Abstract
Despite recent advances in the understanding of the interplay between a dynamic physical environment and phylogeography in Europe, the origins of contemporary Irish biota remain uncertain. Current thinking is that Ireland was colonized post-glacially from southern European refugia, following the end of the last glacial maximum (LGM), some 20 000 years BP. The Leisler's bat (Nyctalus leisleri), one of the few native Irish mammal species, is widely distributed throughout Europe but, with the exception of Ireland, is generally rare and considered vulnerable. We investigate the origins and phylogeographic relationships of Irish populations in relation to those across Europe, including the closely related species N. azoreum. We use a combination of approaches, including mitochondrial and nuclear DNA markers, in addition to approximate Bayesian computation and palaeo-climatic species distribution modelling. Molecular analyses revealed two distinct and diverse European mitochondrial DNA lineages, which probably diverged in separate glacial refugia. A western lineage, restricted to Ireland, Britain and the Azores, comprises Irish and British N. leisleri and N. azoreum specimens; an eastern lineage is distributed throughout mainland Europe. Palaeo-climatic projections indicate suitable habitats during the LGM, including known glacial refugia, in addition to potential novel cryptic refugia along the western fringe of Europe. These results may be applicable to populations of many species.
Keywords: western Europe, cryptic refugia, postglacial colonization, Nyctalus leisleri, Nyctalus azoreum
1. Introduction
The origins of the terrestrial fauna and flora of western Europe in general and Ireland in particular are the subject of prolonged debate [1–4]. Similar to the rest of northern Europe, most of Ireland was covered by ice during the last glacial maximum (LGM) some 20 000 years BP; thus, all endemic Irish species are thought to be descendants from colonists arriving post-glacially from refugia in southern Europe. Ireland is species poor compared with Britain and continental Europe. This is attributed to the ‘steeplechase’ effect [5], as the retreating ice cut off Ireland from Britain around 16 000 years BP and continental Europe around 7800 years BP [6]. However, recent studies (see [7]) examining geophysical models simulating the land mass response to glaciohydro-isostatic adjustment [8] provide no evidence for land-bridges between Ireland and Britain during the Holocene, reopening the debate as to the origin of the terrestrial Irish fauna. Biogeographic studies of species such as the Kerry slug (Geomalacus maculosus) and the strawberry tree (Arbutus unedo) [9] suggest a possible link between Ireland and southwestern Europe. However, phylogeographic studies of Irish flora and fauna have demonstrated that Irish populations are genetically distinct and diverse in relation to their European counterparts [10]. The current consensus is that there are still too few comprehensive phylogeographical studies involving Irish populations to finally resolve the origins of the fauna and flora of western Europe. Here, we use a combination of analytical approaches, including phylogeographic analysis of mitochondrial and nuclear DNA, in addition to palaeo-climatic species distribution modelling (SDM), to investigate the impact of the Pleistocene LGM on the past and present-day distribution of the Leisler's bat Nyctalus leisleri (Kuhl, 1817).
The Leisler's bat is distributed across most of Europe, western Asia as far as the Urals, northwest Africa, the Canary Islands and Madeira [11,12]. Despite widespread distribution, it is rare almost everywhere except Ireland, where it is the most common species after the Pipistrellus spp. [13]. The Leisler's bat is the only representative of the Nyctalus genus in Ireland [14], but in the rest of its range, it occurs sympatrically with the common noctule N. noctula (Schreber, 1774). A closely related species, N. azoreum, is endemic to the Azores. The Irish N. leisleri bat population is considered the largest in Europe [15] and of international importance.
Morphological and behavioural differences occur between N. azoreum and continental European N. leisleri [16–19]. Similarly, behavioural differences have been reported between the Leisler's bat in Ireland and elsewhere. Continental N. leisleri are forest dwelling, roosting mainly in treeholes, whereas in Ireland, maternity colonies are more commonly found in buildings [20,21]. The N. leisleri in Ireland have also been reported to have a lower peak-frequency echolocation call than their counterparts elsewhere [14,22], although such comparisons are difficult owing to disparate recording methods. Continental N. leisleri are migratory, with individuals being recorded moving distances over 1500 km between summer and winter roost sites [23–25]. However, in Ireland, it seems N. leisleri remain within their summer range to hibernate [26] (E. S. M. Boston and A. Hopkirk 2004, personal observations).
Genetic divergence and phylogeographic history of Nyctalus in Europe have been examined by Salgueiro et al. [27] using mitochondrial DNA (mtDNA) genes. Their main interest was to establish genetic relationships between N. azoreum and N. leisleri. They found that N. azoreum was characterized by a number of unique mtDNA haplotypes restricted to the Azores. These findings, in addition to a number of morphological, ecological and behavioural differences, were used to argue for full species status for N. azoreum [27,28]. The low levels of genetic divergence between N. azoreum and N. leisleri suggested that the former species originated recently from a European population of N. leisleri, and that the colonization of the Azores is most to have likely occurred at the end of the Pleistocene. No N. leisleri samples from Ireland or Britain were included in this study and, thus, the phylogeography of N. leisleri in western Europe remains far from complete, and the origins of this species in Ireland and Britain remain uncertain. Further, it is not known whether contemporary gene flow occurs between N. leisleri populations from Ireland, Britain and mainland Europe. Despite their obvious dispersal potential as a flying mammal, Castella et al. [29] demonstrated that in Myotis myotis, for instance, large bodies of water can act as effective barriers to gene flow. Thus, any contemporary exchange of genes between island and mainland populations may be restricted to passive (e.g. wind-blown), rather than to active, dispersal.
Here, we aim to determine the origin(s) and relationship of Irish populations with those across the rest of Europe, including the closely related N. azoreum. We test the hypotheses that (i) N. leisleri in Ireland colonized from a single southern European glacial refugia, via Britain, and (ii) there is limited gene flow among recent populations of N. leisleri in continental Europe, Britain and Ireland. Throughout, we examine the systematic and phylogeographic status of N. leisleri in continental Europe and offshore islands, and, in particular, the proposed endemic species N. azoreum.
2. Material and methods
(a). Sampling
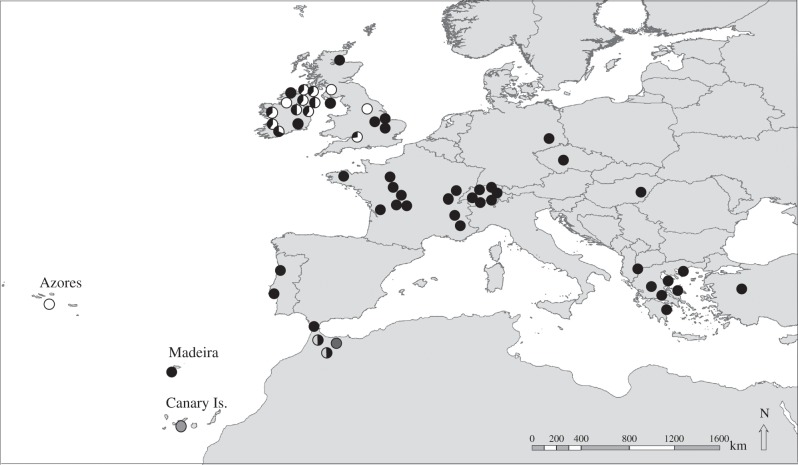
Over two consecutive years, 283 N. leisleri were captured leaving maternity colonies in 12 locations throughout Ireland (n = 138) and at two sites in northeastern France (n = 145). Samples were taken non-destructively, using the sterile wing biopsy technique, as described by Worthington-Wilmer & Barrett [30]. Two 3 mm diameter wing biopsies were taken and placed in 99% molecular grade ethanol. All bats were released immediately after sampling. Biopsy tissue samples from 37 specimens were donated from research colleagues and museums in France, Portugal, Switzerland, Germany, Hungary and Greece, along with 18 faecal samples from Britain (figure 1 and table 1; electronic supplementary material, table A in appendices).
