Abstract
Background
Quorum sensing (QS) is a cell-to-cell communication system used by bacteria to regulate activities such as virulence, bioluminescence and biofilm formation. The most common QS signals in Gram-negative bacteria are N-acyl-homoserine lactones (AHLs). Aliivibrio salmonicida is the etiological agent of cold water vibriosis in Atlantic salmon, a disease which occurs mainly during seasons when the seawater is below 12°C. In this work we have constructed several mutants of A. salmonicida LFI1238 in order to study the LuxI/LuxR and AinS/AinR QS systems with respect to AHL production and biofilm formation.
Results
Using high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) we found that LuxI in A. salmonicida LFI1238 is responsible for producing seven of the different AHLs, whereas AinS is responsible for producing only one. The production of these various AHLs is dependent on both cell density and growth temperature. The AHLs were efficiently produced when wild type LFI1238 was grown at 6 or 12°C, however at 16°C AHL production decreased dramatically, and LFI1238 produced less than 5% of the maximum concentrations observed at 6°C. LitR, the master regulator of QS, was found to be a positive regulator of AinS-dependent AHL production, and to a lesser extent LuxI-dependent AHL production. This implies a connection between the two systems, and both systems were found to be involved in regulation of biofilm formation. Finally, inactivation of either luxR1 or luxR2 in the lux operon significantly reduced production of LuxI-produced AHLs.
Conclusion
LuxI and AinS are the autoinducer synthases responsible for the eight AHLs in A. salmonicida. AHL production is highly dependent on growth temperature, and a significant decrease was observed when the bacterium was grown at a temperature above its limit for disease outbreak. Numerous AHLs could offer the opportunity for fine-tuning responses to changes in the environment.
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-015-0402-z) contains supplementary material, which is available to authorized users.
Keywords: Aliivibrio salmonicida, Quorum sensing, Biofilm, Acyl homoserine lactone, Temperature
Background
Members of the Vibrionaceae family are found in a wide range of aquatic environments, as free-living planktonic cells or attached to surfaces as bacterial aggregates or biofilms. Some members within this family have evolved to become pathogens whereas others have developed symbiotic relationships with their hosts [1]. Aliivibrio salmonicida, previously designated Vibrio salmonicida [2], is the causative agent of cold water vibriosis in Atlantic salmon (Salmo salar), rainbow trout (Oncorhynchus mykiss) and captive Atlantic cod (Gadus morhua) [3-5], a disease which is kept under control due to successful vaccination [6]. The mechanisms of A. salmonicida virulence and pathogenicity are not known in detail, however, temperature-sensitive iron sequestration, motility and flagella activity, as well as quorum sensing (QS) have been suggested to be involved in virulence [7-10]. Moreover, the genome of A. salmonicida LFI1238 encodes putative hemolysins, proteases and several protein secretion systems that may be found to be important in pathogenesis [11].
Bacteria use QS to regulate and coordinate gene expression by secreting and responding to signal molecules in a cell density dependent manner [12]. The most common QS autoinducer signal molecules in Gram-negative bacteria are N-acyl homoserines lactones (AHLs), which consist of a homoserine lactone (HSL) linked to an acyl side chain with 4–18 carbons [13-16]. Several QS systems have been identified in vibrios and aliivibrios, and each species seems to utilize a unique combination of QS systems that can act in parallel, or in a hierarchal manner to regulate species-specific activities including biofilm formation, virulence and colonization factors [17,18]. The well-studied squid symbiont Aliivibrio (Vibrio) fischeri, a close relative of A. salmonicida, regulates bioluminescence and colonization by the QS systems LuxS/LuxPQ, AinS/AinR and LuxI/LuxR, where LuxS, AinS and LuxI are the autoinducer synthases [17,19-21]. AinS in A. fischeri produces N-octanoyl-HSL (C8-HSL) which is recognized by the membrane-bound two-component hybrid sensor kinase AinR [22,23]. LuxS in A. fischeri does not produce AHLs, but instead makes a signal molecule referred to as autoinducer 2 (AI-2) [20] that likely binds the periplasmic receptor LuxP and modulates the activity of the sensor kinase LuxQ, similar to the homologous system in Vibrio harveyi [24]. The LuxS/LuxPQ and AinS/AinR systems in A. fischeri work in parallel, and in the absence of signal molecules LuxPQ and AinR are believed to function as kinases and phosphorylate the response regulator LuxO via LuxU [17,18,20]. Phosphorylated LuxO then activates expression of qrr which encodes a small regulatory RNA. Qrr destabilizes the litR mRNA that encodes the master regulator of QS. In the presence of signal molecules at high cell density, LuxO becomes dephosphorylated leading to suppression of qrr allowing LitR to be expressed [17,25,26]. The third system in A. fischeri, LuxI/LuxR, is activated by the AinS/AinR system, and at medium cell densities LuxR binds AinS-produced C8-HSL and activates transcription of the lux operon (consisting of the luxICDABEG and luxR loci). At higher cell densities, LuxI-produced N-3-oxo-hexanoyl-L-HSL (3-oxo-C6-HSL) binds to LuxR and increases transcription of the luxICDABEG and luxR genes [27,28]. Moreover, the master regulator LitR is a positive regulator of ainS and is indirectly involved in expression of luxI by controlling expression of luxR. Hence, LitR links the AinS/AinR and LuxS/LuxPQ systems to the LuxI/LuxR system in A. fischeri [20,25].
The number and types of AHLs vary between different species of Vibrio and Aliivibrio, as well as between strains of the same species [29-34]. However, the specific autoinducer synthases responsible for the different AHLs are only known for a few bacteria, such as the above mentioned in A. fischeri. In V. anguillarum, another fish pathogen, the AinS homolog VanM is reported to produce N-3-hydroxy-hexanoyl-HSL (3-OH-C6-HSL) and N-hexanoyl-HSL (C6-HSL) [35], whereas the LuxI homolog VanI produces N-3-oxo-decanoyl-HSL (3-oxo-C10-HSL) [36]. In the bioluminescent bacteria V. harveyi the LuxM synthase is reported to produce N-3-hydroxy-butyryl-L-HSL (3-OH-C4-HSL) [37,38]. Thus, while some autoinducer synthases produce only one AHL others are able to produce several.
The genome of A. salmonicida LFl1238 encodes five QS systems: the AinS/AinR, LuxI/LuxR, VarS/VarA, LuxM/LuxN and LuxS/LuxPQ systems. The latter two systems are probably inactive since luxM is absent, and luxN and luxP contain frame-shift mutations [11]. The lux operon of A. salmonicida has a novel organization compared to A. fischeri, with two flanking luxR genes (luxR1 and luxR2) and the luxI gene located outside the luxCDABEG locus [39], and A. salmonicida is only able to produce bioluminescence after addition of decyl aldehyde [40]. Deletion of luxA in the lux operon of A. salmonicida has been associated with decreased virulence [39]. The master regulator of QS, LitR, is also involved in regulation of virulence in A. salmonicida LFl1238 as well as biofilm formation, motility and cryptic bioluminescence [10,41].
In recent work we identified and quantified AHLs in 57 members of the Vibrionaceae family using high-performance liquid chromatography combined with mass spectrometry (HPLC-MS/MS) [34]. Mapping of the resulting AHL profiles onto a host 16S rDNA phylogenetic tree revealed that closely related strains produce similar AHL profiles. One of the 57 isolates included was A. salmonicida strain LFI1238, from which we were able to identify a total of eight different AHLs. The AHLs were analyzed at only one cell density from cultures grown at one temperature in this study [34]. Since in A. salmonicida the regulatory role of LitR on some phenotypes is temperature dependent [10,41] we wanted to analyze the impact of temperature on AHL production. Hence, in the work presented here we have analyzed the AHL profiles of A. salmonicida wild type LFI1238 and a ΔlitR mutant at different cell densities during growth at three different temperatures. We have also inactivated the AHL synthetases AinS and LuxI in order to identify which synthetase makes which AHL(s). Finally, we studied AHL production in luxR mutants, and analyzed the involvement of the AinS/AinR and LuxI/LuxR systems in biofilm formation.
