Abstract
Lentil (Lens culinaris ssp. culinaris) is a nutritious and affordable pulse with an ancient crop domestication history. The genus Lens consists of seven taxa, however, there are many discrepancies in the taxon and gene pool classification of lentil and its wild relatives. Due to the narrow genetic basis of cultivated lentil, there is a need towards better understanding of the relationships amongst wild germplasm to assist introgression of favourable genes into lentil breeding programs. Genotyping-by-sequencing (GBS) is an easy and affordable method that allows multiplexing of up to 384 samples or more per library to generate genome-wide single nucleotide Polymorphism (SNP) markers. In this study, we aimed to characterize our lentil germplasm collection using a two-enzyme GBS approach. We constructed two 96-plex GBS libraries with a total of 60 accessions where some accessions had several samples and each sample was sequenced in two technical replicates. We developed an automated GBS pipeline and detected a total of 266,356 genome-wide SNPs. After filtering low quality and redundant SNPs based on haplotype information, we constructed a maximum-likelihood tree using 5,389 SNPs. The phylogenetic tree grouped the germplasm collection into their respective taxa with strong support. Based on phylogenetic tree and STRUCTURE analysis, we identified four gene pools, namely L. culinaris/L. orientalis/L. tomentosus, L. lamottei/L. odemensis, L. ervoides and L. nigricans which form primary, secondary, tertiary and quaternary gene pools, respectively. We discovered sequencing bias problems likely due to DNA quality and observed severe run-to-run variation in the wild lentils. We examined the authenticity of the germplasm collection and identified 17% misclassified samples. Our study demonstrated that GBS is a promising and affordable tool for screening by plant breeders interested in crop wild relatives.
Introduction
Lentil (Lens culinaris ssp. culinaris) is an annual, herbaceous, self-pollinating grain legume crop. This crop is important in cereal-based cropping systems due to its nitrogen-fixing ability. Lentil has a historical and geographic domestication history [1–3]. Lentil and its wild relatives are naturally distributed in South-west Asia and Mediterranean regions [1]. The genus Lens (2n = 14) is phylogenetically nested within the tribe Vicieae, which are cool-season legumes belonging to sub-family Papilionoideae of family Fabaceae [4]. The genus Lens has seven closely related taxa, namely L. culinaris, L. orientalis, L. tomentosus, L. odemensis, L. lamottei, L. ervoides and L. nigricans. Past taxonomic studies [5–13], based on morphology, cytogenetics, hybridization studies, and/or molecular markers, have frequently disagreed with respect to classification at the species and subspecies level. The most recent classification identified seven taxa grouped into four species, namely L. culinaris ssp. culinaris, L. culinaris ssp. orientalis, L. culinaris ssp. tomentosus, L. culinaris ssp. odemensis, L. ervoides, L. lamottei, and L. nigricans [14, 15]. Despite the taxonomic re-organizations, all studies generally agreed that L. culinaris ssp. orientalis is the most closely related wild progenitor of L. culinaris ssp. culinaris while L. nigricans is the most distant relative.
The ancient domestication history of cultivated lentil has produced bottleneck effects resulting in a narrow genetic basis which has resulted in reduced levels of resistance to biotic and abiotic stresses relative to its wild relatives [16]. Phenotypic variability studies have identified wild lentil germplasm with resistance to anthracnose (Colletotrichum spp.), ascochyta blight (Ascochyta lentis), stemphylium blight (Stemphylium botryosum) and Orobanche spp. root-holoparasitic infection [17–21]. To increase genetic diversity and resistance to biotic and abiotic stresses in new cultivars, introgression of favourable genes from crop wild relatives is necessary. Like many crops, wild relatives can be divided into primary, secondary and tertiary gene pools, according to their relatedness to L. culinaris ssp. culinaris and their ability to produce fertile hybrids when intercrossed with cultivated lentil [22]. Some early studies, based on cross-compatibility and cytogenetic evidence [23–25], placed L. orientalis in the primary gene pool while the secondary gene pool consisted of L. odemensis, L ervoides and L. nigricans [26]. However, the gene pool placement of two more recently identified species L. lamottei and L. tomentosus [27] is inconsistent among studies. These two species were placed in the secondary gene pool by Muehlbauer and McPhee [22] while a later study suggested that more crossing experiments would be necessary to determine their positions [28]. In addition, a more recent study suggested that L ervoides and L. nigricans should be placed in the tertiary gene pool [29]. Nonetheless, the gene pools proposed by all these studies are not in concordance with the species classification from the Ferguson et al. study [14] suggesting more work is needed to clarify these relationships.
Successful and efficient diversification of cultivated lentils through introgression of genes from wild relatives greatly depends on accurate identification of the wild species followed by successful development of fertile hybrid plants. Despite having good standard procedures for the management of plant genetic resources, incorrect classification is not uncommon. Furthermore, human error can be introduced at various stages in a breeding program, e.g., seed handling, storage, harvesting and exchange between plant breeders. Some accessions of the wild progenitor L. orientalis are morphologically almost indistinguishable from the cultivated lentil and the two species are fully inter-fertile [1] making classification difficult. Many studies have employed molecular markers to improve the accuracy of germplasm characterization [30–33], however, the high cost per sample involved in marker discovery and screening has restricted practical application in plant breeding programs. The recent improvements of next-generation sequencing-based genotyping methods has made routine screening of plant germplasm feasible and cost-effective [34].
Genotyping-by-sequencing (GBS) is genome-wide reduced representation of Single Nucleotide Polymorphisms (SNPs) developed for Illumina sequencing technology [35]. Compared to other complexity reduction methods such as Reduced Representation Libraries (RRL) and Restriction site Associated DNA (RAD) sequencing, the GBS method is favoured as a relatively simple and quick method for generating SNP data [36, 37]. The initial protocol was developed using one restriction enzyme [37] and subsequently modified to use two restriction enzymes (a common cutter and a rare cutter) to generate uniform complexity reduction [38]. The two-enzyme approach reduces genome complexity by avoiding sequencing of repetitive regions resulting in more straightforward bioinformatics analysis for large genomes. GBS has already been successfully applied in highly homozygous crops such as maize, rice, soybean, wheat and barley to provide large numbers of SNP markers for association studies and genomic-assisted breeding [37–40]. The low read depth produced by GBS poses a challenge in accurate detection of heterozygous SNPs in mapping populations and crop wild relatives [41, 42]. Nonetheless, recent reports demonstrated the use of GBS in wild crop relatives to resolve phylogenetic relationship and genetic diversity [43–45]. Therefore, the GBS method has great potential for characterization of the large and complex genomes (~4 Gb [46]) of the genus Lens.
