Abstract
Open cell-free translation systems based on Escherichia coli cell lysates have successfully been used to produce antibodies and antibody fragments. In this study, we demonstrate the cell-free expression of functional single-chain antibody variable fragments (scFvs) in a eukaryotic and endotoxin-free in vitro translation system based on Spodoptera frugiperda (Sf21) insect cell extracts. Three scFv candidates with different specificities were chosen as models. The first scFv candidate SH527-IIA4 specifically discriminates between its phosphorylated (SMAD2-P) and nonphosphorylated antigens (SMAD2) (where SMAD is mothers against decapentaplegic homolog 2), whereas the second scFv candidate SH527-IIC10 recognizes both, SMAD2-P and SMAD2. The third scFv candidate SH855-C11 binds specifically to a linear epitope of the CXC chemokine receptor type 5. The translocation of antibody fragments into the lumen of endogenous microsomal vesicles, which are contained in the lysate, was facilitated by fusion of scFv genes to the insect cell specific signal sequence of honeybee melittin. We compared the binding capabilities of scFv fragments with and without melittin signal peptide and detected that translocated scFv fragments were highly functional, whereas scFvs synthesized in the cytosol of the cell extract showed strongly decreased binding capabilities. Additionally, we describe a cell-free protein synthesis method for the incorporation of noncanonical amino acids into scFv molecules in eukaryotic cell lysates. We demonstrate the successful cotranslational labeling of de novo synthesized scFv molecules with fluorescent amino acids, using residue-specific as well as site-specific labeling.
Keywords: Cell-free, ELISA, Insect cell extract, scFv antibody fragment, Sf21 insect lysate
1. Introduction
Antibodies are the key detection elements in research, diagnostics, and therapeutics. Until now, they are the fastest growing class of therapeutic protein agents [1]. Besides manufacturing of the classical IgG antibody format, a rapidly growing number of recombinant antibodies are currently being made available [2]. Antibody fragments are of increasing clinical importance and several methods have evolved to meet these demands [1,3]. Besides classical cell culture production of antibodies and antibody fragments, cell-free methods have shown to provide a suitable platform for the synthesis of disulfide-bonded proteins [4–8]. In contrast to cell-based systems, cell-free methods do not require time- and labor-intensive cloning steps for the generation of appropriate expression vectors, and the transformation of these vectors into host cells [9]. Using cell-free methods, linear DNA templates generated by PCR can be directly applied in the translation system and the metabolic resources of the generated cell lysate are exclusively used for the synthesis of a given target protein [10]. Cell extracts generated from Escherichia coli have been used for the synthesis of antibody fragments as well as antibody fragment fusion proteins [6–8,11–13]. These lysates are usually characterized by high production yields, but only a fraction of synthesized antibodies may be in a functional state [8,14]. Since many proteins tend to accumulate as insoluble aggregates of folding intermediates, the performance of alternative cell-free translation systems, e.g. based on lysates from wheat germs [15] and insect cells [16,17], has been evaluated in the past.
The protein chains of an antibody molecule fold into distinct structural domains, called immunoglobulin domains. Most immunoglobulin domains contain highly conserved cysteines, which allow the formation of one disulfide bond per immunoglobulin fold [18–21]. Thus, the cell-free system used for the expression of properly folded antibody domains needs to fulfill certain requirements in order to allow the formation of disulfide bonds and folding of complex structural domains. In this context, it was already reported that the insect cell-free translation system used in this study is well suited for the synthesis of biologically active disulfide-bonded proteins, such as ice structuring proteins [22], single-chain antibody variable fragments (scFvs) [17], and Fab fragments [16].
ScFv molecules are the smallest recombinant antibody formats still containing the full antigen-binding site, consisting of the variable domain of the heavy antibody chain and variable domain of the light antibody chain, connected by a flexible peptide linker [23,24]. Here, we demonstrate the expression of soluble and functional scFv molecules with different specificities in a eukaryotic cell-free translation system based on cultured Spodoptera frugiperda (Sf21) cells. On the one hand, the cell extract serves as the source for translationally active ribosomes, translation factors, and enzymes [25,26]. On the other hand, the lysate is characterized by the presence of endogenous vesicles, having their origin in the ER of the cultured insect cells, which allow for a cotranslational translocation of target proteins into the lumen of these vesicles, or in the case of membrane proteins, their embedding into microsomal membranes [27,28]. The scFv molecules analyzed in this study were produced under unified redox-optimized reaction conditions and are shown to be highly specific for their corresponding antigen.
2. Materials and methods
2.1. Preparation of insect lysate
Translationally active insect lysates were prepared from cultured Sf21 cells grown in fermenters at 27°C in an animal component free insect cell medium. Cell extracts were prepared as described previously [29]. Translationally active insect lysates were prepared from cultured Sf21 cells grown in fermenters at 27°C in an animal component free insect cell medium (Insect-XPRESS medium, Lonza). At a density of approximately 4 × 106 cells/mL, cells were harvested and collected by centrifugation at 200 × g for 5 min. Cell pellets were washed twice and resuspended in a HEPES-based homogenization buffer (final concentration (f.c.) 40 mM HEPES-KOH, pH 7.5, 100 mM NaOAc). Resuspended Sf21 cells were lysed mechanically by passing the cell suspension through a 20-gauge needle using a syringe. Nuclei and cell debris were removed by centrifugation at 10 000 × g for 10 min. The resulting supernatant was applied to a Sephadex G-25 column (GE Healthcare, Freiburg), which was pre-equilibrated with homogenization buffer. Elution fractions (1 mL each) with the highest RNA/protein ratios were pooled and subsequently treated with S7 nuclease (f.c. 10 U/mL, Roche) and CaCl2 (f.c. 1 mM) in order to remove endogenous messenger RNA (mRNA). The mixture was incubated for 2 min at room temperature (RT) and micrococcal nuclease was subsequently inactivated by addition of EGTA (f.c. 6.7 mM). Aliquots of the Sf21 lysate were immediately shock frozen in liquid nitrogen and subsequently stored at –80°C until further usage.
2.2. Template generation by expression-PCR (E-PCR)
Recombinant human antibodies against SMAD2 (where SMAD2 is nonphosphorylated SMAD2 peptide), SMAD2-P (where SMAD2-P is phosphorylated SMAD2 peptide), and CXC chemokine receptor type 5 (CXCR5) (SH527-IIA4, SH527-IIC10, and SH855-C11, Table1) were selected by phage display using the human naive antibody gene library HAL7/8 [30] as described before [31]. Linear DNA templates of chosen scFv fragments were generated by a two-step E-PCR [32]. In the PCR step one, scFv genes were amplified from their corresponding phage display vectors (pHAL14-SH527-IIA4, pHAL14-SH527-IIC10, and pHAL14-SH855-C11) using gene-specific forward primers and vector-specific reverse primers, both containing 5′ overhangs, which introduced overhang sequences that were complementary for the primers used in PCR step two. In the second PCR step, adapter primers were used, which introduced the regulatory sequences needed for transcription and translation such as a T7 promotor sequence and a ribosome-binding site upstream of the gene's ORF and a T7 terminator sequence downstream of the deduced stop codon. All oligonucleotides used in this study were purchased from IBA GmbH, Göttingen, Germany (Supporting information, Table S1). Furthermore, scFv genes were N-terminally fused to the signal sequence of honeybee melittin (Mel) [33], to promote cotranslational translocation of cell-free synthesized target proteins into microsomal vesicles. Additionally, scFv templates were generated without N-terminal signal sequence. In all cases, scFv templates contained a His-tag and a c-myc-tag sequence at the C-terminus. In order to incorporate fluorescent amino acids in a site-directed manner, additional DNA templates were generated by a two-step E-PCR, bearing the amber stop codon TAG at the C-terminus of the scFv gene right before the c-myc-tag sequence. The codon was introduced by using mismatch primers in the first E-PCR step. scFv genes were amplified using HotStar HiFidelity DNA polymerase (Qiagen) in the first PCR step and Taq DNA polymerase (Thermo Scientific) in the second PCR step. The following PCR conditions were applied during the first PCR step: 5-min initial denaturation at 95°C, 30 cycles comprising 1-min denaturation at 94°C, 1-min annealing at 52°C (SH527-IIA4)/55°C (SH527-IIC10, SH855-C11), 1-min elongation at 72°C, followed by 10-min final extension at 72°C. PCR conditions during the second PCR step: 5-min initial denaturation at 95°C; 30 cycles comprising 1-min denaturation at 94°C, 1-min annealing 45°C, 1-min elongation at 72°C, followed by 10-min final extension at 72°C. Theoretical DNA fragment sizes were calculated in silico. PCR products were analyzed by agarose gel electrophoresis. PCR products of first and second PCR steps were detected as homogenous bands showing the expected sizes (data not shown).
