Piwi-piRNA ribonucleoproteins (piRNPs) enforce retrotransposon silencing, a function critical for preserving genome integrity of germ cells. Vourekas et al. found that MOV10L1 exhibits 5? to 3? directional RNA unwinding activity in vitro and that a point mutation that abolishes this activity causes a failure in primary piRNA biogenesis in vivo. MOV10L1 selectively binds piRNA precursor transcripts and is essential for the generation of intermediate piRNA processing fragments that are subsequently loaded to Piwi proteins.
Keywords: piRNA, Piwi, MOV10L1, Armitage, Argonaute, G quadruplex
Abstract
Piwi–piRNA (Piwi-interacting RNA) ribonucleoproteins (piRNPs) enforce retrotransposon silencing, a function critical for preserving the genome integrity of germ cells. The molecular functions of most of the factors that have been genetically implicated in primary piRNA biogenesis are still elusive. Here we show that MOV10L1 exhibits 5′-to-3′ directional RNA-unwinding activity in vitro and that a point mutation that abolishes this activity causes a failure in primary piRNA biogenesis in vivo. We demonstrate that MOV10L1 selectively binds piRNA precursor transcripts and is essential for the generation of intermediate piRNA processing fragments that are subsequently loaded to Piwi proteins. Multiple analyses suggest an intimate coupling of piRNA precursor processing with elements of local secondary structures such as G quadruplexes. Our results support a model in which MOV10L1 RNA helicase activity promotes unwinding and funneling of the single-stranded piRNA precursor transcripts to the endonuclease that catalyzes the first cleavage step of piRNA processing.
Piwi-interacting RNAs (piRNAs) are a class of small RNAs (23–30 nucleotides [nt]) that bind to Piwi proteins to form piRNPs (Piwi–piRNA ribonucleoproteins), which silence selfish genetic elements in the germline of metazoans (Aravin et al. 2004, 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Vagin et al. 2006; Kuramochi-Miyagawa et al. 2008; Ghildiyal and Zamore 2009; Siomi et al. 2011; Sienski et al. 2012; Huang et al. 2013; Rozhkov et al. 2013). Loss of Piwi genes results in retrotransposon up-regulation, arrest of gametogenesis, and infertility (Cox et al. 1998; Harris and Macdonald 2001; Deng and Lin 2002; Kuramochi-Miyagawa et al. 2004; Carmell et al. 2007; Li et al. 2009). In mice, Mili (Piwil2) and Miwi2 (Piwil4) loaded with retrotransposon-related piRNAs cooperate to establish silencing of long interspersed element-1 (LINE1 or L1) by DNA methylation during prenatal male gonad development (Carmell et al. 2007; Shoji et al. 2009). Postnatally, Mili and Miwi (Piwil1) target L1 RNA for post-transcriptional silencing at the meiotic and post-meiotic stages of spermatogenesis, respectively (De Fazio et al. 2011; Reuter et al. 2011; Di Giacomo et al. 2013). Additionally, Miwi is involved in the cytoplasmic remodeling of spermatids through the formation of the chromatoid body, and Miwi carries out a piRNA-independent stabilization of spermiogenic mRNA in round spermatids (Deng and Lin 2002; Vourekas et al. 2012).
Primary piRNAs are produced from long, single-stranded precursor transcripts in a stepwise process. The Zucchini endonuclease (Zuc, whose mouse homolog, mZuc, is also known as PLD6 or mitoPLD) (Pane et al. 2007; Huang et al. 2011; Watanabe et al. 2011; Ipsaro et al. 2012; Nishimasu et al. 2012) is most likely the nuclease that generates piRNA precursor intermediate fragments (PPIFs) that are loaded onto Piwi proteins, which stabilize their 5′ ends (Pillai and Chuma 2012; Vourekas et al. 2012). Piwi proteins preferentially stabilize PPIFs with a 5′ U, thus generating a nucleotide bias that is inherited by the mature piRNA (Kawaoka et al. 2011; Ipsaro et al. 2012; Nishimasu et al. 2012; Vourekas et al. 2012). High-throughput sequencing after cross-linking and immunoprecipitation (HITS-CLIP or CLIP-seq) provided in vivo evidence for PPIFs loaded to Mili and Miwi (Vourekas et al. 2012). A second processing step is the 3′-to-5′ exonucleolytic trimming of Piwi-loaded PPIFs by an enzymatic activity called Trimmer that has not yet been identified (Kawaoka et al. 2011). It is thought that 3′ trimming proceeds until it is sterically hindered by the Piwi protein, and it is functionally coupled with 2′-O methylation, a 3′ end modification of piRNAs catalyzed by Hen1 (Horwich et al. 2007; Kirino and Mourelatos 2007a,b; Saito et al. 2007; Kamminga et al. 2010; Kawaoka et al. 2011; Montgomery et al. 2012). Recently, it has been shown that a Tudor domain-containing (Tdrd) protein known as Tdrkh promotes the 3′ trimming process (Honda et al. 2013; Saxe et al. 2013). In addition to Piwi proteins and Zuc, other factors essential for piRNA processing, including GASZ (germ cell protein with ankyrin repeats, sterile α motif, and leucine zipper) and Tdrd proteins (Hosokawa et al. 2007; Ma et al. 2009), are localized on the cytoplasmic surface of mitochondria in electron-dense granules known as intermitochondrial cement (ICM) in mammalian spermatocytes or more generally as nuage in Drosophila and other animals (Siomi et al. 2011; Pillai and Chuma 2012).
piRNA precursor genomic loci termed piRNA clusters, especially in Drosophila, contain sequence fragments that are antisense to retrotransposons as a form of molecular memory of past retrotransposon activity (Aravin et al. 2007; Brennecke et al. 2007). Piwi-loaded primary piRNAs derived from such clusters can target, through extensive sequence complementarity, and cleave retroelement-derived RNAs (Saito et al. 2006; Reuter et al. 2011; Di Giacomo et al. 2013). The 3′ cleavage fragments can be processed into secondary piRNAs, which in turn are able to cleave antisense piRNA precursors, thus closing a piRNA amplification loop termed ping-pong (Brennecke et al. 2007; Gunawardane et al. 2007). In mammals, most postnatal piRNAs are produced at the pachytene stage of meiosis from clusters depleted of repeat sequences that in mice are transcribed by A-MYB (Aravin et al. 2004, 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Vagin et al. 2006; Li et al. 2013).
The wealth of genetic information regarding the piRNA pathway is contrasted by scarce knowledge of the molecular functions of piRNA protein factors. MOV10L1 is a predicted RNA helicase that is essential for primary piRNA biogenesis; it is localized in ICM, and its expression starts in prenatal gonocytes, peaks in pachytene spermatocytes, and terminates in post-meiotic spermatids (Frost et al. 2010; Zheng et al. 2010; Zheng and Wang 2012). The Drosophila homolog of MOV10L1, Armitage, is localized to the nuage and is essential for primary piRNA generation (Klattenhoff et al. 2007; Haase et al. 2010; Saito et al. 2010). Furthermore, mice with postnatal deletion of Mov10l1 lack pachytene piRNAs and provide an ideal system to study their functions (Zheng and Wang 2012). By using HITS-CLIP, RNA sequencing (RNA-seq), and computational approaches coupled with in vitro enzymatic assays and in vivo mutagenesis, we uncovered the molecular function of MOV10L1 in piRNA biogenesis. In this model, MOV10L1 selectively binds to piRNA precursors and, by means of its ATP-dependent RNA helicase activity, funnels them to the endonuclease that catalyzes the first cleavage step of piRNA processing to generate piRNA intermediate fragments that are subsequently loaded to Piwi proteins.
