The Arabidopsis AXY9 gene encodes a plant-specific component of a cell wall polysaccharide acetylation pathway.
Abstract
A mutation in the ALTERED XYLOGLUCAN9 (AXY9) gene was found to be causative for the decreased xyloglucan acetylation phenotype of the axy9.1 mutant, which was identified in a forward genetic screen for Arabidopsis (Arabidopsis thaliana) mutants. The axy9.1 mutant also exhibits decreased O-acetylation of xylan, implying that the AXY9 protein has a broad role in polysaccharide acetylation. An axy9 insertional mutant exhibits severe growth defects and collapsed xylem, demonstrating the importance of wall polysaccharide O-acetylation for normal plant growth and development. Localization and topological experiments indicate that the active site of the AXY9 protein resides within the Golgi lumen. The AXY9 protein appears to be a component of the plant cell wall polysaccharide acetylation pathway, which also includes the REDUCED WALL ACETYLATION and TRICHOME BIREFRINGENCE-LIKE proteins. The AXY9 protein is distinct from the TRICHOME BIREFRINGENCE-LIKE proteins, reported to be polysaccharide acetyltransferases, but does share homology with them and other acetyltransferases, suggesting that the AXY9 protein may act to produce an acetylated intermediate that is part of the O-acetylation pathway.
The plant cell wall is a complex composite of polysaccharides, glycoproteins, and polyphenols, with the fine structure and quantity of each varying by species, tissue, and developmental time point (Knox, 2008; Burton et al., 2010). Cellulose, hemicelluloses, and pectic polysaccharides are the three major classes of polysaccharides observed in the wall. Current models of the wall have cellulose microfibrils as the major structural component, with hemicelluloses binding to the microfibrils and pectins as an amorphous matrix in which the cellulose/hemicellulose network is embedded (Pauly et al., 1999a; Somerville et al., 2004; Cosgrove, 2005). Unlike the linear β-1,4-glucan chains making up cellulose microfibrils, hemicelluloses and pectins consist of a diverse set of glycosyl units and linkages as well as other modifications such as methylation and acetylation (Caffall and Mohnen, 2009; Scheller and Ulvskov, 2010; Pauly et al., 2013).
The O-acetyl substitutions on hemicelluloses and pectins occur on a variety of specific glycosyl residues. The hemicellulose xyloglucan (XyG) consists of a β-1,4-glucan backbone with a regular pattern of xylosyl branches, with additional galactosyl, fucosyl, arabinosyl, and/or galacturonosyl substitution depending on the tissue and plant species (Obel et al., 2009; Pauly et al., 2013; Schultink et al., 2014). XyG O-acetylation has been reported on the β-1,4-glucan backbone (Sims et al., 1996; York et al., 1996) as well as on specific galactosyl or arabinosyl side chains (Kiefer et al., 1989; Vierhuis et al., 2001). The hemicellulose xylan is heavily acetylated at positions O2 and O3 of the backbone β-1,4-xylosyl residues, with the degree of acetylation (O-acetyl groups per backbone of xylosyl residue) ranging from approximately 0.4 to 0.6 depending on the species (Teleman et al., 2002; Evtuguin et al., 2003; Prozil et al., 2012; Chong et al., 2014; Lee et al., 2014). The glycosyl substituents of xylan, including glucuronosyl, arabinosyl, and xylosyl groups, have not been reported to be acetylated. The backbone β-1,4-mannosyl residues of the hemicellulosic polysaccharide mannan also can be acetylated (Manna and McAnalley, 1993). The predominant location of O-acetyl groups in pectin has been reported to be on galacturonic acid residues at positions O2 and O3 (Ralet et al., 2005). O-Acetylation of pectin also has been observed on rhamnosyl (Sengkhamparn et al., 2009), fucosyl, and aceric acid residues (Glushka et al., 2003).
The functional significance and biosynthetic pathway of wall polysaccharide O-acetylation are not fully understood. O-Acetylation has been shown to influence the solubility, gelation, and enzymatic accessibility of polysaccharides in vitro (Biely et al., 1986; Huang et al., 2002). These properties are likely to be important for appropriate function in planta. Recently identified Arabidopsis (Arabidopsis thaliana) mutants with polysaccharide O-acetylation deficiencies (reduced wall acetylation [rwa] and trichome birefringence-like [tbl]; Gille and Pauly, 2012) have allowed for testing of the in vivo role of this substituent. The ALTERED XYLOGLUCAN4 (AXY4 [TBL27]) gene from the TBL family was identified in a forward genetic screen of Arabidopsis and is believed to code for a XyG acetyltransferase (Gille et al., 2011). The growth morphology of this mutant, which lacks XyG O-acetylation in leaves, etiolated seedlings, and roots, was not affected under laboratory growth conditions. Arabidopsis mutants deficient for a putative xylan acetyltransferase (TBL29/ESKIMO1 [ESK1]) were reported to have reduced growth and irregular xylem and to be freezing tolerant (Xin et al., 2007; Xiong et al., 2013; Yuan et al., 2013). Arabidopsis mutants deficient for other TBL genes have been reported to exhibit phenotypes such as aberrant trichomes (Bischoff et al., 2010a) and resistance to powdery mildew (Vogel et al., 2004), but polysaccharide acetylation defects have not been demonstrated in these cases. The variation in the morphological phenotypes of different tbl mutants suggests that the function of polysaccharide acetylation is specific to the particular polysaccharide and tissue.
While the TBL gene products seem to affect single wall polysaccharides, Arabidopsis mutants defective for one or more of the four RWA genes have decreased acetylation of multiple polysaccharides and growth phenotypes ranging from mild to severe (Lee et al., 2011; Manabe et al., 2011, 2013). For this reason, and because the RWA proteins are integral membrane proteins with 10 predicted transmembrane domains, it has been hypothesized that they may act as transporters for an activated form of acetate into the Golgi apparatus (Manabe et al., 2011). It has been demonstrated that acetyl-CoA is involved in the pathway of pectin acetylation (Pauly and Scheller, 2000); however, it is not clear if acetyl-CoA is transported into the Golgi or there is an alternative donor substrate that acts as a carrier.
In this study, we report the identification and characterization of AXY9, an additional component of the plant cell wall polysaccharide acetylation pathway.
RESULTS
Identification of the AXY9 Gene
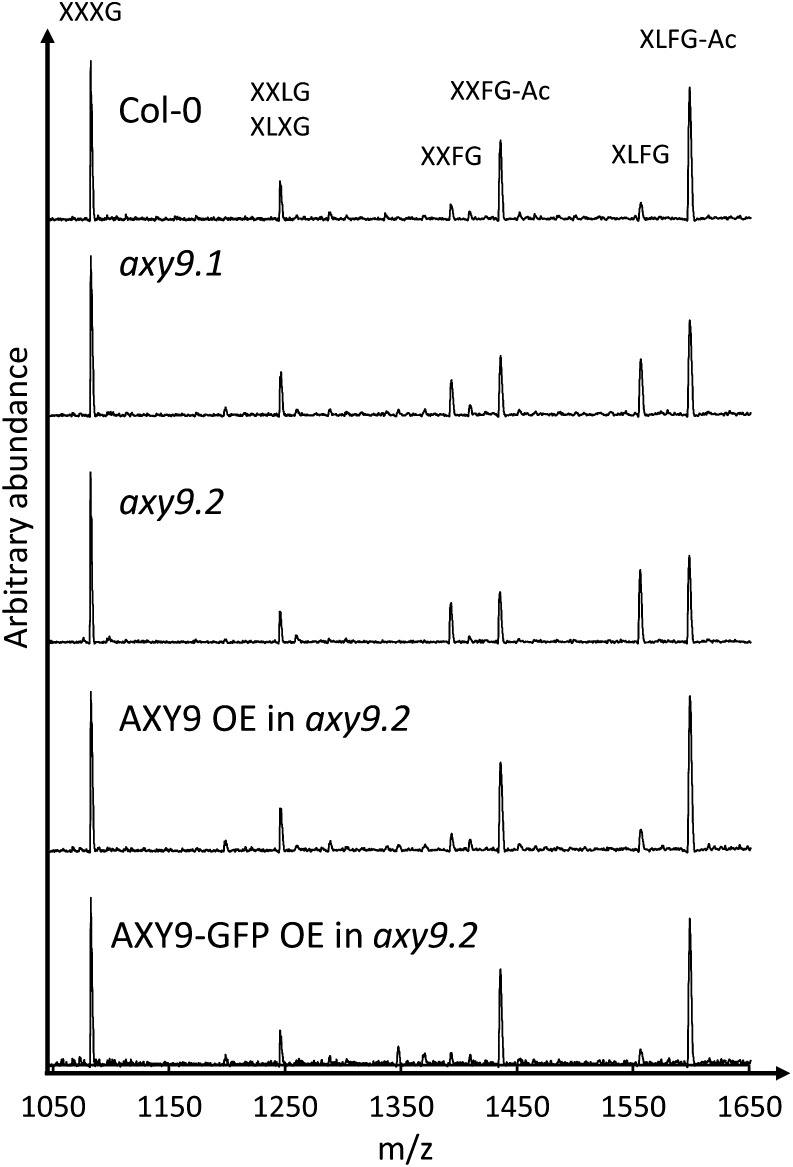
The axy9.1 mutant was identified from a forward genetic screen of an M2 population of ethyl methanesulfonate-mutagenized Arabidopsis by oligosaccharide mass profiling of XyG (Lerouxel et al., 2002). XyG O-acetylation was reduced in cotyledons and etiolated seedlings in the axy9.1 mutant by 25% and 60%, respectively (Fig. 1; Supplemental Fig. S1).
Figure 1.
XyG oligosaccharide profiles of cotyledons from 12-d-old plants. The observed oligosaccharides are labeled using the nomenclature described by Fry et al. (1993). Ac, Acetyl; m/z, mass-to-charge ratio; OE, overexpression.
Genetic linkage analysis was performed utilizing an F2 mapping population from a cross between axy9.1 (Arabidopsis ecotype Columbia-0 [Col-0]) and the Arabidopsis ecotype Landsberg erecta. XyG oligosaccharide mass profiling was performed on leaf tissue of these F2 plants to identify mutants based on the relative abundance of the nonacetylated XyG oligosaccharide XXFG (mass-to-charge ratio = 1,393), which is increased in the mutant. Out of 187 F2 plants phenotyped, 41 mutant and 133 wild-type plants were identified (Supplemental Fig. S2). This segregation ratio is consistent with a single recessive locus based on the χ2 test (P > 0.60). The selected mutants were genotyped at various positions along each chromosome to obtain the Col-0 allele frequency, with enrichment of Col-0 alleles found at the beginning of chromosome 3 (Supplemental Table S1; Supplemental Fig. S3A). Additional markers were used to narrow the genomic region to the first 1.6 Mb of chromosome 3 (Supplemental Table S1; Supplemental Fig. S3B).
