Abstract
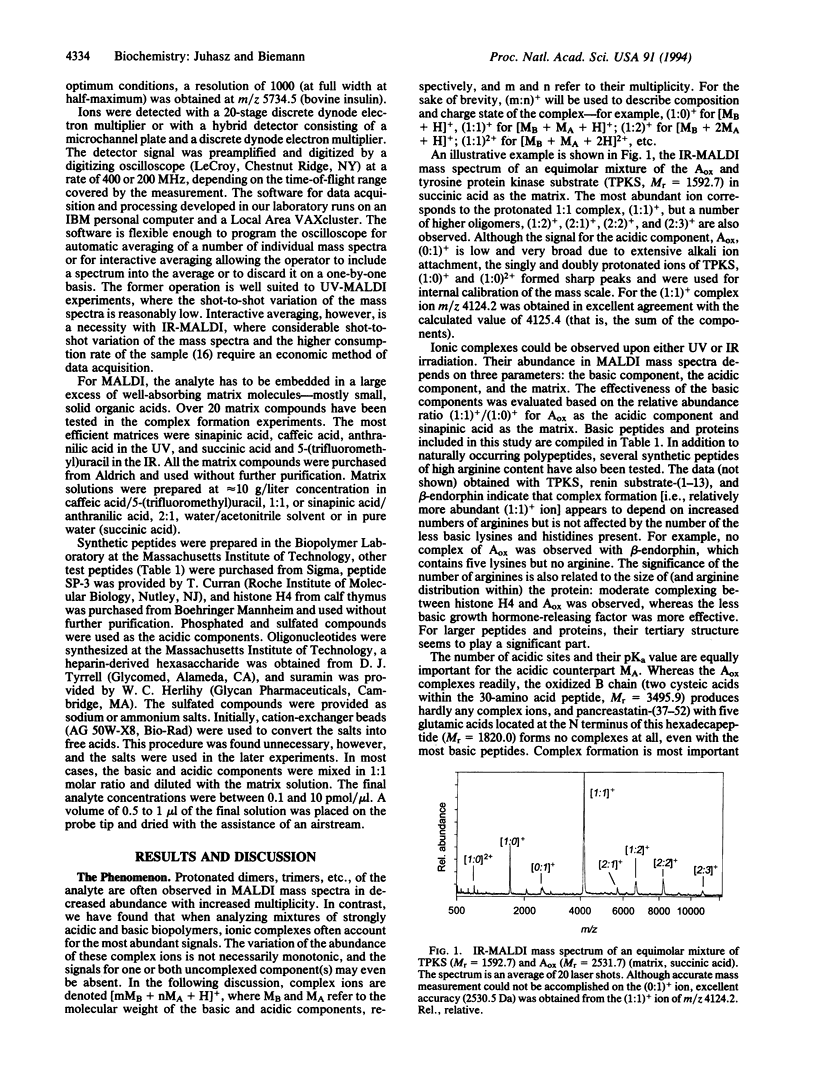
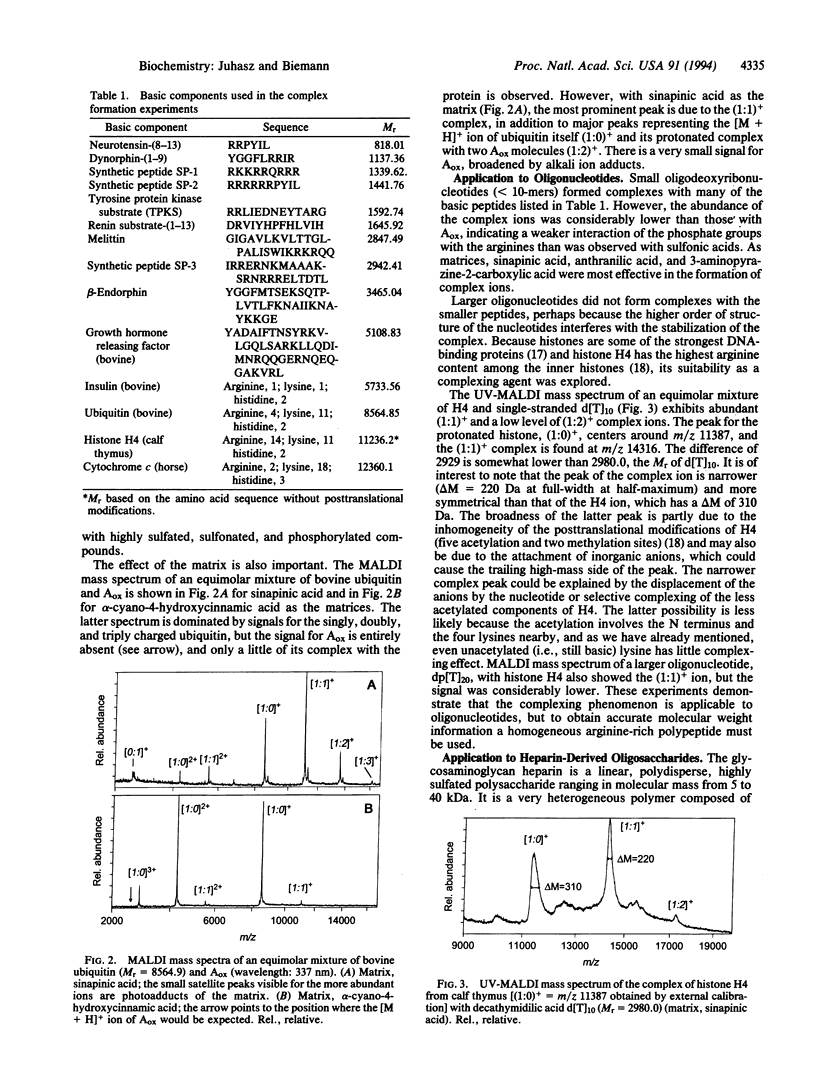
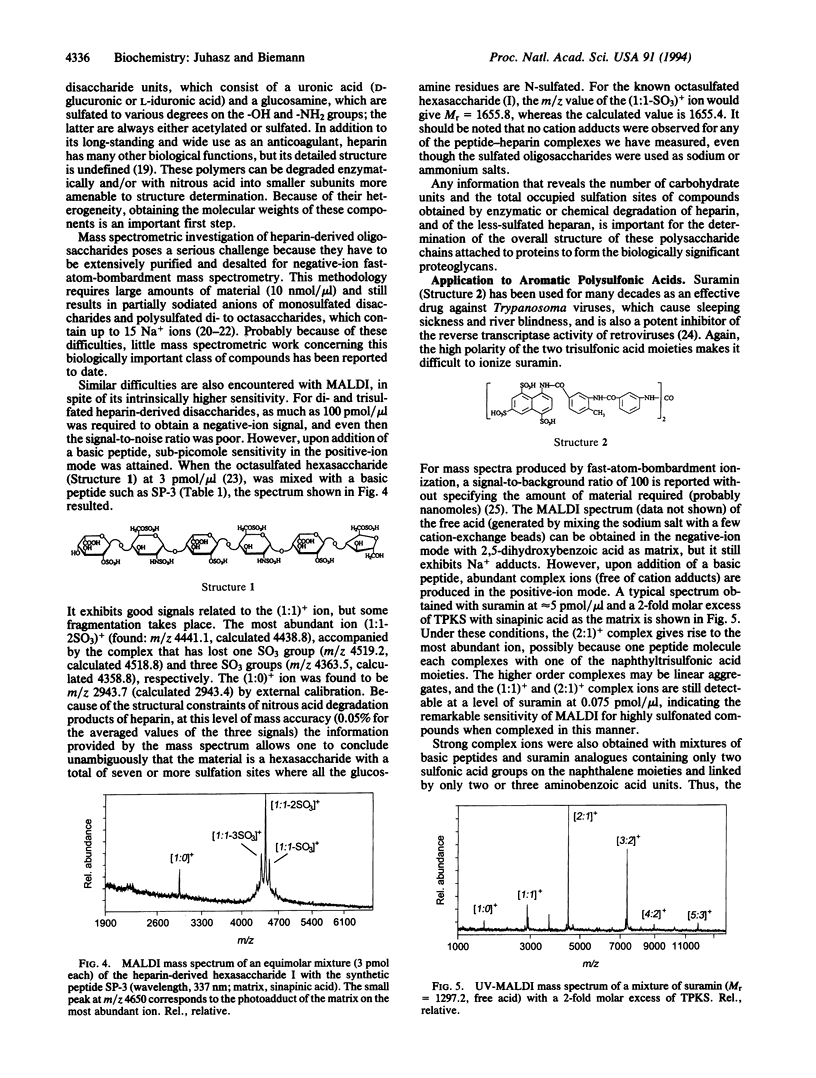
Highly acidic compounds that are difficult to ionize by matrix-assisted laser desorption ionization give excellent spectra when mixed with a basic peptide or protein to form a noncovalent complex. This phenomenon makes it possible to determine the molecular weights of polysulfated, -sulfonated, and -phosphorylated biomolecules such as cysteic acid-containing peptides, oligonucleotides, heparin-derived oligosaccharides, and suramin (a drug containing two trisulfonated naphthalene moieties). Peptides and small proteins rich in arginine were used as the basic components. The extent of complex formation correlates with the number of phosphate and sulfate groups in the acidic component and with the number of arginines in the basic component. Neither the acidic amino acid residue aspartic and glutamic acid nor the basic lysine and histidine contribute to complex formation. For oligonucleotides, histone H4 was found to be the best complexing agent investigated. The analytical utility of the complex formation is demonstrated by the molecular-mass determination of acidic compounds from 500 to 6000 Da at the picomole or sub-picomole level with an accuracy of +/- 0.1% or better and by the absence of alkali cation adducts.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beavis R. C., Chait B. T. High-accuracy molecular mass determination of proteins using matrix-assisted laser desorption mass spectrometry. Anal Chem. 1990 Sep 1;62(17):1836–1840. doi: 10.1021/ac00216a020. [DOI] [PubMed] [Google Scholar]
