Abstract
HSPA9 is located on chromosome 5q31.2 in humans, a region that is commonly deleted in patients with myeloid malignancies [del(5q)], including myelodysplastic syndromes (MDS). HSPA9 expression is reduced by 50% in patients with del(5q)-associated MDS, consistent with haploinsufficient levels. Zebrafish mutants and knockdown studies in human and mouse cells have implicated a role for HSPA9 in hematopoiesis. To comprehensively evaluate the effects of Hspa9 haploinsufficiency on hematopoiesis, we generated an Hspa9 knockout mouse model. While homozygous knockout of Hspa9 is embryonic lethal, mice with heterozygous deletion of Hspa9 (Hspa9+/−) are viable and have a 50% reduction in Hspa9 expression. Hspa9+/− mice have normal basal hematopoiesis and do not develop MDS. However, Hspa9+/− mice have a cell- intrinsic reduction in bone marrow CFU-PreB colony formation without alterations in the number of B-cell progenitors in vivo, consistent with a functional defect in Hspa9+/− B-cell progenitors. We further reduced Hspa9 expression (<50%) using RNAi and observe reduced B-cell progenitors in vivo, indicating that appropriate levels (≥50%) of Hspa9 are required for normal B- lymphopoiesis in vivo. Knockdown of Hspa9 in an IL-7 dependent mouse B-cell line reduced Stat5 phosphorylation following IL-7 receptor stimulation, supporting a role for Hspa9 in Stat5 signaling in B-cells. Collectively, these data implicate a role for Hspa9 in B-lymphopoiesis and Stat5 activation downstream of IL-7 signaling.
Keywords: Hspa9, hematopoiesis, knockout mouse, B-cell, Stat5
INTRODUCTION
HSPA9 (GRP75/mortalin/mtHSP70) is a member of the highly conserved HSP70 family of proteins [1]. Across species, HSP70 family members play an essential role in protein homeostasis and cellular stress response [1,2]. Overexpression of HSP70 proteins results in increased cell survival and is associated with a multitude of cancers, while reduced expression increases apoptosis and susceptibility to cellular stresses [2,3]. Members of this family share a common protein binding function; however, unique expression patterns, subcellular localization and co-chaperone interactions have diversified the functions of the 8 well-described Hsp70 proteins expressed in mouse and human. To date, knockout mice have been described for 5 Hsp70 family members: Hspa1a, Hspa1b, Hspa2, Hspa5 and Hspa8 [1]. Homozygous knockout in mice is embryonic lethal for Hspa5 and Hspa8 [4,5].
Several HSP70 family proteins influence hematopoiesis through a variety of mechanisms, including their regulatory roles in cell signaling, cell cycle and glycolytic metabolism [6–11]. A role for HSPA9, the only HSP70 protein localized in the mitochondria, in hematopoiesis has been highlighted using multiple models. Zebrafish homozygous for a point mutation that results in a hypomorphic allele suffer from severe anemia and have defects in erythroid differentiation with increased apoptosis [12,13]. HSPA9 has also been implicated in erythropoiesis as a downstream mediator of erythropoietin signaling in a primary human CD34+ cell culture system [14]. RNAi-mediated knockdown of HSPA9 in primary human CD34+ cells and Hspa9 in a murine bone marrow transplant model disrupted erythroid differentiation, reduced mature B-cell numbers, reduced cellular proliferation, and increased apoptosis [15]. In a separate study, increased reactive oxygen species and reduced quiescence in hematopoietic stem cells was observed following RNAi-mediated knockdown of Hspa9 in a mouse bone marrow transplant model [16]. Finally, HSPA9 lies within a commonly deleted region affecting one allele of human chromosome 5 (del[5q]) that is lost in patients with myeloid malignancies [17–20]. Consistent with heterozygous loss, HSPA9 mRNA expression is reduced by 50% in del(5q)-associated myelodysplastic syndrome (MDS) compared to non-del(5q) MDS and normal CD34+ cells, suggesting that haploinsufficiency may contribute to disease pathogenesis [21].
In order to precisely model Hspa9 haploinsufficiency, we created a novel Hspa9 knockout mouse model. Homozygous knockout of Hspa9 is embryonic lethal, but heterozygous knockout mice (Hspa9+/−) are viable and express 50% reduced levels of Hspa9 compared to wild-type mice. While Hspa9+/− mice do not develop MDS or leukemia and have normal erythropoiesis, we establish a role for Hspa9 in B-lymphopoiesis through attenuation of Stat5 activation.
MATERIAL AND METHODS
Generation of Hspa9+/− mice
Normal karyotype C57Bl/6N embryonic stem (ES) cell clones containing a gene trap insertion in the third intron of Hspa9 (Hspa9+/Gt(IST14901H6)TIGM) were obtained from the Texas A&M Institute for Genomic Medicine [22]. Mice were maintained on a C57Bl/6N background, unless otherwise indicated. Southern blots were performed using standard methods. PCR genotyping was performed as described previously and genotyping primer sequences are listed in Table S2 [22]. Mouse procedures were performed according to protocols approved by the Washington University Animal Studies Committee.
Western blot and RT-PCR analysis
Western blots were performed as previously described [15,23]. Antibodies used are described in Table S1. RNA was isolated using TRIzol LS reagent (Invitrogen), DNase treated (TURBO DNA-free kit, Ambion), converted to cDNA (SuperScript III First-Strand Synthesis System, Invitrogen), and analyzed using a StepOne Plus Real time PCR System (Applied Biosystems). Individual cDNA samples were normalized according to their levels of Gapdh or β-Actin. The relative standard curve method was used for analysis. Primers used are listed in Table S2.
Flow cytometry
Bone marrow, spleen or peripheral blood cells were isolated by standard methods and red cells were lysed with ACK lysis buffer. Antibodies used are listed in Table S1. For intracellular flow, cells were fixed in 2% paraformaldehyde and permeabilized in 100% methanol prior to intracellular staining. Fluorescent detection was performed by FACScan or Gallios cytometers (BD biosciences) and analyzed using FlowJo software. Cells were isolated using an iCyt Synergy flow sorter (Sony) or Beckman Coulter MoFlo.
Colony forming assays
Bone marrow or spleen cells were harvested, red cell lysed, and plated in methylcellulose media (Stem Cell Technologies) to detect CFU-C/CFU-E/BFU-E (M3434), mature BFU-E (M3234 supplemented with 3 or 6U/mL hEPO), and CFU-PreB (M3630) colonies per standard protocols, unless otherwise indicated. CFU-C and CFU-PreB colonies were scored on day 7–10. BFU-E/mature BFU-E colonies were scored on day 10–11. CFU-E colonies were scored on day 3 following benzidine staining.
Viral preparation, transduction, and bone marrow transplantation
HSPA9 cDNA was cloned from the previously described FLAG-Mortalin-WT vector into an MSCV-IRES-GFP vector [24]. For virus production, HEK293T cells were transfected with MSCV and EcoPack packaging plasmids by calcium phosphate transfection using CalPhos Mammalian Transfection Kit (Clontech) and viral supernatant was collected. Fcy-si control (shLUC) and Hspa9 knockdown (shHspa9 #3) shRNAs were generated as previously described [15]. For HSPA9 overexpression, an MOI of 1 was used. For Fcy-si experiments, lineage-depleted Ly5.2 bone marrow cells were transduced once with an MOI of 20 by spinoculation, as previously described [15]. On the following day, 0.75–1×106 cells were transplanted into lethally irradiated recipients (Ly5.1).
Microarray analysis
Cells were isolated from CFU-PreB media on day 7 and B220+ cells were sorted directly into TRIzol LS Reagent. Samples were hybridized to Mouse Gene 1.0 ST arrays and gene-level normalization and signal summarization was performed for all arrays using Expression Console (Affymetrix). Differentially expressed individual genes were identified using Significance Analysis of Microarrays (SAM) [25]. Pathway analysis was performed on significantly altered genes using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) [26,27]. Microarray data will be deposited in GEO.