The genomic DNA of homozygous axy9.1 plants was subjected to Illumina sequencing and compared with that of other plants from the same mutant population (Gille et al., 2011; Günl et al., 2011; Günl and Pauly, 2011). A total of 148 homozygous, amino acid-changing single-nucleotide polymorphisms (SNPs) were identified as being unique to the axy9.1 mutant, two of which were within the 1.6-Mb region of interest (Supplemental Tables S2 and S3). Transfer DNA (T-DNA) lines containing insertions in the genes of interest were obtained for the two candidate genes (At3g01530 and At3g03210). One of the T-DNA lines, SALK_090612, containing an insertion in the gene At3g03210, was found to have decreased XyG O-acetylation (Fig. 1; Supplemental Fig. S1), and as a second axy9 allele was named axy9.2. The XyG acetylation phenotype of axy9.2 was rescued by overexpression of the coding sequence for this gene (Fig. 1; Supplemental Fig. S1), demonstrating that disruption of At3g03210 is causal for the decreased XyG O-acetylation phenotype. Therefore, the gene At3g03210 was named AXY9.
The SNP identified by Illumina sequencing of axy9.1, resulting in a stop codon at the 276th codon, was confirmed by Sanger sequencing (Supplemental Fig. S4). The precise location of the T-DNA insertion within the AXY9 coding sequence in axy9.2 was identified by sequencing a PCR product spanning the border of the T-DNA insertion. The insertion was found to disrupt the coding sequence of AXY9 starting at the 284th codon and resulted in a predicted polypeptide containing the first 283 amino acids of AXY9 followed by 25 aberrant amino acids coded for by the T-DNA insertion (Supplemental Fig. S4). The presence of AXY9 transcript was checked by reverse transcription (RT)-PCR and quantitative reverse transcription (qRT)-PCR with primer pairs before, spanning, and after the T-DNA insertion (Supplemental Fig. S5). AXY9 transcript was detected in the axy9.2 mutant using the primer pairs before and after the T-DNA insertion but not for the spanning pair (Supplemental Fig. S5), indicating that a partial AXY9 transcript is present in this line.
Distribution and Sequence Conservation of the AXY9 Gene
The AXY9 gene is present as a single copy in Arabidopsis, with no close homologs detectable by BLAST search in the current genome (The Arabidopsis Information Resource [TAIR]10; http://www.arabidopsis.org/). Between one and four putative orthologous AXY9 genes were identified in each of the 35 currently available land plant genomes in the Phytozome database, including those from early-diverging lineages such as the moss Physcomitrella patens and the spikemoss Selaginella moellendorffii (www.phytozome.net). A phylogenetic tree of AXY9 protein sequences is shown in Supplemental Figure S6. An AXY9 ortholog could not be identified in any green algae or other nonland plant species, suggesting that this gene is specific to land plants. The N terminus of the AXY9 protein is highly conserved and contains two hydrophobic stretches separated by four hydrophilic residues (Supplemental Fig. S7). The strong conservation of this domain suggests an important functional role besides simply anchoring the protein in the membrane. Transmembrane domain prediction tools (http://aramemnon.uni-koeln.de/; Schwacke et al., 2003) tend to predict each of these hydrophobic stretches as transmembrane domains, suggesting that the N and C termini would be on the same side of the Golgi membrane. However, experimental evidence (described below) suggests that there is only a single transmembrane domain. While not part of the TBL family, AXY9 does share limited sequence homology with certain parts of these proteins, including a putative GDS active site and a DXXH motif (Supplemental Fig. S8; Akoh et al., 2004; Bischoff et al., 2010b; Gille and Pauly, 2012; Moynihan and Clarke, 2014b). According to the Arabidopsis eFP browser, the AXY9 gene is expressed throughout all examined developmental stages, with moderately elevated expression observed in mature pollen, seeds, and stems (Winter et al., 2007).
The axy9.2 Mutant Has Defects in Growth and Xylem Morphology
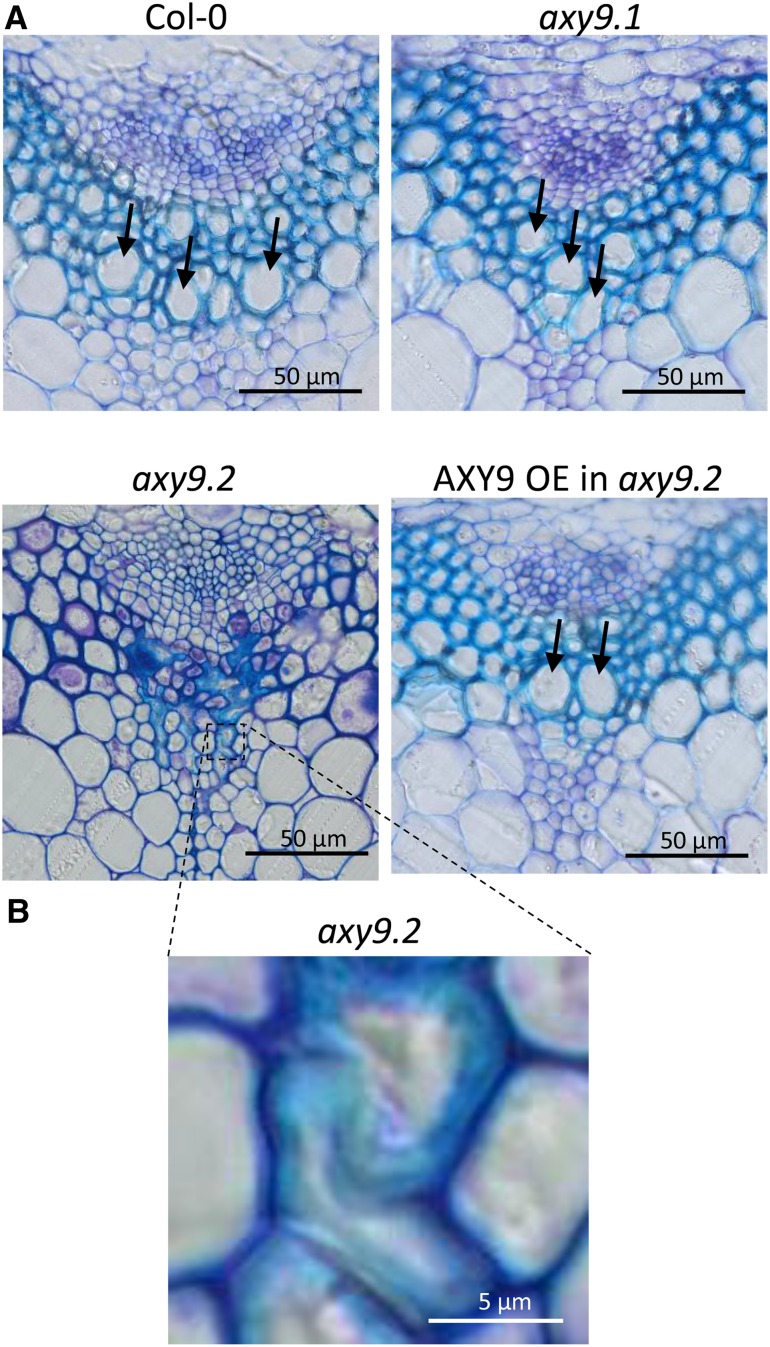
While axy9.2 etiolated seedlings appeared similar to the wild type, soil-grown plants exhibited a severe growth defect (Fig. 2). The leaves of the axy9.2 plants were much smaller and darker green in color, and bolting of the axy9.2 plants was delayed by 4 to 6 weeks compared with the wild type. Analysis of stem cross sections revealed alterations in xylem structure in the axy9.2 mutants (Fig. 3). Large open xylem vessels were observed in Col-0, axy9.1, and the axy9.2 complementation line but not in the axy9.2 mutant, in which the xylem vessels appear to be collapsed.
Figure 2.
Growth phenotype of axy9.2. Two plants from each genotype are shown for Col-0, axy9.1, axy9.2, and AXY9 overexpressed (OE) in the axy9.2 background. A, Seven-day-old etiolated seedlings. B to D, Soil-grown plants at 12 d (B), 21 d (C), and 36 d (D).
Figure 3.
Stem cross sections. A, Stem cross sections of the four genotypes stained with toluidine blue. B, Higher magnification of axy9.2 xylem tissue. Arrows indicate some of the large, open xylem vessels.
Disruption of AXY9 Affects the Acetylation of Multiple Polysaccharides
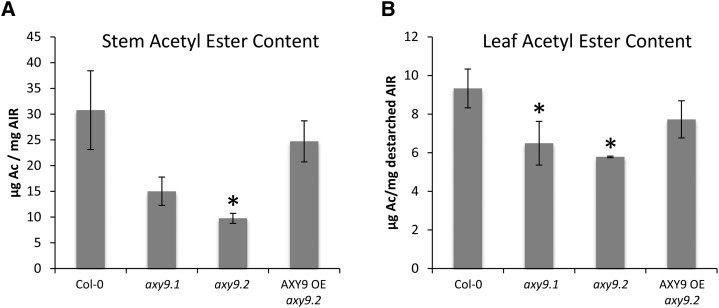
Total wall O-acetyl ester content was measured to determine if polymers other than XyG are affected in axy9 mutants, as XyG provides a relatively minor contribution to total wall acetyl ester content (Gille et al., 2011). For stem tissue, axy9.1 and axy9.2 plants were found to have 50% and 70% decreases, respectively, in overall acetyl ester content relative to Col-0 (Fig. 4A). In destarched leaf material, acetyl ester content was reduced by 30% and 35% in axy9.1 and axy9.2, respectively (Fig. 4B). Two-dimensional (2D) heteronuclear single quantum correlation (HSQC) NMR was performed on stem material to investigate xylan acetylation (Cheng et al., 2013). Total xylan O-acetylation was found to be decreased by 35% in axy9.1 and 80% in axy9.2 (Table I; Supplemental Fig. S9).
Figure 4.
Total wall-bound acetyl ester content for stem (A) and leaf tissue (B). Ac, Acetate; OE, overexpression. Error bars indicate sd (n = 3). Asterisks indicate statistically significant differences from Col-0 (Student’s t test, P < 0.05).
