Abstract
ClpP is a self-compartmentalized proteolytic assembly comprised of two, stacked, heptameric rings that, when associated with its cognate hexameric ATPase (ClpA or ClpX), form the ClpAP and ClpXP ATP-dependent protease, respectively. The symmetry mismatch is an absolute feature of this large energy-dependent protease and also of the proteasome, which shares a similar barrel-shaped architecture, but how it is accommodated within the complex has yet to be understood, despite recent structural investigations, due in part to the conformational lability of the N-termini. We present the structures of Escherichia coli ClpP to 1.9 Å and an inactive variant that provide some clues for how this might be achieved. In the wild type protein, the highly conserved N-terminal 20 residues can be grouped into two major structural classes. In the first, a loop formed by residues 10–15 protrudes out of the central access channel extending ∼ 12–15 Å from the surface of the oligomer resulting in the closing of the access channel observed in one ring. Similar loops are implied to be exclusively observed in human ClpP and a variant of ClpP from Streptococcus pneumoniae. In the other ring, a second class of loop is visible in the structure of wt ClpP from E. coli that forms closer to residue 16 and faces toward the interior of the molecule creating an open conformation of the access channel. In both classes, residues 18–20 provide a conserved interaction surface. In the inactive variant, a third class of N-terminal conformation is observed, which arises from a conformational change in the position of F17. We have performed a detailed functional analysis on each of the first 20 amino acid residues of ClpP. Residues that extend beyond the plane of the molecule (10–15) have a lesser effect on ATPase interaction than those lining the pore (1–7 and 16–20). Based upon our structure–function analysis, we present a model to explain the widely disparate effects of individual residues on ClpP–ATPase complex formation and also a possible functional reason for this mismatch.
Keywords: ClpP, ClpAP, ClpXP, Symmetry mismatch, 26S proteasome, Energy-dependent proteolysis
1. Introduction
Energy-dependent proteases play an essential role in regulating gene expression and eliminating damaged proteins from cells. The five prototypical energy-dependent protease families, FtsH, Lon, Clp, HslUV, and the 26S proteasome, perform similar cellular functions and share a common barrel-shaped architecture (Lupas et al., 1997). In their active form, all possess both ATPase and proteolytic activities that can be present on either the same (FtsH, Lon) or distinct (Clp, HslUV, and 26S proteasome) polypeptide chains. The ATPase components of these enzymes, such as ClpX and ClpA in the case of the Clp family, are hexameric members of the AAA+ ATPase (ATPase associated with various cellular activities) superfamily that transduce the energy of ATP binding and hydrolysis to unfold their protein substrates and present them to the proteolytic components for degradation. By contrast, the proteolytic components do not have a common fold, hydrolytic mechanism or oligomeric state. Despite these divergent properties, all family members maintain a conserved architecture in which their proteolytic active sites are compartmentalized within a central chamber in the oligomer. Structural studies suggest that access to the proteolytic chamber is controlled by narrow entrance/exit pores (∼10–20 Å in diameter) that are too small to allow entry of the native proteins without the aid of their cognate ATPases to unfold and feed them into the chamber (Lupas et al., 1997).
A consequence of this common architecture is that their mechanism of degradation is also likely to be similar, since the physical dimensions of the protease machinery dictate that substrates entering the proteolytic chamber are in a largely extended conformation, and that the products exiting are small peptides that can readily diffuse out of the chamber (Hoskins et al., 2000; Ishikawa et al., 2001; Kim et al., 2000; Ortega et al., 2000; Singh et al., 2000; Weber-Ban et al., 1999). However, one important difference between these machines is that they do not have an identical oligomeric state: the proteolytic components of FtsH, Lon, and HslUV (HslV) have 6-fold symmetry whereas the proteolytic complexes of Clp (ClpP) and the 26S proteasome (20S) have 7-fold symmetry. Since the ATPase components are all hexameric, the first set of proteolytic machines (FtsH, Lon, and HslUV) has a symmetry match whereas the second set has a symmetry mismatch between the ATPase and proteolytic components. This raises the question: how do we reconcile the observation that the oligomeric state of the proteases is not conserved with the contention that these machines perform similar functions and operate with a similar overriding mechanism?
To address this question, we have chosen to focus our attention on the Clp protease as a model system. In the Clp protease, a homo-hexameric ATPase component, ClpX or ClpA in Escherichia coli, stacks axially on one or both ends of the tetradecameric proteolytic component, ClpP. This latter complex is comprised of two apposed heptameric rings enclosing 14 serine-proteolytic active sites within a large proteolytic chamber (Maurizi et al., 1990a,b; Ortega et al., 2004; Wang et al., 1997). The ClpAP and XP proteases degrade protein substrates processively, such that only the intact protein and final peptide products are observed throughout the time course of the reaction (Thompson et al., 1994). The lack of intermediates implies that substrate unfolding and translocation by the ATPases is tightly coupled to transfer of the substrate into the proteolytic chamber where it is degraded. Currently, little is known about how the diverse activities of these symmetry mismatched components are coupled, with the exception that all ClpP-binding AAA+ ATPases contain a tripeptide signature sequence (IGF motif) which appears to be required for assembling functional Clp complexes that are capable of degrading folded substrates (Kim et al., 2001). Although there is currently no high-resolution structure for an assembled hexameric ClpP-binding ATPase, available structures of ClpX and ClpA monomers and electron microscopy (EM) generated models of the intact ATPase have allowed a biochemically consistent model to be generated. Furthermore, EM studies of functional ClpAP and ClpXP complexes show that a central pore on the ATPase aligns with axial entry/exit pores in the 7-fold symmetric ClpP to form a continuous channel through the entire length of the complex allowing protein substrates' access to the proteolytic chamber and product peptides' subsequent release (Beuron et al., 1998; Ortega et al., 2000). To maintain the integrity of this channel, it is presumed that residues at the protease:ATPase interface interact and may play a role in coupling substrate translocation from the ATPase components into the proteolytic chamber. A composite model is emerging to account for how the ATPase and protease components interact. In this model, the IGF loop of the ATPase is predicted to be on the ClpP-binding face near the periphery of the oligomer, ∼35 Å from the 6-fold axis where it could interact with deep hydrophobic grooves on the surface of ClpP, although how it accommodates the symmetry mismatch is not clear. The inferred flexibility of this motif in the ClpA and ClpX monomers may provide a mechanism for allowing multiple ATPase subunits to simultaneously contact the heptameric ClpP face (Guo et al., 2002; Kim and Kim, 2003). An appealing detail of this model is that the IGF motif lies near the proposed sites of substrate binding and ATP hydrolysis in ClpA and ClpX, and thus interaction of this motif with ClpP may play a role in the functional coupling observed in the holo-complex (Joshi et al., 2004; Kim et al., 2001). An extension of this model was recently proposed in which the N-terminal 16 residues in the protease form an axial loop that extends all seven termini in the heptameric ring to the ATPase surface. However, there has been considerable difficulty in visualizing portions of the N-terminus that have been assumed to sit outside of the plane of the pore since they have been uninterpretable in the electron density and hence unmodeled in the deposited structures. More concerning is the fact that this model fails to convincingly account for how the 6–7 symmetry mismatch is accommodated, making only a passing remark that only 6 of the 7 loops would be in contact with the ATPase, but all would have the axial loop conformation. How this loop could be accommodated without contacting the ATPase was not clear. Independent of this work, we have identified biochemically residues within the first 20 amino acids that control the functional binding states of ClpP. Furthermore, we present two structures, one that reflects the functional ATP-binding state and a second which is a biochemically characterized mutant that reflects a non-functional binding state. In addition, we propose a model for ClpAP/ClpXP interaction that not only accounts for the symmetry mismatch but additionally provides a possible reason for a biological function of the mismatch.
2. Materials and methods
2.1. Alanine scanning mutagenesis and protein expression
The expression plasmid, pET9-ClpP, containing the full-length clpP gene (Maurizi et al., 1990a,b) (N-terminal 14-residue leader + residues 1–193 in the mature ClpP sequence; alanine-1 denotes the first residue in the mature sequence, while 14-Methionine is the last residue in the leader peptide) was used to construct a panel of single site ClpP variants where the first 20 residues of the mature sequence were replaced, in turn, with alanine (residues 2–20), or in the case of residue A1, phenylalanine. Substitutions in codons 1–11, 13–16, and 18–20 were constructed using a cassette mutagenesis protocol. The substitutions R12A, F17A, and F112A were obtained by the previously described PCR-based megaprimer method. In all cases, the entire ClpP coding region, including the leader sequence, was sequenced to confirm the presence of the desired mutation alone. For protein expression, pET9a-wtClpP and each of the substituted ClpP constructs were transformed into the E. coli expression host BL21(DE3)1146D, which lacks the chromosomal copy of clpP. In tryptone-containing media, wt ClpP is constitutively expressed in this construct due to an unknown inducer present in all of the batches of tryptone that we examined. With some batches, the level of induction is so high that a significant fraction of the expressed ClpP is unprocessed and found in inclusion bodies. For experiments designed to compare the effects of substitutions on leader peptide processing, CircleGrow media (BIO101) supplemented with 0.2% glucose was used. Under these conditions, 80–90% of wt ClpP is processed and accumulates to ∼100 mg/L of culture during an overnight growth. Similar levels of expression were observed with the entire set of single site ClpP variants, with the exception of ClpP-P4A that expressed at lower levels.
2.2. Purification of wt ClpP and ClpP variants
Wild type ClpP and all of the variants were purified by a streamlined protocol, as described previously (Wang et al., 1997). In all instances, the final purified protein samples were >90% pure as judged by SDS–PAGE analysis and were tetradecameric as determined by size exclusion chromatography. The concentrations of ClpA, ClpX, and ClpP are given with respect to the assembled hexamers and tetradecamers, respectively.
2.3. Crystallography
For the wt and V6A variant ClpP, crystals were grown in sitting drops by the vapor diffusion technique from a reservoir contained 55% MPD, in 100mM MES at pH 6.2. Drops containing 5 μL of protein at a concentration of 16 mg/mL in 200 mM NaCl and 10mM MES at pH 6.5 and 5 μL of reservoir solution. Crystals were cooled directly into a stream of liquid nitrogen gas at 99 K using an Oxford cryostream. Diffraction data to 1.85 Å (wt) and 2.6 Å (V6A) resolution, respectively, were each collected from a single crystal on station X25, NSLS, Upton, NY, using a Brandeis 4 cell CCD detector. The crystals have space group C2 (wt) andP21 (V6A), respectively, with the unit cell dimensions shown in Table 1. Data were processed using the HKL package (Otwinowski and Minor, 1997). The Rfree set was chosen at random within resolution shells due to the 14-fold non-crystallographic symmetry in the crystals (Dodson, 1996).