Cell culture
B7 (Ba/F3 cells stably transduced with MSCV-mIL7R-IRES-GFP) cells were maintained at a density <2 million cells/mL in media with 10ng/mL mIL-7 (Peprotech), as previously described [28]. B7 cells were electroporated with 1000nM Hspa9-targeting siRNAs (Thermo Scientific: D-057872-03 [siRNA 1] and D-057872-04 [siRNA 2]) or non-targeting control siRNA (Thermo Scientific: D-001206-14) using a Nucleofector Device (Lonza) program X-001 according to manufacturer’s protocols.
Statistical analysis and visualization
Statistical analysis and graphing was performed using Prism (GraphPad).
RESULTS
Generation of Hspa9+/− mice
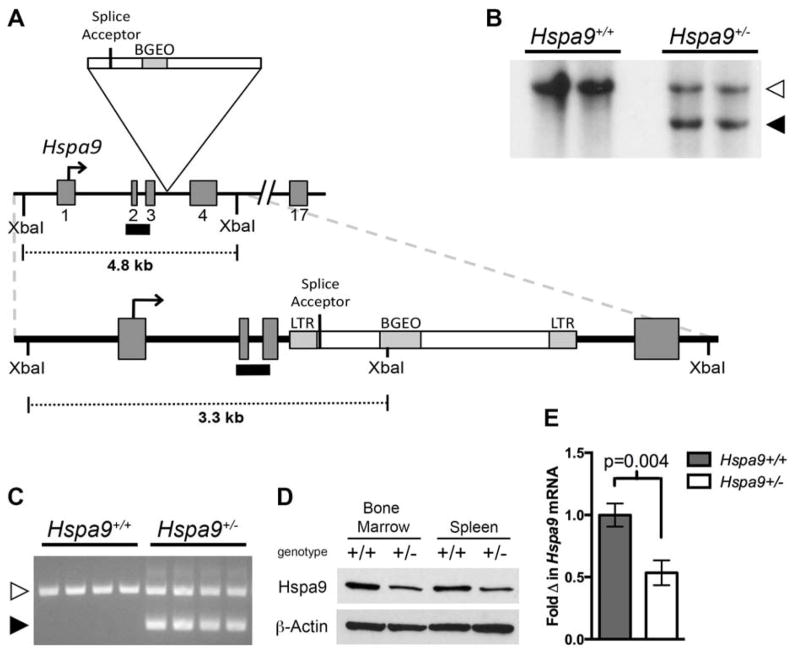
In order to evaluate the effect of Hspa9 deletion on hematopoiesis in vivo, we created a mouse model with heterozygous inactivation of Hspa9 (Hspa9Gt(IST14901H6)TIGM, hereafter referred to as Hspa9+/−) using ES cells containing a gene trap inserted in intron 3 of Hspa9 (Fig. 1A). The gene trap insertion was confirmed by Southern blot and PCR (Fig. 1B and C). Heterozygous loss of Hspa9 resulted in ~50% reduction in Hspa9 protein (Fig. 1D and Fig. S1) and mRNA (Fig. 1E) levels in Hspa9+/− mice compared to wild-type littermates.
Figure 1. Generation of Hspa9+/− mice.
A) Exons 1–4 of the Hspa9 locus with gene trap insertion. A black bar indicates the Southern blot probe. B) Southern blot of tail DNA from Hspa9+/+ and Hspa9+/− mice showing DNA fragments from a wild-type allele (white arrow, 4817 bp) and gene trap-disrupted allele (black arrow, 3303 bp). C) Results of 3 primer PCR amplification of tail DNA from Hspa9+/+ and Hspa9+/− mice showing a band from the wild-type allele (white arrow, 453 bp) and gene trap-disrupted allele (black arrow, 307 bp). D) Expression of Hspa9 in bone marrow and spleen of littermates by C-terminal antibody with β-Actin loading control. E) RT-PCR expression of Hspa9 mRNA in bone marrow of littermates, normalized to Gapdh (N=3/genotype). Statistical analysis by two tailed Student’s t-test. Error bars represent mean ± SD.
Hspa9+/− mice are born at normal Mendelian ratios from Hspa9+/− x Hspa9+/+ matings (N=261) (Table 1). However, intercrossing of Hspa9+/− mice did not produce homozygous mice (Hspa9−/−), suggesting homozygous inactivation of Hspa9 is embryonic lethal, independent of strain (N=73, C57Bl/6N background; N=139, outbred B6129F2 mice) (Table 1). Timed matings from Hspa9+/− x Hspa9+/− intercrosses (pure or mixed strain) failed to identify Hspa9−/− embryos after embryonic day 9.5, preventing analysis of hematopoietic cells from Hspa9−/− fetal livers (Table S3).
Table 1.
Homozygous deletion of Hspa9 is embryonic lethal
| Genotype | Hspa9+/+ x Hspa9+/− | Hspa9+/− x Hspa9+/− † | |
|---|---|---|---|
| Observed (Expected)
|
|||
| Observed (Expected) | C57Bl6/N | B6129F2 | |
| Hspa9+/+ | 121 (130.5) | 30 (18.25) | 52 (34.75) |
| Hspa9+/− | 140 (130.5) | 43 (36.5) | 87 (69.5) |
| Hspa9−/− | 0 (0) | 0 (18.25)* | 0 (34.75)* |
|
| |||
| total | 261 | 73 | 139 |
p<0.0001
C57Bl/6N Hspa9+/− mice were outcrossed to wild-type 129X1/SvJ mice (Jax: 000691) to generate Hspa9+/− B6129F1.
Basal and stress hematopoiesis is normal in Hspa9+/− mice up to 18 months of age
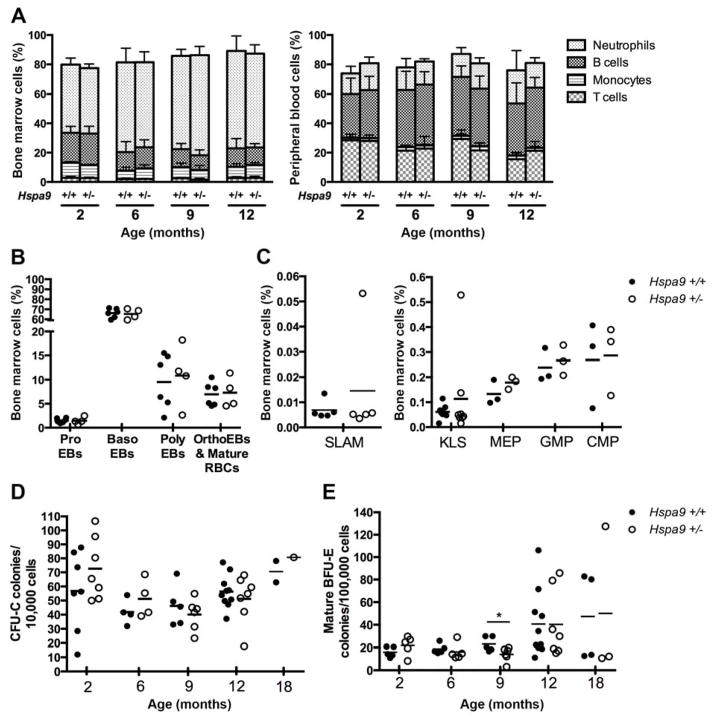
C57Bl/6N Hspa9+/− and Hspa9+/+ littermate mice were evaluated at 2, 6, 9, 12 and 18 months of age. Hspa9+/− mice have normal peripheral blood and bone marrow morphology (data not shown), bone marrow and spleen cellularity (Fig. S2A), and complete blood counts (Fig. S2C). Immunophenotyping of mature and precursor hematopoietic cells in the peripheral blood, bone marrow and spleen of Hspa9+/− mice were normal (Fig. 2A–B and Fig. S3A–B) (see Table S4 for immunophenotypic markers). Immunophenotypic analysis of bone marrow progenitors (CMP, MEP, GMP) and stem cell-enriched populations (KLS, SLAM) between Hspa9+/+ and Hspa9+/− mice at 2, 6 and 12 months of age revealed no difference (Fig. 2C and data not shown). Bone marrow and spleen myeloid (CFU-C) and erythroid progenitors (mature BFU-E/BFU-E/CFU-E), as evaluated by methylcellulose colony-forming assays, were normal up to 18 months of age (Fig. 2D–E and Fig. S4). There was no difference in overall or leukemia-free survival between Hspa9+/+ and Hspa9+/− mice up to 18 months of age (N=35 mice/genotype) (data not shown). Collectively, these results indicate that hematopoiesis in Hspa9+/− mice is largely normal.