- Brown R. S., Gilfrich N. L. Optimizing signal and mass resolution for matrix-assisted laser desorption utilizing a linear time-of-flight mass spectrometer. Rapid Commun Mass Spectrom. 1992 Nov;6(11):697–701. doi: 10.1002/rcm.1290061113. [DOI] [PubMed] [Google Scholar]
- Chait B. T., Wang R., Beavis R. C., Kent S. B. Protein ladder sequencing. Science. 1993 Oct 1;262(5130):89–92. doi: 10.1126/science.8211132. [DOI] [PubMed] [Google Scholar]
- De Clercq E. Suramin: a potent inhibitor of the reverse transcriptase of RNA tumor viruses. Cancer Lett. 1979 Nov;8(1):9–22. doi: 10.1016/0304-3835(79)90017-x. [DOI] [PubMed] [Google Scholar]
- Funk W. D., Pak D. T., Karas R. H., Wright W. E., Shay J. W. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol. 1992 Jun;12(6):2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- Jiménez C. R., van Veelen P. A., Li K. W., Wildering W. C., Geraerts W. P., Tjaden U. R., van der Greef J. Neuropeptide expression and processing as revealed by direct matrix-assisted laser desorption ionization mass spectrometry of single neurons. J Neurochem. 1994 Jan;62(1):404–407. doi: 10.1046/j.1471-4159.1994.62010404.x. [DOI] [PubMed] [Google Scholar]
- Juhasz P., Costello C. E. Generation of large radical ions from oligometallocenes by matrix-assisted laser desorption ionization. Rapid Commun Mass Spectrom. 1993 May;7(5):343–351. doi: 10.1002/rcm.1290070508. [DOI] [PubMed] [Google Scholar]
- Linhardt R. J., Wang H. M., Loganathan D., Lamb D. J., Mallis L. M. Analysis of glycosaminoglycan-derived oligosaccharides using fast-atom-bombardment mass-spectrometry. Carbohydr Res. 1992 Feb 17;225(1):137–145. doi: 10.1016/0008-6215(92)80045-3. [DOI] [PubMed] [Google Scholar]