Figure 1.
Sampling locations of N. leisleri, N. l. verrocosus and N. azoreum used in phylogeographic analysis. Black, white and grey shades in pies represent the proportion of haplotypes belonging to distinct lineages: western (white); eastern (black); Moroccan (Mar1, light grey); Moroccan 2 (Mar2; dark grey); and Canary Islands (Can; medium grey).
Table 1.
ISO country code (map ID), number of samples screened for microsatellites (nn), number of sequences (nmt) and descriptive statistics, including number of haplotypes (nh), number of polymorphic sites, mean number of pairwise differences and haplotype (h ± s.d.) and nucleotide (π ± s.d.) diversities with standard deviation for each sampling location.
| map ID | location | nn | nmt | nh | poly. sites | mean pairwise | h + s.d. | π + s.d. |
|---|---|---|---|---|---|---|---|---|
| IRL | Ireland | 138 | 75 | 12 | 18 | 5.129 | 0.771 ± 0.036 | 0.016 ± 0.009 |
| IOM | Isle of Man | 7 | 4 | 11 | 3.524 | 0.81 ± 0.130 | 0.011 ± 0.007 | |
| GBR | Britain | 21 | 6 | 12 | 4.485 | 0.776 ± 0.059 | 0.014 ± 0.008 | |
| total British Isles | 103 | 15 | 21 | 5.106 | 0.801 ± 0.023 | 0.016 ± 0.009 | ||
| FRA | France | 145 | 27 | 13 | 15 | 2.769 | 0.880 ± 0.044 | 0.009 ± 0.005 |
| PRT | Portugal | 35 | 6 | 8 | 0.947 | 0.603 ± 0.068 | 0.003 ± 0.002 | |
| ESP | Spain | 1 | 1 | 0 | — | — | — | |
| CHE | Switzerland | 7 | 6 | 7 | 2.800 | 1.000 ± 0.094 | 0.009 ± 0.006 | |
| DEU | Germany | 2 | 2 | 4 | 4.000 | 1.000 ± 0.500 | 0.013 ± 0.014 | |
| HUN | Hungary | 2 | 2 | 3 | 3.000 | 1.000 ± 0.500 | 0.009 ± 0.010 | |
| TUR | Turkey | 1 | 1 | 0 | — | — | — | |
| CZE | Czech Rep | 1 | 1 | 0 | — | — | — | |
| MNE | Montenegro | 1 | 1 | 0 | — | — | — | |
| GRC | Greece | 7 | 5 | 9 | 2.571 | 0.952 ± 0.096 | 0.008 ± 0.005 | |
| total Mainland Europe | 84 | 29 | 29 | 2.227 | 0.871 ± 0.024 | 0.007 ± 0.004 | ||
| MAR | Morocco | 4 | 3 | 17 | 10.00 | 0.833 ± 0.222 | 0.031 ± 0.021 | |
| Can. Is. | Canary Islands | 1 | 1 | 0 | — | — | — | |
| Mad. Is. | Madeira | 2 | 1 | 0 | — | — | — | |
| Az. Is. | Azores | 165 | 16 | 17 | 2.762 | 0.837 ± 0.019 | 0.009 ± 0.005 |
(b). DNA extraction, and mitochondrial DNA sequencing and microsatellite screening
Genomic DNA was extracted from tissue biopsies using the DNeasy tissue kit (Qiagen), whereas the 18 faecal samples were extracted using QIAmp DNA stool mini kit (Qiagen) following the modifications from Puechmaille et al. [31], which demonstrates that good-quality DNA can be obtained from bat droppings. A 324 base pair (bp) region of the hypervariable mtDNA control region (D-loop) was amplified for a subset of samples from Ireland (n = 67; an average five per site) and France (n = 15), and all other European samples, by PCR using the universal primers L16517 [32] and 130 sH651 [29]. PCR amplifications were carried out in 50 µl reaction volumes as follows: 1–3 µl of template DNA (approx. 5–25 ng), 2.5 mM MgCl2, 10 pM of each primer, 200 µM dNTPs, 1 U of Taq DNA polymerase (Invitrogen) with 1× of the corresponding PCR buffer. Thermal profiles consisted of an initial denaturation step at 95°C for 3 min, followed by 35 cycles consisting of 45 s at 94°C, 45 s at 50°C, 1 min at 72°C followed by a final extension step of 7 min at 72°C. Amplified products were purified using Roche high purification kits and subsequently sent for commercial sequencing (Macrogen) using the forward primer only. Resulting sequences were imported into BioEdit [33], and aligned using ClustalW [34].
In order to combine our novel mtDNA dataset with existing lineages based on previous studies by Salgueiro et al. [27,28], 46 European N. leisleri were sequenced for the mtDNA control region, resulting in 13 haplotypes (accession nos. DQ887597–DQ887608 and AY756613–AY756615). In addition, sequences representing 16 haplotypes resulting from the screening of 165 specimens of the closely related N. azoreum, and one haplotype from N. l. verrucosus, representing two specimens from Madeira Island (accession nos. DQ887596 and AY756598–AY756612), were included in the analysis.
To account for possible biases of phylogeographic inferences resulting from the analysis of a single mtDNA gene [35], 138 Irish tissue samples and 145 tissue samples from two sites in northeastern France were typed at seven autosomal microsatellite loci (Nleis1, Nleis2, Nleis3, Nleis5, Nleis6, Nleis8, Nleis10) as described by Boston et al. [36]. Genotypic typing was carried out using Gene Profiler v. 4.05 software (Scanalytics, Fairfax, VA). Control samples of known genotypes were used on each run, and the majority of the typing double checked by an independent researcher.
(c). Mitochondrial DNA analysis
Sample diversity statistics were calculated per site in Arlequin v. 3.0 [37]. To evaluate the overall level of population genetic structuring on a large geographical scale, a global exact test for genetic differentiation was carried out among samples grouped by major regions (i.e. Ireland and Britain, mainland Europe and the Azores) using Arlequin v. 3.0, with 20 000 Markov chain steps. Patterns of genetic subdivision both within and among these regions were evaluated using analysis of molecular variance (AMOVA) as implemented in Arlequin. The statistical significance of the AMOVA was tested by 10 000 random permutations. To investigate the phylogeographic origin(s) of N. leisleri in Ireland in relation to Britain and mainland Europe, and to N. azoreum and N. l. verrocosus, a haplotypic network was constructed using the median-joining method [38], implemented in Network v. 4.5.0.2 (Fluxus Technology, www.fluxus-engineering.com).
