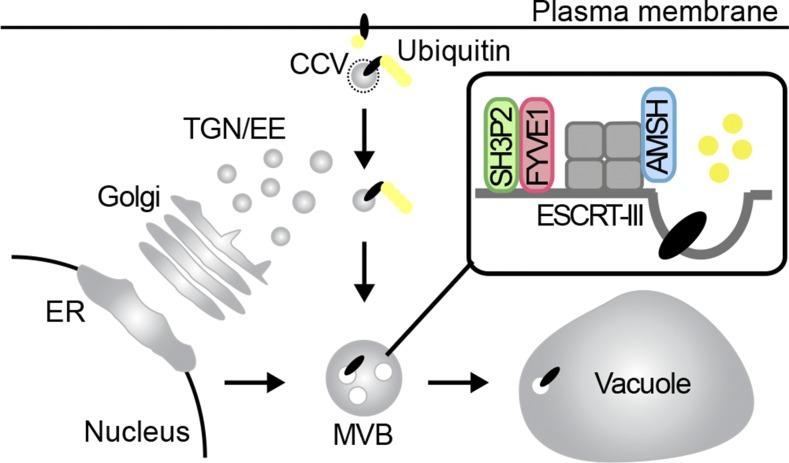
A phospholipid-binding protein regulates intracellular trafficking and vacuole formation.
Abstract
The plant vacuole is a central organelle that is involved in various biological processes throughout the plant life cycle. Elucidating the mechanism of vacuole biogenesis and maintenance is thus the basis for our understanding of these processes. Proper formation of the vacuole has been shown to depend on the intracellular membrane trafficking pathway. Although several mutants with altered vacuole morphology have been characterized in the past, the molecular basis for plant vacuole biogenesis has yet to be fully elucidated. With the aim to identify key factors that are essential for vacuole biogenesis, we performed a forward genetics screen in Arabidopsis (Arabidopsis thaliana) and isolated mutants with altered vacuole morphology. The vacuolar fusion defective1 (vfd1) mutant shows seedling lethality and defects in central vacuole formation. VFD1 encodes a Fab1, YOTB, Vac1, and EEA1 (FYVE) domain-containing protein, FYVE1, that has been implicated in intracellular trafficking. FYVE1 localizes on late endosomes and interacts with Src homology-3 domain-containing proteins. Mutants of FYVE1 are defective in ubiquitin-mediated protein degradation, vacuolar transport, and autophagy. Altogether, our results show that FYVE1 is essential for plant growth and development and place FYVE1 as a key regulator of intracellular trafficking and vacuole biogenesis.
The plant vacuole is the largest organelle in a plant cell in which proteins, metabolites, and ions can be stored or sequestered. The vacuole is essential for plant development and growth and is directly or indirectly involved in various biotic and abiotic stress responses (Zhang et al., 2014). The vacuole is also the central organelle for degradation of endocytic and autophagic protein substrates through the activity of vacuolar proteases. In both degradation pathways, substrates are transported to the vacuole by intracellular membrane trafficking. In endocytic degradation, plasma membrane-localized proteins are targeted to the vacuole for degradation by endosomes (Reyes et al., 2011). This process is important, among others, to control the abundance of plasma membrane receptors and thus downstream signaling events. Autophagic degradation is mainly involved in nutrient recycling. During this process, cytosolic proteins and organelles are either selectively or nonselectively transported by double membrane autophagosomes to the vacuole to be degraded (Liu and Bassham, 2012). Vacuolar transport defines an intracellular transport pathway by which de novo synthesized proteins or metabolic compounds are carried to the vacuole by vesicle transport (Drakakaki and Dandekar, 2013).
In yeast (Saccharomyces cerevisiae), forward genetic screens aimed at finding mutants with defective vacuolar transport or vacuolar morphology have identified more than 30 VACUOLAR PROTEIN SORTING (VPS) and VACUOLAR MORPHOLOGY (VAM) genes (Banta et al., 1988; Raymond et al., 1992; Wada and Anraku, 1992). Closer analyses have shown that many of these mutants have defects both in protein sorting and in vacuole biogenesis, suggesting a close link between these processes. vps and vam mutants were classified into six mutant classes according to their phenotypes. The strategic success of these screens has been confirmed when later studies revealed that many of the genes categorized in the same mutant class were coding for subunits of the same protein complexes. Among them were complexes important for membrane transport and fusion events, such as the endosomal sorting complex required for transport (ESCRT)-I to ESCRT-III (Henne et al., 2011) or the homotypic fusion and vacuole protein sorting (HOPS) complex (Balderhaar and Ungermann, 2013).
Sequence homologs of most yeast VPS genes can be found in the Arabidopsis (Arabidopsis thaliana) genome (Sanderfoot and Raikhel, 2003; Bassham et al., 2008), and some of them were reported to be involved in intracellular trafficking as well as vacuole biogenesis. For example, the Arabidopsis vacuoleless (vcl)/vps16 mutant is embryo lethal and lacks lytic vacuoles (Rojo et al., 2001). VPS16 is a subunit of the HOPS complex, suggesting that membrane fusion events mediated by VCL/VPS16 are also important for plant vacuole biogenesis. Several other Arabidopsis vps mutants were also shown to have altered vacuole morphology at the mature embryo stage (Shimada et al., 2006; Sanmartín et al., 2007; Ebine et al., 2008, 2014; Yamazaki et al., 2008; Zouhar et al., 2009; Shahriari et al., 2010), showing that there is a conserved mechanism regulating vacuolar transport and vacuole biogenesis. However, in contrast to yeast, in which mutants without vacuole or severe biogenesis defects are viable, plant vacuoles seem to be essential for plant development.
We have previously shown that defects in the deubiquitinating enzyme (DUB) ASSOCIATED MOLECULE WITH THE Src homology-3 DOMAIN OF STAM3 (AMSH3) also lead to a severe vacuole biogenesis defect (Isono et al., 2010). AMSH homologs do not exist in budding yeast but are conserved in animals and plants. Our previous studies have shown that AMSH3 can directly interact with ESCRT-III subunits (Katsiarimpa et al., 2013). ESCRT-III is a multiprotein complex that is essential for multivesicular body (MVB) sorting (Winter and Hauser, 2006) and hence for plant growth and development (Haas et al., 2007; Spitzer et al., 2009; Katsiarimpa et al., 2011; Cai et al., 2014). AMSH proteins regulate intracellular trafficking events, including endocytic degradation, vacuolar transport, and autophagic degradation through its interaction with ESCRT-III (Isono et al., 2010; Katsiarimpa et al., 2011, 2013, 2014). Prior to our characterization of the amsh3 mutant, AMSH proteins had not been implicated in vacuole biogenesis. Thus, we reasoned that there might be additional, yet unidentified, factors important for regulating vacuole biogenesis in plants. Further, we reasoned that other mutants with a defect in vacuole biogenesis, analogous to amsh3, might also exhibit seedling lethality.
Thus, with the goal to identify and characterize these factors, we carried out a two-step mutant screen. We first selected seedling lethal mutants from an ethyl methansulfonate (EMS)-mutagenized population and then examined the vacuole morphology in these mutants. The isolated mutants were designated vacuolar fusion defective (vfd). vfd1 is affected in the expression of a functional Fab1, YOTB, Vac1, and EEA1 (FYVE) domain-containing FYVE1 protein. FYVE1 was originally identified in silico as one of 16 FYVE domain-containing proteins in Arabidopsis with no apparent homologs in yeast and mammals (van Leeuwen et al., 2004). FYVE domains bind phosphatidylinositol 3-P, a phospholipid that is a major constituent of endosomal membranes. Hence, FYVE domain-containing proteins are implicated in intracellular trafficking (van Leeuwen et al., 2004; Wywial and Singh, 2010). In a previous work, we have shown that a null mutant of FYVE1, fyve1-1, is defective in IRON-REGULATED TRANSPORTER1 (IRT1) polarization and that FYVE1 is essential for plant growth and development (Barberon et al., 2014). A very recent publication describing the same mutant has shown that FYVE1/FYVE domain protein required for endosomal sorting1 (FREE1) is also important for the early and late endosomal trafficking events (Gao et al., 2014). In this study, we show that FYVE1 is also regulating ubiquitin-dependent membrane protein degradation, vacuolar transport, autophagy, and vacuole biogenesis. Altogether, our results point toward FYVE1 being a key component of the intracellular trafficking machinery in plants.
RESULTS
vfd1 Shows Seedling Lethality and Has Defects in Central Vacuole Formation
To identify additional essential factors that are involved in vacuolar biogenesis and intracellular trafficking processes in Arabidopsis, we conducted a forward genetic screen. For this purpose, we EMS-mutagenized seeds of a tonoplast marker TONOPLAST INTRINSIC PROTEIN (δTIP)-expressing ecotype Columbia (Col-0) line (Cutler et al., 2000). M1 plants were then self-pollinated, and seeds of approximately 3,500 M1 plants were collected in pools containing five plants each. Next, we carried out a two-step phenotype screen on M2 seedlings. We first searched for a seedling lethal phenotype similar to amsh3, a mutant of the ESCRT-III-associated DUB AMSH3, that we have previously characterized (Isono et al., 2010; Katsiarimpa et al., 2014). Subsequently, we examined the identified mutant candidates under the confocal microscope for altered vacuole morphology, typically for a smaller vacuole size in a confocal section. We named these mutants vfd after the apparent defect in vacuolar fusion to form a large central vacuole in the mutants.
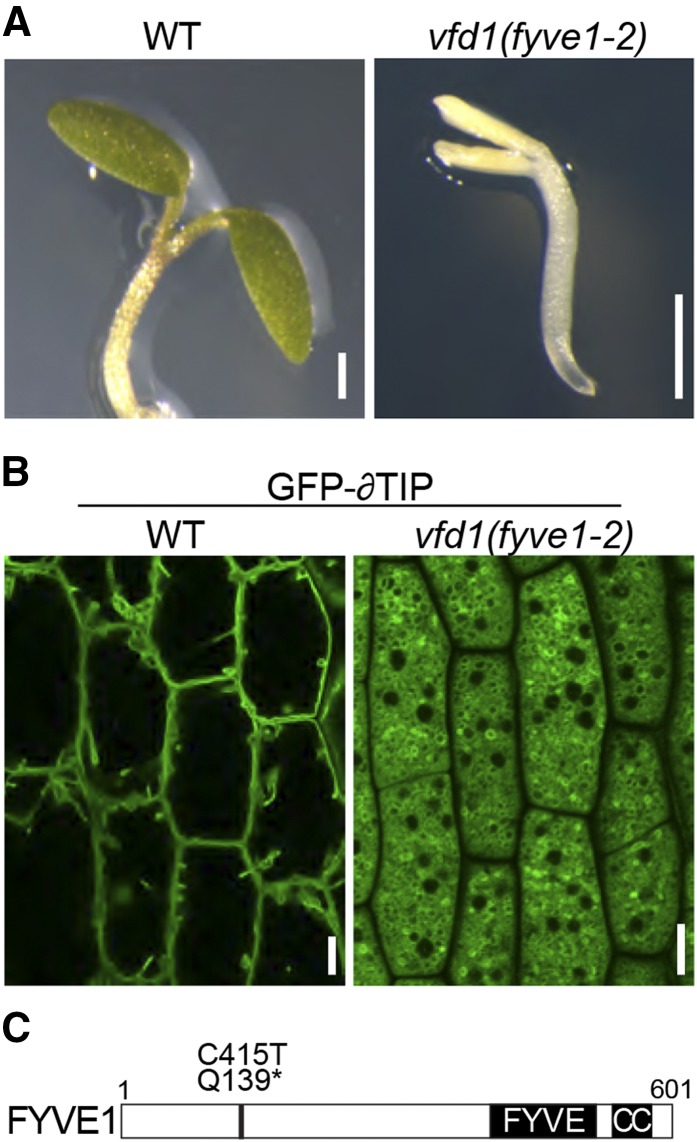
vfd1 is one of the thus identified mutants and shows recessive inheritance of the mutant phenotype. Homozygous vfd1 mutants either did not germinate or showed seedling lethality after germination (Fig. 1A). vfd1 vacuoles appeared to be much smaller compared with the wild type (Fig. 1B) when visualized with the tonoplast marker GFP-δTIP, indicating that vfd1 shares the characteristic phenotype of the previously described amsh3 mutant.
