Abstract
Human mutations in PQBP1, a molecule involved in transcription and splicing, result in a reduced but architecturally normal brain. Examination of a conditional Pqbp1-knockout (cKO) mouse with microcephaly failed to reveal either abnormal centrosomes or mitotic spindles, increased neurogenesis from the neural stem progenitor cell (NSPC) pool or increased cell death in vivo. Instead, we observed an increase in the length of the cell cycle, particularly for the M phase in NSPCs. Corresponding to the developmental expression of Pqbp1, the stem cell pool in vivo was decreased at E10 and remained at a low level during neurogenesis (E15) in Pqbp1-cKO mice. The expression profiles of NSPCs derived from the cKO mouse revealed significant changes in gene groups that control the M phase, including anaphase-promoting complex genes, via aberrant transcription and RNA splicing. Exogenous Apc4, a hub protein in the network of affected genes, recovered the cell cycle, proliferation, and cell phenotypes of NSPCs caused by Pqbp1-cKO. These data reveal a mechanism of brain size control based on the simple reduction of the NSPC pool by cell cycle time elongation. Finally, we demonstrated that in utero gene therapy for Pqbp1-cKO mice by intraperitoneal injection of the PQBP1-AAV vector at E10 successfully rescued microcephaly with preserved cortical structures and improved behavioral abnormalities in Pqbp1-cKO mice, opening a new strategy for treating this intractable developmental disorder.
Introduction
Microcephaly is a group of multiple disorders including PQBP1 gene mutations1, 2, 3, 4, 5 and classified by the presence or absence of architectural change of cortex. The mechanism of neuronal production is one of the most critical factors affecting brain size. According to the classical radial unit hypothesis, brain volume (cortical thickness × surface area) is determined by neuronal production from neural stem cells (NSCs) per radial glia unit.6 Neurons in the neocortex are generated over a 6-day neuronogenesis interval (E11-E17) that comprises 11 cell cycles of NSC self-renewal and neurogenesis.7 In this model, excessive neurogenesis during cell division depletes the NSC pool, decreases the vertical expansion (radial growth) of the pool as well as the final production of neurons, and leads to microcephaly.8 However, lateral expansion (tangential growth), which should be also related to brain size, has not been explained well.
The discovery of new subsets in the neural stem progenitor cell (NSPC) pool has made the story more complex. First, radial glia were shown to be identical to NSCs and apical progenitor cells (AP) in the ventricular zone (VZ).9, 10, 11, 12 Next, the Tbr2-positive basal progenitor cell (BP) in the subventricular zone (SVZ) was identified to be the primary source of neurons and assumed to regulate tangential (horizontal) and radial (vertical) expansion of the cortex.12, 13, 14, 15 Moreover, basal radial glia (bRG), Pax6-positive cells on the basal side of the SVZ, were discovered to be another source of neuron production.8,16, 17, 18 Therefore, the hypothesis was revised to state that the amounts of BPs/bRGs relative to NSCs/APs regulate cortical thickness, although the details of the contribution of the two basal progenitors to corticogenesis remained unknown. Two recent reports have provided important lines of evidence to this question. Analysis of Trnp1, a DNA-associated protein whose exact molecular function is not yet known, revealed that overexpression of Trnp1 increased the self-renewal of radial glia and expanded the NSPC pool laterally, while knockdown (KD) of Trnp1 increased the number of BPs/bRGs and increased the radial expansion and gyrification of the cortex.19 Another group simultaneously reported that the overexpression of Cdk4 and CyclinD1 (4D) in transgenic mice increased BP/bRG numbers and gyrification.20 Importantly, no abnormality was detected in the cortical layer structure of the folded cortex in 4D transgenic mice.20 Therefore, the combination of lateral expansion (tangential growth) of the NSPC pool, which is affected by neurogenesis ratio, and vertical expansion (radial growth) to produce BP/bRG and neurons is now believed to determine the surface area, thickness and gyration of the cortex.19, 20, 21
In this study, we report that cell cycle elongation of NSPCs by the NSPC-specific depletion of Pqbp1, which simply delays the entire process, finally causes brain downsizing in vivo that mimics the primary microcephaly (PM)-like phenotype of human patients with PQBP1-linked intellectual disability (ID). Although neuron and BP production ratios are unchanged, cell cycle elongation in NSPCs decreases the AP and BP pools equally, and results in the proportional reduction of both tangential and radial expansion. Cell death is not increased in vivo during these processes except in the extreme case of shRNA-mediated KD. The molecular mechanism, elongation of cell cycle time, is due to aberrant transcription and RNA splicing of cell cycle-regulating genes. Moreover, we have developed a new therapy, the peritoneal injection of PQBP1-adenoassociated virus (AAV) to pregnant mice, that rescues the microcephaly of the offspring.
Materials and methods
For more detailed information, please refer to ‘Full methods' in Supplementary Information.
Generation of conditional knockout of murine Pqbp1
To generate the targeting vector, three Pqbp1 genomic fragments were PCR amplified from a murine bacterial artificial chromosome (BAC) library (ID: RP23-404N15). A 3.6-kb 5′ fragment containing exons 1 and 2 was inserted upstream of a neomycin resistance cassette flanked by Flp recognition target sites. A 3.9-kb fragment containing exons 3 to 7 was inserted between two LoxP sites and a 4.1-kb non-coding fragment was added 3′, with the diphtheria toxin A gene to prevent random insertion. After electroporation into ES cells (C57BL/6), and G418 selection (Sigma, St Louis, MO, USA—200 mg/ml), clones were analysed by genomic DNA PCR using the following primers: fwd, 5′-AATCTTGGAGTTAGTAATGGTGCTT-3′, and rev, 5′-AATCTCATGTAATTGACGAGACAGAG-3′. Selected ES clones were corroborated by Southern blot analyses of genomic DNA digested with EcoRI or EcoRV. Probes for Southern blot analyses were prepared by PCR from the BAC clone using the following primers (locations given in Supplementary Fig 2): 5′ probe (408 bp), fwd: AAAGTGAACCTGCATTAGAGGAAC-3′, rev: TCAGTGAGATACTCTGACTTCCACA; 3′ probe (462 bp), fwd: GTCAATAAGCATTCAAGGACTCACT, rev: TCAGAATACTCTTGGAACTCCCTTA. Chimeric mice were generated by injecting the recombinant ES cells into C57BL/6 blastocysts, subsequently crossed with C57BL/6 mice to generate the targeted allele. The neomycin resistance cassette was removed by crossing with CAG-FLPe recombinase transgenic mice.22 The resultant Pqbp1-floxed heterozygous female mice were further crossed with Nestin-Cre transgenic male mice (B6.Cg-Tg (Nes-cre) 1Kln/J; The Jackson Laboratory, Bar Harbor, ME, USA) and Synapsin1-Cre transgenic male mice (B6.Cg-Tg (Syn1-Cre)671Jxm/J; The Jackson Laboratory) to generate the Pqbp1 conditional knockout.
Cumulative labeling
The method for analyzing cell cycle parameters in the neuroepithelium23 was employed with minor modifications. BrdU (Sigma; 100 mg/kg of body weight) was injected intraperitoneally into pregnant mice at E14. Cumulative labeling was performed by repeated injections (at 3-h intervals) into pregnant mice, which were killed 1, 1.5, 2, 3.5, 6.5, 15.5 and 24.5 h after the first BrdU injection. Embryonic brains were fixed with 4% paraformaldehyde and paraffin embedded. Sections were made at 3 mm intervals, deparaffinized, rehydrated and then microwaved in 10 mM of citrate buffer, pH 6.0, for 15 min. Antibody incubations were done with the mouse anti-BrdU antibody (1:200, BD Biosciences, Franklin Lakes, NJ, USA) and rabbit anti phospho-histone H3 (pH3) antibody, a marker for M-phase cells (1:500, Millipore, Billerica, MA, USA) at 4 °C overnight. Secondary antibody incubations were done with Alexafluor-488 or Cy3 conjugates (1:500, Invitrogen, Carlsbad, CA, USA). The ratio of BrdU/pH3-double-positive cells to pH3-positive cells in the ventricular zone was calculated at 1, 1.5 and 2 h after a single injection of BrdU to determine the length of the G2/M phase. A straight-line graph of the labeling index values (LIs) at 1, 1.5, 2, 3.5 and 6.5 h allowed us to extrapolate to a y-axis intercept (the LI at 0 h) and calculate the slope. Since the growth fraction (the ratio of proliferating cells) is nearly 1.0 in the ventricular zone of wild mice, the LI at 0 h and slope represent the ratio of S-phase to total cell cycle (Ts/Tc) and the reciprocal of total cell cycle (1/Tc), respectively. Ts and Tc denote the length of the S-phase and total cell cycle, respectively. From these values (Ts/Tc and 1/Tc), Ts and Tc were calculated.
Exon array-based analysis of alternative splicing
We analyzed wild type (WT) and PQBP1 conditional knock-out (cKO) mouse brain samples using Affymetrix GeneChip Mouse Exon 1.0 ST array (exon array) (http://www.affymetrix.com) to find significantly changed genes in terms of potential alternative splicing. The exon array contains over five million probes representing about 1.4 million probesets that are designed based on the genomic regions of known genes and exons to measure both gene-level and exon-level expression in samples. The sequences of the probes and the probesets were downloaded at the Affymetrix website (http://www.affymetrix.com/support/technical/whitepapers/exon_probeset_trans_clust_whitepaper.pdf). To summarize the probes into the exon-level probesets, the PLIER algorithm (http://www.affymetrix.com/analysis/index.affx) was applied to the probe signals of our samples.