Results
MOV10L1 specifically binds piRNA precursors
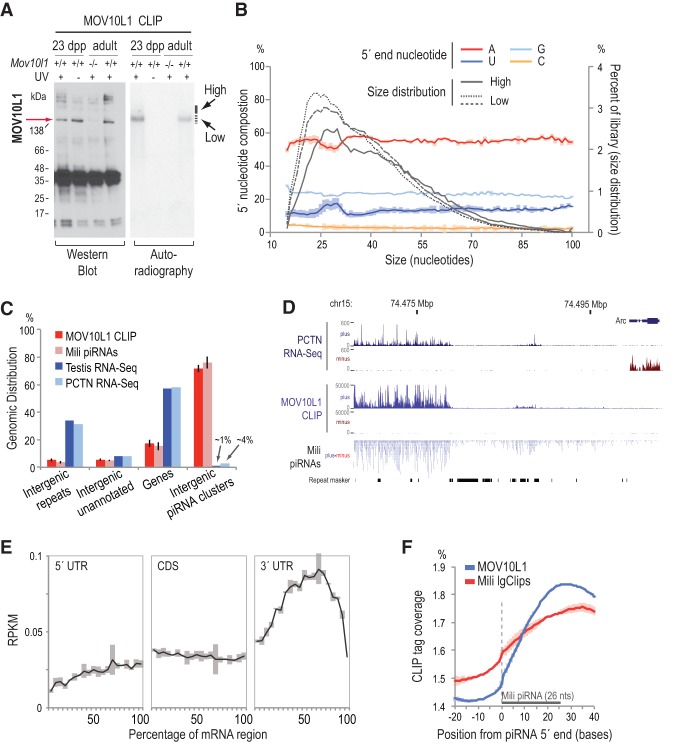
We performed MOV10L1 HITS-CLIP in testes from adult and 23-d post-partum (dpp) wild-type mice, as described previously for Mili and Miwi (Vourekas et al. 2012; Vourekas and Mourelatos 2014), without addition of exogenous nuclease to the cross-linked lysate. We also performed solid support directional (SSD) RNA-seq (Vourekas et al. 2012) of total RNA depleted of ribosomal RNA. By CLIP, we detected specific MOV10L1–RNA protein complexes (indicating direct binding of MOV10L1 to RNA) that are more pronounced in 23-dpp testes, which are enriched in pachytene spermatocytes that express high levels of MOV10L1 (Fig. 1A; Zheng et al. 2010). We extracted RNAs and created three cDNA libraries: two from the main radioactive signal and one from a higher position (Fig. 1A,B; Supplemental Table S1). The size distribution of the mapped reads reveals a similar size profile for all libraries (Fig. 1B). The identity of the 5′ end nucleotide and the genomic distribution are unimodal in all three libraries across the size range of reads (Fig. 1B; Supplemental Fig. S1A). More than 70% of MOV10L1 CLIP tags map within the previously described intergenic piRNA clusters (IPCs) (Aravin et al. 2006; Vourekas et al. 2012; Li et al. 2013), which produce the overwhelming majority of pachytene piRNAs (Fig. 1C,D). IPC coordinates can be found in Supplemental Table S1 (see also the Supplemental Material). Extremely high correlation between the three libraries and within IPCs was observed, and therefore the three libraries were considered replicates (Supplemental Fig. S1B). Standard RNA immunoprecipitation was performed to independently verify the strong enrichment of piRNA precursor transcripts in MOV10L1 immunoprecipitation compared with control rabbit serum immunoprecipitation (Supplemental Fig. S1C,D).
Figure 1.
Transcriptome-wide identification of MOV10L1 RNA targets by CLIP. (A) Western blot and autoradiography of MOV10L1–RNA complexes from CLIP. Noncross-linked and Mov10l1−/− testes served as negative controls. Separate libraries were prepared from RNA extracted from the main radioactive signal (Low, dashed line) and the higher position (High, solid line). (B) Size distribution (right axis) and average 5′ end nucleotide composition (left axis) for the three MOV10L1 CLIP-seq libraries. Shaded areas represent one standard deviation (SD). n = 3. (C) Genomic distribution of reads from three MOV10L1 CLIP-seq libraries and RNA-seq libraries from whole testis and isolated pachytene spermatocytes (PCTN). (D) Genome browser view of MOV10L1 CLIP tags, PCTN RNA-seq reads, and Mili piRNAs mapped within a pachytene piRNA cluster (chr15_cl266) that lies close to an unrelated gene (Arc). This view is an example of MOV10L1-binding specificity toward piRNA precursors. (E) Density of MOV10L1 CLIP tags mapped on mRNAs. Shaded areas represent one SD. n = 3. (F) Relative coverage of MOV10L1 (blue line) and Mili (red line) CLIP tags (Vourekas et al. 2012) mapping within −20/+40 bases from 5′ ends (0, X-axis) of piRNAs. For Mili, the large CLIP tag (lgClip) subpopulation was used, as it contains the PPIFs. The area typically occupied by a piRNA molecule is marked with a gray bar. Shaded areas represent one SD. n = 3.
The genomic distribution is strikingly similar to a typical pachytene piRNA library (Fig. 1C) even though only ∼6% of total MOV10L1 CLIP tags represent mature piRNA sequences (Supplemental Table S2). While 70% of MOV10L1 CLIP tags map within IPCs, only ∼1% of RNA-seq tags from whole testis and ∼4% of RNA-seq tags from purified wild-type pachytene spermatocytes map within IPCs, which again demonstrates the remarkable specificity of our CLIP experiments (Fig. 1C). The correlation between piRNA and MOV10L1 CLIP tags mapping within piRNA clusters was very high (Supplemental Fig. S2A). Moreover, we found a complete and reciprocal overlap at the nucleotide level between Mili-bound piRNAs (Vourekas et al. 2012) and MOV10L1 CLIP tags on all genomic elements. Specifically, even after removing putative piRNA sequences from MOV10L1 libraries, on average, 85.3% of MOV10L1 CLIP tags overlap with Mili piRNAs. The reciprocal overlap is 89.2%. Furthermore, ∼66% of MOV10L1 CLIP tags overlap with Mili piRNAs that map on mRNAs; the reciprocal overlap is 80%. MOV10L1 CLIP tag density shows a strong enrichment within mRNA 3′ untranslated regions (UTRs) (Fig. 1E), and the abundance of MOV10L1 CLIP tags is positively correlated with piRNA abundance in these areas (Supplemental Fig. S2B), in direct accord with the observation that many genic piRNAs are preferentially derived from the 3′ UTRs of mRNAs (Robine et al. 2009; Saito et al. 2009; Vourekas et al. 2012). The above findings demonstrate that MOV10L1 specifically binds virtually all piRNA precursors in vivo.
Binding of MOV10L1 to piRNA precursors precedes Piwi loading
We hypothesized that the positioning of MOV10L1 CLIP tags relative to piRNAs arising from neighboring sites may provide insights into the function of MOV10L1. Instead of a random distribution, the relative nucleotide coverage of MOV10L1 CLIP tags reveals a remarkable preference for binding downstream from and within ∼50 nt of both Mili and Miwi piRNA 5′ ends (Supplemental Fig. S2C), implying that MOV10L1 binds the 5′ cleaved piRNA precursor. By comparing the MOV10L1 CLIP and Mili large CLIP tag (lgClip) (Vourekas et al. 2012) coverage around 5′ ends of piRNAs, we found that the two plots show similar patterns (Fig. 1F), further supporting the above notion. Critically, the 5′ ends of MOV10L1 CLIP tags lie downstream from the 5′ ends of Mili-bound, stabilized PPIFs and piRNAs; we observed markedly lower coverage by MOV10L1 CLIP tags (compared with Mili CLIP tags) within a 9-nt distance downstream from 5′ ends of piRNAs (Fig. 1F). We verified this observation by measuring the 5′–5′ distances of MOV10L1 CLIP tags and mature piRNAs. We found a higher incidence of MOV10L1 CLIP tag 5′ ends downstream from 5′ ends of mature piRNAs and within 25 nt (Supplemental Fig. S2D). Since the 5′ ends of MOV10L1 CLIP tags are located within the piRNA body, this suggests that they represent 5′ cleaved piRNA precursors that were further attacked by a separate nucleolytic activity (Fig. 1F; Supplemental Fig. S2C). MOV10L1 CLIP tags show A/(G) nucleotide bias at the 5′ end and U/(C) bias one position upstream (which is not part of the CLIP tag); the 3′ end has a similar bias (Supplemental Fig. S2E). Mili- and Miwi-bound PPIFs have a 5′ bias for uridine (Vourekas et al. 2012). We carefully analyzed the nucleotide biases of MOV10L1 and Piwi CLIP tags using lysates with or without exogenous nucleases, as described in detail in the Supplemental Material and Supplemental Figure S2E–H. We show that the 5′ ends of Piwi-bound piRNA intermediate fragments are protected from nucleolytic activities, while the 5′ ends of MOV10L1-bound piRNA precursors are not. The sensitivity of the 5′ ends of MOV10L1 CLIP tags to nucleases indicates that the 5′ cleaved piRNA precursor, while bound by MOV10L1, is not bound and protected by a Piwi protein. Overall, the above findings indicate that MOV10L1 binds piRNA precursors close to the site of the endonucleolytic cleavage that will define the 5′ end of PPIFs before PPIFs are loaded to a Piwi protein that will protect their 5′ ends.
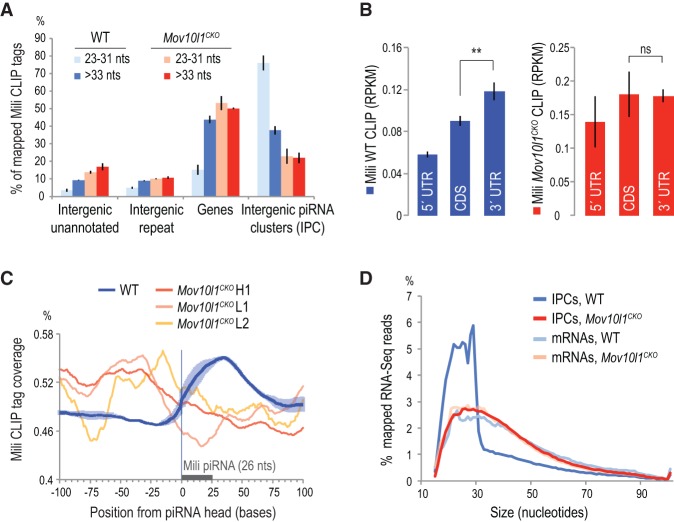
MOV10L1 is required for production of piRNA intermediate processing fragments
To determine whether pachytene piRNA precursors are processed in the absence of MOV10L1, we performed Mili CLIP using testes from Mov10l1fl/− Neurog3-Cre (Mov10l1CKO) mice. Prenatal and early postnatal testis development is normal in these animals, but upon conditional deletion of Mov10l1 after postnatal day 7, they exhibit a deficiency in pachytene piRNA biogenesis and post-meiotic arrest of spermatogenesis (Zheng and Wang 2012). Furthermore, to identify putative transcriptome changes, we performed RNA-seq (Vourekas et al. 2012) using total RNA extracted from wild-type and Mov10l1CKO whole testes and from highly enriched populations of pachytene spermatocytes and round spermatids (Supplemental Table S1).