Figure 1.
The vfd1(fyve1-2) mutation causes seedling lethality and defects in central vacuole formation. A and B, Phenotypes of vfd1(fyve1-2). A, Seven-day-old seedlings of vfd1(fyve1-2) are shown compared with wild-type (WT) seedlings at the same age. B, Confocal microscopy images of the vacuolar membrane marker GFP-δTIP in 7-d-old wild-type and vfd1(fyve1-2) mutant epidermis cells. C, Schematic presentation of the FYVE1 domain structure. Position of the vfd1 mutation (C415T) leading to a premature stop codon (Q139*) is shown. CC, Coiled-coil domain. Bars = 0.5 mm.
VFD1 Encodes the FYVE Domain-Containing FYVE1 Protein
To identify the locus that was responsible for the vfd1 phenotype, we performed small sequence length polymorphism (SSLP)-based mapping. Marker analysis has revealed that the vfd1 mutation is located within a 6-Mb region on the short arm of chromosome 1. We next performed whole-genome sequencing to analyze mutations within the identified region. Sequencing analysis revealed 12 nonsynonymous typical EMS transitions in 12 genes within the rough-mapped region. To identify the gene causing the vfd1 phenotype, we next carried out a complementation assay in vfd1. Only 35Spro:FYVE1 (At1g20110) could complement the seedling-lethal mutant phenotype of vfd1, indicating that a nonsense mutation (C415T) in FYVE1 is responsible for the vfd1 phenotype (Fig. 1C; Supplemental Fig. S1). FYVE1 is a plant-specific FYVE domain-containing protein that is regulating the polarity of the iron transporter IRT1 and MVB sorting (Barberon et al., 2014; Gao et al., 2014).
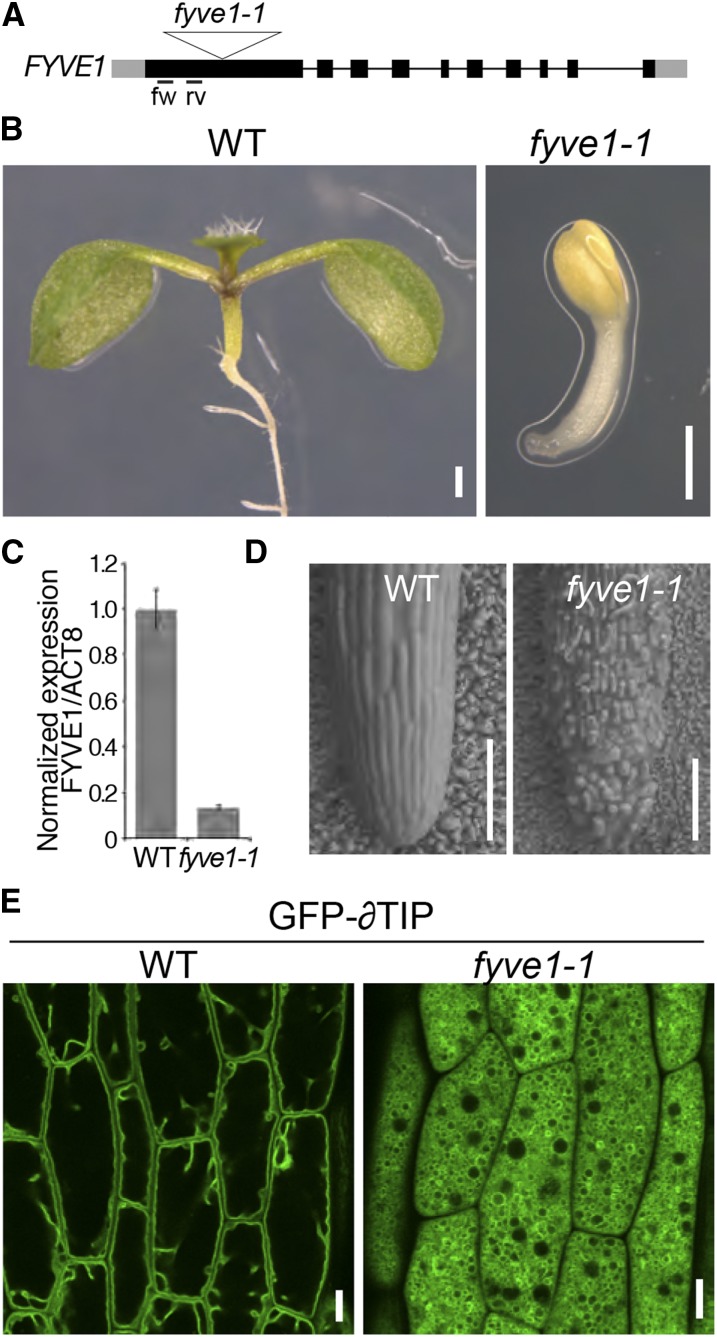
To confirm that dysfunction of FYVE1 causes the vfd1 phenotype, we analyzed a transposon insertion line of FYVE1 (Fig. 2A). We designated this mutant allele fyve1-1 (Barberon et al., 2014). The same line was later referred to as free1 by Gao et al. (2014). fyve1-1 showed a seedling-lethal phenotype similar to vfd1 (Fig. 2B). Quantitative real-time (qRT)-PCR analysis has shown that fyve1-1 has only residual transcript levels of the truncated FYVE1 gene (Fig. 2C). In addition to its seedling lethality, fyve1-1 also shows degenerated root surface structures as seen in Figure 2D, indicating that FYVE1 is essential for proper root development. To verify that fyve1-1 and vfd1 are allelic, we crossed a heterozygous fyve1-1 with a heterozygous vfd1 plant and analyzed the F1 progeny. The seedling phenotypes of transheterozygotes were identical to that of fyve1-1 or vfd1 (Supplemental Fig. S2M), indicating that fyve1-1 and vfd1 are allelic to each other. Thus, we designated vfd1 as fyve1-2.
Figure 2.
The transposon insertion line fyve1-1 shows a similar phenotype as vfd1. A, Schematic presentation of the transposon insertion site in fyve1-1. Gray boxes, Untranslated regions; black boxes, exons; lines, introns. Forward and reverse primer positions used in C are indicated. B, A 7-d-old seedling of fyve1-1 is shown compared with a wild-type (WT) seedling at the same age. C, qRT-PCR analysis of fyve1-1. Expression of FYVE1 in wild-type and fyve1-1 seedlings was analyzed with FYVE1- and ACTIN8 (ACT8)-specific primers. D, Root morphology of fyve1-1. Scanning electron micrographs of wild-type and fyve1-1 root tips. E, Confocal microscopy images of the vacuolar membrane marker GFP-δTIP in 5-d-old wild-type and vfd1(fyve1-2) epidermis cells. Bars = 0.5 mm (B), 100 µm (D), and 10 µm (E).
To examine whether fyve1-1 shows altered vacuole morphology like fyve1-2, we crossed GFP-δTIP into a fyve1-1 background and examined the vacuole phenotype in wild-type and fyve1-1 homozygous mutant seedlings. Instead of a large central vacuole that can be visualized in a wild-type epidermis cell, fyve1-1 mutants accumulate seemingly smaller vacuoles in the cytosol (Fig. 2E), corroborating that FYVE1 is crucial for proper vacuole biogenesis. Furthermore, both the seedling and vacuole phenotype of fyve1 mutants resemble the previously characterized amsh3 mutant (Supplemental Fig. S2, A–L), suggesting that FYVE1 and AMSH3 might function in the same cellular pathway.
Vacuoles of fyve1-1 Show Altered Morphology
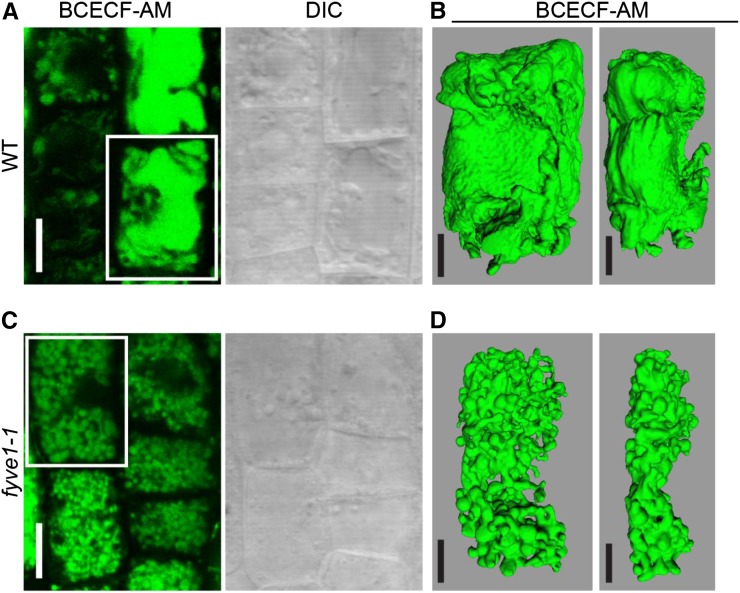
To further establish the vacuole phenotype of fyve1-1, we analyzed 2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein (BCECF)-acetoxymethyl ester (AM)-stained vacuoles of wild-type and fyve1-1 seedlings by confocal microscopy. For the comparison, 2-d-old wild-type seedlings were chosen due to the similar size with the fyve1-1 mutant (Supplemental Fig. S2, B, C, and I). On a normal confocal image, vacuoles of fyve1-1 seem to be fragmented compared with the wild type, and it appears as if many small vacuoles are filling the cytosol of fyve1-1 cells (Fig. 3, A and C). To analyze the vacuole morphology in more details, we performed a three-dimensional reconstruction analysis by surface rendering on Z-stack images of wild-type and fyve1-1 vacuoles from cells of approximately the same size. Surface rendering suggests that the small vacuoles in fyve1-1 are, in fact, interconnected with each other, forming a complex tubular structure (Fig. 3, B and D). This result suggests that FYVE1 is important for the determination of vacuolar morphology.
Figure 3.
Vacuoles in fyve1-1 show tubular-like interconnected structures. A and C, Vacuole morphology in 2-d-old wild-type (WT; A) and 5-d-old fyve1-1 (C) root epidermis cells visualized by BCECF staining. White boxes, The cells used for surface rendering. DIC, Differential interference contrast image. B and D, Representative three-dimensional surface renderings of wild-type (B) and fyve1-1 (D) vacuoles in root epidermis cells. Views from the front (left) and the side (right) are shown. Bars = 10 µm (A and C) and 5 µm (B and D).