Methods
Chemicals and AHL standards
The following AHL standards were purchased from University of Nottingham, UK: N-3-oxo-butyryl-L-homoserine lactone (3-oxo-C4-HSL), N-3-hydroxy-butyryl-L-homoserine lactone (3-OH-C4-HSL), N-3-hydroxy-hexanoyl-L-homoserine lactone (3-OH-C6-HSL), N-3-hydroxy-octanoyl-L-homoserine lactone (3-OH-C8-HSL), N-3-hydroxy-decanoyl-L-homoserine lactone (3-OH-C10-HSL). Standards purchased from Sigma-Aldrich were: N-butyryl-DL-homoserine lactone (C4-HSL), N-hexanoyl-L-homoserine lactone (C6-HSL), N-3-oxo-hexanoyl-L- homoserine lactone (3-oxo-C6-HSL), N-octanoyl-L-homoserine lactone (C8-HSL), N-3-oxo-octanoyl-L-homoserine lactone (3-oxo-C8-HSL), N-decanoyl-DL-homoserine lactone (C10-HSL), N-3-oxo-decanoyl-L-homoserine lactone (3-oxo-C10-HSL), N-dodecanoyl-DL-homoserine lactone (C12-HSL), N-3-oxo-dodecanoyl-L-homoserine lactone (3-oxo-C12-HSL), and N-3-hydroxy-dodecanoyl-DL-homoserine lactone (3-OH-C12-HSL). HPLC grade acetonitrile and formic acid were purchased from Merck.
Strains and culture conditions
Bacterial strains and plasmids used in this study are listed in Table 1. The A. salmonicida strains were grown at 12°C in either Lysogeny Broth (LB) or Tryptic Soy Broth (TSB) (Difco, BD Diagnostics) supplemented with 2.5% or 1.5% NaCl, respectively. The Escherichia coli strains S17.1 and DH5α were cultured in LB medium, whereas DH5αλpir was cultivated in Brain Heart Infusion (BHI) (Oxoid, Cambridge, UK) medium at 37°C. The plasmids pDM4 and pNQ705 were propagated in S17.1 cells [42,43] and pEVS122 and pEVS104 were propagated in DH5αλpir and DH5α, respectively [44,45].
Table 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Description a | Source or reference | |
|---|---|---|---|
| A. salmonicida | |||
| LFI1238 | Wild type, isolated from Atlantic cod | [11] | |
| ΔlitR | LFI1238 containing an in-frame deletion of litR | [10] | |
| ΔainS | LFI1238 containing an in-frame deletion of ainS | This study | |
| ΔainS/luxI ˉ | ΔainS with insertional disruption of luxI; Cmr | This study | |
| luxI ˉ | LFI1238 with insertional disruption of luxI; Cmr | This study | |
| luxR1ˉ | LFI1238 with insertional disruption of luxR1 gene, Ermr | This study | |
| luxR2ˉ | LFI1238 with insertional disruption of luxR2 gene, Ermr | This study | |
| E. coli | |||
| S17-1 | Donor strain for conjugation, λ-pir | [42] | |
| DH5αλ pir | Donor strain for conjugation | [45] | |
| DH5α | Helper strain for conjugation | Invitrogen | |
| Plasmids | |||
| pDM4 | Cmr; suicide vector with an R6K origin (λ-pir requiring) and sacBR | [43] | |
| pNQ705 | Cmr; suicide vector with an R6K origin (λ-pir requiring) | [43] | |
| pEVS122 | Ermr; suicide vector with an R6Kγ oriV, oriT RP4, lacZα,cosN, loxP, incD | [45] | |
| pEVS104 | Helper plasmid | [44] | |
| pDM4ΔainS | pDM4 containing a fragment of ainS harbouring an internal deletion | This study | |
| pNQ705luxI | pNQ705 containing a 245 bp fragment of luxI | This study | |
| pEVSluxR1 | pEVS122 containing a 224 bp fragment of luxR1 | This study | |
| pEVSluxR2 | pEVS122 containing a 280 bp fragment of luxR2 | This study | |
aCmr, Chloremphenicol resistance gene. Ermr, Erythromycin resistance gene.
E. coli carrying pNQ705 and pDM4 constructs were grown on media containing 25 μg/ml chloramphenicol, whereas 100 μg/ml kanamycin or 250 μg/ml erythromycin were used in the media for propagating E. coli carrying pEVS104 and pEVS122. A. salmonicida transconjugants were selected on LB agar plates containing 2.5% NaCl and 2 μg/ml chloramphenicol, or on TSB agar plates containing 1.5% NaCl and 25 μg/ml erythromycin.
DNA extraction, PCR and sequencing
Extraction of DNA, recombinant DNA techniques and transformations were performed according to standard protocols [46]. Restriction enzyme digestion, ligation and plasmid purification were performed as recommended by the manufacturers (NEB Biolabs, OMEGA Bio-Tek and Invitrogen). PCR using Phusion (FinnZyme) or Taq polymerase (Biolabs) as well as Big Dye sequencing (Applied Biosystems) were performed with custom made primers (Sigma, Operon and Medprobe). The primers used for PCR and sequencing are listed in Additional file 1: Table S1.
Construction of A. salmonicida LFI1238 mutants
Construction of the ΔlitR in-frame mutant has been described elsewhere [10]. Similarly, the ainS gene was deleted in A. salmonicida by allelic exchange. In brief, the sequences flanking ainS were amplified by PCR from genomic DNA of LFI1238. The A and B primers were used for amplification of the region upstream ainS, and primers C and D for amplification of the downstream region (Additional file 1: Table S1). Primers B and C contain complementary sequences that enable fusion of the upstream and the downstream PCR products by an overlap PCR using the outermost primers A and D. This results in a removal of 357 codons (including the start codon) from the ainS open reading frame, and hence, only 40 codons remain. The primers A and D contain SpeI and XhoI restriction enzyme sites, respectively, in their 5’end. The overlapping PCR product was digested with SpeI and XhoI, ligated into the corresponding sites of the suicide vector pDM4 and transformed directly into E. coli S17-1 cells.
The luxI gene was inactivated by plasmid insertion in the wild type A. salmonicida LFI1238 as well as in the ΔainS mutant. The plasmid used for insertional inactivation was made by amplifying an internal part of luxI (245 bp) by PCR. Adenine overhangs were added to the PCR products using a Taq polymerase before being ligated into the pGEM T-Easy vector (Invitrogen) and transformed into E. coli DH5α. The resulting plasmid was purified, digested with SpeI and XhoI and finally the re-purified PCR products were cloned into the corresponding sites of the suicide vector pNQ705. Similarly, internal fragments of luxR1 (224 bp) and luxR2 (280 bp) were PCR amplified and cloned into a pCR™4-TOPO® TA cloning vector according to the manufacturer’s instructions (Invitrogen). The luxR1 and luxR2 fragments were cut from the resulting plasmids using BamHI and cloned into the corresponding sites of pEVS122.
The pDM4 and pNQ705 constructs were transformed into E. coli S17-1 and used as donors in mating experiments with their respective parental A. salmonicida strain as described previously [10]. For inactivation of luxR1 and luxR2 tri-parental mating was performed [39,45]. To this end, pEVSluxR1 or pEVSluxR2 was transferred from E. coli DH5αλpir to A. salmonicida LFI1238 using pEVS104 contained in E. coli DH5α as helper plasmid. Briefly, DH5αλpir, DH5α and LFI1238 were grown to their stationary phases. In this experiment the LFI1238 was grown at 15°C in TSB containing 1.5% NaCl. The volume of each culture was adjusted to account for a 1:1:1 ratio of cells. The cells were pelleted by centrifugation, washed twice and combined by re-suspension in chilled TSB before being spotted onto chilled blood agar plates with 2.5% NaCl. The plates were incubated at 21°C for 5–6 hours followed by incubation at 15°C for 16 hours. The resulting confluent growth of cells was re-suspended in chilled TSB and cultivated for 24 hours at 12°C with 200 rpm. Finally, the suspension was plated onto TSB agar plates with 1.5% NaCl and erythromycin. After 5–7 days of growth, erythromycin resistant luxR1 and luxR2 mutants were isolated.
