and 14). Next, to further reduce amplification biases, we developed a coverage-based method to quantify the reads within bins. This coverage-based method significantly reduces bin-to-bin technical noise (see Supplementary Note, Fig 2f and Supplementary Figs. 15,16 for details). The reads are then corrected for GC bias25. The corrected read distribution is then used to identify breakpoints using the circular binary segmentation (CBS) algorithm26. Finally, the median read counts for each segment are used to call copy number variations in single cells (Supplementary Note).
DNA FISH
SK-BR-3 cells were grown on coverglasses and fixed in Methanol:Acetic Acid solution (3:1 vol) for 10 minutes at RT upon reaching confluency. They were washed with PBS/0.1% Triton X-100 and treated with 100 μg/mL of RNAseA in PBS for 1 h at 37°C. They were then washed with PBS and dehydrated with ethanol series (70% ethanol, followed by 85%, followed by 100%) followed by over night air-drying. The next day they were denatured in 70% formamide/2xSSC buffer at 75°C for 5 minutes and placed in 70% ethanol afterwards for 2 minutes, followed by a 2 minute incubation in 85% ethanol and 2 minute incubation in 100% ethanol. They were then air-dried for 30 minutes and during this time the probes were denatured at 75°C for 5 minutes. The hybridization was set up with HD FISH probes resuspended in the CEP buffer (Abbott) (100 ng/20 μL). After sealing the coverglasses on microscope slides with a rubber cement they were incubated at 37°C over night. The next day the coverglasses were removed from the slides and washed 3 times in 2xSSC followed by two washes in 0.2xSSC/0.2% Tween20 at 56°C for 7 minutes each. Afterwards they were rinsed with 4xSSC/0.2% Tween20 and washed once with 2xSSC for 5 minutes. They were then incubated with 50 ng/mL DAPI/2xSSC for 5 minutes at RT and mounted in the mounting solution containing 2xSSC, 10 mM Tris, 0.4% glucose, 100 μg/mL catalase, 37 μg/mL glucose oxidase, 2 mM Trolox. The probes were designed using the www.hdfish.eu database. For CCDC40, HTT and FHIT genes, the HD-FISH probes were prepared by PCR as previously described27. For ZMIZ1, 40-mer oligonucleotides with a 3′ functional amino group were synthesized by Biosearch Technologies Inc., and coupling to Cy5 (GE Healthcare) was performed in-house.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Chenghang Zong (Harvard University) for sharing details of MALBAC and Dominic Grün (Hubrecht Institute) for assistance in data analysis. We would also like to thank Nicola Crosetto (Hubrecht Institute) for assistance in preparing DNA FISH probes. This work was supported by the European Research Council Advanced grant (ERC-AdG 294325-GeneNoiseControl), the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO) Vici award and by the Human Frontier Science Program (to M.B).
Footnotes
Accession Code. Sequencing data is available at Gene Expression Omnibus (GEO) (GSE62952) (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE62952).
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keane TM, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheltzer JM, Torres EM, Dunham MJ, Amon A. Transcriptional consequences of aneuploidy. Proc. Natl. Acad. Sci. U.S.A. 2012;109:12644–12649. doi: 10.1073/pnas.1209227109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falconer E, et al. DNA template strand sequencing of single-cells maps genomic rearrangements at high resolution. Nat. Methods. 2012;9:1107–1112. doi: 10.1038/nmeth.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hou Y, et al. Genome analyses of single human oocytes. Cell. 2013;155:1492–1506. doi: 10.1016/j.cell.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 10.Evrony GD, et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McConnell MJ, et al. Mosaic copy number variation in human neurons. Science. 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 13.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Shalek AK, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue Z, et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500:593–597. doi: 10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Picelli S, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 17.Islam S, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods. 2014;11:163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- 18.Wu AR, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat. Methods. 2014;11:41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng Q, Ramsköld D, Reinius B, Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- 20.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grün D, Kester L, van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nat. Methods. 2014;11:637–640. doi: 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
- 22.Junker JP, van Oudenaarden A. Every cell Is special: Genome-wide studies add a new dimension to single-cell biology. Cell. 2014 doi: 10.1016/j.cell.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 23.Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 24.Baker SC, et al. The External RNA Controls Consortium: a progress report. Nat. Methods. 2005;2:731–734. doi: 10.1038/nmeth1005-731. [DOI] [PubMed] [Google Scholar]
- 25.Zhang C, et al. A single cell level based method for copy number variation analysis by low coverage massively parallel sequencing. PLoS ONE. 2013;8:e54236. doi: 10.1371/journal.pone.0054236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 27.Bienko M, et al. A versatile genome-scale PCR-based pipeline for high-definition DNA FISH. Nat. Methods. 2013;10:122–124. doi: 10.1038/nmeth.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.