Table 1. X-ray crystallographic data collection and refinement statistics.
| Native | V6A | |
|---|---|---|
| Data collection | ||
| Unit cell | a = 192.5 Å, b = 103.3 Å, c = 159.0 Å, β = 98.4° | a = 94.8 Å, b = 161.0 Å, c = 186.6 Å, β = 90.3° |
| Space group | C2 | P21 |
| Resolution (Å) | 30–1.85 | 35.0–2.6 |
| Unique observations | 247 428 | 152 303 |
| Redundancy | 5.5 (2.1) | 3.9(2.4) |
| Rmerge (%) | 8.8 (23.3) | 5.8 (29.0) |
| Completeness (%) | 95.9 (76.4) | 88.7 (81.9) |
| I/σI | 30 (2) | 14 (3) |
| Overall B Wilson plot Å3 | 27.8 | 51.4 |
| Refinement statistics | ||
| Resolution limits (Å) | 30.0–1.9 | 30.0–2.6 |
| No. of reflections I/σI > 0 in work set | 233 080 | 147 121 |
| No. of reflections in free set | 1245 | 1591 |
| No. of protein atoms | 20 735 | 38 820 |
| No. of water molecules | 1791 | 0 |
| No. of MPD atoms | 112 | 0 |
| R-factor (%) | 22.1 | 22.6 |
| Rfree (%) | 25.1 | 26.7 |
| Geometric parameters (rms) | ||
| Bond length (Å) | 0.011 | 0.012 |
| Bond angle (°) | 1.8 | 1.9 |
Residues 20–193 from 1TYS.pdb were used as a template and subject to rigid body refinement followed by a simulated annealing protocol applying non-crystallographic symmetry (NCS) restraints. In the later steps of the refinement of the wt structure, NCS restraints were relaxed, as determined appropriate by monitoring the behavior of the Rfree with a set of refinement protocols. Refinement, punctuated by rounds of model building, was performed until the structure could not be improved as judged by a reduction in Rfree. We refer to this as a structure rather than the more typical usage of model so that it is not confused with the modeling studies that are performed later. The current structures of wt and V6A have R-factors of 22.1% (Rfree 25.1% for 1245 reflections) and 22.0% (Rfree 26.7% for 1591 reflections), respectively, with good geometry. The wild type structure contains the following residues: chain A, B, D, E, F, and G, residues 1–193; chain C, residues 8–193; chain H–N, residues 12–193. The structure of the V6A variant contains residues 15–193 for all 28 chains. Coordinates have been deposited in the PDB with accession codes 1YG6 and 1YG8.
2.4. Determination of subunit mass and amino-terminal residues for ClpP variants
Mass spectrometric analyses for each of the proteins used in this study were carried out on an Applied Biosystems Voyager-DE Biospectrometry Workstation using a 25 kV accelerating voltage. The mass spectra were acquired by adding the individual spectra from 150 laser shots, in the linear mode. The protein solutions ∼10 μM were diluted (1:1) by spotting 1 μL onto the sinapinic acid matrix (10mg/ mL in 50% CH3CN in 0.05% TFA), followed by the application of another 1 μL aliquot of the matrix. The samples were allowed to air-dry on the sample target before analysis. Horse heart myoglobin (16953 Da, Sigma) and wild type ClpP were used as external standards. Samples whose mass from MALDI agreed within 5–10 Da of the expected mass were not submitted for further mass analysis. However, samples of ClpP-P4A, -V6A, -I7A, -D18A, I19A, and Y20A only showed significant peaks of smaller than expected masses. ClpP-P4A, -V6A, and -I7A were submitted for amino-terminal analysis to confirm the identity of the mature amino-terminus. For P4A and V6A variants, the amino-terminal residues were M5 and I7, respectively, while the amino-termini of I7A were a mixture of A1 and E8 consistent with the masses of the predominant peaks in the MALDI-TOF spectra. ClpP-D18A, -I19A, and -Y20A showed a spectrum of smaller peaks, each of which was present in quantities too small to be characterized by N-terminal sequencing.
2.5. Assay of ClpP peptidase activity
Peptidase activity was measured in a continuous spectrophotometric-based assay that exploits the difference between the absorbance spectrum of AMC in the peptide substrate N-succinyl-Leu-Tyr-AMC (Suc-LY-AMC) and free in solution. The substrate, Suc-LY-AMC (Sigma, S-1153), was dissolved in 50mM Tris–HCl, pH 8.0, 100 mM KCl, 1 mM DTT, and 17.6% DMSO to a final concentration of ∼15mM, its solubility limit. The actual concentration of the substrate stock solution was calculated assuming a molar extinction coefficient of 16,000 at 324 nm. The peptidase assays were conducted in 1-mm path-length cuvettes maintained at 37 °C. Each reaction mixture (200 μL) contained 50 mM Tris–HCl, pH 8.0, 100mM KCl, 1 mM DTT, and 9.5% DMSO (final concentration), and varying amounts of substrate ranging from 0.25 to 5 mM. The substrate-containing solutions were pre-incubated at 37 °C for 5 min and the reaction was initiated by addition of wt ClpP, or one of the ClpP variants, to a final concentration of 0.72 μM (ClpP14). The solutions were then quickly transferred to a pre-warmed quartz cuvette and the change in absorbance at 360 nm was recorded for 3 min. The initial rate of peptide hydrolysis was determined from the slope of the curve and converted to the μM of AMC released per second using an AMC standard curve. The parameters Km and Vmax were calculated from the dependence of the initial rate on substrate concentration by fitting the data to the Michaelis–Menten equation using the non-linear regression analysis implemented in the SigmaPlot data analysis package (ISS).
2.6. Assay of ClpAP complex formation by size exclusion chromatography
The extent of complex formation between hexameric ClpA and tetradecameric ClpP was examined by size exclusion chromatography. Purified ClpA or ClpP was chromatographed on a Superose 12 column (Amersham Pharmacia) in the presence of 0.5 mM ATPγS and the absorbance of the eluent at 229 nm was monitored. Prior to chromatography, the ClpA (3 μM) and ClpP (3 μM) were incubated with 1 mM ATPγS in assembly buffer (50 mM Hepes–NaOH, 0.3 M NaCl, 20 mM MgCl2, and 10% glycerol) for 20 min. Aliquots of these reactions were then analyzed by SEC in reaction buffer containing 0.5 mM ATPγS.
2.7. Assay of ClpXP and ClpAP complex formation by ultrafiltration
Briefly, reactions containing ClpA alone, ClpP alone, or mixtures of ClpA and ClpP (wild type and variants) were added to the retentate cup of a UF300 ultrafiltration device (Milipore) and weighed. The device was then assembled and centrifuged at 4 °C for 2 min as described by the manufacturer. At the end of the run, the retentate cup was again weighed and its weight adjusted by addition of buffer to that of the sample before centrifugation. Any protein that was weakly adsorbed to the membrane was resuspended by repeated pipetting of the sample in the retentate cup. Equivalent aliquots from the total assembly mixture taken before centrifugation, the filtrate, and adjusted retentate were then assayed by SDS–PAGE analysis followed by Coomassie staining to visualize the proteins.
2.8. Assay ATP-dependent proteolytic activity
The ATP-dependent hydrolysis of α-casein by the ClpAP protease was followed in a continuous assay that monitored the change in fluorescence anisotropy of BODIPY-labeled α-casein substrate during degradation. The reaction mixtures contained 80 mM Tris–HCl at pH 8.0, 10mM MgCl2, 1mM DTT, 1mM EDTA, 21 mM KCl, 4mM ATP, 1 μg/mL BODIPY-labeled α-casein, and 100 μg/mL unlabeled α-casein, and varying concentrations of wt ClpP or ClpP variant. All the samples were pre-incubated at 37 °C in an Eppendorf tube and the reaction started by adding 10 nM ClpA to the tube. The mixture was quickly transferred to a 0.5 cm path-length quartz cuvette that had been pre-incubated at 37 ° C. Anisotropy measurements were taken every 10 s in an ISS PC1 spectrofluorometer. The instrument was configured with 475 and 535 nm excitation and emission filters, respectively, with a 2 s integration time. The rate of the reaction was calculated from the slope of the linear region of the curve, typically from 100 to 500 s. ATP-dependent degradation of ssrA-GFP by the ClpXP protease was determined as previously described (Weber-Ban et al., 1999).
2.9. Assay for ClpP-dependent inhibition of ClpX ATPase activity
ATP hydrolysis by ClpX was measured in a coupled reaction as described previously (Kim et al., 2001). Briefly, 50 nM ClpX6 alone or in the presence of 3 μM wild type ClpP or a ClpP variant was incubated in 25 mM Hepes–KOH, pH 7.6, 5mM KCl, 5mM MgCl2, 0.03% NP-40, 10% glycerol, 2.5 mM ATP, 1 mM NADH, 7.5 mM phosphoenolpyruvate, 0.05mg/mL pyruvate kinase, and 0.025 mg/mL lactate dehydrogenase. After a 3 min pre-incubation at 25 °C, ATP hydrolysis was monitored by following the decrease in absorbance by NADH at 340 nm. Hydrolysis was calculated using the extinction coefficient of 6230 μM−1 cm−1 for NADH.