Two types of analysis based on statistical hypothesis testing, exon–exon and variance analyses, were performed on the exon-level probesets to detect changes in alternative splicing. Before these tests, the signals of each exon-level probeset in one gene were normalized by their total signals to eliminate the effects of different numbers of exons among compared genes. The exon–exon analysis was performed to compare each exon's signals between WT and cKO mice to find the significant change in exon level that could be induced by the change of alternatively splicing or transcription. The difference was examined by Student's t-test based on the null hypothesis that the means of the two samples are the same. The variance analysis was performed on exon-level probesets in each gene of WT and cKO mice to detect changes in relative expression levels among exons by an F-test for the null hypothesis that these two samples have the same pattern and the variance.
The smallest P-value was selected as the significance level of the gene to find alternatively spliced exons and it was significant with P-value <0.05.
Genes showing significantly different expression between WT and cKO mice in exon–exon and/or variance analysis were listed to apply to PANTHER analysis (http://www.pantherdb.org/). In Panther analysis, statistical overrepresentation test was performed to determine whether the genes were enriched or deprived for specific biological processes.
Three groups of significantly changed genes, (A) Nestin-cKO-NSC specific genes, (B) co-occurrence genes between the NSC and cortex of Nestin-cKO mice and (C) co-occurrence genes of the three types of cKO samples, were tested individually by PANTHER analysis to be compared with all the genes detected in the assay and then pie-charts of protein function classification were formed.
In vivo rescue experiment of Apc4
In utero electroporation and determination of Pial-to-Apical surface area experiments were performed as described previously.16 Full-length murine Apc4 cDNA (Genbank accession number NM_024213) was obtained by RT-PCR using fwd (5′-GGGCTCGAGACCATGGGAATGCTGCGCTTTCCGACCTGTTT-3′) and rev (5′-GGCGGATCCCTATTATTTGTCGTCATCATCC-3′) primers. The product was inserted into XhoI/BamHI cut pIRES2-EGFP (Clontech) to generate pApc4-IRES2-EGFP. For the Apc4 rescue experiments, pAPC4-IRES2-EGFP or pIRES2-EGFP was electroporated into the ventricular zone of E13 embryos. Brain tissues of E18 embryos were fixed, embedded in 3% agarose, and sectioned on the rostral-to-caudal axis as 50-μm-thick sections using a vibratome. Brain sections were incubated with mouse anti-green fluorescent protein (GFP) monoclonal antibody (1:500, Millipore) at 4 °C overnight, followed by treatment with Alexafluor-488 labeled secondary antibodies. Sections were analyzed with a confocal fluorescence microscope (Olympus FV10i, Tokyo, Japan) and the ratio of pial-to-apical surface determined as described previously.8
Gene therapy with PQBP1-AAV vector
The AAV vector plasmids contained an expression cassette, consisting of a human cytomegalovirus immediate-early promoter (CMV promoter), followed by cDNA encoding human PQBP1 or human PQBP1-EGFP, and a simian virus 40 polyadenylation signal sequence (SV40 poly (A)) between the inverted terminal repeats of AAV 3 genome. The recombinant AAV vectors were produced by transient transfection of HEK293 cells using the vector plasmid, an AAV2 rep and AAV1 vp expression plasmid, and an adenoviral helper plasmid, pHelper (Agilent Technologies, Santa Clara, CA, USA). The recombinant viruses were purified by isolation from two sequential CsCl gradients, and the viral titers were determined by qRT-PCR. For in vivo administration of AAV vectors, C57BL/6J pregnant mice (E10) were injected with AAV-PQBP1 vector (2.0 × 1011 genome copies) by intraperitoneal administration.
Results
Human PQBP1 mutations cause PM
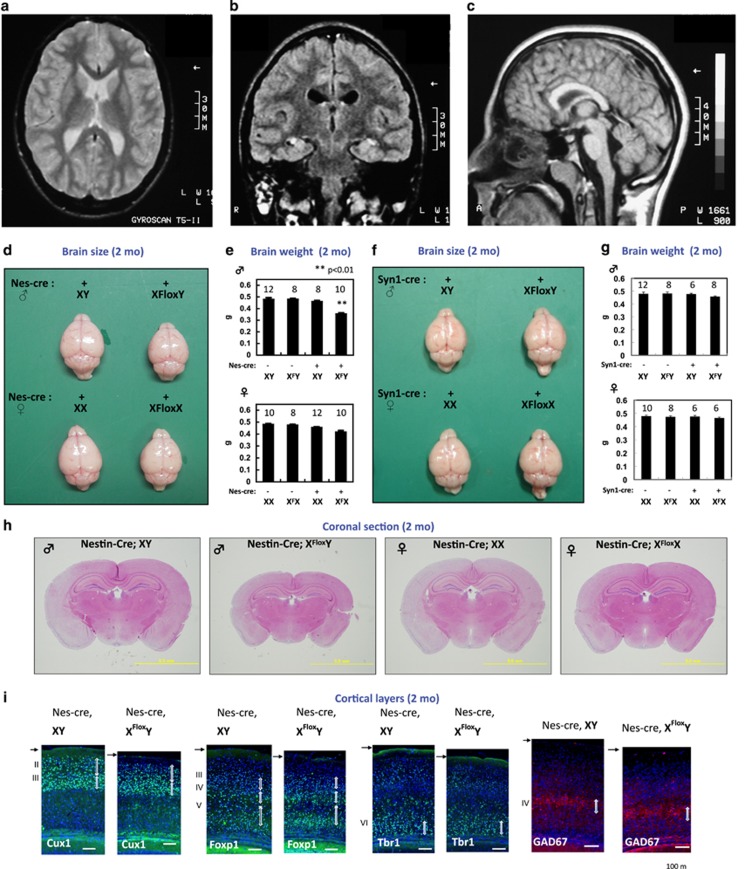
First, we defined the characteristics of PQBP1-linked microcephaly in human patients. Microcephaly is generally defined as an innate non-progressive small brain sized less than 4 standard deviations (s.d.); a milder form of microcephaly (less than 3 or 2 s.d.) has been also reported in some patients. In PQBP1-linked microcephaly, the brain size ranges from less than 6 s.d. to less than 2 s.d. Although one PQBP1-linked microcephaly patient with periventricular heterotopias (PH) was reported,24 PH is likely to be a rare clinical feature as it was not observed in an affected sibling with the identical mutation,24 or in a further 13 microcephalic patients with PQBP1 mutations.25 By magnetic resonance imaging, we confirmed the presence of well-preserved cortical architecture in two previously described but unrelated patients with mutated PQBP12,26 (Figures 1a–c). A slight dilation of the ventricles was observed in these patients having mutated PQBP1, as seen in the autosomal recessive PM (MCPH) cases. All these considerations revealed that PQBP1-linked microcephaly is quite similar to PM in morphology.
Figure 1.
Microcephaly with normal cortical structures in PQBP1 patients and nestin-Cre Pqbp1-cKO (conditional Pqbp1-knockout) mice. (a–c) Magnetic resonance imaging of a PQBP1-mutated patient showed normal cortical structures with no periventricular heterotopia: (a) horizontal, (b) coronal and (c) sagittal sections. (d) Macroscopic images of the brain at the age of 2 months. Male nestin-Cre Pqbp1-cKO mouse brains (Nes-Cre; XFloxY) were consistently the smallest among the littermates. (e) Pqbp1-cKO mice showed reduction of brain weight at 2 months. The bar graph shows the mean+s.e.m. for each genotype with the number of mice used indicated above. The mean and s.e.m. values are provided in the text. Asterisks indicate significance (P<0.01) in one-way analysis of variance with post hoc Bonferroni test. (f) Macroscopic images of the brain at the age of 2 months. Male synapsin 1-Cre Pqbp1-cKO mouse brains (synapsin1-Cre; XFloxY) were not different from the littermates in size. (g) The brain weight of the male synapsin-1-Cre Pqbp1-cKO mouse was not different from that of the background control. (h) Coronal sections of adult brains of nestin-Cre Pqbp1-cKO mouse and littermates (2 months, at −1.82 mm from the bregma in background mice). (i) Staining for layer-specific markers, Cux1, Foxp1 and Tbr1, together with GAD67, shows preservation of cortical layers in Pqbp1-cKO mice at 2 months.
Pqbp1-cKO in NSCs mimics the microcephaly of human patients
Next, we aimed to generate a model of PQBP1-linked microcephaly. Pqbp1 is highly expressed in NSPCs (Sox2-positive AP cells and RC2-positive radial glia in the VZ and SVZ) and Sox2 transcriptionally regulates Pqbp1 in NSPCs.27 Pqbp1 is also expressed in differentiated neurons at a lower level.5,27,28 We therefore generated two types of conditional KO (cKO) mice using nestin-Cre and synapsin-1-Cre (Figures 1d–g, and Supplementary Figure 1A). As expected, the expression level of the Pqbp1 protein was remarkably decreased in the total brain tissue of nestin-Cre cKO mice, although a faint band was detected due to the non-neural tissues in the brain (Supplementary Figure 1B). Immunohistochemistry of the cortical tissue of synapsin-1-Cre cKO mice also confirmed the depletion of Pqbp1 in neurons (Supplementary Figure 1B).
We expected that comparing the two models could distinguish the effect of Pqbp1-KO from NSPCs from that of Pqbp1-KO after neuronal differentiation on brain development.29 Male nestin-Cre-derived Pqbp1-cKO mice had obvious microcephaly. In female hemizygous cKO mice, the brains were slightly smaller than the control (Figures 1d and e). Surprisingly, the brain size was not changed in synapsin-1-Cre derived Pqbp1-cKO mice (Figures 1f and g). This discrepancy indicated that Pqbp1 function in NSPCs but not in neurons is responsible for microcephaly. The PM-like morphological features of the brain indicated that the nestin-Cre cKO mouse is a good model for PQBP1-linked microcephaly.