To test for signals of expansion in the recent demographic history of N. leisleri and N. azoreum, mismatch distribution analysis, using Arlequin, was carried out under a model of sudden (demographic) expansion. The significance of these observed deviations from expectation (i.e. evidence for population growth) was evaluated using the F-statistics proposed by Fu & Li [39]. The peak of the mismatch distribution provides an estimate of τ (tau), the starting time of the expansion in units of 1/(2μ) generations. From τ, it is possible to estimate the time (t) since the most recent expansion using the equation τ = 2 μt [40], where μ is the mutation rate per locus (i.e. the product of the mutation rate per site and the sequence length). We used a range of mutation rates for the control region proposed by Petit et al. [41] for the congeneric N. noctula, with a generation time of 2 years, a lower rate of 6.3%, a higher rate of 25.2%, with an average of 20% Myr−1. The latter value was used in several other studies, including Salgueiro et al. [27] and Chen et al. [42]. The approximate Bayesian computation (ABC) approach, implemented in the DIYABC software [43], was also used to test for alternative scenarios for the colonization of Britain and Ireland by N. leisleri following the LGM using both microsatellite and mtDNA data (details of this analysis are provided in the electronic supplementary material).
(d). Microsatellite analysis
The Bayesian approach implemented in Structure v. 2.3.3 was used to examine the extent of concurrent genetic partitioning between Irish and French populations [44]. Structure was run using an admixture model with correlate allelic frequencies. A burn-in of 5 × 103 was followed by 106 iterations over 20 replicated runs for each K (1–20). Structure Harvester v. 0.6.1 [45] was used to combine the results files from Structure and determine the optimal value of K using the estimated log-likelihood of K according to Pritchard et al. [44]. In order to estimate the level of divergence between Irish and French nursery colonies, pairwise FST estimates were calculated using diveRsity v. 1.7.0 [46].
(e). Species distribution modelling
To ascertain broadly suitable climatic conditions for N. leisleri for both the present day and during the LGM, SDM in conjunction with palaeo-climatic reconstruction was conducted using the maximum entropy approach as implemented in MaxEnt v. 3.3.2 [47]. Data for Leisler's bat occurrence, filtered to include only records found within the known distribution range of the species [48], were obtained from the Global Biodiversity Information Facility (GBIF). In total, occurrence data were obtained from 1679 point localities throughout the range of the species. Data were not filtered by season, and thus included some sites used in summer or for hibernation only. Both current (1950–2000) and LGM (Community Climate System Model) global 19 Bioclim variable layers (2.5 arc-min resolution) were downloaded from WordClim—Global Climate Data (www.worldclim.org) and from the Paleoclimate Modelling Intercomparison Project Phase II (https://pmip2.lsce.ipsl.fr), respectively. Model performance was evaluated using a randomly selected sample comprising 25% of the occurrence data points. The presence threshold was based on the sensitivity–specificity sum maximization approach [49].
3. Results
(a). Genetic diversity
Levels of mtDNA haplotypic diversity for N. leisleri from mainland Europe (0.871 ± 0.024) and the British Isles (0.801 ± 0.023), and those from N. azoreum (0.837 ± 0.019), were very similar (table 1). However, nucleotide diversity was almost twofold higher in N. leisleri populations from Ireland and Britain than N. azoreum (0.016 ± 0.009 versus 0.009 ± 0.005, respectively). All seven microsatellite loci screened were found to be polymorphic with 3–26 alleles per locus.
(b). Mitochondrial sequence analysis
A 324 bp stretch of the D-loop was sequenced for 147 individual bats. These, in addition to 28 equivalent haplotypic sequences downloaded from GenBank, representing 212 bats, resulted in a total of 359 individual sequences available for analysis. A total of 60 haplotypes were identified in our dataset, with 48% of these (n = 29) reported for the first time; these data were submitted to GenBank (see data accessibility information at end of paper). These haplotypes were defined by 64 variable sites, of which 48 were transitions, seven transversions and nine indels. The 22 bp indel found in N. azoreum [27] was absent in all N. leisleri sequences examined in this study.
Overall, only eight haplotypes (13%) were found in two or more locations, whereas four (Po1, A7, Ir1 and Po2) were relatively common (i.e. found in three or more locations). One (Po1) was widespread throughout Europe, being found in 22% of the samples, and it was the only haplotype to occur in continental Europe, Ireland and Britain. Overall, 87% of haplotypes were restricted to single geographical locations. Surprisingly, N. azoreum haplotypes, presumed to be species-specific, were also found in N. leisleri from Ireland and Britain. Thus, the A7 haplotype previously known only in N. azoreum from the Azores [28], also occurred in N. leisleri from Ireland (7 of 75 individuals), the Isle of Man (three of seven individuals) and Britain (6 out of 21 individuals). Eleven haplotypes (18% of all haplotypes), of which 10 were from Ireland (Ir1, Ir3, Ir4, Ir5, Ir6, Ir7, Ir8, Ir9, Ir10 and Ir11) and one from Britain (En2), were restricted to these islands. The remaining 33 haplotypes (55%) were found in Europe (including Madeira and the Canary Islands) and northern Africa only, whereas 15 additional haplotypes (29%) were restricted to the Azores.
(c). Phylogeographic analysis
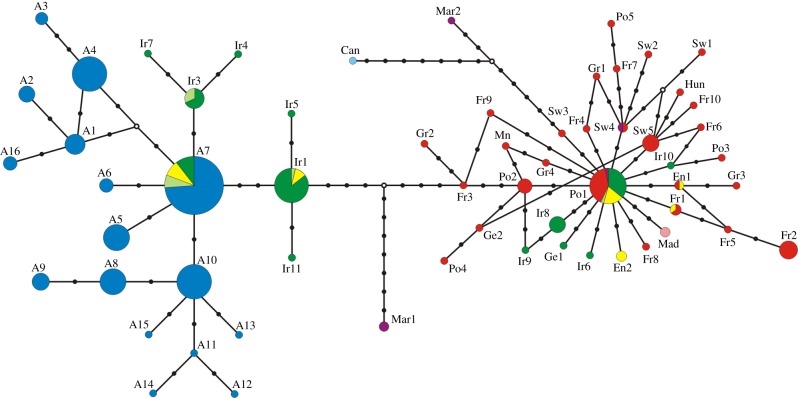
The resulting median-joining haplotypic network (figure 2) revealed two very distinct mtDNA haplotypic lineages. One of the lineages comprised haplotypes restricted to the Azores, Ireland and Britain, whereas the second lineage comprised haplotypes which were widespread throughout the rest of Europe, including Ireland and Britain (figure 1). These two major lineages, henceforth referred to as ‘western’ and ‘eastern’ lineages, respectively, differ by 2% sequence divergence (i.e. an average of nine nucleotide differences between both lineages), and are characterized by four fixed mutational changes, with a minimum divergence of 1.8% (six nucleotide differences). Haplotype divergence within the eastern lineage was 0.7% (average number of nucleotide differences within lineage = 2.3), whereas for the western lineage, this value was 0.8% (average number of nucleotide differences within lineage = 2.7).
