Abstract
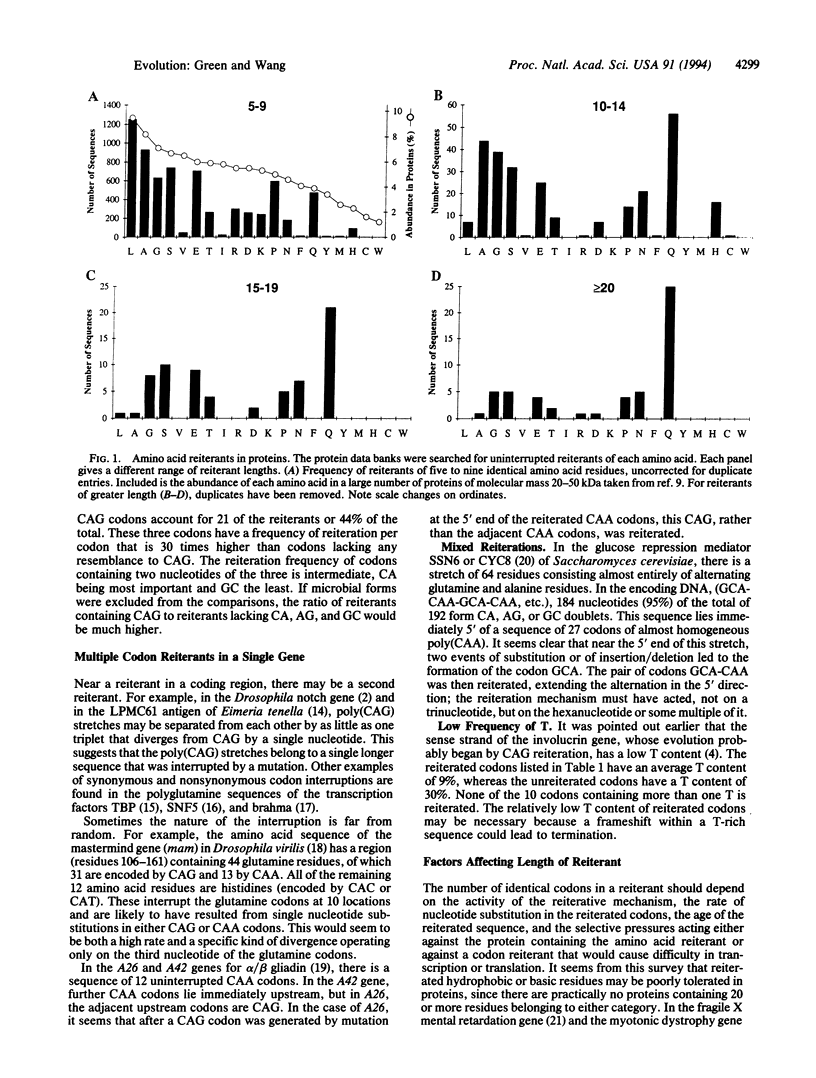
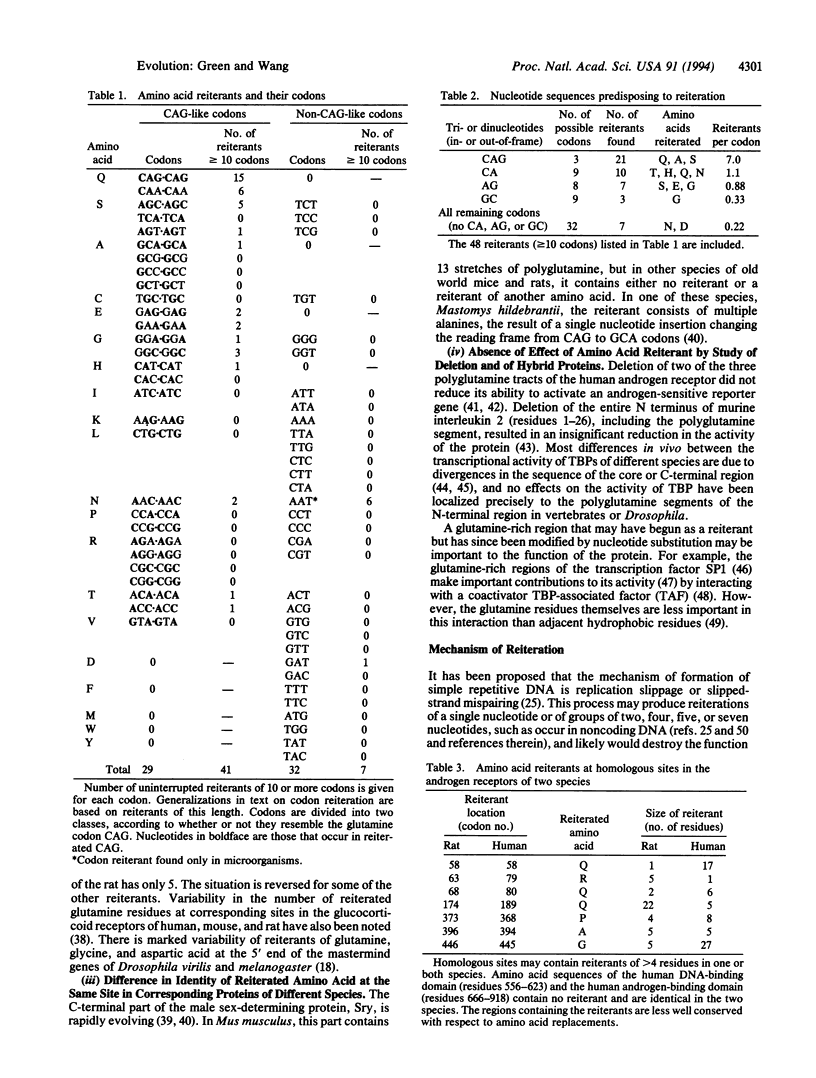
Sequence data banks have been searched for proteins possessing uninterrupted reiterations of any amino acid. Hydrophilic amino acids, and particularly glutamine, account for a large proportion of the longer reiterants. In the genes for these proteins, the most common reiterants are those that contain poly(CAG), even out-of-frame or, to a lesser degree, those that contain repeated doublets of CA, AG, or GC. The preferential generation of such reiterants requires that DNA strand-specific signals predispose to reiteration and thus to the extension of coding regions.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Brook J. D., McCurrach M. E., Harley H. G., Buckler A. J., Church D., Aburatani H., Hunter K., Stanton V. P., Thirion J. P., Hudson T. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3' end of a transcript encoding a protein kinase family member. Cell. 1992 Feb 21;68(4):799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- Caskey C. T., Pizzuti A., Fu Y. H., Fenwick R. G., Jr, Nelson D. L. Triplet repeat mutations in human disease. Science. 1992 May 8;256(5058):784–789. doi: 10.1126/science.1589758. [DOI] [PubMed] [Google Scholar]
- Cavallini B., Faus I., Matthes H., Chipoulet J. M., Winsor B., Egly J. M., Chambon P. Cloning of the gene encoding the yeast protein BTF1Y, which can substitute for the human TATA box-binding factor. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9803–9807. doi: 10.1073/pnas.86.24.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. S., Kokontis J., Liao S. T. Structural analysis of complementary DNA and amino acid sequences of human and rat androgen receptors. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7211–7215. doi: 10.1073/pnas.85.19.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack B. P., Strubin M., Ponticelli A. S., Struhl K. Functional differences between yeast and human TFIID are localized to the highly conserved region. Cell. 1991 Apr 19;65(2):341–348. doi: 10.1016/0092-8674(91)90167-w. [DOI] [PubMed] [Google Scholar]
- Courey A. J., Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988 Dec 2;55(5):887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- Djian P., Phillips M., Easley K., Huang E., Simon M., Rice R. H., Green H. The involucrin genes of the mouse and the rat: study of their shared repeats. Mol Biol Evol. 1993 Nov;10(6):1136–1149. doi: 10.1093/oxfordjournals.molbev.a040069. [DOI] [PubMed] [Google Scholar]