At the macroscopic level, brain sections showed normal cortical, subcortical and brain stem structures in both types of Pqbp1-cKO mice (Figure 1h). At the histological level, cortical layer structures were well preserved in both adult (Figure 1i, and Supplementary Figures 1C and D) and embryonic brains of nestin-Cre Pqbp1-cKO mice (Supplementary Figures 2A–D). Quantitative analyses revealed that the layer thickness was decreased in adult (Supplementary Figures 1C and D) and embryonic brains (Supplementary Figures 2A, C and D). Notably, the thickness of the telencephalon was already smaller at E10 (Supplementary Figure 2B) when proliferation of the NSC pool and neurogenesis began.
Asymmetric cell division is not changed in Pqbp1-cKO mice
At the cell level, we checked pathologies in the NSPCs of nestin-Cre cKO mice. APs facing the ventricular surface of the Pqbp1-cKO mouse did not show abnormal centrosomes or mitotic spindles in vivo and in vitro, even though we examined more than 300 APs (Supplementary Figure 3A). We could not detect detachment of the centrosome from the mitotic spindle pole as observed in Cdk5Rap2-mutant cell lines30 or abnormal spindle poles as seen in ASPM-mutated Drosophila cells.31,32 We also examined the centrosome and mitotic spindle using primary fibroblasts from human patients, but did not detect abnormal findings in these structures (Supplementary Figure 3B).
We analyzed the mitotic plane angle of APs at E10.5, E14.5 and E15.5 (Supplementary Figure 3C), because spindle orientation plays a key role in NSPC differentiation in MCPH. Throughout these embryonic stages, the mitotic plane angles of cKO mice were unchanged (Supplementary Figure 3C). Furthermore, we evaluated the ratio of asymmetric division of APs at E14.5 by immunohistochemistry with an antibody against N-cadherin (Supplementary Figure 3D), which serves as a marker for cell fate through the distribution of the apical membrane to sister cells. The data showed no change in the percentage of asymmetric cell division (Supplementary Figure 3D).
Cell cycle time elongates in NSCs of Pqbp1-cKO
At the cellular function level, the cell cycle plays a crucial role in the tangential and radial expansion of the cortex. The Calegari group reported previously that in utero electroporation of 4D into NSPCs, which shortened G1, resulted in enlarged cortical thickness generated from transfected NSPCs.33 The Huttner group revealed that G1 is longer in BPs than in Aps, and that S-phase shortening occurs both in APs and in BPs during commitment to differentiation.34 Studies of MCPH revealed the role of the M phase in corticogenesis. The causative disease gene products of MCPH1–7 (microcephalin, WDR62, Cdk5Rap2, CEP152, ASPM, CENPJ and STIL)35, 36, 37, 38, 39, 40, 41 are localized to the centrosome-spindle pole of NSPCs38,42,43 and their defective function causes abnormal centrosome-spindle pole structures.44 A recent report showed that shRNA-mediated KD of Cdk5Rap2 actually increased neurogenesis and the BP pool but reduced the AP pool.45 Intriguingly, despite large amounts of accumulated data, this series of cell cycle abnormalities have not yet been sufficiently integrated into the newest hypothesis of corticogenesis.
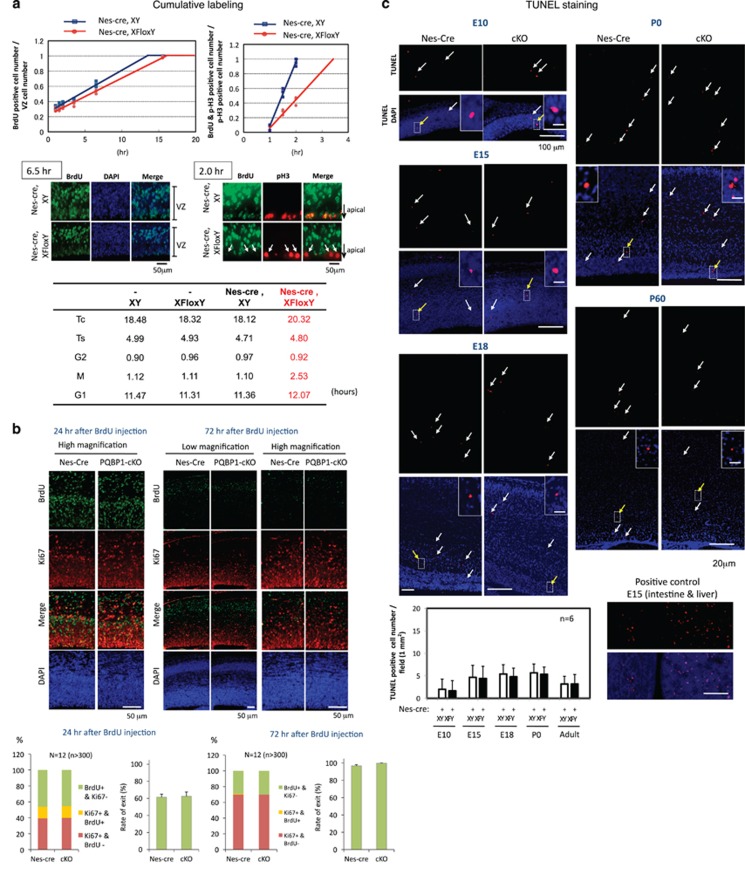
Hence, we first evaluated the cell cycle time of NSPCs in vivo using the cumulative labeling method.23 The data showed that the total cell cycle time (Tc) was increased (+2.2 h, +12%) (Figure 2a). The S phase (Ts) was unaltered compared to the strict control nestin-Cre XY mice (Figure 2a). The G1 phase was slightly longer (Figure 2a). However, G1 elongation in the Pqbp1-cKO was minimal (5.2%) compared to other reports of cell cycle length regulating brain size.33,34 G2 phase time was not changed (Figure 2a). In contrast to the G1, S, and G2 phases, the M phase was remarkably increased (Figure 2a) and the increase in Tc was principally due to a longer M phase (+1.4 h, +67%). This cell cycle change in Pqbp1-cKO mice is quite unique among various disease models.
Figure 2.
Nestin-Cre Pqbp1-cKO (conditional Pqbp1-knockout) delays the cell cycle but does not affect neurogenesis of neural stem progenitor cells (NSPCs). (a) The M phase was specifically elongated in NSPCs of Pqbp1-cKO mice (Nes-Cre; XFloxY) in vivo. Cumulative labeling of NSPCs at E14 in vivo showed an increase in the total cell cycle length (Tc) of +2.2 h, +12% (upper left panel). G2/M phase time was evaluated using phosphorylated histone H3 (upper right panel). The middle panels show immunostaining data corresponding to the upper panels. pH3+/BrdU+ cells were reduced in number in cKO embryos (arrows), indicating elongation of the G2/M phase (middle right panel). The summary of cumulative labeling and G2/M analyses is shown in the lower table. A significant extension of the G2/M phase (+67%) and a slight extension of G1 (+6%) were observed. The crossing point between the plot line and the x-axis indicates the length of G2 phase (upper right panel). Thus, the M phase was remarkably elongated while the length of the G2 phase was unchanged in Pqbp1-cKO mice. (b) Neurogenesis from the stem cell pool was analyzed by co-staining for BrdU and Ki67. At 12 or 72 h after intraperitoneal injection of BrdU, E15 embryonic brains were analyzed to calculate the numbers of cells after neurogenesis (BrdU+/Ki67−), cells remaining in the stem cell pool (BrdU+/Ki67+) and non-labeled stem/progenitor cells (BrdU−/Ki67+). The bar graphs show the relative percentages of the three groups (left graph) and the neurogenesis percentage of BrdU-labeled cells (right graph). No difference was detected at 24 and 72 h after BrdU injection by the Student's t-test or Welch's t-test. (c) Levels of cell death in the cerebral cortex were evaluated by terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) staining at E10, E15, E18, P0 and P60 of Pqbp1-cKO mice (Nes-Cre; XFloxY) and nestin-Cre mice (Nes-Cre; XY). Quantitative analysis of apoptotic cells did not reveal any differences (Student's t-test).
Neurogenesis and apoptosis of NSCs are unchanged in Pqbp1-cKO
M phase arrest or elongation has been linked to increased neurogenesis.35, 36, 37, 38, 39, 40, 41, 42, 43 G1 shortening is related to increased brain size through the delay of neurogenesis.33 Therefore, we asked whether neurogenesis from the stem cell pool is altered in Pqbp1-cKO mouse embryos (Figure 2b). We labeled proliferating cells at the S phase with BrdU, and examined the brain after an interval to check whether they exited from the proliferating stem cell pool, by co-staining with the proliferation marker Ki67. At 24 and 72 h following labeling, the ratios of the remaining stem cells to differentiated cells were not largely different between nestin-Cre XY littermates and Pqbp1-cKO mice (Figure 2b, graphs). In addition, in utero electroporation of a GFP-expressing vector into the embryonic brain of the Pqbp1-cKO mouse was performed at E14 and the embryos were dissected at E16. The neurogenesis rate, calculated from the ratio of GFP+/ki67− versus total GFP+ cells, was also unaltered (data not shown).
Further, to test the effect of Pqbp1 on neurogenesis, we performed in vivo KD using short hairpin RNA against Pqbp1 (Pqbp1-shRNA). We generated plasmids expressing Pqbp1-shRNAs (Supplementary Figure 4A) that successfully suppressed Pqbp1 in P19 cells, as evidenced by western blot (Supplementary Figure 4B), and dramatically reduced Pqbp1 in NSPCs in vitro or in vivo (Supplementary Figures 4C–E). In all these cases, Pqbp1 was decreased to nearly 10%. Pqbp1-KD by in utero electroporation of Pqbp1-shRNA-ZsGreen into normal mice delayed the exit of transfected cells from the stem cell layer VZ/SVZ (Supplementary Figures 5A and B) and delayed their entrance into the differentiating cell layer IZ (Supplementary Figure 5C). These results supported the conclusion from Pqbp1-cKO mice that neurogenesis is not increased in Pqbp1 deficiency.