3. Results and discussion
3.1. Two general conformations exist for the N-termini in the wild type E. coli ClpP oligomer
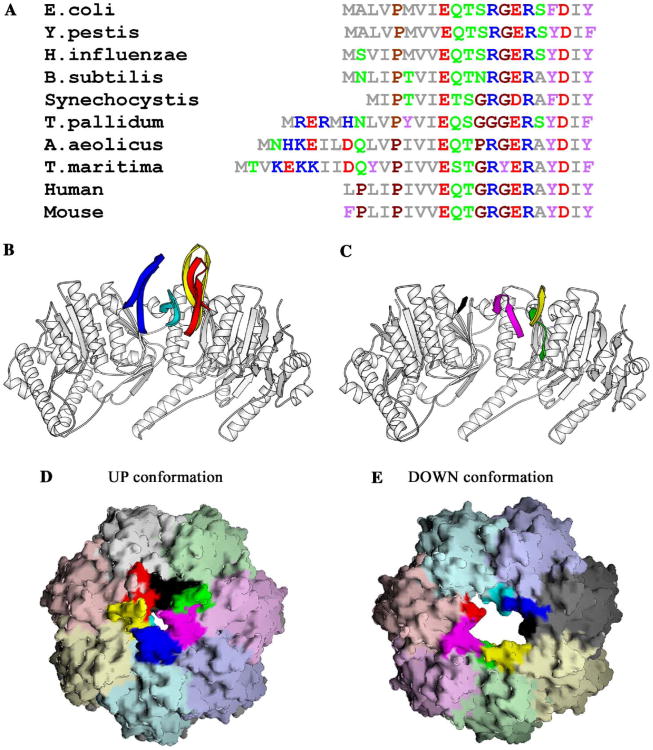
The amino acid sequence of the amino-terminal 20 residues of the mature ClpP is highly conserved across species (Fig. 1 A), yet they exhibit conformational flexibility in their crystal structures. We have determined the structure of ClpP at 1.9 Å resolution which has revealed the existence of two general conformational states for these amino-terminal residues, one populated by 6 monomers and the other by 8 monomers. The protein crystallized with a tetradecamer in the asymmetric unit; in all 14 copies, the basic fold of the N-terminus is characterized by a two-stranded β-sheet connected by a turn; however, the register of the strands differ, which has the effect of altering the residues forming the turn. In all cases, though interactions with residues 18–20 are conserved independent of the conformation of the remainder of the N-terminus, closer inspection reveals that the conformations of the amino-termini can be classed into two major groups. In the first, a portion of the N-terminus extends out of the access pore (∼ 10–15 Å), in a manner consistent with structures available in the PDB. This conformation will be referred as ‘up’ and is present in six of the subunits in ring-1 (molecules A, B, and D–G) (Fig. 1B). In the second conformational class, which will be referred to as ‘down,’ the N-terminal residues are contained entirely within the access pore since the turn occurs further from the N-terminus. This conformation is found in all of the subunits in ring-2, and subunit C in ring-1 (Fig. 1C) and has not been observed previously. The difference between the conformations of the amino-termini in the two rings is probably due to subtle differences in the crystal-packing environment of each ring. This is further suggested by inspecting the packing of molecules in the structures of ClpP from humans and Streptococcus pneumoniae, which each contain a heptamer in the asymmetric unit, constraining each ring of the tetradecamer to be identical. By contrast, in the structure of wild type ClpP presented here, there is a tetradecamer in the asymmetric unit, creating the possibility that the two rings could be different. In general we found that residues 21–193 were very similar when all 14 monomers were compared, however, we observed significant differences in the N-termini of the two rings. In this crystal, the majority of crystal contacts involve residues at the side surfaces of the ClpP cylinder, however, a few contacts are observed with the end surfaces of the oligomer. Specifically, in ring-1, residues 8–11 of chain B are involved in crystal contacts with residues on the side surfaces of an adjacent oligomer (chain K: E56, K57, D58, K84, D86, and K110). Conversely, in ring-2, residues 8–11 in all seven monomers are disordered and not involved in crystal contacts and in particular, the side surfaces in ring-1 equivalent to chain K are not involved in crystal contacts. Not surprisingly, the amino-terminus of chain B is the best ordered of the seven N-termini in ring-1. The conversion of the last N-termini in ring-1 to the up conformation may be prevented by steric considerations imposed by the size of the access pore giving entrance pore in this ring a local “pseudo”-6-fold symmetry (Figs. 1D and E).
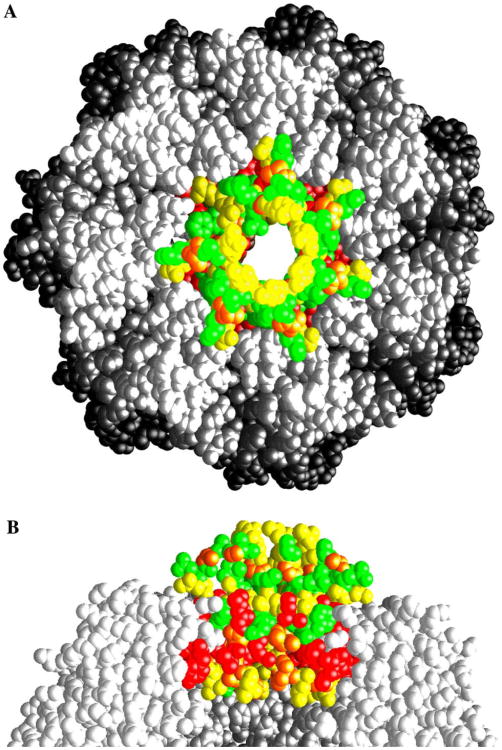
Fig. 1.
The amino-terminal residues of ClpP are conserved across species, yet adopt distinct conformations in each monomer. (A) Amino acid sequence alignment of the N-terminal residues of mature ClpP sequences from a representative set of species: E. coli, Yersinia pestis, Haemophilus influenzae, Bacillus subtilis, Synechocystis, Treponema pallidum, Aquifex aeolicus, Thermotoga maritima, human, and mouse. The amino acids are colored as follows: M, A, L, I, and V: gray; P and G: brown; E and D: red; H, R, and K: blue; Q, N, T, and S: green; F, W, and Y: purple. (B) The amino-terminal residues of ring-1 extend out of the pore. Monomers A–D are shown as ribbon drawings: residues 21–193 are colored white. Residues upto 20, when observed in the electron density, are colored uniquely for each chain: red, yellow, cyan, and blue, respectively. (C) The ordered amino-terminal residues of ring-2 do not extend out of the pore. Monomers H, I, J, and N are shown as ribbon drawings: residues 21–193 are colored white. Residues upto 20, where observed in the electron density, in chains H, I, J, and N are colored green, yellow, magenta, and black, respectively. (D) Molecular surface of the X-ray structure of ring-1. Each monomer is shown in a distinct color, with observed residues upto residue 20 in a deeper hue. (E) Molecular surface of the X-ray structure of ring-2 colored in the same manner as (D). Panels (B) and (C) were drawn using the program Molscript (Kraulis, 1991), and panels (D) and (E) were drawn using the program GRASP (Nicholls et al., 1991).
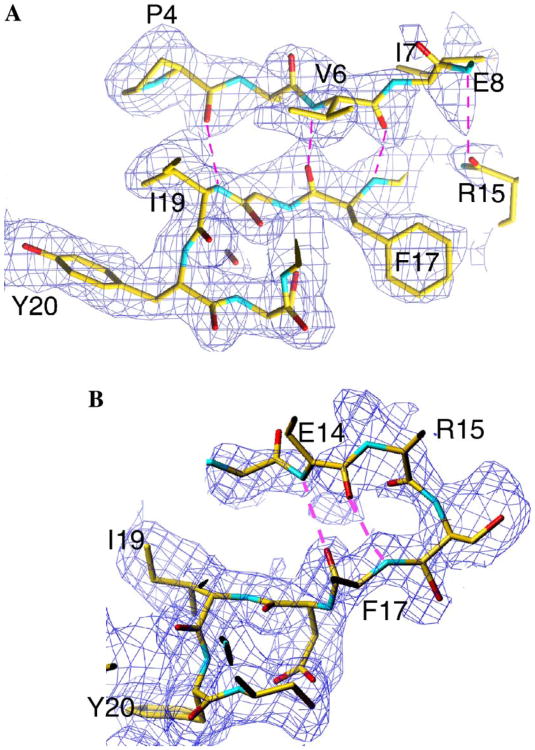
For all six of the monomers in the ‘up’ conformation, clear density is observed for both the backbone and side chain atoms of residues 1–8 and 16–19, which comprise the N-terminal two β-strands. Weaker density is observed in most cases for residues within the turn. The network of hydrogen bonds involving the carbonyl oxygen atoms of P4, V6, F17, and R15 and the amide nitrogen atoms of I19, F17, V6, and E8, respectively, are consistent with the presence of a two-stranded antiparallel β-sheet (Fig. 2A). In this arrangement, the carbonyl atoms of I7, R12, and E14 and the amide nitrogen atoms of S11 and S16 are solvent accessible and present unsatisfied hydrogen-bonding potential towards the center of the access pores where they could form hydrogen bonds with incoming unfolded protein substrates. The amino-terminal β-sheets in this conformation are only loosely tethered to the heptameric ring by a hydrogen bond between S21Oγ (A:G, B:D, D:E, E:F, and F:G) and the amide nitrogen of M5 in an adjacent subunit along the ring. In addition, atoms within residues P4, V6, E18, I19, and Y20 pack against those of the conserved residues of R22, N41, L42, and F49 from the adjacent monomer. The loop regions of the six N-termini in the ‘up’ conformation in ring-1 extend out of the central access pore and over the substrate access pore to close it (Fig. 1D). This has the effect of giving one end of the ClpP oligomer a more pronounced dome-like architecture that is roughly complimentary to the concave ClpP-binding faces that have been inferred for the ClpA and ClpX oligomers (Guo et al., 2002; Kim and Kim, 2003). As a result of the conformation of subunit B, which is involved in crystal contacts, the pore in the ‘up’ conformation is closed (it should be noted that if the N-terminal 20 residues of subunit B are replaced with the analogous region from any of the other five subunits in the up conformation, a ∼12Å wide pore is generated, the same as in ring-2) is strikingly similar to the conformation of the N-termini in the outer α-subunits of the yeast 20S proteasome that close the pore in this oligomer (Groll et al., 1997).
Fig. 2.
Section of |Fo| – |Fc| electron density simulated annealing omit map corresponding to (A) one loop in the up conformation and (B) one loop in the down conformation. The electron density is shown as a blue chicken wire and contoured at a sigma level of 0.7. The residues are shown in ball-and-stick representation, colored according to their atom type: carbon, yellow; oxygen, red; nitrogen, blue; sulfur, green. Hydrogen bonds are shown as a broken magenta line. The figure was produced from a snapshot image taken directly from the screen, using the program TURBO to display the electron density map. Labeling was performed in Adobe Photoshop. Electron density corresponding to F17 is visible in both conformations, however, in (B), the side chain electron density lies outside of the plane of the section.