The objective of this study was to characterize a collection of wild and cultivated Lens spp. germplasm currently used in the University of Saskatchewan lentil breeding program to gain a better understanding of taxon and gene pool classification. Here, we explore the use of GBS using a two-enzyme system to discover and genotype genome-wide SNPs. We developed an automated GBS pipeline to facilitate SNP detections from a large number of samples. To improve the understanding of our existing germplasm, we examined the phylogenetic relationships and population structure of the genus Lens. We also verified the authenticity of all the accessions and their biological replicates and evaluated the reproducibility of GBS results in species classification and accession identification.
Materials and Methods
Plant materials
The genetic diversity panel of the genus Lens consisted of 83 samples which originated from 60 diverse varieties and landraces (subsequently known as accessions). As several seed sources may be available for an accession, each seed source represented a biological replicate and was subsequently referred to as a sample. The number of accessions ranged from 5 to 15 for each of the seven taxa. The information about their accession numbers, species classification, seed source and geographical location is available in S1 Table. To obtain plant materials from these accessions, a single plant of each sample was selfed to produce sufficient seeds for the study. About 3–5 seeds were germinated and the seedlings were grown for 2–3 weeks in a controlled environment growth chamber. Genomic DNA extraction was carried out using pooled fresh leaf tissues using a modified CTAB method [47]. The quantity and quality of the genomic DNA was checked on a 1% agarose gel and determined using Quant-iT PicoGreen dsDNA assay kit (Life Technologies, USA) on a FLUOstar Omega fluorometer (BMG Labtech, Germany).
Library preparation for genotyping-by-sequencing
Two GBS libraries were constructed based on a modified protocol from Poland et al. [38] using a two-enzyme system, PstI (rare cutter) and MspI (common cutter). The protocol consisted of three main steps: restriction digestion, ligation, and PCR amplification. Each GBS library was a 96-plex library consisting of 48 samples in two technical replicates. We first prepared one library consisting of 36 samples from the lentil diversity panel and 12 samples from another experiment. A second GBS library was constructed to include samples from the first library which produced low number of reads and additional samples to increase representation of some species. Each sample was labelled with its accession number as prefix and suffixes representing the seed source (a/b/c/A/S/R) and GBS libraries (r1/r2). See S2 Table for information on GBS libraries.
For each sample, a total of 200 ng of genomic DNA was digested using 8 U of Pst1-HF and 8 U MspI (NEB, USA) in a 20 μl reaction mixture, by incubating at 37 ºC for 2 h followed by denaturation of restriction enzyme at 65 ºC for 20 min. Ligation was performed in a 40 μl reaction volume containing the digested DNA, 0.02 μM (0.1 pM) of Adapter 1 (containing barcode sequence), 3 μM (15 pM) of Adapter 2 and ligation master mix (1X NEB buffer 4, 1 mM ATP and 200 U T4 DNA ligase). The ligation mixture was incubated at 22 ºC for 2 h followed by inactivation of the enzyme at 65 ºC for 20 min. An aliquot of 10 μl of adapter-ligated DNA from each sample was pooled and adjusted to a final volume of 500 μl. About 400 μl of the pooled DNA was purified using QIAquick PCR purification kit (Qiagen, Germany) as per manufacturer's guidelines and eluted into a total volume of 120 μl using EB buffer.
In the amplification step, 8 PCR reactions were set up for each library. Each PCR reaction contained 10 μl of eluted DNA, 1X NEB Master Mix, 0.8 μM Illumina Primer 1 (barcoded adapter with PstI overhangs) and 0.8 μM Illumina Primer 2 (common Y-adapter) prepared in a final volume of 25 μl. PCR amplification was performed using an initial denaturation of 95 ºC for 30 s, followed by 16 cycles of 95 ºC for 30 s, 62 ºC for 20 s and 68 ºC for 30 s and a final elongation step of 72 ºC for 5 min. All eight PCR reactions were pooled and purified using QIAquick PCR purification kit (Qiagen, Germany) and the purified library was eluted in 30 μl EB. The quality and quantity of the library was measured using a Bioanalyzer DNA 1000 Chip (Agilent, USA) and Qubit High Sensitivity dsDNA kit (Invitrogen, USA). The libraries were diluted to 2 nM and sequenced on an Illumina Hiseq 2500 (2 x100 bp) at the DNA Sequencing Laboratory, NRC-Saskatoon. The raw sequencing reads were deposited in NCBI SRA (Accession numbers: SRX703778-SRX703943).
Read mapping and SNP calling
Raw Illumina reads were de-multiplexed and the barcode sequences removed. Any sequences not containing the expected restriction sites for both enzymes were removed. Subsequently, the reads were filtered and trimmed using recommended settings in Trimmomatic-0.17 [48]. Due to uneven read distribution between the two technical replicates for each sample, both reactions were merged before variant calling. The filtered reads for each sample were aligned to the draft lentil genome (version 0.6) of CDC Redberry (Lens culinaris) using Bowtie2-2.1.0 [49] allowing only end-to-end matches. Variant-calling was performed with Samtools-0.1.18 [50] and output in VCF format [51]. The SNP results from all the samples were merged into one large file using custom Perl scripts. This pipeline (Fig 1) is publicly available at http://knowpulse2.usask.ca/pulse-binfo/software/GBS-Pipeline. In silico prediction of ApeKI and PstI-MspI fragments was performed on the draft lentil genome (version 0.6) using a custom Python script. In addition, the scaffolds from which the SNPs originated were searched against M. truncatula genome version Mt4.0 [52] using NCBI-BLAST-2.2.28+[53] to predict genome distribution of SNPs in lentil genome.
Fig 1. Detailed steps performed by automated GBS pipeline.
(A) De-multiplex samples: Raw paired-end Illumina reads are assigned to a sample using barcode sequences, which are subsequently trimmed. (B) Trim and filter reads: De-multiplexed paired-end reads are trimmed for base quality and Illumina adaptors. (C) Align reads to a reference genome. (D) Raw SNP calls: Every position in each sample’s alignment is scanned to determine the probability of a variant.