- Dover G. Molecular drive: a cohesive mode of species evolution. Nature. 1982 Sep 9;299(5879):111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Eckert R. L., Green H. Structure and evolution of the human involucrin gene. Cell. 1986 Aug 15;46(4):583–589. doi: 10.1016/0092-8674(86)90884-6. [DOI] [PubMed] [Google Scholar]
- Edwards A., Hammond H. A., Jin L., Caskey C. T., Chakraborty R. Genetic variation at five trimeric and tetrameric tandem repeat loci in four human population groups. Genomics. 1992 Feb;12(2):241–253. doi: 10.1016/0888-7543(92)90371-x. [DOI] [PubMed] [Google Scholar]
- Fu Y. H., Pizzuti A., Fenwick R. G., Jr, King J., Rajnarayan S., Dunne P. W., Dubel J., Nasser G. A., Ashizawa T., de Jong P. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992 Mar 6;255(5049):1256–1258. doi: 10.1126/science.1546326. [DOI] [PubMed] [Google Scholar]
- Gill G., Pascal E., Tseng Z. H., Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci U S A. 1994 Jan 4;91(1):192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H., Djian P. Consecutive actions of different gene-altering mechanisms in the evolution of involucrin. Mol Biol Evol. 1992 Nov;9(6):977–1017. doi: 10.1093/oxfordjournals.molbev.a040775. [DOI] [PubMed] [Google Scholar]
- Green H. Human genetic diseases due to codon reiteration: relationship to an evolutionary mechanism. Cell. 1993 Sep 24;74(6):955–956. doi: 10.1016/0092-8674(93)90718-6. [DOI] [PubMed] [Google Scholar]
- Hancock J. M. Evolution of sequence repetition and gene duplications in the TATA-binding protein TBP (TFIID). Nucleic Acids Res. 1993 Jun 25;21(12):2823–2830. doi: 10.1093/nar/21.12.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto S., Fujita H., Hasegawa S., Roeder R. G., Horikoshi M. Conserved structural motifs within the N-terminal domain of TFIID tau from Xenopus, mouse and human. Nucleic Acids Res. 1992 Jul 25;20(14):3788–3788. doi: 10.1093/nar/20.14.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoey T., Dynlacht B. D., Peterson M. G., Pugh B. F., Tjian R. Isolation and characterization of the Drosophila gene encoding the TATA box binding protein, TFIID. Cell. 1990 Jun 29;61(7):1179–1186. doi: 10.1016/0092-8674(90)90682-5. [DOI] [PubMed] [Google Scholar]
- Hoey T., Weinzierl R. O., Gill G., Chen J. L., Dynlacht B. D., Tjian R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993 Jan 29;72(2):247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- Hoffman A., Sinn E., Yamamoto T., Wang J., Roy A., Horikoshi M., Roeder R. G. Highly conserved core domain and unique N terminus with presumptive regulatory motifs in a human TATA factor (TFIID). Nature. 1990 Jul 26;346(6282):387–390. doi: 10.1038/346387a0. [DOI] [PubMed] [Google Scholar]