We prepared three Mili CLIP libraries from Mov10l1CKO: two from the main radioactive signal and one from larger Mili–RNA complexes (Supplemental Fig. S3A). The profiles of these libraries are dramatically different from Mili CLIP libraries from wild-type mice (Vourekas et al. 2012), and a robust piRNA population was not detected in any of the three Mov10l1CKO libraries (Supplemental Fig. S3, cf. B and C), which is in agreement with our previous report (Zheng and Wang 2012). Contrary to Mili CLIP from wild-type testes, all three Mili CLIP libraries from Mov10l1CKO have a uniform genomic distribution for smaller- and larger-sized CLIP tags, and there are fewer Mili CLIP tags mapped within precursor piRNA transcripts in the absence of MOV10L1 (Fig. 2A). We specifically interrogated whether these libraries contained 23- to 31-nt-sized CLIP tags that had common 5′ ends with pachytene and/or prepachytene Mili-bound piRNAs from standard immunoprecipitation libraries (Aravin et al. 2007; Kirino et al. 2009). While a typical piRNA-enriched Mili CLIP library (Vourekas et al. 2012) from wild-type mice contains ∼65% piRNA-matching sequences in this size range, Mili CLIP libraries from Mov10l1CKO have only ∼10% matches while containing a similar amount of putative prepachytene piRNAs with wild-type libraries (Supplemental Table S2). Collectively, these data demonstrate a severe defect in piRNA processing upon conditional deletion of MOV10L1.
Figure 2.
Mili CLIP-seq reveals a lack of PPIF production in Mov10l1CKO. (A) Genomic distribution of replicate Mili CLIP libraries from wild-type (WT; blues) and Mov10l1fl/− Neurog3-Cre testes (Mov10l1CKO; reds). In contrast to wild-type, Mili CLIP tags from Mov10l1CKO exhibit a uniform genomic distribution regardless of their sizes. Error bars represent one SD. n = 3. (B) Abundance of Mili wild-type and Mov10l1CKO CLIP tags (reads per kilobase per million [RPKM]) mapping within mRNA untranslated and coding regions. Error bars represent one SD. n = 3. (C) Mili CLIP tag coverage within ±100 nt from piRNA 5′ ends (0, X-axis) in wild-type and Mov10l1CKO. The area occupied by a typical piRNA is marked with a gray bar. Shaded areas represent one SD. n = 3. (D) Size distribution of RNA-seq reads from adult wild-type and Mov10l1CKO testis. The percentage of total mapped tags per read size for each genomic category is plotted. Reads mapping to mRNAs or piRNA clusters (IPCs) were plotted separately. The majority of RNA-seq reads mapping to IPCs in wild-type testis are piRNA-sized or smaller. Although some mRNAs, especially 3′ UTRs, are processed into piRNAs, the overwhelming majority is not; thus mRNA read size distributions are identical.
In addition, the enrichment of Mili CLIP tags in 3′ UTRs (Vourekas et al. 2012) is lost in the absence of MOV10L1 (Fig. 2B), consistent with a piRNA biogenesis defect. Mili CLIP tags mapped within precursor piRNA transcripts in the absence of MOV10L1 are fewer (Fig. 2A, IPC) and lack a 5′ uridine bias (Supplemental Fig. S3D), the hallmark of Piwi-stabilized piRNA intermediates. In fact, they carry the same bias as described above for 5′ ends of MOV10L1 CLIP tags and 3′ ends of Miwi-bound PPIFs (Supplemental Fig. S2E,F, respectively), suggesting that the 5′ ends of these RNAs were not protected by Mili and were generated after cell lysis. Additionally, Mili CLIP tag coverage relative to 5′ ends of piRNAs (Fig. 2C) in Mov10l1CKO reveals a complete loss of the characteristic binding downstream from piRNA 5′ ends, strongly suggesting that these Mili-bound transcript fragments from Mov10l1 mutant testes do not represent bona fide PPIFs.
To investigate these observations further, we examined RNA-seq tags mapping within piRNA clusters. The size profile of IPC reads from wild-type mice shows that piRNA processing of precursor transcripts is detectable in RNA-seq libraries (Fig. 2D) even though the total RNA is fragmented before library preparation. In contrast, the IPC read size profile in Mov10l1CKO testis is identical to that of mRNA reads in wild-type and conditional knockout genotypes, strongly suggesting the absence of piRNAs and piRNA intermediate fragments in mice lacking MOV10L1. Therefore, RNA-seq and Mili CLIP in Mov10l1CKO testis support the notion that MOV10L1 function is essential for the endonucleolytic cleavage that gives rise to intermediate piRNA fragments.
The uncovering of piRNA processing products in wild-type RNA-seq libraries has a broader implication for transcriptome analysis. Long transcripts that are processed and give rise to mature products whose stability and turnover are regulated by different mechanisms than for the precursor transcripts cannot be accurately quantified by a technique that uses short reads such as RNA-seq because the short reads cannot be unambiguously assigned to the precursor or the mature RNA. Therefore, we used quantitative RT–PCR (qRT–PCR) to verify and measure the levels of piRNA precursors, selected mRNAs, and retrotransposons. As previously shown (Zheng and Wang 2012), pachytene piRNA precursors are increased significantly in the Mov10l1 mutant testes, while retrotransposons are only slightly increased (less than twofold) (Supplemental Fig. S3E). Interestingly, Miwi (Piwil1) mRNA is modestly up-regulated, while its protein levels are notably down-regulated (Supplemental Fig. S3F). Quantification of mRNA levels by qRT–PCR is in agreement with RNA-seq results (Supplemental Fig. S3G). We noticed a decrease of spermiogenic mRNA levels despite virtually unchanged levels of CREM, the master spermiogenic transcriptional regulator (Blendy et al. 1996). This finding is consistent with a proposed role of Miwi in post-transcriptional stabilization of spermiogenic mRNAs (Vourekas et al. 2012). It is unclear why the Miwi protein abundance is reduced in Mov10l1CKO testis.
piRNA precursor secondary structure elements are enriched in MOV10L1 footprints
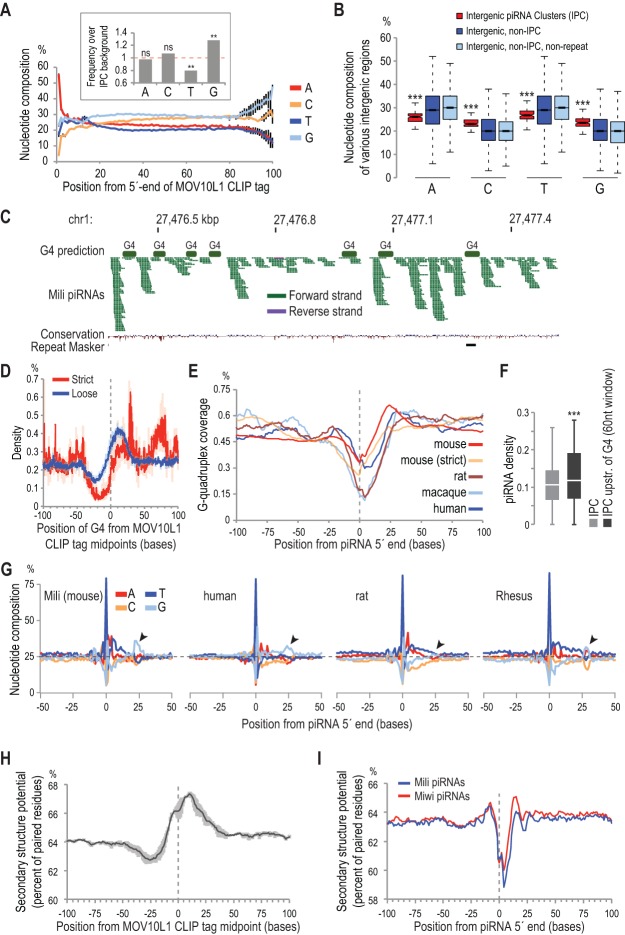
We next analyzed MOV10L1 CLIP tags to identify potential sequence-binding preferences. Intriguingly, we discovered a statistically significant enrichment in G (∼28%) and depletion of U (∼20%) residues in heavily bound areas of the piRNA clusters compared with the entire cluster sequence (P < 0.05) (Fig. 3A). A genome-wide nucleotide composition analysis for intergenic areas revealed that, remarkably, piRNA clusters are already significantly enriched in G residues compared with other intergenic areas, regardless of the presence of repeat elements (Fig. 3B).