Ubiquitinated Membrane-Associated Proteins Accumulate in fyve1-1
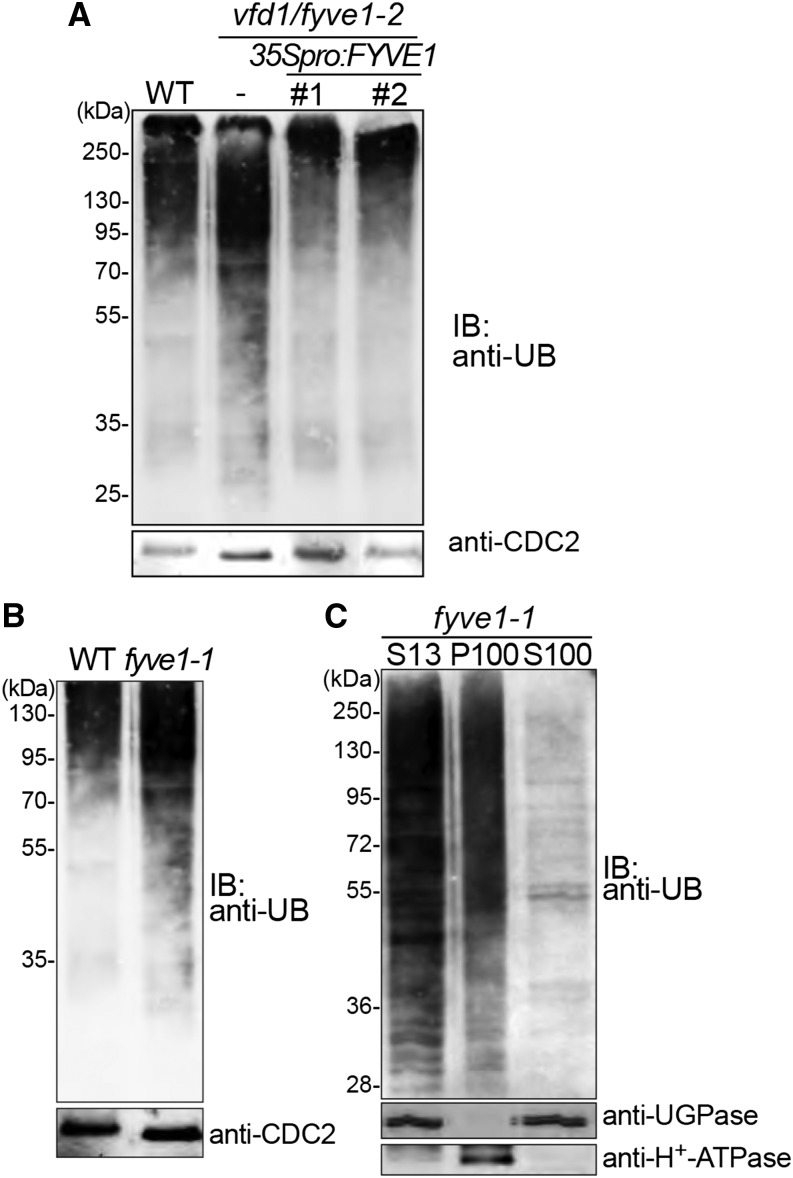
It was previously shown that mutations in ESCRT-III subunits and AMSH proteins or treatment with trafficking inhibitors lead to the accumulation of ubiquitinated proteins (Isono et al., 2010; Katsiarimpa et al., 2013, 2014; Cai et al., 2014) and that most of the accumulated proteins are found in the microsomal fraction (Katsiarimpa et al., 2014). To establish whether a defect in the FYVE1 protein also affects degradation of ubiquitinated proteins, we conducted immunodetection assays with an ubiquitin antibody on total cell extracts of wild-type, fyve1-1, and fyve1-2 seedlings. Both fyve1 mutants accumulated ubiquitinated proteins at a higher level than in the wild type (Fig. 4, A and B), indicating that FYVE1 is necessary for the degradation of ubiquitinated substrates. Accumulation of ubiquitinated proteins is restored to the wild-type level in a 35Spro:FYVE1-expressing plant (Fig. 4A), indicating that FYVE1 is affecting the global ubiquitin profile. As previously reported (Gao et al., 2014), fyve1-1 mutants accumulated ubiquitinated proteins at high levels (Fig. 4B). To examine whether the accumulated ubiquitinated proteins are membrane bound, we performed fractionation by ultracentrifugation to gain microsomal (P100) and soluble (S100) fractions. Immunoblotting showed that the majority of the ubiquitinated proteins were detected in the microsomal fraction (P100; Fig. 4C). Together, these results suggest that FYVE1 is involved in the removal of ubiquitinated proteins that are associated with endomembranes.
Figure 4.
fyve1 mutants accumulate ubiquitinated proteins in the membrane fraction. A and B, Immunoblots with an anti-ubiquitin P4D1 antibody from total protein extracts. A, Total extracts of the 7-d-old wild type (WT) and vfd1(fyve1-2) together with two complemented lines of vfd1(fyve1-2) were subjected to immunoblotting with anti-ubiquitin (anti-UB) antibody. CELL DIVISION CYCLE2 (CDC2) was used as a loading control. B, Seven-day-old fyve1-1 mutants compared with wild-type seedlings of the same age were analyzed with an antiubiquitin immunoblot. CDC2 was used as a loading control. C, Total extracts (S13) of 7-d-old fyve1-1 mutants were fractionated by ultracentrifugation to separate the microsomal (P100) and soluble fraction (S100) and subjected to immunoblotting using an antiubiquitin antibody. Note that the majority of ubiquitinated proteins accumulate in the membrane (P100) fraction. Anti-UDP-glucose pyrophosphorylase (UGPase) and anti-H+-adenosine triphosphatase (ATPase) antibodies were used for controls for the soluble and membrane fractions, respectively. IB, Immunoblot.
fyve1-1 Is Defective in Vacuolar Transport
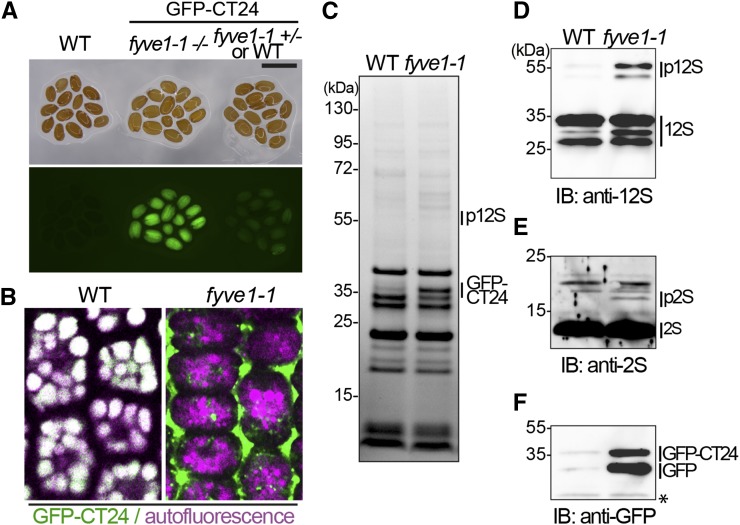
In addition to the endosomal trafficking of ubiquitinated proteins, vacuolar protein sorting is another pathway that transports vesicle cargos to the vacuole. To investigate whether fyve1-1 is defective in protein transport to the vacuole, we examined the behavior of the model vacuolar cargo GFP-CT24 (Nishizawa et al., 2003). GFP-CT24 contains the signal peptide for vacuolar targeting in seeds and was previously shown to cause green seed fluorescence in mutants defective in vacuolar transport (Fuji et al., 2007). We therefore analyzed GFP-CT24-containing seeds from wild type and heterozygous fyve1-1 plants and observed seeds with strong green fluorescence in the fyve1-1 heterozygous line (Fig. 5A). Seeds with strong fluorescence corresponded to fyve1-1 homozygous mutants as verified by PCR-based genotyping (Supplemental Fig. S3). For the following experiments, we used this phenotype to distinguish between fyve1-1 homozygous and nonhomozygous seeds. In wild-type or heterozygous embryos, signals of GFP-CT24 completely overlapped with the autofluorescence of the seed storage vacuoles, whereas in fyve1-1 homozygous embryos, GFP-CT24 signals were observed exclusively in the intercellular spaces (Fig. 5B). Autofluorescence of the seed storage vacuoles was still observed in the homozygous fyve1-1 mutants, and the altered morphology of seed storage vacuoles was apparent (Fig. 5B). This result shows that FYVE1 is involved in vacuole biogenesis already at the mature embryo stage.
Figure 5.
fyve1-1 mutants are impaired in 12S globulin and 2S albumin processing. A, Seeds of the wild type (WT; left) together with GFP-CT24-expressing fyve1-1 homozygous (middle) or fyve1-1 heterozygous and wild-type seeds (right). Seeds were photographed with a binocular microscope (top) or an epifluorescent microscope (bottom). Genotyping results are shown in Supplemental Figure S3. B, GFP-CT24 in mature embryos of the wild type and fyve1-1. Note that whereas in the wild type, GFP-CT24 signals (green) overlap with the autofluorescence of the seed storage vacuoles (magenta), in fyve1-1, GFP-CT24 is secreted into the intercellular spaces. C, A Coomassie Blue-stained gradient gel of total protein extracts of the wild type and fyve1-1 carrying GFP-CT24. p12S, 12S globulin precursors. D to F, Immunoblots of the same extracts using anti-12S globulin, anti-2S albumin, and anti-GFP antibodies to detect 12S globulin subunits and 12S globulin precursors (D), 2S albumin and 2S albumin precursors (p2S; E), and GFP-CT24 (F). Asterisk indicates an unspecific band. Bar = 1 mm. IB, Immunoblot.
To further investigate the vacuolar transport of seed storage proteins, we next analyzed the protein abundance of endogenous seed storage proteins and chose two major seed storage proteins, 2S albumin and 12S globulin. In total extracts derived from seeds, precursor proteins of both 2S albumin and 12S globulin accumulated in fyve1-1 (Fig. 5, C–E). These precursor proteins are readily processed in the wild type upon delivery to the vacuole, thus suggesting that fyve1-1 is affected in proper sorting of these storage proteins to the vacuole. The abundance of GFP-CT24 was also examined at the protein level. When compared with the wild type, fyve1-1 also accumulated higher amounts of GFP-CT24 (Fig. 5, C and F), which is probably due to the increased stability of GFP-CT24 in the intercellular spaces (Fuji et al., 2007). As the accumulation of precursor proteins is a strong indication of defective vacuolar processing, these data indicate that FYVE1 is involved in proper vacuolar transport of seed storage proteins.
fyve1-1 Has Defects in the Autophagy Pathway
Recent reports have shown that intracellular membrane trafficking is closely related to autophagy (Katsiarimpa et al., 2013; Kulich et al., 2013), a pathway that eliminates and recycles cellular components. In autophagic degradation, the cytosol or selected cargo proteins marked with ubiquitin are sequestered by double membranes to form autophagosomes. Autophagosomal membranes contain AUTOPHAGY8 (ATG8), an ubiquitin-like protein that is activated by a cascade of enzymes similar to the ubiquitin pathway (Yoshimoto et al., 2004). After engulfing of contents and closure of the double membrane, autophagosomes are targeted to the vacuole, where upon fusion with the vacuolar membrane, the content is released and degraded by vacuolar proteases. ATG8 is a constitutive structural component of the autophagosome and therefore an ideal marker to monitor autophagy. In amsh3 mutants, ATG8 has been shown to accumulate (Isono et al., 2010). Because fyve1 mutants are phenotypically similar to amsh3 mutants, we wondered whether fyve1 mutants are also altered in the autophagy pathway.
To test this possibility, we analyzed the steady-state level of ATG8 and NEIGHBOR OF BRCA1 (for BREAST CANCER SUSCEPTIBILITY GENE1) GENE1 (NBR1). Whereas ATG8 is present in most types of autophagosomes, NBR1 is specific to selective autophagy and acts as a cargo adapter protein (Svenning et al., 2011). Both autophagy markers accumulated in autophagy-defective mutant seedlings of atg7-2 (Hofius et al., 2009) and atg10-1 (Phillips et al., 2008). atg7-2 and atg10-1 are mutants of the E1-activating and E2-conjugating enzymes of the autophagic pathway, respectively. Similar to the atg mutants, ATG8 and NBR1 also accumulated in fyve1-1 (Fig. 6), indicating that fyve1-1 is also defective in the autophagy pathway. Because not only ATG8 but also NBR1 accumulated in fyve1-1, FYVE1 is involved in both selective and nonselective autophagy pathway.