Increased apoptosis of NSPCs is another plausible mechanism for PM. Indeed, apoptosis was found to be increased in human microcephaly with preserved brain structure caused by mutations in polynucleotide kinase 3′-phosphatase, which is essential for many DNA repair pathways including base-excision repair and non-homologous end joining.46 However, repeated analyses by terminal deoxynucleotidyl transferase dUTP nick end labeling staining failed to demonstrate elevated apoptosis in embryonic (E10–E18), neonatal (P0) and adult brains of Pqbp1-cKO mice (Figure 2c).
Single-cell volume of neuron is unchanged in Pqbp1-cKO
One may suspect that Pqbp1 deficiency causes brain size reduction by decreasing the single-cell volume of neurons. However, this is not plausible because synapsin-1-Cre cKO mice did not show microcephaly, as mentioned previously (Figures 1f and g). In addition, we examined the space occupied by a single neuron (cell volume) and the total dendrite length of a neuron (retrosplenial dysgranular cortex, layer V) in adult nestin-Cre and synapsin-1-Cre Pqbp1 cKO mice (P90) by two-photon microscopy (Supplementary Figures 6A and B). The results revealed no significant changes in total neuronal cell volume in nestin-Cre Pqbp1 cKO mice that had microcephaly, whereas the volume was decreased in synapsin-1-Cre Pqbp1 cKO mice that did not show microcephaly (Supplementary Figures 6A and B), indicating that the single-cell volume does not account for the microcephaly. Unexpectedly, total dendrite length was not changed either in nestin-Cre or in synapsin-1-Cre Pqbp1 cKO mice in comparison to control mice (Supplementary Figures 6A and B). However, the dendrite diameter tended to be thinner than in the control (data not shown).
Our previous collaborative study revealed the localization of Pqbp1 in the cilia of differentiated hippocampal neurons and its essential role in the maintenance of cilia.28 Given that cilia receive and transfer environmental signals like Shh to the cell body47 and that NSPCs also possess cilia, the function of Pqbp1 in cilia might be related to the proliferation of NSPCs.48 However, we could not detect any differences in the morphology of cilia on the ventricular surface of cortex where the cilia of NSPCs are located (Supplementary Figure 6C), and also could not localize the Pqbp1 protein to the centrosome of NSPCs during mitosis (Supplementary Figures 7A and B). These results suggested that the role of PQBP1 in promoting cilia is specific to differentiated neurons but not directly linked to the proliferation of NSPCs. The decreased cell volume of adult neurons in synapsin-1-Cre Pqbp1 cKO mice (Supplementary Figures 6A and B) was consistent with the change in the Pqbp1/cilia-derived signal in differentiated neurons.28 The cell volume change was compensated during development when Pqbp1 was depleted from NSPCs.
A unique mechanism of microcephaly in Pqbp1-cKO
Collectively, our intensive analyses failed to show the typical pathomechanisms of PM in Pqbp1-cKO mice, instead of the morphological similarities. The results from this study do not support the existence of the centrosome-spindle pathology (Supplementary Figures 3A and B), increased asymmetric cell division (Supplementary Figures 3C and D), increased neurogenesis from the stem cell pool (Figure 2b, Supplementary Figures 5A–C), increased cell death (Figure 2c), or decreased neuronal cell volume (Supplementary Figure 6A).
The only remaining possibility is the decrease in the stem cell pool due to elongated cell cycle time. Immunohistochemistry of Pqbp1-cKO mouse embryos revealed that the size of the stem cell pool is already small at E10, when neurogenesis has not yet started (Supplementary Figure 2B). After E11, when neuroepithelial cells start to switch from symmetric proliferative to asymmetric neurogenic division, delayed cell cycle time (Figure 2a) keeps both AP and BP stem cell pools equally reduced at E15 (Supplementary Figures 2C and D), and the reduction of the stem cell pool leads to a decrease in the number of differentiated neurons (Supplementary Figure 2A). These results support a simple hypothesis of microcephaly based on delayed cell proliferation and reduced expansion of the stem cell pool, which is independent of unequal changes in the production of neurons or BPs.
Systems biology analyses reveal the molecular mechanism of microcephaly
Next, we investigated the molecular mechanisms underlying the cell cycle dysregulation of NSPCs. PQBP1 is a factor coupling transcription and splicing through two protein-binding motifs, the WW and C-terminal domains. Currently, mutations of the human PQBP1 gene are classified into two types. The first category causes frame shifts that introduce premature stop codons before the C-terminal domain of PQBP-1.2, 3, 4 The second category targets a conserved amino-acid residue in the WW domain.49,50 These defects relate to a failure of PQBP1 to interact with its partner proteins RNA polymerase II51 and WBP11/SIPP152,53 via the WW domain, and U5-15kD54,55 via the C-terminal domain.
Therefore, mutant PQBP1 is believed to disturb multiple cellular functions through impairment of RNA splicing and/or transcription. Wang et al.56 screened genes affected by aberrant splicing in mouse embryonic neurons and identified neural cell adhesion molecule (NCAM)-140, a splicing isoform of NCAM1 involved in neurite extension, as a target of PQBP1-mediated aberrant splicing. Moreover, the Bonni group recently reported that PQBP1 interacts with the GTPase dynamin2 and affects ciliary morphogenesis in postmitotic neurons.28 However, the splicing target of Pqbp1 in proliferating NSPCs might be different from targets in non-dividing neurons, and even in neurons splicing might be influenced by primary culture and could be different in vivo, where neurons are supported by multiple circumstantial factors.
Therefore, we rescreened the target genes of PQBP1 in NSPCs by a combination of an ordinary mRNA gene chip to evaluate gene expression levels, and an exon array to estimate alternative splicing patterns. Regarding the gene expression levels, we first summarized data from the ordinary gene chip with primary cultured NSPCs prepared from nestin-Cre Pqbp1-cKO mice (E15), and extracted functional gene groups sensitive to Pqbp1 deficiency (reactomes) using Gene Set Enrichment Analysis57 (Supplementary Figure 8A and Supplementary Table 1). This revealed that, in any combination, nearly a half of the reactive gene groups (reactomes) were related to the M phase (red in Supplementary Figure 8A and Supplementary Table 1) or another cell cycle phase (yellow in Supplementary Figure 8A and Supplementary Table 1), and supporting cell cycle genes were the main targets of Pqbp1 in NSPCs at the transcriptional level that are affected by Pqbp1 deficiency. In particular, M phase-related reactomes were significantly downregulated (Supplementary Figure 8B and Supplementary Table 1), consistent with the elongation of the M phase seen in vivo (Figure 2a). Reactomes in a dPQBP1 mutant fly58 showed related changes, specifically in the M phase (data not shown).
Further, to identify critical genes whose expression was altered by Pqbp1 deficiency in NSPCs, we merged genes selected by Gene Set Enrichment Analysis with nodes in the protein–protein interaction network database around PQBP1 (Supplementary Figure 8C). All the genes showed a densely packed interaction pattern, as expected. Several genes, including Apc2 (ANAPC2) and RBMXL1, were linked to more than 25 genes downregulated in NSPCs from Pqbp1-cKO mice (Supplementary Table 2), suggesting their role as hubs for executing the role of Pqbp1 in NSPCs. It is of note that centrosomal proteins were excluded from the network upon increasing the threshold (Supplementary Figure 8D), indicating that their role is not so significant in the case of Pqbp1-linked microcephaly.
Interestingly, the protein–protein interaction network revealed a link from PQBP1 to APC4 via U5-15kD (Supplementary Figure 8E), a spliceosomal component that is transiently incorporated into the U5 spliceosome when the U5 complex recognizes the exon–intron junction.59,60 In addition, Apc4 further links to Apc2. As is well known, Apc2 is the major player in anaphase-promoting complex/cyclosome, which controls the M phase. Given the physical interaction between PQBP1 and U5-15kD,54,55 the synchronized dynamics of PQBP1 and U5-15kD in the spliceosome59,60 and the genetic interaction between U5-15kD/dim1 and Apc4/lid1,61,62 Apc4 was considered to be the best candidate for the target gene of PQBP1 in NSPCs, which could transduce the effect of Pqbp1 deficiency into cell cycle elongation.