Preparation of bacterial supernatants for AHL measurements
Two or more biological replicates were used for all A. salmonicida strains, with three technical replicates of each sample collected at different time points. The primary cultures (2 ml) were grown from individual colonies in LB medium with 2.5% NaCl at 12°C with 220 rpm. After 48 hours, a secondary culture was made by diluting the primary culture 1:20. The secondary cultures were grown 24 hours before being diluted to OD600 = 0.001 or 0.050 (optical density measured at 600 nm) in a total volume of 60 ml LB with 2.5% NaCl. The cultures were grown further in 250 ml baffled flask at 220 rpm. Samples (1 ml) were harvested at regular intervals and centrifuged at 17000 g for 1 minute (Heraeus Fresco 21, Thermo Scientific). The supernatants were acidified before ethyl acetate extraction as previously described [34]. The ethyl acetate phase was dried using a rotary vacuum centrifuge (PH40-11, Savant Instruments Inc.) and re-dissolved in 150 μl of 20% acetonitrile containing 0.1% formic acid and 775 nM of the internal standard 3-oxo-C12-HSL.
Samples from three separate experiments were harvested and prepared as described above. In the first experiment, the secondary cultures of LFI1238 and ΔlitR were diluted to an OD600 of 0.050 and grown at 6, 12 and 16°C. Here, the samples were harvested at eight different time points starting from an OD600 of ~ 0.5 through to the stationary phase. In the second experiment, cultures of LFI1238, ΔainS, luxIˉ and ΔainS/luxIˉ were started at an OD600 of 0.001, grown at 12°C and harvested after 50 hours. In the third experiment the different cultures (LFI1238, luxR1ˉ and luxR2ˉ) were started at an OD600 of 0.001 and grown at 12°C before samples were collected at an OD600 of 1.8 (approx. 36 hours). The AHL molecules were separated by HPLC before being identified using MS/MS with selective reaction monitoring (SRM) (first and second experiments) or using a Full Scan High Resolution (HR) MS method (see below).
Detection of AHL profiles using HPLC-MS/MS analysis
The HPLC-MS/MS analysis was performed as previously described [34]. In brief, HPLC was performed using a Hypersil GOLD C18 reverse phase column (50 × 2.1 mm, 1.9 μm particle size, Thermo Scientific) and eluted with a 162 second gradient of 5-95% acetonitrile in 0.1% formic acid at a flow rate of 500 μl/min. The LTQ (Linear Ion Trap Quadrupole) part of the LTQ orbitrap XL (Thermo Scientific) was used in SRM mode for detection of fragment m/z 102 from parent ions. The retention time was divided into 6 segments of varying lengths with two to three scan events for each segment. The data was collected from 101 to 103 m/z.
Detection of AHL profiles using HPLC with Full Scan HR-MS analysis
HPLC with Full Scan HR-MS was performed using the LTQ Orbitrap XL and Accela Autosampler (Thermo Scientific). The samples (20 μl) were injected onto the same reverse phase column as described above. The elution procedure was performed with an acetonitrile gradient in 0.1% formic acid, and consisted of 5% acetonitrile for 18 seconds, followed by a linear gradient up to 90% acetonitrile over 402 seconds, and finally 90% acetonitrile for 150 seconds. The column was re-equilibrated for 150 seconds with 5% acetonitrile in 0.1% formic acid before the next sample was injected. The flow rate was 200 μl/min for all steps. The separated compounds were detected under positive ion conditions by electrospray ionization using the following settings: sheath gas flow rate 35, auxiliary gas flow rate 20, sweep gas flow rate 0, spray voltage +4.50 kV, capillary temperature 300°C, capillary voltage 47 V, and tube lens 80–90 V. The orbitrap was operated in the full scan mode from m/z 165–450 at a resolution of 15.000 with target setting of 5 × 105 ions per scan and collection of data in the profile mode. The maximum ion injection time was 500 ms. Lock mass was enabled for correction of background ions from caffeine (m/z 195.0877) and diisooctyl phthalate (m/z 391.2843 and m/z 413.2662). The system was calibrated with a mixture of 15 AHLs including the internal standard 3-oxo-C12-HSL, and the ion chromatograms were analyzed using the Xcalibur v. 2.0.7 software package. The mass window was set to 15 parts per million. The limit of detection (LOD) and the limit of quantification (LOQ) for the different AHLs were calculated as previously described [34] and are shown in Additional file 2: Table S2. This method does not allow quantification of C4 chained AHLs.
Biofilm assay
The biofilm assay was performed mainly as described elsewhere [41]. In brief, the different strains were grown in LB with 2.5% NaCl at 12°C before being diluted to OD600 = 0.1 in SWT medium (5 g/l Bacto Peptone (BD), 3 g/l Yeast Extract (Sigma), 28 g/l Marine Sea Salt (Tetra)). A total volume of 300 μl of each dilution was added to wells in a flat bottom non tissue culture treated Falcon 24-well tray (BD Biosciences). The plates were incubated statically at 4°C for 3 days before being stained with 0.1% crystal violet. The air-dried biofilms were dissolved in 96% ethanol (500 μl/well) before the absorbance was read at 590 nm (Vmax Kinetic Microplate Reader, Molecular Devices). Three biological replicates were used for each strain, and the experiments were repeated several times.
Statistical analysis
Student’s t test was performed to calculate statistical significance (p-values) using the Microsoft Excel 2010 software.
Results
AHL signal production in A. salmonicida is cell density and temperature dependent
We recently established a method for detection of AHLs using HPLC-MS/MS and reported that A. salmonicida LFI1238 produces 8 different AHLs: 3-oxo-C4-HSL, C4-HSL, C6-HSL, 3-oxo-C6-HSL, C8-HSL, 3-oxo-C8-HSL, 3-oxo-C10-HSL and 3-OH-C10-HSL. In this previous study the bacterium was grown at 12°C and the supernatants were harvested and analyzed after the bacterium had reached the stationary phase [34]. Under laboratory conditions, A. salmonicida has a growth optimum between 12-16°C in liquid cultures [4]. However, a seawater temperature below 12°C is normally a prerequisite for A. salmonicida to cause cold water vibrosis in Atlantic salmon [47], as well as for siderophore production and iron-regulated outer membrane protein expression [7]. Moreover, growth temperatures below 14-16°C are required for the bacteria to express a number of host-bacterium related phenotypes such as adhesion, colony morphology and biofilm formation [10,41]. Therefore, to investigate whether AHL production is also affected by growth temperature, supernatants from the wild type LFI1238 were harvested at different cell densities during growth at 6, 12 and 16°C.
As shown in Figure 1, the different AHLs had reached detectable levels when the measurements started (OD600 ~ 0.5), and increased thereafter in a cell density dependent manner. For most AHLs, the highest concentrations were observed near the stationary phase, after which they decreased. Of the dominating AHLs, 3-oxo-C6-HSL reached its maximum concentration (22 μM) after 38 hours growth at 12°C, whereas C6-HSL and 3-oxo-C8-HSL, which were both detected in significantly lower concentrations, reached maxima (2.1 μM and 0.95 μM) after 30 and 38 hours growth respectively, at 12°C. The production of AHLs was highly dependent on temperature, being dramatically lower during growth at 16°C. At this temperature, the maximum concentrations of 3-oxo-C6-HSL, C6-HSL, 3-oxo-C8-HSL and 3-OH-C10-HSL were less than 5% of those produced after growth at 6°C. Only small amounts of 3-oxo-C4-HSL, C4-HSL, C8-HSL and 3-oxo-C10-HSL could be detected at 16°C, and these were at concentrations below the LOQ [34]. Aside from C6-HSL and 3-OH-C10-HSL, the different AHLs were produced at approximately similar levels during growth at 6 and 12°C. The concentration of C6-HSL produced at 12°C was approximately twice that compared to at 6°C. Interestingly, the opposite was observed for 3-OH-C10-HSL where the highest concentration was produced at 6°C, with the concentration rising steadily throughout the experiment reaching a final value of 610 nM compared to only 140 nM after growth at 12°C. Thus, the decrease in growth temperature to 6°C resulted in a four-fold increase of the 3-OH-C10-HSL concentration.