Table 1.
Model antibody fragments used in this study.
| Antibody | Target | VH |
VL |
|||
|---|---|---|---|---|---|---|
| V | D | J | V | J | ||
| SH527-IIA4 | SMAD2-P | IGHV3–30*03 | IGHD3–22*01 | IGHJ4*02 | IGLV2–14*02 | IGLJ3*02 |
| SH527-IIC10 | SMAD2, SMAD2-P | IGHV5–51*01 | IGHD2–15*01 | IGHJ3*02 | IGLV1–50*01 | IGLJ3*02 |
| SH855-C11 | CXCR5 | IGHV1–69*01 | IGHD3–10*01 | IGHJ4*02 | IGLV1–47*02 | IGLJ3*01 |
The most similar human germline genes were identified by VBASE 2 (http://www.vbase2.org).
VH = variable domain of the heavy antibody chain; VL = variable domain of the light antibody chain; V = variable gene segment; D = diversity gene segment; J = Joining gene segment
2.3. Cell-free protein synthesis
2.3.1. Cell-free protein synthesis based on insect lysate
Synthesis of scFv molecules was performed using the batch-formatted insect cell-free translation system. Transcription and translation were carried out as independent and subsequently performed reactions, separated by an intermediate gel filtration step (linked mode) [25]. In vitro transcription reactions based on T7 RNA polymerase (f.c. 1 U/μL) were performed using the EasyXpress Insect Kit II (Qiagen) according to the manufacturer's instructions. Obtained mRNA samples were purified by gel filtration (DyeEx spin columns, Qiagen; illustra NAP-5 columns, GE Healthcare) and analyzed qualitatively by agarose gel electrophoresis. mRNA templates were detected as homogenous bands showing the expected size (data not shown). In vitro transcription reactions were initiated by addition of purified scFv mRNA templates (f.c. 240–280 μg/mL).
Cell-free translation reactions were performed using 40% v/v insect lysate supplemented with HEPES-KOH (f.c. 30 mM, pH 7.6; Merck), Mg(OAc)2 (f.c. 2.5 mM; Merck), KOAc (f.c. 75 mM; Merck), amino acids (complete 200 μM f.c.; Merck), spermidine (f.c. 0.25 mM; Serva), creatine-phosphate (f.c. 20 mM; Roche), and energy-regenerating components (f.c. 1.75 mM ATP, 0.3 mM GTP; Roche). Synthesis of scFv fragments without signal sequence was performed in presence of reduced glutathione (GSH) (f.c. 0.5 mM; Roth) and oxidized glutathione (GSSG) (f.c. 2.5 mM; Roth), which was directly added to the cell-free reaction. To monitor protein quality and quantity, translation mixtures (TMs) were supplied with14C-labeled leucine (f.c. 60 μM; PerkinElmer) yielding a specific radioactivity of 46.15 dpm/pmol. To monitor background translational activity in every experiment, a translation reaction without the addition of mRNA was run in parallel, in the following designated as “no template control” (NTC). Batch reactions were performed in a 100 μL reaction volume in a thermomixer (Thermomixer comfort, Eppendorf) at 25°C for up to 4 h with gentle agitation at 750 rpm.
2.3.2. Residue-specific and site-specific labeling of scFv molecules
In order to label scFv molecules in an amino acid selective (phenylalanine, Phe) manner, fluorescent BODIPY-TMR-lysine-tRNAGAA (f.c. 2 μM, RiNA GmbH) was added to the open cell-free translation reaction. Using this residue-specific labeling technique, the fluorescent amino acid is incorporated at multiple sites in the protein. Based on its anticodon, the precharged tRNA decodes the TTC codon, which results in a competition of endogenous Phe-tRNAGAA with precharged tRNAs, leading to the incorporation of the fluorescent amino acid in a stochastic manner. Site-specific labeling of scFv molecules was facilitated by using linear DNA templates bearing the amber stop codon TAG and by addition of BODIPY-TMR-lysine-tRNACUA (f.c. 5 μM, RiNA GmbH). To enhance labeling efficiency in de novo synthesized proteins, while decreasing the fraction of unspecifically labeled proteins synthesized from endogenous mRNA, fluorescent amino acids were added to the cell-free translation reaction 10 min after starting the reaction. Batch reactions were performed in a 20 μL reaction volume in a thermomixer (Thermomixer comfort, Eppendorf) at 25°C for up to 4 h with gentle agitation at 750 rpm.
2.3.3. Cell-free protein synthesis based on lysates from E. coli
Cell-free expression in E. coli cell lysates was performed using the EasyXpress Disulfide E. coli Kit (Qiagen) following the manufacturer's instructions. For qualitative and quantitative analysis of de novo synthesized target proteins, reactions were supplemented with 14C-labeled leucine (60 μM; PerkinElmer) yielding a specific radioactivity of 9.5 dpm/pmol.
2.4. Release of melittin-fused scFv molecules from insect vesicles
Prior to functional analysis, TMs of cell-free synthesized scFv molecules were separated into distinct lysate fractions. To release translocated and trapped scFv molecules from the lumen of the insect vesicles, TMs were centrifuged at 16 000 × g and 4°C for 10 min. During centrifugation, insect vesicles accumulated as a defined but soluble pellet, designated as vesicular fraction 1 (VF1). After centrifugation, the supernatant 1 (SN1) was separated from VF1, followed by resuspension of VF1 in PBS supplemented with 0.2% n-dodecyl-β-maltoside (DDM) (Qiagen) and agitation for at least 45 min at RT. Subsequently, this solution was centrifuged (16 000 × g, 10 min, 4°C) and the resulting supernatant 2 (SN2) was separated from the vesicular fraction 2 (VF2). Lysate fractions SN1 and SN2 were directly applied for functional analysis of cell-free synthesized scFv molecules. Lysate fractions were stored on ice (short-term storage for up to 2 days) or frozen at –20°C until further usage.
2.5. Qualitative analysis of cell-free produced scFv molecules
14C-leucine-labeled scFv molecules were analyzed qualitatively by SDS-PAGE and autoradiography. Aliquots of 5 μL were precipitated in precooled acetone and incubated on ice for a minimum of 15 min. Afterwards, samples were centrifuged (16 000 × g, 10 min, 4°C) and the resulting protein pellet was dried for 1 h at 45°C, whereas the supernatant containing soluble and nonincorporated 14C-leucine was discarded. Dried protein pellets were subsequently resuspended in 20 μL of 1× sample buffer (NuPAGE® LDS Sample Buffer; Life technologies) supplemented with 50 mM DTT (Merck) by intense agitation on a vibrax mixer. Samples were subjected to 10% SDS-PAGE using precast gels (NuPAGE®, 10% Bis-Tris Gel with MES SDS buffer; Life technologies). SDS-PAGE gels were run at 200 V for 35 min. After gel electrophoresis, gels were washed with demineralized water for three times in a microwave and subsequently dried (Unigeldryer 3545D, Uniequip) for 1 h at 70°C. 14C-leucine-labeled proteins were visualized using a phosphoimager system (Typhoon TRIO+ Imager, GE Healthcare). scFv molecules labeled with fluorescent amino acids were analyzed by in-gel fluorescence. After gel electrophoresis, SDS-PAGE gels were incubated in a solution of 50% methanol/50% water for 30 min at RT. Labeled proteins were subsequently visualized by using a phosphoimager system (excitation 532 nm, emission filter 580 nm BP, Typhoon TRIO+ Imager, GE Healthcare).