Figure 6.
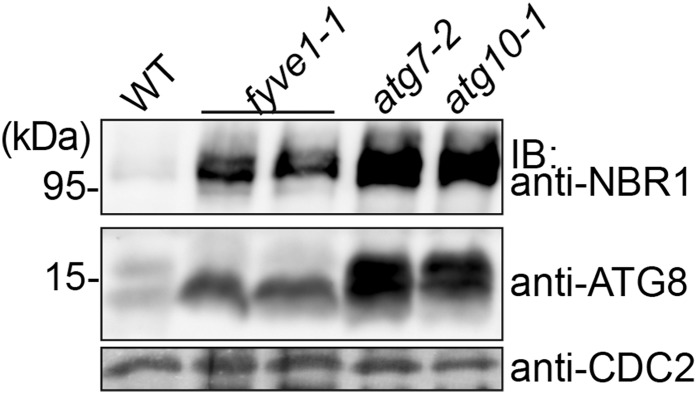
fyve1-1 accumulates autophagosome markers. Total extracts from wild-type (WT), fyve1-1, atg7-2, and atg10-1 seedlings grown under long-day conditions were subjected to immunoblotting using an anti-NBR1 and an anti-ATG8 antibody. CDC2 was used as a loading control. IB, Immunoblot.
FYVE1 Is Localized to the Cytosol and to Late Endosomes
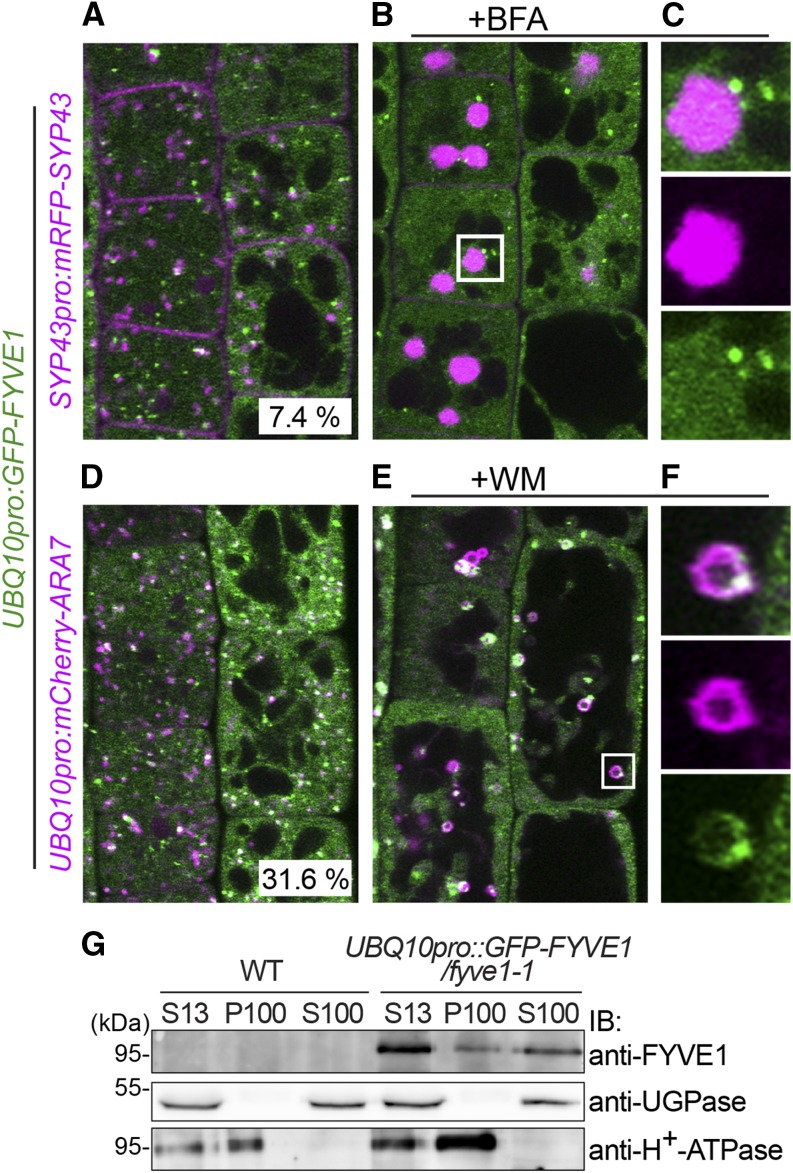
FYVE1 was previously shown to bind to phosphoinositides and localize to late endosomal vesicles together with the 2×FYVE marker and two late endosome (LE)/MVB markers Rha1 and VACUOLE SORTING RECEPTOR2 (Barberon et al., 2014; Gao et al., 2014). We analyzed the subcellular localization of a functional UBQUITIN10 promotor (UBQ10):GFP-FYVE1 construct (Barberon et al., 2014) with further markers. As previously reported, GFP-FYVE1 signals appeared in the cytosol as well as in small vesicles in the cytosol. Among the total of 257 GFP-FYVE1 vesicles, 31.6% colocalized with the LE/MVB marker mCherry-ARABIDOPSIS RAT SARCOMA-RELATED PROTEINS IN BRAIN (RAB) GTPase7 (ARA7; Geldner et al., 2009), whereas only 7.4% of the GFP-FYVE1 vesicles showed overlap with the early endosome (EE)/trans-Golgi network (TGN) marker SYNTAXIN OF PLANTS43 (SYP43)-mRFP (Ebine et al., 2008; Fig. 7, A and D). To further analyze this localization, we treated the seedlings with the inhibitor of guanine nucleotide exchange factor for ADP-ribosylation factor brefeldin A (BFA) and the inhibitor of phosphatidylinositol 3-kinase (PI3K) and PI4K Wortmannin (WM). After 45 min of 50 µm BFA treatment, mRFP-SYP43 accumulated in BFA bodies (Fig. 7, B and C), while the localization of GFP-FYVE1 remained unaffected. By contrast, after 45 min of 33 µm WM treatment, both mCherry-ARA7 and GFP-FYVE1 were localized to the WM-induced ring-like structure (Fig. 7, D and E), corroborating the observation that FYVE1 is associated with LE/MVBs.
Figure 7.
GFP-FYVE1 colocalizes with ARA7. A, Localization of GFP-FYVE1 and mRFP-SYP43 in 5-d-old root epidermis cells. The percentage of GFP-FYVE1 vesicle colocalizing with mRFP-SYP43 is indicated at bottom. Total number of GFP-FYVE1 vesicle counted was n = 257. B and C, Localization of GFP-FYVE1 and mRFP-SYP43 upon 45 min of BFA treatment. Magnification of a BFA body in the area indicated in B is shown in C. Note that mRFP-SYP43, but not GFP-FYVE1, localizes to BFA bodies. D, Localization of GFP-FYVE1 and mCherry-ARA7 in 5-d-old root epidermis cells. The percentage of GFP-FYVE1 vesicle colocalizing with mCherry-ARA7 is indicated at the bottom. The total number of GFP-FYVE1 vesicle counted was n = 253. E and F, Localization of GFP-FYVE1 and mCherry-ARA7 and upon 45 min of WM treatment. Magnification of a WM compartment in the area indicated in E is shown in F. Note that both proteins localize to WM compartments. G, Total extracts (S13) from the wild type (WT) and complementing GFP-FYVE1 line were subjected to ultracentrifugation to separate soluble (S100) and microsomal (P100) fractions. Proteins from each fraction were subjected to immunoblotting using anti-FYVE1. Anti-UGPase and anti-H+-ATPase antibodies were used for controls for soluble and membrane fractions, respectively. IB, Immunoblot.
To investigate the membrane association of the FYVE1 biochemically, we generated an anti-FYVE1-specific antibody. The antibody recognized overexpressed GFP-FYVE1 but failed to detect the endogenous FYVE1 protein in plant total extracts (data not shown). Therefore, we used the GFP-FYVE1 line for fractionation and immunodetection analysis. Total extracts (S13) of wild-type and GFP-FYVE1 seedlings were subjected to ultracentrifugation to obtain membrane (P100) and soluble (S100) fractions. GFP-FYVE1 was detected in both soluble and membrane fractions (Fig. 7D), suggesting that GFP-FYVE1 is not exclusively associated with membranes. This result is in accordance with the observation that GFP-FYVE1 is localized in both vesicles and the cytosol and also suggests the possibility that GFP-FYVE1 recycles between the cytosol and the membrane.
FYVE1 Interacts with SH3P2 and SH3P3
Arabidopsis has 16 FYVE domain-containing proteins whose molecular and biological functions are diverse (van Leeuwen et al., 2004; Wywial and Singh, 2010; Hirano et al., 2011; Serrazina et al., 2014). The FYVE domain serves to bind membranes, but it is the interaction with other proteins that is important in defining the role of a given FYVE domain protein. Therefore, to better understand the molecular framework surrounding FYVE1 function, we wanted to identify interacting proteins of FYVE1. To this end, we performed a yeast two-hybrid (YTH) screen against 12,000 Arabidopsis proteins encoded by sequence-verified open reading frames (ORFs; Arabidopsis Interactome Mapping Consortium, 2011; Weßling et al., 2014) using FYVE1 as a bait. We used a truncated variant of FYVE1 lacking the FYVE domain, FYVE1(∆FYVE), because we wanted to rule out the possibility that FYVE1 might also be targeted to membranes in yeast and thereby interfere with the efficiency of the YTH screen. The YTH screen, in which the bait was screened in a binary test against all individual ORFs in the collection, was carried out independently three times. To ensure reliability and reproducibility of the interactions, only interactor candidates that appeared at least in two of the three independent experiments were further considered. Because fyve1 mutants showed severe phenotypes in intracellular trafficking, we decided to first focus on proteins with a known function in this pathway. One of these was Src homology-3-containing protein3 (SH3P3), which has been described as a protein implied in intracellular trafficking in Arabidopsis (Lam et al., 2001).