Table I. Analysis of O-acetylation status of xylan in inflorescence stem by 2D HSQC NMR.
The values are percentages of acetylated hydroxyl groups out of the total detected. The full spectra are shown in Supplemental Figure S9. Boldface values indicate significance compared with the wild type (P < 0.01, n = 3).
| O-Acetylation Status | Wild Type | axy9.1 | axy9.2 | AXY9 Overexpressed in axy9.2 |
|---|---|---|---|---|
| O2-Acetate | 23.0 ± 0.8 | 14.2 ± 0.5 | 4.4 ± 0.1 | 19.5 ± 0.4 |
| O3-Acetate | 25.7 ± 2.1 | 18.2 ± 0.7 | 4.7 ± 0.1 | 25.1 ± 0.5 |
| O2,3-Acetate | 8.1 ± 0.4 | 4.1 ± 0.7 | 1.8 ± 0.2 | 8.1 ± 0.6 |
| Total | 56.9 ± 2.4 | 36.4 ± 1.9 | 11.0 ± 0.3 | 53.0 ± 0.8 |
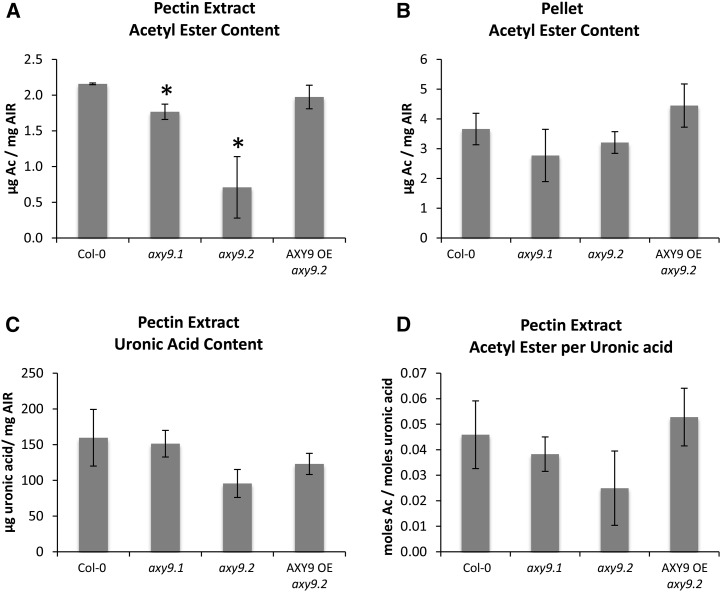
To investigate the effect of disrupting the AXY9 gene on pectin acetylation, we examined the acetyl ester content of etiolated seedlings, as seedlings contain significant amounts of pectin and, unlike later in development, the axy9.2 plants do not have a strong morphological defect at this stage (Fig. 2). Cell wall material was treated with pectinases, and the acetyl ester contents of the released pectin fraction (Fig. 5A) and the remaining pellet (Fig. 5B) were measured. Decreases of 18% and 67% were observed for the axy9.1 and axy9.2 pectin fractions, respectively. No statistically significant difference was observed for the remaining pellet. To determine whether the observed decrease in acetyl ester content of the pectinase-released fraction was due to decreased pectin acetylation or a decrease in the amount of enzymatically released pectin, the uronic acid content of the pectin fraction was determined (Fig. 5C) and the molar ratio of acetyl esters per uronic acid was calculated (Fig. 5D). A statistically significant difference (P < 0.05) in the ratio of acetyl ester per uronic acid was not observed between Col-0 and either of the axy9 mutant lines, indicating that pectin O-acetylation status is not affected as determined with the experimental setup used here.
Figure 5.
A and B, Total acetic ester content of a pectin extract (A) and the remaining pellet (B) of etiolated seedling material. C and D, The uronic acid content was measured for the pectin extract (C), and the molar ratio of acetic acid to uronic acid was calculated (D). Ac, Acetate; OE, overexpression. Error bars indicate sd (n = 3). Asterisks indicate statistically significant differences from Col-0 (Student’s t test, P < 0.05).
AXY9 Localization and Topology
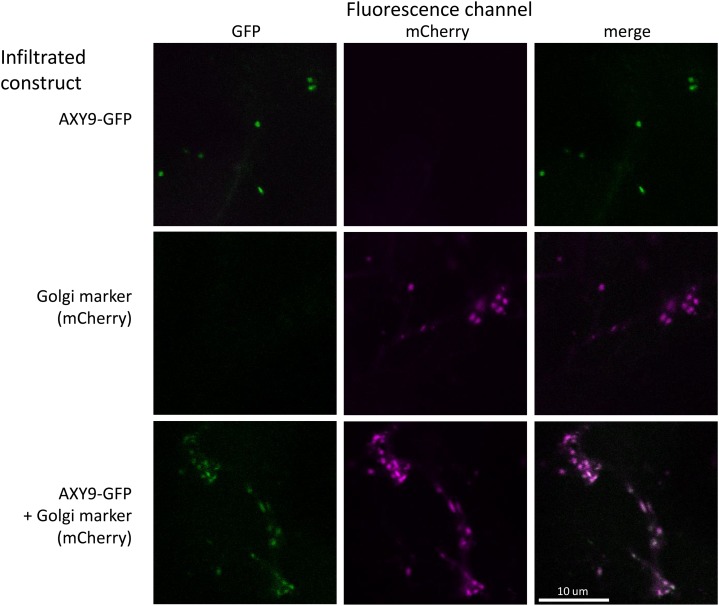
The subcellular localization of the AXY9 protein was investigated by generating a C-terminal GFP fusion construct that was transiently expressed in Nicotiana benthamiana. A GFP signal was observed in small mobile vesicles and colocalized with a known Golgi marker (Nelson et al., 2007; Fig. 6). The functionality of the AXY9-GFP fusion protein was tested by transformation of the construct into the axy9.2 background, where it fully or partially rescued the XyG acetylation phenotype of the mutant (Fig. 1; Supplemental Fig. S1). A GFP signal was observed in these stable lines in small mobile vesicles, consistent with the localization of other Golgi-associated proteins (Supplemental Fig. S10; Nelson et al., 2007; Chiu et al., 2012; Jensen et al., 2012).
Figure 6.
Subcellular localization of AXY9. A fusion construct of AXY9-GFP was transiently expressed in N. benthamiana with or without a Golgi marker (N-terminal fragment of a mannosidase from soybean [Glycine max] fused to mCherry; Nelson et al., 2007). Leaves of the infiltrated plants were examined by confocal microscopy.
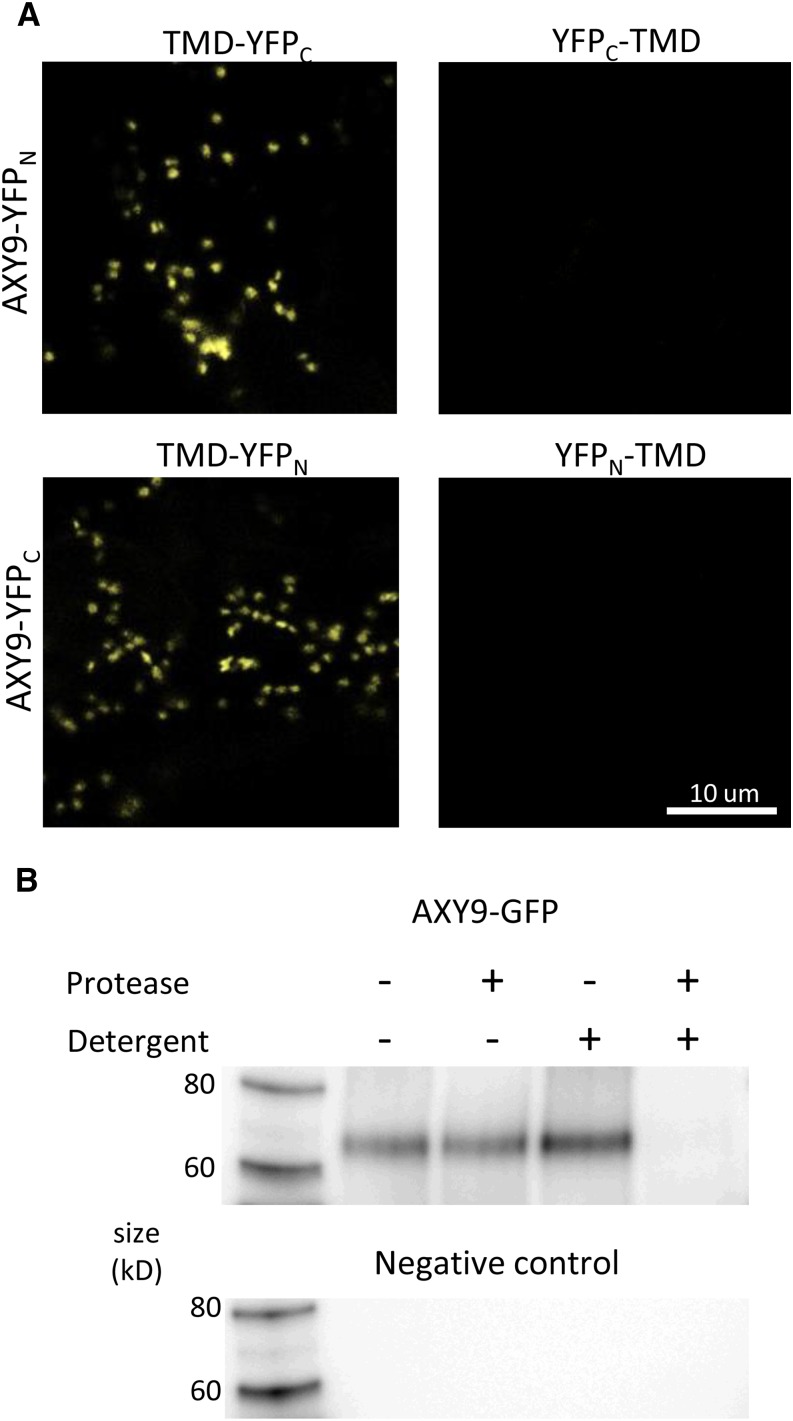
The topology of the AXY9 protein within the Golgi membrane was determined using both a split yellow fluorescent protein (YFP) assay (Søgaard et al., 2012) and a protease protection assay (Akiyama and Ito, 1987; Boyd et al., 1987). In the split YFP assay, the N- and C-terminal portions of the YFP protein are expressed as separate polypeptides and will form a functional YFP protein only if both peptides are present in the same compartment of the cell. Constructs containing a transmembrane domain (TMD) of a truncated sialyltransferase from rat fused to the N- or C-terminal portion of YFP (YFPN or YFPC, respectively) were used as controls (Søgaard et al., 2012). When a YFP domain is fused to the N or C terminus of TMD, the YFP domain localizes outside or inside the Golgi, respectively (Søgaard et al., 2012). When the split YFP TMD control constructs were transiently expressed in N. benthamiana, a YFP signal was observed only in the combinations TMD-YFPN with TMD-YFPC and YFPN-TMD with YFPC-TMD (Supplemental Fig. S11). A signal was not observed for YFPN-TMD with TMD-YFPC or TMD-YFPN with YFPC-TMD, indicating that the constructs are specific for orienting the YFPN/C peptide either inside or outside of the Golgi. For TMD-YFPN and TMD-YFPC, the YFP signal was observed specifically in Golgi-like small mobile puncta, whereas in YFPC-TMD and YFPN-TMD, the YFP signal was observed both in small mobile puncta and long filamentous structures reminiscent of endoplasmic reticulum-like structures (Supplemental Fig. S11). The failure of these peptides to be restricted to the Golgi in the latter case may have been an artifact of overexpression; however, the topology of the peptides appeared to be preserved despite the mislocalization.