Phylogenetic tree construction
The phylogenetic tree was initially constructed using a relatively easy and quick distance-matrix method until a pipeline with more advanced methods and bootstrapping is available to handle large SNP datasets. For the first GBS run, a pairwise distance matrix was generated based on unfiltered calls across our preliminary test run. After removing samples with a low number of reads, a dendrogram was constructed using the Neighbour-Joining method in Phylip [54] based on this matrix. When the second set of GBS data became available, the phylogenetic tree for both GBS runs was constructed using SNPhylo [55]. The SNP datasets were first filtered for SNPs which fell in repetitive regions detected by Repeatmasker-4.0.3[56] using Repbase-18.11[57] to remove any potential false positive SNPs caused by mismapping. The pipeline was used to construct a maximum-likelihood tree with bootstrap values by performing the following steps: 1) filter the SNP datasets using minor allele frequency of 10% and 20% missing data; 2) remove redundant SNPs based on linkage disequilibrium information (cutoff threshold 0.8) using SNPRelate [58]; 3) construct multiple sequence alignment of the SNP dataset using MUSCLE [59]; 4) construct the maximum-likelihood tree using DNAML from Phylip [54]; 5) perform 1000 bootstraps using Phangorn [60]. The phylogenetic tree was drawn and visualized using iTOL [61].
Determination of population structure
Determination of population structure was performed on a filtered SNP dataset of all individuals used in maximum-likelihood tree construction using STRUCTURE-2.3.4 [62] based on admixture and the USEPOPINFO model. Linkage disequilibrium was assumed to be absent in the filtered SNP dataset after SNPRelate filtering in the SNPhylo pipeline. The estimation of clusters (K) was performed in five replications using K of 1 to 10. An additional STRUCTURE analysis (K of 1 to 5) was performed on a filtered SNP dataset of individuals from L. odemensis, L. lamottei, L. ervoides and L. nigricans to evaluate population substructure within these four species. The analysis was optimized using higher burnin period and MCMC Reps after burnin until standard deviations of L(K) were low. The best K value was chosen based on Evanno’s methods [63] using STRUCTURE HARVESTER [64] and visualized using Distruct v1.1 [65].
Results
The sequencing of the first GBS library consisting of 36 samples resulted in a total of 90,218,745 raw de-multiplexed reads with an average 2.5 million reads per sample. We successfully mapped between 59 and 94% of reads per technical replicate to the reference genome. After excluding two technical replicates from IG_110813_r1 and IG_110810_r1 that yielded less than 1,000 reads, the estimated site coverage per technical replicate ranged from 0.055 to 14.637 x.
As the use of the two-enzyme approach in GBS library construction was selected based on previous reports from other crops which represents a priori knowledge in lentil, we evaluated the one-enzyme and two-enzyme GBS approach in our preliminary lentil draft genome assembly using in silico prediction of restriction sites. We predicted the restriction sites of ApeKI, MspI and PstI in the lentil draft genome assembly and calculated the number of genomic fragments containing ApeKI-ApeKI (one-enzyme approach) and MspI-PstI (two-enzyme approach) cuts. We found that the two-enzyme MspI-PstI approach is more favourable as it should produce 17% fewer fragments than the ApeKI one-enzyme method. We also evaluated the distribution of 5,389 SNPs used in final phylogenetic tree which were represented in 2,120 scaffolds of the L. culinaris genome assembly (v0.6) and found them to be evenly distributed across Medicago truncatula genome version Mt4.0.
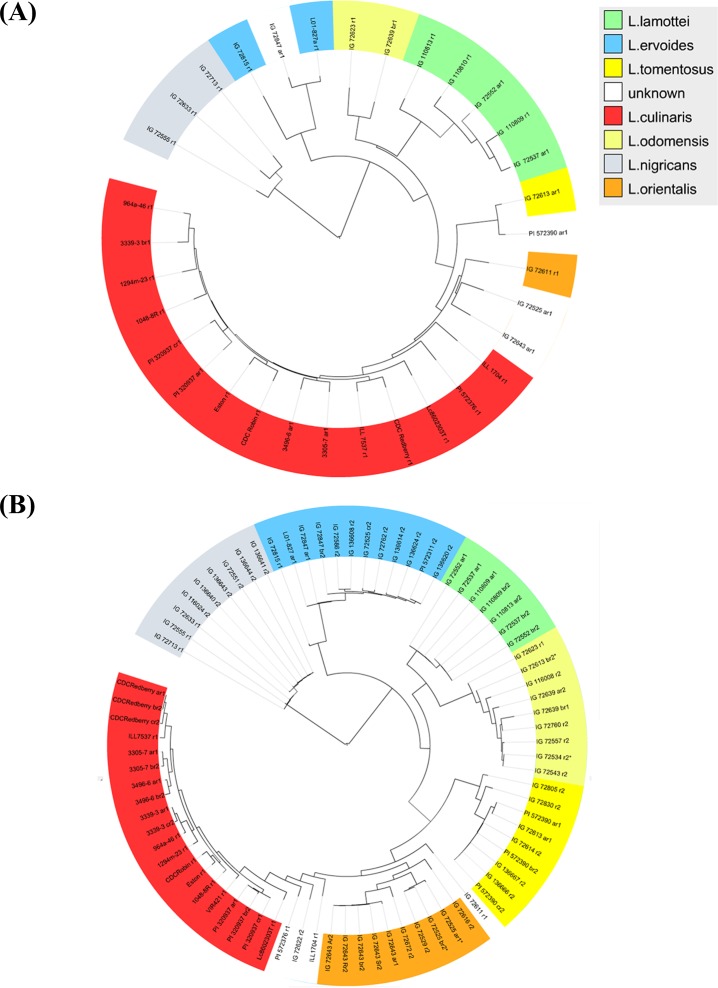
A preliminary analysis using Neighbour-Joining tree based on 353,656 unfiltered SNPs classified the diverse germplasm into four major groups; namely L. nigricans, L. ervoides, L. odemensis/L. lamottei, and L. culinaris/L. orientalis/L. tomentosus (Fig 2A). All but four of the samples fell within their respective groups as anticipated. IG 72847 was classified as L. orientalis, however, it grouped with L. ervoides IG 72815. Fiala et al. [66] reported that the original seed source of IG 72847 was a seed mixture of L. orientalis and L. ervoides since L01-827A, a single-plant selection from IG 72847, also grouped in L. ervoides and therefore, the sample we used is likely originated from a L. ervoides plant. PI 572390 was listed as L. orientalis but it appeared to be L. tomentosus. Two samples, IG 72525 and IG 72643, grouped with a L. orientalis accession (i.e. IG 72611) despite being classified as L. ervoides and L. tomentosus, respectively. The sterility issues that arose in two populations made from putative intra-specific crosses: IG 72611 x PI 572390 (supposedly a L. orientalis cross) and IG 72643 x IG 72613 (supposedly a L. tomentosus cross) [67] provided further evidence that there had been mislabeling or misclassification of the parental accessions IG 72643 and PI 572390.
