Abstract
Global folding of bacterial chromosome requires the activity of condensins. These highly conserved proteins are involved in various aspects of higher order chromatin dynamics in a diverse range of organisms. Two distinct superfamilies of condensins have been identified in bacteria. The SMC-ScpAB proteins bear significant homology to eukaryotic condensins and cohesins and are found in most of presently sequenced bacteria. This review focuses on the MukBEF/MksBEF superfamily, which is broadly distributed across diverse bacteria and is characterized by low sequence conservation. The prototypical member of this superfamily, the Escherichia coli condensin MukBEF, continues to provide critical insights into the mechanism of the proteins. MukBEF acts as a complex molecular machine that assists in chromosome segregation and global organization. The review focuses on mechanistic analysis of DNA organization by MukBEF with the emphasis on its involvement in formation of chromatin scaffold and plausible other roles in chromosome segregation.
Keywords: condensins, MukB, MksB, condensin, bacterial chromosome, chromosome structure, chromosome scaffold, Pseudomonas aeruginosa
Preface: Chromatin scaffold and global folding of the chromosome
The problem of chromatin folding is often described as the need to pack a very long chromosomal DNA into the confines of a tiny cell. Indeed, a single human cell carries 2 meters of DNA, whereas a typical bacterial chromosome, whilst admittedly shorter, about few mm, fits inside a proportionally smaller 1 μm cell. The resulting DNA packing density is about the same in bacteria and eukaryotic cells suggesting that at least some key features of chromosome organization are, perhaps, also conserved across kingdoms of life.
Besides bulk packing, chromosome structure must also meet even more pressing challenges of maintaining at least partial order in DNA without compromising its accessibility to information processing machinery. This must be accomplished in the face of thermal motion, which randomizes spatial arrangement of DNA, and DNA unwinding activity of various polymerases, which introduce supercoils and promote intertwining of DNA strands [Alexandrov et al., 1999; Schvartzman and Stasiak, 2004].
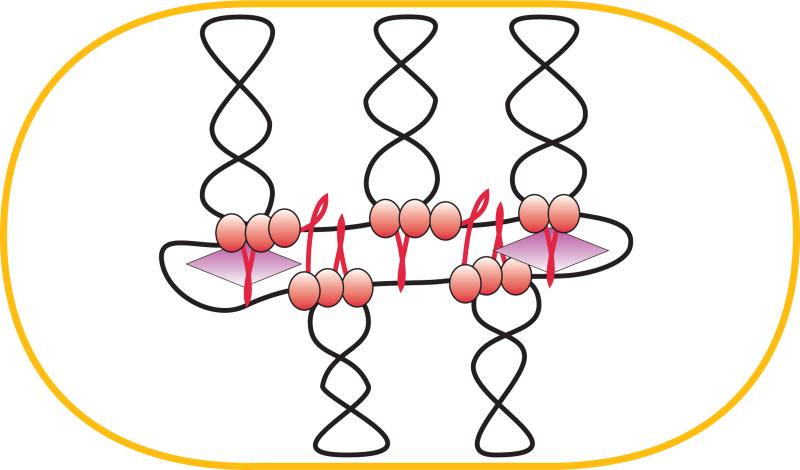
The scaffold model of the chromosome (Fig. 1) seeks to meet these demands by postulating that the chromosome is split into a series of giant loops, which are anchored to proteinaceous matrix at their base [Johnson et al., 2005; Pettijohn, 1996; Saitoh et al., 1995]. The model was initially inspired by electron microscopy observations of gently lysed bacterial [Kavenoff and Bowen, 1976] and eukaryotic [Paulson and Laemmli, 1977] cells and later substantiated by cell biology and genetic tests. Notable among those were the findings that isolated bacterial chromosomes require multiple DNA breaks for complete relaxation and that supercoiling-driven assembly of pre-recombination complexes occurs only for rather closely spaced DNA fragments [Higgins et al., 1996; Sinden and Pettijohn, 1981; Worcel and Burgi, 1972]. This line of reasoning builds upon realization that supercoiling is a global property of DNA and, therefore, supercoils should readily equilibrate throughout the entire molecule unless some DNA fragments are attached to a larger structure. By anchoring bases of DNA loops to a scaffold, the molecule can be subdivided into a set of topologically insulated fragments, known as topological domains. The most reliable current estimates reveal that the E. coli chromosome is split into multiple, about 10 kb domains that are dynamically and stochastically distributed through the chromosome [Deng et al., 2004; Postow et al., 2004].
Figure 1.
A scaffold model of the chromosome. The giant loop architecture of the chromosome is stabilized by multiple attachments to the cellular matrix. The matrix does not have to be continuous but is likely to be linked to the chromosome segregation machinery. The chromosome is further stabilized by DNA supercoiling and numerous nucleoid associated proteins, NAPs, which bend and bridge DNA (not shown).
The scaffold model offers an attractive balance of order and disorder. Whereas most of the DNA remains accessible to information processing enzymes, the sparsely set scaffold attachment sites should suffice when a force needs to be applied to DNA during chromosome segregation. This arrangement also offers the most effective way to control the size of the chromosome or separate topological domains [Marko and Trun, 1998]. The biggest challenge of the model is to explain location of the scaffold attachment sites.
The issue seems trivial at the first glance. Indeed, numerous proteins display DNA bridging activity and are expected to stabilize DNA loops inside the cell (reviewed in [Browning et al., 2010; Dillon and Dorman, 2010; Johnson et al., 2005; Luijsterburg et al., 2006; Rimsky and Travers, 2011]). Should the reaction follow the random collision mechanism, however, the size of the loops would be small, around few hundred bp, with chances of obtaining larger loops rapidly decaying with increasing DNA separation [Levene et al., 2013]. This pattern is at odds with the predictions of the scaffold model. This suggests a contribution of nonrandom mechanisms to selection of scaffold attachment regions, SARs. An easy way to accomplish that would be by employing a sequence specific DNA binding protein whose binding sites are dispersed throughout the chromosome. This idea, however, does not readily agree with the dynamic and stochastic nature of SARs. Moreover, sequencing of eukaryotic SARs did not reveal any conserved motifs beyond these regions being intergenic and AT-rich [Mirkovitch et al., 1984]. Apparently, a novel mechanism needs to be devised to explain the sparse organization of the SARs. A plausible such mechanism is beginning to emerge from studies of condensins.
Condensins were discovered over the span of few years in several diverse organisms [Cobbe and Heck, 2004; Graumann and Knust, 2009; Gruber, 2011; Reyes-Lamothe et al., 2012; Rybenkov, 2009]. Distinct lines of inquiry led to their discovery. One of those lines employed fractionation of the poorly soluble frog chromatin scaffold, which revealed DNA topoisomerase II and condensins as the major protein components of the scaffold [Earnshaw et al., 1985; Saitoh et al., 1994]. The first discovered condensin was MukBEF, which emerged from an elegant screen for E. coli proteins involved in partitioning of chromosomes but not necessarily plasmids [Hiraga et al., 1989; Niki et al., 1991]. Independently, condensins and cohesins were discovered in Saccharomyces ceverisiae as the proteins required for chromosome segregation [Strunnikov et al., 1995; Strunnikov et al., 1993]. Yet another line of research identified condensins in frog oocytes as the soluble factors responsible for chromosome condensation during cell division [Hirano and Mitchison, 1994]. The convergence of the multiple approaches likely reflects the central role of condensins in many aspects of the higher order chromosome dynamics and must be kept in mind when deducing their mechanism.
Architecture of MukBEF
Three families of condensins have been identified so far in bacteria. The SMC-ScpAB condensins are found in vast majority of sequenced bacteria. The core SMC (structural chromosome maintenance) subunit of the complex shares high degree of homology to archeal and eukaryotic condensins [Britton et al., 1998; Cobbe and Heck, 2004; Mascarenhas et al., 2002; Soppa et al., 2002]. The second family, MukBEF, is found in enterobacteria and several other related orders of γ-Proteobacteria [Hiraga, 2000; Niki et al., 1991; Yamanaka et al., 1996]. The name of the complex stands for mukaku, anucleate, to mark its phenotype. Recently, a third family of condensins, MksBEF (MukBEF-like SMCs), was identified in diverse proteobacteria [Petrushenko et al., 2011]. These proteins have the same operon organization as MukBEF but display virtually no sequence homology to it. One or more members of MksBEF family typically coexist with SMC-ScpAB or MukBEF and augment rather than substitute their function. These last two families will be the focus of this review. Bacteria also harbor other SMC-like proteins, including SbcCD [Connelly et al., 2003] and RecN [Reyes et al., 2010], which function in DNA recombination and repair but do not seem to contribute to chromosome partitioning.