This split YFP system was utilized to gain insights into the orientation of AXY9. Chimeric constructs were made with the AXY9 coding sequence fused to that of either YFPN or YFPC at the 5′ or 3′ end. A YFP signal was observed for AXY9-YFPN/C coexpressed with the complementary TMD-YFPN/C but not with YFPN/C-TMD, indicating that the C terminus of the AXY9 protein is present in the Golgi lumen (Fig. 7A). A YFP signal was observed for YFPC-TMD expressed with YFPN-AXY9, indicating that the N terminus of AXY9 resides outside of the Golgi (Supplemental Fig. S12).
Figure 7.
Protein topology of AXY9. A, Split YFP protein fusions, AXY9-YFPN and AXY9-YFPC, were coexpressed with the complementary half of the YFP expressed inside (TMD-YFPN/C) or outside (YFPN/C-TMD) of the Golgi in N. benthamiana and examined using confocal microscopy. B, Protease protection assay using microsomes prepared from N. benthamiana transiently expressing the AXY9-GFP fusion protein. A western blot using an anti-GFP antibody was used to detect the fusion peptide after digestion with (+) or without (−) protease and detergent.
A protease protection assay was performed to independently determine the topology of the AXY9 protein. Protein fusion constructs (AXY9-GFP, YFP-TMD, and TMD-YFP) were transiently expressed in N. benthamiana, and microsomal preparations were generated from leaves. The microsomes were then treated with or without protease and detergent. The YFP-TMD protein, detected by western blot, was sensitive to protease with or without detergent, confirming a cytosolic orientation of the YFP domain (Supplemental Fig. S13). The TMD-YFP protein was protected from protease alone, but treatment with protease and detergent together resulted in protein degradation, indicating a lumenal localization for the YFP domain (Supplemental Fig. S13). These results are consistent with those reported previously and serve as controls that the assay is working as expected (Søgaard et al., 2012). The AXY9-GFP fusion polypeptide was detected intact near its expected mass of 72 kD by western blot with an anti-GFP antibody at similar intensities without or with protease in the absence of detergent (Fig. 7B). Coincubation with both detergent and protease resulted in a loss of detection of the AXY9-GFP fusion protein, indicating degradation of the polypeptide under these conditions. The protease protection results thus confirm the interpretation of the split-YFP assays that the C terminus of the AXY9 protein is located in the Golgi lumen.
DISCUSSION
Functionality of the axy9 Alleles
The small stature, delayed flowering, dark-green coloration, and collapsed xylem vessels that represent the severe growth phenotype of the axy9.2 mutant can be rescued by genetic complementation, indicating that a defect in the AXY9 gene is indeed responsible for these phenotypes (Figs. 2 and 3). As the axy9.1 plants do not have a strong growth phenotype like axy9.2 and show less of a defect in XyG and xylan O-acetylation (Fig. 1; Table I; Supplemental Fig. S1), the mutation in axy9.1 appears to be a weak allele. The stop codon in axy9.1 occurs before the conserved DXXH motif, which as a conserved feature is likely to be important for protein function. However, limited activity of a bacterial O-acetyltransferase was retained despite mutation of the Asp and His residues in the corresponding motif, suggesting that the motif may in fact be dispensable (Moynihan and Clarke, 2014b). The axy9.2 mutant still contains acetylated XyG, suggesting that either there is an AXY9-independent pathway for XyG acetylation or that axy9.2 also represents a weak allele rather than a knockout. As the T-DNA insertion present in the AXY9 gene in axy9.2 occurs after the point mutation present in the axy9.1 allele, the predicted protein in axy9.2 contains all of the amino acids present in the truncated AXY9 protein produced in the axy9.1 mutant (Supplemental Fig. S4). This suggests that axy9.2 has the potential to retain limited AXY9 functionality, if axy9.1 is indeed a weak allele.
Polymer O-acetylation defects were observed across several tissues and polymers in axy9.1 and axy9.2, and while axy9.2 tended to have a larger deficiency than axy9.1, the precise magnitude of the defect was variable. For example, a 60% decrease in XyG acetylation was observed in axy9.1 etiolated seedlings, whereas only a 25% decreased was observed in cotyledons (Supplemental Fig. S1). Such differences also were observed for some of the rwa mutants (Manabe et al., 2013) and may be influenced by differing rates of O-acetyl ester deposition into the cell wall between tissues and developmental stages.
Growth Phenotype of the axy9.2 Mutant
Arabidopsis mutants lacking XyG O-acetylation have not been reported to display any growth phenotype (Gille et al., 2011), demonstrating that the reduction in XyG O-acetylation present in the axy9.2 mutant cannot account for the severe growth phenotype observed in these plants. However, there are several mutants deficient for xylan biosynthesis that have been demonstrated to have growth phenotypes reminiscent of that of the axy9.2 mutant with varying degrees of severity. These include knockouts of a xylan acetyltransferase (TBL29/ESK1; Xiong et al., 2013; Yuan et al., 2013), the RWA quadruple mutant (Manabe et al., 2013), as well as several glycosyltransferases (Brown et al., 2007). As these mutants tend to have irregular or partially collapsed xylem vessels, it is thought that the aberrant xylan produced in these mutants leads to a weaker xylem wall that is unable to withstand the negative pressure of the xylem vessels during water transport (Turner and Somerville, 1997; Turner et al., 2001; Persson et al., 2007). It is likely, therefore, that the decreased degree of xylan acetylation observed in the axy9.2 mutant contributes to the severe growth phenotype of the plant. Differences in the staining of the xylem parenchyma and interfascicular regions also were observed between the axy9.2 mutant and the other genotypes (Fig. 3); however, these differences may result from developmental or pleiotropic effects, given the severe growth phenotype of the axy9.2 mutant. While mutants defective for cellulose synthase and xylan backbone biosynthesis have obvious implications for wall integrity, the connection between wall integrity and xylan O-acetylation is less clear. Mutants lacking xylan side chains such as glucuronosyl (Mortimer et al., 2010), arabinosyl (Anders et al., 2012), or xylosyl (Chiniquy et al., 2012) residues have not been reported to have such a strong phenotype. This indicates that the acetyl substituent is unique in having such a large impact on the functional properties of xylan. O-Acetylation of xylan was recently demonstrated to occur primarily on alternating xylosyl residues, allowing for the adsorption of xylan onto cellulose microfibrils, with the acetyl groups facing outward and increasing the hydrophobicity of the microfibrils (Busse-Wicher et al., 2014). Interestingly, expression of a fungal xylan acetylesterase in Arabidopsis was reported to result in a 50% decrease in stem O-acetyl content, a similar reduction to that in the tbl29/esk1 mutant, without resulting in a growth phenotype for the plants (Pogorelko et al., 2013; Xiong et al., 2013). This suggests that the timing or distribution of xylan O-acetylation depletion has functional implications for the wall.
The collapsed xylem phenotype of the axy9.2 mutant is likely to result in the disruption of water transport, as has been reported for the tbl29/esk1 mutant, which has less severe but significant xylem and growth phenotypes (Lugan et al., 2009; Lefebvre et al., 2011). Such a disruption in water transport may ultimately lead to the overall growth phenotype (decreased size and delayed flowering) of the axy9.2 mutant when grown on soil. It seems likely that abscisic acid (ABA) signaling plays a role in this response, as ABA is increased in tbl29/esl1 (Lefebvre et al., 2011) and the transcript profile of tbl29/esk1 appears similar to that of wild-type plants treated with ABA or drought stressed (Xin et al., 2007; Bouchabke-Coussa et al., 2008). The dark-green coloration of axy9.2 is very evident and also can be observed in tbl29/esk1 as well as other mutants with severe defects in vascular tissue (Jones et al., 2001; Brown et al., 2005; Chen et al., 2005; Xiong et al., 2013). A similar phenotype has been reported for mutants defective for GA biosynthesis (Koornneef et al., 1990). As ABA signaling is typically antagonistic to GA signaling (Razem et al., 2006), a possible explanation for this similarity is that the likely ABA-mediated stress response in the axy9.2 mutant suppresses either the production or perception of GA, leading to the axy9.2 mutants having a similar phenotype to that of mutants defective for GA biosynthesis.
Involvement of AXY9 in the Polysaccharide Acetylation Pathway
The TBL proteins have been reported previously to be putative polysaccharide acetyltransferases acting in the Golgi apparatus, with specificity for particular acceptor polysaccharides and positions (Gille et al., 2011; Gille and Pauly, 2012; Xiong et al., 2013; Yuan et al., 2013). In the case of TBL29/ESK1, xylan O-acetyltransferase activity has been demonstrated in vitro (Urbanowicz et al., 2014). The AXY9 protein shares homology with the TBL proteins, having approximately 32% sequence similarity and containing the conserved GDS and DXXH motifs (Supplemental Fig. S8), suggesting that it also functions as an acetyltransferase. The TBL proteins, while believed to be acetyltransferases, do have sequence homology to known acetylesterases (Bischoff et al., 2010b), raising the possibility that AXY9 could function as either an esterase or a transferase. However, when disrupted, a polysaccharide acetylesterase would be expected to result in increased polysaccharide acetyl ester content (de Souza et al., 2014), as opposed to the decreased acetyl ester content observed in the axy9 mutants. Although an indirect effect of an acetylesterase activity cannot be excluded, the phenotype data suggest that AXY9 plays a role in the O-acetyltransferase process of various wall polymers upstream from the polysaccharide-specific acetyltransferases (TBLs; Bischoff et al., 2010b; de Souza et al., 2014).