Figure 2. Immunophenotyping of bone marrow and peripheral blood cells in Hspa9+/− mice are normal compared to Hspa9+/+ littermates.
A) No difference was observed in bone marrow (left panel) or peripheral blood (right panel) of Hspa9+/− and Hspa9+/+ littermates analyzed by flow cytometry for immunophenotypic markers for (bars labeled top to bottom) neutrophils (Gr1+/CD115−), B-cells (B220+), monocytes (Gr1lo/CD115+) and T-cells (CD3e+) (N=3–6/genotype at each time point). B) Red blood cell precursors evaluated by flow cytometry (CD71+/−/Ter119+) in bone marrow of Hspa9+/− and Hspa9+/+ littermates at 12 months of age showing no difference between genotypes. C) Bone marrow cells from 12-month-old Hspa9+/− and wild-type littermates were stained with immunophenotypic markers for SLAM (Lin−Sca+cKit+CD150+CD48−), KLS (cKit+, Lin−, Sca+), megakaryocyte-erythrocyte progenitors (MEP, Lin−Sca−cKit+FcγRloCD34−), granulocyte-monocyte progenitors (GMP, Lin−Sca−cKit+FcγRhiCD34+), and common myeloid progenitors (CMP, Lin−Sca−cKit+FcγRloCD34+). Bone marrow cells were isolated at indicated time points from Hspa9+/+ and Hspa9+/− mice, D) plated in CFU-C media (10,000 cells/plate) and counted on day 7 or E) plated in mature BFU-E media containing only erythropoietin (100,000 cells/plate) and counted on day 10–11. (Hspa9+/+, filled circles; Hspa9+/−, open circles) ProEBs, proerythroblasts; BasoEBs, basophilic erythroblasts; PolyEBs, polychromatic erythroblasts; OrthoEBs, orthochromatic erythroblasts. Statistical analysis by two tailed Student’s t-test. Error bars represent mean ± SD. *p<0.05
Hspa9 is up-regulated in response to a number of cellular stresses to provide a cytoprotective effect [14,29–31]. Next, we tested recovery of Hspa9+/− and Hspa9+/+ littermate mice following the induction of three different types of hematopoietic stress. First, we evaluated survival following weekly doses of 5-fluorouracil, which significantly reduces hematopoietic progenitors in normal mice (150mg/kg) (Fig. S5A) [32]. Second, recovery of red blood cells in the peripheral blood of older mice was evaluated for 15 days following phenylhydrazine-induced hemolytic anemia (2 doses of 30mg/kg) (Fig. S5B). Finally, radiation treatment was previously described to induce up-regulation of Hspa9 protein levels; therefore, we evaluated peripheral blood cell count recovery following a single dose of sublethal irradiation (500 Rads) (Fig. S5C) [33]. No differences in hematopoietic stress recovery were observed between Hspa9+/− and Hspa9+/+ littermates for these assays.
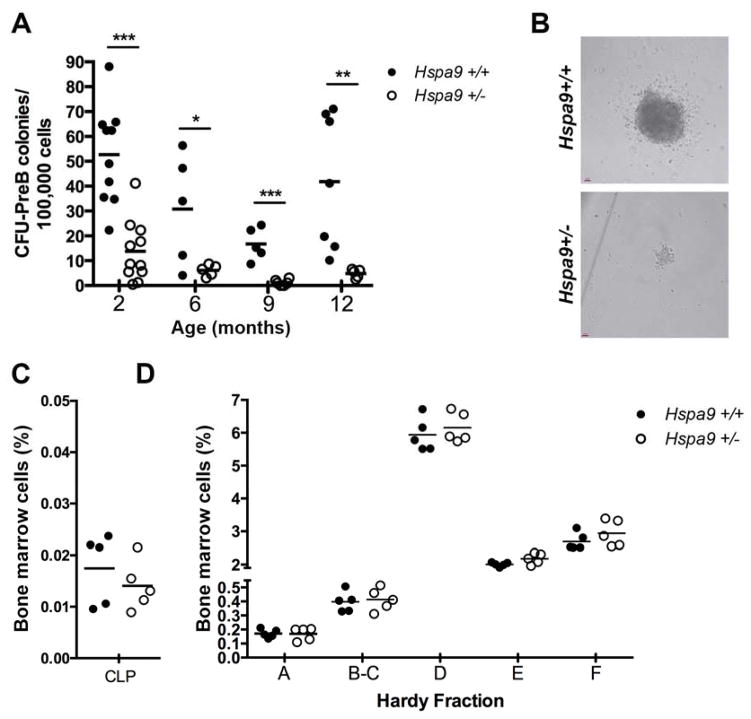
CFU-PreB colonies are reduced in Hspa9+/− mice
As early as 2 months of age, the number of bone marrow CFU-preB methylcellulose colonies was significantly reduced in Hspa9+/− mice compared to Hspa9+/+ littermates (Fig. 3A). The Hspa9+/− CFU-PreB colonies observed were also typically smaller in size (Fig. 3B). The same results were observed using CFU-PreB media containing recombinant mouse IL-7 (Fig. S6). To confirm the CFU-PreB phenotype was Hspa9-dependent, we expressed HSPA9 in Hspa9+/− or Hspa9+/+ bone marrow cells using a retrovirus. We observed an increase in the number of CFU-preB colonies only in Hspa9+/− bone marrow expressing HSPA9 (Fig. S7).
Figure 3. Colony forming ability of B-cell progenitors is significantly reduced in Hspa9+/− compared to Hspa9+/+ mice.
A) The number of Hspa9+/− CFU-PreB colonies counted on day 7 were significantly reduced compared to colonies from Hspa9+/+ littermate bone marrow at all ages evaluated. B) Representative images of CFU-PreB colonies. Percentage of bone marrow cells that are common lymphoid progenitors (C) and Hardy fractions (D) were not significantly different between Hspa9+/− and Hspa9+/+ littermate mice at 4–5 months of age. (Hspa9+/+, filled circles; Hspa9+/−, open circles) CLP, common lymphoid progenitor. Statistical analysis by two tailed Student’s t-test. *p<0.05, **p<0.01, ***p<0.001
We next measured bone marrow B-cell progenitors and precursors using flow cytometry to determine whether the CFU-PreB colony reduction was due to a reduction in the total number of progenitors added to the media. There was no significant difference in the percent and absolute number of common lymphoid progenitors (CLP) or Hardy fractions A, B/C, D, E, or F in Hspa9+/− mice, indicating the reduction in colony number was not due to fewer numbers of Hspa9+/− progenitors (N=5–10/genotype) (Fig. 3C and D).