MukBEF is the sole condensin in E. coli [Hiraga, 2000; Petrushenko et al., 2011]. All three subunits of the protein are encoded in the operon smtA-mukF-mukE-mukB together with the unrelated gene smtA [Yamanaka et al., 1995, 1996]. Mutational inactivation of MukBEF results in chromosome disorganization, decondensation and cutting, anucleate cell formation (even at permissive temperatures) and severe decline in colony formation at temperatures above 25°C [Niki et al., 1991; Sawitzke and Austin, 2000; Wang et al., 2006; Yamanaka et al., 1996] presumably due to chromosome decondensation [Sawitzke and Austin, 2000; Weitao et al., 1999]. Similar defects are observed in condensin-deficient strains of Bacillus subtilis and Caulobacter crescentus indicating that these proteins play the same role inside the cell [Britton et al., 1998; Jensen and Shapiro, 1999]. The severity of mukB phenotype is notable given its modest copy number, about 400 MukBEFs per growing cell [Kido et al., 1996; Petrushenko et al., 2006b]. For comparison, the copy number of “traditional” nucleoid proteins, such as HU, HNS or FIS, often exceeds 10,000 [Johnson et al., 2005]. This observation lends support to the notion that MukBEF serves as a part of the chromosome scaffold rather than a bulk DNA packing protein [Saitoh et al., 1995].
At the heart of MukBEF is MukB, a member of the SMC protein family [Cobbe and Heck, 2004; Graumann and Knust, 2009; Gruber, 2011; Rybenkov, 2009]. MukB consists of two globular, the N- and C-terminal domains connected by two long α-helices with a hinge region in between (Fig. 2A and [Melby et al., 1998; Niki et al., 1992]). The N- and C-terminal domains, which contain the Walker A and Walker B motifs, respectively, fold into a single globular head domain with the ATP binding site located on its surface [Woo et al., 2009]. In solution, MukB dimerizes via the hinge domain to form a distinctive V-shaped molecule where two long coiled-coils protruding from the hinge terminate in globular head domains [Matoba et al., 2005; Melby et al., 1998; Niki et al., 1992]. The head domains themselves can associate via the shared ABC-type ATP binding site leading to ATP-modulated formation of protein rings or macromolecular assemblies [Matoba et al., 2005; Woo et al., 2009]. The DNA binding site is located on the positively charged hinge-proximal side of the head domain and spreads over its sides ([Woo et al., 2009] and Fig 3A).
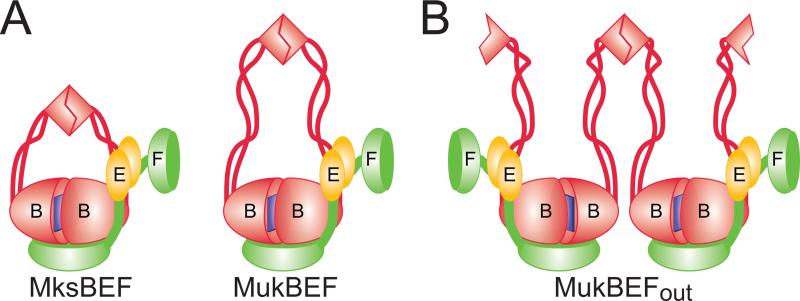
Figure 2. Architecture of MukBEF.
(A) Organization of MukBEF and MksBEF. ATP-mediated dimerization of MukB head creates a high affinity DNA binding site. The head can accommodate a single C-terminal WHD of MukF. N-terminal WHD of MukF is prone to further dimerization. With the exception of the shorter coiled coils, MksBEF is predicted to form a similar structure to MukBEF.
(B) A possible organization of MukBEF if dimerization interfaces of its hinge and head domains face away from each other.
Figure 3.
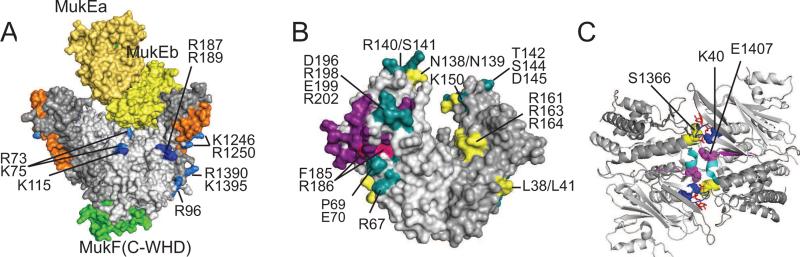
Architecture of crystallized MukBEF fragments. (A) Crystal structure of dimerized MukB head in complex with a fragment of MukF and a dimer of MukE (PDB 3EUK). The N- and C-terminal domains of MukB are shown in light and darker grey, respectively, MukF is green and the two MukEs are in two shades of yellow. The stems of the long coiled coils are shown in orange. Amino acids involved in DNA binding are blue (essential) or light blue (reduced affinity). Amino acids are numbered according to their position in the E. coli MukB. Note that no MuB-MukE contacts hold MukE above MukB head, where it blocks the DNA binding site.
(B) Crystal structure of MukE (PDB 3EUH) with known mutations. The two MukE monomers are shown in two shades of gray. Purple marks the interface with MukF. Also shown are non-essential amino acids (yellow), essential amino acids found on the MukE-MukF interface (red) and essential amino acids elsewhere (green).
(C) Organization of the ATPase site (PDB 3EUK). The dimeric MukB head viewed from the top with two bound ATPγS (red). Shown are the conserved ATPases regions, including Walker A (blue), C-motif (signature; yellow), Walker B (magenta) and D-loop (cyan). The N- and C-terminal domains are shown in light and darker shades of grey.
Because of the sheer length of the coiled-coil region (~ 50 nm in MukB), no crystal structures of a full length condensin has emerged so far. This leaves open the question about connectivity within the complex. It is generally assumed that the ATP-binding sites of MukB heads face each other within the V-shaped dimer and, by extension, the protein acts as an ATP-modulated protein ring. The opposite arrangement, however, is also conceivable, in which case formation of multimeric structures would be expected (Fig. 2B). Notably, experimental evidence strongly supports existence of multimeric MukBEFs in vitro and in vivo [Badrinarayanan et al., 2012b; Petrushenko et al., 2006b] and, furthermore, biochemical studies suggest that multimerization of MukBEF is essential for its function [Cui et al., 2008].
The other two subunits, MukE and MukF, form a stable complex with each other and dynamically associate with MukB [Petrushenko et al., 2006b; Woo et al., 2009; Yamazoe et al., 1999]. MukEF does not show any DNA binding activities of its own but modulates MukB-DNA interactions [Cui et al., 2008; Petrushenko et al., 2006b]. MukF serves as a kleisin that interacts with MukB heads and links MukE to the complex [Fennell-Fezzie et al., 2005; Woo et al., 2009]. Both purified and reconstituted MukEF form an elongated complex MukE4F2 [Gloyd et al., 2007; Petrushenko et al., 2006b]. MukF and MukE also form a complex MukE2F2 but only at substoichiometric levels of MukE [Petrushenko et al., 2006b].
The structural basis for this ambiguity lies in the unusual organization of MukF. MukF consists of two winged-helix domains (WHD) connected by a long unstructured linker peptide [Woo et al., 2009]. The N-terminal WHD provides a dimerization interface whereas the C-terminal WHD associates with MukB head via mostly hydrophobic interactions on its hinge-distal side. The long unstructured linker serves as a docking site for MukB and MukE and is perfectly structured in their presence. The MukE binding stretch of the linker can asymmetrically bind two MukE monomers. Within the crystal, MukE occupies space immediately atop of the DNA binding site of MukB (Fig. 3A) which could conceivably lead to DNA displacement. However, no direct contacts between MukE and MukB were observed, indicating that the entire assembly could swivel away.
In this arrangement, MukF acts essentially as a tether that ensures proximity of all globular domains but does not impose a rigid structure, Therefore, the complex can assemble and be at least partially functional even in the absence of some of the subunits. It remains unclear whether the two MukEs in the MukE2F2 complex are bound to the same or different MukFs. However, the former possibility seems more likely given the propensity of MukE for self-association. Notably, a recent study revealed essential mutations in MukE that seem unrelated to DNA binding activities of MukB (Fig. 3B), suggesting that the protein plays a distinct role of its own [She et al., 2013]. Although some of these mutations were proposed to fall on a novel MukE-MukE interface [Gloyd et al., 2011], others face solvent in all available crystal structures of MukBEF.