2.6. Quantitative analysis of cell-free produced scFv molecules
Yields of 14C-leucine-labeled proteins were determined by liquid scintillation counting after hot trichloroacetic acid (TCA) precipitation. After synthesis, aliquots from the TM (5 μL) were precipitated in 3 mL of 10% TCA (Roth) supplemented with 2% casein hydrolysate (Merck) and incubated for 15 min at 80°C. Afterwards, samples were incubated on ice for a minimum of 30 min. The solutions were filtered using a vacuum filtration system (Hoefer). 14C-leucine-labeled proteins were retained on filter papers (MN GF-3, Machery-Nagel), which were subsequently washed two times with 5% TCA and dried by rinsing the filters twice with acetone. Dried filter papers were transferred to scintillation tubes (Zinsser Analytic), soaked with 3 mL of scintillation cocktail (Quicksafe A, Zinsser Analytic), and agitated for at least 1 h on an orbital shaker. Incorporation of 14C-leucine was measured by liquid scintillation counting using the LS6500 Multi-Purpose scintillation counter (Beckman Coulter).
2.7. Functional analysis by ELISA
Functional analysis of cell-free synthesized scFv molecules was performed by ELISA. Ninety-six well microtiter plates (Costar 96, Sigma-Aldrich) were coated with streptavidin (0.74 μg/mL in PBS, 270 μL/well; Serva) for 1 h at RT, followed by 2 × 5 min washing in PBS supplemented with 0.05% Tween-20 (PBST). Wells were subsequently blocked with 2% BSA (Sigma-Aldrich) in PBS (270 μL/well) for 1 h at RT, followed by 3 × 5 min washing with PBST. Afterwards, wells were coated with the biotinylated antigen (2 μg/mL in PBS, 100 μL/well), which was incubated for 1 h at RT or overnight at 4°C. The following antigens were purchased from Peps4LS GmbH (Heidelberg, Germany): Phosphorylated SMAD2 peptide (Biotin-PEG-(GGS)2 GPLQWLDKVLTQMGSPSVRCSpSMpS); nonphosphorylated SMAD2 peptide (Biotin-PEG-(GGS)2 GPLQWLDKVLTQMGSPSVRCSSMS); human CXCR5 chemokine receptor peptide (Biotin-PEG-(GGS)2SLVENHLCPATEGPLMASFKAVFVP). After 3 × 5 min washing with PBST, wells were incubated with cell-free synthesized scFvs. scFv molecules without signal sequence were analyzed in lysate fraction SN1, whereas scFv molecules with melittin signal sequence were analyzed in lysate fraction SN2. NTCs were treated likewise and were analyzed in parallel. A total of 100 μL of lysate fraction SN1 or SN2, serially diluted in 1% BSA in PBST, was added to the wells and incubated for at least 1.5 h at RT, followed by 3 × 5 min washing with PBST. For detection of bound scFv molecules, wells were incubated with the monoclonal anti-c-myc-tag antibody 9E10 as primary antibody (Hybrotec; dilution 1:1000 in 1% BSA in PBST, 100 μL/well) and anti-mouse-IgG-HRP-linked antibody (Cell signaling) (dilution 1:3000 in 1% BSA in PBST, 100 μL/well) for 1.5 h at RT, followed by three PBST washing cycles. Finally, substrate solution (3,3′,5,5′-tetramethylbenzidine, 100 μL/well; Life technologies) was added to the wells and reactions were stopped after an incubation of approximately 30 min by addition of 0.5 M sulfuric acid (100 μL/well). Plates were measured at 450 and 620 nm (reference) using a microplate reader (FLUOstar Omega, BMG LABTECH). Values measured at 620 nm (scattered light) were subtracted from the values obtained at 450 nm. Furthermore, signals obtained from identically treated NTCs (SN1 and SN2) were subtracted from the values measured from scFv-containing lysate fractions.
3. Results
3.1. Generation of recombinant human antibodies against SMAD2 and CXCR5
The selection of recombinant antibodies was performed according to Schirrmann and Hust with modifications [31]. Antibody fragments were selected against peptides using the human naive antibody gene library HAL7/8 [30]. Monoclonal binders were identified by antigen ELISA using soluble scFv fragments (data not shown). scFv fragments were produced in microtiter plates and monoclonal binders were identified as described before [34]. These binders were sequenced to identify unique binders and analyzed using VBASE2 (http://www.vbase2.org) [35]. Antigen binding was verified by titration ELISA.
3.2. Cell-free synthesis of scFv candidates
The scFv molecules SH527-IIA4, SH527-IIC10, and SH855-C11 were chosen as test candidates, in order to evaluate the performance of the insect cell-free translation system for the synthesis of functional antibody fragments. Two different kinds of DNA templates were generated: scFv genes fused to the melittin signal sequence at the N-terminus and scFv genes without a signal sequence. The honeybee melittin signal peptide enables efficient translocation of target proteins into the lumen of the ER, as has been demonstrated in vivo [33] using Sf cells and in vitro using a cell-free translation system generated from Sf21 cells [25]. Translocation of proteins into the ER is the prerequisite for their posttranslational modification, such as formation of disulfide bonds, glycosylation, and lipidation [36]. Therefore, scFv genes were N-terminally fused to the melittin signal sequence in order to test if the translocation of de novo synthesized scFv molecules might have a positive effect on their functionality. To analyze expression, scFv candidates were synthesized in presence of 14C-leucine. De novo synthesized antibody fragments reached protein yields of 10–15 μg/mL and were detected as distinct and homogeneous bands in the autoradiograph, showing the expected migration pattern (Fig.1).
Figure 1.
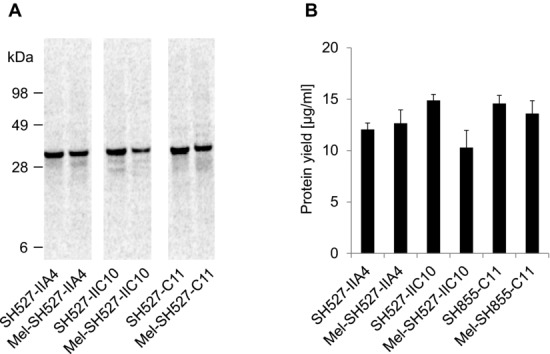
Qualitative (A) and quantitative (B) analyses of cell-free synthesized and 14C-leucine-labeled scFv molecules. Cell-free synthesis was performed using the insect cell-free expression system. (A) Autoradiograph of 14C-leucine-labeled scFvs. (B) Diagram showing total protein yields of scFvs in the translation mixture. Error bars show SDs calculated from triplicate analysis (n = 3).
Next, we investigated the influence of the melittin signal peptide on the translocation efficiency of cell-free expressed scFv molecules into the endogenous vesicles contained in the insect lysate. After translation of scFv constructs with or without signal sequence, reaction mixtures were separated into four different lysate fractions by centrifugation. Expression and localization of scFv molecules in the insect lysate were subsequently monitored by SDS-PAGE followed by autoradiography. As expected, scFv molecules without signal sequence were mainly detected in the TM and supernatant fraction (SN1) after centrifugation, indicating that these proteins were synthesized in soluble and nonaggregated form (Fig.2A). scFv molecules with N-terminally fused signal sequence were detected mainly in the vesicular fraction (VF1), which was gained after centrifugation of the TM, and to some extent in SN1 (Fig.2B). Most probably, proteins localized in SN1 represented nontranslocated scFv molecules, whereas those detected in VF1 indicated the translocation of target proteins into the lumen of the insect vesicles. scFv molecules present in VF1 were most efficiently released from the vesicles by resuspension of the vesicular fraction using 0.2% DDM in PBS, as could be seen by the detection of soluble scFv molecules in SN2 rather than vesicular fraction 2. These results were consistent with published data for a cell-free synthesized and fluorescein-binding scFv [37,38], which was released by using 0.1% DDM in PBS [17].