The reduction of CFU-PreB colonies is hematopoietic-cell intrinsic
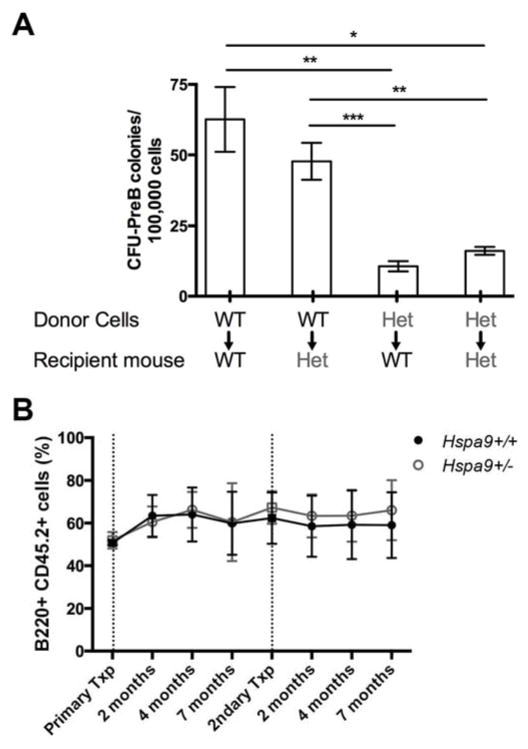
We tested whether the reduction in CFU-PreB colonies was a hematopoietic cell-intrinsic phenotype or due to a defect in non-hematopoietic stromal cells by performing non-competitive bone marrow transplants of Hspa9+/− and Hspa9+/+ cells into wild-type (Hspa9+/+) or Hspa9+/− recipients. After long-term engraftment was established (>6 months), the number of CFU-PreB colonies from wild-type mice that received Hspa9+/− bone marrow was significantly reduced compared to mice that received Hspa9+/+ bone marrow (N=7–9 mice/genotype, p=0.002) (Fig. 4A). There was no reduction in CFU-PreB colonies following transplantation of wild-type donor bone marrow cells into Hspa9+/− recipients (N=5) (Fig. 4A). These results indicate that the reduction in B-cell progenitors in Hspa9+/− mice is hematopoietic cell-intrinsic.
Figure 4. The reduction in Hspa9+/− CFU-PreB colony formation is hematopoietic cell-intrinsic.
A) Donor bone marrow from Hspa9+/+ (WT) or Hspa9+/− (HET) mice was transplanted into lethally irradiated Hspa9+/+ (WT) or Hspa9+/− (HET) recipients. Bone marrow was harvested 6 months after transplant and plated in CFU-PreB promoting methylcellulose (N=4–9 mice/genotype). B) A ratio of 1:1 Hspa9+/+ (black lines) or Hspa9+/− (grey lines) test cells (Ly5.2) and competitor bone marrow (Ly5.1/5.2) were transplanted into lethally irradiated recipients (Ly5.1). Mice were bled at intervals indicated after transplant and relative chimerism of B220+ peripheral blood cells were evaluated in recipients. Following long-term engraftment, bone marrow from recipients were pooled and transplanted into lethally irradiated secondary recipients. Data represents pooled results from two independently transplanted cohorts (N=10–15 mice/genotype). Txp, transplant. Statistical analysis by two tailed Student’s t-test and an ANOVA. Error bars represent mean ± SD. *p<0.05, **p<0.01, ***p<0.001
Next, we tested whether Hspa9+/− lymphoid progenitors are at a functional disadvantage compared to progenitors from control mice in vivo by performing competitive repopulation studies. Pools of donor bone marrow from C57Bl/6 (Ly5.2) Hspa9+/− or Hspa9+/+ mice were mixed at a 1:1 ratio with a competitor bone marrow pool from wild-type C57Bl/6 (Ly5.1/Ly5.2) mice, and transplanted into wild-type C57Bl/6 recipient mice (Ly5.1). Analysis of donor-derived cells (Ly5.2) following competitive transplant revealed no significant difference in the number or percent of mature B-cells (Fig. 4B), CLPs, or Hardy fractions in primary or secondary recipients (data not shown). Collectively, the data indicate that in vivo homeostatic regulation may compensate for a functional defect in Hspa9+/− B-cell progenitors.
B-cell progenitors are reduced in Hspa9 knockdown mice
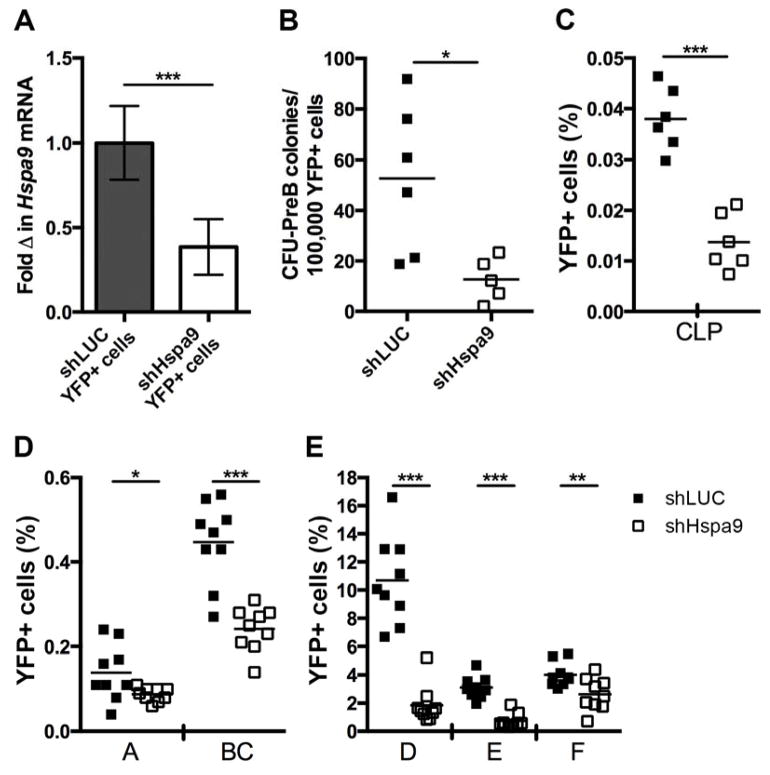
In vivo B-cell differentiation and proliferation are regulated by a variety of extracellular signals. In contrast, cells in an in vitro CFU-PreB assay are exposed to few stimulatory signals, which may sensitize cells to the effects of Hspa9 loss. In an attempt to overcome compensation and sensitize cells in vivo, we reduced Hspa9 levels >50% acutely in adult bone marrow cells. Because homozygous knockout of Hspa9 was lethal prior to fetal liver formation, preventing analysis of Hspa9−/− B-cell progenitors in the fetal liver, we knocked down Hspa9 levels below the 50% level using RNAi. Bone marrow cells transduced with virus (YFP+) were sorted from recipient mice 8–14 weeks post transplant and the mRNA expression level of Hspa9 in cells that received anti-Hspa9 shRNA (shHspa9) was 37% of the level measured in cells that received the control knockdown construct (shLUC) (i.e., 63% knockdown of Hspa9) (Fig. 5A). Consistent with Hspa9+/− mice, YFP+ Hspa9 knockdown (shHspa9) bone marrow cells had significantly reduced CFU-PreB colony formation compared to YFP+ control (shLUC) cells (p=0.017, N=5–6 mice/genotype) (Fig. 5B).
Figure 5. B-cell progenitors are significantly reduced in mice following >50% knockdown of Hspa9.
A) Expression of Hspa9 analyzed by RT-PCR in YFP+ bone marrow cells sorted from wild-type mice that received bone marrow transduced with shLUC or shHspa9 lentiviral constructs (N=7–8 mice/group, normalized to Gapdh). B) YFP+ bone marrow cells sorted from shLUC (closed squares) or shHspa9 (open squares) mice 10–12 weeks post-transplant were plated in CFU-PreB methylcellulose medium. CFU-PreB colonies were counted on day 7 and significantly reduced in shHspa9 bone marrow. The percent of CLPs (C) and Hardy fractions A–F (D, E) from the YFP+ bone marrow population of recipient mice is reduced. Statistical analysis by two tailed Student’s t-test. Error bars represent mean ± SD. *p<0.05, **p<0.01, ***p<0.001
The percent and absolute number of mature peripheral blood B-cells were significantly reduced, while T-cells and myeloid cells were not, as previously described [15]. Next, we interrogated B-cell progenitors and precursors not previously investigated. Acute knockdown of Hspa9 >50% in adult bone marrow resulted in a significant reduction in the frequencies of common lymphoid progenitors (CLP) and all Hardy fractions versus the control knockdown, with the largest difference occurring in Hardy fraction D (5.75 fold-change, N=9 mice/group, p<0.001) (Fig. 5C–E), indicating Hspa9 levels can affect B-cell progenitors in vivo. The percent of YFP+ Hardy fractions B–F in the spleen cells of mice that received the shHspa9 construct were also significantly reduced (1.7–3.5 fold, N=8 mice/group, p<0.05, Fig. S8B).