- Horikoshi M., Wang C. K., Fujii H., Cromlish J. A., Weil P. A., Roeder R. G. Cloning and structure of a yeast gene encoding a general transcription initiation factor TFIID that binds to the TATA box. Nature. 1989 Sep 28;341(6240):299–303. doi: 10.1038/341299a0. [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T., Carner K. R., Masiarz F. R., Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987 Dec 24;51(6):1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- Kao C. C., Lieberman P. M., Schmidt M. C., Zhou Q., Pei R., Berk A. J. Cloning of a transcriptionally active human TATA binding factor. Science. 1990 Jun 29;248(4963):1646–1650. doi: 10.1126/science.2194289. [DOI] [PubMed] [Google Scholar]
- Ko C., Smith C. K., 2nd, McDonell M. Identification and characterization of a target antigen of a monoclonal antibody directed against Eimeria tenella merozoites. Mol Biochem Parasitol. 1990 Jun;41(1):53–63. doi: 10.1016/0166-6851(90)90096-5. [DOI] [PubMed] [Google Scholar]
- Laughon A., Carroll S. B., Storfer F. A., Riley P. D., Scott M. P. Common properties of proteins encoded by the Antennapedia complex genes of Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1985;50:253–262. doi: 10.1101/sqb.1985.050.01.032. [DOI] [PubMed] [Google Scholar]
- Laurent B. C., Treitel M. A., Carlson M. The SNF5 protein of Saccharomyces cerevisiae is a glutamine- and proline-rich transcriptional activator that affects expression of a broad spectrum of genes. Mol Cell Biol. 1990 Nov;10(11):5616–5625. doi: 10.1128/mcb.10.11.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G., Gutman G. A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987 May;4(3):203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Lubahn D. B., Joseph D. R., Sar M., Tan J., Higgs H. N., Larson R. E., French F. S., Wilson E. M. The human androgen receptor: complementary deoxyribonucleic acid cloning, sequence analysis and gene expression in prostate. Mol Endocrinol. 1988 Dec;2(12):1265–1275. doi: 10.1210/mend-2-12-1265. [DOI] [PubMed] [Google Scholar]
- Mahadevan M., Tsilfidis C., Sabourin L., Shutler G., Amemiya C., Jansen G., Neville C., Narang M., Barceló J., O'Hoy K. Myotonic dystrophy mutation: an unstable CTG repeat in the 3' untranslated region of the gene. Science. 1992 Mar 6;255(5049):1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- Mhatre A. N., Trifiro M. A., Kaufman M., Kazemi-Esfarjani P., Figlewicz D., Rouleau G., Pinsky L. Reduced transcriptional regulatory competence of the androgen receptor in X-linked spinal and bulbar muscular atrophy. Nat Genet. 1993 Oct;5(2):184–188. doi: 10.1038/ng1093-184. [DOI] [PubMed] [Google Scholar]