Figure 2.
Median-joining network of N. leisleri, and N. azoreum mtDNA haplotypes. Pie sizes are proportional to the number of individuals showing a particular haplotype. Black full circles represent mutational steps; white circles represent inferred haplotypes (not observed in dataset); different colours represent broad geographical haplotype locations (blue, Azores; green, Ireland; pale green, Isle of Man; yellow, England; red, continental Europe; purple, Morocco; light blue, Canary Islands; pink, Madeira) Reticulations for unresolved links between haplotypes are shown.
Most N. leisleri haplotypes found in Ireland and Britain (Ir1, Ir3, Ir4, Ir5, Ir7 and Ir11) were closely related to haplotypes associated with N. azoreum, with one haplotype (A7) occurring both in N. azoreum and in the N. leisleri from Ireland and Britain. Within the network, Po1 was placed in the centre of a typical star-shaped topology of the European lineage, and had the highest frequency and widest geographical distribution, which is indicative of its ancestral status in relation to other haplotypes. Four haplotypes unique to Ireland (Ir6, Ir8, Ir9 and Ir10), and an additional haplotype unique to Britain (En2), belonged to the eastern mtDNA lineage, and thus were closely related to the Po1 haplotype. Two further haplotypes of the eastern lineage were shared between France and Britain, En1 and Fr1. The single haplotype (Mad) representing the subspecies N. l. verrocosus from Madeira also belonged to the eastern N. leisleri lineage. The most divergent haplotypes were found in Morocco (Mar1 and Mar2), and the Canary Islands (Can1). Interestingly, Mar1 and Mar2 represented two highly divergent haplotypes with 11 fixed differences and 3% sequence divergence between them.
(d). Population structuring
The global exact test for genetic differentiation of mtDNA haplotypes among major regions was highly significant (p < 0.001), confirming a considerable degree of genetic structuring among regions. Pairwise FST comparisons revealed that the majority of this differentiation was observed between regions (i.e. Ireland and Britain, continental Europe and the Azores).
AMOVA between Ireland and Britain, continental Europe and the Azores shows that 45.9% of the observed genetic variation was explained by differences between regions, whereas 38.72% of variation occurred within regions. These findings are supported by the analysis of nuclear microsatellite data between Ireland and France (FST = 0.023, 95% CI 0.015–0.031). The highest probability of the data (i.e. higher estimated log-likelihood) was observed at K = 2, clearly separating Irish and French samples (data not shown).
(e). Demographic history
Mismatch distribution demonstrated that both the western (τ = 2.71) and eastern (τ = 2.42) lineages were consistent with a model of sudden demographic expansion. Fu's test was found to be significant for both western (Fs = −6.256, p = < 0.05) and eastern (Fs = −27.386, p = <0.01) lineages. Assuming an average mutation rate of 20% Myr−1 (i.e. μ = 2 × 10−7 × 324 bp = 6.48 × 10−5), the times elapsed from the most recent expansion, using this estimate, were approximately 21 000 (95% CI 10 500–32 000) and approximately 18 600 (95% CI 3000–32 000) years for the western and eastern lineages, respectively. The lower-bound estimates (i.e. 6.3% Myr−1) for the expansion times for the eastern and western lineages were approximately 69 600 years BP (95% CI 35 000–107 000) and approximately 62 000 years BP (95% CI 10 200–107 500), respectively. The upper-bound estimate (25.2% Myr−1) was approximately 16 500 (95% CI 8300–25 500) for the western lineage and approximately 15 000 (95% CI 2400–25 600) for the eastern lineage. Results from the ABC analyses were not entirely consistent. Scenario 1 (i.e. Britain and Ireland diverged from a common ancestor) and scenario 2 (i.e. western and eastern lineages diverged from the common ancestor, whereas Britain and Ireland diverged more recently from the western lineage) had low probability, whereas both scenario 3 (i.e. the Britain and Ireland group diverged more recently from the western lineage) and scenario 4 (i.e. the Britain and Ireland group is the result of an admixture event between the western and eastern lineages) showed the highest probability. Close examination of type I and II errors (data not shown), however, suggests that the current data are insufficient to reliably discriminate among the competing hypotheses.
(f). Species distribution modelling
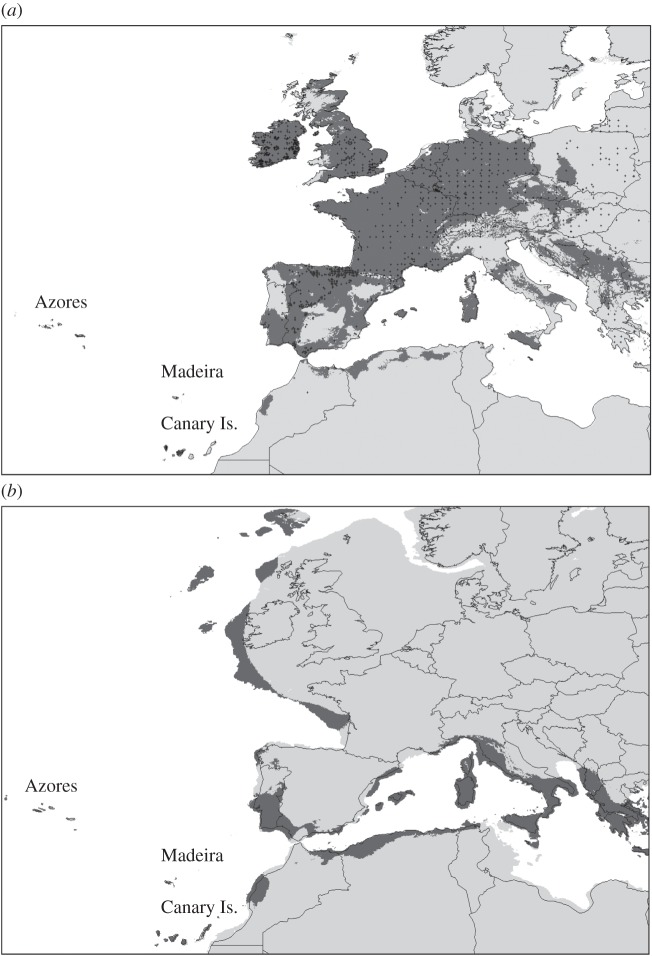
The predicted geographical distribution for the present day and LGM based on SDM are shown in figure 3. Despite recent criticism [50], MaxEnt is thought be a robust and useful approach for predicting relative occurrence probability [51,52]. Overall, the present-day modelled distribution N. leisleri (figure 3a) is in accordance with the known distribution of the species [48]. Discrepancies are most likely to reflect poor data on species occurrence/distribution in eastern Europe. Among the 19 Bioclim variable layers, the best predictors for habitat suitability (i.e. accounting for 92.8% of the total contribution to the MaxEnt model) were bio19 (precipitation of coldest quarter—21.4%), bio3 (isothermality—17.8%), bio7 (temperature annual range—15.6%), bio6 (minimum temperature of coldest month—15.6%), bio8 (mean temperature of wettest quarter—12.7%), bio4 (temperature seasonality—6.6%) and bio10 (mean temperature of warmest quarter—3.1%).