The mRNA expression of B-cell proliferation and activation genes is reduced in Hspa9+/− CFU-PreB colonies
Both cytokine availability and cytokine receptor expression can affect the ability of cells to respond to extracellular signals, and may contribute to in vivo compensation. Several cytokines, including IL-7 and Flt3-ligand, drive early B-cell proliferation and differentiation in vivo; however, IL-7 is the only cytokine supplement added to the methylcellulose media to promote CFU-PreB colony formation [34–36]. Levels of IL-7Rα expressed on the cell surface of Hardy fractions A–D were not significantly different between Hspa9+/− and Hspa9+/+ mice as measured by flow cytometry (N=5/genotype) (data not shown). To measure cytokine expression in vivo, we extracted RNA from femurs by flushing them with TRIzol. We observed no difference in mRNA expression levels of IL-7 or Flt3-ligand between Hspa9+/− and wild-type littermates as measured by qRT-PCR (3 months old, N=3/genotype, data not shown).
To investigate whether downstream signaling is disrupted by heterozygous loss of Hspa9, we performed gene expression array profiling on colonies produced by Hspa9+/− bone marrow and compared them to wild-type colonies. Day 7 CFU-PreB colonies were collected and sorted for B220+ cells (N=5 mice/genotype) (Fig. S9A and B). Two hundred forty-two genes were significantly down-regulated and 169 were significantly up-regulated in Hspa9+/− colonies compared to controls in a supervised analysis (FDR<0.05). Hspa9 expression was reduced in Hspa9+/− versus Hspa9+/+ colonies, as expected (p=0.01) (Fig. S9C). Pathway analysis of down-regulated genes in Hspa9+/− colonies using DAVID identified high enrichment scores (>3 fold enrichment) in pathways that promote lymphocyte proliferation and activation. No pathway enrichment was observed in up-regulated genes in Hspa9+/− colonies (>1.5 fold enrichment) (Table 2). Microarray results suggest that the reduction in CFU-PreB colonies may be due to a blunted response to proliferation signals in vitro.
Table 2.
Pathways significantly down-regulated in Hspa9+/− CFU-PreB colonies
| Annotation Cluster 1: Enrichment Score 5.02 | ||||
|---|---|---|---|---|
| Term | # genes | P Value | Benjamini | |
| GO:0045321 | leukocyte activation | 13 | 5.56E-06 | 6.28E-03 |
| GO:0046649 | lymphocyte activation | 12 | 8.65E-06 | 4.89E-03 |
| GO:0001775 | cell activation | 13 | 1.80E-05 | 6.77E-03 |
| Annotation Cluster 2: Enrichment Score 3.48 | ||||
|---|---|---|---|---|
| Term | # genes | P Value | Benjamini | |
| GO:0051249 | regulation of lymphocyte activation | 9 | 1.94E-04 | 5.36E-02 |
| GO:0032944 | regulation of mononuclear cell proliferation | 7 | 2.51E-04 | 5.53E-02 |
| GO:0050670 | regulation of lymphocyte proliferation | 7 | 2.51E-04 | 5.53E-02 |
| GO:0070663 | regulation of leukocyte proliferation | 7 | 2.87E-04 | 5.28E-02 |
| GO:0002694 | regulation of leukocyte activation | 9 | 3.07E-04 | 4.85E-02 |
| GO:0050865 | regulation of cell activation | 9 | 3.35E-04 | 4.64E-02 |
| GO:0050863 | regulation of T cell activation | 7 | 1.24E-03 | 1.31E-01 |
Knockdown of Hspa9 reduces proliferation and Stat5 activation in an IL-7 dependent mouse B-cell line
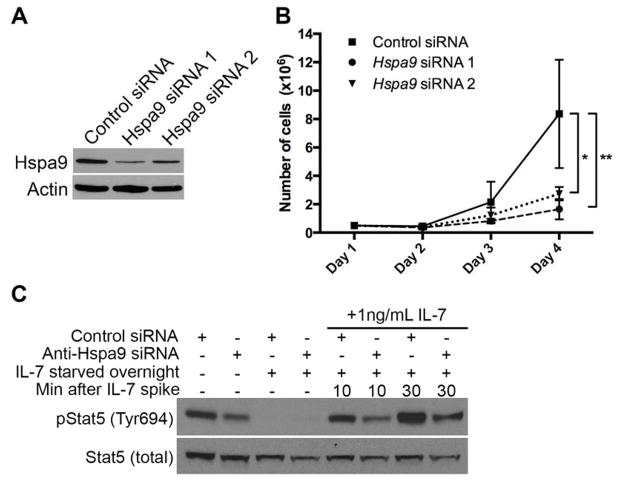
Because expression levels of the IL-7 receptor and its activating ligand were not different, we determined whether knockdown of Hspa9 altered the downstream cellular response to IL-7 stimulation. For this, we utilized the IL-7-dependent mouse B7 cell line. Knockdown of Hspa9 using Hspa9-targeting siRNAs in B7 cells causes a reduction in cell numbers over time compared to control siRNA-treated B7 cells grown in 10 ng/mL of IL-7 (Fig. 6B). A similar reduction in cell numbers was observed when B7 cells are grown in low concentrations (≤1ng/mL) of IL-7 (Fig. S10).
Figure 6. Knockdown of Hspa9 inhibits growth and IL-7 receptor mediated Stat5 phosphorylation in B7 cells.
B7 cells were electroporated with a non-targeting control siRNA or siRNA targeting Hspa9. The following day (day 1), cells were counted and plated at the same cell concentration. A) Hspa9 expression levels were evaluated by Western blot on day 2. B) Growth of B7 cells maintained in 10ng/mL IL-7 is significantly inhibited by knockdown of Hspa9 by two independent Hspa9-targeting siRNAs (N=3/group; Control siRNA, solid line; siRNA 1, dashed line; siRNA 2, dotted line). C) Western blot of lysates from B7 cells treated with Hspa9-targeting siRNA 1 or non-targeting control were analyzed for total and phosphorylated (Tyr694) Stat5. Cells were grown in 10mg/mL IL-7 for 4 days (Lane 1 and 2). Cells were starved overnight starting on day 3 (Lane 3 and 4). Cells starved overnight were stimulated on day 4 with 1ng/mL IL-7 and lysates were collected after 10 minutes (Lane 5 and 6) and 30 minutes (Lane 7 and 8). Stat5 phosphorylation was reduced in cells treated with Hspa9-targeting siRNA. Representative data is shown from 4 biological replicates. Error bars represent mean ± SD. *p<0.05, **p<0.01, ***p<0.001
We next investigated signaling downstream of the IL-7 receptor. Stat5 is indispensible for B-cell development and is rapidly activated by phosphorylation following IL-7 stimulation of the IL-7 receptor [37]. B7 cells were electroporated with Hspa9-targeting siRNAs, rested for two days, starved of IL-7 overnight, and stimulated with different concentrations of IL-7 (0.1, 1 or 10ng/mL). Cells were collected at 5, 10, 15 and 30 minutes following IL-7 stimulation and analyzed for Stat5 phosphorylation by flow cytometry. Following IL-7 stimulation, phosphorylation of Stat5 was reduced in B7 cells treated with Hspa9 siRNA compared to control siRNA at all time-points and concentrations evaluated by flow cytometry (Fig. S11). These results were confirmed by Western blotting of B7 cells collected following 10 and 30 minutes of stimulation with 1ng/mL IL-7 (Fig. 6C).