Figure 1.
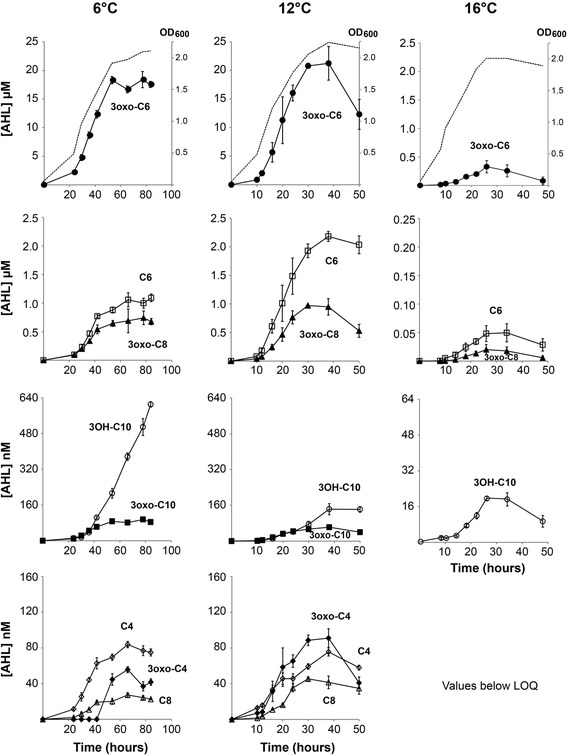
AHLs produced by A. salmonicida LFI1238 at different temperatures and cell densities. Supernatants were harvested at different time points during growth at 6, 12 and 16°C. The AHL concentrations were determined by HPLC-MS/MS and are shown with respect to time (hours) and cell density (OD600). Symbols indicate the different AHLs: 3-oxo-C6-HSL (●), C6-HSL (□), 3-oxo-C8-HSL (▲), 3-OH-C10-HSL (○), 3-oxo-C10-HSL (■), C4-HSL (◇), 3-oxo-C4-HSL (◆) and C8-HSL (∆). Each value represents the mean of triplicates from three biological replicates. The error bars represent the standard deviations. The dotted lines in the top panels display the growth curves of LFI1238 at different temperatures.
Identification of AHLs produced by LuxI and AinS
Comparison of the genomes of A. salmonicida and A. fischeri, which are ~65% identical, allowed identification of the genes encoding the AHL synthases LuxI and AinS in A. salmonicida LFI1238 [11]. To determine which AHLs are produced by which synthases in LFI1238, the ainS and luxI genes were interrupted by allelic exchange and plasmid insertion respectively, giving rise to the ∆ainS and the luxIˉ mutants. A double mutant (∆ainS/luxIˉ) was also made by interrupting both genes. The wild type LFI1238 and all three mutants were grown at 12°C for 50 hours (OD600 ~ 2.2) before samples were collected and analyzed. As shown in Figure 2, LFI1238 produced 8 different AHLs which were identified by their retention times (RT) and mass-to-charge ratio (m/z). The ∆ainS mutant was unable to produce 3-OH-C10-HSL but the remaining seven AHLs were still detected; whereas the luxIˉ mutant produced only 3-OH-C10-HSL, and the double mutant ∆ainS/luxIˉ showed no AHL production. Thus, the AinS synthase is responsible for the 3-OH-C10-HSL production whereas LuxI is responsible for production of 3-oxo-C4-HSL, C4-HSL, 3-oxo-C6-HSL, C6-HSL, C8-HSL, 3-oxo-C8-HSL and 3-oxo-C10-HSL.
Figure 2.
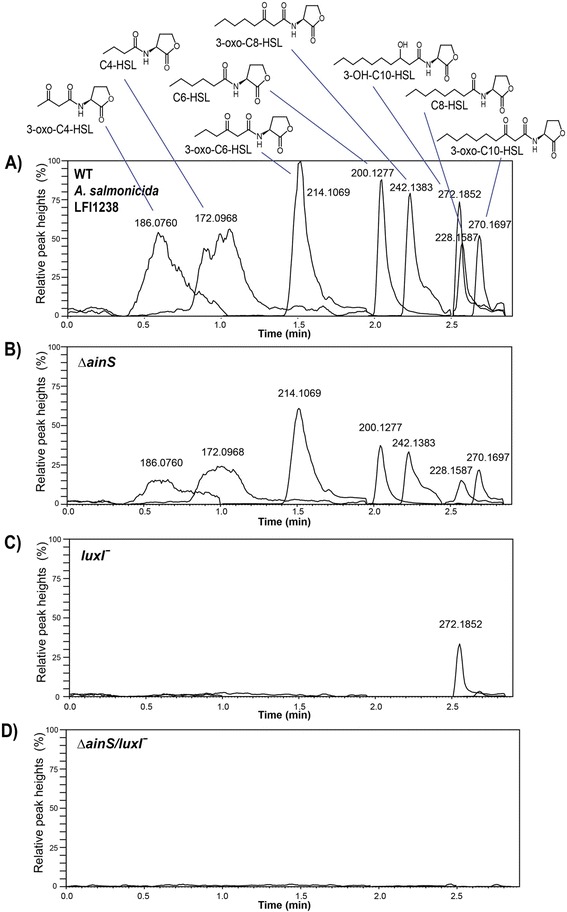
HPLC-MS/MS ion chromatograms of AHLs produced by A. salmonicida LFI1238 and QS mutants. (A) The wild type LFI1238 produces eight AHLs those being the C4-HSL, 3-oxo-C4-HSL, C6-HSL, 3-oxo-C6-HSL, C8-HSL, 3-oxo-C8-HSL, and 3-oxo-C10-HSL and 3-OH-C10-HSL. (B) The ΔainS mutant lacks 3-OH-C10-HSL production, and the chromatogram shows the seven remaining AHLs. (C) The luxIˉ mutant produces only 3-OH-C10-HSL and (D) no AHLs were detected in the supernatants harvested from the double mutant ΔainS/ luxIˉ. The peaks of the different AHLs were scaled so that the different AHLs could be shown in the same chromatogram. The scaling factors were as follows: 1.6 × 103 for 3-oxo-C4-HSL, 6.0 × 103 for C4-HSL, 9.0 × 105 for 3-oxo-C6-HSL, 2.7 × 105 for C6-HSL, 5.4 × 104 for 3-oxo-C8-HSL, 1.5 × 104 for C8-HSL, 1.9 × 104 for 3-OH-C10-HSL and 1.0 x 104 for 3-oxo-C10-HSL. The same scaling was used for all four chromatograms (A, B, C and D). The numbers above the peaks are mass-to-charge ratios (m/z).