Fig 2. Phylogenetic relationship within genus Lens.
(A) Dendrogram generated using Neighbour-Joining model based on results from first GBS library. (B) A maximum-likelihood tree based on combined results from two GBS libraries.
To verify the authenticity of these four samples, we constructed a second GBS library using old and fresh DNA samples (from different seed sources if available) of 15 accessions from the first GBS library and included 27 additional accessions to improve separation and classification of the wild species during phylogenetic tree construction. Both GBS libraries produced an average of 2.4 million raw de-multiplexed reads per sample. Although the range of reads that mapped to reference genome remained the same, the estimated site coverage was increased to the range of 0.55 to 31.80 x after removing three samples with low number of reads (IG_110813_r1, IG_110810_r1 and IG_72847_cr2). After removing SNPs that were present in repetitive regions of the genome, we detected a total of 266,356 SNPs with 11,581–116,510 SNPs per sample. Removal of SNPs and samples with more than 20% missing SNP data resulted in a final dataset of 32,019 SNPs from 80 samples. Only 0.57% of the SNPs had heterozygous genotypes, mainly found in L. nigricans and L. ervoides samples.
A major problem we observed from the sequencing results of both GBS libraries was the large variation in the number of reads assigned to a given barcode adaptor in each library. By having two technical replicates per sample and each technical replicate multiplexed with a different barcode adaptor, we were able to examine whether technical replicate and barcode sequence influenced the number of reads per sample. We found no significant difference in the number of reads per technical replicate between both runs, suggesting that this bias was not caused by run-to-run variation (paired t-test, p>0.05). We also observed that the number of reads were the same between the two technical replicates (paired t-test, p<0.05). Assuming the number of reads between technical replicates was the same, the huge variation of number of reads must be sample dependent and thus, the DNA quantity and quality of the samples was likely to be the main contributing factor to the observed biases.
After removing 893 low quality and 25,737 redundant SNPs, the number of SNPs was reduced to 5,389 and subsequently used to construct a maximum-likelihood tree (Fig 2B). The phylogenetic tree provided 100% bootstrap support for the paraphyletic relationship among the four major Lens groups; namely L. nigricans, L. ervoides, L. lamottei/L.odemensis, and L. culinaris/L. orientalis/L. tomentosus. As expected, L. nigricans exhibited the greatest genetic distance from L. culinaris followed by L. ervoides, L. lamottei/L. odemensis and finally L. tomentosus/ L. orientalis was the closest. The L. lamottei/L. odemensis group was further separated into their respective taxa. The L. culinaris/L. orientalis/L. tomentosus clade is paraphyletic in which L. tomentosus first branched off with good bootstrap support followed by the branching of L. orientalis from L. culinaris with weak bootstrap support. The species boundary between L. culinaris and its wild progenitor L. orientalis was difficult to distinguish due to this paraphyletic relationship. L. culinaris accessions ILL1704, IG 72622 and PI 572376 were located at this species boundary. In addition, IG 72611 was classified as L. orientalis, however, it was the earliest diverging member of the L. orientalis/L. culinaris clade. We conclude that these four samples are natural hybrids between L. orientalis and L. culinaris due to contradictory morphological and phylogenetic evidence.
Next, we examined the reproducibility of GBS results based on phylogenetic positioning since about 25% of the accessions had two or more biological replicates. Our hypothesis was that biological replicates of the same accession should be closely related and shared minimal genetic distance with each other. We found that the biological replicates of accessions from L. culinaris and L. orientalis were closely grouped together, however, this was not observed in other species especially in L. nigricans, L. ervoides and L. lamottei where an accession was more closely related to other accessions from the same GBS run than to their biological replicates from a different GBS run. This suggested that run-to-run variation was present in most wild lentils accessions and had a profound effect on the groupings of accessions and their biological replicates.
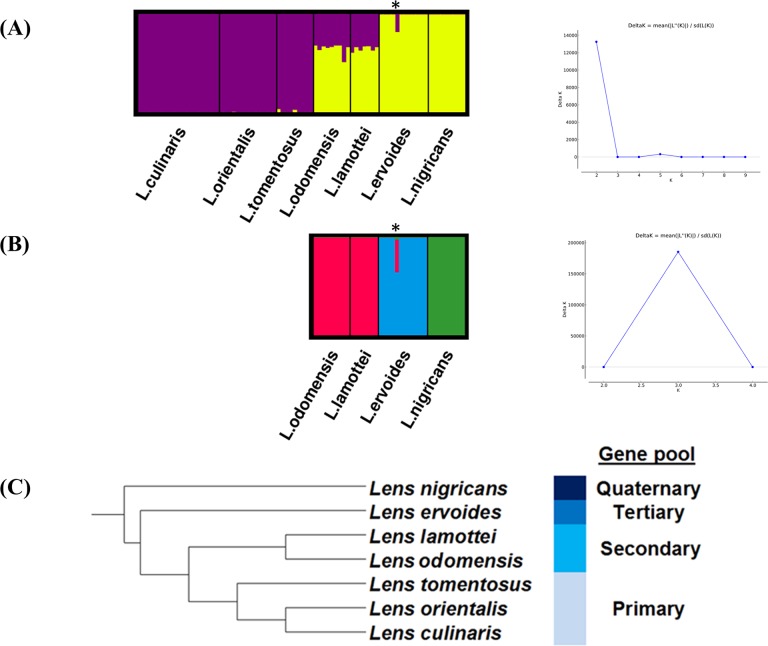
The Bayesian STRUCTURE analysis based on all individuals (Fig 3A) led to the observation that L. culinaris/L. orientalis/L. tomentosus and L. ervoides/L. nigricans each belong to one cluster while L. lamottei and L. odemensis showed mixed ancestry with major proportion in L. ervoides/L. nigricans cluster. This finding was in agreement with phylogenetic analysis except the fact that L. ervoides and L. nigricans were placed in the same cluster. The failure to reveal strong level of genetic divergence between two distinct populations suggested that hierarchical clustering plays a role in masking population substructure as reported by [68]. Additional STRUCTURE analysis using only individuals from L. odemensis, L. lamottei, L. ervoides and L. nigricans indicated three population substructures within these four species (Fig 3B). L. odemensis and L. lamottei belonged to one cluster while L. ervoides and L. nigricans each formed a distinct cluster. Based on these findings, we proposed a new gene pool classification for the genus Lens (Fig 3C). Both analyses revealed potential admixture between populations in one L. ervoides accession (IG 72815). This finding was consistent with the observation that IG 72815 shared shorter genetic distance compared to other accessions from the same species in the phylogenetic tree. Since this accession is from multiple plants, it is not clear if the observed admixture occurred due to natural hybridization or contaminated seed sources from another species.