Figure 3.
Sequence analysis of MOV10L1 CLIP tags and piRNA precursors implicates elements of secondary structures in piRNA processing. (A) Nucleotide composition per position along MOV10L1 CLIP tags. Error bars represent one SD. n = 3. (Inset) Nucleotide composition of MOV10L1 CLIP tags mapped within IPCs over nucleotide composition of IPCs. Significance test was performed using permutations (see the Supplemental Material). (ns) Nonsignificant; (**) P-value < 0.05. (B) Notched box and whisker plots showing nucleotide occurrence within the indicated genomic areas. (***) P-value < 10−15. (C) Distribution of predicted G quadruplexes (G4s) and mapped piRNAs in a piRNA cluster on chromosome 1. (D) Density plot of G4 midpoints relative to MOV10L1 CLIP tag midpoints (0, X-axis). Strict and loose G4 predictions are plotted. Shaded areas represent one SD (n = 3). Notable enrichment of G4 occurrence is observed downstream from MOV10L1 CLIP tag midpoints. (E) Density plot of G4s within mammalian piRNA genomic loci (5′ ends of piRNAs at position 0, X-axis). Strict and loose G4 predictions are plotted for mouse piRNAs, and loose prediction is plotted for rat, macaque, and human piRNAs. (F) Box and whisker plot of piRNA densities (piRNA counts/sequence length) of whole piRNA clusters (light gray) and of 60-nt windows upstream of 5′ ends of G4s (loose prediction) within the same clusters (dark gray). (***) P-value < 10−4. (G) Genomic nucleotide composition within 50 bases from 5′ ends of mammalian piRNAs for the indicated species. Arrowheads indicate local increase of Gs toward the 3′ ends of piRNAs. (H) Secondary structure potential around MOV10L1 CLIP tag midpoints within IPCs. The gray area represents one SD. n = 3. (I) Secondary structure potential in the genomic context of Mili and Miwi piRNAs.
These findings raised the possibility that elements of secondary structures known as G quadruplexes (G4s) (Biffi et al. 2014) may play a role in piRNA processing and prompted us to perform genome-wide prediction of G4s using strict and loose prediction algorithms (see the Supplemental Material), which we overlaid with MOV10L1 CLIP tags and piRNAs (Fig. 3C,D). Notably, G4 elements are enriched within MOV10L1 CLIP tags, downstream from their mid-points (Fig. 3D). Our analysis revealed a paucity of G4 density on and around 5′ ends of piRNAs from four mammalian species followed by a relative increase downstream from piRNA 5′ ends (Fig. 3E). Additionally, Mili piRNA density within a 60-nt window upstream of G4 5′ ends is significantly higher than overall piRNA density within IPCs (P < 10−4, paired t-test) (Fig. 3F, see also Supplemental Table S3). Subsequently, we asked whether G4 relative positioning and frequency created a specific nucleotide “signature” around piRNA loci. Examining the genomic nucleotide composition around piRNA 5′ ends, we found a notable increase in G residues downstream from position 15 (Fig. 3G). Strikingly, this pattern is present within human, Rhesus, mouse, and rat piRNA genomic loci (Fig. 3G) despite the absence of piRNA sequence conservation, suggesting a conserved role for G4s in mammalian piRNA precursor processing. We performed G4 prediction within piRNA cluster sequences for all four mammalian species and found significantly more G4s than would be expected by chance (P < 10−4) (Supplemental Table S3). The above findings suggest that G4s within piRNA precursors are not an indirect consequence of guanosine enrichment but a result of selective pressure and may have been co-opted as landmarks for piRNA biogenesis.
Additionally, we performed a secondary structure prediction of intergenic piRNA precursors using RNAfold (Hofacker et al. 1994) and plotted it relative to MOV10L1 CLIP tags and piRNAs. The base-pairing potential is remarkably increased close to the 3′ end of MOV10L1 CLIP tags, in concordance with G4 density in the same areas, and shows a local minimum around piRNA 5′ ends (Fig. 3H,I, respectively). Collectively, these results support the notion that processing of piRNA precursor transcripts is intimately coupled with local secondary structures: 5′ ends are generated more often in areas of lesser secondary structure potential, and MOV10L1 is associated with structured regions (G4 or otherwise) of precursor transcripts.
To examine the formation of G4s within piRNA precursor transcripts in vivo, we used the highly specific BG4 antibody (Biffi et al. 2014). BG4 antibody specifically immunoprecipitated piRNA precursors from testis lysates (Fig. 4A), especially in Mov10l1CKO, strongly suggesting the formation of G4s within piRNA precursor transcripts in vivo. The degree of enrichment of piRNA precursors in BG4 immunoprecipitations from Mov10l1CKO lysates compared with wild type (10-fold to 75-fold) greatly exceeded the increase of their overall levels in total RNA (twofold to sixfold) (Fig. 4B). Therefore, the dramatic precursor RNA enrichment in BG4 immunoprecipitations from Mov10l1CKO testis lysates is only partially attributed to precursor transcript abundance. The increased antigenicity of piRNA precursors from Mov10l1CKO lysates in this assay indicates the increased presence of G4 structures within these transcripts in Mov10l1 mutant testis and thus a role for MOV10L1 in the resolution of these thermostable secondary structures.
Figure 4.
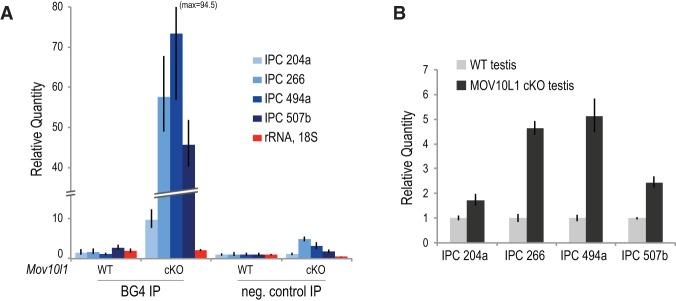
G4 antigenicity of piRNA precursor transcripts increases in Mov10l1 mutant mice. (A) RNA immunoprecipitation of piRNA precursor transcripts using the BG4 antibody that specifically recognizes RNA G4s using testis lysates from wild-type (WT) and Mov10l1CKO mice. Relative quantity of the indicated RNAs extracted from the immunoprecipitation beads was determined by qRT–PCR. (B) Determination of steady-state levels of piRNA precursor transcripts by qRT–PCR using wild-type and Mov10l1CKO testis total RNA.
MOV10L1 exhibits 5′-to-3′ RNA-unwinding activity in vitro
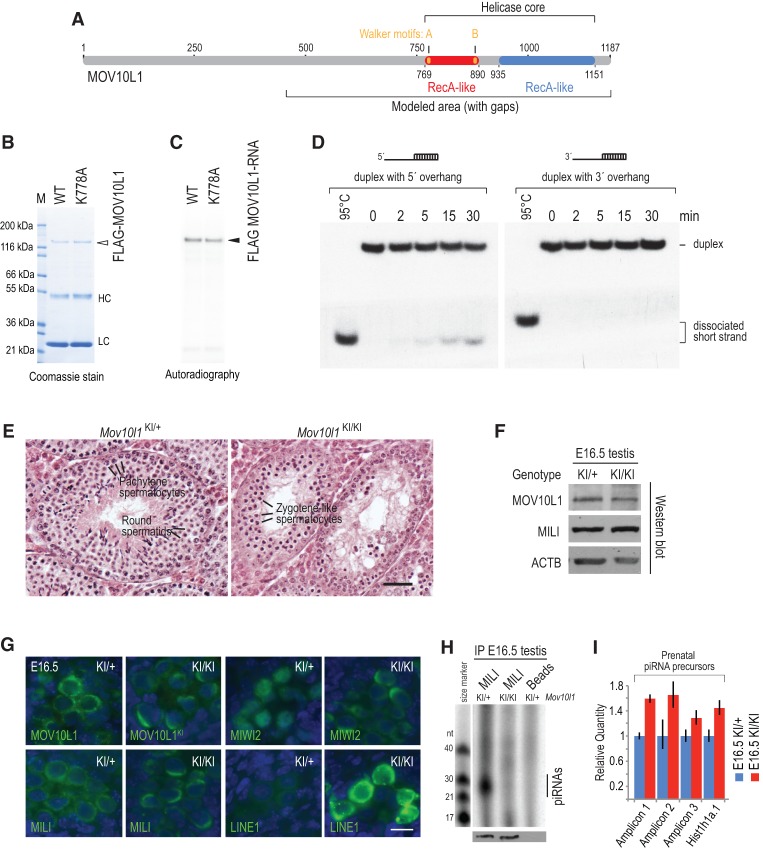
MOV10L1 belongs to the superfamily I (SF I) of DNA/RNA helicases (Fig. 5A; Fairman-Williams et al. 2010), and one of the closest homologs of MOV10L1 is UPF1, an RNA helicase involved in nonsense-mediated RNA decay (Supplemental Fig. S4A; Kervestin and Jacobson 2012). Tertiary structure modeling showed extensive similarities between the helicase domains of MOV10L1 and UPF1 (Supplemental Fig. S4A–E; Supplemental Material), suggesting that MOV10L1 may use its conserved helicase core to translocate the piRNA precursor transcript, analogous to the translocation of UPF1 on mRNAs with a 5′-to-3′ directionality (Franks et al. 2010). Recently, MOV10, a ubiquitously expressed RNA helicase that has not been implicated in the piRNA pathway, was found to possess RNA-unwinding activity and assist UPF1 in mRNA degradation (Gregersen et al. 2014).