Figure 2.
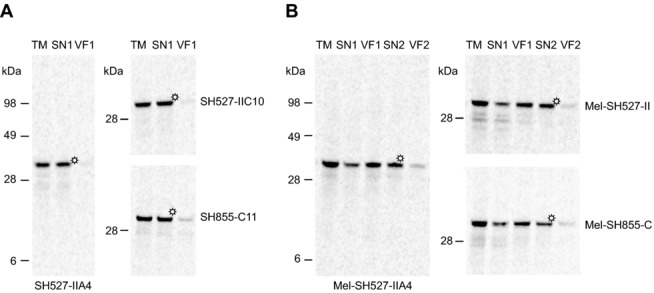
Qualitative analysis of scFv molecules in different lysate fractions using autoradiography. Cell-free synthesis of scFvs was performed in presence of 14C-leucine using the insect cell-free expression system. Immediately following the protein synthesis reaction, translation mixtures (TM) were centrifuged and the supernatant fraction (SN1) was separated from the vesicular fraction (VF1). VF1 was resuspended in PBS supplemented with 0.2% DDM. After an additional centrifugation step, SN2 was separated from the remaining VF2, resulting in altogether five lysate fractions that were subsequently analyzed by SDS-PAGE and autoradiography. (A) Detection of scFv molecules without signal sequence (SH527-IIA4, SH527-IIC10, SH855-C11) in TM, SN1, and VF1. (B) Detection of scFv molecules with melittin signal sequence (Mel-SH527-IIA4, Mel-SH527-IIC10, Mel-SH855-C11) in TM, SN1, VF1, SN2, and VF2. Asterisks indicate lysate fractions that were directly used for functional analysis of scFv antibody fragments by ELISA.
3.3. Residue-specific and site-specific labeling of scFv molecules with fluorescent amino acids
Incorporation of 14C-leucine is an efficient method to quantify the amount of de novo synthesized antibody fragments. However, nonradioactive detection methods of cell-free synthesized proteins have their own advantages since no isotope lab and occupational safety measures are required. The open nature of cell-free reactions allows the addition of supplements in order to obtain de novo synthesized target proteins with modifications and even advanced functions. To address this issue, cell-free synthesized scFv fragments were labeled with fluorescent amino acids. For proof of principle, Mel-SH527-IIA4 was labeled in a residue-specific and site-specific manner using the precharged fluorescent amino acids BODIPY-TMR-lysine-tRNAGAA and BODIPY-TMR-lysine-tRNACUA, respectively. As a prerequisite for site-specific labeling, the amber stop codon TAG was introduced by E-PCR at the 3′-end of the ORF encoding the scFv fragment. Labeled scFv molecules were successfully visualized by in-gel fluorescence as distinct and homogenous bands showing the expected migration pattern (Fig.3). In addition, protein bands with lower molecular weight (∼17 kDa) became visible, which are supposed to originate from BODIPY-TMR-charged tRNAs. Taken together, the cotranslational labeling of scFv molecules with fluorescent amino acids allows for an easy-to-handle and nonradioactive detection method of de novo synthesized scFvs. Fluorescence-labeled scFv molecules were detected in the complex TM without the need for prior purification. Moreover, cell-free synthesized scFv molecules could be equipped with additional functions for various downstream applications using other modified amino acids.
Figure 3.
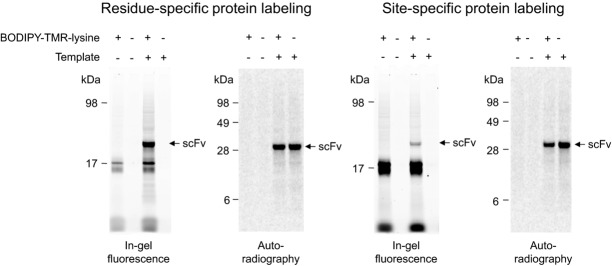
Residue-specific and site-specific labeling of Mel-SH527-IIA4 with BODIPY-TMR-lysine. Cell-free synthesis was performed in presence of 14C-leucine using the insect cell-free expression system. De novo synthesized scFv antibody fragments were visualized by in-gel fluorescence (extinction 532 nm, emission filter 580 nm bandpass, Typhoon TRIO+ Imager, GE Healthcare) and autoradiography.
3.4. Functional analysis of scFv molecules by ELISA
Antigen binding of cell-free synthesized scFv fragments was analyzed by ELISA. Biotinylated peptide antigens SMAD2, SMAD2-P, and CXCR5 were immobilized on streptavidin-coated microtiter plates. First, binding properties of scFv molecules with and without melittin signal sequence were analyzed. Lysate fractions containing Mel-SH527-IIA4, Mel-SH527-IIC10, and Mel-SH855-C11 were found to bind to their corresponding antigens (Fig.4). As expected, the phospho-specific scFv Mel-SH527-IIA4 showed specific binding to SMAD2-P, but not to SMAD2 or the unrelated antigen CXCR5. On the other hand, the phospho-unspecific scFv Mel-SH527-IIC10 was shown to bind to both, SMAD2 and SMAD2-P, but as expected, no binding to CXCR5 was detected. Likewise, Mel-SH855-C11 showed a specific binding to CXCR5 only, while no interaction with SMAD2 and SMAD2-P was measured. Although scFv antibody fragments without signal sequence were synthesized in presence of GSH and GSSG, no binding or very weak binding signals were detected by ELISA, indicating a remarkable difference in the binding capabilities of scFv candidates synthesized in the cytosol (without signal peptide) or translocated into microsomal vesicles.
Figure 4.
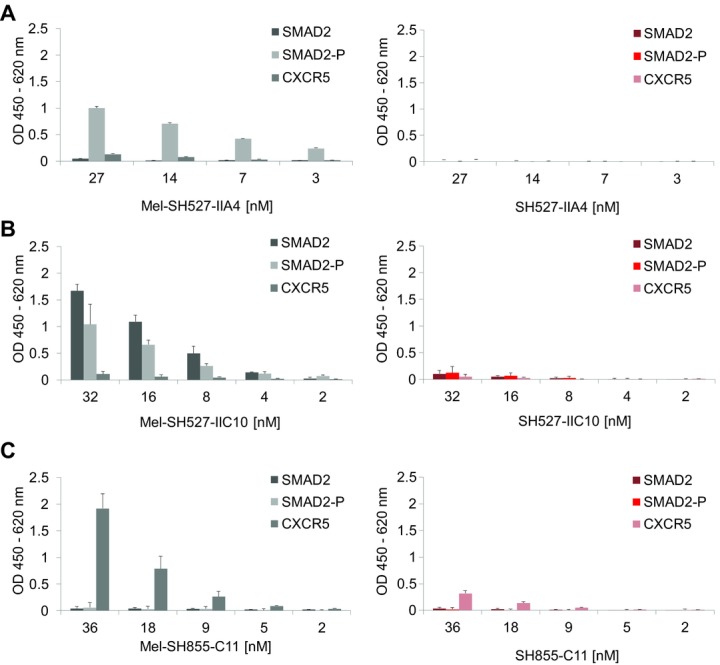
Functional analysis of cell-free synthesized scFv fragments by ELISA. Cell-free synthesis was performed using the vesicle-containing insect cell-free expression system. scFv molecules were tested for binding to their peptide antigens SMAD2, SMAD2-P, and CXCR5. (A) Binding of Mel-SH527-IIA4 vs. SH527-IIA4, (B) binding of Mel-SH527-IIC10 vs. SH527-IIC10, and (C) binding of Mel-SH855-C11 vs. SH855-C11. Lysate fractions containing scFv fragments with melittin signal sequence (Mel-SH527-IIA4, Mel-SH527-IIC10, Mel-SH855-C11) show strong binding to their corresponding antigen, whereas scFv molecules without signal sequence (SH527-IIA4, SH527-IIC10, SH855-C11) only show weak binding or no binding at all. Error bars show SDs calculated from triplicate analysis (n = 3).