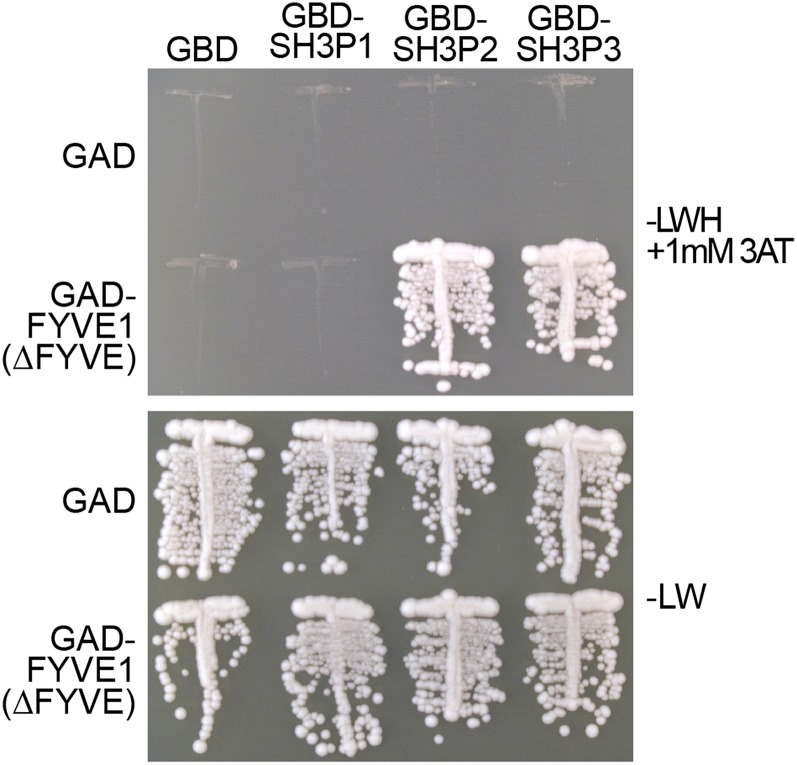
SH3P1, SH3P2, and SH3P3 are the three SH3 domain-containing proteins that were previously reported to be involved in clathrin-mediated endocytosis (Lam et al., 2001). Among the three, SH3P2 and SH3P3 share the highest sequence homology. To establish whether SH3P1 and SH3P2 can also interact with FYVE1, we performed a targeted YTH analysis. To increase the confidence in the interaction, we subcloned the FYVE1(∆FYVE) into another YTH vector and analyzed the interaction between FYVE1(∆FYVE) and SH3P1, SH3P2, and SH3P3. SH3P2 and SH3P3, but not SH3P1, showed interaction with FYVE1, indicating that they share a common interaction partner in FYVE1 (Fig. 8; Supplemental Fig. S4). Moreover, as the ∆FYVE variant was able to interact with these proteins, the FYVE domain is not involved in the interaction between these proteins.
Figure 8.
FYVE1 interacts with SH3P2 and SH3P3. YTH analysis of GAD-FYVE1(∆FYVE) with GBD-fused SH3P2 and SH3P3. Transformants were grown on media lacking Leu and Trp (−LW) or Leu, Trp, and His (−LWH) with the indicated amount of 3-amino-1,2,4-triazole (3AT) to test their auxotrophic growth. The expression of all fusion proteins was verified by immunoblotting (Supplemental Fig. S4).
FYVE1 Colocalizes with SH3P2 on Class E Compartments
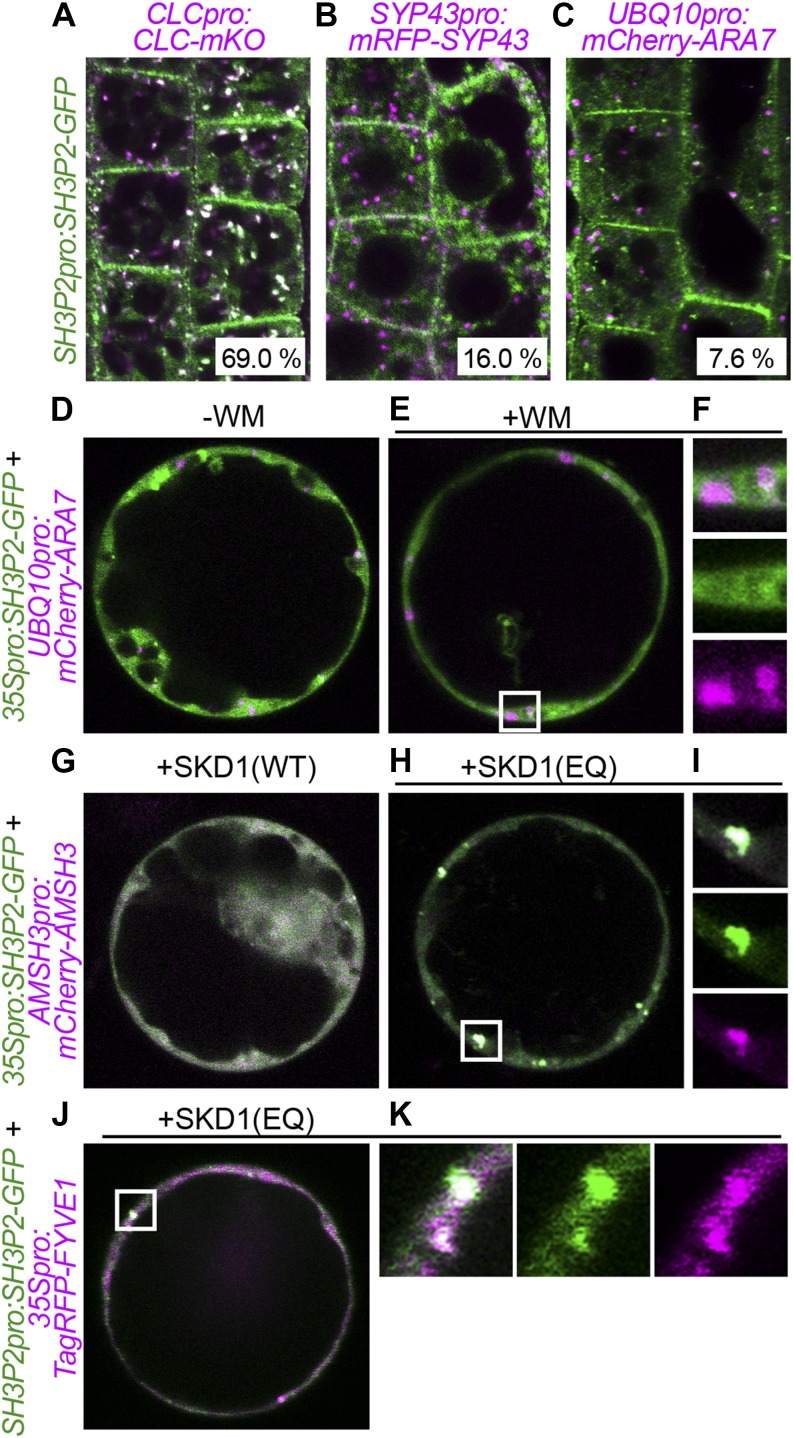
We next examined whether these proteins function on the same cellular compartments. SH3P1 was previously shown to localize on clathrin-coated vesicles by immunoelectron microscopy (Lam et al., 2001). To examine the localization of SH3P2, we generated a SH3P2pro:SH3P2-GFP plant line. We noticed that SH3P2-GFP localizes on vesicles, on the plasma membrane, and in the cytosol. To analyze the nature of these vesicles, we crossed the SH3P2-GFP-expressing line with CLATHRIN LIGHT CHAIN (CLC)-mKO-, mRFP-SYP43-, and mCherry-ARA7-expressing plants (Ito et al., 2012). SH3P2-GFP signals showed strong overlap with CLC-mKO (69.0%, n = 619; Fig. 9A). We detected also partial colocalization of SH3P2-GFP with the EE/TGN marker mRFP-SYP43 (16.0%, n = 162) and with the LE/MVB marker mCherry-ARA7 (7.2%, n = 158; Fig. 9, B and C), albeit with much lower frequency. These results show that the majority of SH3P2 molecules functions on CLC-positive vesicles, though a fraction of SH3P2 molecules localize to the cytosol and to other vesicles.
Figure 9.
FYVE1 colocalizes with SH3P2 on SKD1(EQ)-induced class E compartments. A to C, GFP-SH3P2-expressing plants were crossed with CLC-mKO-expressing (A), mRFP-SYP43-expressing (B), and mCherry-ARA7-expressing (C) lines. Colocalization was analyzed in root epidermis cells of 5-d-old seedlings. The percentage of GFP-SH3P2 vesicle colocalizing with each marker is presented at bottom. Total numbers of GFP-SH3P2 vesicle counted were n = 619 for A, n = 162 for B, and n = 158 for C. D and F, Coexpression analysis of SH3P2-GFP with mCherry-ARA7 in Arabidopsis cell culture-derived protoplasts. Cells in E were treated with WM for 120 min before observation. Note that whereas mCherry-ARA7 accumulates in WM-induced compartments, GFP-SH3P2 does not. Magnification of a WM compartment in the area indicated in E is shown in F. G to I, Coexpression of SH3P2-GFP and mCherry-AMSH3 with SKD1(WT; G) or SKD1(EQ; H) in Arabidopsis cell culture-derived protoplasts. Note that a fraction of GFP-SH3P2 colocalizes with mCherry-AMSH3 in SKD1(EQ)-induced class E compartments. Magnification of a class E compartment in the area indicated in H is shown in I. J and K, Localization of GFP-SH3P2 with TagRFP-FYVE1 upon coexpression of SKD1(EQ) in an Arabidopsis root-derived protoplast. Magnification of a class E compartment in the area indicated in J is shown in K.
Because GFP-FYVE1 showed colocalization with ARA7 (Fig. 7) and because we detected interactions between FYVE1 and SH3P2, we further investigated the possibility that SH3P2 can transiently associate with LE/MVBs together with FYVE1. To this end, we further examined the localization of SH3P2-GFP upon WM treatment and upon inhibition of ESCRT-III function. WM is an inhibitor of PI3 and PI4 kinases and thereby inhibits endosomal trafficking. One hundred twenty minutes of WM treatment caused accumulation of the LE/MVB marker mRFP-ARA7 to WM-induced compartments. However, under this condition, the localization of SH3P2-GFP remained unaffected (Fig. 9, D–F). This result confirms that SH3P2, unlike ARA7, is not a stable component of LE/MVBs.
To test whether the localization of SH3P2 is affected by the inhibition of ESCRT-III function, we next examined the behavior of SH3P2-GFP together with the ESCRT-III-associated mCherry-AMSH3 upon coexpression of an inactive SUPPRESSOR OF K+ TRANSPORT GROWTH DEFECT1 (SKD1). SKD1 is an AAA-ATPase that is essential for the disassembly of the ESCRT-III complex. Upon expression of the ATPase-inactive SKD1(EQ) variant, ESCRT-III disassembly is inhibited and enlargement of the MVB occurs (Haas et al., 2007), leading to the formation of so-called class E compartments.
Because the expression of SKD1(EQ) is toxic in planta, we investigated the effect of SKD1(EQ) in Arabidopsis cell culture-derived protoplasts. This system was successfully used to stabilize and visualize transient interaction between ESCRT-III and its interacting AMSH3 protein, both of which show uniform cytosolic localization that was not affected by WM treatment (Katsiarimpa et al., 2011). To verify the specificity of the effect caused by SKD1(EQ), we analyzed the coexpression of SKD1(WT) and SKD1(EQ) with CLC-mKO, mRFP-SYP41, and mRFP-ARA7. As a control, ESCRT-III subunits VPS2.1 and VPS24.1 were cotransformed. SKD1(EQ) induction specifically affected the localization of the LE/MVB marker ARA7 causing ARA7 to accumulate in class E compartments together with ESCRT-III subunits (Supplemental Fig. S5). By contrast, CLC and the EE/TGN marker SYP43 did show accumulation on class E compartments (Supplemental Fig. S5). This experiment shows that at least in this experimental system, SKD1(EQ) induction specifically accumulates ARA7- and ESCRT-III-related proteins to class E compartments.
When SH3P2-GFP and mCherry-AMSH3 were coexpressed with SKD1(EQ), but not with SKD1(WT), we observed that SH3P2-GFP accumulated together with mCherry-AMSH3 on class E compartments (Fig. 9, G–I). Because AMSH3 was shown to interact directly with ESCRT-III (Katsiarimpa et al., 2011) and because ESCRT-III was shown to localize on LE/MVBs (Scheuring et al., 2011), this result suggests that there must be at least a transient interaction of SH3P2-GFP with LE/MVB. SH3P2-GFP also colocalized with TagRFP-FYVE1 (Fig. 9, J and K) on SKD1(EQ)-induced class E compartments, suggesting that they can function together on a (late-)endosomal compartment sensitive to SKD1(EQ).