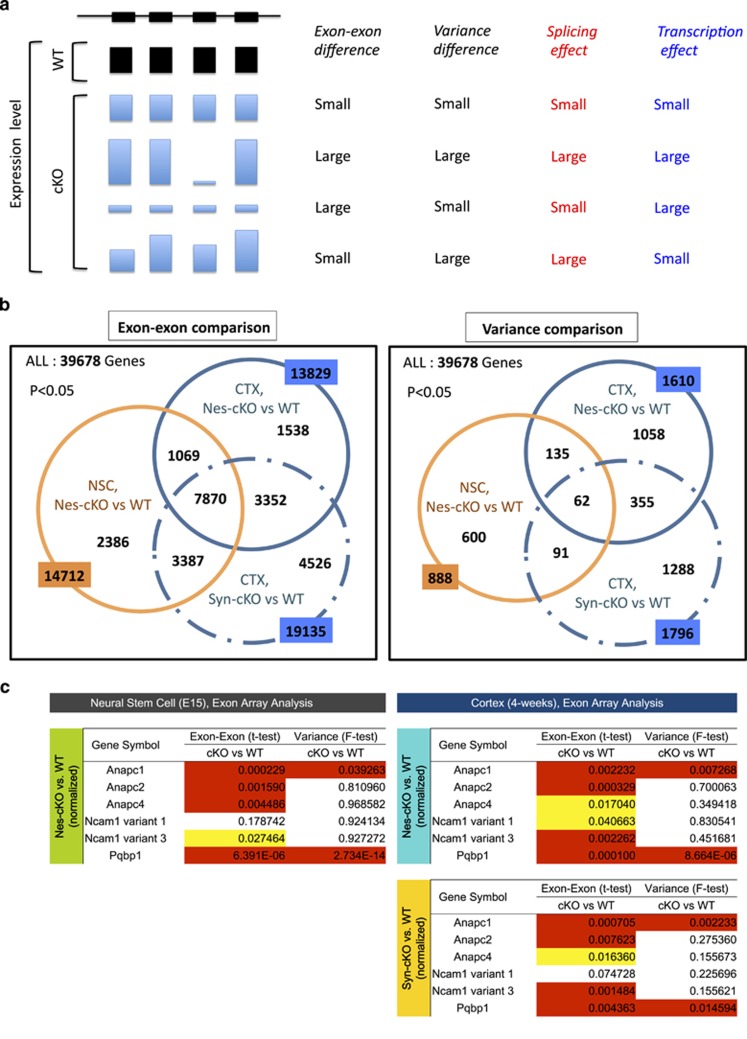
To identify the target genes of Pqbp1 that were affected by aberrant splicing under Pqbp1 deficiency, we next performed two independent analyses using the results of exon-array gene chips (Figure 3a). In the first analysis, we normalized all the exon signals by the total signals in an array, and calculated the ratios of exon signals to the integrated signal of each gene as described in the methods. We then compared the ratio of each exon between the nestin-Cre cKO and the background B6 mice (male) (n=3) or between the synapsin-1-Cre cKO mice and B6 mice (male) (n=3) in all genes (Supplementary Table 3). In this exon–exon comparison, we employed the lowest P-value as the representative of the gene for ranking when multiple exons were significantly changed in a gene (Supplementary Table 3). Genes ranked at a high position were considered to be remarkably affected with respect to their splicing and transcription by Pqbp1 deficiency.
Figure 3.
Analyses of genes affected by aberrant splicing in neurogenesis of neural stem progenitor cells (NSPCs) from nestin-Cre conditional Pqbp1-knockout (cKO) mice. (a) Representative patterns of exon array signals in a gene are shown. The expected results in the two analyses (exon–exon and variance) of exon array data and the contribution of splicing/transcription are correlated. (b) Affected genes (P<0.05) were selected from the exon array results of NSPCs from nestin-Cre cKO mice, the cortex of nestin-Cre cKO mice and the cortex of synapsin-1-Cre cKO mice by comparison with wild-type mice (B6). The selected genes were further compared among the three genotypes as shown in the Venn diagram. (c) Results from the two analyses (exon–exon and variance) of exon array data are shown for APC1, APC2, APC4, NCAM (variants 1 and 3) and Pqbp1. When a gene possessed multiple exon probes, the lowest P-value was used as the representative. APC1 and Pqbp1 were remarkably affected in both analyses in all genotypes. APC4 was significantly affected in neural stem cells (NSCs) but not strongly in the cortex. From the speculation in (a), the effect of transcription was relatively large on APC4 while both transcription and splicing affect APC1 and Pqbp1. NCAM1 (especially variant 3) was affected in the cortex of two types of cKO mice, but the change in NSCs was not so significant.
In the second approach, we calculated the ‘variance' of multiple exon signals from the same gene, and compared it between background mice and cKO mice (Supplementary Table 4). A P-value of <0.05 in the F-test was used for judgment that the ‘variance' was different between background and cKO mice. The significant change in ‘variance' was considered to reflect the aberrant splicing. We used the results from these two approaches to evaluate whether the gene was changed due to aberrant splicing and/or transcription, as shown schematically (Figure 3a).
Next, we compared the three sets of results. In the first ‘exon–exon comparison' analysis, among the 39 678 gene transcripts on the exon array gene chip, 14 712 were affected by Pqbp1 deficiency in NSPCs, 13 829 genes were affected in the cortical tissues of nestin-Cre cKO mice (Figure 3b, left panel), and 19 135 genes were affected in the cortical tissues of synapsin-1-Cre cKO mice (Figure 3b, left panel). Half of the genes affected in the NSPCs of nestin-Cre cKO mice (7870 genes) were also affected in the cortical tissues of both types of cKO mice, while nearly 16% of them (2386 genes) were unique to nestin-Cre cKO mice (Figure 3b, left panel; Supplementary Table 5). It is of note that the ratio of tissue- or model-unique genes was highest in the cortical tissue of synapsin-1-Cre cKO mice (4526 genes) (Figure 3b, left panel). In the second analysis, we calculated the variances of exon signals and compared them between mouse models (Supplementary Table 4). First, the numbers of changed genes based on the variance analysis were far smaller than that selected by exon–exon comparison in all groups (Figure 3b, right panel), suggesting that Pqbp1 deficiency affects gene expression more widely through transcription than through splicing. The comparison among the three arrays also showed that the ratios of genes in the intersection groups were smaller (Figure 3b, right panel; Supplementary Table 6).
Data from the ordinary gene chip suggested that anaphase-promoting complex/cyclosome proteins would be a downstream target of Pqbp1 deficiency in NSPCs (Supplementary Figure 8). The new results from exon arrays consistently supported this idea. Apc1, which links to Apc4, was significantly affected with respect to splicing in NSPCs and the cortex (Figure 3c, Supplementary Table 7). Both splicing and transcription affected the expression of Apc1 judging from the results of ‘exon–exon comparison' and ‘variance' analyses. Apc2 expression was also remarkably changed in NSPC and the cortex, but the effect of splicing on Apc2 did not seem large (Figures 3a and c, and Supplementary Table 7). On the other hand, Apc4 was altered specifically in NSPCs of nestin-Cre cKO mice (Figure 3c and Supplementary Table 7), suggesting a relatively large effect of Apc4 on NSPCs and on microcephaly. Interestingly, Pqbp1 was also affected by itself more strongly through splicing than through transcription (Figure 3c and Supplementary Table 7).
With regard to NCAM1, which was previously reported as a target of aberrant splicing by Pqbp1 KD,56 we confirmed aberrant splicing of the NCAM1 gene in the cortical tissue of nestin-Cre and synapsin-1-Cre Pqbp1-cKO mice in exon–exon signal comparison (Figure 3c and Supplementary Table 7). The discrepancy between normal dendrite length in vivo (Supplementary Figure 6) and the aberrant splicing of NCAM1 (Figure 3 and Wang et al.56) suggests that certain compensatory mechanism(s) recovered the neurite length in vivo. The aberrant splicing of NCAM1 was not significant in NSPCs under Pqbp1 deficiency (Figure 3c and Supplementary Table 7).
Apc4 is a target gene of Pqbp1 that links to microcephaly
By carrying out reverse transcription polymerase chain reaction (RT-PCR) with Southern blot, we further tested whether Pqbp1 affected the splicing of Apc4. Pqbp1-KD by verified shRNA (Supplementary Figure 4) increased the amount of intron-containing pre-mRNAs of Apc4 in primary NSPCs (Supplementary Figure 9A). In addition, we found that the Pqbp1 protein was essential for the splicing of Apc4 pre-mRNA because the addition of anti-PQBP1 antibodies specifically inhibited the in vitro splicing of Apc4 but not of crystallin (Supplementary Figure 9B). Nonsense RNA decay dependent on Upf1 was expected to be the mechanism for the decrease of Apc4 in NSPCs induced by Pqbp1-KD, (Supplementary Figure 9C). Co-transfection of Upf1-shRNA into NSPCs increased the number of unspliced introns of Apc4 between exons 22 and 23 or exons 6 and 7 (Supplementary Figure 9C) and supported nonsense RNA decay.
In addition, we confirmed that human APC4 mRNA is normally recognized as a splicing target by a spliceosome complex containing PQBP1, by showing the co-precipitation of PQBP1 and APC4 mRNA in human HEK-293 cells (Supplementary Figures 9D and E). Blocking nonsense-mediated mRNA decay by cycloheximide after PQBP1 KD resulted in the preservation of aberrant isoforms of APC4 pre-mRNA containing intron 23 in human HEK-293 cells (Supplementary Figures 9F–H). The levels of APC4 mRNAs measured by quantitative PCR were also reduced in PQBP1-KD HEK-293 cells (Supplementary Figures 9F and G). Taken together with NSPC-specific suppression of Apc4 (Figure 3c), these results suggested that Apc4 is a specific target of Pqbp1 deficiency in NSPCs that represses NSPC proliferation through its aberrant transcription and splicing.