Fig 3. Gene pool classification of lentil based on STRUCTURE results.
(A) Graphical representation of STRUCTURE results indicates two clusters (K = 2) in genus Lens based on the highest delta K score. (B) Additional STRUCTURE analysis revealed substructures (K = 3) within individuals of L. odemensis, L. lamottei, L. ervoides and L. nigricans. (C) The new gene pool classification proposed in this study is shown next to a simplified maximum-likelihood tree of genus Lens. Accession IG 72815 is marked with asterisk.
Lastly, we re-examined the authenticity of our existing germplasm collection based on results from both GBS runs and identified 17% misclassified samples (See S1 Table). In the first GBS run, we identified discrepancies in taxon classification between genebank records and the phylogenetic tree in four samples and subsequently, re-sequenced those samples from a different seed source. Results from the second GBS run confirmed that the taxon classification based on the first phylogenetic tree was correct and it is consistent with the evidence based on morphological traits and crossability of mapping populations. We also identified four mislabeled samples which were likely caused by human error during sample and library preparation. One sample of IG 72613 was mislabeled as IG 72623, IG 72534 was suspected to be IG 72543 while two samples of IG 72529 were mislabeled as IG 72525. Besides identifying mislabeled accessions, we were able to validate the authenticity of some accessions and characterize the genetic differences within the same accession. One example is L. orientalis IG 72643. Our germplasm collection contains several seed sources of IG 72643 including one seed source which had red cotyledons (IG_72643_Sr2) instead of the expected yellow cotyledons. Our observation that all the seed sources formed a clade with minimal genetic distance in the phylogenetic tree suggested that they were authentic and no mislabeling or contamination has occurred and rather there is residual genetic variability within this accession.
Discussion
Our comparison of one-enzyme vs. two-enzyme approach using a draft lentil genome assembly suggested that the two-enzyme approach can reduce overall genome complexity more than the single-cutter approach. Even genome distribution of GBS SNPs was observed based on the M. truncatula genome suggested that the SNPs are likely distributed evenly across the lentil genome as the genomes of both species share high levels of conserved synteny[69]. Since an automated GBS pipeline to map paired-end reads was not available at the start of this study, we developed a pipeline to reproduce the workflow from processing of raw reads through to SNP calling based on commonly-used open-source bioinformatics tools such as Bowtie2[49] and Samtools[50]. This enabled us to combine and re-use previously written codes to analyze our GBS data. A total of four commands are called to process raw Illumina paired-end reads through de-multiplexing, trimming and filtering, alignment to a reference genome and SNP calling. Our pipeline has several advantages compared to other GBS pipelines such as Tassel-GBS v3.0[70] and its reference-free pipeline UNEAK[71]. Our pipeline allows the paired-end reads to be more accurately mapped to a reference genome based on expected fragment sizes and has no upper limit on read length compared to the single-end mode and 64 bp tag length limitation in Tassel-GBS v3.0 and UNEAK. Our pipeline allowed us to simplify customization and alteration of all parameters from barcode sequence to mapping and variant calling parameters for future analysis while also providing a simplified user interface for more standardized runs. Through automatic generation of summaries at the end of each step, the user is given the opportunity to evaluate data integrity and tweak parameters accordingly.
Our assessment of technical reproducibility of GBS results identified several factors affecting the reproducibility which were not reported in previous GBS studies. One major problem with GBS for large genomes is low coverage sequencing data resulting in large numbers of missing data and false positive SNP calls. Our results suggested that the quality of the DNA samples is likely to be the main determinant of the number of sequencing reads. Firstly, sequencing bias is observed between samples but not found between technical replicates of a given sample, suggesting that sequencing bias is sample dependent. Assuming minimal methodological and sequencing machine variation, this bias is likely to be caused by DNA quality. This is not surprising as it is generally known that DNA quality affects digestion of restriction enzymes and ligation of adaptors. PCR amplification further escalates sequencing bias resulting in overrepresentation of PCR fragments from good quality samples. Therefore, we recommend paying particular attention to the quality of the DNA samples to ensure even sequencing coverage of a GBS run. Other known factors causing sequencing bias are fragment length bias, GC bias, unequal pooling of primers and DNA samples in multiplex reaction [72, 73], any or all of which may also have played a role in the uneven sequence coverage.
Our observation that an accession shared minimal genetic distance with other accessions from the same run in the final phylogenetic tree compared to its biological replicates of a different run suggested that run-to-run variation affects GBS results. This is not surprising as run-to-run variation has already been reported [74, 75]. One explanation is that run-to-run variation is caused by low sequencing coverage resulting in non-uniform sequencing of genomic regions between runs and thus, different SNPs are detected and used in phylogenetic tree construction. To reduce this effect, repeat sequencing with selected samples and increasing sequencing coverage has been recommended [74]. Interestingly, the effect of run-to-run variation was observed in all lentil samples except those of L. culinaris and L. orientalis, suggesting that ascertainment bias is likely playing a role. It is generally accepted that GBS eliminates most sources of marker ascertainment biases by discovering and genotyping markers simultaneously [34]. However, ascertainment bias can be re-introduced by using a reference genome from one species to map reads from its relatives. This can result in reduced read mapping and subsequent inability to detect rare alleles from genomic regions which are absent in the reference genome due to structural variation and differences in genome content. The inability to capture the real level of variation among individuals of a wild species made it appear as if there is very little genetic variability within and between species. To more accurately reflect the diversity within a wild species, it would be necessary to either carry out a de novo assembly or non-reference mapping based on the results of individual species. Either of these options would require additional sequencing to increase the depth of coverage sufficiently, which would add to the cost considerably. If the goal is simply to classify accessions to species, however, this additional work is not necessary.