LitR is a positive regulator of AinS AHL production
In A. fischeri the master regulator LitR is a positive regulator of the LuxI/LuxR and the AinS/AinR systems [20,25]. We therefore wanted to determine if LitR has a similar role in A. salmonicida. The ∆litR mutant was grown at different temperatures as described above and samples were collected throughout the growth curve. In contrast to a previous report [10] the ΔlitR mutant did not reach to the same cell densities as the wild type LFI1238 in this experiment (Figure 3), which may be due to different culturing conditions or media; however AHL production was still affected by deletion of litR. Compared to the wild type LFI1238 (Figure 1) the maximum concentrations of 3-OH-C10-HSL (AinS product) produced by ΔlitR were only 11% at 6°C and 14% at 12°C (P values < 0.05) (Figure 3), while the amounts of 3-OH-C10-HSL detected after growth at 16°C were below the quantification limit [34]. Similarly, inactivation of litR resulted in decreased production of 3-oxo-C6-HSL, C6-HSL and 3-oxo-C8-HSL (LuxI products), and at 12°C the maximum concentrations reached by the ∆litR mutant were only 50-60% of those reached by LFI1238 (P values < 0.05). The opposite situation was observed when the bacteria were grown at 16°C, where in the absence of litR the concentrations of 3-oxo-C6-HSL, C6-HSL and 3-oxo-C8-HSL were approximately two-three times higher than in the wild type. Still, these levels were much lower than those of LFI1238 at 6°C (less than13%). No significant differences in the production of 3-oxo-C4-HSL, C4-HSL, C8-HSL and 3-oxo-C10-HSL were detected between LFI1238 and ΔlitR at any of the conditions.
Figure 3.
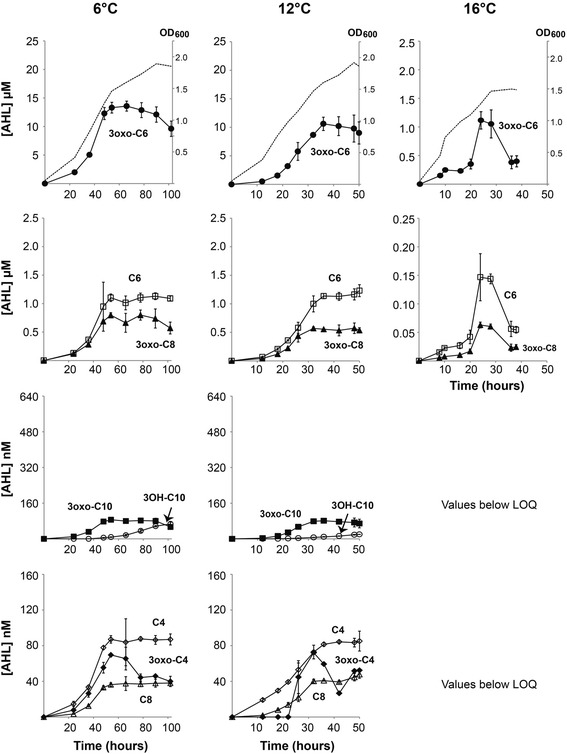
AHLs produced by the A. salmonicida ΔlitR mutant at different temperatures and cell densities. Supernatants were harvested at different time points during growth at 6, 12 and 16°C. The AHL concentrations were measured by HPLC-MS/MS and are shown with respect to time (hours) and cell density (OD600). Symbols indicate the different AHLs; 3-oxo-C6-HSL (●), C6-HSL (□), 3-oxo-C8-HSL (▲), 3-OH-C10-HSL (○), 3-oxo-C10-HSL (■), C4-HSL (◇), 3-oxo-C4-HSL (◆) and C8-HSL (∆). Each value represents the mean of triplicates from three biological replicates. The error bars represent the standard deviations. The dotted lines in the top panels display the growth curves of the ΔlitR mutant at the different temperatures.
LuxR1 and LuxR2 are required for production of LuxI AHLs
In A. fischeri, LuxR and autoinducer activate transcription the lux operon encoding the AHL synthetase LuxI and proteins required for bioluminescence [27,28]. We therefore inactivated both copies of luxR (luxR1 and luxR2) in A. salmonicida LFI1238 by plasmid insertion and analyzed the AHL profiles of the resulting mutants (luxR1ˉ and luxR2ˉ) using HPLC with Full Scan HR-MS. The wild type and mutants were grown at 12°C and samples collected in the stationary phase. As shown in Table 2, inactivation of either copy caused a significant decrease in the production of LuxI-dependent AHLs, with mutants producing only 5.6% C6-HSL, 3.6% 3-oxo-C6-HSL and 2.7% 3-oxo-C8-HSL relative to wild type LFI1238 concentrations. Small amounts of C8-HSL and 3-oxo-C10-HSL were detected but in concentrations close to or below the quantification limit (Additional file 2: Table S2). Finally, production of the AinS signal (3-OH-C10-HSL) was not affected by mutations in luxR1 or luxR2. It should be noted that the AHL concentrations produced by the wild type were much lower here (Table 2) than detected in the experiment above (see Figure 1). A reason for this could be that the cultures were started at OD600 = 0.001 and not at OD600 = 0.050 as above. The concentrations of 3-oxo-C4-HSL and C4-HSL were not determined in this experiment.
Table 2.
AHL production in A. salmonicida LFI1238 and luxRˉ mutants a
| Bacterial strain | C6 (nM) | 3-oxo-C6 (nM) | C8 (nM) | 3-oxo-C8 (nM) | 3-oxo-C10 (nM) | 3-OH-C10 (nM) |
|---|---|---|---|---|---|---|
| LFI1238 | 359 ± 17 | 5021 ± 173 | 15 ± 2 | 332 ± 27 | 32 ± 2 | 53 ± 6 |
| luxR1ˉ | 20 ± 3 | 180 ± 51 | 1.9 ± 0.4 | 8 ± 1 | * | 60 ± 1 |
| luxR2ˉ | 19 ± 5 | 139 ± 75 | * | 9 ± 2 | * | 52 ± 5 |
aEach value represents the mean of triplicates from two biological replicates harvested at OD600 ~ 1.8.
The samples were analyzed by the HPLC Full Scan HR-MS method. The concentrations of C4-HSL or 3-oxo-C4-HSL were not determined.
*Below the limit of quantification (LOQ) given in Additional file 2: Table S2.
The AinS/AinR and LuxI/LuxR pathways regulate biofilm formation
LitR is a negative regulator of biofilm in A. salmonicida LFI1238, hence deletion of litR results in increased biofilm formation. The biofilm formation is both temperature- and medium-dependent [10,41]. To establish the impact of AHLs in this process, we analyzed the biofilm formation capabilities of the different mutants. As shown in Figure 4, disruption of either ainS or luxI alone did not alter biofilm formation, however interruption of both genes simultaneously resulted in a phenotype similar to ΔlitR. Although luxR1 and luxR2 are necessary for manufacture of LuxI-produced AHLs, interruption of these genes did not affect biofilm formation in the assays performed here.
Figure 4.
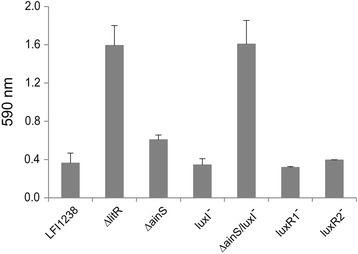
Biofilm formation of A. salmonicida LFI1238 and QS mutants. The biofilms were grown statically for 72 hours at 4°C before being stained with crystal violet. The absorbance was read at 590 nm. The error bars represent the standard deviation of three biological replicates.
Discussion
Although A. salmonicida is no longer an immediate threat to the aquaculture industry it would be beneficial to understand factors involved in its virulence. Disruption of bacterial communication or QS may present a way to reduce virulence, with the usual strategies being destruction of signals by enzymes, or inactivation by chemicals or natural compounds with antagonistic activities [48].