In the ‘down’ conformation, residues 12–20 are generally interpretable and the turn between the two β-strands involves residues 13–17 (Fig. 2B). In ring-2, the packing of the N-termini produces an open access pore, with an ∼ 12 Å diasmeter (Fig. 1E) and is reminiscent of the open access pores in the 20S proteasome from the archaebacterium Thermoplasma acidophilum (Lowe et al., 1995), and HslV (Bochtler et al., 1997), the proteolytic component of the HslUV ATP-dependent protease (Bochtler et al., 2000; Rohrwild et al., 1996). This pore size can accommodate the passage of unfolded polypeptides in an extended conformation. In the down conformation, the electron density for residues 1–11 is not readily interpretable; however, diffuse density is observed leading from residue 12 down the access pore and into the proteolytic chamber suggesting a path for these residues. In this conformation, the visible residues of the N-termini make few contacts with the remainder of the molecule; however, in general, I19 and Y20 pack against R22, N41, and L42. Further, a rotation around the Cβ–Cγ bond of N41 results in the creation of a hydrogen bond between Nδ1 of N41 and OH of Y20 being made in the subunits in ring-2 that is generally not present in the up conformation. In addition, a hydrogen bond between S21Oγ in one subunit with the side chain of R12 from the next subunit along in the ring is also observed. Conversion between the ‘up’ and ‘down’ conformations can thus be thought of as a translocation of residues 1–11 into the access pore, with S21 acting as the lynchpin that stabilizes each conformation. Translocation of the N-terminus into the pore may occur in much the same way that we envision the entry of an unfolded substrate, for example comparing the position of R12 in ring-1 and ring-2, a displacement of ∼10 Å is observed. Interestingly, a conformational change between the closed and open states of pores in the yeast 20S proteasome is required for function (Kohler et al., 2001a,b), thus the two conformations of the N-termini in ClpP may also be mechanistically important. The structures of ClpP available in the PDB have utilized the structure of E. coli ClpP as a template: human ClpP (1TG6) was determined using a polyalanine model derived from E. coli ClpP and S. pneumoniae used a combination of SAD phasing and molecular replacement (1Y7O), again using E. coli ClpP as a template without removing the mismodeled regions. However, the authors of the E. coli ClpP structure determined to 2.3 Å resolution were concerned about the correctness of the N-terminal 3–4 residues due to the method utilized to phase the molecule (Wang et al., 1997). With this in mind, we collected a dataset from crystals of selomethionine-derivatized E. coli ClpP at the peak selenium wavelength. Due to the large number of selenium atoms in this molecule (140), we calculated an anomalous difference Fourier map using the coordinates of residues 20–193 to obtain phases while attempting to locate the selenomethionine atoms independently. E. coli ClpP has a methionine residue at position 5 in the N-terminus allowing unambiguous positioning of this residue in the anomalous difference Fourier map, if ordered. In the region of the N-terminus, we observed only six Se anomalous difference peaks, the positions of which superimposed onto the sulphur atoms of Met5 in the up conformation in the structure of the wild type enzyme and no peaks in the ring that contains N-termini in the down conformation, consistent with our observation that the N-termini in the down conformation are not visible before residue 12. This provided direct evidence that the amino-termini in E. coli ClpP do not adopt identical conformations in the molecule, as previously suggested (Gribun et al., 2005; Kang et al., 2004; Wang et al., 1997).
Is it possible that the conformation observed in ring-1 of E. coli ClpP, in which six of the N-termini are in the up conformation and the seventh is in the down conformation, might be a general feature of ClpP? If so, how can we reconcile the differences in the N-termini observed in the three ClpP structures? In order to address this, we looked in detail at the conformations and temperature factors of the amino-termini for the structures of human (PDB code 1TG6) and S. pneumoniae ClpP (PDB code 1Y7O). We noticed that the temperature factors in these two structures were unusual. In the structure of S. pneumoniae ClpP, the temperature factors for residues 1–6 in the N-termini of chain F are relatively higher (approximately twice the value of the remaining chains) suggesting that this loop is more mobile. By contrast, the average temperature factor for the remainder of chain F is similar to other chains, alternatively suggesting that the relatively higher temperature factors could also indicate that residues 1–6 of chain F might be otherwise modeled. Since the structure was determined to 2.5 Å resolution with reduced data completeness (79% after the omission of more than 4600 reflections for the free set), the required NCS restraints would tend to weight the model towards a more 7-fold symmetric molecule. Since the structure factors have been deposited, we calculated a series of electron density maps including a 2fo – fc map and a simulated annealing omit map although we were unable to calculate any anomalous maps as the required data was not available in PDB. The calculated maps did not contain clear density for some regions, however, the density for at least one N-termini suggested that a shorter loop could also be modeled. It is not within the scope of this paper, however, to rerefine a structure, nor would the authors feel comfortable doing so. This region will presumably become clearer when higher-resolution data become available. For human ClpP, again, the refined temperature factors are somewhat odd, for example, the temperature factor for residue 17 is unphysical (B = ∼107& Å3 compared to residues 18–20 which vary from ∼39 to ∼43& Å3). More worryingly for residues 1–7, the temperature factor of an individual atom has the same value in each monomer, suggesting that the temperature factors had been NCS-constrained, which would be unreasonable since their conformations are different (for example, the nitrogen atom of Pro1 is 57.68 Å3 in each monomer). By contrast, in the remainder of the chain, identical atoms have different temperature factors in each monomer, as expected when individual temperature factors are refined. These observations may indicate that only six of the amino-termini are in the up conformation in all of the structures solved to date.
3.2. Residues at positions 4, 6–8, and 18–20 in the mature N-terminus affect the proper maturation of ClpPγS
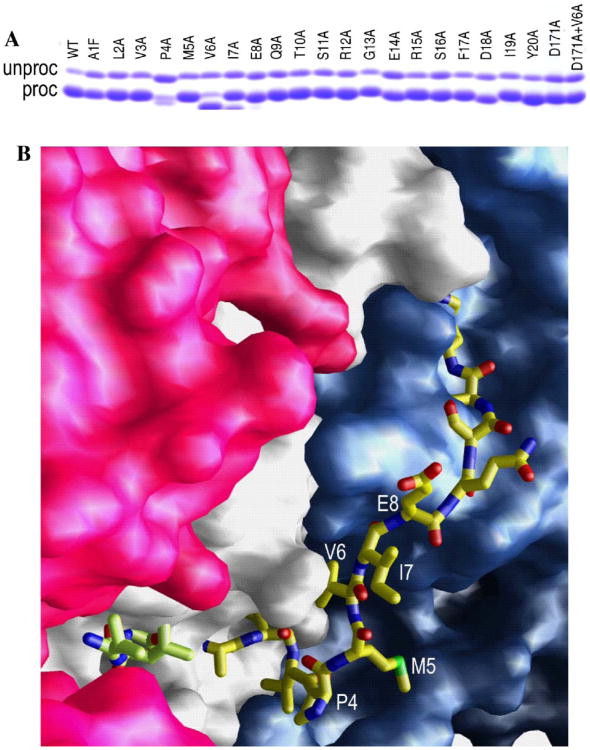
Based on the structure that is described here, particularly noting the conservation of hydrogen bonds to the backbone of residues 18–20, we reasoned that residues 1–20 would be important in the proper functioning of ClpP. In order to fully investigate the roles of each of these residues, we created a panel of single amino acid variants of ClpP that span residues 1–20, using scanning alanine mutagenesis (Table 2). Wild type protein and each of the ClpP variants in this panel, with the exception of ClpP-P4A, are expressed at high levels and efficiently processed (routinely 90% of the expressed ClpP is processed under our standard test induction conditions, Fig. 3A). The unprocessed material is found exclusively in the insoluble fraction and thus only processed subunits are found in purified preparations of these proteins (data not shown). By contrast, the ClpP-P4A variant showed different behavior: both the expression and processing were markedly reduced relative to wt, and both full-length and processed ClpP-P4A were found in the soluble fraction. For this variant, propeptide processing continued slowly throughout purification, suggesting that the rate of processing is altered. A possible explanation for this can be visualized by modeling residues 1–12 of the amino-terminus in the down conformation by eye along the internal groove that lies close to the subunit interface (Fig. 3B). Modeling the path of residues 1–12 in this way suggests that the limited conformational space that proline can adopt might constrain the path of the leader peptide and the first four residues of the mature amino-terminus, helping to orient the propeptide appropriately in the active site cleft. Further support for this is given by Gribun et al., who reported that substituting P4 in E. coli ClpP with a glycine residue resulted in protein that was uncleaved (Gribun et al., 2005). Given the wider conformational flexibility of glycine, it is likely that the amino-terminus never adopt the appropriate conformation.
Table 2.
| Variant | Mature N-terminus | Peptidase Suc-LY-AMC | Protease α-casein | ClpAP complex formation | |||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| Km (mM)a | Kcat (s−1)a | Relative kcat/Kma | Kd (nM)b | Vmax (ng/s) | Relative Kapp | SECc | ClpP retainedd | ||
| WT | A1 | 4.4 | 2.4 | 1.0 | 0.5 | 76 | 1.0 | + | ++++ |
| A1F | F1 | 5.1 | 2.5 | 0.9 | 0.3 | 62 | 1.0 | + | ++++ |
| L2A | A1 | 3.9 | 2.7 | 1.3 | 211 | 50 | 422 | nd | +++ |
| V3A | A1 | 4.3 | 2.9 | 1.2 | 2606 | 49 | 5212 | nd | − |
| P4A | M5e | 5.0 | 1.9 | 0.7 | na | 0 | >100000 | − | − |
| M5A | A1 | 3.6 | 2.4 | 1.2 | 4303 | 42 | 8606 | nd | − |
| V6A | I7 | 5.8 | 2.9 | 0.9 | na | 0 | >100000 | − | − |
| V6S | A1 | 4.6 | 2.7 | 1.1 | na | 0 | >100000 | nd | nd |
| I7A | A1 | 5.0 | 2.5 | 0.9 | na | 0 | >100000 | nd | − |
| I7S | A1 | 4.8 | 2.5 | 0.9 | na | 0 | >100000 | nd | nd |
| E8A | A1/Q9f | 5.0 | 2.5 | 0.9 | 751 | 31 | 1502 | nd | ++ |
| Q9A | A1 | 4.4 | 2.1 | 0.9 | 3 | 37 | 6 | nd | ++++ |
| T10A | A1 | 4.6 | 2.7 | 1.1 | 3784 | 34 | 7568 | nd | − |
| S11A | A1 | 4.4 | 2.2 | 1.0 | 0.5 | 48 | 1 | nd | ++++ |
| R12A | A1 | 4.1 | 2.2 | 1.0 | 637 | 28 | 1374 | nd | ++ |
| G13A | A1 | 3.4 | 2.5 | 1.3 | 4485 | 40 | 8960 | nd | − |
| E14A | A1 | 4.1 | 2.5 | 1.1 | 9 | 51 | 18 | nd | ++++ |
| R15A | A1 | 4.1 | 2.4 | 0.9 | 493 | 27 | 986 | nd | +++ |
| S16A | A1 | 4.7 | 2.2 | 0.9 | 2 | 52 | 4 | nd | ++++ |
| F17A | A1 | 4.5 | 2.7 | 1.1 | 24 | 32 | 48 | nd | ++++ |
| D18A | A1g | 3.8 | 2.7 | 1.3 | na | 0 | >100000 | − | − |
| I19A | A1g | 4.4 | 2.6 | 1.1 | na | 0 | >100000 | nd | − |
| Y20A | A1g | 4.4 | 2.1 | 0.9 | na | 0 | >100000 | nd | − |
| F112A | A1 | 4.2 | 2.2 | 0.9 | 2300 | 65 | 4600 | nd | + |
| D171A | A1 | — | — | — | 10 | — | 20 | + | ++++ |
The symbol — in this column denotes inactive in hydrolyzing Suc-LY-AMC, a fluorogenic peptide substrate of ClpP.
na in this column denotes not active meaning no degradation of α-casein was observed with 10 μM ClpP mutant.