3.5. Comparison of scFv fragments synthesized in insect and E. coli cell lysates
Cell-free systems based on lysates generated from E. coli have shown their potential to produce functional scFv and Fab fragments [6–8,14] as well as IgG antibodies [11]. Therefore, a comparison of the insect cell-free system with these well-established prokaryotic systems might help to evaluate the benefits and also drawbacks of the eukaryotic system applied in this study. The performance of the insect cell-free expression system used in this study was compared to a commercially available cell-free translation system based on E. coli cell lysates (EasyXpress Disulfide E. coli Kit, Qiagen). Interestingly, significant differences were observed regarding total protein yields of antibody fragments with and without signal sequence. Each of the tested scFv candidates exhibited highest yields when synthesized with signal sequence. As a result, protein yields of around 200–300 μg/mL (Fig.5A and B) were reached. In contrast, expression of the corresponding scFv candidates without signal sequence resulted in a 10- to 15-fold decrease, with total protein yields of approximately 20 μg/mL (Fig.5C). Due to these findings, melittin-fused scFv constructs were chosen as the preferred templates for expression in the redox-optimized E. coli cell-free translation system. The three model scFv fragments were expressed using the insect cell-free expression system and E. coli based cell-free expression system. Lysate fractions SN2 (insect) and SN1 (E. coli) containing soluble scFv molecules were diluted to the same scFv concentration and analyzed in parallel by ELISA. Significant differences in antigen-binding function were observed between scFv fragments made in the different cell-free translation systems. scFv fragments synthesized in E. coli cell extracts showed very weak binding or no binding at all (Fig.6). In contrast, scFv fragments synthesized in insect lysate were shown to bind to their corresponding antigens with high specificity. To exclude the possibility that the melittin signal peptide might have a negative influence on protein functionality, scFv fragments without signal sequence were synthesized in the E. coli cell-free translation system and subsequently tested for their binding capabilities. As expected, also these antibody fragments were not able to detect their corresponding antigen in the ELISA experiment (data not shown).
Figure 5.
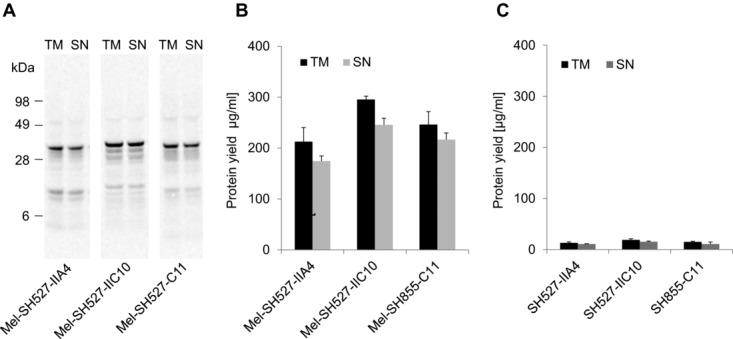
Qualitative and quantitative analyses of cell-free synthesized and 14C-leucine-labeled scFv molecules. Cell-free synthesis was performed using a commercially available cell-free transcription–translation system based on E. coli cell extracts (EasyXpress Disulfide E. coli Kit, Qiagen). (A) Autoradiograph of 14C-leucine-labeled scFvs. (B) Diagram showing total protein yields of scFv antibody fragments with melittin signal sequence in the translation mixture (TM) and supernatant fraction (SN) after centrifugation. (C) Diagram showing total protein yields of scFv antibody fragments without melittin signal sequence in the TM and supernatant fraction (SN) after centrifugation. Error bars show SDs calculated from triplicate analysis (n = 3).
Figure 6.
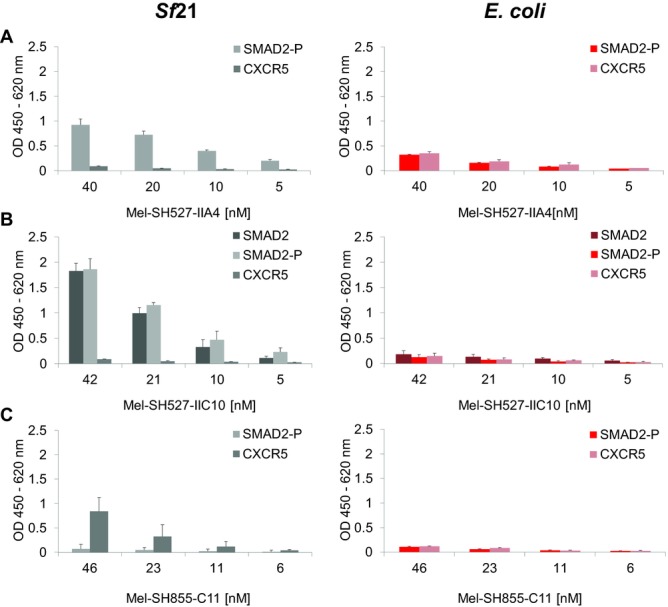
ELISA analysis of scFv molecules synthesized using the insect cell-free expression system or a commercially available E. coli cell-free expression system (EasyXpress Disulfide E. coli Kit, Qiagen). scFv molecules were tested for binding to their peptide antigens SMAD2, SMAD2-P, and CXCR5. (A) Binding of Mel-SH527-IIA4, (B) binding of Mel-SH527-IIC10, and (C) binding of Mel-SH855-C11. scFv molecules synthesized in insect cell lysate recognize their corresponding antigens with high specificity. Error bars show SDs calculated from triplicate analysis (n = 3).
4. Discussion
We demonstrate the expression of functional scFv antibody fragments in a eukaryotic in vitro translation system based on lysates derived from cultured Sf21 cells. Cell-free synthesized scFv molecules were produced rapidly within a total incubation time of 4 h in small-scaled batch reactions. Translation of target proteins was initiated by the addition of PCR products coding for the individual scFv candidates. In this way, recombinant human scFv genes were successfully expressed in the Sf21 lysate without the need for codon optimization and cloning procedures. Three model antibody fragments, each with and without Mel signal sequence, were expressed as nonaggregated and soluble proteins with production yields (∼10–15 μg/mL) sufficient for research applications and functional analysis. As expected, melittin-fused scFv molecules showed a different distribution in the insect lysate compared to non-melittin-fused scFvs, indicating their signal peptide induced translocation into the lumen of the endogenous vesicles present in the insect lysate. The translocation and enrichment of melittin-fused scFv molecules has previously been demonstrated for a fluorescein-binding scFv antibody fragment, C-terminally fused to the enhanced yellow fluorescent protein, by fluorescence imaging and confocal laser scanning microscopy [17].
In order to facilitate disulfide bond formation in de novo synthesized target proteins, all buffers and the insect lysate itself were produced in the absence of the reducing agent DTT. The open nature of cell-free reactions allows the addition of further supplements, such as defined mixtures of GSH and GSSG. scFv fragments without signal sequence were expressed in presence of 2.5 mM GSSG and 0.5 mM GSH, in order to support the formation of intramolecular disulfide bonds. This particular ratio has been shown to be well-suited to allow the production of a functional scFv antibody fragment binding to fluorescein and fluorescein derivatives [17]. The antibody fragments SH527-IIA4, SH527-IIC10, and SH855-C11 were expressed applying similar conditions, but binding to their corresponding antigens was not detected. In contrast, scFv fragments fused to a melittin signal sequence were found to be highly functional as determined by antigen ELISA. Functional analysis of scFv molecules was possible without the need for further purification. On the basis of the presented autoradiographs (Fig.1A), it can be assumed that the melittin sequence of the analyzed scFv antibody fragments may not be cleaved off by endogenous signal peptidases, because scFv fragments with melittin signal sequence appear to have a slightly higher apparent molecular weight compared to their counterparts without signal sequence. However, since these antibody fragments were shown to be highly functional, a potential negative influence of the melittin signal peptide (if not cleaved off) on protein functionality can be excluded.