DISCUSSION
The present study addresses the role of Hspa9 in hematopoiesis and highlights the effects of Hspa9 expression levels on B-lymphopoiesis. While basal and stress hematopoiesis was normal in heterozygous knockout mice (Hspa9+/−), we observed a significant reduction in CFU-PreB colony numbers in Hspa9+/− mice. Embryonic lethality prevented the study of Hspa9 homozygous null mice; however, reduction of Hspa9 expression >50% by RNAi-mediated knockdown led to a reduction in B-cell progenitors and precursors in vivo. Our results suggest that reduced levels of Hspa9 impair Stat5 activation following IL-7 stimulation of B-cells. Collectively, these results indicate reduced Hspa9 expression alters B-lymphopoiesis.
We report for the first time that homozygous knockout of Hspa9 is embryonic lethal in mice, similar to knockout of Hspa5 and Hspa8. This is consistent with other genetic models of Hspa9 orthologs and indicates the importance of Hspa9 during embryogenesis [12,38–41]. Based on the altered hematopoiesis observed in Hspa9 mutant zebrafish, murine bone marrow shRNA transduction/transplantation models, and altered differentiation observed in human CD34+ cells, we predicted that Hspa9 heterozygous knockout mice would have abnormal hematopoiesis, specifically affecting erythroid and B-cells[12,15,16]. However, constitutive heterozygous deletion of Hspa9 did not alter erythroid or mature B-cell numbers, suggesting that >50% reduction of Hspa9 is necessary to perturb hematopoiesis in mice. It remains possible that the differences in phenotypes between these models are due to the level of Hspa9 expression and/or the constitutive deletion of Hspa9 in the knockout model versus the acute reduction of Hspa9 that occurs using RNAi-mediated methods. Compensation for Hspa9 deletion in the knockout model may occur during development. Similarly, acute RNAi-mediated knockdown of HSPA9 in primary human CD34+ cells may induce a phenotype not seen in the knockout mice, in addition to species-specific differences. Creation of an Hspa9 conditional knockout mouse will be necessary to test this possibility.
We postulate that the reduction in CFU-PreB colony formation in Hspa9+/− mice is mediated through Stat5 activation. Our microarray analysis did not reveal dysregulation of the SOCS or phosphatase family of genes that could explain altered Stat5 activation. Alternatively, we propose that Hspa9 may regulate Stat5 activation through its co-chaperone Dnaja3, a negative regulator of Stat5b [42]. Hspa9 is an Hsp70 protein that relies on interactions with co-chaperones such as J-proteins for its normal functions. Dnaja3 is the J-protein associated with the non-mitochondrial import functions of Hspa9 and is responsible for recruiting substrates to Hspa9 [13]. Overexpression of Dnaja3 results in dose-dependent inhibition of Stat5 phosphorylation, cell growth, and expression of the Stat5 target Bcl-xl in two IL-7 responsive B-cell lines (human 697 pre-B and mouse mIL-7R expressing Ba/F3 cells) [42]. Hspa9 is expressed in excess of Dnaja3, a ratio that is important for normal chaperone and cellular functions [43,44]. Reducing the abundance of Hspa9 may increase the amount of unbound Dnaja3, increasing the Dnaja3 available to bind and inhibit the phosphorylation of Stat5b. Hspa9 and Dnaja3 have also been established as regulators of p53, through a similar mechanism [45–47].
Although CFU-PreB colonies were reduced in vitro from Hspa9+/− mice, there were no alterations in basal B-lymphopoiesis in vivo. This suggests that homeostatic regulation of B-cell numbers may account for in vivo compensation. IL-7 is the only supportive cytokine added to CFU-PreB media even though other cytokines such as Scf, Flt3-ligand and Cxcl12 regulate B-cell differentiation and proliferation in vivo [34–36]. Relying solely on the IL-7 signaling cascade in CFU-PreB culture conditions in the absence of these other cytokines may sensitize cells to subtle alterations in IL-7 signaling caused by heterozygous Hspa9 loss. While we did not detect differences in bulk bone marrow IL-7 expression, these results do not exclude the possibility that smaller, local bone marrow niche changes in IL-7 concentration exist. Additionally, increased expression of other B-cell supportive cytokines may exist. Genetic loss of Hspa9 throughout the entire lifespan of the animal may allow compensation to occur and account for the difference between chronic heterozygous (knockout model) and acute (RNAi knockdown model) loss of Hspa9 in mice. Indeed, further reduction of Hspa9 (>50%) does disrupt B-lymphopoiesis in vivo; indicating a threshold level of Hsap9 is required for normal B-cell development to proceed.
HSPA9 protects cells from a variety of stresses. Although Hspa9+/− mice respond similarly to Hspa9+/+ littermates when challenged with hematopoietic stress inducers, it is possible that heterozygous loss of Hspa9 impairs response to cellular stresses that we did not evaluate. Increased levels of reactive oxygen species (ROS) have been implicated in MDS pathogenesis [48,49]. HSPA9 haploinsufficiency in MDS patients could alter cellular function as increased levels of ROS occur in hematopoietic cells from Hspa9 mutant zebrafish and knockdown mice [12,16]. In other cellular systems, loss of Hspa9 has been associated with increased ROS generation while Hspa9 overexpression has been shown to protect from oxidative damage [29,31,50]. In addition, deletion of RPS14, a gene commonly deleted with HSPA9 on del(5q), results in cellular stress due to ribosomal deficiency and p53 activation [51,52]. HSPA9 haploinsufficiency may limit a cell’s ability to appropriately regulate these stress states and may ultimately contribute to the ineffective hematopoiesis observed in del(5q) MDS. It is possible that an impaired ability of Hspa9+/− cells to respond to ex vivo cellular stresses may explain the observed reduction in CFU-PreB colony formation from Hspa9+/− bone marrow in the absence of in vivo reductions in B-cell progenitors.
HSPA9 is located in a commonly deleted region in myelodysplastic syndrome (MDS), del(5q), one of the most recurrent genetic abnormalities associated with the disease [17]. No single gene can explain the full spectrum of clinical phenotypes observed in MDS, suggesting that multiple del(5q) genes may contribute to pathogenesis [53]. Our studies suggest that HSPA9 haploinsufficiency may contribute to B-cell progenitor alterations observed in patients with MDS. Studies of B-cells in MDS are limited, but alterations in B-cell progenitors have been described, including reductions in B-cell progenitors, increased levels of B-cell apoptosis, and reduced expression of B-cell signaling pathways [54–56]. Data we provide here indicate that loss of HSPA9 may contribute to these B-cell alterations in patients with del(5q). Additional studies are needed to better understand how loss of Hspa9 attenuates Stat5 activation and B-cell development; however, this model provides evidence that Hspa9 loss may alter B- lymphopoiesis. This model could be used for future studies investigating how simultaneous deletion of multiple genes contributes to myeloid disease pathogenesis.
Supplementary Material
HIGHLIGHTS.