The relationship between AHL production and cell density is thoroughly documented for a variety of Gram-negative bacteria [18,27]. Similarly, in the study presented here we show that the eight AHLs identified in A. salmonicida LFI1238 are also produced in a cell density-dependent manner. A. salmonicida was initially reported to produce only two AHLs: C6-HSL and 3-oxo-C6-HSL [30], and reasons for this discrepancy are likely due to different detection methods, strains, growth medium and cultivation temperatures between the different studies. However, by inactivation of two putative AHL synthases in A. salmonicida LFI1238 we found that LuxI produces seven AHLs (3-oxo-C4-HSL, C4-HSL, 3-oxo-C6-HSL, C6-HSL, C8-HSL, 3-oxo-C8-HSL and 3-oxo-C10-HSL) whereas AinS only produces one AHL (3-OH-C10-HSL). LuxI homologues in other bacteria are known to catalyze the acylation and lactonization reactions between the substrates S-adenosylmethionine and the acylated ACP (acyl carrier protein) [16,28], thus it is reasonable to assume that LuxI perform the same reaction in A. salmonicida. The ability of LuxI to produce an array of different AHLs is intriguing and suggests that this enzyme in A. salmonicida LFI1238 does not discriminate well between different substrates, and can accept ACP carrying acyl chains of different lengths as well as acyl chains without or with a keto substitution in the third position (3-oxo). The ability of LuxI homologues to produce a broad spectrum of AHLs is not restricted to A. salmonicida. Indeed, the human pathogenic bacteria Yersinia pseudotuberculosis has been reported to produce a far greater range of AHLs. This bacterium possesses two LuxI homologous, YpsI amd YtbI, which are responsible for producing over 20 different AHLs containing acyl chains of both odd and even numbers of carbons [49].
The majority of vibrios and aliivibrios produce multiple types of AHLs suggesting that it provides some biological advantages [34]. This was shown for A. fischeri where C8-HSL binds and activates both AinR and LuxR at intermediate cell densities, and at higher cell densities LuxR binds 3-oxo-C6-HSL [19]. This sequential activation allows specific regulation of early colonization factors by AinS/AinR, whereas late colonization factors and luminescence are preferentially regulated by LuxI/LuxR. Both systems however are required for persistent colonization of the squid host [21]. Thus, it is possible that A. salmonicida exploits its AHL diversity to regulate specific activities in response to temperature or other environmental changes, or during transmission from the seawater into the host and vice versa.
Cold-water vibriosis in Atlantic salmon occurs mainly in late autumn, winter and early spring when the water temperature is below 12°C [47]. We have recently shown that LitR regulates a number of phenotypes, including virulence, in A. salmonicida LFI1238 and that this regulation was stronger at lower temperatures [10,41]. Similarly, as shown here, the growth temperature of wild type LFI1238 was found to have a significant impact on AHL production, and increasing the temperature from 12 to 16°C resulted in a drastic decrease of all eight AHLs. This decline cannot be explained by differences in growth rate, since the LFI1238 cultures all reached stationary phase at similar cell densities (OD600 ~ 2) irrespective of their growth temperatures. The LuxI AHLs were produced at comparable or higher levels after growth at 12°C compared to 6°C. On the other hand, the AinS signal concentration produced at 6°C was approximately four times higher than at 12°C in LFI1238. This is interesting considering that LitR was found to regulate production of the AinS signal 3-OH-C10-HSL, and the ΔlitR mutant produced 3-OH-C10-HSL corresponding to ≤ 14% of the wild type concentrations at 6 and 12°C. This finding strengthens the suggestion that LitR exerts its regulatory function(s) more strongly at low temperatures.
Similar to LitR in A. fischeri [25], our results suggest that LitR also connects the LuxI/LuxR systems to the AinS/AinR and LuxS/LuxPQ systems in A. salmonicida (Figure 5). This conclusion is based on the finding that; (i) deletion of litR influenced production of LuxI AHLs, (ii) inactivation of ainS and luxI simultaneously was needed to produce a biofilm similar to the ΔlitR mutant, as well as our previous results showing that (iii) the ∆litR mutant produces up to 20-fold less cryptic bioluminescence compared to the wild type LFI1238 [10]. Although LitR is involved in regulation of the lux operon, deletion of litR does not completely prevent AHL production through LuxI, and the ΔlitR mutant still produced the LuxI-dependent AHLs 3-oxo-C6-HSL, C6-HSL and 3-oxo-C8-HSL at 50-60% of wild type levels at 6 and 12°C. This suggests that LitR is not essential for luxR transcription and that other regulators may be involved in the transcriptional regulation of the two luxR genes and hence luxI. Autoregulation of luxR [50,51], and LitR independent activation of luxR have been demonstrated in A. fischeri. One of those LitR independent mechanisms involves cAMP and cAMP receptor protein (CRP) [52,53], and more recently the regulatory RNA-binding protein carbon storage regulator A (CsrA) has been shown to increase transcription of luxR independently of LitR [54]. A. salmonicida LFI1238 encodes the genes for CsrA and CRP [11] and similar mechanisms may therefore explain why AHL production of LuxI is only reduced when litR is deleted. On the other hand, our study shows that both LuxRs in A. salmonicida LFI1238 are necessary for producing LuxI-AHLs, and inactivation of either luxR1 or luxR2 reduced LuxI-produced AHLs significantly without affecting the AinS signal production. A possible explanation for this is that LuxR1 and LuxR2 function as a heterodimer in order to activate transcription of luxI, however further studies are needed to elucidate this.
Figure 5.

Illustration of the proposed model of the QS system in A. salmonicida LFI1238. The autoinducer synthases LuxS, LuxI and AinS, produce the different AHLs and AI-2 which are transported across the inner membrane (IM) and the outer membrane (OM). Their respective receivers are believed to be LuxPQ, a LuxR1-LuxR2 heterodimer, and AinR. The LuxS/LuxPQ pathway may be inactive due to a frame shift mutation within luxP. It is unknown which AHLs bind the LuxRs (illustrated with a question mark). At low cell density, a phosphorylation cascade is believed to start from the receivers LuxPQ and AinR, and proceed downstream to LuxO via LuxU (illustrated with dashed arrows). LuxO probably regulates expression of Qrr, which in turn controls the expression of the master regulator LitR. When the autoinducer concentrations are high, LitR is expressed and regulates the production of the AinS AHL, as well as activities such as motility, biofilm, adhesion, virulence and bioluminescence [10]. Both LitR and the LuxRs are probably involved in regulation of the lux operon as illustrated.
In our work we have studied the role of different QS genes using gene inactivation. It should be pointed out that although our observations with the different mutants are coherent complementation studies for the different mutants would be required to unambiguously prove the function of the inactivated genes.
Conclusions
In this study we have shown that the AHL autoinducer synthases LuxI and AinS in A. salmonicida LFI1238 produced the eight different AHLs. Their production is dependent on both the cell density and cultivation temperature of the bacteria. Production of numerous AHLs suggests that the QS signaling cascade is complex and probably important to fine-tune activities such as virulence, biofilm and other adaption processes to respond to changes in the environment. The AinS/AinR and LuxR/LuxI systems are connected to, and needed for down-regulation of biofilm formation. However, further investigations are needed to understand the regulation and complexity of QS in A. salmonicida.
Ethics statement
The work presented in this paper does not involve human subjects, and we see no ethical issues.
Acknowledgements
This work was financed by The Norwegian Research Council, UiT The Arctic University of Norway and the Norwegian Structural Biology Centre. We thank Dr. Debra Milton (Umeå University) for the pDM4 and pNQ705 plasmids, and Dr. Pat M. Fidopiastis (Cal Poly) for the pEVS122 and pEVS104 plasmids which were originally constructed by Dr. Eric V. Stabb (University of Georgia). We also thank Adele Kim Williamson (UiT The Arctic University of Norway) for proofreading of the manuscript.