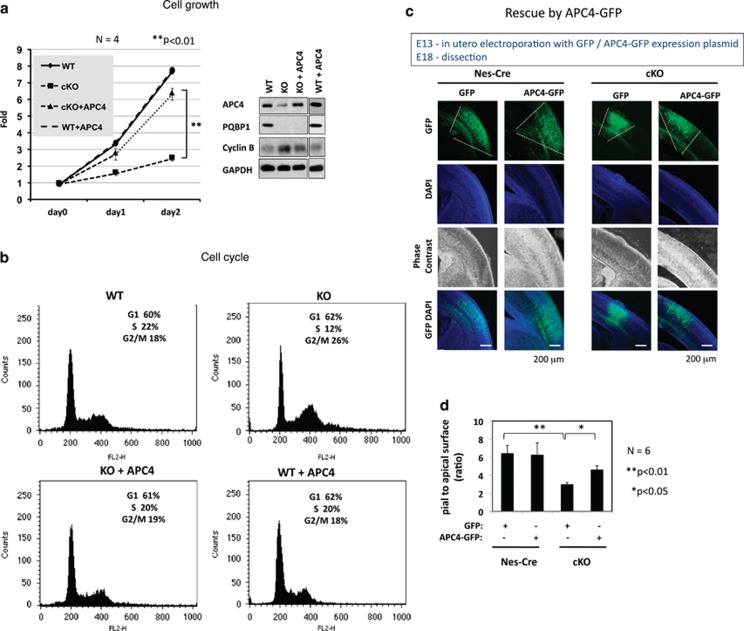
To test whether Apc4 is a major contributor to microcephaly in Pqbp1 deficiency, we performed rescue experiments in vitro and in vivo. First, we transfected the Apc4 expression vector into NSPCs from nestin-Cre cKO mice (Figures 4a and b). Overexpression of Apc4 recovered the decreased Apc4 protein level in NSPCs from cKO mice and normalized the cell growth speed (Figure 4a). Consistent with this observation, Apc4 recovered the accumulation of NSPCs at the G2/M phase, as seen with fluorescence-activated cell sorting (Figure 4b). The effect of Apc4 was also tested in vivo by in utero electroporation of the APC4 expression vector into the brains of nestin-Cre cKO mouse embryos (Figures 4c and d). We transfected either a GFP or a GFP-Apc4 plasmid into control (nestin-Cre) or Pqbp1-cKO (nestin-Cre; Floxed) mouse embryos at E13 by in utero electroporation, and dissected them at E18, following a previously reported method to quantify cortical expansion from NSPCs.33 The ratio of pial-to-apical surface area was calculated from serial consecutive vibratome sections (50 μm thick) through the entire rostral-to-caudal axis of E18 brains, as described.33 The decreased pial-to-apical surface area ratio in Pqbp1-cKO mice was partially rescued by electroporation of GFP-Apc4 plasmid but not by GFP in controls (Figures 4c and d). All the results revealed the cascade from Pqbp1 deficiency to microcephaly by cell cycle elongation via Apc4 and related molecules that were decreased via aberrant transcription and splicing.
Figure 4.
Apc4 plays a main role in the proliferation and horizontal expansion of neural stem progenitor cells (NSPCs). (a) Proliferation of NSPCs from E14 nestin-Cre conditional Pqbp1-knockout (cKO) embryos was retarded in primary culture but rescued by transfection of Apc4 (n=4). **P<0.01 in one-way analysis of variance (ANOVA) with post hoc Tukey's test. (b) Fluorescence-activated cell sorting analysis showed G2/M accumulation and a mild increase in the G1 population in NSPCs derived from E14 cKO embryos. Apc4 transfection rescued delayed proliferation and prevented cyclin B accumulation in NSPCs from cKO embryos. (c) The pial-to-apical surface area ratio reflects the proliferation of in-utero transfected EGFP-positive cells after a defined time period. pApc4-IRES-hrGFPII or pIRES-hrGFPII were electroporated into the ventricular zone of E13 embryos (nestin-Cre control and cKO mice), and the brains were analyzed at E18. (d) Pial-to-apical ratio was deduced from 3D reconstruction of the rostral-to-caudal axis serial sections of six embryos in each genotype. The ratio was decreased in Pqbp1-cKO embryos, reflecting the decreased cell cycle times of NSPCs, but it was rescued by Apc4 expression. **P<0.01 or *P<0.05 in one-way ANOVA with post hoc Tukey's test.
PQBP1 rescues microcephaly of nestin-Cre cKO mice
Finally, we performed therapeutic trials for Pqbp1 deficiency-induced microcephaly using AAV expressing human PQBP1 (Figure 5). The viral vector was similar to the one used previously for human gene therapy of a young girl who suffered from aromatic L-amino acid decarboxylase deficiency63 and for experimental therapy of a mouse model of spinal and bulbar muscular atrophy (Kennedy's disease) with miR-196.64 The viral vector was supplied to mother mice by peritoneal injection at E10, the earliest time point at which we could confirm pregnancy.
Figure 5.
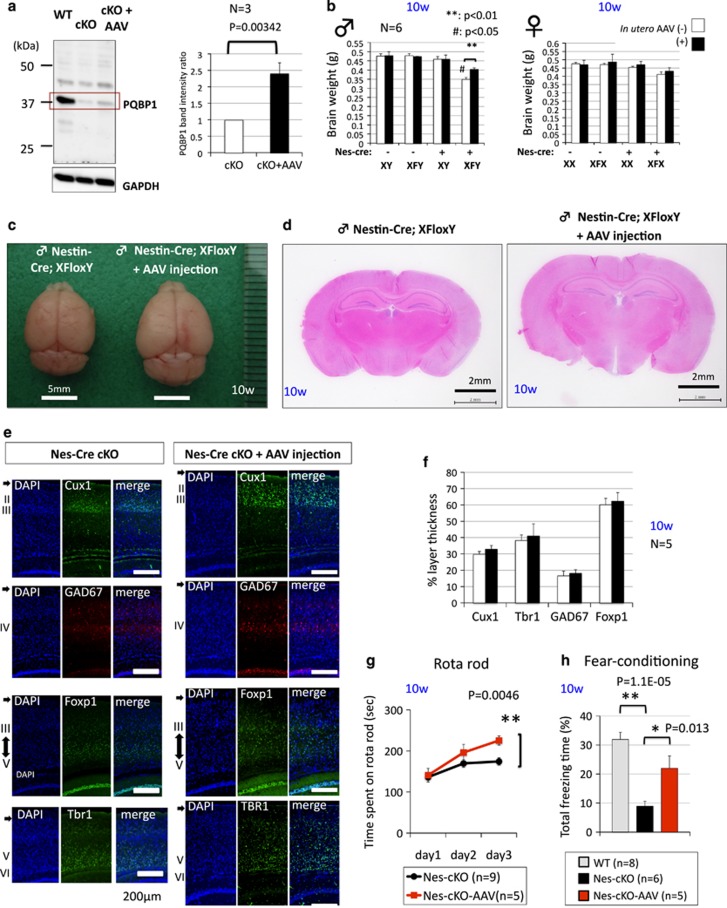
Peritoneal injection of PQBP1-AAV recovers the microcephaly and behaviors of nestin-Cre Pqbp1-cKO (conditional Pqbp1-knockout) mice. (a) Confirmation of the increase of Pqbp1 protein level in nestin-Cre Pqbp1-cKO mice after injection of PQBP1-AAV vector. Western blot analysis revealed a 2.5-fold increase. (b) The brain weight of nestin-Cre Pqbp1-cKO mice was recovered by PQBP1-AAV (*P<0.01 in Student's t-test). In multiple-group comparison with Tukey's test, the change was also confirmed (#P<0.05). (c) Macroscopic comparison of brain morphology between non-treated and AAV-injected male nestin-Cre Pqbp1-cKO mice at 2.5 months (10 weeks). (d) Comparison of coronal sections of the brain at the exactly same position of non-treated and AAV-injected male nestin-Cre Pqbp1-cKO mice at 2.5 months (10 weeks) revealed recovery of brain size by the PQBP1-AAV vector. (e) Staining for layer-specific markers, Cux1, Foxp1, and Tbr1, together with GAD67, shows the preservation of cortical layers in the rescue of nestin-Cre Pqbp1-cKO mice by the PQBP1-AAV vector at 2.5 months. (f) Quantitative analysis of the relative thickness of each layer to total thickness of the cortex. No difference was detected between AAV-injected and non-injected nestin-Cre Pqbp1-cKO mice. (g) The decline of nestin-Cre Pqbp1-cKO mice in the rotarod test was recovered at 3 months after birth by in utero gene therapy with the PQBP1-AAV vector. **P<0.01 in ANOVA with post hoc Tukey's test. (h) The decline of nestin-Cre Pqbp1-cKO mice in fear-conditioned memory was recovered at 3 months by in utero gene therapy with the PQBP1-AAV vector. **P<0.01 or *P<0.05 in ANOVA with post hoc Tukey's test.
The effect of the AAV vector is known to be sustained for more than a year.63 Consistent with this, we found that the expression level of the Pqbp1/PQBP1 proteins, whose sizes are exactly similar on western blot due to their high homology, was increased to 2.5-fold at 10 weeks after birth (Figure 5a) though the expression level was still far lower than that of the background control mice (Figure 5a). Interestingly, even this limited recovery of Pqbp1/PQBP1 expression resulted in a significant increase in brain weight (Figure 5b). The macroscopic and microscopic brain architectures of nestin-Cre cKO mice were normal and proportionally enlarged (Figures 5c and d). Immunohistological analyses with layer markers also revealed that the cortical layer structure was well preserved (Figure 5e), and this conclusion was supported by quantitative analyses of the thickness of each layer (Figure 5f).
Recovery by PQBP1 supplementation was confirmed not only for brain morphology but also for behavioral phenotypes (Figures 5g and h). We performed multiple behavior tests with nestin-Cre and synapsin-1-Cre Pqbp1 cKO mice (Supplementary Figures 10 and 11). Interestingly, while the fear conditioning and rotarod tests showed similarly abnormal results in both types of cKO mice, the open field, light-dark box, elevated plus maze, and water maze tests revealed some differences in the results from the different mouse models (Supplementary Figures 10 and 11). In brief, synapsin-1-Cre cKO mice were typically careless and insensitive to fear. On the other hand, the phenotypes of nestin-Cre cKO mice seemed more complex, which might be a result of developmental compensation for Pqbp1 deficiency in this model. Among such behavior tests, we observed significant recovery of nestin-Cre Pqbp1 cKO mice in rotarod and fear conditioning tests, which were abnormal in both types of cKO mice (Supplementary Figures 10 and 11) at 10 weeks after birth, following the intra-peritoneal injection of PQBP1-AAV to mother mice (Figures 5g and h).