MukEF and MukB interact with each other in a dynamic ATP-modulated manner both in vitro and in vivo [Badrinarayanan et al., 2012b; Petrushenko et al., 2006b; Woo et al., 2009]. Association between MukB and MukEF is stimulated in the presence of calcium and magnesium [Yamazoe et al., 1999]. When reconstituted in vitro, MukB and MukEF produce two complexes, MukB2-Muk(E2F) and MukB2-Muk(E2F)2 [Petrushenko et al., 2006b]. Whereas MukB2-Muk(E2F) is stable under a range of conditions, MukB2-Muk(E2F)2 can be only observed at low salt and in the presence of magnesium. The MukB2-Muk(E2F)2 complex is similarly disrupted in the presence of ATP [Woo et al., 2009]. The structural basis for this dynamics lies in steric hindrances that develop upon ATP-mediated dimerization of MukB heads. Whilst each MukB monomer can associate with a C-terminal WHD of MukF the room for only one remains upon MukB dimerization [Woo et al., 2009].
Notably, the MukB2-Muk(E2F)2 complex proved to be completely inert in DNA binding whereas DNA remodeling activities of MukB2-Muk(E2F) seem indistinguishable from those of MukB2 [Petrushenko et al., 2006b]. This suggests that alterations in the composition of the complex could be coupled to loading of the protein onto DNA. This idea is consistent with the observation that the ATPase activity of MukB is stimulated by MukEF but not DNA [She et al., 2013; Woo et al., 2009]. This reveals that MukEF but not DNA helps stabilize the transition state of the MukB ATPase and thereby activate conformational changes, which, in turn, modulate interaction of the protein with DNA. Furthermore, real time single molecule fluorescence studies of MukBEF in live cells indeed revealed two populations of the complexes [Badrinarayanan et al., 2012b]. The presumably DNA-bound stationary population consists of oligomers of MukB4-Muk(E2F)2 complexes, whereas MukB2-Muk(E2F)2 complexes were mobile and apparently DNA-free. The generality of this phenomenon remains unclear since MukBEF is so far the only condensin for which variation in composition was reported. For comparison, the B. subtilis condensin operates at SMC2-ScpAB composition [Burmann et al., 2013].
A lingering issue in this context is the oligomeric state of the active MukBEF. Hydrodynamic methods clearly show that the protein exists as a MukB2-Muk(E2F) complex [Petrushenko et al., 2006b]. This, however, is at odds with high dimerization propensity of MukEF [Petrushenko et al., 2006b; Woo et al., 2009]. Indeed, purified MukEF always exists in its dimeric form, Muk(E2F)2. What precludes MukF dimerization when it is a part of MukBEF? Several possibilities must be considered. Perhaps the dimeric complex has a short life time or its dissociation constant could be too high in vitro. The complex could conceivably be stabilized inside the cell through its interaction with DNA or other cofactors. Alternatively, the N-terminal WHD domain of MukF could be engaged in the as yet unidentified secondary binding site on MukBEF. Such arrangement would not be unlike that for the SMC-ScpAB complex, where the N- and C-terminal domains of the kleisin ScpA associate, respectively, with the neck and head regions of the SMC subunit [Burmann et al., 2013].
The hinge domain of MukB is a rather small compact globule from which two coiled coils emanate at almost a 90° angle to each other [Ku et al., 2010; Li et al., 2010a]. The two halves of the hinge could conceivably be separated if a twisting or peeling stress is applied to it from the head via coiled coils. The coiled coils of MukB are not contiguous but include several interruptions [Weitzel et al., 2011]. In these so called knuckle regions, one of the α-helices of the coiled coil buckles out of the register to produce a short three-helix bundle. These knuckles might serve as bends or flexure points; however, their functional significance is unknown.
Perhaps the most striking feature of the hinge is its association with ParC, a subunit of DNA topoisomerase IV [Hayama and Marians, 2010; Li et al., 2010b]. The interface is located on the outward side of the hinge and is comprised mostly of charged amino acids [Vos et al., 2013]. Although this association is rather weak, it seems essential. Mutations in ParC or MukB that disrupt their interface are detrimental for the cell. In fact, one of the long known temperature sensitive mutations in ParC, G725D, maps close to this interface [Vos et al., 2013]. The interaction is specific to the C-terminal domain (CTD) of ParC; no association with DNA gyrase is detected. Because the interaction is fairly weak, it is unclear whether ParC should be viewed as a part of MukBEF complex or a transient cofactor. The presence of MukB stimulates intramolecular but not intermolecular reactions of topo IV [Hayama et al., 2013] by neutralizing inhibitory effects of CTD on some of the topo IV reactions [Vos et al., 2013]. Specifically, MukB stimulates topo IV-catalyzed relaxation of negatively supercoiled DNA but has no effect on relaxation of positive supercoils or decatenation and promotes DNA knotting [Hayama et al., 2013]. How topo IV affects reactions of MukBEF is yet to be determined.
ATP plays a central role in coordinating MukB interactions with its accessory subunits and DNA [Cui et al., 2008; Woo et al., 2009]. The ATPase site of MukB is homologous to that of other SMC proteins with readily identifiable Walker A and B regions, the signature motif and the D-loop (Fig. 3C). A number of mutations in the ATPase site have been described. Some of them are particularly useful in conformational analysis. In particular, the E1407Q mutation disrupts ATP hydrolysis but not binding and stabilizes the ATP-sandwiched MukB head [Woo et al., 2009]. In contrast, the D1406A mutation is deficient in ATP binding and head engagement and S1366R (in C-motif) precludes head engagement but not ATP binding or association with MukEF [Hirano et al., 2001; Woo et al., 2009]. The Walker A K40I mutation is expected to fail in ATP binding [Hirano et al., 2001; Woo et al., 2009], whereas the K40D S41G was deficient in DNA bridging but not binding [Petrushenko et al., 2010].
A recurrent theme in these studies is that ATP serves as a conformational switch (Fig. 4). ATP binding promotes MukB loading onto DNA, whereas ATP hydrolysis helps to displace it. Accordingly, ATPase deficient mutant E1407Q displayed markedly longer life time within stationary foci in live cells than its ATPase proficient counterpart whereas the ATP binding mutant D1406A failed to associate with foci [Badrinarayanan et al., 2012b]. The effect is even more pronounced with the P. aeruginosa MukBEF-like protein MksBEF. MksB binding to DNA significantly declines in the presence of ATP, whereas ATPase rate is negatively affected by DNA [Petrushenko et al., 2011].
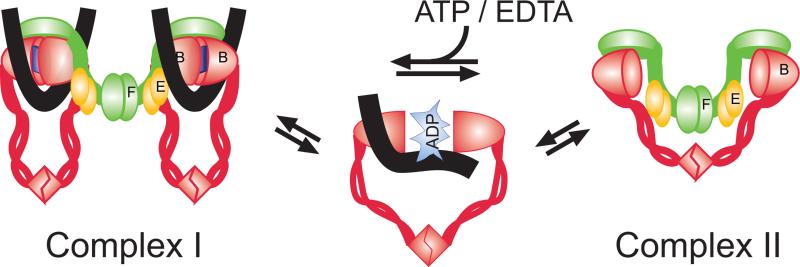
Figure 4.
A schematic view on ATP-mediated regulation of DNA binding and architecture of MukBEF. ATP binding promotes engagement of MukB heads, which creates a high affinity DNA binding site and is accompanied bybinding of DNA (black lines) and formation of the MukB2-Muk(E2F) complex (Complex I). ATP hydrolysis promotes dissociation of the heads (or perhaps another functionally identical conformational change), which disrupts the DNA binding site and forces dissociation of DNA and promotes formation of the MukB2-Muk(E2F)2 complex (Complex II).
In contrast to its functional role, structural perturbations caused by ATP are yet to be established. Clearly, ATP binding and hydrolysis lead to major conformational changes within the dimeric MukB head. What is less clear is whether or not these changes involve disassociation of the dimer. The evidence in favor of disengagement is based upon electron microscopy studies, which reveal both V- and I-shaped MukB molecules [Matoba et al., 2005; Melby et al., 1998] as well as sizing studies of ATPase mutants [Hirano et al., 2001; Woo et al., 2009]. The argument against such disengagement stems from the comparison with the ABC transporters, the only other known class of ABC type ATPases. The ATPase unit of the transporters is believed to stay intact throughout the enzymatic cycle using, instead, the generated energy to agitate the transmembrane domain [Oldham et al., 2008]. This view recently found strong support from crystallography studies of the SMC-like protein Rad50 [Lammens et al., 2011]. ATP hydrolysis was shown to cause a massive transversal motion within the dimeric Rad50 head that was tilting the coiled coils but did not disrupt the interface between the two heads. Such tilting motion could conceivably be occurring in MukB and used to transmit the conformational change along the coiled coils. Whether or not this change leads to conformational rearrangement at the hinge or its disassociation is unknown.