Figure 3.
Species distribution model projections of (a) present-day distributions of N. leisleri and (b) the distribution during the LGM. Black crosses in (a) indicate N. leisleri occurrence data, obtained from the Global Biodiversity Information Facility (GBIF). Dark grey areas in (b) show suitable habitats within the ecological niche of N. leisleri, whereas the light grey areas indicate unsuitable habitat.
Projection of the present-day modelled distribution of N. leisleri onto reconstructed palaeo-climatic conditions at the LGM (figure 3b) indicates the presence of a number of locations with suitable habitats where the species could have persisted. In addition to well-known glacial refugia (i.e. the south of the Iberian Peninsula, Italy, south of the Alps and the Balkans), other putative regions included parts of northern Spain, the Balearics, northern Africa, Madeira, the Canary Islands and the Azores. Potential terrestrial glacial refuge areas were also identified in now submersed land south of the Bay of Biscay in France, towards the west/southwest of Ireland, as well as sites probably suitable for hibernation only and as far north as western Scotland and the Faroe Islands.
4. Discussion
(a). Taxonomy and phylogeography
The present results reveal new insights into the taxonomy and evolutionary relationship of the Leisler's bat (N. leisleri) and the Azorean bat (N. azoreum). Once considered only a subspecies of N. leisleri [16], N. azoreum has more recently proposed as a good species [17,28]. Analysis of the new molecular data generated in this study, in combination with previous data by Salgueiro et al. [27,28], unambiguously demonstrates that the majority of Irish and British N. leisleri mtDNA individuals (67%) are represented by haplotypes from the western lineage (i.e. more closely related to N. azoreum haplotypes than to any other haplotypes from N. leisleri from continental Europe). Thus, from an mtDNA perspective, N. azoreum does not represent a monophyletic group, and hence does not meet the phylogenetic species criterion.
We demonstrate that both the western and eastern mtDNA haplotypic groups represent distinct evolutionary lineages, which probably diverged in allopatry in separate glacial refugia. Based on the results of the network analyses, Ireland and Britain are likely to comprise a zone of secondary contact and admixture between these two lineages. While this is not supported by the ABC analysis, this approach, as applied to our dataset (i.e. combination of samples and molecular data available for analyses), did not discriminate among competing colonization scenarios. The presence of unique derived haplotypes representing both lineages in Ireland and Britain only suggests that this secondary contact is of some antiquity and no longer occurs. Thus, excluding the common eastern lineage Po1, the putative ancestral haplotype, and two further haplotypes that occur in Britain (En1 and Fr1), none of the other eastern haplotypes found in Ireland and Britain (Ir6, Ir8, Ir9, Ir10 and En2) are found in mainland Europe. While this could be partially attributed to small sample size, the occurrence of geographically restricted haplotypes is also compatible with new mutations occurring since the time of expansion, which is estimated at between 10 516–32 114 years BP for the western lineage and between 3070–32 268 years BP for the eastern lineage using the average mutation rate of 20% Myr−1, coinciding with the LGM, which peaked some 20 000 years BP. Furthermore, N. leisleri is characterized by long migrations in mainland Europe. Thus, even considering small sample sizes per location, our sample size (n = 186) over all of Europe is sufficient to argue against sample size bias. Colonization of the Azores archipelago by early ancestors of N. azoreum, in agreement with Salgueiro et al. [27], is most likely to have occurred from a single source of colonists, given the widespread distribution of the haplotype A7 throughout Ireland, Britain and the Azores. An additional colonization event took place subsequently in the British Isles, involving ancestors of the eastern European lineage (probably represented by the Po1 haplotype).
The importance of the Balkans, Italy and the Iberian Peninsula as glacial refugia for northern temperate species during the LGM has been well documented [53,54], and is supported here by SDM of the past climatic conditions for N. leisleri. Fossil remains of N. leisleri from the middle and late Pleistocene have been found in southern Spain [55], whereas fossil remains of N. noctula and M. myotis have been found in the Balkans [56,57]. Thus, given the current geographical distribution of the eastern lineage, with no strong signal in the current dataset, N. leisleri may have inhabited any one of these refugia during the LGM. The SDM results also suggest the possibility of refugia in northern Africa, northern parts of Italy, Spain and the Balearics. The current distribution of the western lineage, however, is hard to explain by invoking any of these three accepted glacial refugia, given the apparent absence of haplotypes from this lineage in continental Europe. The Iberian Peninsula, being the most westerly of these refugia, would be the most probable location. In support of this, the ‘Lusitanian flora and fauna’ in Ireland proposed by a number of authors [9] links some 15–20 extant species between southwest Ireland and Iberia. In this study, however, no haplotypes of the western lineage were found in Iberia, despite a good sample size (n = 36) and geographical coverage of the area, arguing against this region as the refuge for this lineage.
One possible explanation for the absence of this lineage in western Europe is replacement by the expanding eastern lineage. A hypothesis was proposed by Martínková et al. [10] investigating the phylogeography of the Irish stoat (Mustela erminea hibernicus). These authors suggested that a cold tolerant lineage that initially colonized Britain and Ireland was replaced in Britain by an expanding lineage from Europe, which was assumed to have a fitness advantage under warmer environmental conditions. Piertney et al. [58] and Searle et al. [59] have used the same mtDNA replacement hypothesis to explain the Celtic fringe distribution seen in many British mammals, including the pygmy shrew (Sorex minutus), bank vole (Clethrionomys glareolus), field vole (Microtus agrestis) and water vole (Arvicola terrestris), which is strikingly similar to that described for the Celtic people [60]. In this study, however, the coexistence of both western and eastern lineages of N. leisleri in Ireland and Britain, and the fact that according to microsatellite analysis they constitute a single population, argues against mtDNA replacement to explain the absence of the western lineage in continental Europe.
Alternatively, there are several potential histories involving cryptic glacial refugia for the western lineage of N. leisleri during the LGM which are independently supported by palaeo-climatic SDM. It is possible that the Irish and British populations originated from populations that diverged in an Azorean refugium where suitable habitat was available according to the SDM. While active dispersal from the Azores to Ireland and Britain is unlikely given the over-water distances involved, it is possible that passive dispersal may have occurred as a result of the prevailing winds in the North Atlantic.
Furthermore, suitable habitats along the coastal regions of western Europe in areas now submerged were identified by SDM methods. The influence of ice loading on the land masses [61] would have meant that ice-free, dry land extended several hundred kilometres into the Atlantic Ocean and Celtic Sea. Exposed land along the coast of western Europe including southern France, the Bay of Biscay and land to the southwest of Ireland, according to our model, would have provided suitable habitat for N. leisleri throughout the LGM. In support of this hypothesis, both southern France and the Bay of Biscay have been proposed as potential refugia by a recent study modelling the past geographical distribution of another European bat species, Plecotus austriacus [62]. The existence of refugia in northwestern Europe is also supported by the presence of several tree species [63] in the region during the LGM. Furthermore, Mitchell [64] suggests that some tree species colonized Ireland as early as 9500 years BP, supporting the existence of a nearby, forest glacial refugium. Another clue comes from the study of the sibling species N. noctula by Petit et al. [41]. In their analysis of the mtDNA Dloop of N. noctula populations in Europe, they reported on the occurrence of highly differentiated haplotypes (i.e. long-branch haplotypes) among specimens collected in both the UK and Germany. Although their sample size was small, the presence of these haplotypes suggested the existence of highly divergent lineages among the mtDNA Dloop of N. noctula. These lineages restricted to western Europe lend weight to the hypothesis of novel cryptic western European refugia for bats. A northwesterly refugia, particularly along the Atlantic coast, could also help to explain why many Irish flora and fauna are genetically distinct and diverse from British and continental European counterparts, including the stoat [10], the natterjack toad [65] and the brown trout [66,67].