- Muchardt C., Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993 Nov;12(11):4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhich M. L., Iida C. T., Horikoshi M., Roeder R. G., Parker C. S. cDNA clone encoding Drosophila transcription factor TFIID. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9148–9152. doi: 10.1073/pnas.87.23.9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newfeld S. J., Smoller D. A., Yedvobnick B. Interspecific comparison of the unusually repetitive Drosophila locus mastermind. J Mol Evol. 1991 May;32(5):415–420. doi: 10.1007/BF02101281. [DOI] [PubMed] [Google Scholar]
- Okita T. W., Cheesbrough V., Reeves C. D. Evolution and heterogeneity of the alpha-/beta-type and gamma-type gliadin DNA sequences. J Biol Chem. 1985 Jul 5;260(13):8203–8213. [PubMed] [Google Scholar]
- Poole S. J., Kauvar L. M., Drees B., Kornberg T. The engrailed locus of Drosophila: structural analysis of an embryonic transcript. Cell. 1985 Jan;40(1):37–43. doi: 10.1016/0092-8674(85)90306-x. [DOI] [PubMed] [Google Scholar]
- Poon D., Schroeder S., Wang C. K., Yamamoto T., Horikoshi M., Roeder R. G., Weil P. A. The conserved carboxy-terminal domain of Saccharomyces cerevisiae TFIID is sufficient to support normal cell growth. Mol Cell Biol. 1991 Oct;11(10):4809–4821. doi: 10.1128/mcb.11.10.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggins G. J., Lokey L. K., Chastain J. L., Leiner H. A., Sherman S. L., Wilkinson K. D., Warren S. T. Human genes containing polymorphic trinucleotide repeats. Nat Genet. 1992 Nov;2(3):186–191. doi: 10.1038/ng1192-186. [DOI] [PubMed] [Google Scholar]
- Schlötterer C., Tautz D. Slippage synthesis of simple sequence DNA. Nucleic Acids Res. 1992 Jan 25;20(2):211–215. doi: 10.1093/nar/20.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Cowe E., Higgins D. G., Shields D. C., Wolfe K. H., Wright F. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 1988 Sep 12;16(17):8207–8211. doi: 10.1093/nar/16.17.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simental J. A., Sar M., Lane M. V., French F. S., Wilson E. M. Transcriptional activation and nuclear targeting signals of the human androgen receptor. J Biol Chem. 1991 Jan 5;266(1):510–518. [PubMed] [Google Scholar]
- Simon M., Green H. The glutamine residues reactive in transglutaminase-catalyzed cross-linking of involucrin. J Biol Chem. 1988 Dec 5;263(34):18093–18098. [PubMed] [Google Scholar]