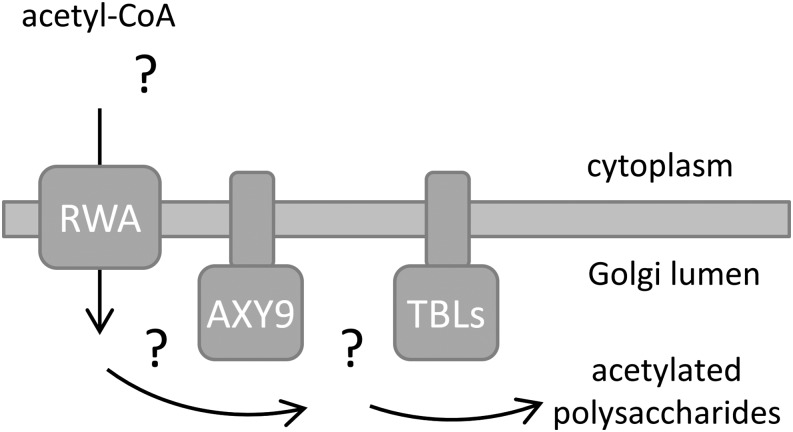
The protease protection and split YFP assays demonstrate that the active site of the AXY9 protein is in the Golgi lumen, implying that AXY9 acts downstream from the RWA proteins, which have been proposed to transport an activated form of acetate into the Golgi (Manabe et al., 2011). Acetyl-CoA has been shown to be an upstream donor of acetyl groups for the acetylation of pectin; however, it has not been shown whether acetyl-CoA itself is transported into the Golgi or if there is an alternative intermediate carrier molecule (Pauly and Scheller, 2000). Therefore, it appears that RWA transports an activated form of acetate, possibly acetyl-CoA, into the Golgi for use directly or indirectly by AXY9 and/or the TBL proteins (Fig. 8). AXY9 may act to produce an acetyl donor substrate used by some or all of the TBL proteins. Recently, acetyl-CoA was shown to be a viable donor for the acetyltransferase activity of TBL29 in vitro (Urbanowicz et al., 2014). However, TBL29 and a bacterial O-acetyltransferase are able to utilize multiple acetyl-donor substrates in vitro, including the artificial substrate p-nitrophenyl acetate (Moynihan and Clarke, 2014a; Urbanowicz et al., 2014). Therefore, the physiological utilization of an alternative donor substrate in vivo cannot be excluded. Future work is required to unravel the existence and identity of the hypothetical acetylated intermediate(s) and whether there is a single or multiple acetyl donor substrates for the polysaccharide acetyltransferases.
Figure 8.
A hypothetical model of plant wall polysaccharide acetylation. The RWA proteins have been hypothesized to act as transporters for an acetyl donor molecule, possibly acetyl-CoA. The TBL proteins have been reported to likely represent polysaccharide acetyltransferases, but their donor substrate is unknown. The AXY9 protein, with its active site in the Golgi lumen, likely acts downstream of the RWA proteins but upstream of the polysaccharide acetyltransferases, as disruption of this gene affects multiple polysaccharides. As a putative acetyltransferase, the AXY9 protein may act to produce a hitherto unknown acetyl donor substrate used by some or all of the TBL polysaccharide acetyltransferases.
Acetylation of carbohydrates is not restricted to land plants and also has been observed in fungi (Cryptococcus neoformans; glucoronoxylomannan [Cherniak and Sundstrom, 1994]), mammals (sialic acids [Hutchins et al., 1988]), and bacteria (peptidoglycan [Clarke and Dupont, 1992]; alginate [Skjåk-Braek et al., 1986], and cellulose [Spiers et al., 2003]). These nonplant organisms contain acetylation machinery that has homology to the RWA and TBL proteins, sometimes as parts of a single polypeptide (Gille and Pauly, 2012). Additional components of the O-acetylation machinery such as AXY9 in plants have not been reported in mammalian or fungal systems. As nonland plant species do not have a putative ortholog of AXY9, it appears that this protein represents a plant-specific component of the polysaccharide acetylation pathway. Given that plants incorporate a large amount of acetyl esters into the wall, this additional step may provide a mechanism to more effectively shuttle activated acetyl equivalents into the Golgi lumen. Alternatively, AXY9 could serve as a point of regulation either for the total amount of acetyl esters being added to the wall or, if this protein is only required for the acetylation of a subset of polysaccharides, to particular wall polysaccharides.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The axy9.1 mutant was identified from an ethyl methanesulfonate-treated population of Arabidopsis (Arabidopsis thaliana) of the Col-0 ecotype (Berger and Altmann, 2000; http://paulylab.berkeley.edu/axy-mutants.html). The axy9.2 mutant is also from the Col-0 background and was obtained from the Arabidopsis Biological Resource Center (stock no. SALK_090612).
For growth of Arabidopsis on plates, the seeds were surface sterilized and plated onto one-half-strength Murashige and Skoog (Murashige and Skoog, 1962) medium with Suc (1%, w/v), agar (0.8%, w/v), and MES (0.06%, w/v) adjusted to pH 5.6 with KOH. Etiolated seedlings were grown in the dark for 7 d at 22°C following 2 d of stratification at 4°C and 6 h of light exposure to induce germination. Soil-grown plants and light-grown plants on plates were grown at 22°C under long-day conditions (16 h of light/8 h of dark) with a light intensity of 130 to 140 μmol m−2 s−1.
For analysis of stem material, the inflorescence stem was collected starting from immediately above the rosette leaves to immediately below the first silique of the main stem and all secondary stems. Stem material was collected when the stems had reached mature height but prior to senescence. For the wild type, axy9.1, and the complementation line, this was at 6 weeks of growth. For axy9.2, which grows much slower, material was collected between 10 and 12 weeks.
Sequencing and Data Analysis
Homozygous axy9.1 seeds were grown on plates under long-day conditions for 4 weeks. Genomic DNA was extracted using the DNeasy genomic DNA extraction kit (Qiagen). The genomic DNA was submitted to the Vincent J. Coates DNA Sequencing Facility at the University of California, Berkeley, for library preparation and sequencing. The DNA was sequenced using one lane of 100-bp single-end reads on a HiSeq 2000 (Illumina). The reads were mapped onto a Col-0 reference genome (TAIR; www.arabidopsis.org), and putative SNPs were identified using the Genomics Workbench software (CLC Bio).
Phylogenetic Analysis
Putative orthologs of AXY9 were obtained from Phytozome (www.phytozome.net) using a BLAST search. The results were manually curated for correctness of the gene models and exclusion of nonorthologous genes. The protein sequences were aligned using ClustalO (Sievers et al., 2011), and a phylogenetic tree was constructed using PhyML (Guindon et al., 2010).
Cloning and Transformation
New plasmids generated for this work along with the method, plasmid backbones, primer sequences, and restriction enzymes used for the construction are listed in Supplemental Tables S4 and S5. The strong constitutive ENTCUP2 promoter was used to drive AXY9 expression for complementation (Coutu et al., 2007). Phusion DNA Polymerase (Thermo Scientific) was used to amplify the DNA sequences for assembly using the manufacturer’s recommendations. The PCR products were purified by gel extraction (Qiagen) following agarose gel electrophoresis and used for Gateway BP reactions (Life Technologies) or Gibson Assembly (Gibson et al., 2009) using a reaction mix from New England Biolabs following the suggested reaction conditions with the appropriate plasmid backbones. The final constructs were verified by restriction digest and sequencing of the gene inserts and border regions.
Genotyping for Mapping and Arabidopsis Lines
Genotyping of the F2 plants of the axy9.1 (Col-0) × Landsberg erecta cross for genetic linkage analysis was performed using simple sequence length polymorphism and cleaved-amplified polymorphic sequence (CAPS) PCR-based markers. The simple sequence length polymorphism markers were obtained from the TAIR Web site and are listed in Supplemental Table S1. For fine mapping, CAPS markers were obtained by using a custom python script to search the Arabidopsis Col-0 reference genome for restriction sites disrupted by mutations present in the Landsberg erecta ecotype (TAIR; www.arabidopsis.org). Candidate marker locations were checked for specificity using an automated BLAST search (Altschul et al., 1997), and primers avoiding overlap with polymorphisms were designed by Primer3 (Rozen and Skaletsky, 1999). The CAPS markers used are listed in Supplemental Table S1. Genotyping of the Arabidopsis axy9.2, AXY9-GFP, and AXY9 overexpression was performed using primers specific to each (Supplemental Table S4). The PCRs were performed with JumpStart RedTaq ReadyMix (P0982; Sigma) following the manufacturer’s recommendations in a total reaction volume of 20 μL. For restriction digest of the CAPS markers, 10× EcoRI buffer (2.5 μL), EcoRI (0.4 μL), and water (2.1 μL) were added following the PCR, and the reactions were incubated for 2 h at 37°C. Agarose gel electrophoresis was used to visualize the genotyping results.
Transcript Analysis of Arabidopsis Lines
For RT-PCR and qRT-PCR analysis, total RNA was extracted from 2-week-old Arabidopsis leaves using the Plant RNeasy kit (Qiagen) according to the manufacturer’s protocol and treated DNase using the TURBO DNA-free kit (AM1907; Life Technologies). RNA (500 ng) was used for complementary DNA (cDNA) synthesis using M-MLV Reverse Transcriptase (Life Technologies) following the manufacturer’s recommendations. For RT-PCR, cDNA (2 µL) was used as a template for PCR with JumpStart RedTaq ReadyMix (Sigma). For qRT-PCR, cDNA (1 µL) was used with Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific) in the StepOnePlus Real-Time PCR System (Applied Biosciences). Relative transcript abundance was calculated using the ACTIN2 gene (At3g18780) as an internal control. For primer sequences, see Supplemental Table S4.
Analysis of XyG Structure
Analysis of XyG was performed as described previously (Lerouxel et al., 2002). In brief, alcohol-insoluble residue (AIR) of plant material was digested with a XyG-specific endoglucanase (Pauly et al., 1999b) in a buffer containing 50 mm ammonium formate at pH 4.5. Following digestion overnight at 37°C, the digest was desalted with cation-exchange beads (Bio-Rex MSZ 501; Bio-Rad) and spotted onto 2,5-dihydroxybenzoic acid matrix for analysis with AXIMA Performance matrix-assisted laser-desorption ionization time of flight mass spectrometry (Shimadzu) in linear positive mode with an accelerating voltage of 20,000 V.