- Mallis L. M., Wang H. M., Loganathan D., Linhardt R. J. Sequence analysis of highly sulfated, heparin-derived oligosaccharides using fast atom bombardment mass spectrometry. Anal Chem. 1989 Jul 1;61(13):1453–1458. doi: 10.1021/ac00188a030. [DOI] [PubMed] [Google Scholar]
- Nordhoff E., Ingendoh A., Cramer R., Overberg A., Stahl B., Karas M., Hillenkamp F., Crain P. F. Matrix-assisted laser desorption/ionization mass spectrometry of nucleic acids with wavelengths in the ultraviolet and infrared. Rapid Commun Mass Spectrom. 1992 Dec;6(12):771–776. doi: 10.1002/rcm.1290061212. [DOI] [PubMed] [Google Scholar]
- Reinhold V. N., Carr S. A., Green B. N., Petitou M., Choay J., Sinaÿ P. Structural characterization of sulfated glycosaminoglycans by fast atom bombardment mass spectrometry: application to heparin fragments prepared by chemical synthesis. Carbohydr Res. 1987 Apr 1;161(2):305–313. doi: 10.1016/s0008-6215(00)90088-0. [DOI] [PubMed] [Google Scholar]
- Turner B. M., O'Neill L. P., Allan I. M. Histone H4 acetylation in human cells. Frequency of acetylation at different sites defined by immunolabeling with site-specific antibodies. FEBS Lett. 1989 Aug 14;253(1-2):141–145. doi: 10.1016/0014-5793(89)80947-0. [DOI] [PubMed] [Google Scholar]
- Tyrrell D. J., Ishihara M., Rao N., Horne A., Kiefer M. C., Stauber G. B., Lam L. H., Stack R. J. Structure and biological activities of a heparin-derived hexasaccharide with high affinity for basic fibroblast growth factor. J Biol Chem. 1993 Mar 5;268(7):4684–4689. [PubMed] [Google Scholar]
- Wu K. J., Steding A., Becker C. H. Matrix-assisted laser desorption time-of-flight mass spectrometry of oligonucleotides using 3-hydroxypicolinic acid as an ultraviolet-sensitive matrix. Rapid Commun Mass Spectrom. 1993 Feb;7(2):142–146. doi: 10.1002/rcm.1290070206. [DOI] [PubMed] [Google Scholar]