Presence of ClpP co-eluting with ClpA in size exclusion chromatography determined by SDS–PAGE analysis of column fractions. + denotes complex formation, − denotes no complex formation, and nd denotes not determined.
The amount of ClpP14 retained by the 300kDa cutoff membrane in the presence of 700nM ClpA6 and 1 mM ATPγS is denoted as – (≤5%), + (5–25%), ++ (26–50%), +++ (51–75%), or ++++ (76–100%). In the absence of ClpA ≤5% of WT ClpP14 is retained, while >95% is retained in the presence of 1 mM ATPγS and ClpA.
Additional degradation occurs during purification to give this single shorter species.
Mixed processing yielding a mixture of subunit having A1 and Q9 at their N-termini.
Approximately, 10–15% heterogeneous population of smaller products corresponding to N-termini I6, V7, E8, and Q9.
Fig. 3.
The effects of residues in the mature amino-terminus on propeptide processing. (A) SDS gel of wt protein, each of the amino terminal mutants, the D171 active site mutant and the double mutant V6A + D171A showing the degree of propeptide processing. (B) Surface representation residues 12–193 of chains L and M and residues 13–193 of chain M in a ClpP heptamer, as observed in the electron density (colored red, gray, and purple, respectively). Residues 1–12 of chain M have been modeled by eye along the internal groove that runs approximately parallel to the subunit interface. It is drawn as a ball-and-stick representation and colored according to atom type: blue, nitrogen; red, oxygen; yellow, carbon; green, sulfur. The position of the competitive inhibitor, acetyl-LLM, is also shown in ball-and stick representation.
During the course of these experiments, we observed that seven of our variants (P4A, V6A, I7A, E8A, D18A, I19A, and Y20A) were, to varying degrees, processed to smaller species than authentic wild type ClpP, as judged by SDS–PAGE and MALDI-TOF analysis of the purified proteins (data not shown, and Table 2). The percentage of the smaller species ranged from ∼90 to 100% for the P4A and V6A variants to <10% for the D18A, I19A, and Y20A variants; although the exact values were dependent on the culture conditions (data not shown). For the P4A, V6A, I7A, and E8A variants, the single smaller species corresponded to subunits with deletions of 4, 6, 7, or 8 residues from their amino-terminus, respectively, as shown by N-terminal analysis. By contrast, D18A, I19A, and Y20A yielded multiple discrete species in quantities too small to be characterized by N-terminal sequencing directly (2–4% of the total expressed ClpP). However, the masses of the minor peaks in the MALDI-TOF spectra of these variants are consistent with deletions of 5–8 residues from the mature N-terminus (Table 2). It is interesting to note at this point that the residues most disruptive to the proper maturation of ClpP form a belt of residues in the up conformation that line the entrance to the proteolytic chamber and are disordered (P4, V6, and I7) in the down conformation. This will be discussed here later in the context of additional functional studies.
Previous studies have demonstrated that authentic leader peptide processing requires the catalytic serine (S97) and histidine (H122) residues observed in the native serine-proteolytic triad suggesting that processing is autocatalytic (Thompson et al., 1994). Similarly, the internal cleavage of the mature N-terminus that was observed in some variants also requires the catalytic serine. This is demonstrated by the ClpP-V6A+S97A variant for which only the unprocessed protein accumulated. Moreover, even when the leader peptide was removed genetically, the catalytically inactive and leaderless construct (−l)ClpP-V6A + S97A was not cleaved within the mature sequence whereas the equivalent catalytically active construct (−l)ClpP-V6A was cleaved to the shorter form having I7 at its N-terminus (data not shown). Unexpectedly, we observed that the third member of the catalytic triad in the native ClpP oligomer, D171, is not required for leader peptide processing since ClpP-D171A is processed efficiently (Fig. 3A, Table 2). However, D171 is required for all of the other proteolytic functions of the enzyme, including the alternate processing associated with the V6A substitution since ClpP-V6A + D171A is correctly processed (Fig. 3A, and Table 2). This observation may indicate that the region around D171 has not adopted its correct conformation at the time of propeptide cleavage. It is currently not clear whether propeptide cleavage requires a serine dyad (Ser-His) or if another residue serves this function in this reaction. Experiments are currently underway to further define the catalytic center involved in propeptide processing. Together, these data indicate that propeptide processing and the cleavage of the mature polypeptide observed for several of the variants in our panel occur at distinct points along the maturation pathway of the ClpP subunit because both reactions have different requirements for active site residues. Consistent with this interpretation, the slow processing ClpP-P4A variant has an internally cleaved species beginning at M5 which accumulates at the expense of the authentically processed species during purification. Furthermore, in cells that are actively expressing ClpP-V6A, a faint band was observed corresponding to the authentically processed species that is not present in the cells at stationary phase where only the N-terminally truncated form is found. The simplest explanation for these observations is that these mutations affect the folding/stability of the amino-terminus during the maturation process, allowing promiscuous cleavage of the mature protein sequence.
3.3. Residues within the pore of ClpP are essential to ClpAP and ClpXP complex formation
To further define the role(s) of the amino-terminal residues in the function of mature ClpP, we examined whether our panel of N-terminal single site variants affected the catalytic activity of ClpP in degrading short (<10 residue) peptide and protein substrates (Hwang et al., 1988; Katayama et al., 1988; Woo et al., 1989). Peptide substrates, such as the fluorogenic substrate Suc-LY-AMC, can be hydrolyzed by ClpP alone, presumably because they can diffuse unaided into the proteolytic chamber (Thompson and Maurizi, 1994) where they are cleaved by the serine-proteolytic active sites. Analysis of the kinetics of Suc-LY-AMC hydrolysis by purified wt and the 20 single site N-terminal ClpP variants revealed that none of the N-terminal substitutions significantly altered this reaction (Table 2) indicating that they have little effect on peptide bond hydrolysis or the free diffusion of small substrates into the proteolytic chamber.
By contrast, the degradation of protein substrates by ClpP requires the formation of a complex with one of its cognate ATPase subunits (ClpA or ClpX) and hydrolysis of ATP (Grimaud et al., 1998; Maurizi et al., 1998; Singh et al., 1999). The resultant ClpAP and ClpXP complexes are capable of unfolding protein substrates and feeding them into the proteolytic chamber of ClpP, where they are degraded. These latter steps presumably require a tight coupling between the ATPase and ClpP complexes. To determine whether the mature N-terminus of ClpP is required for the functional coupling between the ATPase and proteolytic components, we tested the ability of each member in the panel of N-terminal ClpP variants to form functional complexes with ClpA and degrade the model unfolded substrate, α-casein (Fig. 4 and Table 2).
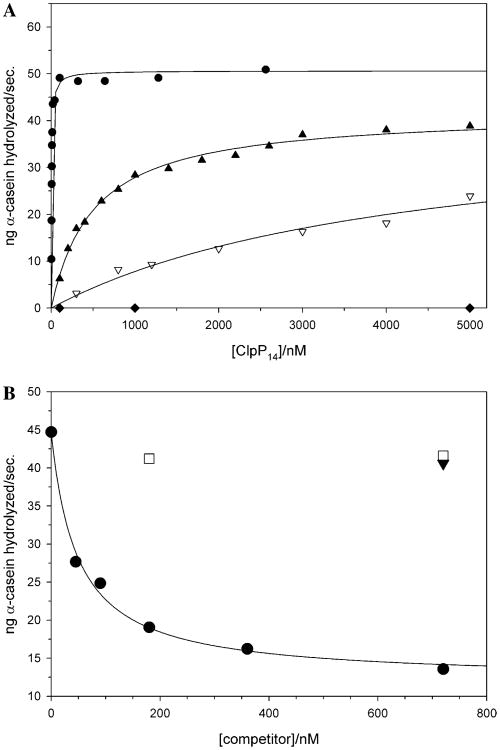
Fig. 4.
Kinetic analysis of the ATP-dependent degradation of α-casein by ClpAP, using either wild type or variant ClpP oligomers. (A) The effects of representative N-terminal alanine substitutions in ClpP on the ATP and ClpA-dependent hydrolysis of α-casein, WT (●), R15A (▲), G13A (▽), and V6A (◆). (B) Effects of increasing concentrations of inactive three ClpP variants on the ATP-dependent degradation of α-casein by wt ClpAP (ClpP-D171A (●), ClpP-V6A (■), and ClpP-V6A + D171A (▼).
Wild type ClpAP complexes rapidly degrade α-casein (kcat ∼ 10min−1) in an ATP-dependent manner to short 3- to 14-residue peptides. The apparent dissociation constant (Kapp) of these ClpAP complexes, determined kinetically, is ≤1 nM (Table 2). This value is ∼5- to 10-fold lower than that obtained by equilibrium centrifugation in the absence of proteolytic substrates (Maurizi et al., 1998), and may reflect the communication between the bound ATPase and proteolytically engaged active sites within ClpP that has recently been shown to increase the stability of the ClpXP complex (Joshi et al., 2004). Consistent with this idea, the catalytically inactive ClpP-D171A variant is 10- to 20-fold weaker than wt ClpP in this assay (Table 2 and below). Comparison of the kinetics of ClpA and ATP-dependent α-casein hydrolysis by the 20 amino-terminal variants revealed that most of the N-terminal single site variants displayed reduced activity in this assay (Fig. 4, Table 2). Their effects were reflected primarily as a decreased affinity for ClpA (increased Kapp) since only relatively small changes in Vmax were observed, in the cases for which it could be determined. The substitutions A1F, Q9A, S11A, and S16A had close to wild type activity (< 10-fold change in Kapp), while E14A and F17A variants displayed modest effects (∼20- to 50-fold) on Kapp. The L2A, V3A, M5A, E8A, T10A, R12A, G13A, and R15A substitutions showed more dramatic effects, decreasing the affinity of the resultant ClpP molecules for ClpA by 400- to 10 000-fold in this reaction indicating that they play a critical role in complex formation.