Stem Sectioning
For visualization of the xylem tissue, 7-week-old Arabidopsis stems were cut into approximately 1-mm sections and fixed in neutral buffered formalin (4% [w/v] paraformaldehyde, 1.88% [w/v] sodium dihydrogen phosphate, and 0.43% [w/v] sodium hydroxide, heated at 80°C until clear) for 45 min at 37°C. The material was dehydrated by sequential incubations (15 min) with ethanol solutions (50%, 70%, 95%, and twice 100% [v/v]). The tissue was infiltrated with Spurr’s embedding medium (Electron Microscopy Sciences) following the manufacturer’s recommendations and heat cured (20 h, 60°C). Sections (10 μm) were made using a rotary microtome and stained by incubation with 0.02% (w/v) toluidine blue. Imaging was performed with a light microscope (DM5000 B; Leica Microsystems).
Enzymatic Destarching of Cell Wall Material
AIR material (approximately 10 mg) was suspended in McIlvaine buffer (0.1 m Na2HPO4 and 0.05 m citric acid, pH 5; 1 mL), incubated for 20 min at 80°C, and cooled on ice. To this suspension, a reaction buffer was added resulting in a final concentration of 0.067 μg mL−1 sodium azide, 0.67 μg mL−1 α-amylase (A6380; Sigma), and 10.27 units mL−1 pullulanase M2 (Megazyme). The mixture was incubated overnight (37°C, 230 rpm, 15 h) and then terminated by high-temperature incubation (99°C, 10 min). The samples were centrifuged (15 min, 21,000g), the supernatant was discarded, and the pellet was washed three times with water and once with 70% (v/v) ethanol. The pellet was dried for subsequent analysis of acetic acid content.
Pectin Extraction from Etiolated Seedlings
AIR material (6 mg) from 7-d-old etiolated seedlings were treated with pectinases in a digestion mixture containing 50 mm ammonium formate, pH 4.5, 0.2 μg mL−1 sodium azide, 2 units mL−1 endopolygalacturonase M2 (Megazyme), and 0.04 units mL−1 pectin methyl esterase (Novozymes) for 17.5 h at 37°C in a shaker at 230 rpm. The reaction was stopped by incubating at 80°C for 20 min. Following centrifugation (10 min, 21,000g), the supernatant was analyzed for acetic acid and uronic acid content, and the pellet was washed three times with water and then dried.
Total Acetic Acid Content
Cell wall preparations and pectic fractions (10 mg sample mL−1) were saponified through the addition of an equal volume of 1 m NaOH. Reactions were incubated 1 h at 25°C with 600 rpm shaking. The reaction was neutralized through the addition of an equal volume of 1 m HCl. The material was pelleted (14,000 rpm for 10 min), and total acetic acid content of the supernatant was determined using the Acetic Acid Kit (Megazymes).
Hydroxybiphenyl Microtiter Plate Assay for Uronic Acid Content
Supernatant fractions from pectinase digest of etiolated seedling cell wall preparations were saponified (see above). Material was pelleted, and supernatant fractions were used for biphenyl plate assay. Saponified pectic extract (5 μL) was diluted eight times with water, treated with 200 μL of 0.012 m sodium tetraborate in 96% sulfuric acid, and incubated 1 h at 80°C. Absorbance was measured at 525 nm before and after the addition of 40 μL of hydroxybiphenyl mix (2 μg mL−1 3-phenylphenol, 2% [v/v] dimethyl sulfoxide [DMSO], and 78% [v/v] sulfuric acid). Uronic acids were quantified by comparison of absorbance with a standard curve of galacturonic acid.
HSQC NMR Method
2D HSQC NMR experiments were performed on AIR material of stem tissue as described previously (Cheng et al., 2013). In brief, freeze-dried stem tissue from numerous plants was pooled and then split into three equal aliquots. Ball-milled AIR material (25 mg) was prepared from each aliquot and dissolved in 0.75 mL of DMSO-d6 and 10 µL of 1-ethyl-3-methylimidazolium acetate-d14. The 2D 13C-1H HSQC NMR spectra were acquired on a Bruker AVANCE 600-MHz NMR spectrometer equipped with a 5-mm TXI 1H/13C/15N cryoprobe using the pulse sequence hsqcetgpsisp.2. The experiments were carried out at 25°C with the following parameters: spectral width of 12 ppm in F2 (1H) dimension with 4,096 data points (TD1) and 160 ppm in F1 (13C) dimension with 256 data points (TD2); scan number of 200; interscan delay of 1 s. The chemical shifts were referenced to the DMSO solvent peak (δC 39.5 ppm, δH 2.49 ppm). The NMR data processing and analysis were performed on Bruker’s Topspin 3.1 software. The NMR data were quantified as described previously (Chong et al., 2014). In brief, signals in the anomeric/aliphatic region (H1-C1 signals of 2-O-acetyl-xylose [Ac-Xyl], 3-O-Ac-Xyl, 2,3-O-Ac-Xyl, Xyl, and reducing ends of xylan [α/β-Xyl-R]) were summed up to 100%, and the signal in the aliphatic region was integrated separately to calculate the relative content of each form of O-acetyl-xylan unit. The relative contents of 3-O-acetyl-xylan and 2,3-O-acetyl-xylan units were calculated from the H3-C3 signals, and that of the 2-O-acetyl unit was calculated from the H2-C2 signal.
Transient Protein Expression in Nicotiana benthamiana
For the protease protection assays, the coding sequence of interest was cloned into pICH31070 (Icon Genetics) along with the appropriate accessory constructs (a plasmid with p19 for the fluorescence microscopy [Voinnet et al., 2003] and plasmids pICH20111 [Kalthoff et al., 2010] and pICH14011 [Giritch et al., 2006] for the protease protection assays). The plasmids containing the desired coding sequences were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. The A. tumefaciens harboring the desired vectors was grown for approximately 36 h (30°C, shake flasks) in LB medium with kanamycin (60 μg mL−1), gentamycin (25 μg mL−1), and rifampicin (10 μg mL−1). The cells were collected by centrifugation (5,000g, 10 min) and resuspended in infiltration buffer (10 mm MES, 10 mm MgCl2, pH 5.6, with KOH) to a final optical density at 600 nm of 0.2 for fluorescence microscopy or 0.02 for the production of material for the protease protection assays. The A. tumefaciens solution was vacuum infiltrated into leaves of 6-week-old N. benthamiana plants.
Stable Transformation of Arabidopsis
A. tumefaciens (strain GV3101) containing the desired vectors was grown and used for floral dip transformation as described previously (Clough and Bent, 1998). Transformants were selected by growing T1 seeds on one-half-strength Murashige and Skoog agar plates (described above) supplemented with 25 μg mL−1 hygromycin. Resistant plants were picked after 2 weeks of growth under long-day conditions and moved to soil for seed production. The presence of the transgenes was confirmed by genotyping PCR as described above.
Confocal Microscopy
A laser scanning confocal microscope (LSM 710; Carl Zeiss Microscopy) was used to image the N. benthamiana and Arabidopsis leaves used for transient and stable expression of the fluorescent proteins, respectively. Excitation of GFP, YFP, and mCherry was performed using lasers at 488, 514, and 594 nm, respectively. The emission filter was 519 to 621 nm for YFP, 599 to 696 nm for mCherry, and either 489 to 546 nm (Arabidopsis) or 493 to 578 nm (N. benthamiana) for GFP. Chlorophyll was excited at 488 nm and detected at 593 to 735 nm to image chloroplasts as a visual reference. Four-week old Arabidopsis leaves expressing AXY9-GFP and N. benthamiana leaves 3 d after infiltration were used for imaging. The Golgi mCherry marker G-rk (CD3-967) obtained from the Arabidopsis Biological Resource Center was used as a control (Nelson et al., 2007).
Protease Protection Assay
Microsome preparation was performed as described previously with modifications (Abas and Luschnig, 2010). Leaf material was collected and snap frozen in liquid nitrogen at 10 d after infiltration for AXY9-GFP and 7 d after infiltration for YFP-TMD and TMD-YFP. Leaf material was thawed on ice, mixed with cold extraction buffer (100 mm Tris-HCl, pH 7.5, 25% [w/v] Suc, 5% [v/v] glycerol, 10 mm EDTA, 5 mm KCl, and 1% [w/v] polyvinylpolypyrrolidone) with Halt protease inhibitor mix (78429; Thermo Scientific), and homogenized with a mortar and pestle at 0°C. Cell debris was removed by centrifugation (three times, 600g, 3 min), and the final supernatant was diluted two times with water at 4°C. The solution was divided into aliquots of 200 μL and centrifuged (21,000g, 90 min, 4°C) with 50 μL of cushion buffer (50 mm Tris-HCl, pH 7.5, 5 mm EDTA, 1.8 m Suc, 2.5% [v/v] glycerol, 5 mm EDTA, and 2.5 mm KCl) added to the bottom of the tube. The microsomal layer (on top of the cushion buffer) was gently resuspended in 200 μL wash buffer (20 mM Tris-HCl, pH 7.5, 5 mM EDTA) at 4°C and centrifuged (21,000g, 45 min, 4°C) with cushion buffer (50 μL). The microsomal fraction (on top of the cushion buffer) was gently suspended in storage buffer (20 mm Tris-HCl, pH 7.5, 5 mm EDTA, and 20% [v/v] glycerol) and stored at −80°C. Microsomal fractions containing YFP-TMD and TMD-YFP were diluted 30- and 3-fold, respectively, with storage buffer before use to obtain a signal on the western blot of similar intensity to that of AXY9-GFP.
For the protease assay, microsomes (25 μL) were treated with or without detergent (0.07% [v/v] Triton X-100) and with or without protease (4.4 ng μL−1 proteinase K [P6556; Sigma]) in a total reaction volume of 28.5 μL for 90 min at 37°C. Phenylmethanesulfonyl fluoride (2 μL of 100 mm in isopropanol) was added to the reactions, which were then incubated for 10 min at room temperature to inactivate the protease. The proteins were denatured and reduced by the addition of 4× LDS NuPage loading buffer (Life Technologies) and 10× NuPage reducing agent (Life Technologies) and incubation at 70°C for 10 min. The proteins were separated by SDS-PAGE and then transferred to a polyvinylidene fluoride membrane for immunoblotting. A rabbit anti-GFP polyclonal antibody (A11122; Life Technologies) was used at a 1:4,000 dilution as the primary antibody, and a goat anti-rabbit horseradish peroxidase (SC2054; Santa Cruz Biotech) was used at a 1:2,000 dilution as the secondary antibody.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Relative amount of XyG O-acetylation.