Using phylogenetic and population structure analysis based on 5,389 good quality and non-redundant SNPs, we propose that the genus Lens should be separated into four gene pools, namely L. culinaris/L. orientalis/L. tomentosus, L. lamottei/L. odemensis, L. ervoides and L. nigricans. There is no doubt that L. tomentosus and L. orientalis belong to the primary gene pool as they are most closely related to L. culinaris and they can easily produce seeds following interspecific crossing. The second most closely related group to L. culinaris is L. lamottei/L. odemensis, placing them in the secondary gene pool. Our observation that L. odemensis is a sister clade to L. lamottei is in agreement with Verma et al. [76]. We propose that L. odemensis should be a separate species instead of a subspecies under L. culinaris as it is clearly distinct from the primary gene pool. As was suggested by Fratini and Ruiz [29], L. ervoides belongs to the tertiary gene pool as evidenced by the successful development of RIL mapping populations following F1 embryo rescue [17, 66]. Considering that interspecific crosses between L. culinaris and L. nigricans have never been reported to be successful beyond the F1 generation and both species formed distinct groups as revealed by phylogenetic and STRUCTURE analysis, L. nigricans should be placed in the quaternary gene pool and probably should be considered a last resort as a source of genetic diversity for cultivated lentil.
These results demonstrate several uses of GBS in the characterization of diverse germplasm. Firstly, GBS can be used as a reliable screening tool for lentil breeders interested in using wild relatives as a source of genetic diversity. GBS results provide good classification at the taxon level. Basing SNPs on comparisons with cultivated lentil however, results in the discriminatory power within species being limited to only L. culinaris and L. orientalis. Therefore, the use of GBS to correct identification of genotypes within wild species is not recommended until further optimization and improvement can be made. Secondly, GBS is an affordable screening tool replacing the need for other technology by discovering and genotyping many markers at once. We estimated that the cost per sample in this study for 48 samples in a 96-plex GBS run is CAD $48. The decreasing cost of sequencing is expected to drive cost per sample below USD $10 [34]. The low cost makes it feasible and practical to screen wild germplasm accessions before use in introgression. This cost would offset the potential larger cost of making a mistake in crossing due to mis-classification. Lastly, GBS is useful for checking the authenticity of germplasm. This has been demonstrated in Labate et al. [43] where a Solanum arcanum accession was re-classified as S. huaylasense based on GBS data. A previous diversity study in lentil germplasm [77] re-classified 10.8% of L. culinaris and L. orientalis germplasm based on phylogenetic tree results suggesting the need to examine the authenticity of lentil germplasm before utilization. Marker information can be used to detect errors in germplasm collections, resulting in more time and resources devoted to varietal development with a higher success rate.
In summary, GBS is a promising technology for plant breeders interested to work with crop wild relatives as an affordable and reliable routine screening of germplasm provided that several technical problems are addressed. The reliability and practically of GBS is likely to increase in the future with the improvement of sample preparation, increased sequencing depth, reduced sequencing cost per base and de novo sequence assembly of wild relatives. This technology offers high potential for screening largely uncharacterized gene pools of non-model crops with few genomic resources.
Supporting Information
(XLS)
(XLS)
(XLS)
Acknowledgments
We would like to acknowledge ICARDA and USDA for supplying seeds of the wild lentil germplasm accessions, Abebe Tullu for providing single plant sources of seeds of some lentil germplasm accessions, Lacey-Anne Sanderson for constructive suggestions on the GBS pipeline, and technical support from Devini De Silva and Shyamali Saha.
Data Availability
Relevant data are available at NCBI's Sequence Read Archive: SRX703778-SRX703943.
Funding Statement
This work was supported by the Saskatchewan Ministry of Agriculture’s Agriculture Development Fund contract # 20110236 (http://www.agriculture.gov.sk.ca); Saskatchewan Pulse Growers contract # BRE1202 (http://www.saskpulse.com/); Natural Sciences and Engineering Research Council of Canada #IRCPJ 395994-09 (http://www.nserc-crsng.gc.ca/index_eng.asp). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Zohary D (1972) The wild progenitor and the place of origin of the cultivated lentil:Lens culinaris . Economic Botany 26: 326–332. [Google Scholar]
- 2. Zohary D, Hopf M (1973) Domestication of pulses in the old world: Legumes were companions of wheat and barley when agriculture began in the Near East. Science 182: 887–894. [DOI] [PubMed] [Google Scholar]
- 3. Vishnumittrre A (1974) The beginnings of agriculture-paleo-botanical evidence in India In: Hutchinson J. B., editor editors. Evolutionary Studies on World Crops: Diversity and Change in the Sub-Continent. Cambridge, UK: Cambridge University Press; pp. 23–24. [Google Scholar]
- 4. Schaefer H, Hechenleitner P, Santos-Guerra A, Menezes de Sequeira M, Pennington RT, Kenicer G, et al. (2012) Systematics, biogeography, and character evolution of the legume tribe Fabeae with special focus on the middle-Atlantic island lineages. BMC Evol Biol 12: 250 10.1186/1471-2148-12-250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mayer MS, Soltis PS (1994) Chloroplast DNA phylogeny of Lens (Leguminosae): origin and diversity of the cultivated lentil. Theor Appl Genet 87: 773–781. 10.1007/BF00221128 [DOI] [PubMed] [Google Scholar]
- 6. Sharma SK, Knox MR, Ellis THN (1996) AFLP analysis of the diversity and phylogeny of Lens and its comparison with RAPD analysis. Theor Appl Genet 93: 751–758. 10.1007/BF00224072 [DOI] [PubMed] [Google Scholar]
- 7. Sonnante G, Galasso I, Pignone D (2003) ITS sequence analysis and phylogenetic inference in the genus Lens mill. Ann Bot 91: 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mayer MS, Bagga SK (2002) The phylogeny of Lens (Leguminosae): new insight from ITS sequence analysis. Plant Systematics and Evolution 232: 145–154. [Google Scholar]