Altogether, our results indicate that FYVE1 is essential for ubiquitin-mediated protein degradation, vacuolar transport, autophagy, and proper vacuole biogenesis and/or maintenance. FYVE1 localizes on LEs and, like the ESCRT-III-interacting DUB AMSH3, is recruited together with its interactor SH3P2 to ESCRT-III-positive class E compartments. Through the interaction with other proteins that are involved in intracellular trafficking, as illustrated by the interaction with SH3P2, FYVE1 could play a central role in the regulation of these key cellular processes and hence is pivotal for plant growth and development (Fig. 10).
Figure 10.
FYVE1 localizes to LEs, regulates intracellular trafficking and vacuole biogenesis and interacts with SH3P2. FYVE1 function together with SH3P2 or other interactors is essential for endocytic protein degradation, vacuolar protein transport, autophagy, and vacuole biogenesis. Lack of FYVE1 causes seedling lethality, indicating that FYVE1 is essential for plant growth and development. ER, Endoplasmic reticulum.
DISCUSSION
FYVE1 as an Essential Factor in Vacuole Biogenesis and Endomembrane Trafficking
With the goal to identify new proteins involved in vacuole biogenesis, we used a forward genetics approach and screened an EMS-mutagenized population for mutants with altered vacuole morphology. In our study, we report the identification and characterization of the vfd1(fyve1-2) and fyve1-1 mutants and show that FYVE1 is an essential component in vacuole biogenesis. FYVE1 was recently reported as an interactor and regulator of the polarity of the iron transporter IRT1, and it was also shown that overexpression of FYVE1 leads to hypersensitivity toward low-iron conditions (Barberon et al., 2014). In another recent publication, it was shown that the fyve1-1 mutant (named free1 by the authors) is defective in MVB sorting and ubiquitin-dependent protein degradation in Arabidopsis (Gao et al., 2014). Our characterization of the fyve1 mutants revealed that, in addition to the vacuole biogenesis defect, they are also affected in the degradation of ubiquitinated proteins associated with membranes, vacuolar transport, and autophagy, indicating that FYVE1 has a pleiotropic role in endomembrane trafficking.
Vacuoles in Arabidopsis root cells have been shown to change their morphology dynamically during cell elongation (Viotti et al., 2013). fyve1-1 cells do not contain a large central vacuole but instead have a complex tubular structure of vacuoles that is reminiscent of wild-type vacuoles in the meristematic cells (Viotti et al., 2013). The process of central vacuole formation is not well understood yet, but it most certainly requires homotypic membrane fusion events. The HOPS tethering complex, soluble N-ethylmaleimide-sensitive factor attachment receptors, and RAB7 GTPases are described as proteins involved in this process in yeast (Wickner, 2010). The fact that mutants of the Arabidopsis homologs of these factors also display altered vacuole morphology (Rojo et al., 2001; Sanmartín et al., 2007; Ebine et al., 2008, 2014; Zouhar et al., 2009) suggests that the same proteins are involved in vacuolar membrane fusion events in plants. Because it is essential for these proteins to be properly targeted to the correct endomembrane, FYVE1 dysfunction and the defect in intracellular trafficking as a consequence might impact the transport of these tethering/fusion factors to the vacuolar membrane. This, in turn, might cause defects in vacuolar membrane fusion, causing altered vacuolar morphology in fyve1 mutants.
Role of FYVE1 in Vacuolar Protein Transport
Different cargos of the vacuolar sorting pathway employ distinct transport routes to the vacuole in a RAB5 and RAB7 activity-dependent process (Cui et al., 2014; Ebine et al., 2014; Singh et al., 2014). Storage protein sorting also relies on the proper function of vacuolar sorting receptors (Fuji et al., 2007), and the fact that the fyve1-1 mutant missorted seed storage proteins indicates that FYVE1 function might affect the localization, transport, or activity of these proteins. Alternatively, the vacuole biogenesis defect in fyve1-1 might alter the vacuolar membrane lipid or protein composition in the first place. Therefore, in fyve1-1, regulators of vacuolar protein transport might not be able to recognize the tonoplast, their target membrane, causing defects in vacuolar protein transport.
FYVE1 harbors a FYVE domain, a well-characterized domain that can bind to phospholipids, especially PI3P (van Leeuwen et al., 2004). In contrast to yeast and mammals, in which PI3P is found also in EEs, PI3P was reported to be enriched in LEs in plants (Vermeer et al., 2006). This is in accordance with the localization of FYVE1 on LEs as well as its reported function in MVB sorting (Gao et al., 2014). The presence of GFP-FYVE1 in the cytosol suggests that the FYVE domain alone is not enough for a constitutive binding of FYVE1 to vesicles. Depending on the protein structure outside of the FYVE domain and the presence of catalytic domains, FYVE domain-containing proteins can play variable roles in endomembrane-related pathways. FYVE1 does not contain obvious domains with catalytic activity, implying that it probably executes its cellular function by interacting with other proteins of the pathway, for example, by serving as a scaffold to recruit other proteins onto the endosomal membrane. Both fyve1 mutants used in this work cannot produce transcripts with a functional FYVE domain. Thus, even if the truncated proteins are stably expressed, they would not be able to bind to endomembranes, which may render the protein nonfunctional.
FYVE1 Function in Different Intracellular Trafficking Processes
Although it is evident from recently published studies (Barberon et al., 2014; Gao et al., 2014) and the data presented in this study that FYVE1 is key to many membrane trafficking processes, the molecular function of FYVE1 still remains to be elucidated. Because FYVE1 does not have an apparent functional domain except for the FYVE domain and a C-terminal coiled-coil domain, it is informative to identify interacting proteins of FYVE1 to better understand its molecular function. We therefore carried out a YTH screen using FYVE1 as a bait and identified two SH3 domain-containing proteins SH3P2 and SH3P3 as interactors of FYVE1.
The Arabidopsis genome contains three genes that code for SH3 domain-containing proteins SH3P1, SH3P2, and SH3P3. SH3P1 was originally reported to localize on clathrin-coated vesicles and interact with the clathrin disassembling factor Auxillin (Lam et al., 2001). Our results show that although almost 70% of vesicle-localized SH3P2 are on CLC-positive vesicles, a small portion of SH3P2 can localize to the LE/MVB. Though the interaction of FYVE1 and SH3P2 may be of a transient nature, they may regulate membrane transport together at the LE/MVB. In addition to the function at LE/MVB, as assumed from its localization, FYVE1 might also play a role in different types of endosomes. A recent publication has shown that FYVE1 can interact with the ESCRT-I component VPS23.1 in vitro and in vivo (Gao et al., 2014). The ability of FYVE1 to interact with VPS23.1 and with ubiquitin led to the assumption that FYVE1 acts as an ESCRT-I-interacting ubiquitin adapter protein. Although we could detect only a little overlap between GFP-FYVE1 and the EE/TGN marker, we noticed that GFP-FYVE1 is also localized in the cytosol. This might suggest that the cytosolic fraction of FYVE1 can transiently interact with early endosomal components and regulates endocytosis in association with these. The fact that the fyve1 mutant is defective in a plethora of intracellular trafficking pathways supports this hypothesis.
fyve1-1 showed accumulation of ATG8 that is a structural component of autophagosomes, indicating that the mutant has defects in the autophagic pathway. In addition, it also showed stabilization of NBR1, indicating that not only bulk autophagy but also selective autophagy is inhibited in this mutant. It has been shown that Arabidopsis mutants in the ESCRT-III and ESCRT-III-related amsh mutants (Isono et al., 2010; Katsiarimpa et al., 2013) as well as an Exocyst mutant (Kulich et al., 2013) have defects in autophagy. These reports indicate that there is a strong correlation between the functionality of basic intracellular trafficking and autophagic degradation. Because FYVE1 seems to play a central role in different aspects of endomembrane trafficking as described in this report, we can assume that these defects in turn could affect the autophagy pathway. Interestingly, SH3P2 was reported to interact with ATG8 proteins, to localize on autophagosomes, and to regulate autophagy (Zhuang et al., 2013). Thus, in regard to the interaction of FYVE1 with SH3P2, FYVE1 could also be involved directly in the regulation of autophagic degradation.
fyve1 mutants phenocopy the amsh3 mutant in regard to their general seedling morphology, vacuolar structure, defects in ubiquitinated cargo degradation, vacuolar protein sorting, and autophagy. Both proteins are predicted to function at the LEs, although, in contrast to AMSH3 (Katsiarimpa et al., 2011), it was shown that FYVE1 does not interact with ESCRT-III subunits (Gao et al., 2014). It might still be possible that AMSH3 and FYVE1 act together in the regulation of ubiquitin-mediated proteolysis in the endosomal pathway. It is our future subject to investigate the relationship between FYVE1 and AMSH3 and to elucidate the molecular framework surrounding FYVE1 function in intracellular trafficking to decipher its role in plant growth and development.
MATERIALS AND METHODS
Plant Material and Medium
All plant experiments were performed with Arabidopsis (Arabidopsis thaliana). Wild-type Col-0 and ecotype Nossen were used as controls for experiments with vfd1/fyve1-2 and fyve1-1, respectively. Transposon insertion line of FYVE1 in ecotype Nossen, designated fyve1-1 (RIKEN pst18264), was described previously (Barberon et al., 2014). fyve1-1 was genotyped with primers EI511, EI512, and Ds5-2a. Sequences of primers used for genotyping are listed in Supplemental Table S1. Genotyping PCR products were analyzed using the QIAxcel Advanced System (Qiagen). Plant transformations were performed using the floral dip method (Clough and Bent, 1998). Seedlings were grown in continuous light at 110 μmol m–2 s–1 light intensity. Standard Murashige and Skoog growth medium (Duchefa Biochemie) supplemented with 1% (w/v) Suc (2.15 g L–1 Murashige and Skoog and 2.3 mm MES, pH 5.7) was used to grow seedlings, and adult plants were grown in soil. Arabidopsis root cell culture was used to produce protoplasts for transient transformation and localization analysis as described previously (Katsiarimpa et al., 2011).
Identification of the VFD1/FYVE1 Gene
To map the VFD1/FYVE1 locus, an F2-mapping population was generated by crossing a vfd1 heterozygous plant (Col-0) with a wild-type ecotype Landsberg erecta plant. F1 plants were allowed to self-pollinate, and F2 progenies were screened for the vfd1 phenotype. Genomic DNA was isolated and subjected to SSLP-based marker analysis. SSLP analysis restricted the vfd1 map interval to a 6-Mb region defined by markers F16J7-TRB and NGA392. To identify the region carrying the causative mutation, genomic DNA was pooled from approximately 100 F2 mutant seedlings that had been obtained from a single backcross of vfd1 to Col-0. A paired-end sequencing library was prepared using the NEBNext DNA Sample Prep Reagent Set for Illumina (New England Biolabs) according to the manufacturer’s instructions. The library was subjected to 100-bp paired-end whole-genome sequencing using a single lane on an Illumina Hiseq2000 machine. The resulting reads were mapped against the Col-0 reference genome (The Arabidopsis Information Resource 9) using GenomeMapper (Schneeberger et al., 2009a), and single nucleotide polymorphisms (SNPs) were called using SHORE (Ossowski et al., 2008). SNPs were filtered by read support (minimum 10), concordance (>80%), uniqueness, and typical EMS transitions (C > T and G > A), resulting in 216 potential EMS-induced mutations on the whole genome. Using plotting functions of SHOREmap (Schneeberger et al., 2009b), we found that almost one-half of them clustered on the left arm of chromosome 1, indicating low recombination in this region. Annotating the SNPs resulted in 12 nonsynonymous mutations within the previously rough-mapped region.