Homozygous knockout of Hspa9 in mice is embryonic lethal
Haploinsufficiency of Hspa9 causes a significant reduction in CFU-PreB colony formation
Knockdown of Hspa9 >50% significantly reduces B-cell progenitor numbers
Knockdown of Hspa9 attenuates Stat5 activation following IL-7 stimulation of a mouse B-cell line
Acknowledgments
We thank Deepta Bhattacharya for kindly providing the IL-7Rα antibody as well as helpful scientific discussions, and Charles Mullighan for kindly providing the B7 cells. Research reported in this publication was supported by the National Cancer Institute and the National Heart Lung Blood Institute of the National Institutes of Health under Award Number F31CA165702 (K.K.) and Research Project Grant R01HL109336 (M.J.W.), respectively, as well as a Howard Hughes Medical Institute Physician-Scientist Early Career Award (M.J.W.) and a Siteman Cancer Biology Pathway Fellowship (K.K). Technical assistance was provided by the Alvin J. Siteman Cancer Center Flow Cytometry Core, which provided cell sorting; Tissue Procurement Core, which provided RNA processing for microarray analysis; Biostatistics Core, which provided informatics support; and GTAC for microarray hybridization, and are supported by an NCI Cancer Center Support Grant (P30CA91842). Additional technical assistance was provided by the Hope Center Viral Vectors Core, which is supported by a Neuroscience Blueprint Interdisciplinary Center Core Award (P30NS057105).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Daugaard M, Rohde M, Jäättelä M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007 Jul 31;581(19):3702–10. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 2.Murphy ME. The HSP70 family and cancer. Carcinogenesis. 2013 Jun;34(6):1181–8. doi: 10.1093/carcin/bgt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hampton CR, Shimamoto A, Rothnie CL, Griscavage-Ennis J, Chong A, Dix DJ, et al. HSP70.1 and -70.3 are required for late-phase protection induced by ischemic preconditioning of mouse hearts. Am J Physiol Heart Circ Physiol. 2003 Aug;285(2):H866–74. doi: 10.1152/ajpheart.00596.2002. [DOI] [PubMed] [Google Scholar]
- 4.Luo S, Mao C, Lee B, Lee AS. GRP78/BiP is required for cell proliferation and protecting the inner cell mass from apoptosis during early mouse embryonic development. Mol Cell Biol. 2006 Aug;26(15):5688–97. doi: 10.1128/MCB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Florin L, Becker KA, Sapp C, Lambert C, Sirma H, Müller M, et al. Nuclear translocation of papillomavirus minor capsid protein L2 requires Hsc70. J Virol. 2004 Jun;78(11):5546–53. doi: 10.1128/JVI.78.11.5546-5553.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo F, Sigua C, Bali P, George P, Fiskus W, Scuto A, et al. Mechanistic role of heat shock protein 70 in Bcr-Abl-mediated resistance to apoptosis in human acute leukemia cells. Blood. 2005 Feb 1;105(3):1246–55. doi: 10.1182/blood-2004-05-2041. [DOI] [PubMed] [Google Scholar]
- 7.Ribeil J-A, Zermati Y, Vandekerckhove J, Cathelin S, Kersual J, Dussiot M, et al. Hsp70 regulates erythropoiesis by preventing caspase-3-mediated cleavage of GATA-1. Nature. 2007 Jan 4;445(7123):102–5. doi: 10.1038/nature05378. [DOI] [PubMed] [Google Scholar]
- 8.Zou P, Yoshihara H, Hosokawa K, Tai I, Shinmyozu K, Tsukahara F, et al. p57(Kip2) and p27(Kip1) cooperate to maintain hematopoietic stem cell quiescence through interactions with Hsc70. Cell Stem Cell. 2011 Sep 2;9(3):247–61. doi: 10.1016/j.stem.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 9.Miharada K, Karlsson G, Rehn M, Rörby E, Siva K, Cammenga J, et al. Cripto Regulates Hematopoietic Stem Cells as a Hypoxic-Niche-Related Factor through Cell Surface Receptor GRP78. Cell Stem Cell Elsevier. 2011 Oct;9(4):330–44. doi: 10.1016/j.stem.2011.07.016. [DOI] [PubMed] [Google Scholar]
- 10.Mjahed H, Girodon F, Fontenay M, Garrido C. Heat shock proteins in hematopoietic malignancies. Exp Cell Res. 2012 Sep;318(15):1946–58. doi: 10.1016/j.yexcr.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 11.Matsui H, Asou H, Inaba T. Cytokines direct the regulation of Bim mRNA stability by heat-shock cognate protein 70. Mol Cell. 2007 Jan 12;25(1):99–112. doi: 10.1016/j.molcel.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Craven SE, French D, Ye W, de Sauvage F, Rosenthal A. Loss of Hspa9b in zebrafish recapitulates the ineffective hematopoiesis of the myelodysplastic syndrome. Blood. 2005 May 1;105(9):3528–34. doi: 10.1182/blood-2004-03-1089. [DOI] [PubMed] [Google Scholar]
- 13.Goswami AV, Chittoor B, D’Silva P. Understanding the Functional Interplay between Mammalian Mitochondrial Hsp70 Chaperone Machine Components. J Biol Chem. 2010 Jun 11;285(25):19472–82. doi: 10.1074/jbc.M110.105957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohtsuka R, Abe Y, Fujii T, Yamamoto M, Nishimura J, Takayanagi R, et al. Mortalin is a novel mediator of erythropoietin signaling. Eur J Haematol. 2007 Aug;79(2):114–25. doi: 10.1111/j.1600-0609.2007.00870.x. [DOI] [PubMed] [Google Scholar]
- 15.Chen TH-P, Kambal A, Krysiak K, Walshauser MA, Raju G, Tibbitts JF, et al. Knockdown of Hspa9, a del(5q31.2) gene, results in a decrease in hematopoietic progenitors in mice. Blood. 2011 Feb 3;117(5):1530–9. doi: 10.1182/blood-2010-06-293167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tai-Nagara I, Matsuoka S, Ariga H, Suda T. Mortalin and DJ-1 coordinately regulate hematopoietic stem cell function through the control of oxidative stress. Blood. 2014 Jan 2;123(1):41–50. doi: 10.1182/blood-2013-06-508333. [DOI] [PubMed] [Google Scholar]
- 17.Haase D, Germing U, Schanz J, Pfeilstöcker M, Nösslinger T, Hildebrandt B, et al. New insights into the prognostic impact of the karyotype in MDS and correlation with subtypes: evidence from a core dataset of 2124 patients. Blood. 2007 Dec 15;110(13):4385–95. doi: 10.1182/blood-2007-03-082404. [DOI] [PubMed] [Google Scholar]
- 18.Zhao N, Stoffel A, Wang PW, Eisenbart JD, Espinosa R, Larson RA, et al. Molecular delineation of the smallest commonly deleted region of chromosome 5 in malignant myeloid diseases to 1–1.5 Mb and preparation of a PAC-based physical map. Proc Natl Acad Sci USA. 1997 Jun 24;94(13):6948–53. doi: 10.1073/pnas.94.13.6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walter MJ, Shen D, Shao J, Ding L, White BS, Kandoth C, et al. Clonal diversity of recurrently mutated genes in myelodysplastic syndromes. Leukemia. 2013 Jun;27(6):1275–82. doi: 10.1038/leu.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jerez A, Gondek LP, Jankowska AM, Makishima H, Przychodzen B, Tiu RV, et al. Topography, clinical, and genomic correlates of 5q myeloid malignancies revisited. J Clin Oncol. 2012 Apr 20;30(12):1343–9. doi: 10.1200/JCO.2011.36.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graubert TA, Payton MA, Shao J, Walgren RA, Monahan RS, Frater JL, et al. Integrated genomic analysis implicates haploinsufficiency of multiple chromosome 5q31. 2 genes in de novo myelodysplastic syndromes pathogenesis. PLoS ONE Public Library of Science. 2009;4(2):e4583. doi: 10.1371/journal.pone.0004583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zambrowicz BP, Abuin A, Ramirez-Solis R, Richter LJ, Piggott J, BeltrandelRio H, et al. Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc Natl Acad Sci USA. 2003 Nov 25;100(24):14109–14. doi: 10.1073/pnas.2336103100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magee JA, Ikenoue T, Nakada D, Lee JY, Guan K-L, Morrison SJ. Temporal changes in PTEN and mTORC2 regulation of hematopoietic stem cell self-renewal and leukemia suppression. Cell Stem Cell. 2012 Sep 7;11(3):415–28. doi: 10.1016/j.stem.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma Z, Izumi H, Kanai M, Kabuyama Y, Ahn NG, Fukasawa K. Mortalin controls centrosome duplication via modulating centrosomal localization of p53. Oncogene. 2006 Apr 17;25(39):5377–90. doi: 10.1038/sj.onc.1209543. [DOI] [PubMed] [Google Scholar]