Surprisingly, several single amino acid substitutions (P4A, V6A, I7A, D18A, I19A, and Y20A) resulted in tetradecameric complexes that were completely inactive in degrading α-casein even when present at 10μM, under the conditions tested (Fig. 4A, Table 2). These six variants all had, to varying extents, truncated N-termini, thus their effects on α-casein degradation may reflect the truncation. To explore this possibility, we exploited the doubly substituted ClpP-V6A+D171A variant as a model for the authentically processed species and compared its behavior to the two singly substituted variants (ClpP-V6A and ClpP-D171A). As expected, the replacement of the catalytic aspartate residue rendered ClpP-D171A and ClpP-V6A + D171A catalytically inactive even for the hydrolysis of the short peptide substrate, Suc-LY-AMC (ClpP-V6A is fully active against this substrate). All three variants (ClpP-V6A, ClpP-D171A, and ClpP-V6A+D171A) were unable to support ClpA- and ATP-dependent hydrolysis of α-casein. However, in competition assays the ClpP-D171A variant acted as a competitive inhibitor of wt ClpP in degrading α-casein (Fig. 4B) and thus is capable of forming stable but proteolytically inactive ClpAP complexes. By contrast, ClpP-V6A and ClpP-V6A+D171A did not compete with wt ClpP even when present at 3 μM (Fig. 4A and data not shown) indicating that they do not form stable complexes with ClpA under these conditions, confirming that the V6A substitution in ClpP is sufficient to block binding of ClpA. Similarly, the substitutions in the five other “inactive” variants (P4A, I7A, D18A, I19A, and Y20A) also are likely to compromise ClpAP complex formation since they are also catalytically inactive in α-casein degradation, independent of the relative amounts of authentically cleaved protein (Table 2). This is borne out by size exclusion chromatography and ultrafiltration analyses of several “inactive” variants that confirm that they do not form stable complexes with ClpA (Table 2 and Supplemental Figs. 1–3). Further, these “inactive” variants were unable to support degradation of ssrA-tagged GFP by the ClpXP complex or inhibit its basal ATPase activity suggesting that they are also essential for ClpXP complex formation (Table 3 and Supplemental Fig. 4). Similar analyses of the remaining panel of ClpP variants showed that variants with more modest effects upon α-casein hydrolysis (relative Kapp > 1000) also showed significantly reduced affinity for ClpA in the ultrafiltration experiments (Table 2) and lower affinity in ClpX-dependent assays (Table 3). Taken together, these results point towards an essential role for the first 20 residues of the mature amino-terminus of ClpP in stabilizing the ClpAP and ClpXP complexes. Recently, two groups have implicated this region in stabilizing the interactions of ClpP with its cognate ATPase (Gribun et al., 2005; Kang et al., 2004). However, our more systematic analyses of the role of this first 20 residues now provides strong support for a model in which the N-termini of ClpP interact with ClpA and ClpX in the “up” conformation.
Table 3. Summary of the effects of substitutions in ClpP on the ATPase and proteolytic activities of the resultant ClpXP complexes.
| ClpP | Relative Kapp ClpAPa | GFP-ssrA degradation vClpXPmut/vClpXPwtb | Inhibition of ClpX ATPase activity (%)c |
|---|---|---|---|
| WT | 1.0 | 1.0 ± 0.1 | 41 ± 4 |
| A1F | 1.0 | 1.0 | 40 |
| L2A | 422 | 0.3 | 37 |
| V3A | 5212 | 0 | 5 |
| P4A | >100 000 | 0 | 0 |
| M5A | 8606 | 0 | 0 |
| V6A | >100 000 | 0 | 0 |
| V6S | >100 000 | 0 | 0 |
| I7A | >100 000 | 0 | 0 |
| I7S | >100 000 | 0 | 0 |
| E8A | 1502 | 0.1 | 24 |
| Q9A | 6 | 1.1 | 42 |
| T10A | 7568 | 0 | 0 |
| S11A | 1 | 1.0 | 39 |
| R12A | 1374 | 0.2 | 27 |
| G13A | 8960 | 0 | 0 |
| E14A | 18 | 1.1 | 41 |
| R15A | 986 | 0.1 | 31 |
| S16A | 4 | 0.9 | 38 |
| F17A | 48 | 0.9 | 39 |
| D18A | >100 000 | 0 | 0 |
| I19A | >100 000 | 0 | 0 |
| Y20A | >100 000 | 0 | 0 |
| F112A | 4600 | 0 | 9 |
| V6A + D171A | >100 000 | 0 | 0 |
| D171A | 20 | 0 | 43 |
Kapp values for ClpAP-dependent α-casein degradation are taken from Table 2.
Relative rate of GFP-ssrA ClpXP-variant/ClpXPwt degradation. Degradation experiments were performed as described in the legend to Supplemental Fig. 4A.
Effect of ClpP (wild type and variants) on the basal ATPase activity of ClpX6. ATPase assays were performed as described in the legend to Supplemental Figs. 4B and C.
3.4. The structure of V6A shows a change in conformation of the N-terminus and provides an explanation for the roles of P4, V6, I7, D18, I19, and Y20 in the conformation of the N-terminus
Having established that the amino-termini are required for stabilizing ClpAP and ClpXP complexes, we tried to reconcile the structure with the genetic and biochemical data. Based simply on mutational data, it would appear that the residues P4, V6, I7, D18, I19, and Y20, which exist within the pore in either conformation, play a dominant role in ClpAP and ClpXP complex formation (Fig. 5, Table 2). Moreover, during maturation of the ClpP oligomer, these residues appear to play a structural role in stabilizing an amino-terminal conformation that is resistant to auto-proteolysis during maturation. Thus, one possibility is that they are required for stabilizing a local conformation of the amino terminus that interacts with its cognate ATPases. As an initial test of this hypothesis, the structure of the V6A variant of ClpP was determined to be 2.6 Å resolution. Interestingly, each monomer, including the visible region of the N-terminus, is similar in every copy, despite the presence of a tetradecamer in the asymmetric unit. The first residue for which there is clear density is R15. Two lines of evidence suggest that this is not a resolution-dependent artifact. First, the structures of ClpP solved with several tripeptide inhibitors at this resolution and refined using the same protocols show the asymmetric amino-termini (unpublished results). Second, the conformation of residues 15–17, which have clear density in the structure of ClpP-V6A, differs from that observed in either the ‘up’ or ‘down’ conformations found in wild type ClpP, suggesting that the V6A substitution, and the accompanying deletion of six residues, stabilizes a novel structure for the amino-termini (Fig. 6). Further support for this is found in our preliminary analysis of a ClpP-V6A + D171A variant at 2.9 Å resolution with 54 monomers in the asymmetric unit which shows that F17 adopts a rotomer conformation similar to that observed in the structure of V6A. Like ClpP-V6A, ClpP-V6A + D171A does not interact with ClpA, but has an authentic N-terminus. Since the conformation of F17 is similar in both structures, we suggest that the disorder observed for this region in the ClpP-V6A crystal form is due to the substitution rather than the truncation of the first five residues at its amino-terminus. Higher-resolution data for the ClpP-V6A + D171A variant will provide a clearer answer. However, when all of the data for both the V6A and V6A-D171A variants are taken together, the results suggest that V6 contributes to the conformational stability of the N-terminus.
Fig. 5.
The most deleterious mutations lie within the inner surface of the pore of ring-1. Symmetrized model of ClpP where the monomer of chain B was rotated onto the positions of chain A, D–G, respectively. Chain C is shown as it exists in the crystal structure. Space filling representation of ClpP with residues 1–20 colored according to the kinetic data in Table 2. Red represents inactive variants, orange represents a 5000- to 10 000-fold decrease in Kapp values, yellow represents a 400- to 1500-fold reduction in ClpA binding, and green represents wild type-like activity. Residues 21–193 are colored white. (A) A top-view of the ClpP heptamer. The top of the pore is lined with residues (T10, R12, and R15) that have significant effects on ClpA binding. (B) Side view of four of the ClpP monomers. The central pore contains a ring of residues that abrogate ClpA binding, protected from solution by a layer of residues that have little or no effect upon ClpA binding.
Fig. 6.
Phe17 has a different rotomer conformation in the structure of the V6A variant compared to that of the wild type. Ribbon diagram for the ClpP monomer. Residues 25–193 are colored in gray and the amino terminal 24 residues, where visible in the electron density, are colored according to their structure. Representative conformations of the wild type protein, corresponding to the up and down conformations, are colored green and blue, respectively. The conformations of the loops in the structures of V6A and V6A + D171A superimpose and are colored in red. In each case, the location of F17 is shown in a ball-and-stick representation. This figure was drawn using Bobscript (Esnouf, 1991) and Raster3D (Merritt and Bacon, 1997).
3.5. Phenylalanine-112 in the hydrophobic depression also contributes to ClpAP complex stability
Our current results indicate that residues of the N-terminus of ClpP play a crucial role in complex formation with ClpA and ClpX. However, previous studies have identified a tripeptide (IGF) motif in the ClpA and ClpX ATPases that is responsible for interacting with ClpP (Kim et al., 2001). Based upon models for the ClpA and ClpX hexamers (Guo et al., 2002; Kim and Kim, 2003), it has been proposed that this motif interacts with a hydrophobic depression (Y60, Y62, F82, I90, F112, and L189) near the outer edge of the ClpP oligomer and not the N-terminus. As an initial test of this hypothesis, we constructed the F112A variant and tested its ability to hydrolyze model peptide and protein substrates. As with the N-terminal variants, the peptidase activity of F112A was unaffected, however, the relative Kapp for this variant is ∼4000 (Table 2) and is consistent with the substitution significantly reducing the stability of the ClpAP complex. Further support for this interpretation is provided by the results of the ultrafiltration experiments, which show that this substitution markedly decreases the retention of the resultant ClpP construct in the presence of ATPγS and ClpA (Supplemental Table 2). This indicates that F112 is also critical for ClpA binding; however, since it is ∼40 Å from the closest residue in the amino-terminus, it is likely that both the amino-terminus and the hydrophobic depression in ClpP interact with different surfaces on the ATPases. Coincidentally, in the structure of the symmetric, ATP-bound HslUV complex, the regions of contact are distributed along two annuli (Sousa et al., 2000) with a similar spacing. Thus, despite the difference in symmetry of the protease component in each case and the structural dissimilarity of their folds, the similarity in the distribution of interacting residues is striking.