The incomplete recovery in behavioral tests (Supplementary Figure 10) can be simply explained by the insufficient expression of the PQBP1/Pqbp1 protein by AAV vector (Figure 5a). However, it might suggest that brain size recovery is not sufficient for functional recovery. If this is the case, two explanations are possible. The first one is that the levels of Pqbp1 protein necessary for architectural recovery and functional recovery might be different. A higher level of Pqbp1 might be necessary for synapse dynamism. The second possibility is that the AAV-mediated delivery of Pqbp1 was too late. There might be a critical period before E10 for some downstream functions. Further investigations of these possibilities will be necessary to develop this technique towards human gene therapy of the ID of PQBP1-mutated patients.
Discussion
This study provides a unique mechanism of brain size regulation that depends largely on cell cycle elongation of NSPCs but not on unequally changed neurogenesis or decreased BP production ratio. Our findings might be also discussed from the viewpoint of the hypothesis that the cell division times of NSPCs determine neuronal subtypes.65,66 Given that the cell division number of NSPCs was expected to decrease roughly from 11 to 10 in Pqbp1-deficient cKO embryos, our findings might suggest that extrinsic factors rather than intrinsic determination of NSPCs control the cortical layer to produce from the NSPC pool at a developmental time point. However, since the decrease of cell division number is small, it might also be explained by assuming that cortical layer formation was compensated in nestin-Cre cKO embryos by overlapped time spans of cortical layers. This is a question for further investigation.
The results in this study, especially from a newly developed cKO mouse model that faithfully mimics the magnetic resonance images of human PQBP1-linked microcephaly, elucidated the molecular mechanism of Pqbp1-dependent brain size regulation. Systems biology analyses revealed candidate molecules that are possibly involved in the mechanism of microcephaly. Especially, Apc4 is a critical downstream target of Pqbp1 for microcephaly, and both aberrant splicing and transcription contribute to the downregulation of Apc4 by Pqbp1.
In the other categories of developmental disorders such as enzyme deficiencies or cystic fibrosis, in utero gene therapy has been considered.64,67 However, it has not been considered for normalizing brain tissues or whole brains that were impaired by genetic defects in microcephaly. Therefore, this is the first study to prove that brain size disorder could be another objective of in utero gene therapy. This study is just an example, but the method could be applied to the other microcephalies by targeting each causative gene. Future advance in genetic diagnosis will help extremely early initiation of gene therapy especially in the case of pregnancy of high-risk mothers with the patients in her or husband's family.
Technically, gene transfer efficiency, virulence, tumorigenesis and inflammation would be the next issues to tackle in order to adapt this technique to human patients, as shown in the application of gene therapy for treating cystic fibrosis and other diseases.68,69 With regard to expression level, this study revealed that low-level PQBP1 expression was still effective for treating microcephaly. Regarding the toxicity of AAV, it has been already used in clinical trials of multiple diseases like cystic fibrosis, hemophilia B, muscular dystrophy, Parkinson's disease, Canavan's disease and Alzheimer's disease, and side effects have rarely been observed.70 Tumorigenesis or inflammation has not been reported thus far. Therefore, a similar approach would be a potential therapeutic approach to treat human patients with PQBP1-linked microcephaly if further intensive studies in higher animals can exclude such side effects.
Naturally, ethical consideration is also essential before clinical application to human patients. The timing of administration of AAV vector and the route of gene delivery (intravenous, amniotic fluid, or cerebrospinal fluid administration; delivery to mother or embryo) would be the issues in such a case. However, this is the first step for the possibility of gene therapy in microcephaly, and the issues raised above will be cleared in the future by technical advances. It would be worth discussing and investigating on it further.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research on Innovative Areas ‘Foundation of Synapse and Neurocircuit Pathology' (22110001, 22110002), Strategic Research Program for Brain Sciences (SRPBS) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS) (18390254), CREST from Japan Science and Technology Agency (JST) to HO, the Deutsche Forschungsgemeinschaft, SFB577 to VMK and HHR, research grants from the Department of Health, Pennsylvania, USA, RFA60707 and RFA09200903 to MS and NIH grant NS084393 to AB. We thank Drs Kazuhiko Tagawa, Takuya Tamura, Masaki Sone, Naoyuki Kataoka, Miho Soma, Koichi Tanaka, and Ms Tayoko Tajima (TMDU) for technical assistance and critical discussion. We appreciate the technical support of Drs Yoshiaki V Nishimura, Daijiro Konno, Shigeaki Kanatani and Kazunori Nakajima (Keio University), Mss Mami Terao and Mimi Adachi (Kyoto University), and critical advice from Dr Shigetaka Kitajima (TMDU) and continuous encouragement from Professor Ichiro Kanazawa (National Center for Neurology and Psychiatry).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- Waragai M, Lammers CH, Takeuchi S, Imafuku I, Udagawa Y, Kanazawa I, et al. PQBP-1, a novel polyglutamine tract-binding protein, inhibits transcription activation by Brn-2 and affects cell survival. Hum Mol Genet. 1999;8:977–987. doi: 10.1093/hmg/8.6.977. [DOI] [PubMed] [Google Scholar]
- Kalscheuer VM, Freude K, Musante L, Jensen LR, Yntema HG, Gécz J, et al. Mutations in the polyglutamine binding protein 1 gene cause X-linked mental retardation. Nat Genet. 2003;35:313–315. doi: 10.1038/ng1264. [DOI] [PubMed] [Google Scholar]
- Stevenson RE, Bennett CW, Abidi F, Kleefstra T, Porteous M, Simensen RJ, et al. Renpenning syndrome comes into focus. Am J Med Genet A. 2005;134:415–421. doi: 10.1002/ajmg.a.30664. [DOI] [PubMed] [Google Scholar]
- de Brouwer AP, Yntema HG, Kleefstra T, Lugtenberg D, Oudakker AR, de Vries BB, et al. Mutation frequencies of X-linked mental retardation genes in families from the EuroMRX consortium. Hum Mutat. 2007;28:207–208. doi: 10.1002/humu.9482. [DOI] [PubMed] [Google Scholar]
- Qi Y, Hoshino M, Wada Y, Marubuchi S, Yoshimura N, Kanazawa I, et al. PQBP-1 is expressed predominantly in the central nervous system during development. Eur J Neurosci. 2005;22:1277–1286. doi: 10.1111/j.1460-9568.2005.04339.x. [DOI] [PubMed] [Google Scholar]
- Rakic P. A small step for the cell, a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends Neurosci. 1995;18:383–388. doi: 10.1016/0166-2236(95)93934-p. [DOI] [PubMed] [Google Scholar]
- Caviness VS, Goto T, Tarui T, Takahashi T, Bhide PG, Nowakowski RS. Cell output, cell cycle duration and neuronal specification: a model of integrated mechanisms of the neocortical proliferative process. Cereb Cortex. 2003;13:592–598. doi: 10.1093/cercor/13.6.592. [DOI] [PubMed] [Google Scholar]
- Fietz SA, Huttner WB. Cortical progenitor expansion, self-renewal and neurogenesis-a polarized perspective. Curr Opin Neurobiol. 2011;21:23–35. doi: 10.1016/j.conb.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Malatesta P, Hartfuss E, Gotz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Okano H, Ogawa M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 2001;31:727–741. doi: 10.1016/s0896-6273(01)00420-2. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci USA. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- Shitamukai A, Konno D, Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J Neurosci. 2011;31:3683–3695. doi: 10.1523/JNEUROSCI.4773-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reillo I, de Juan Romero C, Garcia-Cabezas MA, Borrell V. A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb Cortex. 2011;21:1674–1694. doi: 10.1093/cercor/bhq238. [DOI] [PubMed] [Google Scholar]
- Stahl R, Walcher T, De Juan Romero C, Pilz GA, Cappello S, Irmler M, et al. Trnp1 regulates expansion and folding of the mammalian cerebral cortex by control of radial glial fate. Cell. 2013;153:535–549. doi: 10.1016/j.cell.2013.03.027. [DOI] [PubMed] [Google Scholar]
- Nonaka-Kinoshita M, Reillo I, Artegiani B, Martínez-Martínez MÁ, Nelson M, Borrell V, et al. Regulation of cerebral cortex size and folding by expansion of basal progenitors. EMBO J. 2013;32:1817–1828. doi: 10.1038/emboj.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki H, Suzuki H, Itohara S. High-efficiency CAG-FLPe deleter mice in C57BL/6J background. Exp Anim. 2006;55:137–141. doi: 10.1538/expanim.55.137. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Nowakowski RS, Caviness VS., Jr. Cell cycle parameters and patterns of nuclear movement in the neocortical proliferative zone of the fetal mouse. J Neurosci. 1993;13:820–833. doi: 10.1523/JNEUROSCI.13-02-00820.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen VL, Torres AR, Du X, Barry B, Walsh CA, Kimonis VE. Mutation in PQBP1 is associated with periventricular heterotopia. Am J Med Genet A. 2010;152:2888–2890. doi: 10.1002/ajmg.a.33507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germanaud D, Rossi M, Bussy G, Gérard D, Hertz-Pannier L, Blanchet P, et al. The Renpenning syndrome spectrum: new clinical insights supported by 13 new PQBP1-mutated males. Clin Genet. 2011;79:225–235. doi: 10.1111/j.1399-0004.2010.01551.x. [DOI] [PubMed] [Google Scholar]
- Fichera M, Falco M, Lo Giudice M, Castiglia L, Guarnaccia V, Calì F, et al. Skewed X-inactivation in a family with mental retardation and PQBP1 gene mutation. Clin Genet. 2005;67:446–447. doi: 10.1111/j.1399-0004.2005.00436.x. [DOI] [PubMed] [Google Scholar]