Supplemental Figure S2. Selection of mutants for genetic linkage analysis.
Supplemental Figure S3. Mapping of axy9.1.
Supplemental Figure S4. Gene and protein models of AXY9.
Supplemental Figure S5. RT-PCR and qRT-PCR of AXY9 in the axy9 mutants.
Supplemental Figure S6. Phylogenetic tree of AXY9 protein sequences.
Supplemental Figure S7. Alignment of AXY9 protein sequences.
Supplemental Figure S8. Alignment and similarity of AXY9 and TBL protein sequences.
Supplemental Figure S9. 2D HSQC NMR spectra of dissolved stem tissue.
Supplemental Figure S10. Subcellular localization of AXY9-GFP in Arabidopsis.
Supplemental Figure S11. Split YFP controls.
Supplemental Figure S12. Split YFP AXY9 N-terminal fusions.
Supplemental Figure S13. Protease protection assay of internal and external Golgi controls.
Supplemental Table S1. Markers used for genetic linkage analysis of axy9.1.
Supplemental Table S2. Genomic sequencing of axy9.1.
Supplemental Table S3. Candidate SNPs for the basis of the axy9.1 phenotype within the mapped region.
Supplemental Table S4. Primer sequences used for genotyping and plasmid construction.
Supplemental Table S5. Plasmid construction.
Supplementary Material
Acknowledgments
We thank Kirk Schnorr (Novozymes) for the generous gift of the XyG endoglucanase enzyme.
Glossary
- XyG
xyloglucan
- Col-0
Columbia-0
- SNP
single-nucleotide polymorphism
- T-DNA
transfer DNA
- qRT
quantitative reverse transcription
- RT
reverse transcription
- TAIR
The Arabidopsis Information Resource
- HSQC
heteronuclear single quantum correlation
- 2D
two-dimensional
- TMD
transmembrane domain
- ABA
abscisic acid
- CAPS
cleaved-amplified polymorphic sequence
- cDNA
complementary DNA
- AIR
alcohol-insoluble residue
- DMSO
dimethyl sulfoxide
Footnotes
This work was supported by the Fred Dickinson Chair of Wood Science and Technology Endowment (to A.S. and M.P.) and a National Institutes of Health National Research Service Award Trainee appointment (grant no. GM007127 to A.S.).
Articles can be viewed without a subscription.
References
- Abas L, Luschnig C (2010) Maximum yields of microsomal-type membranes from small amounts of plant material without requiring ultracentrifugation. Anal Biochem 401: 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama Y, Ito K (1987) Topology analysis of the SecY protein, an integral membrane protein involved in protein export in Escherichia coli. EMBO J 6: 3465–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akoh CC, Lee GC, Liaw YC, Huang TH, Shaw JF (2004) GDSL family of serine esterases/lipases. Prog Lipid Res 43: 534–552 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders N, Wilkinson MD, Lovegrove A, Freeman J, Tryfona T, Pellny TK, Weimar T, Mortimer JC, Stott K, Baker JM, et al. (2012) Glycosyl transferases in family 61 mediate arabinofuranosyl transfer onto xylan in grasses. Proc Natl Acad Sci USA 109: 989–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger D, Altmann T (2000) A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev 14: 1119–1131 [PMC free article] [PubMed] [Google Scholar]
- Biely P, MacKenzie C, Puls J, Schneider H (1986) Cooperativity of esterases and xylanases in the enzymatic degradation of acetyl xylan. Nat Biotechnol 4: 731–733 [Google Scholar]
- Bischoff V, Nita S, Neumetzler L, Schindelasch D, Urbain A, Eshed R, Persson S, Delmer D, Scheible WR (2010a) TRICHOME BIREFRINGENCE and its homolog AT5G01360 encode plant-specific DUF231 proteins required for cellulose biosynthesis in Arabidopsis. Plant Physiol 153: 590–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff V, Selbig J, Scheible WR (2010b) Involvement of TBL/DUF231 proteins into cell wall biology. Plant Signal Behav 5: 1057–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchabke-Coussa O, Quashie ML, Seoane-Redondo J, Fortabat MN, Gery C, Yu A, Linderme D, Trouverie J, Granier F, Téoulé E, et al. (2008) ESKIMO1 is a key gene involved in water economy as well as cold acclimation and salt tolerance. BMC Plant Biol 8: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd D, Manoil C, Beckwith J (1987) Determinants of membrane protein topology. Proc Natl Acad Sci USA 84: 8525–8529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Goubet F, Wong VW, Goodacre R, Stephens E, Dupree P, Turner SR (2007) Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant J 52: 1154–1168 [DOI] [PubMed] [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Gidley MJ, Fincher GB (2010) Heterogeneity in the chemistry, structure and function of plant cell walls. Nat Chem Biol 6: 724–732 [DOI] [PubMed] [Google Scholar]
- Busse-Wicher M, Gomes TCF, Tryfona T, Nikolovski N, Stott K, Grantham NJ, Bolam DN, Skaf MS, Dupree P (2014) The pattern of xylan acetylation suggests xylan may interact with cellulose microfibrils as a twofold helical screw in the secondary plant cell wall of Arabidopsis thaliana. Plant J 79: 492–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffall KH, Mohnen D (2009) The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res 344: 1879–1900 [DOI] [PubMed] [Google Scholar]
- Chen Z, Hong X, Zhang H, Wang Y, Li X, Zhu JK, Gong Z (2005) Disruption of the cellulose synthase gene, AtCesA8/IRX1, enhances drought and osmotic stress tolerance in Arabidopsis. Plant J 43: 273–283 [DOI] [PubMed] [Google Scholar]
- Cheng K, Sorek H, Zimmermann H, Wemmer DE, Pauly M (2013) Solution-state 2D NMR spectroscopy of plant cell walls enabled by a dimethylsulfoxide-d6/1-ethyl-3-methylimidazolium acetate solvent. Anal Chem 85: 3213–3221 [DOI] [PubMed] [Google Scholar]
- Cherniak R, Sundstrom JB (1994) Polysaccharide antigens of the capsule of Cryptococcus neoformans. Infect Immun 62: 1507–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiniquy D, Sharma V, Schultink A, Baidoo EE, Rautengarten C, Cheng K, Carroll A, Ulvskov P, Harholt J, Keasling JD, et al. (2012) XAX1 from glycosyltransferase family 61 mediates xylosyltransfer to rice xylan. Proc Natl Acad Sci USA 109: 17117–17122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu TY, Christiansen K, Moreno I, Lao J, Loqué D, Orellana A, Heazlewood JL, Clark G, Roux SJ (2012) AtAPY1 and AtAPY2 function as Golgi-localized nucleoside diphosphatases in Arabidopsis thaliana. Plant Cell Physiol 53: 1913–1925 [DOI] [PubMed] [Google Scholar]
- Chong SL, Virkki L, Maaheimo H, Juvonen M, Derba-Maceluch M, Koutaniemi S, Roach M, Sundberg B, Tuomainen P, Mellerowicz EJ, et al. (2014) O-Acetylation of glucuronoxylan in Arabidopsis thaliana wild type and its change in xylan biosynthesis mutants. Glycobiology 24: 494–506 [DOI] [PubMed] [Google Scholar]
- Clarke AJ, Dupont C (1992) O-Acetylated peptidoglycan: its occurrence, pathobiological significance, and biosynthesis. Can J Microbiol 38: 85–91 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Coutu C, Brandle J, Brown D, Brown K, Miki B, Simmonds J, Hegedus DD (2007) pORE: a modular binary vector series suited for both monocot and dicot plant transformation. Transgenic Res 16: 771–781 [DOI] [PubMed] [Google Scholar]
- de Souza A, Hull PA, Gille S, Pauly M (2014) Identification and functional characterization of the distinct plant pectin esterases PAE8 and PAE9 and their deletion mutants. Planta 240: 1123–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evtuguin DV, Tomás JL, Silva AM, Neto CP (2003) Characterization of an acetylated heteroxylan from Eucalyptus globulus Labill. Carbohydr Res 338: 597–604 [DOI] [PubMed] [Google Scholar]
- Fry SC, York WS, Albersheim P, Darvill A, Hayashi T, Joseleau JP, Kato Y, Lorences EP, Maclachlan GA, Mcneil M, et al. (1993) An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol Plantarum 89: 1–3 [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345 [DOI] [PubMed] [Google Scholar]
- Gille S, de Souza A, Xiong G, Benz M, Cheng K, Schultink A, Reca IB, Pauly M (2011) O-Acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL and DUF231 domain. Plant Cell 23: 4041–4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S, Pauly M (2012) O-Acetylation of plant cell wall polysaccharides. Front Plant Sci 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giritch A, Marillonnet S, Engler C, van Eldik G, Botterman J, Klimyuk V, Gleba Y (2006) Rapid high-yield expression of full-size IgG antibodies in plants coinfected with noncompeting viral vectors. Proc Natl Acad Sci USA 103: 14701–14706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glushka JN, Terrell M, York WS, O’Neill MA, Gucwa A, Darvill AG, Albersheim P, Prestegard JH (2003) Primary structure of the 2-O-methyl-alpha-L-fucose-containing side chain of the pectic polysaccharide, rhamnogalacturonan II. Carbohydr Res 338: 341–352 [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Günl M, Neumetzler L, Kraemer F, de Souza A, Schultink A, Pena M, York WS, Pauly M (2011) AXY8 encodes an α-fucosidase, underscoring the importance of apoplastic metabolism on the fine structure of Arabidopsis cell wall polysaccharides. Plant Cell 23: 4025–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günl M, Pauly M (2011) AXY3 encodes a α-xylosidase that impacts the structure and accessibility of the hemicellulose xyloglucan in Arabidopsis plant cell walls. Planta 233: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Takahashi R, Kobayashi S, Kawase T, Nishinari K (2002) Gelation behavior of native and acetylated konjac glucomannan. Biomacromolecules 3: 1296–1303 [DOI] [PubMed] [Google Scholar]
- Hutchins JT, Reading CL, Giavazzi R, Hoaglund J, Jessup JM (1988) Distribution of mono-, di, and tri-O-acetylated sialic acids in normal and neoplastic colon. Cancer Res 48: 483–489 [PubMed] [Google Scholar]