3.6. Structural model for the ClpAP and ClpXP complexes
Based simply on the structures of ClpP and the available mutational data, a unique conformation of the N-termini in the ClpAP complex cannot be identified, if indeed a single conformational state exists. However, several lines of evidence provide important clues to possible modes of interaction of ClpP with its cognate ATPases. For example, in the recent structure of the 20S proteasome in complex with its non-ATPase regulator, PA26, the exposed loop formed by the N-termini of the outer α-subunits in the proteasome interacts with PA26 and pulls the remainder of the largely disordered N-termini out of the pore into a large chamber formed in PA26 (Whitby et al., 2000). Removing the N-terminal residues from the pore region in the proteasome opens the access pores and the authors speculate that this may promote the egress of products. The interacting loop in the proteasome is analogous to the surface exposed residues 10–15 in the ‘up’ conformation in ClpP that has been shown here and elsewhere contributes significantly to the stability of the ClpAP complex. However, unlike the 20S proteasome–PA26 complex, the N-termini in ClpP are unlikely to be pulled out of the access channel into the ClpA ATPase, since EM studies of ClpAP complexes formed with unprocessed ClpP indicate that the propeptides (196 residues in total) occupy the proteolytic chamber, blocking the translocation of substrates (Ishikawa et al., 2001). This observation is difficult to reconcile with a model in which residue 2 of the N-terminus is out of the pore and interacting with the ATPase subunits. In addition, the geometry of the ATPase subunits is distinctly different to the open chamber in PA26: the putative substrate channel in the ATPase complexes is narrow therefore extension of even a single N-termini of ClpP into this channel would effectively block it, preventing substrate passage (Sousa et al., 2000; Wang et al., 2001). By extension, it is also unlikely that regions in the ATPase structure project into the substrate pore in ClpP to contact buried regions of the N-termini, since this pore is an equivalent size.
If the first 20 residues are not pulled out of the molecule to make direct contacts with the ATPase complex, why are the V6A, I7A, D18A, I19A, and Y20A ClpP variants unable to form stable ClpAP complexes? The structure of the V6A variant provides one potential answer to this question. Residues 7–14 in this structure are disordered and residues 15–17 adopt a conformation that is distinct from that of the wild type protein; consequently, the amino acid residues implicated in ClpA binding are either buried or disordered. Although this provides an explanation for the lack of ClpA binding, the extent of the destabilization for the inactive class of mutants is still striking. For example, the Kapp for ClpAP complexes involving wild type ClpP is ∼0.5 nM and corresponds to a free energy change, ΔG0, of ∼13kcal/mol (ΔG0 = RTlnKapp) for the complex. Based upon the absence of detectable ClpAP complex formation (rates of casein degradation <1% of wild type degradation) with the P4A, V6A, I7A, D18A, I19A, and Y20A ClpP variants, at 10 μM concentrations, the Kapp values for their complexes must be >100 μM, corresponding to a ΔG0 for complex stabilization of 6kcal/mol, in the most favorable case. The sheer magnitude of this change in binding free energy (ΔΔG0>7kcal/mol) argues against their effects being exclusively due to a loss in their interacting surface with the ATPase. Instead, it seems likely that a major effect of the P4A, V6A, I7A, D18A, I19A, and Y20A substitution is to destabilize the folding core of the β-hairpin structure of the N-terminus in the ‘up’ conformation and thereby preventing any of the residues in this structure from interacting with the ATPase. Consistent with this idea, the β-hairpin conformation of the N-terminus is disrupted in the structure of the V6A and V6A + D171A variants.
With this in mind, a simple docking model of ClpXP was generated based on the structures of their components, ClpX and ClpP (Fig. 7). Since similar residues in ClpP are required for stabilizing the ClpAP and ClpXP complexes (Table 2 and 3), and the IGF motif in the structure of ClpX is ordered whereas in ClpA it is not, we used the coordinates of ClpX to generate a docking model. The structure of ClpX contains a monomer in the asymmetric unit, therefore the hexameric structure of HslU (PDB code 1DO0) (Bochtler et al., 2000) was used as a template to generate a hexameric ClpX. In agreement with EM reconstructions, the central pore of the ATPase was aligned with that of the protease, and translated along this axis. The general features of the two surfaces made chemical sense—there was overall charge-complementarity on the interacting surfaces and the hydrophobic surfaces of each molecule colocalized. Arranged in this way, 2–3 of the ClpX IGF loops could interact with the hydrophobic depressions on the surface of the ClpP oligomer (Fig. 7). This docking model is consistent with the recent biochemical evidence from the Baker and Sauer laboratories which conclude that 2–3 IGF loops on ClpX interact simultaneously with ClpP (Joshi et al., 2004). Furthermore, additional loops could be made to interact by small movements of the IGF loop which is entirely consistent with their observation that the IGF-loop is sensitive to the nucleotide state of ClpX (and by analogy ClpA) altering its affinity for ClpP (Joshi et al., 2004). Since some of the loops in the structure of ClpX are disordered, this may have an effect upon the detailed interactions between ClpX and ClpP. Even so, this analysis does provide insight into the gross characteristics of the interface. A key difference between this model and that proposed by Gribun et al. is in the number of loops in the up conformation. In the Gribun model, all loops are placed in the up conformation, with one loop not being engaged in ClpX binding. However, the atoms in this loop still need to be somehow accommodated. In our model presented here, supported by the high-resolution structure of E. coli ClpP, only six loops adopt the up conformation, with the remaining loop in the down conformation perhaps engaged in translocating substrate to the proteolytic chamber or in ejecting products.
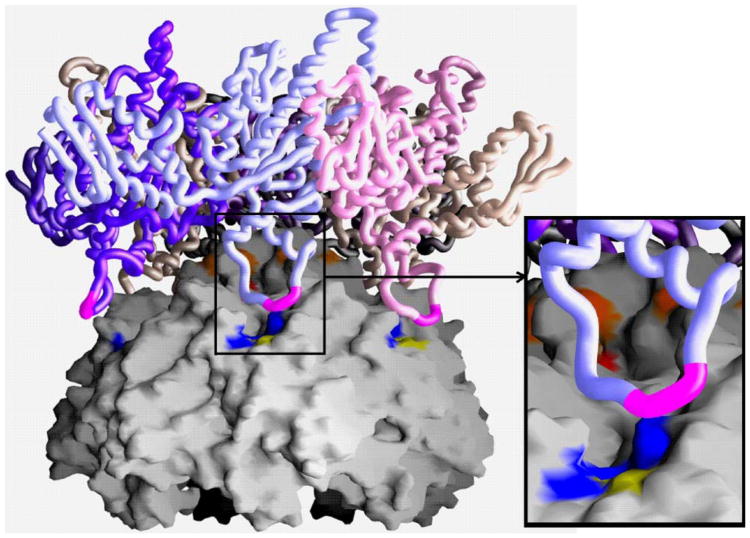
Fig. 7.
Docked model of the hexameric ClpX onto the heptameric ClpP. The molecular surface of a symmetrized ClpP heptamer is shown and a worm of the ClpX hexamer. The dome surface generated by the N-termini of ClpP provides a complementary docking surface for the interaction at the pore of ClpX. The IGF loop of ClpX is colored magenta. The hydrophobic groove on the surface of ClpP is colored blue with the exception of F112, which has been characterized biochemically, is colored yellow. The inset shows this groove in more detail.
Based upon the observed physical data, and the theoretical docking model, we propose a schematic model for the interaction of the assembled ATP-dependent protease (Fig. 8). In the functional ClpAP or ClpXP complexes, the access pores must adopt an open conformation to allow substrate passage. Based upon the location of mutations that abolish ClpAP complex formation, the ‘up’ conformation of the N-termini in ClpP is important for interacting with the ATPase components, however, it is possible that their structure in the complex might be slightly different to that found in ring-1 in our current model. One possibility that is consistent with the observed flexibility of residues 13–16 in all structures determined to date is that the surface exposed loop in the ‘up’ conformation might act as a flap that opens when ClpP interacts with ClpA or ClpX. This possibility is attractive because the N-termini at the center of ClpP are ideally placed to interact with all six ATPase subunits simultaneously in contrast to the hydrophobic grooves near the edge of the oligomer that can only reasonably interact with, at most, three subunits in the ATPases. With the N-termini arranged in this way, interactions would be made between the pseudo-6-fold symmetric N-termini and the hexameric ATPase subunits reminiscent of those in the HslUV complex (Kwon et al., 2003). This model would predict that the interacting conformation of the amino-termini in which they have a perfect local 6-fold symmetric arrangement would only be observed in the presence of the ATPase, since interactions with the ATPase would presumably stabilize this conformation. The presence of the seventh non-interacting N-terminus introduces the possibility of ATPase gated substrate translocation through the ∼15 Å long access pores in ClpP. Substrate translocation into the proteolytic chamber in ClpP may be catalyzed by the ATPase components using the ATP hydrolysis induced transition of one N-terminus from the ‘up’ to the down conformation during ATP hydrolysis. An alternative possibility is that the seventh amino-terminus is actually in the up conformation in the complex creating a 6–7 symmetry mismatch between the ATPase and proteolytic subunits. This arrangement could facilitate the rotational movement of the ATPase during substrate translocation in a manner analogous to virus entry portals, and V- and F-type ATPases (Stock et al., 1999; Tao et al., 1998). However, there is no evidence for ClpAP, ClpXP, and the 26S proteasome that the ATPase component rotates during the proteolytic cycle. Moreover, the HslUV protease which is 6-fold symmetric, and the FtsH and Lon proteases in which the six ATPase and protease subunits are encoded within a single polypeptide chain suggest that rotational motion is not a necessary step in the ATP-dependent proteolysis of substrates. Based upon these considerations, we favor the pseudo-symmetric model for the Clp proteases presented above.