- Tamura T., Sumita K., Fujino I., Aoyama A., Horikoshi M., Hoffmann A., Roeder R. G., Muramatsu M., Mikoshiba K. Striking homology of the 'variable' N-terminal as well as the 'conserved core' domains of the mouse and human TATA-factors (TFIID). Nucleic Acids Res. 1991 Jul 25;19(14):3861–3865. doi: 10.1093/nar/19.14.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T., Matsui H., Fujita T., Takaoka C., Kashima N., Yoshimoto R., Hamuro J. Structure and expression of a cloned cDNA for human interleukin-2. Nature. 1983 Mar 24;302(5906):305–310. doi: 10.1038/302305a0. [DOI] [PubMed] [Google Scholar]
- Treier M., Pfeifle C., Tautz D. Comparison of the gap segmentation gene hunchback between Drosophila melanogaster and Drosophila virilis reveals novel modes of evolutionary change. EMBO J. 1989 May;8(5):1517–1525. doi: 10.1002/j.1460-2075.1989.tb03536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumbly R. J. Cloning and characterization of the CYC8 gene mediating glucose repression in yeast. Gene. 1988 Dec 15;73(1):97–111. doi: 10.1016/0378-1119(88)90316-2. [DOI] [PubMed] [Google Scholar]
- Tucker P. K., Lundrigan B. L. Rapid evolution of the sex determining locus in Old World mice and rats. Nature. 1993 Aug 19;364(6439):715–717. doi: 10.1038/364715a0. [DOI] [PubMed] [Google Scholar]
- Wharton K. A., Yedvobnick B., Finnerty V. G., Artavanis-Tsakonas S. opa: a novel family of transcribed repeats shared by the Notch locus and other developmentally regulated loci in D. melanogaster. Cell. 1985 Jan;40(1):55–62. doi: 10.1016/0092-8674(85)90308-3. [DOI] [PubMed] [Google Scholar]
- White S. H. Amino acid preferences of small proteins. Implications for protein stability and evolution. J Mol Biol. 1992 Oct 20;227(4):991–995. doi: 10.1016/0022-2836(92)90515-l. [DOI] [PubMed] [Google Scholar]
- Whitfield L. S., Lovell-Badge R., Goodfellow P. N. Rapid sequence evolution of the mammalian sex-determining gene SRY. Nature. 1993 Aug 19;364(6439):713–715. doi: 10.1038/364713a0. [DOI] [PubMed] [Google Scholar]
- Wong J. M., Liu F., Bateman E. Cloning and expression of the Acanthamoeba castellanii gene encoding transcription factor TFIID. Gene. 1992 Aug 1;117(1):91–97. doi: 10.1016/0378-1119(92)90494-a. [DOI] [PubMed] [Google Scholar]
- Yokota T., Arai N., Lee F., Rennick D., Mosmann T., Arai K. Use of a cDNA expression vector for isolation of mouse interleukin 2 cDNA clones: expression of T-cell growth-factor activity after transfection of monkey cells. Proc Natl Acad Sci U S A. 1985 Jan;82(1):68–72. doi: 10.1073/pnas.82.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski S. M., Zurawski G. Identification of three critical regions within mouse interleukin 2 by fine structural deletion analysis. EMBO J. 1988 Apr;7(4):1061–1069. doi: 10.1002/j.1460-2075.1988.tb02914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]