Abbreviations
- AHLs
N-acyl-homoserine lactones
- 3-OH-C4-HSL
N-3-hydroxy-butyryl HSL
- 3-OH-C6-HSL
N-3-hydroxy-hexanoyl HSL
- 3-OH-C8-HSL
N-3-hydroxy-octanoyl HSL
- 3-OH-C10-HSL
N-3-hydroxy-decanoyl HSL
- 3-OH-C12-HSL
N-3-hydroxy-dodecanoyl HSL
- 3-oxo-C4-HSL
N-3-oxo-butyryl HSL
- 3-oxo-C6-HSL
N-3-oxo-hexanoyl HSL
- 3-oxo-C8-HSL
N-3-oxo-octanoyl HSL
- 3-oxo-C10-HSL
N-3-oxo-decanoyl HSL
- 3-oxo-C12-HSL
N-3-oxo-dodecanoyl HSL
- C4-HSL
N-butyryl HSL
- C6-HSL
N-hexanoyl HSL
- C8-HSL
N-octanoyl HSL
- C10-HSL
N-decanoyl HSL
- C12-HSL
N-dodecanoyl HSL
- HPLC-MS/MS
High-performance liquid chromatography tandem mass spectrometry
- HR
High Resolution
- m/z
Mass-to-charge ratio
- HSL
Homoserine lactone
- LB
Lysogeny broth
- min
Minutes
- OD600
Optical density measured at 600 nm
- PCR
Polymerase chain reaction
- QS
Quorum sensing
- rpm
Rounds per minute
- SRM
Selective reaction
- TSB
Tryptic Soy Broth
Additional files
The table lists primers used in this study.
The table lists AHL detection limits and quantification parameters for the HPLC Full Scan HR-MS method.
Footnotes
Hilde Hansen and Amit Anand Purohit contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
HH, AAP and NPW conceived and designed the experiments. HH, AAP and AMB constructed the mutants. AAP, SJK and JAJ performed the AHL measurements. HH performed the biofilm assay. All authors were involved in analyzing the data. HH, AAP, HKSL and NPW wrote the paper. All authors read and approved the final manuscript.
Contributor Information
Hilde Hansen, Email: hilde.hansen@uit.no.
Amit Anand Purohit, Email: amitpurohit21@outlook.com.
Hanna-Kirsti S Leiros, Email: hanna-kirsti.leiros@uit.no.
Jostein A Johansen, Email: jostein.johansen@uit.no.
Stefanie J Kellermann, Email: stefanie.kellermann@uni-muenster.de.
Ane Mohn Bjelland, Email: ane.mohn.bjelland@nmbu.no.
Nils Peder Willassen, Email: nils-peder.willassen@uit.no.
References
- 1.Thompson FL, Austin B, Swings J. The biology of vibrios. Washington, D.C.: ASM Press; 2006. [Google Scholar]
- 2.Urbanczyk H, Ast JC, Higgins MJ, Carson J, Dunlap PV. Reclassification of Vibrio fischeri, Vibrio logei, Vibrio salmonicida and Vibrio wodanis as Aliivibrio fischeri gen. nov., comb. nov., Aliivibrio logei comb. nov., Aliivibrio salmonicida comb. nov. and Aliivibrio wodanis comb. nov. Int J Syst Evol Microbiol. 2007;57(Pt 12):2823–9. doi: 10.1099/ijs.0.65081-0. [DOI] [PubMed] [Google Scholar]
- 3.Egidius E, Andersen K, Clausen E, Raa J. Cold-water vibriosis or “Hitra disease” in Norwegian salmonid farming. J Fish Dis. 1981;4(4):353–4. doi: 10.1111/j.1365-2761.1981.tb01143.x. [DOI] [Google Scholar]
- 4.Holm KO, Strøm E, Stensvag K, Raa J, Jørgensen T. Characteristics of a Vibrio sp. associated with the “Hitra disease” of Atlantic salmon in Norwegian fish farms. Fish Pathology. 1985;20(2–3):125–9. doi: 10.3147/jsfp.20.125. [DOI] [Google Scholar]
- 5.Egidius E, Wiik R, Andersen K, Hoff KA, Hjeltnes B. Vibrio salmonicida sp. nov., a new fish pathogen. Int J Syst Evol Microbiol. 1986;36(4):518–20. [Google Scholar]
- 6.Helsesituasjonen hos laksefisk [http://www.vetinst.no/nor/Forskning/Publikasjoner/Fiskehelserapporten/Fiskehelserapporten-2009]
- 7.Colquhoun DJ, Sørum H. Temperature dependent siderophore production in Vibrio salmonicida. Microb Pathog. 2001;31(5):213–9. doi: 10.1006/mpat.2001.0464. [DOI] [PubMed] [Google Scholar]
- 8.Karlsen C, Paulsen SM, Tunsjø HS, Krinner S, Sørum H, Haugen P, et al. Motility and flagellin gene expression in the fish pathogen Vibrio salmonicida: Effects of salinity and temperature. Microb Pathog. 2008;45(4):258–64. doi: 10.1016/j.micpath.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Bjelland AM, Johansen R, Brudal E, Hansen H, Winther-Larsen HC, Sørum H. Vibrio salmonicida pathogenesis analyzed by experimental challenge of Atlantic salmon (Salmo salar) Microb Pathog. 2012;52(1):77–84. doi: 10.1016/j.micpath.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Bjelland AM, Sørum H, Tegegne DA, Winther-Larsen HC, Willassen NP, Hansen H. LitR of Vibrio salmonicida is a salinity-sensitive quorum-sensing regulator of phenotypes involved in host interactions and virulence. Infect Immun. 2012;80(5):1681–9. doi: 10.1128/IAI.06038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hjerde E, Lorentzen MS, Holden MT, Seeger K, Paulsen S, Bason N, et al. The genome sequence of the fish pathogen Aliivibrio salmonicida strain LFI1238 shows extensive evidence of gene decay. BMC Genomics. 2008;9:616. doi: 10.1186/1471-2164-9-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–46. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 13.Krick A, Kehraus S, Eberl L, Riedel K, Anke H, Kaesler I, et al. A marine Mesorhizobium sp produces structurally novel long-chain N-acyl-L-homoserine lactones. Appl Environ Microbiol. 2007;73(11):3587–94. doi: 10.1128/AEM.02344-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pomini AM, Cruz PL, Gai C, Araujo WL, Marsaioli AJ. Long-chain acyl-homoserine lactones from Methylobacterium mesophilicum: synthesis and absolute configuration. J Nat Prod. 2009;72(12):2125–9. doi: 10.1021/np900043j. [DOI] [PubMed] [Google Scholar]
- 15.Decho AW, Frey RL, Ferry JL. Chemical challenges to bacterial AHL signaling in the environment. Chem Rev. 2011;111(1):86–99. doi: 10.1021/cr100311q. [DOI] [PubMed] [Google Scholar]
- 16.Churchill ME, Chen L. Structural basis of acyl-homoserine lactone-dependent signaling. Chem Rev. 2011;111(1):68–85. doi: 10.1021/cr1000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milton DL. Quorum sensing in vibrios: complexity for diversification. Int J Med Microbiol. 2006;296(2–3):61–71. doi: 10.1016/j.ijmm.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 18.Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lupp C, Urbanowski M, Greenberg EP, Ruby EG. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol Microbiol. 2003;50(1):319–31. doi: 10.1046/j.1365-2958.2003.t01-1-03585.x. [DOI] [PubMed] [Google Scholar]
- 20.Lupp C, Ruby EG. Vibrio fischeri LuxS and AinS: comparative study of two signal synthases. J Bacteriol. 2004;186(12):3873–81. doi: 10.1128/JB.186.12.3873-3881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lupp C, Ruby EG. Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. J Bacteriol. 2005;187(11):3620–9. doi: 10.1128/JB.187.11.3620-3629.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuo A, Callahan SM, Dunlap PV. Modulation of luminescence operon expression by N-octanoyl-L-homoserine lactone in ainS mutants of Vibrio fischeri. J Bacteriol. 1996;178(4):971–6. doi: 10.1128/jb.178.4.971-976.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanzelka BL, Parsek MR, Val DL, Dunlap PV, Cronan JE, Jr, Greenberg EP. Acylhomoserine lactone synthase activity of the Vibrio fischeri AinS protein. J Bacteriol. 1999;181(18):5766–70. doi: 10.1128/jb.181.18.5766-5770.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, Jeffrey PD, et al. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126(6):1095–108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fidopiastis PM, Miyamoto CM, Jobling MG, Meighen EA, Ruby EG. LitR, a new transcriptional activator in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol Microbiol. 2002;45(1):131–43. doi: 10.1046/j.1365-2958.2002.02996.x. [DOI] [PubMed] [Google Scholar]