Figure 5.
MOV10L1 RNA helicase activity is essential for piRNA biogenesis. (A) Schematic representation of the MOV10L1 protein domain structure. (B) Immunopurification of Flag-tagged wild-type (WT) and ATP-binding-deficient point mutant (K778A) MOV10L1 proteins from 293T cells. (C) UV cross-linking (365 nm) of wild-type and K778A MOV10L1 proteins with a radiolabeled RNA oligo (38 nt) bearing an internal 4SU residue. (D, left panel) 5′-to-3′ directional RNA duplex-unwinding activity of wild-type MOV10L1. MOV10L1 was incubated with an RNA duplex bearing a 5′ or 3′ overhang in the presence of Mg2+ and ATP for the indicated times. A heat-treated duplex (95°C) shows the migration pattern of the fully dissociated short oligonucleotide. (Right panel) 3′-to-5′ activity was not detected. (E) Histological analysis of testes from adult (8-wk-old) Mov10l1KI/+ and Mov10l1KI/KI mice. Mov10l1KI/+ seminiferous tubules contain spermatogenic cells in all stages of differentiation, whereas Mov10l1KI/KI tubules exhibit meiotic arrest, with the most differentiated cells being zygotene-like spermatocytes. Bar, 50 μm. (F) Western blots for the indicated proteins from embryonic day 16.5 (E16.5) Mov10l1KI/+ and Mov10l1KI/KI testes. (G) Immunofluorescence (green) of MOV10L1, MILI, MIWI2, and LINE1orf1p in E16.5 Mov10l1KI/+ and Mov10l1KI/KI testes. DNA was stained with DAPI (blue). Bar, 10 μm. (H) MILI is depleted of piRNAs in E16.5 Mov10l1KI/KI testes. Ten pairs of embryonic testes per genotype were used for immunoprecipitation. Ninety percent of the immunoprecipitated material was used for analysis of associated RNAs, whereas the remaining 10% was used for detection of MILI by Western blot (WB). Lysate incubated with uncoupled beads served as a background control. (I) Up-regulation of piRNA precursors in E16.5 Mov10l1KI/KI testes. Total RNA extracted from Mov10l1KI/− and Mov10l1KI/KI testes was analyzed by qRT–PCR. Genomic locations of E18 Mili-bound piRNAs (Xiol et al. 2012) and of the amplicons detected in this experiment are shown in Supplemental Figure S5C.
To test whether MOV10L1 can unwind RNA in vitro, we adapted the RNA duplex-unwinding assay (Jankowsky and Putnam 2010). We immunopurified Flag-tagged wild-type and ATP-binding-deficient point mutant MOV10L1 (K778A) from transfected 293T cells (Fig. 5B). Lys778 is conserved among SF I RNA helicases (Supplemental Fig. S4A) and is essential for ATP binding and therefore RNA helicase activity of UPF1 (Weng et al. 1996). Both wild-type and mutant MOV10L1 can be cross-linked efficiently and specifically to a 38-nt-long RNA bearing a 4-thio-uridine residue (Fig. 5C), indicating that the point mutation does not impair the RNA-binding capacity of MOV10L1, similar to the mutation of the homologous residue in UPF1 (Weng et al. 1996). Wild-type MOV10L1 shows time-dependent, 5′-to-3′ (Fig. 5D, left panel) but not 3′-to-5′ (Fig. 5D, right panel) RNA-unwinding activity when using a partial RNA duplex with a single-stranded overhang, showing that MOV10L1 is indeed an RNA helicase. In the same assay, the K778A point mutant is catalytically inactive (Supplemental Fig. S5A). These results, coupled with the CLIP analysis, indicate that MOV10L1 may play a role in directional, 5′-to-3′ translocation of the precursor transcripts and the remodeling of their secondary structures during piRNA biogenesis.
A point mutation in the MOV10L1 RNA helicase core abolishes piRNA biogenesis
To determine the requirement of MOV10L1 RNA helicase activity for piRNA biogenesis in vivo, we generated a Mov10l1 knock-in (Mov10l1KI) allele harboring the K778A amino acid change in the ATP-binding motif through gene targeting in embryonic stem (ES) cells (Supplemental Fig. S5B). Heterozygous Mov10l1KI/+ mice were viable and fertile, suggesting that the knock-in allele is not dominant negative. In contrast, while Mov10l1KI/KI females were fertile, Mov10l1KI/KI males were sterile and exhibited meiotic arrest at the zygotene-like stage of prophase I (Fig. 5E), a phenotype similar to Mov10l1 knockout mice (Zheng et al. 2010). The MOV10L1K778A protein was expressed in gonocytes in embryonic day 16.5 (E16.5) Mov10l1KI/KI testis at a level comparable with MOV10L1 in Mov10l1KI/+ testis (Fig. 5F,G). MOV10L1 localized throughout the cytoplasm in a punctate pattern in the Mov10l1KI/+ gonocytes but exhibited an abnormal polar cytoplasmic localization in the Mov10l1KI/KI gonocytes (Fig. 5G). Miwi2 was present in the nucleus in Mov10l1KI/+ gonocytes but excluded from the nucleus in Mov10l1KI/KI gonocytes, similar to the Mili knockout phenotype (Shoji et al. 2009; Zheng et al. 2010). Furthermore, both Mili and Miwi2 exhibited a similar polar cytoplasmic localization in Mov10l1KI/KI gonocytes, suggesting that the piRNA biogenesis pathway is perturbed by the point mutation in the MOV10L1 RNA helicase domain.
We next determined whether transposon silencing and piRNA biogenesis were affected. Immunofluorescence analysis showed that LINE1 retrotransposons were highly derepressed in Mov10l1KI/KI gonocytes (Fig. 5G). Mili was associated with piRNAs in E16.5 Mov10l1KI/+ testes but depleted of piRNAs in E16.5 Mov10l1KI/KI testes while maintaining wild-type expression levels (Fig. 5H). Additionally, using qRT–PCR, we detected up-regulation of piRNA precursor transcripts in E16.5 Mov10l1KI/KI testes (Fig. 5I; see also Supplemental Fig. S5C). Taken together, these results indicate that MOV10L1 RNA helicase activity is essential for the processing of the precursor transcripts during piRNA biogenesis in vivo.
Discussion
Our data provide a transcriptome-wide view of primary piRNA biogenesis in unprecedented detail and describe the biochemical role of MOV10L1 in this process. We found that MOV10L1 binds to the piRNA precursor with remarkable specificity. The in vitro enzymatic assays and the generation of an ATPase point mutant knock-in mouse of MOV10L1 establish the requirement of the MOV10L1 ATP-dependent RNA helicase activity for piRNA biogenesis in vivo. In vitro systems that recapitulate primary piRNA biogenesis have not been generated, likely due to the numerous factors and activities involved and the complexity of their intricate network of interactions. Nevertheless, we were able to show that MOV10L1 exhibits a 5′-to-3′ directional RNA-unwinding activity in vitro. PPIFs were identified biochemically by Piwi HITS-CLIP (Vourekas et al. 2012) and genetically in the Tdrkh mutant mice (Saxe et al. 2013). MOV10L1 and the endonuclease, which is most likely a mZuc dimer (Haase et al. 2010; Ipsaro et al. 2012; Nishimasu et al. 2012; Voigt et al. 2012), are both required for the generation of PPIFs, indicating that these activities are functionally linked. We envision that MOV10L1 helicase activity is essential for proper piRNA precursor “feeding” to the endonuclease for cleavage. In this model, PPIFs are generated by consecutive cleavages of the 5′ end of the precursor, facilitated by the directional translocation of the precursor by MOV10L1 toward the endonuclease (Fig. 6). The directional translocation is supported by our in vitro duplex-unwinding assay and the pronounced downstream relative positioning of MOV10L1 CLIP tags with respect to the piRNA 5′ ends. Fittingly, the shape of the relative coverage by MOV10L1 CLIP tags with respect to piRNAs (sharp leading edge with a long tail) (Supplemental Fig. S2C) is not consistent with a static type of binding that forms a symmetrical distribution of CLIP tags around a central point, such as Ago binding of mRNAs by microRNA (miRNA)-mediated recognition of a sequence element (Chi et al. 2009). We found increased secondary structure potential toward the 3′ end of MOV10L1 CLIP tags, consistent with a temporary stalling or slowing of the precursor translocation when highly stable secondary structures, such as G4s (Zhang et al. 2011), reach the helicase core. Accordingly, piRNA density is higher upstream of G4s. We speculate that such translocation “stalling” provides an opportunity for the endonuclease to cleave the piRNA precursor. This relative positioning pattern of G4s creates a prominent G bias toward the 3′ ends of mammalian piRNAs, supporting a conserved role in mammalian piRNA biogenesis for G4s, which appear to be selected at the level of secondary structures and are not simply a consequence of G enrichment within piRNA cluster sequences. To our knowledge, this is the first time that local secondary structures of the precursor have been implicated in piRNA processing. Our data using anti-G4 antibodies indicate the increased presence of G4s within piRNA precursors in Mov10l1CKO testis, in turn supporting the notion that G4s—and most likely other elements of secondary structure as well—are resolved by MOV10L1 helicase activity to promote piRNA biogenesis. MOV10L1 may also have an RNP remodeling activity.