DNA organization: binding, bridging and condensation
MukBEF displays a complex set of DNA reshaping activities that befit its role in global chromosome organization. We review these activities below with the focus on the presumed ability of MukBEF to produce chromatin scaffold while also pointing out observations that do not quite fit into this scheme and suggest an even richer mechanism.
All DNA binding activities of the complex reside in its SMC subunit, MukB, whereas MukEF regulates it and does not bind DNA on its own [Petrushenko et al., 2006b]. MukB binds linear and circular DNA equally well and has only slight preference, if at all, to supercoiled DNA [Petrushenko et al., 2006a]. This underscores the importance of physical association between the protein and DNA. Curiously, MukB binds phage single stranded DNA with high affinity. The significance of this interaction has not been explored. Perhaps it reflects preferred binding to DNA crossings which has been also reported for other SMC proteins [Akhmedov et al., 1998; Kimura and Hirano, 1997]. Fluorescence microscopy and cell biology studies find MukB in complex with the chromosome [Badrinarayanan et al., 2012b; She et al., 2007], which reveals that its primary substrate is double stranded DNA.
When overproduced in living cells [Wang et al., 2006] or mixed in vitro with long phage DNA [Chen et al., 2008], MukB induces DNA condensation. Such condensation cannot be explained by DNA binding alone and implies through-space DNA bridging events. Similarly, condensation of single DNAs stretched using magnetic tweezers involves DNA looping [Cui et al., 2008]. Notably, DNA condensation proceeds slowly on single DNAs, suggesting that this is not the primary mechanism of DNA compaction. In contrast, DNA bridging events between separate DNAs are highly efficient and might be limited by collision frequency [Petrushenko et al., 2010]. Moreover, a single MukB can apply only a weak force, < 0.4 pN, to DNA during condensation, which further argues for the random collision mechanism of DNA condensation.
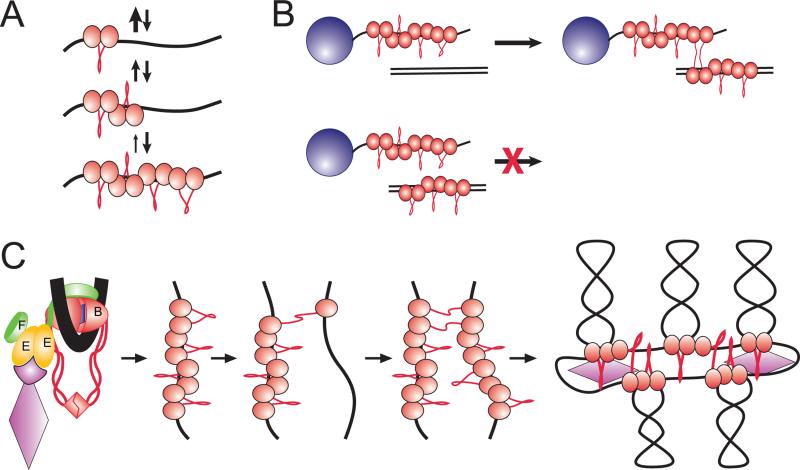
A single MukB has a short life time on DNA. However, it can be stabilized by further protein binding [Cui et al., 2008]. MukB binding to DNA is highly cooperative, with the Hill coefficient of 3. As a result, the protein binds DNA according to the zipper mechanism, much like for DNA annealing, when the slow nucleation step is followed by a fast propagation of the protein cluster (Fig 5A). DNA condensed by such clusters displays significant resilience to applied forces, which might become handy if the protein is indeed involved in chromosome segregation.
Figure 5. DNA scaffolding activity of MukBEF.
(A) Nucleation-propagation mechanism of DNA binding. A single MukB forms only a short-lived complex with DNA. The binding, however, is highly cooperative, and each subsequent bound protein stabilizes the cluster through nearest neighbor interactions.
(B) Bridging occurs between DNA bound MukB clusters and naked DNA.
(C) Proposed mechanism of chromosome organization. The protein is postulated to be recruited to its initial binding site with the help of the regulatory subunits. The protein binds the target site via the clamping mechanism described in (A). Once the clamp is formed, the MukB cluster attempts bridging with randomly colliding DNA segments. Finding a sufficiently long stretch of naked DNA allows stable bridging and further spreading of the scaffold.
The nucleation-propagation mechanism could conceivably serve as a sensor for finding a suitable DNA binding site. Indeed, an ideal scaffold protein is expected not to interfere with information processing enzymes but, rather, stay in the shades and bind only silent DNA. Such silent sites are likely to be only sparsely bound by other proteins and would offer a good landing spot for MukB clusters. If, on the other hand, a need arises for the bound DNA, MukB clusters could be readily removed, owing to low stability of the monomers, one protein at a time with the help of chromatin remodeling activity of DNA helicases or polymerases. In this sense, the protein behaves as a macromolecular clamp that associates with sparsely populated DNAs.
Dynamics of DNA bridging is also consistent with this mechanism. Mixing order experiments using magnetic bead pull-down assay [Petrushenko et al., 2010] revealed that DNA binding and bridging are two distinct events in the reaction cycle of MukB and that bridging occurs most efficiently between a preformed, DNA-bound MukB cluster and a fragment of protein-free DNA (Fig. 5B). Similar to the case of DNA condensation, the formed bridges initially display low stability but are quickly strengthened by propagation of the bridge.
Both DNA binding and bridging can occur in the absence of any nucleotide. Both reactions, however, are accelerated in the presence of ATP. Owing to the high cooperativity of the reaction, the effect is quite dramatic, indicating that ATP can be used to regulate association of MukB with DNA. Importantly, ATP plays a role of a conformational switch here rather than an energy cofactor since effects of ATP could be reproduced by chelating magnesium [Cui et al., 2008]. This is not unlike MukB-MukEF interaction, which is similarly affected by inclusion of ATP and removal of magnesium [Petrushenko et al., 2006b; Woo et al., 2009]. Apparently, the two treatments stabilize the same conformation of MukB. This feature remains a poorly understood curiosity of MukBEF. Perhaps it is somehow related to the high affinity of MukF for another divalent cation, calcium [Yamazoe et al., 1999], and reflects a dormant ability of MukBEF to serve as an environmental sensor.
Taken together, the activities of MukB offer a plausible view of how chromatin scaffold could be organized ([Cui et al., 2008] and Fig. 5C). Owing to its DNA clamping activity, MukB is expected to bind random regions of DNA that do not participate in other cellular activities and relocate to a different place when needed. Given its low intracellular level, MukB may not be able to cover the entire chromosome but probably spreads from the initial binding site to the neighboring DNA loops via a series of bridging events. This binding pattern should minimize non-productive use of the protein that does not result in bridges and offers an excellent example of self-organization in complex systems.
Another attractive aspect of this mechanism is the prediction that chromosomes are condensed without any external force, which could be damaging to chromosome integrity. Indeed, the bridges are predicted to form through capture of randomly colliding protein-free DNA fragments. Thus, by activating MukB through the use of cofactors, the cell can modulate the density of bridges and, therefore, the overall compactness of the chromosome. Once formed, however, the bridges become resilient to external forces and can be used to pull chromosomes apart if needed. This scaffolding mechanism appears generic and might be shared by other chromosome organizing proteins.
Chirality of MukB-DNA interactions
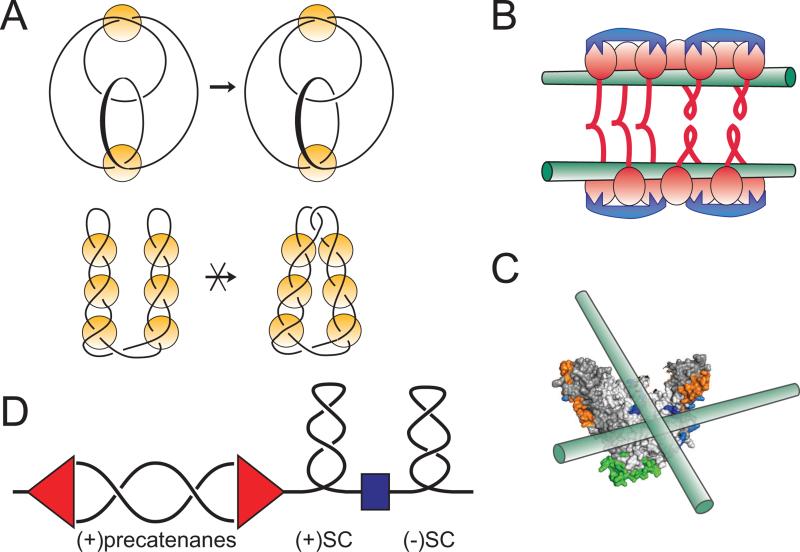
When examined using the topo-2 coupled DNA knotting assay, all tested condensins displayed propensity for stabilization of right handed DNA loops [Kimura et al., 1999; Petrushenko et al., 2006a; Stray et al., 2005]. Importantly, the topology of the generated knots is such that it rules out existence of a plectoneme, i.e. the interwound superhelix between the loops. Such plectoneme spontaneously forms in supercoiled DNA owing to the rules of DNA mechaniscs [Vologodskii and Cozzarelli, 1994] and dramatically alters the topological complexity of the topoisomerase reaction [Wasserman and Cozzarelli, 1991], The fact that such plectonemes were not observed reveals that the proteins do not generate freely diffusing DNA supercoils but rather stabilize right handed DNA crossings at the base of the loops (Fig. 6A). In contrast, the accompanying DNA supercoiling varied from protein to protein. For MukB, the generated supercoils are negative, which is the opposite to what is expected given the right handed chirality of the loops [Petrushenko et al., 2006a]. Apparently, MukB unwinds DNA upon binding, which might explain its high affinity for single stranded DNA.