- 26.Miyashiro T, Wollenberg MS, Cao X, Oehlert D, Ruby EG. A single qrr gene is necessary and sufficient for LuxO-mediated regulation in Vibrio fischeri. Mol Microbiol. 2010;77(6):1556–67. doi: 10.1111/j.1365-2958.2010.07309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176(2):269–75. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaefer AL, Val DL, Hanzelka BL, Cronan JE, Jr, Greenberg EP. Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc Natl Acad Sci U S A. 1996;93(18):9505–9. doi: 10.1073/pnas.93.18.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ravn L, Christensen AB, Molin S, Givskov M, Gram L. Methods for detecting acylated homoserine lactones produced by Gram-negative bacteria and their application in studies of AHL-production kinetics. J Microbiol Methods. 2001;44(3):239–51. doi: 10.1016/S0167-7012(01)00217-2. [DOI] [PubMed] [Google Scholar]
- 30.Bruhn JB, Dalsgaard I, Nielsen KF, Buchholtz C, Larsen JL, Gram L. Quorum sensing signal molecules (acylated homoserine lactones) in Gram-negative fish pathogenic bacteria. Dis Aquat Organ. 2005;65(1):43–52. doi: 10.3354/dao065043. [DOI] [PubMed] [Google Scholar]
- 31.Buchholtz C, Nielsen KF, Milton DL, Larsen JL, Gram L. Profiling of acylated homoserine lactones of Vibrio anguillarum in-vitro and in-vivo: influence of growth conditions and serotype. Syst Appl Microbiol. 2006;29(6):433–45. doi: 10.1016/j.syapm.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 32.Garcia-Aljaro C, Vargas-Cespedes GJ, Blanch AR. Detection of acylated homoserine lactones produced by Vibrio spp and related species isolated from water and aquatic organisms. J Appl Microbiol. 2012;112(2):383–9. doi: 10.1111/j.1365-2672.2011.05199.x. [DOI] [PubMed] [Google Scholar]
- 33.Yang Q, Han Y, Zhang XH. Detection of quorum sensing signal molecules in the family Vibrionaceae. J Appl Microbiol. 2011;110(6):1438–48. doi: 10.1111/j.1365-2672.2011.04998.x. [DOI] [PubMed] [Google Scholar]
- 34.Purohit AA, Johansen JA, Hansen H, Leiros HK, Kashulin A, Karlsen C, et al. Presence of acyl-homoserine lactones in 57 members of the Vibrionaceae family. J Appl Microbiol. 2013;115(3):835–47. doi: 10.1111/jam.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milton DL, Chalker VJ, Kirke D, Hardman A, Camara M, Williams P. The LuxM homologue VanM from Vibrio anguillarum directs the synthesis of N-(3-hydroxyhexanoyl)homoserine lactone and N-hexanoylhomoserine lactone. J Bacteriol. 2001;183(12):3537–47. doi: 10.1128/JB.183.12.3537-3547.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milton DL, Hardman A, Camara M, Chhabra SR, Bycroft BW, Stewart GS, et al. Quorum sensing in Vibrio anguillarum: characterization of the vanI/vanR locus and identification of the autoinducer N-(3-oxodecanoyl)-L-homoserine lactone. J Bacteriol. 1997;179(9):3004–12. doi: 10.1128/jb.179.9.3004-3012.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bassler BL, Wright M, Showalter RE, Silverman MR. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9(4):773–86. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- 38.Cao JG, Meighen EA. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J Biol Chem. 1989;264(36):21670–6. [PubMed] [Google Scholar]
- 39.Nelson EJ, Tunsjø HS, Fidopiastis PM, Sørum H, Ruby EG. A novel lux operon in the cryptically bioluminescent fish pathogen Vibrio salmonicida is associated with virulence. Appl Environ Microbiol. 2007;73(6):1825–33. doi: 10.1128/AEM.02255-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fidopiastis PM, Sørum H, Ruby EG. Cryptic luminescence in the cold-water fish pathogen Vibrio salmonicida. Arch Microbiol. 1999;171(3):205–9. doi: 10.1007/s002030050700. [DOI] [PubMed] [Google Scholar]
- 41.Hansen H, Bjelland AM, Ronessen M, Robertsen E, Willassen NP. LitR is a repressor of syp genes and has a temperature-sensitive regulatory effect on biofilm formation and colony morphology in Vibrio (Aliivibrio) salmonicida. Appl Environ Microbiol. 2014;80(17):5530–41. doi: 10.1128/AEM.01239-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in-vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotech. 1983;1(9):784–91. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 43.Milton DL, O’Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol. 1996;178(5):1310–9. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stabb EV, Ruby EG. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. In: Clark VL, Bavoil PM, editors. Methods in Enzymology. San Diego: Academic; 2002. pp. 413–26. [DOI] [PubMed] [Google Scholar]
- 45.Dunn AK, Martin MO, Stabb EV. Characterization of pES213, a small mobilizable plasmid from Vibrio fischeri. Plasmid. 2005;54(2):114–34. doi: 10.1016/j.plasmid.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 47.Colquhoun DJ, Alvheim K, Dommarsnes K, Syvertsen C, Sørum H. Relevance of incubation temperature for Vibrio salmonicida vaccine production. J Appl Microbiol. 2002;92(6):1087–96. doi: 10.1046/j.1365-2672.2002.01643.x. [DOI] [PubMed] [Google Scholar]
- 48.Natrah FM, Defoirdt T, Sorgeloos P, Bossier P. Disruption of bacterial cell-to-cell communication by marine organisms and its relevance to aquaculture. Mar Biotechnol (NY) 2011;13(2):109–26. doi: 10.1007/s10126-010-9346-3. [DOI] [PubMed] [Google Scholar]
- 49.Ortori CA, Atkinson S, Chhabra SR, Camara M, Williams P, Barrett DA. Comprehensive profiling of N-acylhomoserine lactones produced by Yersinia pseudotuberculosis using liquid chromatography coupled to hybrid quadrupole-linear ion trap mass spectrometry. Anal Bioanal Chem. 2007;387(2):497–511. doi: 10.1007/s00216-006-0710-0. [DOI] [PubMed] [Google Scholar]
- 50.Shadel GS, Baldwin TO. The Vibrio fischeri LuxR protein is capable of bidirectional stimulation of transcription and both positive and negative regulation of the luxR gene. J Bacteriol. 1991;173(2):568–74. doi: 10.1128/jb.173.2.568-574.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shadel GS, Baldwin TO. Positive autoregulation of the Vibrio fischeri luxR gene. LuxR and autoinducer activate cAMP-catabolite gene activator protein complex-independent and -dependent luxR transcription. J Biol Chem. 1992;267(11):7696–702. [PubMed] [Google Scholar]
- 52.Dunlap PV, Greenberg EP. Control of Vibrio fischeri luminescence gene expression in Escherichia coli by cyclic AMP and cyclic AMP receptor protein. J Bacteriol. 1985;164(1):45–50. doi: 10.1128/jb.164.1.45-50.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dunlap PV, Greenberg EP. Control of Vibrio fischeri lux gene transcription by a cyclic AMP receptor protein-LuxR protein regulatory circuit. J Bacteriol. 1988;170(9):4040–6. doi: 10.1128/jb.170.9.4040-4046.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams JW, Ritter AL, Stevens AM. CsrA modulates luxR transcript levels in Vibrio fischeri. FEMS Microbiol Lett. 2012;329(1):28–35. doi: 10.1111/j.1574-6968.2012.02499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