Figure 6.
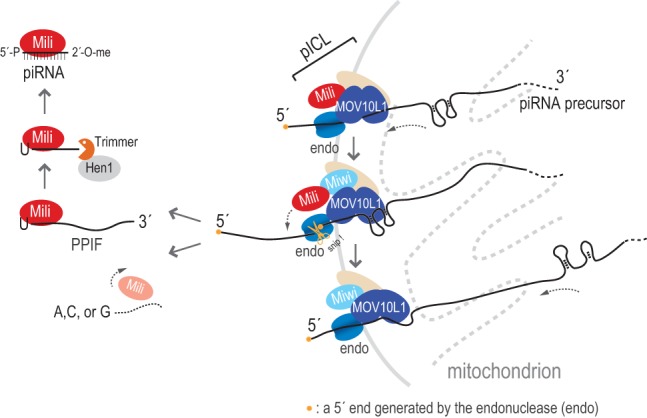
Refined model for piRNA biogenesis. MOV10L1, a Piwi protein, and the endonuclease (endo) are functionally linked within the piRNA intermediate cleavage and loading (pICL) complex, which likely contains additional factors (beige oval), such as GASZ, Tdrds, and chaperones. In this model, MOV10L1 translocates the piRNA precursor transcript in a 5′-to-3′ direction toward the endonuclease. MOV10L1 helicase activity resolves RNA secondary structures and facilitates cleavage that creates PPIFs. Mili binds the newly formed PPIFs, and both are released from MOV10L1. MOV10L1 may also have an RNP remodeling activity in pICL. Mili preferentially binds and stabilizes PPIFs with 5′ U; this is followed by 3′ end trimming and 2′-O methylation, resulting in mature piRNAs. This process may repeat as MOV10L1 in pICL continues piRNA precursor translocation.
There is substantial evidence from genetic and cell culture experiments that Zuc is essential for primary piRNA biogenesis (Pane et al. 2007; Malone et al. 2009; Haase et al. 2010; Saito et al. 2010; Watanabe et al. 2011), and it was recently shown both structurally and biochemically that Zuc is an endonuclease that cleaves ssRNA and leaves a 5′ phosphate end (Ipsaro et al. 2012; Nishimasu et al. 2012; Voigt et al. 2012). Therefore, Zuc fits the description for the endonuclease that generates the 5′ ends of piRNA intermediates, but direct proof of this is lacking. Our results are consistent with an endonuclease that is inhibited by secondary structures and is activated by the secondary structure-resolving helicase activity of MOV10L1. In support of the possibility that Zuc endonuclease is responsible for piRNA intermediate generation in the piRNA intermediate cleavage and loading (pICL) complex (see below), we were able to detect MOV10L1 and mZuc association by coimmunoprecipitation using a heterologous expression system (Supplementary Fig. S5D).
In our model (Fig. 6), cleavage events occur downstream from previous ones and “prune” the longer precursor, creating the 5′ ends of forthcoming PPIFs and releasing intermediate fragments, which are bound by Piwi. The loaded Piwi breaks off from MOV10L1, and the latter may carry on with additional rounds of precursor processing. The 5′ U of PPIFs is stabilized by Piwi, and the 3′ end is trimmed and methylated, leading to generation of mature piRNA. Since MOV10L1 does not bind Piwi-stabilized PPIFs, MOV10L1 RNA-binding activity may not be directly involved in the subsequent PPIF maturation steps. To illustrate the separation of the 5′ and 3′ end maturation processes, we propose the designation of a functional pICL complex, which contains MOV10L1, Piwi, and the endonuclease (Fig. 6). Additional proteins, such as GASZ, Tdrds, and chaperones, are likely components of the pICL complex (Olivieri et al. 2012; Preall et al. 2012; Xiol et al. 2012; Izumi et al. 2013). MOV10L1 does not interact with Tdrkh (Saxe et al. 2013), which is required for 3′ end maturation. Experiments in Drosophila are consistent with our model; RNAs isolated from Armitage by standard immunoprecipitation conditions show characteristics similar to those of piRNA precursor fragments (Saito et al. 2010). A significant number of Armitage-bound RNA fragments mapped within the flamenco locus, which gives rise to long piRNA precursor transcripts (Malone et al. 2009), overlapping with piRNA sequences (Saito et al. 2010). Most of these fragments (which the investigators named piR-ILs) did not match the 5′ ends of mature piRNAs, suggesting that Armitage binds piRNA precursors. Moreover, based on the above, inhibition of endonuclease activity would result in Piwi protein free of RNA, which would be “trapped” in a complex with MOV10L1. This was indeed observed in Drosophila: Upon Zuc knockdown, Piwi was less loaded, and more Piwi could be immunoprecipitated with Armitage (Saito et al. 2010), indicating conserved function of MOV10L1 helicases in piRNA biogenesis.
How primary piRNA precursors that are 5′ capped and 3′ polyadenylated (Li et al. 2013) are selected from other RNA polymerase II transcripts is not known. We speculate that cytoplasmic transcripts that are not translated and enter ICM/nuage can be recognized by pICL and processed into piRNAs. This could explain why the bulk of genic piRNAs are derived from 3′ UTRs and the presence in pachytene piRNA precursors of G4s and other secondary structure elements, which may function to impede translation (Arora et al. 2008; Beaudoin and Perreault 2010). Ultimately, identification of all components of the pICL complex and biochemical characterization with reconstitutions of its activities will be required to further address how piRNA precursors are selected to enter ICM/nuage, the role of G4 and other structural elements, and the precise molecular function of all pICL complex components in piRNA processing.
Materials and methods
MOV10L1 HITS-CLIP
MOV10L1 HITS-CLIP was performed essentially as for Mili and Miwi previously (Vourekas et al. 2012). The protocol was described in exhaustive detail (Vourekas and Mourelatos 2014). Testes from 23-dpp or adult mice were collected, detunicated, disrupted by mild pipetting in ice-cold HBSS, and immediately UV-irradiated three times at 254 nm (400 mJ/cm2). The cells were pelleted and washed with PBS, and the final cell pellet was flash-frozen in liquid nitrogen and kept at −80°C. UV light-treated cells (two testes) were lysed in 300 μL of 1× PMPG with protease inhibitors, 2 U/μL RNasin, and no exogenous nucleases; lysates were treated with DNase (Promega) for 5 min at 37°C and then centrifuged at 90,000g for 30 min at 4°C.
For each immunoprecipitation, ∼5 μg of our anti-MOV10L1 rabbit polyclonal purified total IgG (Zheng et al. 2010) was bound on protein A Dynabeads in antibody-binding buffer (0.1 M Na-phosphate at pH 8, 0.1% NP-40) for 3 h at 4°C; antibody-bound beads were washed three times with 1× PMPG (1x PBS [no Mg2+ and no Ca2+], 2% Empigen).
Antibody beads were incubated with lysates (supernatant of 90,000g) for 3 h at 4°C. Low- and high-salt washes of immunoprecipitation beads were performed with 1× and 5× PMPG (5× PBS, 2% Empigen).
RNA linkers (RL3 and RL5) as well as 3′ adaptor labeling and ligation to CIP (calf intestinal phosphatase)-treated RNA CLIP tags were previously described (Vourekas and Mourelatos 2014).
Immunoprecipitation beads were eluted for 12 min at 70°C using 30 μL of 2× Novex reducing loading buffer. Samples were analyzed by NuPAGE (4%–12% gradient precast gels run with MOPS buffer). Cross-linked RNA–protein complexes were transferred onto nitrocellulose (Invitrogen, LC2001), and the membrane was exposed to film for 1–2 h. Membrane fragments containing the main radioactive signal and fragments up to ∼15 kDa higher were cut (Fig. 1B). RNA extraction, 5′ linker ligation, RT–PCR, and the second PCR step were performed with the DNA primers (DP3 and DP5 or DSFP3 and DSFP5) as described previously (Vourekas and Mourelatos 2014). cDNA from two PCR steps was resolved on and extracted from 3% Metaphor 1× TAE gels stained with ethidium bromide. The size profiles of cDNA libraries prepared from the main radioactive signal and higher molecular weights were similar (Fig. 1B). DNA was extracted with QIAquick gel extraction kit and submitted for deep sequencing. The cDNA libraries were sequenced on an Illumina HiSeq 2500 at 100 cycles.