Various strategies are available to adjust the redox potential in the cytosol, in order to achieve functional scFv fragments even without translocation into ER-derived vesicles. For example, the ratio of reduced and oxidized glutathione could be varied or one could add exogenous enzymes and folding helpers to the lysate as already successfully shown for E. coli-based cell-free translation systems [6,8,14]. In this study, addition of 2.5 mM GSSG and 0.5 mM GSH did not result in positive effects, while the significant differences in the functionality of translocated and nontranslocated antibody fragments illustrated the beneficial effect of insect vesicles on the disulfide bond formation process in vitro, most probably by vastly facilitating the protein folding. During lysate preparation, the ER of the insect cells is disrupted; membrane structures rearrange and form smaller compartments, designated as microsomal vesicles. It can be assumed that ER-resident membrane-associated and luminal proteins (e.g. ER-oxidoreductin-1 and protein disulfide isomerase), which are involved in the formation and reshuffling of disulfide bonds [39], are maintained inside the vesicles in an active state, as well as folding helpers (e.g. binding immunoglobulin protein, BiP) originating from the lumen of the ER.
In this study, scFv antibody fragments were additionally expressed in a commercially available cell-free protein synthesis system derived from E. coli, which was especially optimized for the synthesis of disulfide-bonded proteins. Using this system, expression of scFv fragments with melittin signal sequence resulted in significantly increased protein yields compared to their counterparts without signal sequence. Translation initiation is supposed to be the rate-limiting step in protein synthesis [40,41]. Thus, it can be speculated that the presence of the melittin signal sequence at the 5′ end of the target gene stabilizes the initiation activity and translational processivity, resulting in significantly increased protein yields. This interpretation is also supported by a publication, in which the authors have shown the stimulatory effect of signal sequences on protein synthesis efficiency in an E. coli-based cell-free protein synthesis system [42]. Interestingly, the stimulatory effect of the signal sequence was mainly dependent on the identity of its second codon. In particular, in most effective signal sequences, position +2 was occupied by the codon AAA [42], an observation that also accounts for the melittin signal sequence that was applied in this study [33]. Although melittin-fused antibody fragments could be produced in sufficient amounts, only very weak binding or no binding at all was detected for these scFv molecules. This result was rather unexpected, since others have already shown the production of functional scFv antibody fragments [6,8] and scFv fusion proteins [12,13] in cell-free translation systems generated from E. coli. The failure of the E. coli-based cell-free translation system for the scFv antibody fragments analyzed in this study is not completely understood. It can be speculated that the scFv fragments analyzed in this study may just be an exception and the environment in the E. coli based system was not sufficient for efficient folding. Furthermore, it might be possible that functional scFv antibody fragments have been produced, but the fraction was just too small in comparison to unfolded and nonfunctional antibody fragments to be detected in the ELISA experiment. Since both scFv fragments with and without melittin signal sequence did not show specific binding in the ELISA experiment, a negative influence of the melittin signal peptide on protein functionality could be excluded.
E. coli based cell-free translation systems are very popular and widely used because of several reasons. (i) For instance, the organism itself is well-characterized and thus, biochemical pathways are elucidated and genetic modification approaches are well-understood. (ii) Cell fermentation is inexpensive and lysate preparation is well established and easy. (iii) Cell-free systems based on E. coli lysates are most productive, reaching protein yields from hundreds of micrograms to milligrams per milliliter [43], of course depending on the protein type to be expressed and the reaction format that is used. On the other hand, eukaryotic cell-free systems have some advantages when it comes to the expression of more complex eukaryotic proteins and proteins that require posttranslational modifications [44,45] (for a comparison of state-of-the-art cell-free protein synthesis systems, see [9]). In addition, since eukaryotic translation systems are naturally free of endotoxins, de novo synthesized target proteins could potentially be applied directly in cell-based assays, without the need for time and labor-intensive purification strategies. We addressed this point by using a eukaryotic translation system based on Sf21 cells for the expression of antibody fragments. Of course, scFv antibody fragments represent a rather small antibody format, but taking this type of molecule as advance guard, we may assume the usefulness of the system also for other and more sophisticated formats of recombinant antibodies. This assumption may be strengthened by the fact that the vesicle-containing Sf21 cell-free system has been shown to be capable of producing functional tissue-type plasminogen activator, a complex eukaryotic protein bearing multiple disulfide bonds [46].
The possibility to label proteins cotranslationally is one of the outstanding advantages of cell-free protein synthesis systems [47]. In this study, residue-specific and site-specific labeling of scFv molecules with the fluorescent amino acid BODIPY-TMR-lysine was achieved, demonstrating the potential of the system to cotranslationally equip a protein with an additional chemical function. Residue-specific incorporation of noncanonical amino acids leads to heterogeneously labeled proteins, bearing their labels at varying and multiple positions. This technology is beneficial for the introduction of a high number of, e.g., fluorescence-labeled amino acids per scFv molecule. However, negative effects on the functionality of the protein cannot be excluded, possibly compromising the biological, physical, and pharmacological properties [48]. This can be circumvented by applying site-specific labeling techniques that allow the introduction of a noncanonical amino acid at just one defined position, carefully chosen to avoid negative effects on protein function. The site-specific incorporation of a noncanonical amino acid to the heavy chains of an IgG antibody, and the subsequent conjugation with a chemotherapeutic small molecule, has recently been shown. These experiments were performed using mammalian cells [48] as well as an E. coli based cell-free translation system [49]. Moreover, site-specific labeling approaches by means of an orthogonal amber suppressor tRNA–synthetase pair have also been exploited by using the vesicle-containing insect cell-free translation system, thereby enabling the synthesis of site-specifically labeled erythropoietin [50].
In summary, the presented results demonstrate the potential of the vesicle-containing insect cell-free expression system to serve as a general platform for the production of highly functional antibody fragments. The capability of the vesicle-containing insect cell-free expression system to produce even complex eukaryotic proteins, such as posttranslationally modified proteins and membrane proteins [25,27,28,46,51–53], promises its successful utilization for the synthesis of more complex antibody formats, such as full-length antibodies, and antibody fusion proteins. In addition, the use of batch-based translation reactions offers the particular advantages of a scalable system, amenable for miniaturization and large-scale production (e.g. antibody production in 4 L scale [11] and cytokine production in 100 L scale [54]), as well as automation and high-throughput protein synthesis. Combination of these features with cotranslational labeling approaches results in an extremely potent, rapid, and versatile biochemical engineering and manufacturing technology, which might be the future source for tailor-made, functionalized antibodies.
Practical application
Open cell-free translation systems have shown their potential to produce functionally active target proteins, including recombinant antibody formats. Antibody fragments synthesized in a eukaryotic translation system based on cultured insect cells are shown to be highly functional. Since these proteins are synthesized in an endotoxin-free environment, they could be directly applied in cell-based assays. The combination of cell-free protein synthesis and the cotranslational labeling of target proteins with noncanonical amino acids accelerates the production of antibody fragments with advanced functions. Modified molecules could potentially be applied as versatile detection reagents, e.g. in fluorescence-activated cell sorting, ELISA, and fluorescence microscopy.
Acknowledgments
The authors would like to thank Doreen Wüstenhagen, Conny Mascher, and Dana Wenzel for cell cultivation and insect lysate preparation (Fraunhofer IZI, Potsdam-Golm, Germany). Furthermore, we thank Jörg Schenk (Hybrotec GmbH, UP Transfer GmbH, Potsdam-Golm, Germany) for provision of the monoclonal anti-c-myc-tag antibody 9E10. Moreover, we would like to gratefully acknowledge the contribution of Saskia Helmsing (Technische Universität Braunschweig, Germany) for the production of scFv-Fc binders that were used as positive controls. In addition, we thank Dr. Felix Oden (Max-Delbrück-Center for Molecular Medicine, Berlin, Germany) for his collaboration regarding the CXCR5-binding scFv antibody fragment SH855-C11. Special thanks belong to Marie Burger (Fraunhofer IZI, Potsdam-Golm, Germany) for the careful revision of this manuscript. This research is supported by the German Ministry of Education and Research (BMBF Nos. 0312039 and 0315942) and EU FP7 program “Affinomics.”