Figure 6. Chirality of MukB-DNA interactions.
(A) Knotting patterns expected on looped and interwound DNA substrates in topo-2 coupled assay. Knots are often classified according to the minimal number of crossings in a projection of the knot onto a flat surface [Rolfsen, 1976]. The simplest knots, trefoils, contain three irreducible crossings and can exist as either right- or left-handed enantiomers. Condensins promote formation of (+) trefoils, which reveals right-handed DNA looping (top diagram). In contrast, topo-2 reaction on an interwound superhelix would produce twist family knots with multiple irreducible crossings (bottom diagram). Yellow circles depict proteins that capture DNA crossings that were either spontaneously formed in the interwound DNA superhelix (bottom) or comprise the base of protein-induced DNA loops (top).
(B and C) The high preference of MukB for right handed crossings could be explained by chiral binding of DNA to different MukBs from the same cluster (B) or the same MukB head (C).
(C) Pre-catenane formation around replication fork. Progression of the replication fork (red) induces waves of positive supercoiling, whose propagation is limited by domain boundaries (blue). Negative supercoiling ahead of the fork is restored by DNA gyrase. Failure to achieve that results in positive supercoiling which migrates into the replicated region in the form of pre-catenanes. Recruitment of topo IV to this region might facilitate their removal and further progression of DNA replication.
Magnetic tweezers analysis of DNA bridging revealed high preference of MukB for right handed crossings (Fig. 6B and [Petrushenko et al., 2010]). This is consistent with chirality of DNA loops detected using knotting assay and further implicates DNA bridging as the chirality discriminating reaction step. The structural basis of such discrimination is unclear. Indeed, MukB is a homodimer and, therefore, is expected to be achiral or exist as a racemic mixture of enantiomers. This should result, in turn, in a racemic mixture of DNA loops. The proteins could conceivably acquire chirality due to posttranslational modifications present in one of the subunits of the protein. This explanation, however, seems stretched given that overproduced MukB was employed in the experiments and that chirality of DNA looping is conserved across kingdoms of life. A more plausible explanation postulates that the protein somehow senses the double helical nature of the DNA- perhaps via DNA unwinding- and amplifies it by preferentially stabilizing right handed crossings. Yet another possibility envisions binding of two DNAs to the same MukB head on two distinct surfaces of the protein (Fig. 6C). Such binding would be chiral simply by the virtue of stereochemistry of the binding site.
The chirality of condensin-DNA interactions might impose it onto other aspects of higher order chromosome packing and might be ultimately responsible for chiral appearance of the chromosomes as observed using light microscopy [Hochstrasser et al., 1986]. Evolutionary advantage of such anisotropy, however, is unclear. This anisotropy could become mechanistically significant in the light of the recently discovered interaction between MukB and ParC, a subunit of DNA topoisomerase IV [Hayama and Marians, 2010; Li et al., 2010b]. As outlined below, this interaction might help recruit the protein to a unique DNA substrate.
Beyond scaffold: Recruit and be recruited
The chromatin scaffolding activity of MukB does not recuperate all of its intracellular functions nor does it even require a contribution from MukE and MukF. This suggests other roles for the protein besides maintenance of the chromosomal size. A direct test confirmed this conjecture by revealing that MukEF is essential even in cells with constitutively condensed chromosomes [Wang et al., 2006]. This additional function is likely related to focal subcellular localization of MukBEF.
Similar to other bacterial condensins [Lindow et al., 2002; Minnen et al., 2011; Volkov et al., 2003], MukBEF forms clusters in the middle of the nucleoids [Ohsumi et al., 2001; She et al., 2007]. This location changes from the middle of the short cells to the ¼ and ¾ positions in cells with replicating chromosomes. MukE appears to play the primary role in driving such localization as was revealed in a recent random mutagenesis study. All point loss-of-function mutations in MukE disrupted focal localization of MukBEF but not its interaction with DNA ([She et al., 2013] and Fig. 3B). A similar loss of focal localization was observed upon disruption of the linker region in MukF, which tethers MukE to MukB, and inducer-triggered degradation of MukE in live cells [Shin et al., 2009]. Taken together, these data point to a certain separation of functions within MukBEF. While MukB is responsible for DNA organization, MukE ensures its targeting to specific cellular addresses and MukF links the two together and perhaps coordinates their activities by modulating ATP turnover by MukB (Fig. 5C).
Neither MukB nor MukE can form clusters in the absence of the other components of the complex [Ohsumi et al., 2001; She et al., 2007]. Apparently, both the DNA binding activity of MukB and the positioning activity of MukE are required for their formation. Accordingly, MukBEF found in the clusters consists of multimers of the DNA proficient MukB4-Muk(E2F)2 complex [Badrinarayanan et al., 2012b]. In contrast, the diffused fraction of MukBEF has composition MukB2-Muk(E2F)2. The coexistence of the mobile and stationary fractions was also observed for the B. subtilis condensin SMC-ScpAB, in which case the mobile and stationary fractions of condensin was composed of SMC and SMC-ScpAB complexes, respectively [Kleine Borgmann et al., 2013a; Kleine Borgmann et al., 2013b]. It should be noted that intracellular dynamics of MukBEF composition needs further elucidation, since ultracentrifugation studies of E. coli cell extracts did reveal coexistence of MukB2-Muk(E2F)2 and MukB2 complexes but not of MukB4-Muk(E2F) [She et al., 2013].
Subcellular localization of MukBEF is similar to that of oriC, the origin of chromosomal replication [Danilova et al., 2007]. This similarity is functionally significant since depletion of MukB or MukE leads to the loss of oriC localization whereas repletion of the protein restores the foci [Adachi et al., 2005; Badrinarayanan et al., 2012a]. The loss of oriC localization is accompanied by disorganization of the entire chromosome and the failure to relocate the chromosome arms into the opposite halves of the cell. Thus, recruitment of MukBEF towards the origin region is essential for chromosome segregation in E. coli. Of note, repletion of MukBEF leads to rapid restoration of MukBEF foci followed by slow repositioning of the chromosome, suggesting that formation of the protein clusters could be prerequisite to spatial chromosome organization [Badrinarayanan et al., 2012a].
The force responsible for positioning of MukBEF is unknown. Perhaps it involves a possibly indirect interaction with an origin proximal region as was found for SMC-ScpAB. In B. subtilis and Streptococcus pneumoniae, condensin is enriched in the vicinity of the replication origin and this enrichment is dependent on the presence of the parABS operon [Gruber and Errington, 2009; Minnen et al., 2011]. ParABS encodes a bacterial mitotic apparatus, which consists of two proteins, Par and ParB, and a DNA element, ParS, which serves as a ParB binding site (reviewed in [Mierzejewska and Jagura-Burdzy, 2012; Szardenings et al., 2011]). ParABS is located close to oriC of many bacteria but is completely absent from the genome of E. coli K12. SMC binding is enriched in the vicinity of ParS, and moving the parABS operon away was sufficient to relocate SMC towards chromosomal arms as well. Clearly, ParABS recruits SMC towards the origin region. Which system recruits MukBEF to oriC is unknown.
Besides being recruited to its site of action, MukBEF appears to serve as a recruiter itself. A weak interaction between MukB and ParC was recently discovered using co-immunoprecipitation [Hayama and Marians, 2010; Li et al., 2010b]. This interaction is sufficiently selective to discriminate between topo IV and DNA gyrase [Vos et al., 2013] and sufficiently strong to recruit a subset of topo IVs to the quarter positions inside the cell [Nicolas et al., 2014]. Mutations that disrupt MukB-ParC interface adversely affect such localization and E. coli viability [Hayama and Marians, 2010; Li et al., 2010b].