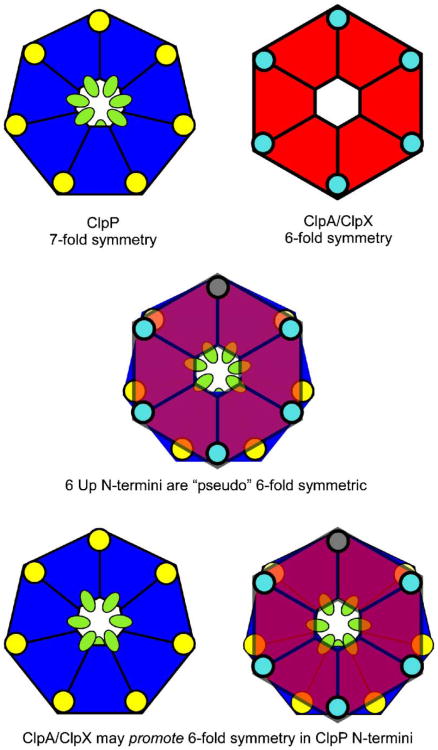
Fig. 8.
Schematic of ClpXP, showing how a local pseudo-6-fold interaction in ClpP allows ClpX to dock. A local pseudo-6-fold symmetric interaction at the central pore allows for a tethering of the molecule. Small changes in the local conformation of the loop, including the IGF motif, are driven by ATP hydrolysis. These movements can be “sensed” by ClpP through the interactions of some of the IGF motifs with the corresponding hydrophobic depressions on the ClpP surface. ClpP is represented as a blue heptamer. The hydrophobic groove, including F112, is represented as a yellow circle and the amino-terminal 20 residues are drawn as green ovals. The ATPase component is drawn as a red hexagon and the IGF residues within the motif are shown as a cyan circle.
Supplementary Material
Acknowledgments
We thank all of our colleagues in the Biology Department at Brookhaven National Laboratory for their support during much of this research. This research was initiated when all authors were members of the Biology Department at Brookhaven National Laboratory, and completed after relocation to Pennsylvania State University College of Medicine in Hershey, PA. In addition, we thank the beam-line managers and night staV at the NSLS on beamlines X12B, X12C, and X25 for their continued support in this and other projects. The macromolecular beamlines X12B, X12C, and X25 at the National Synchrotron Light Source are supported in part by the NSF and NIH Grant 1P41 RR12408-01A1. This work is supported by Grant GM057390 from National Institutes of Health to J.M.F.
Footnotes
Appendix A. Supplementary data: Supplementary data associated with this article can be found, in the online version, at doi:10.1016/ j.jsb.2005.09.011.
References
- Beuron F, Maurizi MR, et al. At sixes and sevens: characterization of the symmetry mismatch of the ClpAP chaperone-assisted protease. J Struct Biol. 1998;123(3):248–259. doi: 10.1006/jsbi.1998.4039. [DOI] [PubMed] [Google Scholar]
- Bochtler M, Ditzel L, et al. Crystal structure of heat shock locus V (HslV) from Escherichia coli. Proc Natl Acad Sci USA. 1997;94(12):6070–6074. doi: 10.1073/pnas.94.12.6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochtler M, Hartmann C, et al. The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature. 2000;403(6771):800–805. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- Dodson E. Report of a workshop on the use of statistical validators in protein X-ray crystallography. Acta Crystallogr D Biol Crystallogr. 1996;52(Pt. 1):228–234. doi: 10.1107/S0907444995010638. [DOI] [PubMed] [Google Scholar]
- Esnouf RM. J Mol Graph. 1991;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- Gribun A, Kimber MS, et al. The ClpP double ring tetradecameric protease exhibits plastic ring–ring interactions, and the N termini of its subunits form Xexible loops that are essential for ClpXP and ClpAP complex formation. J Biol Chem. 2005;280(16):16185–16196. doi: 10.1074/jbc.M414124200. [DOI] [PubMed] [Google Scholar]
- Grimaud R, Kessel M, et al. Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J Biol Chem. 1998;273(20):12476–12481. doi: 10.1074/jbc.273.20.12476. [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, et al. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386(6624):463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Guo F, Maurizi MR, et al. Crystal structure of ClpA, an HSP100 chaperone and regulator of ClpAP protease. J Biol Chem. 2002 doi: 10.1074/jbc.M207796200. [DOI] [PubMed] [Google Scholar]
- Hoskins JR, Singh SK, et al. Protein binding and unfolding by the chaperone ClpA and degradation by the protease ClpAP. Proc Natl Acad Sci USA. 2000;97(16):8892–8897. doi: 10.1073/pnas.97.16.8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang BJ, Woo KM, et al. Protease Ti, a new ATP-dependent protease in Escherichia coli, contains protein-activated ATPase and proteolytic functions in distinct subunits. J Biol Chem. 1988;263(18):8727–8734. [PubMed] [Google Scholar]
- Ishikawa T, Beuron F, et al. Translocation pathway of protein substrates in ClpAP protease. Proc Natl Acad Sci USA. 2001;98(8):4328–4333. doi: 10.1073/pnas.081543698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi SA, Hersch GL, et al. Communication between ClpX and ClpP during substrate processing and degradation. Nat Struct Mol Biol. 2004;11(5):404–411. doi: 10.1038/nsmb752. [DOI] [PubMed] [Google Scholar]
- Kang SG, Maurizi MR, et al. Crystallography and mutagenesis point to an essential role for the N-terminus of human mitochondrial ClpP. J Struct Biol. 2004;148(3):338–352. doi: 10.1016/j.jsb.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Katayama Y, Gottesman S, et al. The two-component, ATP-dependent Clp protease of Escherichia coli Purification, cloning, and mutational analysis of the ATP-binding component. J Biol Chem. 1988;263(29):15226–15236. [PubMed] [Google Scholar]
- Kim DY, Kim KK. Crystal structure of ClpX molecular chaperone from Helicobacter pylori. J Biol Chem. 2003 doi: 10.1074/jbc.M305882200. [DOI] [PubMed] [Google Scholar]
- Kim YI, Burton RE, et al. Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol Cell. 2000;5(4):639–648. doi: 10.1016/s1097-2765(00)80243-9. [DOI] [PubMed] [Google Scholar]
- Kim YI, Levchenko I, et al. Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat Struct Biol. 2001;8(3):230–233. doi: 10.1038/84967. [DOI] [PubMed] [Google Scholar]
- Kohler A, Bajorek M, et al. The substrate translocation channel of the proteasome. Biochimie. 2001a;83(3–4):325–332. doi: 10.1016/s0300-9084(01)01242-1. [DOI] [PubMed] [Google Scholar]
- Kohler A, Cascio P, et al. The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol Cell. 2001b;7(6):1143–1152. doi: 10.1016/s1097-2765(01)00274-x. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- Kwon AR, Kessler BM, et al. Structure and reactivity of an asymmetric complex between HslV and I-domain deleted HslU, a prokaryotic homolog of the eukaryotic proteasome. J Mol Biol. 2003;330(2):185–195. doi: 10.1016/s0022-2836(03)00580-1. [DOI] [PubMed] [Google Scholar]
- Lowe J, Stock D, et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science. 1995;268(5210):533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- Lupas A, Flanagan JM, et al. Self-compartmentalizing proteases. Trends Biochem Sci. 1997;22(10):399–404. doi: 10.1016/s0968-0004(97)01117-1. [DOI] [PubMed] [Google Scholar]
- Maurizi MR, Clark WP, et al. Sequence and structure of Clp P, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J Biol Chem. 1990a;265(21):12536–12545. [PubMed] [Google Scholar]
- Maurizi MR, Clark WP, et al. Clp P represents a unique family of serine proteases. J Biol Chem. 1990b;265(21):12546–12552. [PubMed] [Google Scholar]
- Maurizi MR, Singh SK, et al. Molecular properties of ClpAP protease of Escherichia coli: ATP-dependent association of ClpA and clpP. Biochemistry. 1998;37(21):7778–7786. doi: 10.1021/bi973093e. [DOI] [PubMed] [Google Scholar]
- Merritt EA, Bacon DJ. In: Methods in Enzymology. Carter CW, Sweet RM, editors. Vol. 277. 1997. pp. 505–524. [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, et al. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Ortega J, Lee HS, et al. ClpA and ClpX ATPases bind simultaneously to opposite ends of ClpP peptidase to form active hybrid complexes. J Struct Biol. 2004;146(1–2):217–226. doi: 10.1016/j.jsb.2003.11.023. [DOI] [PubMed] [Google Scholar]
- Ortega J, Singh SK, et al. Visualization of substrate binding and translocation by the ATP-dependent protease, ClpXP. Mol Cell. 2000;6(6):1515–1521. doi: 10.1016/s1097-2765(00)00148-9. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW, Sweet RM, editors. Methods in Enzymology. Vol. 276. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Rohrwild M, Coux O, et al. HslV-HslU: A novel ATP-dependent protease complex in Escherichia coli related to the eukaryotic proteasome. Proc Natl Acad Sci USA. 1996;93(12):5808–5813. doi: 10.1073/pnas.93.12.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Grimaud R, et al. Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proc Natl Acad Sci USA. 2000;97(16):8898–8903. doi: 10.1073/pnas.97.16.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Guo F, et al. ClpA and ClpP remain associated during multiple rounds of ATP-dependent protein degradation by ClpAP protease. Biochemistry. 1999;38(45):14906–14915. doi: 10.1021/bi991615f. [DOI] [PubMed] [Google Scholar]
- Sousa MC, Trame CB, et al. Crystal and solution structures of an HslUV protease-chaperone complex. Cell. 2000;103(4):633–643. doi: 10.1016/s0092-8674(00)00166-5. [DOI] [PubMed] [Google Scholar]
- Stock D, Leslie AG, et al. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286(5445):1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- Tao Y, Olson NH, et al. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell. 1998;95(3):431–437. doi: 10.1016/s0092-8674(00)81773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MW, Maurizi MR. Activity and specificity of Escherichia coli ClpAP protease in cleaving model peptide substrates. J Biol Chem. 1994;269(27):18201–18208. [PubMed] [Google Scholar]
- Thompson MW, Singh SK, et al. Processive degradation of proteins by the ATP-dependent Clp protease from Escherichia coli. Requirement for the multiple array of active sites in ClpP but not ATP hydrolysis. J Biol Chem. 1994;269(27):18209–18215. [PubMed] [Google Scholar]
- Wang J, Hartling JA, et al. The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell. 1997;91(4):447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- Wang J, Song JJ, et al. Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure (Camb) 2001;9(2):177–184. doi: 10.1016/s0969-2126(01)00570-6. [DOI] [PubMed] [Google Scholar]
- Weber-Ban EU, Reid BG, et al. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature. 1999;401(6748):90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- Whitby FG, Masters EI, et al. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408(6808):115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- Woo KM, Chung WJ, et al. Protease Ti from Escherichia coli requires ATP hydrolysis for protein breakdown but not for hydrolysis of small peptides. J Biol Chem. 1989;264(4):2088–2091. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.