Generation of Mov10l1 knock-in mice
To generate the Mov10l1 knock-in targeting construct (Supplemental Fig. S5B), DNA fragments were amplified by high-fidelity PCR using a Mov10l1 BAC clone (RP23-269F24). Codon 778 (exon 17) was mutated from AAG (lysine) to GCG (alanine) by PCR-based mutagenesis. The neomycin selection cassette was flanked by loxP sites for Cre-mediated removal. The targeting construct was sequenced to confirm the introduced mutation. V6.5 hybrid ES cells were electroporated with linearized targeting construct (pUP109/ClaI) and cultured in the presence of 350 μg/mL G418. Five-hundred-seventy-six G418-resistant ES cell clones were screened for homologous recombination. Three targeted ES clones were obtained, and two were injected into blastocysts. The Mov10l1 knock-in allele was transmitted through the germline. To delete the neo selection cassette, Mov10l1KI/+ mice were crossed with Actb-Cre mice that express Cre ubiquitously (Lewandoski and Martin 1997). All of the studies were performed with mice without the neo cassette. The wild-type (439-base-pair [bp]) and knock-in (557-bp) alleles were assayed by PCR with the primers ACCTCCGGGAACTGGAGCGACTGTG and ATCCCAAGCCCGGCTTGCAGTA. PCR primers flanked the remaining loxP site and adjacent vector sequence.
RNA immunoprecipitation using the BG4 antibody
Bacterial expression and purification of Flag/His dual-tagged BG4 antibody were performed as previously described (Biffi et al. 2014). Twenty microliters of M2 agarose beads (Sigma) was washed three times in lysis and immunoprecipitation buffer (50 mM Tris at pH 7.5, 100 mM KCl, 0.1% Igepal CA-360 [NP-40] with EDTA-free protease inhibitors [Roche], 0.5 U/μL rRNasin) and bound with 3 μg of purified BG4 antibody in 600 μL of lysis buffer for 1.5 h at 4°C. One detunicated testis (from 3-mo-old wild-type or Mov10l1CKO animals) was lysed using 1200 μL of lysis buffer with a plastic pestle. The lysate was centrifuged at 16,000g for 20 min, and the supernatant was mixed with the antibody-bound M2 beads or M2 beads alone for 1.5 h at 4°C; the latter served as the negative control. RNA was extracted from the beads using Trizol, precipitated using ethanol, treated with DNase I for 1 h at 37°C, and extracted again using acid phenol. The RNA was spiked with 1 ng of in vitro transcribed Renilla luciferase mRNA. qRT–PCR was performed on a StepOnePlus system (Applied Biosystems) using the primers described above. The Renilla luciferase mRNA was used in place of an endogenous control for calculating relative mRNA quantities.
MOV10L1 protein expression and purification
The nucleotide sequence encoding the 1187 amino acids (Uniprot: Q99MV5) and the K778A mutant were cloned into the pRK5 vector with an in-frame N-terminal Flag tag using Phanta Super-Fidelity DNA polymerase and ClonExpress II (Vazyme). The pRK5 constructs were used to transfect 293T cells with TurboFect transfection reagent (Thermo). 293T cells expressing wild-type and K778A MOV10L1 were lysed by sonication in K150 lysis and immunoprecipitation buffer (50 mM HEPES at pH 7.5, 150 mM KoAc, 1 mM DTT, 0.1% NP-40 [Igepal] with EDTA-free protease inhibitors cocktail [Roche]), and the lysate was centrifuged at 16,000g for 20 min. The cleared lysate was mixed with 50 μL of M2 magnetic bead slurry (Sigma), prewashed with K150, and incubated for 2 h at 4°C. Immunoprecipitation beads were washed three times with K150, once with K150 containing 250 mM NaCl, and three more times with K150. The beads were resuspended in 1 mL of K150 and kept on ice for a maximum of 2 d. Fifty microliters of bead suspension contained 1 μg of MOV10L1.
Cross-linking of MOV10L1 with an RNA oligo bearing a 4-thio-uridine residue
The following oligo was used for cross-linking: U = 4-thio-uridine (4SU), 5′-CAACCACCACUAAACAUUUAGAUGCCACUCUCAACGUU-3′.
Fifty microliters of wild-type or K778A MOV10L1-bound bead slurry (1 μg of MOV10L1 protein) was washed three times with RNA-binding buffer (50 mM HEPES at pH 7.4, 100 mM KoAC, 1 mM DTT, 0.01% NP-40, 0.2 mM MgCl2) and resuspended in 25 μL of the same buffer supplied with 0.5 μL of 40 U/μL rRNasin and 1 μL of 5′ 32P-labeled RNA oligo carrying 4SU residues. The reaction mixture was incubated for 20 min at 37°C with shaking and irradiated with UV (365 nm) for 30 min on ice. The beads were washed twice, and the protein–RNA complexes were eluted with reducing SDS loading buffer and analyzed on a 4%–12% Bis-Tris NuPAGE gel. The gel was dried and exposed to film overnight.
RNA duplex-unwinding assay
We used the RNA duplex-unwinding assay developed by the Jankowsky laboratory, also used recently for Mov10, with some modifications (Jankowsky and Putnam 2010; Gregersen et al. 2014). The following RNA oligos were used: duplex with 5′ end overhang (Top 5′, 5′-GCGUCUUUACGGUGCU-3′ and Bottom 5′, 5′-AAAACAAAACAAAACAAAACAAAAUAGCACCGUAAAGACGC-3′) and duplex with 3′ end overhang (Top 3′, 5′-AGCACCGUAAAGACGC-3′, and Bottom 3′, 5′-GCGUCUUUACGGUGCUUAAAACAAAACAAAACAAAACAAAA-3′). The top oligos were 5′ end-labeled, and all four oligos were gel-purified. The duplexes were prepared in 50 mM MOPS (pH 6.5) and 5 mM EDTA. To detect the duplex-unwinding activity, 50 μL of MOV10L1-bound bead suspension (1 μg of MOV10L1 protein) was washed in reaction buffer (50 mM Tris at pH 7.5, 20 mM KoAc, 0.2 mM MgCl2, 0.01% Igepal [Nonidet P-40], 1 mM DTT); mixed with 20 nM duplex, 1 pmol of cold top oligo, and 20 U of rRNasin in 30 μL of reaction buffer; and incubated for 10 min at 37°C with shaking in a Thermomixer (1000 rpm). Subsequently, ATP was added at a final concentration 0.5 mM, and incubation was resumed for the indicated time. Four-microliter reaction samples were mixed with 4 μL of stop solution and kept on ice. Reaction products were analyzed by electrophoresis on a 12% acrylamide nondenaturing gel and visualized by autoradiography.
SSD RNA-seq, RNA and protein analysis, and bioinformatic analysis
SSD RNA-seq was performed as previously described (Vourekas et al. 2012) using total RNA (depleted of ribosomal RNA) isolated from wild-type and Mov10l1fl/− Neurog3-Cre whole adult testes or from purified pachytene spermatocytes and round spermatids obtained using the STA-PUT procedure (Bellvé et al. 1977) with modifications (Gerton and Millette 1986). qRT–PCR, histological analysis, immunofluorescence, Western blotting, piRNA immunoprecipitation, MOV10L1 and mZuc coimmunoprecipitation, MOV10L1 sequence and structure analysis, and bioinformatic analysis are described in the Supplemental Material. Deep sequencing data were deposited to Sequence Read Archive (SRA), project ID PRJNA230507.
Supplementary Material
Acknowledgments
Many thanks to members of the Mourelatos and Wang laboratories for useful discussions and technical help. We are grateful to S. Balasubramanian and J. McCafferty (University of Cambridge) for the plasmid encoding the BG4 antibody; N.A. Leu for blastocyst injection; W. Chen (Nanjing Medical Univeristy) for preparing the pRK5-Mov10l1 and pcDNA3.1-mZuc constructs; S.L. Martin (University of Colorado) and A. Bortvin (Carnegie Institute) for LINE1-ORF1p antibody; G.L. Gerton and M.L. Vadnais (University of Pennsylvania) for guidance on separation of spermatogenic cell types; F.B. Johnson (University of Pennsylvania) for critical reading of our manuscript; and J. Grubb, J. Schug, and J. Geskes (University of Pennsylvania) for Illumina sequencing. This work is supported by a Brody family fellowship to M.M., National Research Science Award fellowship F31HD081892 (to Q.F.), National Natural Science Foundation of China grant 31471228 and Nanjing Medical University Startup Funding 2013RC02 (to K.Z.), National Institute of Health grants GM072777 (to Z.M.) and HD069592 (to P.J.W.). A.V., K.Z., Q.F., Z.M., and P.J.W. designed the experimental approach. R.S.P. contributed to the design of the knock-in approach. A.V., K.Z., Q.F., and J.M. performed the experiments. M.M. performed bioinformatic analyses with contributions from P.A. A.V., M.M., K.Z., Q.F., Z.M., and P.J.W. analyzed data. A.V. wrote the manuscript with contributions from all authors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.254631.114.