Mechanistic understanding of the benefits of PaC-MukB interaction is still lacking. The primary effect of MukB on topo IV is to derepress its activity on negatively supercoiled DNA [Vos et al., 2013]. It seems tempting to conclude then that MukB helps topo IV in removal of negative supercoils. This idea, however, does not readily explain the findings that suppressors of MukB deficiency map to topoisomerase mutations that increase rather than decrease negative supercoiling [Adachi and Hiraga, 2003; Sawitzke and Austin, 2000]. Among alternative- or, rather, additional- possibilities, we favor the one that envisions recruitment of topo IV to a hazardous, hard-to-find substrate. Such a substrate could be the right handed pre-catenane nodes that might be generated behind replication fork if topoisomerases are unable to keep up with its rapid progression (Fig. 6C and [Postow et al., 2001; Wang, 2009]). Accumulation of such nodes would entangle the daughter chromosomes and could preclude their segregation into opposite halves of the cell potentially leading to a collapse of the segregation machine. Topo IV is the only enzyme capable of resolving such interlinks. Although it performs this reaction efficiently in vitro it could conceivably be slow locating this substrate on its own within the cellular milieu.
In this view, increased negative DNA supercoiling should alleviate the need for topo IV via two mechanisms. First, DNA supercoiling strongly favors DNA unlinking [Alexandrov et al., 1999; Rybenkov et al., 1997] and, thereby, promotes decatenation of newly made sister chromatids. Second, increased negative supercoiling would decrease the rate with which positive supercoils are generated by DNA polymerase and preclude accumulation of pre-catenanes [Alexandrov et al., 1999; Postow et al., 2001].
Concluding remarks
MukBEF functions at the heart of several cellular processes that ensure chromosome replication, segregation and global organization.
MukBEF acts as a mobile, dynamic complex with distinct contribution from its subunits. MukB is responsible for DNA binding and reshaping. MukE is responsible for recruitment of MukBEF to the protein clusters in the middle of nucleoids. MukF physically links MukB and MukE and also coordinates ATP hydrolysis and DNA binding.
MukB displays macromolecular clamping and bridging activities, which, taken together, should result in formation of a chromatin scaffold. These activities could be shared by other chromosome organizing proteins. What makes MukBEF unique is its recruitment and regulation. MukBEF could also serve as a platform for recruitment of additional components. MukB-ParC interaction could be an example of such recruitment.
The nature of MukBEF clusters at the quarter positions remains unknown. A possibility worth exploring is that these clusters are chromosome replication and segregation factories, of which MukBEF is an integral part.
Functional and structural interactions between MukBEF and DNA topoisomerases were reported in numerous, seemingly unrelated studies. These coincidences are difficult to ignore and implicate MukBEF as a key guardian of the topological integrity of the chromosome.
Acknowledgment
This work was supported by Grants 1049755 from the National Science Foundation and AI094124 from the National Institutes of Health.
Footnotes
The authors have no conflict of interests to declare.
References
- Adachi S, Hiraga S. Mutants suppressing novobiocin hypersensitivity of a mukB null mutation. J Bacteriol. 2003;185:3690–3695. doi: 10.1128/JB.185.13.3690-3695.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi S, Kohiyama M, Onogi T, Hiraga S. Localization of replication forks in wild-type and mukB mutant cells of Escherichia coli. Mol Genet Genomics. 2005;274:264–271. doi: 10.1007/s00438-005-0023-6. [DOI] [PubMed] [Google Scholar]
- Akhmedov AT, Frei C, Tsai-Pflugfelder M, Kemper B, Gasser SM, Jessberger R. Structural maintenance of chromosomes protein C-terminal domains bind preferentially to DNA with secondary structure. J Biol Chem. 1998;273:24088–24094. doi: 10.1074/jbc.273.37.24088. [DOI] [PubMed] [Google Scholar]
- Alexandrov AI, Cozzarelli NR, Holmes VF, Khodursky AB, Peter BJ, Postow L, Rybenkov V, Vologodskii AV. Mechanisms of separation of the complementary strands of DNA during replication. Genetica. 1999;106:131–140. doi: 10.1023/a:1003749416449. [DOI] [PubMed] [Google Scholar]
- Badrinarayanan A, Lesterlin C, Reyes-Lamothe R, Sherratt D. The Escherichia coli SMC complex, MukBEF, shapes nucleoid organization independently of DNA replication. Journal of bacteriology. 2012a;194:4669–4676. doi: 10.1128/JB.00957-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badrinarayanan A, Reyes-Lamothe R, Uphoff S, Leake MC, Sherratt DJ. In vivo architecture and action of bacterial structural maintenance of chromosome proteins. Science. 2012b;338:528–531. doi: 10.1126/science.1227126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton RA, Lin DC, Grossman AD. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 1998;12:1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning DF, Grainger DC, Busby SJ. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr Opin Microbiol. 2010;13:773–780. doi: 10.1016/j.mib.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Burmann F, Shin HC, Basquin J, Soh YM, Gimenez-Oya V, Kim YG, Oh BH, Gruber S. An asymmetric SMC-kleisin bridge in prokaryotic condensin. Nature structural & molecular biology. 2013;20:371–379. doi: 10.1038/nsmb.2488. [DOI] [PubMed] [Google Scholar]
- Chen N, Zinchenko AA, Yoshikawa Y, Araki S, Adachi S, Yamazoe M, Hiraga S, Yoshikawa K. ATP-induced shrinkage of DNA with MukB protein and the MukBEF complex of Escherichia coli. J Bacteriol. 2008;190:3731–3737. doi: 10.1128/JB.01863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbe N, Heck MM. The evolution of SMC proteins: phylogenetic analysis and structural implications. Mol Biol Evol. 2004;21:332–347. doi: 10.1093/molbev/msh023. [DOI] [PubMed] [Google Scholar]
- Connelly JC, de Leau ES, Leach DR. Nucleolytic processing of a protein-bound DNA end by the E. coli SbcCD (MR) complex. DNA Repair (Amst) 2003;2:795–807. doi: 10.1016/s1568-7864(03)00063-6. [DOI] [PubMed] [Google Scholar]
- Cui Y, Petrushenko ZM, Rybenkov VV. MukB acts as a macromolecular clamp in DNA condensation. Nat Struct Mol Biol. 2008;15:411–418. doi: 10.1038/nsmb.1410. [DOI] [PubMed] [Google Scholar]
- Danilova O, Reyes-Lamothe R, Pinskaya M, Sherratt D, Possoz C. MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves. Mol Microbiol. 2007;65:1485–1492. doi: 10.1111/j.1365-2958.2007.05881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S, Stein RA, Higgins NP. Transcription-induced barriers to supercoil diffusion in the Salmonella typhimurium chromosome. Proc Natl Acad Sci U S A. 2004;101:3398–3403. doi: 10.1073/pnas.0307550101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Halligan B, Cooke CA, Heck MM, Liu LF. Topoisomerase II is a structural component of mitotic chromosome scaffolds. The Journal of cell biology. 1985;100:1706–1715. doi: 10.1083/jcb.100.5.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennell-Fezzie R, Gradia SD, Akey D, Berger JM. The MukF subunit of Escherichia coli condensin: architecture and functional relationship to kleisins. Embo J. 2005;24:1921–1930. doi: 10.1038/sj.emboj.7600680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloyd M, Ghirlando R, Guarne A. The role of MukE in assembling a functional MukBEF complex. J Mol Biol. 2011;412:578–590. doi: 10.1016/j.jmb.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloyd M, Ghirlando R, Matthews LA, Guarne A. MukE and MukF form two distinct high affinity complexes. The Journal of biological chemistry. 2007;282:14373–14378. doi: 10.1074/jbc.M701402200. [DOI] [PubMed] [Google Scholar]
- Graumann PL, Knust T. Dynamics of the bacterial SMC complex and SMC-like proteins involved in DNA repair. Chromosome Res. 2009;17:265–275. doi: 10.1007/s10577-008-9014-x. [DOI] [PubMed] [Google Scholar]
- Gruber S. MukBEF on the march: taking over chromosome organization in bacteria? Mol Microbiol. 2011;81:855–859. doi: 10.1111/j.1365-2958.2011.07764.x. [DOI] [PubMed] [Google Scholar]