Cloning Procedures
All primers used for cloning and subcloning are listed in Supplemental Table S1. 35Spro:FYVE1 was generated by cloning the ORF of FYVE1 with primers CK40 and CK41 into pGWB414 (Nakagawa et al., 2007) using Gateway technology (Invitrogen). UBQ10pro:GFP-FYVE1 and 35Spro:TagRFP-FYVE1 were generated by cloning the PCR-amplified FYVE1 ORF into pUBN-GFP-DEST (Grefen et al., 2010) and pGWB461 (Nakagawa et al., 2007), respectively. 35Spro:GFP-SH3P2 was obtained by PCR amplifying the ORF of SH3P2 with the primer pair MN53 and MN54 and inserting the resulting PCR product into 35S-GW-GSP (MPI Cologne). SH3P2pro:SH3P2-GFP was generated by amplifying a genomic fragment of SH3P2 with primers SH3P2-up5 fw and SH3P2-dw3 rv and cloning the resulting PCR product into pGWB404 (Nakagawa et al., 2007). GAD-FYVE1 (∆FYVE) for the YTH ORF screen was created by cloning the coding region of FYVE1 without the FYVE domain using primers CK41, CK131, CK140, and CK141 into pDEST-DB (Mukhtar et al., 2011). For the verification of the YTH results, FYVE1 (∆FYVE) was amplified with the primer pair VC11 and VC12 and cloned into pGADT7 (Clontech). ORFs of SH3P1, SH3P2, and SH3P3 were amplified with primer pairs MN215 and MS8, MS11 and MS12, or MS9 and MS10, respectively, and cloned into pGBKT7 (Clontech). AMSH3pro:mCherry-AMSH3 was created by replacing the YFP of AMSH3pro:YFP-AMSH3 (Katsiarimpa et al., 2011) with mCherry. For antigen production, a FYVE1 fragment was amplified with the primer pair EI528 and EI529 and cloned into pET21a (Novagen). 35Spro:SKD1(WT), 35Spro:SKD1(EQ), UBQ10pro:YFP-VPS2.1, UBQ10pro:YFP-VPS24.1 (Katsiarimpa et al., 2011), 35Spro:mRFP-SYP41, and 35Spro:mRFP-ARA7 (Ueda et al., 2004; Uemura et al., 2004) were described previously.
qRT-PCR and Reverse Transcription-PCR
Sequences of primers used for qRT-PCR (EI541 and EI542) and reverse transcription-PCR (CK104 and CK105) are listed in Supplemental Table S1. Total RNA was extracted from 7-d-old seedlings with a NucleoSpin RNA plant kit (Machery-Nagel). One microgram of total RNA was reverse transcribed with an oligo(dT) primer and M-MulV reverse transcriptase (Fermentas) following the manufacturer’s instructions. Quantitative real-time PCR was performed using iQ SYBR Green Supermix (Bio-Rad) in a CFX96 Real-Time System Cycler (Bio-Rad). A 45-cycle two-step amplification protocol (10 s at 95°C and 25 s at 60°C) was used for all measurements.
EMS Mutagenesis
Mutagenesis of GFP-δTIP-expressing Col-0 seeds with EMS was performed as described previously (Weigel and Glazebrook, 2001). Progeny of about 3,500 M1 plants were screened for seedling lethality and for the vfd phenotype.
Protein Extraction, Immunoblotting, and Antibodies
Yeast (Saccharomyces cerevisiae) total proteins were extracted according to a previously described protocol (Kushnirov, 2000). SDS-PAGE and immunoblotting were performed according to standard methods. Plant total protein extracts were prepared in extraction buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5% [v/v] Triton X-100, and protease inhibitor cocktail [Roche]). For seed storage protein analysis, NuPAGE 4% to 12% (w/v) Bis-Tris Gels (Life Sciences) were used. For fractionation, total extracts were centrifuged for 20 min at 13,000g at 4°C, and the supernatant (S13) was further centrifuged for 1 h at 100,000g at 4°C in a Sorvall MTX 500 bench-top ultracentrifuge (Thermo Scientific) to obtain the soluble (S100) and microsomal (P100) fractions. Antibodies used in this study were as follows: anti-ATG8 (Thompson et al., 2005), anti-GFP (Invitrogen), anti-CDC2 (Santa Cruz), anti-GAL4 DNA-binding (GAL4BD; Santa Cruz), anti-HA (Roche), anti-NBR1 (Svenning et al., 2011), anti-UB (P4D1; Santa Cruz), anti-UGPase (Agrisera), anti-proton (H+)-ATPase (Agrisera), anti-2S, and anti-12S (Shimada et al., 2003). An anti-FYVE1 antibody was raised in rabbits (Eurogentec) using a truncated FYVE1 protein (amino acids 36–280) that was purified from bacteria as an antigen.
Microscopy
To visualize the vacuole, seedlings were incubated with 5 μm BCECF-AM (Molecular Probes) for 1 h. WM (Applichem) treatment was performed at a concentration of 33 µm for 120 min at room temperature. To visualize vacuoles, seedlings were incubated with 5 μm BCECF-AM (Molecular Probes, Invitrogen) and 0.02% (w/v) Pluronic F-127 (Molecular Probes, Invitrogen) for 1 h at room temperature in darkness. GFP- or TagRFP/mCherry/mRFP/mKO-fused proteins and BCECF staining were analyzed with an FV-1000/IX81 confocal laser-scanning microscope (Olympus) equipped with GaAsP detectors (Olympus) and a UPlanSApo X60/1.20 (Olympus) objective using the 488-, 559-, and 405-nm laser line, respectively. For surface rendering of wild-type and fyve1-1 vacuoles, 90 Z-stack images with 0.2-µm step size were obtained. Images were subsequently processed using Imaris 7 (Bitplane). Surface rendering was performed with the following parameters. The surface area detail level was set to 0.122 µm, and signal intensity threshold was set to 80. Seeds expressing GFP-CT24 were photographed using the Olympus BZX16 stereomicroscope. For scanning electron microscopy, seedling roots were observed directly with a TM-3000 scanning electron microscope (Hitachi).
YTH Screen and Interaction Analysis
A YTH screen using GAD-FYVE1(∆FYVE; cloned into pDest-AD-CYH2) as bait was performed as described previously (Dreze et al., 2010) against a collection of 12,000 Gal4 DNA-binding domain (GBD)-fused Arabidopsis ORFs (Weßling et al., 2014) using Y8800/Y8930 (MATa/α, leu2-3,112 trp1-901 his3-200 ura3-52 Gal4D gal80D GAL2-ADE2 LYS2::GAL1-HIS3 MET2::GAL7-lacZ cyh2R) strains. The bait was screened by yeast mating against a collection of 12,000 Arabidopsis ORFs fused to the GBD (Weßling et al., 2014). The screen was done as a binary 1:1 screen (i.e. the single bait was screened against 12,000 individual clones in our collection, and growth was assayed using the HIS3 and ADE2 reporters, the former using 1 mm 3-amino-1,2,4-triazole to suppress background growth). This screen was carried out independently three times, and candidates were sequence verified. Only partners that appeared in at least two of the experiments were selected for further analysis. For a targeted analysis of protein pairs, GBD-fused SH3P1, SH3P2, SH3P3, (cloned into pGBKT7), and GAD-fused FYVE1(∆FYVE; cloned into pGADT7) were transformed into Y8800 cells, and double transformants were grown on selective plates to test auxotrophic growth.
Sequence data from this article can be found in the Arabidopsis Genome Initiative Database under the following accession numbers: ACT8 (AT1G49240), AMSH3 (AT4G16144), ATG8a (AT4G21980), ATG8b (AT4G04620), ATG8c (AT1G62040), ATG8d (AT2G05630), ATG8e (AT2G45170), ATG8f (AT4G16520), ATG8g (AT3G60640), ATG8h (AT3G06420), ATG8i (AT3G15580), FYVE1 (At1g20110), NBR1 (AT4G24690), SH3P1 (At1g31440), SH3P2 (AT4g34660), and SH3P3 (At4g18060).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Complementation of vfd1(fyve1-2).
Supplemental Figure S2. Phenotypes of fyve1 compared with amsh3.
Supplemental Figure S3. Protein expression analysis of YTH clones.
Supplemental Figure S4. Genotyping of GFP-CT24-expressing seeds.
Supplemental Figure S5. Effect of SKD1(EQ) expression on endosomal markers.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Detlef Weigel and Markus Schmid (Max Planck Institute for Developmental Biology) for the whole-genome sequencing of vfd1(fyve1-2); Christa Lanz and Jens Riexinger (Max Planck Institute for Developmental Biology) for technical assistance in Illumina sequencing; Jerome Moriniere, Vanessa Chmielewski, and Markus Schneider (Technische Universität München) for assistance in the project; Richard Vierstra (University of Wisconsin, Madison), Tomoo Shimada and Ikuko Hara-Nishimura (Kyoto University), Tomohiro Uemura, Takashi Ueda, and Akihiko Nakano (University of Tokyo), Niko Geldner (Université Lausanne), Tsuyoshi Nakagawa (Shimane University), Christopher Grefen (Tübingen University), and Mike Blatt (University of Glasgow) for kindly sharing published materials; RIKEN-BioResource Center for the fyve1-1 mutant seeds; Emi Ito (University of Tokyo) and Carina Behringer, Balaji Enugutti, Farhah Assaad, and Alexander Steiner (Technische Universität München) for technical advice; and Frédéric Brunner (Tübingen University) for critical reading of the article.
Glossary
- DUB
deubiquitinating enzyme
- MVB
multivesicular body
- EMS
ethyl methansulfonate
- Col-0
ecotype Columbia
- SSLP
small sequence length polymorphism
- qRT
quantitative real-time
- BCECF
27-bis-(2carboxyethyl)-5-(and-6)-carboxyfluorescein
- AM
acetoxymethyl ester
- LE
late endosome
- TGN
trans-Golgi network
- EE
early endosome
- BFA
brefeldin A
- WM
Wortmannin
- YTH
yeast two-hybrid
- ORF
open reading frame
- ATPase
adenosine triphosphatase
- SNP
single nucleotide polymorphism
- H+
proton-translocating
- GBD
Galactose4 DNA-binding domain
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft SFB924 (grant no. A06 to E.I.) and SFB924 (grant no. A10 to P.B.) and the Ministry of Education, Culture, Sports, Science, and Technology, Japan (Scientific Research on Innovative Areas Grant-in-Aid no. 25119720 to M.H.S.).