(b). Genetic diversity and contemporary gene flow
Regional differentiation in mtDNA indicates limited gene flow between populations of N. leisleri across the major geographical areas of Ireland and Britain, mainland Europe and the Azores. This is supported by 42% of the observed genetic variation being explained by differences between these major geographical regions. Results from the nuclear microsatellites amplified for samples from sites in Ireland and France support this, as does the apparent lack of large scale migratory movement observed among populations in Ireland [26], and the timing and number of records of N. leisleri recorded on islands in the eastern North Atlantic [68]. Furthermore, this is supported by the reported morphological and behavioural differences in N. azoreum [18,19]. We suggest that N. azoreum and N. leisleri populations from the Azores and continental Europe comprise at least two distinct evolutionary significant units that display distinct genetic diversity, and possibly adaptive features, whereas Britain and Ireland represent a zone of admixture. Owing to a lack of contemporary gene flow, these three areas should be considered separate management units.
(c). Concluding remarks
We demonstrate that populations of N. leisleri in Ireland and Britain probably recolonized following the LGM from two distinct glacial refugia. One lineage is consistent with the hypothesis of colonization from a southern European refugium, with no strong evidence to support colonization across a land bridge with Britain. The second lineage is inconsistent with a southern European origin, and thus we cannot refute a hypothesis for the existence of cryptic glacial refugia in western Europe. There is limited contemporary gene flow between recent populations of N. leisleri in continental Europe, Britain and Ireland. This indicates that after the LGM, dispersal of lineages of this species was conservative, underscoring the role of a cryptic refugium in the western isles, and explaining the current distribution of lineages. The post-glacial history revealed here with regard to N. leisleri may be applicable to other species that are dependent on deciduous woodland and were able to colonize Britain and Ireland rapidly after the LGM. We also conclude that there is no genetic support for species status for the endemic N. azoreum, but that the genetic variation apparent in its populations may play a vital role in securing its future against the challenge of rapid climatic change, and thus is worthy of independent conservation consideration.
Supplementary Material
Acknowledgements
The authors acknowledge the Department of Education and Learning (DEL) for funding this study and the following people for help with samples: Susie Brown, Austin Hopkirk, Kate McAney, Enda Mullen, Conor Kelleher, Caroline Shiel, Jim Hurley, Tina Aughney, Niamh Roche, the Northern Ireland Bat Group, the National Trust, Stéphane G Roue and his team in France, Freider Mayer, Beatrice Blochlinger at the Naturhistorshes Museum Bern, Laurent Duverge, George Bemment, Forsyth, Sarah Harris and the Veterinary Laboratory Agency (VLA), Wigbert Schortz and Julia Prüger, Richard Selman, John Haddow, Mick Canham, Shiela Wright, Jackie Wedd, Cindy Blaney, Danilo Russo, Eric Petit, Jean-Francois Noblet, Armand Fayard for samples from the Natural History Museum Grenoble, Bruno Fronseca Simões, and Plecotus, Estudos Ambientais, Ld, Sebastièn Puechmaille and Laurent Arthur at the Muséum d'Histoire naturelle de la ville de Bourges. In addition, we are indebted to the advice and comments of several anonymous reviewers.
Ethics statement
All bats were caught and sampled under licence of the Northern Ireland Environment Agency, the National Parks and Wildlife Service and the Republique Francaise (Prefecture de la Côte-d'Or and Prefecture de la Nièvre).
Data accessibility
DNA sequences for each new haplotype identified in this study are available on GenBank (accession nos. KP462690–KP462718). Microsatellite data (Genepop format) are available for download on Dryad (doi:10.5061/dryad.6g6r6).
References
- 1.Stuart AJ. 1995. Insularity and quaternary vertebrate faunas in Britain and Ireland. In Island Britain: a quaternary perspective (ed. Preece RC.), pp. 111–125. London, UK: Geological Society. [Google Scholar]
- 2.Wingfield RTR. 1995. A model of sea-levels in the Irish and Celtic seas during the end-Pleistocene to Holocene transition. In Island Britain: a quaternary perspective (ed. Preece RC.), pp. 209–242. London, UK: Geological Society. [Google Scholar]
- 3.Yalden D. 1999. The history of British mammals. London, UK: Poyser. [Google Scholar]
- 4.Stewart JR, Lister AM. 2001. Cryptic northern refugia and the origins of the modern biota. Trends Ecol. Evol. 16, 608–613. ( 10.1016/S0169-5347(01)02338-2) [DOI] [Google Scholar]
- 5.Mitchell F, Ryan M. 2001. Reading the Irish Landscape. Dublin, Ireland: Town House. [Google Scholar]
- 6.Montgomery WI, Provan J, McCabe AM, Yalden DW. 2014. Origin of British and Irish mammals: disparate post-glacial recolonisation and species introductions. Quat. Sci. Rev. 98, 144–165. ( 10.1016/j.quascirev.2014.05.026). [DOI] [Google Scholar]
- 7.Edwards R, Brooks A. 2008. The island of Ireland, Drowning the myth of an Irish land-bridge? In Mind the gap: postglacial colonization of Ireland (eds Davenport JJ, Sleeman DP, Woodman PC.), pp. 19–34. Dublin, Ireland: Irish Naturalists Journal. [Google Scholar]
- 8.Brooks AJ, Bradley SL, Edwards RJ, Milne GA, Horton B, Shennan I. 2008. Postglacial relative sea-level observations from Ireland and their role in glacial rebound modeling. J. Quat. Sci. 23, 175–192. ( 10.1002/jqs.1119) [DOI] [Google Scholar]
- 9.Vincent P. 1990. The Biogeography of the British Isles. London, UK: Routledge. [Google Scholar]
- 10.Martínková N, McDonald RA, Searle JB. 2007. Stoats (Mustela eriminea) provide evidence of natural overland colonisation of Ireland. Proc. R. Soc. B 274, 1387–1393. ( 10.1098/rspb.2007.0334) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell-Jones AJ, et al. 1999. The atlas of European mammals. London, UK: Academic Press. [Google Scholar]
- 12.Bogdanowicz W, Ruprecht AL. 2004. Nyctalus leisleri (Kuhl, 1817) Kleinabendsegler. In Handbuch der Säugetiere Europas. Band 4/II: Fledertiere (Chiroptera) II (ed. KRAPP F.), pp. 717–756. Wiebelsheim, Germany: AULA. [Google Scholar]
- 13.Roche N, Langton S, Aughney T, Russ J. 2007. The car-based bat monitoring scheme for Ireland, report for 2006. Dublin, Ireland: Bat Conservation Ireland. [Google Scholar]