- 9. Havey MJ, Muehlbauer FJ (1989) Variability for restriction fragment lengths and phylogenies in lentil. Theor Appl Genet 77: 839–843. 10.1007/BF00268336 [DOI] [PubMed] [Google Scholar]
- 10. Abo-Elwafa A, Murai K, Shimada T (1995) Intra- and inter-specific variations in Lens revealed by RAPD markers. Theor Appl Genet 90: 335–340. 10.1007/BF00221974 [DOI] [PubMed] [Google Scholar]
- 11. Sharma SK, Dawson IK, Waugh R (1995) Relationships among cultivated and wild lentils revealed by RAPD analysis. Theor Appl Genet 91: 647–654. 10.1007/BF00223292 [DOI] [PubMed] [Google Scholar]
- 12. Ahmad M, McNeil DL (1996) Comparison of crossability, RAPD, SDS-PAGE and morphological markers for revealing genetic relationships within and among Lens species. Theor Appl Genet 93: 788–793. 10.1007/BF00224077 [DOI] [PubMed] [Google Scholar]
- 13. Ahmad M, McNeil DL, Fautrier AG, Armstrong KF, Paterson AM (1996) Genetic relationships in Lens species and parentage determination of their interspecific hybrids using RAPD markers. Theor Appl Genet 92: 1091–1098. 10.1007/BF00224054 [DOI] [PubMed] [Google Scholar]
- 14. Ferguson ME, Maxted N, Slageren MV, Robertson LD (2000) A re-assessment of the taxonomy of Lens Mill. (Leguminosae, Papilionoideae, Vicieae). Botanical Journal of the Linnean Society 133: 41–59. [Google Scholar]
- 15. Kole C, Gupta D, Ford R, Taylor PJ (2011) Lens Wild Crop Relatives: Genomic and Breeding Resources. Berlin Heidelberg: Springer; pp. 127–139. [Google Scholar]
- 16. Erskine W, Chandra S, Chaudhry M, Malik IA, Sarker A, Sharma B, et al. (1998) A bottleneck in lentil: widening its genetic base in South Asia. Euphytica 101: 207–211. [Google Scholar]
- 17. Vail S, Strelioff JV, Tullu A, Vandenberg A (2012) Field evaluation of resistance to Colletotrichum truncatum in Lens culinaris, Lens ervoides, and Lens ervoides x Lens culinaris derivatives. Field Crops Research 126: 145–151. [Google Scholar]
- 18. Fernández-Aparicio M, Sillero JC, Rubiales D (2009) Resistance to broomrape in wild lentils (Lens spp.). Plant Breeding 128: 266–270. [Google Scholar]
- 19. Tullu A, Banniza S, Tar'an B, Warkentin T, Vandenberg A (2010) Sources of resistance to ascochyta blight in wild species of lentil (Lens culinaris Medik.). Genetic Resources and Crop Evolution 57: 1053–1063. [Google Scholar]
- 20. Tullu A, Buchwaldt L, Lulsdorf M, Banniza S, Barlow B, Slinkard AE, et al. (2006) Sources of resistance to anthracnose (Colletotrichum truncatum) in wild Lens species. Genetic Resources and Crop Evolution 53: 111–119. [Google Scholar]
- 21. Podder R, Banniza S, Vandenberg A (2012) Screening of wild and cultivated lentil germplasm for resistance to stemphylium blight. Plant Genetic Resources 11: 26–35. [Google Scholar]
- 22. Muehlbauer FJ, McPhee KE (2005) Lentil (Lens culinaris Medik.) In: Singh R. J., Jauhar P. P., editors. Genetic Resources, Chromosome Engineering, and Crop Improvement: Grain Legumes. Taylor & Francis; pp. 268–280. [Google Scholar]
- 23. Ladizinsky G (1979) The origin of lentil and its wild genepool. Euphytica 28: 179–187. [Google Scholar]
- 24. Ladizinsky G, Cohen D, Muehlbauer FJ (1985) Hybridization in the genus Lens by means of embryo culture. Theor Appl Genet 70: 97–101. 10.1007/BF00264489 [DOI] [PubMed] [Google Scholar]
- 25. Ladizinsky G, Muehlbauer FJ (1993) Wild lentils. Critical Reviews in Plant Sciences 12: 169–184. [Google Scholar]
- 26. Muehlbauer FJ, Kaiser WJ, Clement SL, Summerfield RJ, Donald LS (1995) Production and breeding of lentil Advances in Agronomy. Academic Press; pp. 283–332. [Google Scholar]
- 27. van Oss H, Aron Y, Ladizinsky G (1997) Chloroplast DNA variation and evolution in the genus Lens Mill. Theor Appl Genet 94: 452–457. [Google Scholar]
- 28. Yadav S, McNeil D, Stevenson P, Mishra SK, Sharma B, Sharma SK (2007) Genetics and cytogenetics of lentil Lentil. Springer Netherlands; pp. 187–208. [Google Scholar]
- 29. Fratini R, Ruiz M (2006) Interspecific hybridization in the genus Lens applying in vitro embryo rescue. Euphytica 150: 271–280. [Google Scholar]
- 30. Wouw MJvd, Treuren Rv, Hintum TJLv (2011) Authenticity of old cultivars in genebank collections: a case study on lettuce. Crop Science 51: 736–746. [Google Scholar]
- 31. Samec D, Bogovic M, Vincek D, Martincic J, Salopek-Sondi B (2014) Assessing the authenticity of the white cabbage (Brassica oleracea var. capitata f. alba) cv. 'Varazdinski' by molecular and phytochemical markers. Food Research International 60: 266–272. [Google Scholar]
- 32. Dossett M, Bassil NV, Finn CE (2012) SSR fingerprinting of black raspberry cultivars shows discrepancies in identification. X International Rubus and Ribes Symposium 946: 49–53. [Google Scholar]
- 33. Kostamo K, Toljamo A, Antonius K, Kokko H, Karenlampi SO (2013) Morphological and molecular identification to secure cultivar maintenance and management of self-sterile Rubus arcticus . Ann Bot 111: 713–721. 10.1093/aob/mct029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Poland JA, Rife TW (2012) Genotyping-by-sequencing for plant breeding and genetics. Plant Gen 5: 92–102. [Google Scholar]
- 35. Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, et al. (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6: e19379 10.1371/journal.pone.0019379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML (2011) Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet 12: 499–510. 10.1038/nrg3012 [DOI] [PubMed] [Google Scholar]
- 37. Sonah H, Bastien M, Iquira E, Tardivel A, Legare G, Boyle B, et al. (2013) An improved genotyping by sequencing (GBS) approach offering increased versatility and efficiency of SNP discovery and genotyping. PLoS One 8: e54603 10.1371/journal.pone.0054603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Poland JA, Brown PJ, Sorrells ME, Jannink JL (2012) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7: e32253 10.1371/journal.pone.0032253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu H, Bayer M, Druka A, Russell JR, Hackett CA, Poland J, et al. (2014) An evaluation of genotyping by sequencing (GBS) to map the Breviaristatum-e (ari-e) locus in cultivated barley. BMC Genomics 15: 104 10.1186/1471-2164-15-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Spindel J, Wright M, Chen C, Cobb J, Gage J, Harrington S, et al. (2013) Bridging the genotyping gap: using genotyping by sequencing (GBS) to add high-density SNP markers and new value to traditional bi-parental mapping and breeding populations. Theor Appl Genet 126: 2699–2716. 10.1007/s00122-013-2166-x [DOI] [PubMed] [Google Scholar]