References
- Aravin AA, Klenov MS, Vagin VV, Bantignies F, Cavalli G, Gvozdev VA. 2004. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol Cell Biol 24: 6742–6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. 2007. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747. [DOI] [PubMed] [Google Scholar]
- Arora A, Dutkiewicz M, Scaria V, Hariharan M, Maiti S, Kurreck J. 2008. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA 14: 1290–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin JD, Perreault JP. 2010. 5′-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Res 38: 7022–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellvé AR, Cavicchia JC, Millette CF, O'Brien DA, Bhatnagar YM, Dym M. 1977. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol 74: 68–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi G, Di Antonio M, Tannahill D, Balasubramanian S. 2014. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat Chem 6: 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blendy JA, Kaestner KH, Weinbauer GF, Nieschlag E, Schütz G. 1996. Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature 380: 162–165. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514. [DOI] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. 2009. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460: 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. 1998. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev 12: 3715–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O'Carroll D. 2011. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480: 259–263. [DOI] [PubMed] [Google Scholar]
- Deng W, Lin H. 2002. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2: 819–830. [DOI] [PubMed] [Google Scholar]
- Di Giacomo M, Comazzetto S, Saini H, De Fazio S, Carrieri C, Morgan M, Vasiliauskaite L, Benes V, Enright AJ, O'Carroll D. 2013. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol Cell 50: 601–608. [DOI] [PubMed] [Google Scholar]
- Fairman-Williams ME, Guenther UP, Jankowsky E. 2010. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol 20: 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Singh G, Lykke-Andersen J. 2010. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell 143: 938–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost RJ, Hamra FK, Richardson JA, Qi X, Bassel-Duby R, Olson EN. 2010. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci 107: 11847–11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton GL, Millette CF. 1986. Stage-specific synthesis and fucosylation of plasma membrane proteins by mouse pachytene spermatocytes and round spermatids in culture. Biol Reprod 35: 1025–1035. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. 2009. Small silencing RNAs: an expanding universe. Nat Rev Genet 10: 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202. [DOI] [PubMed] [Google Scholar]
- Gregersen LH, Schueler M, Munschauer M, Mastrobuoni G, Chen W, Kempa S, Dieterich C, Landthaler M. 2014. MOV10 is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol Cell 54: 573–585. [DOI] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. 2006. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev 20: 1709–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590. [DOI] [PubMed] [Google Scholar]
- Haase AD, Fenoglio S, Muerdter F, Guzzardo PM, Czech B, Pappin DJ, Chen C, Gordon A, Hannon GJ. 2010. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev 24: 2499–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM. 2001. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development 128: 2823–2832. [DOI] [PubMed] [Google Scholar]
- Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P. 1994. Fast folding and comparison of RNA secondary structures. Monatshefte Für Chemie Chem Mon 125: 167–188. [Google Scholar]
- Honda S, Kirino Y, Maragkakis M, Alexiou P, Ohtaki A, Murali R, Mourelatos Z, Kirino Y. 2013. Mitochondrial protein BmPAPI modulates the length of mature piRNAs. RNA 19: 1405–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. 2007. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 17: 1265–1272. [DOI] [PubMed] [Google Scholar]
- Hosokawa M, Shoji M, Kitamura K, Tanaka T, Noce T, Chuma S, Nakatsuji N. 2007. Tudor-related proteins TDRD1/MTR-1, TDRD6 and TDRD7/TRAP: domain composition, intracellular localization, and function in male germ cells in mice. Dev Biol 301: 38–52. [DOI] [PubMed] [Google Scholar]
- Huang H, Gao Q, Peng X, Choi SY, Sarma K, Ren H, Morris AJ, Frohman MA. 2011. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell 20: 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XA, Yin H, Sweeney S, Raha D, Snyder M, Lin H. 2013. A major epigenetic programming mechanism guided by piRNAs. Dev Cell 24: 502–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. 2012. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature 491: 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi N, Kawaoka S, Yasuhara S, Suzuki Y, Sugano S, Katsuma S, Tomari Y. 2013. Hsp90 facilitates accurate loading of precursor piRNAs into PIWI proteins. RNA 19: 896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E, Putnam A. 2010. Duplex unwinding with DEAD-box proteins. Methods Mol Biol 587: 245–264. [DOI] [PubMed] [Google Scholar]
- Kamminga LM, Luteijn MJ, den Broeder MJ, Redl S, Kaaij LJT, Roovers EF, Ladurner P, Berezikov E, Ketting RF. 2010. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J 29: 3688–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka S, Izumi N, Katsuma S, Tomari Y. 2011. 3′ end formation of PIWI-interacting RNAs in vitro. Mol Cell 43: 1015–1022. [DOI] [PubMed] [Google Scholar]
- Kervestin S, Jacobson A. 2012. NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol 13: 700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z. 2007a. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA 13: 1397–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z. 2007b. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol 14: 347–348. [DOI] [PubMed] [Google Scholar]
- Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z. 2009. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol 11: 652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. 2007. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell 12: 45–55. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. 2004. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131: 839–849. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. 2008. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22: 908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. 2006. Characterization of the piRNA complex from rat testes. Science 313: 363–367. [DOI] [PubMed] [Google Scholar]
- Lewandoski M, Martin GR. 1997. Cre-mediated chromosome loss in mice. Nat Genet 17: 223–225. [DOI] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. 2009. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZ, Roy CK, Dong X, Bolcun-Filas E, Wang J, Han BW, Xu J, Moore MJ, Schimenti JC, Weng Z, et al. 2013. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell 50: 67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Buchold GM, Greenbaum MP, Roy A, Burns KH, Zhu H, Han DY, Harris RA, Coarfa C, Gunaratne PH, et al. 2009. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet 5: e1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery TA, Rim YS, Zhang C, Dowen RH, Phillips CM, Fischer SEJ, Ruvkun G. 2012. PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet 8: e1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, et al. 2012. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature 491: 284–287. [DOI] [PubMed] [Google Scholar]
- Olivieri D, Senti KA, Subramanian S, Sachidanandam R, Brennecke J. 2012. The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Mol Cell 47: 954–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schüpbach T. 2007. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell 12: 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Chuma S. 2012. piRNAs and their involvement in male germline development in mice. Dev Growth Differ 54: 78–92. [DOI] [PubMed] [Google Scholar]
- Preall JB, Czech B, Guzzardo PM, Muerdter F, Hannon GJ. 2012. shutdown is a component of the Drosophila piRNA biogenesis machinery. RNA 18: 1446–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS. 2011. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480: 264–267. [DOI] [PubMed] [Google Scholar]
- Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC. 2009. A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Curr Biol 19: 2066–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhkov NV, Hammell M, Hannon GJ. 2013. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev 27: 400–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. 2006. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20: 2214–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. 2007. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev 21: 1603–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. 2009. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299. [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC. 2010. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev 24: 2493–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxe JP, Chen M, Zhao H, Lin H. 2013. Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J 32: 1869–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, et al. 2009. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell 17: 775–787. [DOI] [PubMed] [Google Scholar]
- Sienski G, Dönertas D, Brennecke J. 2012. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151: 964–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258. [DOI] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324. [DOI] [PubMed] [Google Scholar]
- Voigt F, Reuter M, Kasaruho A, Schulz EC, Pillai RS, Barabas O. 2012. Crystal structure of the primary piRNA biogenesis factor Zucchini reveals similarity to the bacterial PLD endonuclease Nuc. RNA 18: 2128–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourekas A, Mourelatos Z. 2014. HITS-CLIP (CLIP-seq) for mouse Piwi proteins. Methods Mol Biol 1093: 73–95. [DOI] [PubMed] [Google Scholar]
- Vourekas A, Zheng Q, Alexiou P, Maragkakis M, Kirino Y, Gregory BD, Mourelatos Z. 2012. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol 19: 773–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, Hoki Y, Fujiyama A, Shibata T, Sado T, et al. 2011. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell 20: 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW. 1996. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol 16: 5477–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiol J, Cora E, Koglgruber R, Chuma S, Subramanian S, Hosokawa M, Reuter M, Yang Z, Berninger P, Palencia A, et al. 2012. A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol Cell 47: 970–979. [DOI] [PubMed] [Google Scholar]
- Zhang AYQ, Bugaut A, Balasubramanian S. 2011. A sequence-independent analysis of the loop length dependence of intramolecular RNA G-quadruplex stability and topology. Biochemistry 50: 7251–7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Wang PJ. 2012. Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity. PLoS Genet 8: e1003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Xiol J, Reuter M, Eckardt S, Leu NA, McLaughlin KJ, Stark A, Sachidanandam R, Pillai RS, Wang PJ. 2010. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc Natl Acad Sci 107: 11841–11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.