- Li C, Ito H, Fujita K, Shiwaku H, Qi Y, Tagawa K, et al. Sox2 transcriptionally regulates PQBP1, an intellectual disability-microcephaly causative gene, in neural stem progenitor cells. PLoS One. 2013;8:e68627. doi: 10.1371/journal.pone.0068627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi Y, de la Torre-Ubieta L, Matsuda T, Steen H, Okazawa H, Bonni A. The XLID Protein PQBP1 and the GTPase dynamin 2 define a signaling link that orchestrates ciliary morphogenesis in postmitotic neurons. Cell Rep. 2013;4:879–889. doi: 10.1016/j.celrep.2013.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Romero MI, Ghosh P, Ye Z, Charnay P, Rushing EJ, et al. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15:859–876. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr AR, Kilmartin JV, Gergely F. CDK5RAP2 functions in centrosome to spindle pole attachment and DNA damage response. J Cell Biol. 2010;189:23–39. doi: 10.1083/jcb.200912163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll P, Pimpinelli S, Valdivia MM, Avila J. A cell division mutant of Drosophila with a functionally abnormal spindle. Cell. 1985;41:907–912. doi: 10.1016/s0092-8674(85)80071-4. [DOI] [PubMed] [Google Scholar]
- Saunders RD, Avides MC, Howard T, Gonzalez C, Glover DM. The Drosophila gene abnormal spindle encodes a novel microtubule-associated protein that associates with the polar regions of the mitotic spindle. J Cell Biol. 1997;137:881–890. doi: 10.1083/jcb.137.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C, Huttner WB, Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5:320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- Arai Y, Pulvers JN, Haffner C, Schilling B, Nüsslein I, Calegari F, et al. Neural stem and progenitor cells shorten S-phase on commitment to neuron production. Nat Commun. 2011;2:154. doi: 10.1038/ncomms1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AP, Eastwood H, Bell SM, Adu J, Toomes C, Carr IM, et al. Identification of microcephalin, a protein implicated in determining the size of the human brain. Am J Hum Genet. 2002;71:136–142. doi: 10.1086/341283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilguvar K, Oztürk AK, Louvi A, Kwan KY, Choi M, Tatli B, et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, Roberts E, Mochida GH, Hampshire DJ, Scott S, Askham JM, et al. ASPM is a major determinant of cerebral cortical size. Nat Genet. 2002;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- Bond J, Roberts E, Springell K, Lizarraga SB, Scott S, Higgins J, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet. 2005;37:353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- Guernsey DL, Jiang H, Hussin J, Arnold M, Bouyakdan K, Perry S, et al. Mutations in centrosomal protein CEP152 in primary microcephaly families linked to MCPH4. Am J Hum Genet. 2010;87:40–51. doi: 10.1016/j.ajhg.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalay E, Yigit G, Aslan Y, Brown KE, Pohl E, Bicknell LS, et al. CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat Genet. 2011;43:23–26. doi: 10.1038/ng.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Girimaji SC, Duvvari MR, Blanton SH. Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. Am J Hum Genet. 2009;84:286–290. doi: 10.1016/j.ajhg.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Jackson AP, Bond J, Woods CG. What primary microcephaly can tell us about brain growth. Trends Mol Med. 2006;12:358–366. doi: 10.1016/j.molmed.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Pfaff KL, Straub CT, Chiang K, Bear DM, Zhou Y, Zon LI. The zebrafish cassiopeia mutant reveals that SIL is required for mitotic spindle organization. Mol Cell Biol. 2007;27:5887–5897. doi: 10.1128/MCB.00175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish JL, Kosodo Y, Enard W, Paabo S, Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci USA. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman JJ, Tseng HC, Zhou Y, Frank CL, Xie Z, Tsai LH. Cdk5rap2 interacts with pericentrin to maintain the neural progenitor pool in the developing neocortex. Neuron. 2010;66:386–402. doi: 10.1016/j.neuron.2010.03.036. [DOI] [PubMed] [Google Scholar]
- Shen J, Gilmore EC, Marshall CA, Haddadin M, Reynolds JJ, Eyaid W, et al. Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat Genet. 2010;42:245–249. doi: 10.1038/ng.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A, Grove EA. Cilia in the CNS: the quiet organelle claims center stage. Neuron. 2011;69:1046–1060. doi: 10.1016/j.neuron.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador-Arjona A, Elliott J, Miller A, Ginbey A, Pazour GJ, Enikolopov G, et al. Primary cilia regulate proliferation of amplifying progenitors in adult hippocampus: implications for learning and memory. J Neurosci. 2011;31:9933–9944. doi: 10.1523/JNEUROSCI.1062-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubs H, Abidi FE, Echeverri R, Holloway L, Meindl A, Stevenson RE, et al. Golabi-Ito-Hall syndrome results from a missense mutation in the WW domain of the PQBP1 gene. J Med Genet. 2006;43:e30. doi: 10.1136/jmg.2005.037556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia VE, Nicolaescu E, McDonald CB, Musi V, Oka T, Inayoshi Y, et al. Y65C missense mutation in the WW domain of the Golabi-Ito-Hall syndrome protein PQBP1 affects its binding activity and deregulates pre-mRNA splicing. J Biol Chem. 2010;285:19391–19401. doi: 10.1074/jbc.M109.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazawa H, Rich T, Chang A, Lin X, Waragai M, Kajikawa M, et al. Interaction between mutant ataxin-1 and PQBP-1 affects transcription and cell death. Neuron. 2002;34:701–713. doi: 10.1016/s0896-6273(02)00697-9. [DOI] [PubMed] [Google Scholar]
- Komuro A, Saeki M, Kato S. Association of two nuclear proteins, Npw38 and NpwBP, via the interaction between the WW domain and a novel proline-rich motif containing glycine and arginine. J Biol Chem. 1999;274:36513–36519. doi: 10.1074/jbc.274.51.36513. [DOI] [PubMed] [Google Scholar]
- Llorian M, Beullens M, Lesage B, Nicolaescu E, Beke L, Landuyt W, et al. Nucleocytoplasmic shuttling of the splicing factor SIPP1. J Biol Chem. 2005;280:38862–38869. doi: 10.1074/jbc.M509185200. [DOI] [PubMed] [Google Scholar]
- Waragai M, Junn E, Kajikawa M, Takeuchi S, Kanazawa I, Shibata M, et al. PQBP-1/Npw38, a nuclear protein binding to the polyglutamine tract, interacts with U5-15kD/dim1p via the carboxyl-terminal domain. Biochem Biophys Res Commun. 2000;273:592–595. doi: 10.1006/bbrc.2000.2992. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lindblom T, Chang A, Sudol M, Sluder AE, Golemis EA. Evidence that dim1 associates with proteins involved in pre-mRNA splicing, and delineation of residues essential for dim1 interactions with hnRNP F and Npw38/PQBP-1. Gene. 2000;257:33–43. doi: 10.1016/s0378-1119(00)00372-3. [DOI] [PubMed] [Google Scholar]
- Wang Q, Moore MJ, Adelmant G, Marto JA, Silver PA. PQBP1, a factor linked to intellectual disability, affects alternative splicing associated with neurite outgrowth. Genes Dev. 2013;27:615–626. doi: 10.1101/gad.212308.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Horiuchi D, Chen YC, Sone M, Miyashita T, Saitoe M, et al. Drosophila PQBP1 regulates learning acquisition at projection neurons in aversive olfactory conditioning. J Neurosci. 2010;30:14091–14101. doi: 10.1523/JNEUROSCI.1319-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, et al. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- Makarova OV, Makarov EM, Urlaub H, Will CL, Gentzel M, Wilm M, et al. A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J. 2004;23:2381–2391. doi: 10.1038/sj.emboj.7600241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry LD, Gould KL. Fission yeast dim1(+) encodes a functionally conserved polypeptide essential for mitosis. J Cell Biol. 1997;137:1337–1354. doi: 10.1083/jcb.137.6.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry LD, Feoktistova A, Wright MD, Gould KL. The Schizosaccharomyces pombe dim1(+) gene interacts with the anaphase-promoting complex or cyclosome (APC/C) component lid1(+) and is required for APC/C function. Mol Cell Biol. 1999;19:2535–2546. doi: 10.1128/mcb.19.4.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwu WL, Muramatsu S, Tseng SH, Tzen KY, Lee NC, Chien YH, et al. Gene therapy for aromatic L-amino acid decarboxylase deficiency. Sci Transl Med. 2012;4:134ra61. doi: 10.1126/scitranslmed.3003640. [DOI] [PubMed] [Google Scholar]
- Miyazaki Y, Adachi H, Katsuno M, Minamiyama M, Jiang YM, Huang Z, et al. Viral delivery of miR-196a ameliorates the SBMA phenotype via the silencing of CELF2. Nat Med. 2012;18:1136–1141. doi: 10.1038/nm.2791. [DOI] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, et al. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- Gaspard N, Bouschet T, Hourez R, Dimidschstein J, Naeije G, van den Ameele J, et al. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature. 2008;455:351–357. doi: 10.1038/nature07287. [DOI] [PubMed] [Google Scholar]
- Driskell RA, Engelhardt JF. Current status of gene therapy for inherited lung diseases. Annu Rev Physiol. 2003;65:585–612. doi: 10.1146/annurev.physiol.65.092101.142426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb BR, Pickles RJ, Ye H, Yankaskas JR, Vick RN, Engelhardt JF, et al. Inefficient gene transfer by adenovirus vector to cystic fibrosis airway epithelia of mice and humans. Nature. 1994;371:802–806. doi: 10.1038/371802a0. [DOI] [PubMed] [Google Scholar]
- Wagner AM, Schoeberlein A, Surbek D. Fetal gene therapy: opportunities and risks. Adv Drug Deliv Rev. 2009;61:813–821. doi: 10.1016/j.addr.2009.04.011. [DOI] [PubMed] [Google Scholar]
- Kaplitt MG, Feigin A, Tang C, Fitzsimons HL, Mattis P, Lawlor PA, et al. Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson's disease: an open label, phase I trial. Lancet. 2007;369:2097–2105. doi: 10.1016/S0140-6736(07)60982-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.