- Jensen JK, Schultink A, Keegstra K, Wilkerson CG, Pauly M (2012) RNA-Seq analysis of developing nasturtium seeds (Tropaeolum majus): identification and characterization of an additional galactosyltransferase involved in xyloglucan biosynthesis. Mol Plant 5: 984–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, Ennos AR, Turner SR (2001) Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis. Plant J 26: 205–216 [DOI] [PubMed] [Google Scholar]
- Kalthoff D, Giritch A, Geisler K, Bettmann U, Klimyuk V, Hehnen HR, Gleba Y, Beer M (2010) Immunization with plant-expressed hemagglutinin protects chickens from lethal highly pathogenic avian influenza virus H5N1 challenge infection. J Virol 84: 12002–12010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer LL, York WS, Darvill AG, Albersheim P (1989) Structure of plant-cell walls. 27. Xyloglucan isolated from suspension-cultured sycamore cell-walls is O-acetylated. Phytochemistry 28: 2105–2107 [Google Scholar]
- Knox JP. (2008) Revealing the structural and functional diversity of plant cell walls. Curr Opin Plant Biol 11: 308–313 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Bosma TD, Hanhart CJ, van der Veen JH, Zeevaart JA (1990) The isolation and characterization of gibberellin-deficient mutants in tomato. Theor Appl Genet 80: 852–857 [DOI] [PubMed] [Google Scholar]
- Lee C, Teng Q, Zhong R, Ye ZH (2011) The four Arabidopsis reduced wall acetylation genes are expressed in secondary wall-containing cells and required for the acetylation of xylan. Plant Cell Physiol 52: 1289–1301 [DOI] [PubMed] [Google Scholar]
- Lee C, Teng Q, Zhong R, Ye ZH (2014) Alterations of the degree of xylan acetylation in Arabidopsis xylan mutants. Plant Signal Behav 9: e27797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, Fortabat MN, Ducamp A, North HM, Maia-Grondard A, Trouverie J, Boursiac Y, Mouille G, Durand-Tardif M (2011) ESKIMO1 disruption in Arabidopsis alters vascular tissue and impairs water transport. PLoS ONE 6: e16645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouxel O, Choo TS, Séveno M, Usadel B, Faye L, Lerouge P, Pauly M (2002) Rapid structural phenotyping of plant cell wall mutants by enzymatic oligosaccharide fingerprinting. Plant Physiol 130: 1754–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugan R, Niogret MF, Kervazo L, Larher FR, Kopka J, Bouchereau A (2009) Metabolome and water status phenotyping of Arabidopsis under abiotic stress cues reveals new insight into ESK1 function. Plant Cell Environ 32: 95–108 [DOI] [PubMed] [Google Scholar]
- Manabe Y, Nafisi M, Verhertbruggen Y, Orfila C, Gille S, Rautengarten C, Cherk C, Marcus SE, Somerville S, Pauly M, et al. (2011) Loss-of-function mutation of REDUCED WALL ACETYLATION2 in Arabidopsis leads to reduced cell wall acetylation and increased resistance to Botrytis cinerea. Plant Physiol 155: 1068–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manabe Y, Verhertbruggen Y, Gille S, Harholt J, Chong SL, Pawar PM, Mellerowicz EJ, Tenkanen M, Cheng K, Pauly M, et al. (2013) Reduced wall acetylation proteins play vital and distinct roles in cell wall O-acetylation in Arabidopsis. Plant Physiol 163: 1107–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna S, McAnalley BH (1993) Determination of the position of the O-acetyl group in a beta-(1→4)-mannan (acemannan) from Aloe barbardensis Miller. Carbohydr Res 241: 317–319 [DOI] [PubMed] [Google Scholar]
- Mortimer JC, Miles GP, Brown DM, Zhang Z, Segura MP, Weimar T, Yu X, Seffen KA, Stephens E, Turner SR, et al. (2010) Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass. Proc Natl Acad Sci USA 107: 17409–17414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan PJ, Clarke AJ (2014a) Substrate specificity and kinetic characterization of peptidoglycan O-acetyltransferase B from Neisseria gonorrhoeae. J Biol Chem 289: 16748–16760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan PJ, Clarke AJ (2014b) The mechanism of action of peptidoglycan O-acetyltransferase B involves a Ser-His-Asp catalytic triad. Biochemistry 53: 6243–6251 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Obel N, Erben V, Schwarz T, Kühnel S, Fodor A, Pauly M (2009) Microanalysis of plant cell wall polysaccharides. Mol Plant 2: 922–932 [DOI] [PubMed] [Google Scholar]
- Pauly M, Albersheim P, Darvill A, York WS (1999a) Molecular domains of the cellulose/xyloglucan network in the cell walls of higher plants. Plant J 20: 629–639 [DOI] [PubMed] [Google Scholar]
- Pauly M, Andersen LN, Kauppinen S, Kofod LV, York WS, Albersheim P, Darvill A (1999b) A xyloglucan-specific endo-β-1,4-glucanase from Aspergillus aculeatus: expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology 9: 93–100 [DOI] [PubMed] [Google Scholar]
- Pauly M, Gille S, Liu L, Mansoori N, de Souza A, Schultink A, Xiong G (2013) Hemicellulose biosynthesis. Planta 238: 627–642 [DOI] [PubMed] [Google Scholar]
- Pauly M, Scheller HV (2000) O-Acetylation of plant cell wall polysaccharides: identification and partial characterization of a rhamnogalacturonan O-acetyl-transferase from potato suspension-cultured cells. Planta 210: 659–667 [DOI] [PubMed] [Google Scholar]
- Persson S, Caffall KH, Freshour G, Hilley MT, Bauer S, Poindexter P, Hahn MG, Mohnen D, Somerville C (2007) The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell 19: 237–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogorelko G, Lionetti V, Fursova O, Sundaram RM, Qi M, Whitham SA, Bogdanove AJ, Bellincampi D, Zabotina OA (2013) Arabidopsis and Brachypodium distachyon transgenic plants expressing Aspergillus nidulans acetylesterases have decreased degree of polysaccharide acetylation and increased resistance to pathogens. Plant Physiol 162: 9–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prozil SO, Costa EV, Evtuguin DV, Lopes LPC, Domingues MRM (2012) Structural characterization of polysaccharides isolated from grape stalks of Vitis vinifera L. Carbohydr Res 356: 252–259 [DOI] [PubMed] [Google Scholar]
- Ralet MC, Cabrera JC, Bonnin E, Quéméner B, Hellìn P, Thibault JF (2005) Mapping sugar beet pectin acetylation pattern. Phytochemistry 66: 1832–1843 [DOI] [PubMed] [Google Scholar]
- Razem FA, Baron K, Hill RD (2006) Turning on gibberellin and abscisic acid signaling. Curr Opin Plant Biol 9: 454–459 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H (1999) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P (2010) Hemicelluloses. Annu Rev Plant Biol 61: 263–289 [DOI] [PubMed] [Google Scholar]
- Schultink A, Liu L, Zhu L, Pauly M (2014) Structural diversity and function of xyloglucan sidechain substituents. Plants 3: 526–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flügge UI, Kunze R (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol 131: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengkhamparn N, Bakx EJ, Verhoef R, Schols HA, Sajjaanantakul T, Voragen AG (2009) Okra pectin contains an unusual substitution of its rhamnosyl residues with acetyl and alpha-linked galactosyl groups. Carbohydr Res 344: 1842–1851 [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims IM, Munro SL, Currie G, Craik D, Bacic A (1996) Structural characterisation of xyloglucan secreted by suspension-cultured cells of Nicotiana plumbaginifolia. Carbohydr Res 293: 147–172 [DOI] [PubMed] [Google Scholar]
- Skjåk-Braek G, Grasdalen H, Larsen B (1986) Monomer sequence and acetylation pattern in some bacterial alginates. Carbohydr Res 154: 239–250 [DOI] [PubMed] [Google Scholar]
- Søgaard C, Stenbæk A, Bernard S, Hadi M, Driouich A, Scheller HV, Sakuragi Y (2012) GO-PROMTO illuminates protein membrane topologies of glycan biosynthetic enzymes in the Golgi apparatus of living tissues. PLoS ONE 7: e31324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, et al. (2004) Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211 [DOI] [PubMed] [Google Scholar]
- Spiers AJ, Bohannon J, Gehrig SM, Rainey PB (2003) Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol Microbiol 50: 15–27 [DOI] [PubMed] [Google Scholar]
- Teleman A, Tenkanen M, Jacobs A, Dahlman O (2002) Characterization of O-acetyl-(4-O-methylglucurono)xylan isolated from birch and beech. Carbohydr Res 337: 373–377 [DOI] [PubMed] [Google Scholar]
- Turner SR, Somerville CR (1997) Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9: 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SR, Taylor N, Jones L (2001) Mutations of the secondary cell wall. Plant Mol Biol 47: 209–219 [PubMed] [Google Scholar]
- Urbanowicz BR, Peña MJ, Moniz HA, Moremen KW, York WS (2014) Two Arabidopsis proteins synthesize acetylated xylan in vitro. Plant J 80: 197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierhuis E, York WS, Kolli VS, Vincken J, Schols HA, Van Alebeek GW, Voragen AG (2001) Structural analyses of two arabinose containing oligosaccharides derived from olive fruit xyloglucan: XXSG and XLSG. Carbohydr Res 332: 285–297 [DOI] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Somerville CR, Somerville SC (2004) Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J 40: 968–978 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z, Mandaokar A, Chen J, Last RL, Browse J (2007) Arabidopsis ESK1 encodes a novel regulator of freezing tolerance. Plant J 49: 786–799 [DOI] [PubMed] [Google Scholar]
- Xiong G, Cheng K, Pauly M (2013) Xylan O-acetylation impacts xylem development and enzymatic recalcitrance as indicated by the Arabidopsis mutant tbl29. Mol Plant 6: 1373–1375 [DOI] [PubMed] [Google Scholar]
- York WS, Kumar Kolli VS, Orlando R, Albersheim P, Darvill AG (1996) The structures of arabinoxyloglucans produced by solanaceous plants. Carbohydr Res 285: 99–128 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Teng Q, Zhong R, Ye ZH (2013) The Arabidopsis DUF231 domain-containing protein ESK1 mediates 2-O- and 3-O-acetylation of xylosyl residues in xylan. Plant Cell Physiol 54: 1186–1199 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.