Glossary
- CXCR5
CXC chemokine receptor type 5
- DDM
n-dodecyl-β-maltoside
- E-PCR
expression-PCR
- f.c.
final concentration
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- Mel
melittin
- mRNA
messenger RNA
- NTC
no template control
- PBST
PBS Tween
- RT
room temperature
- scFv
single-chain antibody variable fragment
- Sf21
Spodoptera frugiperda
- SMAD
mothers against decapentaplegic homolog 2
- SN1
supernatant 1
- SN2
supernatant 2
- TCA
trichloroacetic acid
- TM
translation mixture
- VF1
vesicular fraction 1
The authors have declared no conflict of interest.
Additional supporting information may be found in the online version of this article at the publisher's web-site
5 References
- [1].Weisser NE, Hall JC. Applications of single-chain variable fragment antibodies in therapeutics and diagnostics. Biotechnol. Adv. 2009;27:502–520. doi: 10.1016/j.biotechadv.2009.04.004. [DOI] [PubMed] [Google Scholar]
- [2].Colwill K, Graslund S. A roadmap to generate renewable protein binders to the human proteome. Nat. Meth. 2011;8:551–558. doi: 10.1038/nmeth.1607. [DOI] [PubMed] [Google Scholar]
- [3].Hagemeyer C, von Zur Muhlen C, von Elverfeldt D, Peter K. Single-chain antibodies as diagnostic tools and therapeutic agents. Thromb. Haemost. 2009;101:1012–1019. [PubMed] [Google Scholar]
- [4].Bundy BC, Swartz JR. Efficient disulfide bond formation in virus-like particles. J. Biotechnol. 2011;154:230–239. doi: 10.1016/j.jbiotec.2011.04.011. [DOI] [PubMed] [Google Scholar]
- [5].Kim D-M, Swartz JR. Efficient production of a bioactive, multiple disulfide-bonded protein using modified extracts of Escherichia coli. Biotechnol. Bioeng. 2004;85:122–129. doi: 10.1002/bit.10865. [DOI] [PubMed] [Google Scholar]
- [6].Merk H, Stiege W, Tsumoto K, Kumagai I. Cell-free expression of two single-chain monoclonal antibodies against lysozyme: Effect of domain arrangement on the expression. J. Biochem. (Tokyo) 1999;125:328–333. doi: 10.1093/oxfordjournals.jbchem.a022290. [DOI] [PubMed] [Google Scholar]
- [7].Oh I-S, Lee J-C, Lee M-S, Chung J-H. Cell-free production of functional antibody fragments. Bioproc. Biosyst. Eng. 2010;33:127–132. doi: 10.1007/s00449-009-0372-3. [DOI] [PubMed] [Google Scholar]
- [8].Ryabova LA, Desplancq D, Spirin AS, Plückthun A. Functional antibody production using cell-free translation: effects of protein disulfide isomerase and chaperones. Nat. Biotechnol. 1997;15:79–84. doi: 10.1038/nbt0197-79. [DOI] [PubMed] [Google Scholar]
- [9].Carlson ED, Gan R, Hodgman CE, Jewett MC. Cell-free protein synthesis: Applications come of age. Biotechnol. Adv. 2012;30:1185–1194. doi: 10.1016/j.biotechadv.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rungpragayphan S, Nakano H, Yamane T. PCR-linked in vitro expression: A novel system for high-throughput construction and screening of protein libraries. FEBS Lett. 2003;540:147–150. doi: 10.1016/s0014-5793(03)00251-5. [DOI] [PubMed] [Google Scholar]
- [11].Yin G, Garces ED, Yang J, Zhang J. Aglycosylated antibodies and antibody fragments produced in a scalable in vitro transcription-translation system. MAbs. 2012;4:217–225. doi: 10.4161/mabs.4.2.19202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kanter G, Yang J, Voloshin A, Levy S. Cell-free production of scFv fusion proteins: An efficient approach for personalized lymphoma vaccines. Blood. 2007;109:3393–3399. doi: 10.1182/blood-2006-07-030593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Patel KG, Ng PP, Levy S, Levy R. Escherichia coli-based production of a tumor idiotype antibody fragment—Tetanus toxin fragment C fusion protein vaccine for B cell lymphoma. Protein Expres. Purif. 2011;75:15–20. doi: 10.1016/j.pep.2010.09.005. [DOI] [PubMed] [Google Scholar]
- [14].Jiang X, Ookubo Y, Fujii I, Nakano H. Expression of Fab fragment of catalytic antibody 6D9 in an Escherichia coli in vitro coupled transcription/translation system. FEBS Lett. 2002;514:290–294. doi: 10.1016/s0014-5793(02)02383-9. [DOI] [PubMed] [Google Scholar]
- [15].Kawasaki T, Gouda MD, Sawasaki T, Takai K. Efficient synthesis of a disulfide-containing protein through a batch cell-free system from wheat germ. Eur. J. Biochem. 2003;270:4780–4786. doi: 10.1046/j.1432-1033.2003.03880.x. [DOI] [PubMed] [Google Scholar]
- [16].Merk H, Gless C, Maertens B, Gerrits M. Cell-free synthesis of functional and endotoxin-free antibody Fab fragments by translocation into microsomes. BioTechniques. 2012;53:153–160. doi: 10.2144/0000113904. [DOI] [PubMed] [Google Scholar]
- [17].Stech M, Merk H, Schenk J, Stöcklein W. Production of functional antibody fragments in a vesicle-based eukaryotic cell-free translation system. J. Biotechnol. 2012;164:220–231. doi: 10.1016/j.jbiotec.2012.08.020. [DOI] [PubMed] [Google Scholar]
- [18].Alzari P, Lascombe M, Poljak R. Three-dimensional structure of antibodies. Annu. Rev. Immunol. 1988;6:555–580. doi: 10.1146/annurev.iy.06.040188.003011. [DOI] [PubMed] [Google Scholar]
- [19].Davies D, Padlan E, Sheriff S. Antibody-antigen complexes. Annu. Rev. Biochem. 1990;59:439–473. doi: 10.1146/annurev.bi.59.070190.002255. [DOI] [PubMed] [Google Scholar]
- [20].Glockshuber R, Schmidt T, Plückthun A. The disulfide bonds in antibody variable domains: Effects on stability, folding in vitro, and functional expression in Escherichia coli. Biochemistry. 1992;31:1270–1279. doi: 10.1021/bi00120a002. [DOI] [PubMed] [Google Scholar]
- [21].Goto Y, Hamaguchi K. The role of the intrachain disulfide bond in the conformation and stability of the constant fragment of the immunoglobulin light chain. J. Biochem. 1979;86:1433–1441. doi: 10.1093/oxfordjournals.jbchem.a132661. [DOI] [PubMed] [Google Scholar]
- [22].Brödel AK, Raymond JA, Duman JG, Bier FF. Functional evaluation of candidate ice structuring proteins using cell-free expression systems. J. Biotechnol. 2013;163:301–310. doi: 10.1016/j.jbiotec.2012.11.001. [DOI] [PubMed] [Google Scholar]
- [23].Bird R, Hardman K, Jacobsen J, Johnson S. Single-chain antigen-binding proteins. Science. 1988;242:423–426. doi: 10.1126/science.3140379. [DOI] [PubMed] [Google Scholar]
- [24].Huston J, Levinson D, Mudgett H, Tai M. Protein engineering of antibody binding sites: Recovery of specific activity in an anti-digosin single-chain Fv analogue produced in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1988;85:5879–5883. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kubick S, Gerrits M, Merk H, Stiege W. In vitro synthesis of posttranslationally modified membrane proteins. In: DeLucas L, editor. “Membrane Protein Crystallization” Current Topics in Membranes. Burlington: Elsevier; 2009. pp. 25–49. in: (Ed.), et al.,, Chapter 2. [Google Scholar]