References
- Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderhaar HJ, Ungermann C (2013) CORVET and HOPS tethering complexes: coordinators of endosome and lysosome fusion. J Cell Sci 126: 1307–1316 [DOI] [PubMed] [Google Scholar]
- Banta LM, Robinson JS, Klionsky DJ, Emr SD (1988) Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J Cell Biol 107: 1369–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon M, Dubeaux G, Kolb C, Isono E, Zelazny E, Vert G (2014) Polarization of IRON-REGULATED TRANSPORTER 1 (IRT1) to the plant-soil interface plays crucial role in metal homeostasis. Proc Natl Acad Sci USA 111: 8293–8298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC, Brandizzi F, Otegui MS, Sanderfoot AA (2008) The Secretory System of Arabidopsis. American Society of Plant Biologists, Rockville, MD [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Zhuang X, Gao C, Wang X, Jiang L (2014) The Arabidopsis endosomal sorting complex required for transport III regulates internal vesicle formation of the prevacuolar compartment and is required for plant development. Plant Physiol 165: 1328–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cui Y, Zhao Q, Gao C, Ding Y, Zeng Y, Ueda T, Nakano A, Jiang L (2014) Activation of the Rab7 GTPase by the MON1-CCZ1 complex is essential for PVC-to-vacuole trafficking and plant growth in Arabidopsis. Plant Cell 26: 2080–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR (2000) Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G, Dandekar A (2013) Protein secretion: how many secretory routes does a plant cell have? Plant Sci 203-204: 74–78 [DOI] [PubMed] [Google Scholar]
- Dreze M, Monachello D, Lurin C, Cusick ME, Hill DE, Vidal M, Braun P (2010) High-quality binary interactome mapping. Methods Enzymol 470: 281–315 [DOI] [PubMed] [Google Scholar]
- Ebine K, Inoue T, Ito J, Ito E, Uemura T, Goh T, Abe H, Sato K, Nakano A, Ueda T (2014) Plant vacuolar trafficking occurs through distinctly regulated pathways. Curr Biol 24: 1375–1382 [DOI] [PubMed] [Google Scholar]
- Ebine K, Okatani Y, Uemura T, Goh T, Shoda K, Niihama M, Morita MT, Spitzer C, Otegui MS, Nakano A, et al. (2008) A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell 20: 3006–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuji K, Shimada T, Takahashi H, Tamura K, Koumoto Y, Utsumi S, Nishizawa K, Maruyama N, Hara-Nishimura I (2007) Arabidopsis vacuolar sorting mutants (green fluorescent seed) can be identified efficiently by secretion of vacuole-targeted green fluorescent protein in their seeds. Plant Cell 19: 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Luo M, Zhao Q, Yang R, Cui Y, Zeng Y, Xia J, Jiang L (2014) A unique plant ESCRT component, FREE1, regulates multivesicular body protein sorting and plant growth. Curr Biol 24: 2556–2563 [DOI] [PubMed] [Google Scholar]
- Geldner N, Dénervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J (2009) Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C, Donald N, Hashimoto K, Kudla J, Schumacher K, Blatt MR (2010) A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J 64: 355–365 [DOI] [PubMed] [Google Scholar]
- Haas TJ, Sliwinski MK, Martínez DE, Preuss M, Ebine K, Ueda T, Nielsen E, Odorizzi G, Otegui MS (2007) The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. Plant Cell 19: 1295–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD (2011) The ESCRT pathway. Dev Cell 21: 77–91 [DOI] [PubMed] [Google Scholar]
- Hirano T, Matsuzawa T, Takegawa K, Sato MH (2011) Loss-of-function and gain-of-function mutations in FAB1A/B impair endomembrane homeostasis, conferring pleiotropic developmental abnormalities in Arabidopsis. Plant Physiol 155: 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NH, Mattsson O, Jørgensen LB, Jones JD, Mundy J, Petersen M (2009) Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137: 773–783 [DOI] [PubMed] [Google Scholar]
- Isono E, Katsiarimpa A, Müller IK, Anzenberger F, Stierhof YD, Geldner N, Chory J, Schwechheimer C (2010) The deubiquitinating enzyme AMSH3 is required for intracellular trafficking and vacuole biogenesis in Arabidopsis thaliana. Plant Cell 22: 1826–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito E, Fujimoto M, Ebine K, Uemura T, Ueda T, Nakano A (2012) Dynamic behavior of clathrin in Arabidopsis thaliana unveiled by live imaging. Plant J 69: 204–216 [DOI] [PubMed] [Google Scholar]
- Katsiarimpa A, Anzenberger F, Schlager N, Neubert S, Hauser MT, Schwechheimer C, Isono E (2011) The Arabidopsis deubiquitinating enzyme AMSH3 interacts with ESCRT-III subunits and regulates their localization. Plant Cell 23: 3026–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsiarimpa A, Kalinowska K, Anzenberger F, Weis C, Ostertag M, Tsutsumi C, Schwechheimer C, Brunner F, Hückelhoven R, Isono E (2013) The deubiquitinating enzyme AMSH1 and the ESCRT-III subunit VPS2.1 are required for autophagic degradation in Arabidopsis. Plant Cell 25: 2236–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsiarimpa A, Muñoz A, Kalinowska K, Uemura T, Rojo E, Isono E (2014) The ESCRT-III-interacting deubiquitinating enzyme AMSH3 is essential for degradation of ubiquitinated membrane proteins in Arabidopsis thaliana. Plant Cell Physiol 55: 727–736 [DOI] [PubMed] [Google Scholar]
- Kulich I, Pečenková T, Sekereš J, Smetana O, Fendrych M, Foissner I, Höftberger M, Zárský V (2013) Arabidopsis exocyst subcomplex containing subunit EXO70B1 is involved in autophagy-related transport to the vacuole. Traffic 14: 1155–1165 [DOI] [PubMed] [Google Scholar]
- Kushnirov VV. (2000) Rapid and reliable protein extraction from yeast. Yeast 16: 857–860 [DOI] [PubMed] [Google Scholar]
- Lam BC, Sage TL, Bianchi F, Blumwald E (2001) Role of SH3 domain-containing proteins in clathrin-mediated vesicle trafficking in Arabidopsis. Plant Cell 13: 2499–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bassham DC (2012) Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol 63: 215–237 [DOI] [PubMed] [Google Scholar]
- Mukhtar MS, Carvunis AR, Dreze M, Epple P, Steinbrenner J, Moore J, Tasan M, Galli M, Hao T, Nishimura MT, et al. (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333: 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nishizawa K, Maruyama N, Satoh R, Fuchikami Y, Higasa T, Utsumi S (2003) A C-terminal sequence of soybean β-conglycinin α′ subunit acts as a vacuolar sorting determinant in seed cells. Plant J 34: 647–659 [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Clark RM, Lanz C, Warthmann N, Weigel D (2008) Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res 18: 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AR, Suttangkakul A, Vierstra RD (2008) The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178: 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH (1992) Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell 3: 1389–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes FC, Buono R, Otegui MS (2011) Plant endosomal trafficking pathways. Curr Opin Plant Biol 14: 666–673 [DOI] [PubMed] [Google Scholar]
- Rojo E, Gillmor CS, Kovaleva V, Somerville CR, Raikhel NV (2001) VACUOLELESS1 is an essential gene required for vacuole formation and morphogenesis in Arabidopsis. Dev Cell 1: 303–310 [DOI] [PubMed] [Google Scholar]
- Sanderfoot AA, Raikhel N (2003) The Secretory System of Arabidopsis. American Society of Plant Biologists, Rockville, MD [Google Scholar]
- Sanmartín M, Ordóñez A, Sohn EJ, Robert S, Sánchez-Serrano JJ, Surpin MA, Raikhel NV, Rojo E (2007) Divergent functions of VTI12 and VTI11 in trafficking to storage and lytic vacuoles in Arabidopsis. Proc Natl Acad Sci USA 104: 3645–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuring D, Viotti C, Krüger F, Künzl F, Sturm S, Bubeck J, Hillmer S, Frigerio L, Robinson DG, Pimpl P, et al. (2011) Multivesicular bodies mature from the trans-Golgi network/early endosome in Arabidopsis. Plant Cell 23: 3463–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K, Hagmann J, Ossowski S, Warthmann N, Gesing S, Kohlbacher O, Weigel D (2009a) Simultaneous alignment of short reads against multiple genomes. Genome Biol 10: R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jørgensen JE, Weigel D, Andersen SU (2009b) SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods 6: 550–551 [DOI] [PubMed] [Google Scholar]
- Serrazina S, Dias FV, Malhó R (2014) Characterization of FAB1 phosphatidylinositol kinases in Arabidopsis pollen tube growth and fertilization. New Phytol 203: 784–793 [DOI] [PubMed] [Google Scholar]
- Shahriari M, Keshavaiah C, Scheuring D, Sabovljevic A, Pimpl P, Häusler RE, Hülskamp M, Schellmann S (2010) The AAA-type ATPase AtSKD1 contributes to vacuolar maintenance of Arabidopsis thaliana. Plant J 64: 71–85 [DOI] [PubMed] [Google Scholar]
- Shimada T, Koumoto Y, Li L, Yamazaki M, Kondo M, Nishimura M, Hara-Nishimura I (2006) AtVPS29, a putative component of a retromer complex, is required for the efficient sorting of seed storage proteins. Plant Cell Physiol 47: 1187–1194 [DOI] [PubMed] [Google Scholar]
- Shimada T, Yamada K, Kataoka M, Nakaune S, Koumoto Y, Kuroyanagi M, Tabata S, Kato T, Shinozaki K, Seki M, et al. (2003) Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana. J Biol Chem 278: 32292–32299 [DOI] [PubMed] [Google Scholar]
- Singh MK, Krüger F, Beckmann H, Brumm S, Vermeer JE, Munnik T, Mayer U, Stierhof YD, Grefen C, Schumacher K, et al. (2014) Protein delivery to vacuole requires SAND protein-dependent Rab GTPase conversion for MVB-vacuole fusion. Curr Biol 24: 1383–1389 [DOI] [PubMed] [Google Scholar]
- Spitzer C, Reyes FC, Buono R, Sliwinski MK, Haas TJ, Otegui MS (2009) The ESCRT-related CHMP1A and B proteins mediate multivesicular body sorting of auxin carriers in Arabidopsis and are required for plant development. Plant Cell 21: 749–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenning S, Lamark T, Krause K, Johansen T (2011) Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7: 993–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD (2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Uemura T, Sato MH, Nakano A (2004) Functional differentiation of endosomes in Arabidopsis cells. Plant J 40: 783–789 [DOI] [PubMed] [Google Scholar]
- Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH (2004) Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct Funct 29: 49–65 [DOI] [PubMed] [Google Scholar]
- van Leeuwen W, Okrész L, Bögre L, Munnik T (2004) Learning the lipid language of plant signalling. Trends Plant Sci 9: 378–384 [DOI] [PubMed] [Google Scholar]
- Vermeer JE, van Leeuwen W, Tobeña-Santamaria R, Laxalt AM, Jones DR, Divecha N, Gadella TW Jr, Munnik T (2006) Visualization of PtdIns3P dynamics in living plant cells. Plant J 47: 687–700 [DOI] [PubMed] [Google Scholar]
- Viotti C, Krüger F, Krebs M, Neubert C, Fink F, Lupanga U, Scheuring D, Boutté Y, Frescatada-Rosa M, Wolfenstetter S, et al. (2013) The endoplasmic reticulum is the main membrane source for biogenesis of the lytic vacuole in Arabidopsis. Plant Cell 25: 3434–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Anraku Y (1992) Genes for directing vacuolar morphogenesis in Saccharomyces cerevisiae. II. VAM7, a gene for regulating morphogenic assembly of the vacuoles. J Biol Chem 267: 18671–18675 [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2001) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- Weßling R, Epple P, Altmann S, He Y, Yang L, Henz SR, McDonald N, Wiley K, Bader KC, Gläßer C, et al. (2014) Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe 16: 364–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W. (2010) Membrane fusion: five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol 26: 115–136 [DOI] [PubMed] [Google Scholar]
- Winter V, Hauser MT (2006) Exploring the ESCRTing machinery in eukaryotes. Trends Plant Sci 11: 115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wywial E, Singh SM (2010) Identification and structural characterization of FYVE domain-containing proteins of Arabidopsis thaliana. BMC Plant Biol 10: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M, Shimada T, Takahashi H, Tamura K, Kondo M, Nishimura M, Hara-Nishimura I (2008) Arabidopsis VPS35, a retromer component, is required for vacuolar protein sorting and involved in plant growth and leaf senescence. Plant Cell Physiol 49: 142–156 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y (2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Hicks GR, Raikhel NV (2014) Plant vacuole morphology and vacuolar trafficking. Front Plant Sci 5: 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Wang H, Lam SK, Gao C, Wang X, Cai Y, Jiang L (2013) A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 25: 4596–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouhar J, Rojo E, Bassham DC (2009) AtVPS45 is a positive regulator of the SYP41/SYP61/VTI12 SNARE complex involved in trafficking of vacuolar cargo. Plant Physiol 149: 1668–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.