- Gruber S, Errington J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 2009;137:685–696. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- Hayama R, Bahng S, Karasu ME, Marians KJ. The MukB-ParC interaction affects the intramolecular, not intermolecular, activities of topoisomerase IV. The Journal of biological chemistry. 2013;288:7653–7661. doi: 10.1074/jbc.M112.418087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Marians KJ. Physical and functional interaction between the condensin MukB and the decatenase topoisomerase IV in Escherichia coli. Proc Natl Acad Sci U S A. 2010;107:18826–18831. doi: 10.1073/pnas.1008140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins NP, Yang X, Fu Q, Roth J. Surveying a supercoil domain by using the gamma-delta resolution system in Salmonella typhimurium. Journal of Bacteriology. 1996;178:2825–2835. doi: 10.1128/jb.178.10.2825-2835.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S. Dynamic localization of bacterial and plasmid chromosomes. Annu Rev Genet. 2000;34:21–59. doi: 10.1146/annurev.genet.34.1.21. [DOI] [PubMed] [Google Scholar]
- Hiraga S, Niki H, Ogura T, Ichinose C, Mori H, Ezaki B, Jaffe A. Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J Bacteriol. 1989;171:1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Anderson DE, Erickson HP, Hirano T. Bimodal activation of SMC ATPase by intraand inter-molecular interactions. Embo J. 2001;20:3238–3250. doi: 10.1093/emboj/20.12.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ. A heterodimeric coiled-coil protein required for mitotic chromsome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M, Mathog D, Gruenbaum Y, Saumweber H, Sedat JW. Spatial organization of chromosomes in the salivary gland nuclei of Drosophila melanogaster. The Journal of cell biology. 1986;102:112–123. doi: 10.1083/jcb.102.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Shapiro L. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc Natl Acad Sci U S A. 1999;96:10661–10666. doi: 10.1073/pnas.96.19.10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RC, Johnson LM, Schmidt JW, Gardner JF. Major nucleoid proteins in the structure and function of the Escherichia coli chromosome. In: Higgins NP, editor. The bacterial chromosome. ASM Press; Washington, D. C.: 2005. pp. 65–132. [Google Scholar]
- Kavenoff R, Bowen BC. Electron Microscopy of Membrane-Free Folded Chromosomes from Escherichia coli. Chromosoma. 1976;59:89–101. doi: 10.1007/BF00328479. [DOI] [PubMed] [Google Scholar]
- Kido M, Yamanaka K, Mitani T, Niki H, Ogura T, Hiraga S. RNase E polypeptides lacking a carboxyl-terminal half suppress a mukB mutation in Escherichia coli. J Bacteriol. 1996;178:3917–3925. doi: 10.1128/jb.178.13.3917-3925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- Kimura K, Rybenkov V, Crisona N, Hirano T, Cozzarelli N. 13S Condensin Actively Reconfigures DNA by Introducing Global Positive Writhe: Implications for Chromosome Condensation. Cell. 1999;98:239–248. doi: 10.1016/s0092-8674(00)81018-1. [DOI] [PubMed] [Google Scholar]
- Kleine Borgmann LA, Hummel H, Ulbrich MH, Graumann PL. SMC condensation centers in Bacillus subtilis are dynamic structures. Journal of bacteriology. 2013a;195:2136–2145. doi: 10.1128/JB.02097-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine Borgmann LA, Ries J, Ewers H, Ulbrich MH, Graumann PL. The bacterial SMC complex displays two distinct modes of interaction with the chromosome. Cell Rep. 2013b;3:1483–1492. doi: 10.1016/j.celrep.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Ku B, Lim JH, Shin HC, Shin SY, Oh BH. Crystal structure of the MukB hinge domain with coiled-coil stretches and its functional implications. Proteins. 2010;78:1483–1490. doi: 10.1002/prot.22664. [DOI] [PubMed] [Google Scholar]
- Lammens K, Bemeleit DJ, Mockel C, Clausing E, Schele A, Hartung S, Schiller CB, Lucas M, Angermuller C, Soding J, Strasser K, Hopfner KP. The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell. 2011;145:54–66. doi: 10.1016/j.cell.2011.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene SD, Giovan SM, Hanke A, Shoura MJ. The thermodynamics of DNA loop formation, from J to Z. Biochem Soc Trans. 2013;41:513–518. doi: 10.1042/BST20120324. [DOI] [PubMed] [Google Scholar]
- Li Y, Schoeffler AJ, Berger JM, Oakley MG. The crystal structure of the hinge domain of the Escherichia coli structural maintenance of chromosomes protein MukB. J Mol Biol. 2010a;395:11–19. doi: 10.1016/j.jmb.2009.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Stewart NK, Berger AJ, Vos S, Schoeffler AJ, Berger JM, Chait BT, Oakley MG. Escherichia coli condensin MukB stimulates topoisomerase IV activity by a direct physical interaction. Proc Natl Acad Sci U S A. 2010b;107:18832–18837. doi: 10.1073/pnas.1008678107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow JC, Kuwano M, Moriya S, Grossman AD. Subcellular localization of the Bacillus subtilis structural maintenance of chromosomes (SMC) protein. Mol Microbiol. 2002;46:997–1009. doi: 10.1046/j.1365-2958.2002.03235.x. [DOI] [PubMed] [Google Scholar]
- Luijsterburg MS, Noom MC, Wuite GJ, Dame RT. The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J Struct Biol. 2006;156:262–272. doi: 10.1016/j.jsb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Marko J, Trun N. Architecture of a bacterial chromosome. ASM News. 1998;64:276–283. [Google Scholar]
- Mascarenhas J, Soppa J, Strunnikov AV, Graumann PL. Cell cycle-dependent localization of two novel prokaryotic chromosome segregation and condensation proteins in Bacillus subtilis that interact with SMC protein. Embo J. 2002;21:3108–3118. doi: 10.1093/emboj/cdf314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba K, Yamazoe M, Mayanagi K, Morikawa K, Hiraga S. Comparison of MukB homodimer versus MukBEF complex molecular architectures by electron microscopy reveals a higher-order multimerization. Biochem Biophys Res Commun. 2005;333:694–702. doi: 10.1016/j.bbrc.2005.05.163. [DOI] [PubMed] [Google Scholar]
- Melby TE, Ciampaglio CN, Briscoe G, Erickson HP. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J Cell Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzejewska J, Jagura-Burdzy G. Prokaryotic ParA-ParB-parS system links bacterial chromosome segregation with the cell cycle. Plasmid. 2012;67:1–14. doi: 10.1016/j.plasmid.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Minnen A, Attaiech L, Thon M, Gruber S, Veening JW. SMC is recruited to oriC by ParB and promotes chromosome segregation in Streptococcus pneumoniae. Mol Microbiol. 2011;81:676–688. doi: 10.1111/j.1365-2958.2011.07722.x. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J, Mirault ME, Laemmli UK. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984;39:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Nicolas E, Upton AL, Uphoff S, Henry O, Badrinarayanan A, Sherratt D. The SMC complex MukBEF recruits topoisomerase IV to the origin of replication region in live Escherichia coli. MBio. 2014;5:e01001–01013. doi: 10.1128/mBio.01001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H, Imamura R, Kitaoka M, Yamanaka K, Ogura T, Hiraga S. E.coli MukB protein involved in chromosome partition forms a homodimer with a rod-and-hinge structure having DNA binding and ATP/GTP binding activities. Embo J. 1992;11:5101–5109. doi: 10.1002/j.1460-2075.1992.tb05617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H, Jaffe A, Imamura R, Ogura T, Hiraga S. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 1991;10:183–193. doi: 10.1002/j.1460-2075.1991.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi K, Yamazoe M, Hiraga S. Different localization of SeqA-bound nascent DNA clusters and MukF-MukE- MukB complex in Escherichia coli cells. Mol Microbiol. 2001;40:835–845. doi: 10.1046/j.1365-2958.2001.02447.x. [DOI] [PubMed] [Google Scholar]
- Oldham ML, Davidson AL, Chen J. Structural insights into ABC transporter mechanism. Current opinion in structural biology. 2008;18:726–733. doi: 10.1016/j.sbi.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson JR, Laemmli UK. The structure of histone-depleted metaphase chromosomes. Cell. 1977;12:817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Petrushenko ZM, Cui Y, She W, Rybenkov VV. Mechanics of DNA bridging by bacterial condensin MukBEF in vitro and in singulo. Embo J. 2010;29:1126–1135. doi: 10.1038/emboj.2009.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrushenko ZM, Lai CH, Rai R, Rybenkov VV. DNA reshaping by MukB. Right-handed knotting, left-handed supercoiling. J Biol Chem. 2006a;281:4606–4615. doi: 10.1074/jbc.M504754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrushenko ZM, Lai CH, Rybenkov VV. Antagonistic interactions of kleisins and DNA with bacterial condensin MukB. J Biol Chem. 2006b;281:34208–34217. doi: 10.1074/jbc.M606723200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrushenko ZM, She W, Rybenkov VV. A new family of bacterial condensins. Mol Microbiol. 2011;81:881–896. doi: 10.1111/j.1365-2958.2011.07763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettijohn DE. The nucleoid. In: Neidhardt FC, editor. Escherichia coli and Salmonella typhimurium. ASM Press; Washington: 1996. [Google Scholar]
- Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR. Topological challenges to DNA replication: conformations at the fork. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8219–8226. doi: 10.1073/pnas.111006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Hardy CD, Arsuaga J, Cozzarelli NR. Topological domain structure of the Escherichia coli chromosome. Genes Dev. 2004;18:1766–1779. doi: 10.1101/gad.1207504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Lamothe R, Nicolas E, Sherratt DJ. Chromosome replication and segregation in bacteria. Annual review of genetics. 2012;46:121–143. doi: 10.1146/annurev-genet-110711-155421. [DOI] [PubMed] [Google Scholar]
- Reyes ED, Patidar PL, Uranga LA, Bortoletto AS, Lusetti SL. RecN is a cohesin-like protein that stimulates intermolecular DNA interactions in vitro. The Journal of biological chemistry. 2010;285:16521–16529. doi: 10.1074/jbc.M110.119164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimsky S, Travers A. Pervasive regulation of nucleoid structure and function by nucleoid-associated proteins. Curr Opin Microbiol. 2011;14:136–141. doi: 10.1016/j.mib.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Rolfsen D. Knots and Links. Publish or Perish, Inc.; Berkeley, CA: 1976. [Google Scholar]
- Rybenkov VV. Towards the architecture of the chromosomal architects. Nat Struct Mol Biol. 2009;16:104–105. doi: 10.1038/nsmb0209-104. [DOI] [PubMed] [Google Scholar]
- Rybenkov VV, Vologodskii AV, Cozzarelli NR. The effect of ionic conditions on the conformations of supercoiled DNA. II. Equilibrium catenation. J Mol Biol. 1997;267:312–323. doi: 10.1006/jmbi.1996.0877. [DOI] [PubMed] [Google Scholar]
- Saitoh N, Goldberg I, Earnshaw WC. The SMC proteins and the coming of age of the chromosome scaffold hypothesis. Bioessays. 1995;17:759–766. doi: 10.1002/bies.950170905. [DOI] [PubMed] [Google Scholar]
- Saitoh N, Goldberg IG, Wood ER, Earnshaw WC. ScII: an abundant chromosome scaffold protein is a member of a family of putative ATPases with an unusual predicted tertiary structure. J Cell Biol. 1994;127:303–318. doi: 10.1083/jcb.127.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawitzke JA, Austin S. Suppression of chromosome segregation defects of Escherichia coli muk mutants by mutations in topoisomerase I. Proc Natl Acad Sci U S A. 2000;97:1671–1676. doi: 10.1073/pnas.030528397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman JB, Stasiak A. A topological view of the replicon. EMBO reports. 2004;5:256–261. doi: 10.1038/sj.embor.7400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She W, Mordukhova E, Zhao H, Petrushenko ZM, Rybenkov VV. Mutational analysis of MukE reveals its role in focal subcellular localization of MukBEF. Mol Microbiol. 2013;87:539–552. doi: 10.1111/mmi.12112. [DOI] [PubMed] [Google Scholar]
- She W, Wang Q, Mordukhova EA, Rybenkov VV. MukEF is required for stable association of MukB with the chromosome. J Bacteriol. 2007;189:7062–7068. doi: 10.1128/JB.00770-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HC, Lim JH, Woo JS, Oh BH. Focal localization of MukBEF condensin on the chromosome requires the flexible linker region of MukF. FEBS J. 2009;276:5101–5110. doi: 10.1111/j.1742-4658.2009.07206.x. [DOI] [PubMed] [Google Scholar]
- Sinden RR, Pettijohn DE. Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc Natl Acad Sci U S A. 1981;78:224–228. doi: 10.1073/pnas.78.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppa J, Kobayashi K, Noirot-Gros MF, Oesterhelt D, Ehrlich SD, Dervyn E, Ogasawara N, Moriya S. Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB. Mol Microbiol. 2002;45:59–71. doi: 10.1046/j.1365-2958.2002.03012.x. [DOI] [PubMed] [Google Scholar]
- Stray JE, Crisona NJ, Belotserkovskii BP, Lindsley JE, Cozzarelli NR. The Saccharomyces cerevisiae SMC2/4 condensin compacts DNA into (+) chiral structures without net supercoiling. J Biol Chem. 2005;280:34723–34734. doi: 10.1074/jbc.M506589200. [DOI] [PubMed] [Google Scholar]
- Strunnikov A, Hogan E, Koshland D. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev. 1995;9:587–599. doi: 10.1101/gad.9.5.587. [DOI] [PubMed] [Google Scholar]
- Strunnikov AV, Larionov VL, Koshland D. SMC1: an essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J Cell Biol. 1993;123:1635–1648. doi: 10.1083/jcb.123.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szardenings F, Guymer D, Gerdes K. ParA ATPases can move and position DNA and subcellular structures. Current opinion in microbiology. 2011;14:712–718. doi: 10.1016/j.mib.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Volkov A, Mascarenhas J, Andrei-Selmer C, Ulrich HD, Graumann PL. A prokaryotic condensin/cohesin-like complex can actively compact chromosomes from a single position on the nucleoid and binds to DNA as a ring-like structure. Mol Cell Biol. 2003;23:5638–5650. doi: 10.1128/MCB.23.16.5638-5650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vologodskii AV, Cozzarelli NR. Conformational and thermodynamic properties of supercoiled DNA. Annu Rev Biophys Biomol Struct. 1994;23:609–643. doi: 10.1146/annurev.bb.23.060194.003141. [DOI] [PubMed] [Google Scholar]
- Vos SM, Stewart NK, Oakley MG, Berger JM. Structural basis for the MukB-topoisomerase IV interaction and its functional implications in vivo. The EMBO journal. 2013;32:2950–2962. doi: 10.1038/emboj.2013.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC. A journey in the world of DNA rings and beyond. Annual review of biochemistry. 2009;78:31–54. doi: 10.1146/annurev.biochem.78.030107.090101. [DOI] [PubMed] [Google Scholar]
- Wang Q, Mordukhova EA, Edwards AL, Rybenkov VV. Chromosome condensation in the absence of the non-SMC subunits of MukBEF. J Bacteriol. 2006;188:4431–4441. doi: 10.1128/JB.00313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman SA, Cozzarelli NR. Supercoiled DNA-directed knotting by T4 topoisomerase. J Biol Chem. 1991;266:20567–20573. [PubMed] [Google Scholar]
- Weitao T, Nordstrom K, Dasgupta S. Mutual suppression of mukB and seqA phenotypes might arise from their opposing influences on the Escherichia coli nucleoid structure. Mol Microbiol. 1999;34:157–168. doi: 10.1046/j.1365-2958.1999.01589.x. [DOI] [PubMed] [Google Scholar]
- Weitzel CS, Waldman VM, Graham TA, Oakley MG. A repeated coiled-coil interruption in the Escherichia coli condensin MukB. Journal of molecular biology. 2011;414:578–595. doi: 10.1016/j.jmb.2011.10.028. [DOI] [PubMed] [Google Scholar]
- Woo JS, Lim JH, Shin HC, Suh MK, Ku B, Lee KH, Joo K, Robinson H, Lee J, Park SY, Ha NC, Oh BH. Structural Studies of a Bacterial Condensin Complex Reveal ATP-Dependent Disruption of Intersubunit Interactions. Cell. 2009;136:85–96. doi: 10.1016/j.cell.2008.10.050. [DOI] [PubMed] [Google Scholar]
- Worcel A, Burgi E. On the structure of the folded chromosome of Escherichia coli. J Mol Biol. 1972;71:127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]
- Yamanaka K, Ogura T, Niki H, Hiraga S. Characterization of the smtA gene encoding an S-adenosylmethionine- dependent methyltransferase of Escherichia coli. FEMS Microbiol Lett. 1995;133:59–63. doi: 10.1111/j.1574-6968.1995.tb07861.x. [DOI] [PubMed] [Google Scholar]
- Yamanaka K, Ogura T, Niki H, Hiraga S. Identification of two new genes, mukE and mukF, involved in chromosome partitioning in Escherichia coli. Mol Gen Genet. 1996;250:241–251. doi: 10.1007/BF02174381. [DOI] [PubMed] [Google Scholar]
- Yamazoe M, Onogi T, Sunako Y, Niki H, Yamanaka K, Ichimura T, Hiraga S. Complex formation of MukB, MukE and MukF proteins involved in chromosome partitioning in Escherichia coli. EMBO J. 1999;18:5873–5884. doi: 10.1093/emboj/18.21.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]