- 25.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001 Apr 17;98(9):5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009 Jan;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Mullighan CG, Collins-Underwood JR, Phillips LAA, Loudin MG, Liu W, Zhang J, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009 Nov;41(11):1243–6. doi: 10.1038/ng.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Liu W, Song X-D, Zuo J. Molecular and cellular biochemistry. 1–2. Vol. 268. Springer; 2005. Effect of GRP75/mthsp70/PBP74/mortalin overexpression on intracellular ATP level, mitochondrial membrane potential and ROS accumulation following glucose deprivation in PC12 cells; pp. 45–51. [DOI] [PubMed] [Google Scholar]
- 30.Williamson CL, Dabkowski ER, Dillmann WH, Hollander JM. Mitochondria protection from hypoxia/reoxygenation injury with mitochondria heat shock protein 70 overexpression. Am J Physiol Heart Circ Physiol. 2008 Jan;294(1):H249–56. doi: 10.1152/ajpheart.00775.2007. [DOI] [PubMed] [Google Scholar]
- 31.Orsini F, Migliaccio E, Moroni M, Contursi C, Raker VA, Piccini D, et al. The life span determinant p66Shc localizes to mitochondria where it associates with mitochondrial heat shock protein 70 and regulates trans-membrane potential. J Biol Chem. 2004 Jun 11;279(24):25689–95. doi: 10.1074/jbc.M401844200. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Wong J, Klinger M, Tran MT, Shannon KM, Killeen N. MLL5 contributes to hematopoietic stem cell fitness and homeostasis. Blood. 2009 Feb 12;113(7):1455–63. doi: 10.1182/blood-2008-05-159905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadekova S, Lehnert S, Chow TY. Induction of PBP74/mortalin/Grp75, a member of the hsp70 family, by low doses of ionizing radiation: a possible role in induced radioresistance. Int J Radiat Biol. 1997 Dec;72(6):653–60. doi: 10.1080/095530097142807. [DOI] [PubMed] [Google Scholar]
- 34.Dolence JJ, Gwin K, Frank E, Medina KL. Threshold levels of Flt3-ligand are required for the generation and survival of lymphoid progenitors and B-cell precursors. Eur J Immunol. 2011 Feb;41(2):324–34. doi: 10.1002/eji.201040710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Funk PE, Varas A, Witte PL. Activity of stem cell factor and IL-7 in combination on normal bone marrow B lineage cells. J Immunol. 1993 Feb 1;150(3):748–52. [PubMed] [Google Scholar]
- 36.Egawa T, Kawabata K, Kawamoto H, Amada K, Okamoto R, Fujii N, et al. The earliest stages of B cell development require a chemokine stromal cell-derived factor/pre-B cell growth-stimulating factor. Immunity. 2001 Aug;15(2):323–34. doi: 10.1016/s1074-7613(01)00185-6. [DOI] [PubMed] [Google Scholar]
- 37.Goetz CA, Harmon IR, O’Neil JJ, Burchill MA, Farrar MA. STAT5 activation underlies IL7 receptor-dependent B cell development. J Immunol. 2004 Apr 15;172(8):4770–8. doi: 10.4049/jimmunol.172.8.4770. [DOI] [PubMed] [Google Scholar]
- 38.Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, et al. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999 Sep;153(1):135–77. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Craig EA, Kramer J, Kosic-Smithers J. SSC1, a member of the 70-kDa heat shock protein multigene family of Saccharomyces cerevisiae, is essential for growth. Proc Natl Acad Sci USA. 1987 Jun;84(12):4156–60. doi: 10.1073/pnas.84.12.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimura K, Tanaka N, Nakamura N, Takano S, Ohkuma S. Knockdown of mitochondrial heat shock protein 70 promotes progeria-like phenotypes in caenorhabditis elegans. J Biol Chem. 2007 Feb 23;282(8):5910–8. doi: 10.1074/jbc.M609025200. [DOI] [PubMed] [Google Scholar]
- 41.Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci USA. 2004 Aug 31;101(35):12792–7. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dhennin-Duthille I, Nyga R, Yahiaoui S, Gouilleux-Gruart V, Régnier A, Lassoued K, et al. The tumor suppressor hTid1 inhibits STAT5b activity via functional interaction. J Biol Chem. 2011 Feb 18;286(7):5034–42. doi: 10.1074/jbc.M110.155903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finka A, Goloubinoff P. Proteomic data from human cell cultures refine mechanisms of chaperone-mediated protein homeostasis. Cell Stress Chaperones. 2013 Sep;18(5):591–605. doi: 10.1007/s12192-013-0413-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laufen T, Mayer MP, Beisel C, Klostermeier D, Mogk A, Reinstein J, et al. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc Natl Acad Sci USA. 1999 May 11;96(10):5452–7. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahn BY, Trinh DLN, Zajchowski LD, Lee B, Elwi AN, Kim S-W. Tid1 is a new regulator of p53 mitochondrial translocation and apoptosis in cancer. Oncogene. 2010 Feb 25;29(8):1155–66. doi: 10.1038/onc.2009.413. [DOI] [PubMed] [Google Scholar]
- 46.Kaul S, Reddel RR, Mitsui Y, Wadhwa R. An N-terminal region of mot-2 binds to p53 in vitro. Neoplasia. 2001 Mar;3(2):110–4. doi: 10.1038/sj.neo.7900139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanai M, Ma Z, Izumi H, Kim S-H, Mattison CP, Winey M, et al. Physical and functional interaction between mortalin and Mps1 kinase. Genes Cells. 2007 Jun;12(6):797–810. doi: 10.1111/j.1365-2443.2007.01091.x. [DOI] [PubMed] [Google Scholar]
- 48.Rassool FV, Gaymes TJ, Omidvar N, Brady N, Beurlet S, Pla M, et al. Reactive oxygen species, DNA damage, and error-prone repair: a model for genomic instability with progression in myeloid leukemia? Cancer Res. 2007 Sep 15;67(18):8762–71. doi: 10.1158/0008-5472.CAN-06-4807. [DOI] [PubMed] [Google Scholar]
- 49.Sallmyr A, Fan J, Rassool FV. Genomic instability in myeloid malignancies: increased reactive oxygen species (ROS), DNA double strand breaks (DSBs) and error-prone repair. Cancer Lett. 2008 Oct 18;270(1):1–9. doi: 10.1016/j.canlet.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 50.EQ, Liu X, Liu Y, Liu W, Zuo J. Over-expression of GRP75 inhibits liver injury induced by oxidative damage. Acta Biochim Biophys Sin (Shanghai) 2013 Feb;45(2):129–34. doi: 10.1093/abbs/gms098. [DOI] [PubMed] [Google Scholar]
- 51.Dutt S, Narla A, Lin K, Mullally A, Abayasekara N, Megerdichian C, et al. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2011 Mar 3;117(9):2567–76. doi: 10.1182/blood-2010-07-295238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barlow JL, Drynan LF, Hewett DR, Holmes LR, Lorenzo-Abalde S, Lane AL, et al. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q– syndrome. Nat Med. 2009 Nov 22;16(1):59–66. doi: 10.1038/nm.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ebert BL. Molecular dissection of the 5q deletion in myelodysplastic syndrome. Semin Oncol. 2011 Oct;38(5):621–6. doi: 10.1053/j.seminoncol.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sternberg A, Killick S, Littlewood T, Hatton C, Peniket A, Seidl T, et al. Evidence for reduced B-cell progenitors in early (low-risk) myelodysplastic syndrome. Blood. 2005 Nov 1;106(9):2982–91. doi: 10.1182/blood-2005-04-1543. [DOI] [PubMed] [Google Scholar]
- 55.Amin HM, Jilani I, Estey EH, Keating MJ, Dey AL, Manshouri T, et al. Increased apoptosis in bone marrow B lymphocytes but not T lymphocytes in myelodysplastic syndrome. Blood. 2003 Sep 1;102(5):1866–8. doi: 10.1182/blood-2003-01-0221. [DOI] [PubMed] [Google Scholar]
- 56.Pellagatti A, Cazzola M, Giagounidis A, Perry J, Malcovati L, Porta Della MG, et al. Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells. Leukemia. 2010 Apr;24(4):756–64. doi: 10.1038/leu.2010.31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.