- 14.Buckley D, Puechmaille S, Roche N, Teeling E. 2011. A critical assessment of the presence of Barbastella barbastellus and Nyctalus noctula in Ireland with a description of N. leisleri echolocation calls from Ireland. Hystrix, Italian J. Mammal. 22, 111–127. [Google Scholar]
- 15.McAney C, Fairley J. 1990. Activity of Leisler's bat (Nyctalus leisleri Kuhl, 1818) at a summer roost in Ireland. Myotis 28, 83–91. [Google Scholar]
- 16.Palmeirim JM. 1991. A morphometric assessment of the systematic position of the Nyctalus from the Azores and Madeira Mammalia, Chiroptera. Mammalia 55, 381–388. ( 10.1515/mamm.1991.55.3.381) [DOI] [Google Scholar]
- 17.Speakman JR, Webb PI. 1993. Taxonomy, status and distribution of the Azorean bat (Nyctalus azoreum). J. Zool. 231, 27–38. ( 10.1111/j.1469-7998.1993.tb05350.x) [DOI] [Google Scholar]
- 18.Rainho A, Marques JT, Palmeirim JM. 2002. Os morcegos dos arquipélagos dos Açores e da Madeira, um contributo para a sua conservação. Lisbon, Portugal: Instituto da Conservação da Natureza. [Google Scholar]
- 19.Skiba R. 2003. Europäische Fledermäuse. Kennzeichen, Echoortung und Detektoranwendung, pp. 212 Hohenwarsleben, Germany: Westarp Wissenschaften. [Google Scholar]
- 20.Ruczyński I, Bogdanowicz W. 2005. Roost cavity selection by Nyctalus noctula and N. leisleri (Vespertilionidae, Chiroptera) in the Białowieża Primeval Forest, eastern Poland. J. Mammal. 86, 921–930. ( 10.1644/1545-1542(2005)86[921:RCSBNN]2.0.CO;2) [DOI] [Google Scholar]
- 21.Harris S, Yalden DW. 2008. Mammals of the British Isles: Handbook, 4th edn London, UK: Mammal Society. [Google Scholar]
- 22.O'Neill JK. 2000. Aspects of the ecology and conservation of Leisler's bats (Nyctalus leisleri) in Northern Ireland. Unpublished PhD thesis, Queen's University Belfast, Ireland. [Google Scholar]
- 23.Ohlendorf B, Hecht B, Strassburg D, Agirre-Mendi PT. 2000. Fernfund eines Kleinabendseglers Nyctalus leisleri in Spanien. Nyctalus 7, 239–242. [Google Scholar]
- 24.Wohlgemuth R, Devrient I, García A, Hutterer R. 2004. Long distance flight of a Lesser noctule (Nyctalus leisleri) after rehabilitation. Myotis 42–42, 69–73. [Google Scholar]
- 25.Dondini G, Rutowski T, Vergari S, Wojtaszyn G. 2012. Long distance migration of female Leisler's bat (Nyctalus leisleri) from Italy to Poland. Hystrix, Ital. J. Mammal. 23, 93–94. [Google Scholar]
- 26.Shiel CB, Shiel RE, Fairley JS. 1999. Seasonal changes in the foraging behaviour of Leisler's bats (Nyctalus leisleri) in Ireland as revealed by radiotelemetry. J. Zool. Lond. 249, 195–214. ( 10.1111/j.1469-7998.1999.tb00770.x) [DOI] [Google Scholar]
- 27.Salgueiro P, Coehlo MM, Palmeirim JM, Ruedi M. 2004. Mitochondrial DNA variation and population structure of the island endemic Azorean bat (Nyctalus azoreum). Mol. Ecol. 13, 3357–3366. ( 10.1111/j.1365-294X.2004.02354.x) [DOI] [PubMed] [Google Scholar]
- 28.Salgueiro P, Ruedi M, Coelho M, Palmeirim M. 2007. Genetic divergence and phylogeography in the genus Nyctalus (Mammalia, Chiroptera): implications for population history of the insular bat Nyctalus azoreum. Genetica 130, 169–181. ( 10.1007/s10709-006-9004-x) [DOI] [PubMed] [Google Scholar]
- 29.Castella V, Ruedi M, Excoffier L. 2001. Contrasted patterns of mitochondrial and nuclear structure among nursery colonies of the bat Myotis myotis. J. Evol. Biol. 14, 708–720. ( 10.1046/j.1420-9101.2001.00331.x) [DOI] [Google Scholar]
- 30.Worthington-Wilmer J, Barrett E. 1996. A non-lethal method of tissue sampling for genetic studies of chiropterans. Bat Res. News 37, 1–3. [Google Scholar]
- 31.Puechmaille S, Mathy G, Petit E. 2007. Good DNA from bat droppings. Acta Chiropterologica 91, 269–276. ( 10.3161/1733-5329(2007)9[269:GDFBD]2.0.CO;2) [DOI] [Google Scholar]
- 32.Fumagalli L, Hausser J, Taberlet P, Giely L, Stewart DT. 1996. Origin and evolution of homologous repeated sequences in the mitochondrial DNA control region of shrews. Mol. Biol. Evol. 13, 31–46. ( 10.1093/oxfordjournals.molbev.a025568) [DOI] [PubMed] [Google Scholar]
- 33.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Series 41, 95–98. [Google Scholar]
- 34.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W, improving the sensitivity of progressive multiple sequence alignments through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. ( 10.1093/nar/22.22.4673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ballard JW, Whitlock MC. 2004. The incomplete natural history of mitochondria. Mol. Ecol. 13, 729–744. ( 10.1046/j.1365-294X.2003.02063.x) [DOI] [PubMed] [Google Scholar]
- 36.Boston ESM, Montgomery I, Prodöhl PA. 2009. Development and characterization of 11 polymorphic compound tri- and tetranucleotide microsatellite loci for the Leisler's bat, Nyctalus leisleri (Vespertilionidae, Chiroptera). Conserv. Genet. 10, 1501–1504. ( 10.1007/s10592-008-9768-x) [DOI] [Google Scholar]
- 37.Excoffier L, Laval G, Schneider S. 2005. Arelequin (v. 3.0): an integrated software package for population genetics analysis. Evol. Bioinf. Online 1, 47–50. [PMC free article] [PubMed] [Google Scholar]
- 38.Bandelt H, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogeneis. Mol. Biol. Evol. 16, 37–48. ( 10.1093/oxfordjournals.molbev.a026036) [DOI] [PubMed] [Google Scholar]
- 39.Fu YX, Li WH. 1993. Statistical tests of neutrality of mutations. Genetics 133, 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogers AR, Harpending H. 1992. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9, 552–569. [DOI] [PubMed] [Google Scholar]
- 41.Petit E, Excoffier L, Mayer F. 1999. No evidence of bottle neck in the postglacial recolonisation of Europe by the noctule bat (Nyctalus noctula). Evolution 53, 1247–1258. ( 10.2307/2640827) [DOI] [PubMed] [Google Scholar]
- 42.Chen S-F, Rossiter SJ, Faulkes CG, Jones G. 2006. Population genetic structure and demographic history of the endemic Formosan lesser horseshoe bat (Rhinolophus monoceros). Mol. Ecol. 15, 1643–1656. ( 10.1111/j.1365-294X.2006.02879.x) [DOI] [PubMed] [Google Scholar]
- 43.Cornuet J-M, Ravigné V, Estoup A. 2010. Inference on population history and model checking using DNA sequence and microsatellite data with the software DIYABC (v. 1.0). BMC Bioinformatics 11, 401 ( 10.1186/1471-2105-11-401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 1552, 945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Earl DA. 2009. Structure Harvester v. 0.3 See http://taylor0.biology.ucla.edu/struct_harvest/.