- [26].Kubick S, Schacherl J, Fleischer-Notter H, Royall E. In vitro translation in an insect-based cell-free system. In: Swartz JR, editor. Cell-Free Protein Expression. Berlin, Heidelberg/New York: Springer; 2003. pp. 209–217. in: (Ed.), et al. [Google Scholar]
- [27].Sachse R, Wüstenhagen D, Šamalíková M, Gerrits M. Synthesis of membrane proteins in eukaryotic cell-free systems. Eng. Life Sci. 2013;13:39–48. [Google Scholar]
- [28].Stech M, Brödel AK, Quast RB, Sachse R. Advances in Biochemical Engineering/Biotechnology. Berlin Heidelberg: Springer; 2013. pp. 67–102. [DOI] [PubMed] [Google Scholar]
- [29].Brödel A, Sonnabend A, Roberts L, Stech M. IRES-mediated translation of membrane proteins and glycoproteins in eukaryotic cell-free systems. PLoS One. 2013;8:e82234. doi: 10.1371/journal.pone.0082234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hust M, Meyer T, Voedisch B, Rülker T. A human scFv antibody generation pipeline for proteome research. J. Biotechnol. 2011;152:159–170. doi: 10.1016/j.jbiotec.2010.09.945. [DOI] [PubMed] [Google Scholar]
- [31].Schirrmann T, Hust M. Construction of human antibody gene libraries and selection of antibodies by phage display. Methods Mol. Biol. 2010;651:177–209. doi: 10.1007/978-1-60761-786-0_11. [DOI] [PubMed] [Google Scholar]
- [32].Merk H, Meschkat D, Stiege W. Expression-PCR: From gene pools to purified proteins within 1 day. In: Swartz JR, editor. Cell-Free Protein Expression. Berlin Heidelberg/New York: Springer; 2003. pp. 15–23. in: (Ed.) [Google Scholar]
- [33].Tessier DC, Thomas DY, Khouri HE, Laliberié F. Enhanced secretion from insect cells of a foreign protein fused to the honeybee melittin signal peptide. Gene. 1991;98:177–183. doi: 10.1016/0378-1119(91)90171-7. [DOI] [PubMed] [Google Scholar]
- [34].Hust M, Steinwand M, Al-Halabi L, Helmsing S. Improved microtitre plate production of single chain Fv fragments in Escherichia coli. New Biotechnol. 2009;25:424–428. doi: 10.1016/j.nbt.2009.03.004. [DOI] [PubMed] [Google Scholar]
- [35].Mollova S, Retter I, Hust M, Dübel S. Analysis of single chain antibody sequences using the VBASE2 Fab analysis tool. In: Kontermann R, editor. Antibody Engineering. Berlin Heidelberg: Springer; 2010. pp. 3–10. in: (Ed.), et al. [Google Scholar]
- [36].Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: Coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- [37].Micheel B, Jantscheff P, Böttger V, Scharte G. The production and radioimmunoassay application of monoclonal antibodies to fluorescein isothiocyanate (FITC) J. Immunol. Methods. 1988;111:89–94. doi: 10.1016/0022-1759(88)90063-4. [DOI] [PubMed] [Google Scholar]
- [38].Schenk JA, Sellrie F, Böttger V, Menning A. Generation and application of a fluorescein-specific single chain antibody. Biochimie. 2007;89:1304–1311. doi: 10.1016/j.biochi.2007.06.008. [DOI] [PubMed] [Google Scholar]
- [39].Tu T, Weissman J. Oxidative protein folding in eukaryotes mechanisms and consequences. J. Cell Biol. 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kierzek AM, Zaim J, Zielenkiewicz P. The effect of transcription and translation initiation frequencies on the stochastic fluctuations in prokaryotic gene expression. J. Biol. Chem. 2001;276:8165–8172. doi: 10.1074/jbc.M006264200. [DOI] [PubMed] [Google Scholar]
- [41].Sonenberg N, Hinnebusch A. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ahn J-H, Hwang M-Y, Lee K-H, Choi C-Y. Use of signal sequences as an in situ removable sequence element to stimulate protein synthesis in cell-free extracts. Nucleic Acids Res. 2007;35:e21. doi: 10.1093/nar/gkl917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kigawa T, Yabuki T, Yoshida Y, Tsutsui M. Cell-free production and stable-isotope labeling of milligram quantities of proteins. FEBS Lett. 1999;442:15–19. doi: 10.1016/s0014-5793(98)01620-2. [DOI] [PubMed] [Google Scholar]
- [44].Tarui H, Murata M, Tani I, Imanishi S. Establishment and characterization of translation/glycosylation in insect cell (Spodoptera frugiperda 21) extract prepared with high pressure treatment. Appl. Microbiol. Biotechnol. 2001;55:446–453. doi: 10.1007/s002530000534. [DOI] [PubMed] [Google Scholar]
- [45].Vinarov DA, Lytle BL, Peterson FC, Tyler EM. Cell-free protein production and labeling protocol for NMR-based structural proteomics. Nat. Meth. 2004;1:149–153. doi: 10.1038/nmeth716. [DOI] [PubMed] [Google Scholar]
- [46].Stech M, Quast RB, Sachse R, Schulze C, Wüstenhagen DA, Kubick S. A continuous-exchange cell-free protein synthesis system based on extracts from cultured insect cells. PLoS One. 2014:e96635. doi: 10.1371/journal.pone.0096635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Shimizu Y, Kuruma Y, Ying B-W, Umekage S. Cell-free translation systems for protein engineering. FEBS J. 2006;273:4133–4140. doi: 10.1111/j.1742-4658.2006.05431.x. [DOI] [PubMed] [Google Scholar]
- [48].Axup JY, Bajjuri KM, Ritland M, Hutchins BM. Synthesis of site-specific antibody-drug conjugates using unnatural amino acids. Proc. Natl. Acad. Sci. 2012;109:16101–16106. doi: 10.1073/pnas.1211023109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zimmerman ES, Heibeck, Gill TH, Li A. Production of site-specific antibody-drug conjugates using optimized non-natural amino acids in a cell-free expression system. Bioconjugate Chem. 2014;25:351–361. doi: 10.1021/bc400490z. X. [DOI] [PubMed] [Google Scholar]
- [50].Quast RB, Claussnitzer I, Merk H, Kubick S. Synthesis and site-directed fluorescence labeling of azido proteins using eukaryotic cell-free orthogonal translation systems. Anal. Biochem. 2014;451:4–9. doi: 10.1016/j.ab.2014.01.013. [DOI] [PubMed] [Google Scholar]
- [51].Kumar Dondapati S, Kreir M, Quast RB, Wüstenhagen DA. Membrane assembly of the functional KcsA potassium channel in a vesicle-based eukaryotic cell-free translation system. Biosens. Bioelectron. 2014;59:174–183. doi: 10.1016/j.bios.2014.03.004. [DOI] [PubMed] [Google Scholar]
- [52].Fenz SF, Sachse R, Schmidt T, Kubick S. Cell-free synthesis of membrane proteins: Tailored cell models out of microsomes. Biochim. Biophys. Acta. 2014;1838:1382–1388. doi: 10.1016/j.bbamem.2013.12.009. [DOI] [PubMed] [Google Scholar]
- [53].Shaklee PM, Semrau S, Malkus M, Kubick S. Protein incorporation in giant lipid vesicles under physiological conditions. ChemBioChem. 2010;11:175–179. doi: 10.1002/cbic.200900669. [DOI] [PubMed] [Google Scholar]
- [54].Zawada JF, Yin G, Steiner AR, Yang J. Microscale to manufacturing scale-up of cell-free cytokine production—A new approach for shortening protein production development timelines. Biotechnol. Bioeng. 2011;108:1570–1578. doi: 10.1002/bit.23103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