- 41. Nielsen R, Paul JS, Albrechtsen A, Song YS (2011) Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet 12: 443–451. 10.1038/nrg2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Uitdewilligen JG, Wolters AM, D'Hoop B B, Borm TJ, Visser RG, van Eck HJ (2013) A next-generation sequencing method for genotyping-by-sequencing of highly heterozygous autotetraploid potato. PLoS One 8: e62355 10.1371/journal.pone.0062355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Labate JA, Robertson LD, Strickler SR, Mueller LA (2014) Genetic structure of the four wild tomato species in the Solanum peruvianum s.l. species complex. Genome 57: 169–180. 10.1139/gen-2014-0003 [DOI] [PubMed] [Google Scholar]
- 44. Escudero M, Eaton DA, Hahn M, Hipp AL (2014) Genotyping-by-sequencing as a tool to infer phylogeny and ancestral hybridization: A case study in Carex (Cyperaceae). Mol Phylogenet Evol 79C: 359–367. 10.1016/j.ympev.2014.06.026 [DOI] [PubMed] [Google Scholar]
- 45. Girma G, Hyma KE, Asiedu R, Mitchell SE, Gedil M, Spillane C (2014) Next-generation sequencing based genotyping, cytometry and phenotyping for understanding diversity and evolution of guinea yams. Theor Appl Genet 127: 1783–1794. 10.1007/s00122-014-2339-2 [DOI] [PubMed] [Google Scholar]
- 46. Hamwieh A, Udupa SM, Choumane W, Sarker A, Dreyer F, Jung C, et al. (2005) A genetic linkage map of Lens sp. based on microsatellite and AFLP markers and the localization of fusarium vascular wilt resistance. Theor Appl Genet 110: 669–677. [DOI] [PubMed] [Google Scholar]
- 47. Doyle JJ (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13–15. [Google Scholar]
- 48. Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. (2011) The variant call format and VCFtools. Bioinformatics 27: 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tang H, Krishnakumar V, Bidwell S, Rosen B, Chan A, Zhou S, et al. (2014) An improved genome release (version Mt4.0) for the model legume Medicago truncatula . BMC Genomics 15: 312 10.1186/1471-2164-15-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. (2009) BLAST+: architecture and applications. BMC Bioinformatics 10: 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Felsenstein J (1989) PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 5: 164–166. [Google Scholar]
- 55. Lee TH, Guo H, Wang X, Kim C, Paterson AH (2014) SNPhylo: a pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics 15: 162 10.1186/1471-2164-15-162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smit AFA, Hubley R, Green P (1996) RepeatMasker Open-3.0.
- 57. Jurka J (2000) Repbase update: a database and an electronic journal of repetitive elements. Trends Genet 16: 418–420. [DOI] [PubMed] [Google Scholar]
- 58. Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS (2012) A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28: 3326–3328. 10.1093/bioinformatics/bts606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schliep KP (2011) phangorn: phylogenetic analysis in R. Bioinformatics 27: 592–593. 10.1093/bioinformatics/btq706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Letunic I, Bork P (2011) Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39: W475–478. 10.1093/nar/gkr201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- 64. Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4: 359–361. [Google Scholar]
- 65. Rosenberg NA (2004) distruct: a program for the graphical display of population structure. Molecular Ecology Notes 4: 137–138. [Google Scholar]
- 66. Fiala JV, Tullu A, Banniza S, Séguin-Swartz G, Vandenberg A (2009) Interspecies transfer of resistance to anthracnose in lentil (Lens culinaris Medic.). Crop Sci 49: 825–830. [Google Scholar]
- 67. Suvorova G (2014) Hybridization of cultivated lentil Lens culinaris Medik. and wild species Lens tomentosus Ladizinsky. Czech J Genet Plant Breed 50: 130–134. [Google Scholar]
- 68. Schreier AD, Mahardja B, May B (2012) Hierarchical patterns of population structure in the endangered Fraser River white sturgeon (Acipenser transmontanus) and implications for conservation. Canadian Journal of Fisheries and Aquatic Sciences 69: 1968–1980. [Google Scholar]
- 69. Sharpe AG, Ramsay L, Sanderson LA, Fedoruk MJ, Clarke WE, Li R, et al. (2013) Ancient orphan crop joins modern era: gene-based SNP discovery and mapping in lentil. BMC Genomics 14: 192 10.1186/1471-2164-14-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, et al. (2014) TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One 9: e90346 10.1371/journal.pone.0090346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lu F, Lipka AE, Glaubitz J, Elshire R, Cherney JH, Casler MD, et al. (2013) Switchgrass genomic diversity, ploidy, and evolution: novel insights from a network-based SNP discovery protocol. PLoS Genet 9: e1003215 10.1371/journal.pgen.1003215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Dabney J, Meyer M (2012) Length and GC-biases during sequencing library amplification: a comparison of various polymerase-buffer systems with ancient and modern DNA sequencing libraries. Biotechniques 52: 87–94. 10.2144/000113809 [DOI] [PubMed] [Google Scholar]
- 73. Sint D, Raso L, Traugott M (2012) Advances in multiplex PCR: balancing primer efficiencies and improving detection success. Methods Ecol Evol 3: 898–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ge Y, Schimel JP, Holden PA (2014) Analysis of run-to-run variation of bar-coded pyrosequencing for evaluating bacterial community shifts and individual taxa dynamics. PLoS One 9: e99414 10.1371/journal.pone.0099414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Daley T, Smith AD (2013) Predicting the molecular complexity of sequencing libraries. Nat Meth 10: 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Verma P, Sharma T, Srivastava P, Abdin MZ, Bhatia S (2014) Exploring genetic variability within lentil (Lens culinaris Medik.) and across related legumes using a newly developed set of microsatellite markers. Molecular Biology Reports: 1–19. [DOI] [PubMed] [Google Scholar]
- 77. Alo F, Furman BJ, Akhunov E, Dvorak J, Gepts P (2011) Leveraging genomic resources of model species for the assessment of diversity and phylogeny in wild and domesticated lentil. J Hered 102: 315–329. 10.1093/jhered/esr015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(XLS)
(XLS)
Data Availability Statement
Relevant data are available at NCBI's Sequence Read Archive: SRX703778-SRX703943.