- 46.Keenan K, McGinnity P, Cross T, Crozier WW, Prodöhl PA. 2013. diveRsity: an R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol. Evol. 8, 782–788. ( 10.1111/2041-210X.12067) [DOI] [Google Scholar]
- 47.Philips SJ, Anderson RP, Schapire RE. 2006. Maximum entropy modelling of species geographic distributions. Ecol. Model. 190, 231–259. ( 10.1016/j.ecolmodel.2005.03.026) [DOI] [Google Scholar]
- 48.Dietz C, von Helversen O, Nill D. 2009. The handbook of the bats of Britain, Europe and North Africa. London, UK: A&C Black. [Google Scholar]
- 49.Cantor SB, Sun CC, Tortolero-Luna G, Richards-Kortum R, Follen M. 1999. A comparison of C/B ratios from studies using receiver operating characteristic curve analysis. J. Clin. Epidemiol. 53, 885–892. ( 10.1016/S0895-4356(99)00075-X) [DOI] [PubMed] [Google Scholar]
- 50.Royle JA, Chandler RB, Yackulic C, Nichols JD. 2012. Likelihood analysis of species occurrence probability from presence-only data for modelling species distributions. Methods Ecol. Evol. 3, 545–554. ( 10.1111/j.2041-210X.2011.00182.x) [DOI] [Google Scholar]
- 51.Elith J, Phillips SJ, Hastie T, Dudík M, Chee YE, Yates CJ. 2011. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 17, 43–57. ( 10.1111/j.1472-4642.2010.00725.x) [DOI] [Google Scholar]
- 52.Merow C, Silander JA. 2014. A comparison of Maxlike and Maxent for modelling species distributions. Methods Ecol. Evol. 5, 215–225. ( 10.1111/2041-210X.12152) [DOI] [Google Scholar]
- 53.Hewitt GM. 1999. Post-glacial re-colonization of European biota. Mol. Genet. Anim. Ecol. 68, 87–112. [Google Scholar]
- 54.Taberlet P, Fumagalli L, Wust-Saucy AG, Cossons JF. 1998. Comparative phylogeography and post-glacial colonization routes in Europe. Mol. Ecol. 7, 453–464. ( 10.1046/j.1365-294x.1998.00289.x) [DOI] [PubMed] [Google Scholar]
- 55.Sevilla P. 1989. Quaternary fauna of bats in Spain, Paleoecologic and biogeographic interest. In European bat research 1987 (eds Hanak V, Horacek I, Gaisler J.), pp. 349–355. Prague, Czech Republic: Charles University Press. [Google Scholar]
- 56.Popov V, Ivanova T. 1995. Morphoecological analysis and late Quaternary history of a bat community in a karstic landscape of North Bulgaria. Myotis 32–33, 21–32. [Google Scholar]
- 57.Bogdanowicz W, Van Den Bussche RA, Gajewska M, Postawa T, Harutyunyan M. 2009. Ancient and contemporary DNA sheds light on the history of mouse-eared bats in Europe and the Caucasus. Acta Chiropterologica 11, 289–305. ( 10.3161/150811009X485530) [DOI] [Google Scholar]
- 58.Piertney SB, Stewart WA, Lambin X, Telfer S, Aars J, Dallas JF. 2005. Phylogeographic structure and postglacial evolutionary history of the water voles (Arvicola terrestris) in the United Kingdom. Mol. Ecol. 14, 1435–1444. ( 10.1111/j.1365-294X.2005.02496.x) [DOI] [PubMed] [Google Scholar]
- 59.Searle JB, Kotlik P, Rambau RV, Marková S, Herman JS, Mc Devitt AD. 2009. The Celtic fringe of Britain, insights from small mammal phylogeography. Proc. R. Soc. B 276, 4287–4294. ( 10.1098/rspb.2009.1422) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sykes B. 2006. The blood of the Isles. Ealing, UK: Bantam Press. [Google Scholar]
- 61.Lowe JJ, Walker MJC. 1997. Reconstructing quaternary environments, 2nd edn London, UK: Longman. [Google Scholar]
- 62.Razgour O, et al. 2013. The shaping of genetic variation in edge-of-range populations under past and future climate change. Ecol. Lett. 16, 1258–1266. ( 10.1111/ele.12158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Svenning JC, Normand S, Kageyama M. 2008. Glacial refugia of temperate trees in Europe, insights from species distribution modelling. J. Ecol. 966, 1117–1127. ( 10.1111/j.1365-2745.2008.01422.x) [DOI] [Google Scholar]
- 64.Mitchell FJG. 2006. Where did Ireland's trees come from? Biology and the Environment. Proc. Royal Irish Acad. B 106, 251–259. ( 10.3318/BIOE.2006.106.3.251) [DOI] [Google Scholar]
- 65.Rowe G, Harris DJ, Beebee TJC. 2006. Lusitania revisited, A phylogeographic analysis of the natterjack toad (Bufo calamita) across its entire biogeograpical range. Mol. Phylogenet. Evol. 39, 335–346. ( 10.1016/j.ympev.2005.08.021) [DOI] [PubMed] [Google Scholar]
- 66.Ferguson A. 2004. The importance of identifying conservation units, brown trout and pollan biodiversity in Ireland. Biol. Environ. Proc. Royal Irish Acad. B 104, 33–41. ( 10.3318/BIOE.2004.104.3.33) [DOI] [Google Scholar]
- 67.McKeown NJ, Hynes RA, Duguid RA, Ferguson A, Prodöhl PA. 2010. Phylogeographic structure of brown trout (Salmo trutta) in Britain and Ireland, glacial refugia, post-glacial colonisation, and origins of sympatric populations. J. Fish Biol. 76, 319–347. ( 10.1111/j.1095-8649.2009.02490.x) [DOI] [PubMed] [Google Scholar]
- 68.Petersen A, Jensen J-K, Jenkins P, Dorete B, Ingimarsson F. 2014. A review of the occurrence of bats (Chiroptera) on islands in the North East Atlantic and on North Sea installations. Acta Chiropterologica 16, 169–195. ( 10.3161/150811014X683381) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences for each new haplotype identified in this study are available on GenBank (accession nos. KP462690–KP462718). Microsatellite data (Genepop format) are available for download on Dryad (doi:10.5061/dryad.6g6r6).