Significance
In the heart, phospholamban regulates Ca2+-ATPase function, controlling cardiac output. A single deletion (R14del) in the phospholamban recognition sequence kinase A is linked to the progression of familial dilated cardiomyopathy, a leading cause of death worldwide. Here, we provide the molecular mechanism for the sluggish phosphorylation of R14del by protein kinase A. We found that the R14 deletion affects the organization of the active site, which remains partially open and quite dynamic, preventing the formation of catalytically committed complex. We conclude that well-tuned structural and dynamic interplay between kinase and substrate is crucial for efficient phosphorylation. These results provide new structural basis to understand for the reduced phosphorylation levels of the R14del phospholamban and impaired Ca2+ transport in heart muscle.
Keywords: phospholamban, phosphorylation, conformational dynamics, NMR, calcium regulation
Abstract
The dynamic interplay between kinases and substrates is crucial for the formation of catalytically committed complexes that enable phosphoryl transfer. However, a clear understanding on how substrates modulate kinase structural dynamics to control catalytic efficiency is still missing. Here, we used solution NMR spectroscopy to study the conformational dynamics of two complexes of the catalytic subunit of the cAMP-dependent protein kinase A with WT and R14 deletion phospholamban, a lethal human mutant linked to familial dilated cardiomyopathy. Phospholamban is a central regulator of heart muscle contractility, and its phosphorylation by protein kinase A constitutes a primary response to β-adrenergic stimulation. We found that the single deletion of arginine in phospholamban’s recognition sequence for the kinase reduces its binding affinity and dramatically reduces phosphorylation kinetics. Structurally, the mutant prevents the enzyme from adopting conformations and motions committed for catalysis, with concomitant reduction in catalytic efficiency. Overall, these results underscore the importance of a well-tuned structural and dynamic interplay between the kinase and its substrates to achieve physiological phosphorylation levels for proper Ca2+ signaling and normal cardiac function.
Phosphorylation by cAMP-dependent protein kinase A (PKA) is a central signaling pathway in cardiomyocytes, where it modulates the activity of several Ca2+ handling proteins (1, 2). In particular, PKA targets phospholamban (PLN), a small membrane protein embedded in the sarcoplasmic reticulum that binds and regulates the activity of the sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA). The SERCA/PLN complex plays a critical role in cardiac contractility, because it is responsible for Ca2+ reuptake in the sarcoplasmic reticulum, thereby controlling muscle relaxation, or diastole (1). PLN regulation of SERCA keeps Ca2+ flux within a physiological window (3). Unphosphorylated PLN reduces SERCA’s apparent affinity for Ca2+, whereas phosphorylation at Ser16 reverses PLN’s inhibitory effect, increasing Ca2+ flux and enhancing cardiac muscle relaxation (4). PKA phosphorylation of PLN is a primary response to β-adrenergic stimulation in the heart, affecting cardiac output directly (5). Altered levels of PLN phosphorylation (i.e., hypo- or hyperphosphorylation) cause SERCA’s function to be outside this physiological window. Specifically, aberrant PLN phosphorylation depresses Ca2+ cycling, with an attenuation of Ca2+ transients in both amplitude and frequency, resulting in cardiac disease (6, 7). Six naturally occurring mutations of PLN have been linked to early and/or late onset dilated cardiomyopathy (DCM), a leading cause of morbidity and mortality worldwide (8, 9). First reported by Kranias et al. (10, 11), the R14del mutation of PLN () was found in several DCM patients (10) and was more recently identified in 14% of a cohort of 354 patients diagnosed with either DCM or arrhythmogenic right ventricular cardiomyopathy (12). It has been proposed that the dysfunctional effects of are due to chronic suppression of SERCA activity, with a synergistic inhibition by the mutant and the WT PLN () (10, 13). Recent studies, however, show that PLN phosphorylation by PKA-C is significantly hindered in R14del transgenic mice (∼7% of WT levels), causing Ca2+ mishandling and progression of DCM (13). Although the phenotype and genotype of the R14 deletion have been identified, the molecular mechanisms for its aberrant phosphorylation are still unknown.
The R14 deletion is located at the recognition sequence for the catalytic subunit of PKA (PKA-C) (Fig. 1) (10). From the X-ray structure of the PKA-C/ATPγN/ complex [Protein Data Bank (PDB) ID code 3O7L], the PLN consensus sequence for PKA-C (R-R-X1-S-X2, where X denotes a variable amino acid) adopts an extended conformation within the binding groove (14, 15). The side chain of R14 (P-2 site) points toward the large lobe and interacts with Y204, E230, and E203, whereas the side chain of R13 (P-3 site) interacts with S51, the ribose ring, and hydroxyl group of Y330, clamping on the small lobe (Fig. 1 A and B) (14). Based on these structural features, we hypothesized that the R14 deletion disrupts intermolecular interactions between enzyme and substrate, affecting the motions responsible for opening and closing the active cleft and ultimately enzyme turnover. To test this hypothesis, we compared the kinetics of phosphorylation, thermodynamics of binding, and enzyme structural dynamics of the PKA-C/ complex to PKA-C/ using steady-state kinetic measurements, isothermal titration calorimetry (ITC), NMR spectroscopy, and molecular dynamic (MD) simulations. We found that, thermodynamically and kinetically, exhibits characteristics of a poor substrate compared with . More importantly, deletion of R14 prevents the formation of a catalytically committed Michaelis complex, causing dysfunctional conformational dynamics that concur with the reduction of catalytic efficiency. These findings explain the molecular mechanisms for the sluggish phosphorylation kinetics, leading to the Ca2+ mishandling associated with DCM. Overall, our study emphasizes the importance of well-tuned dynamic interplay between enzyme and substrate to achieve optimal catalysis.
Fig. 1.
Molecular interactions between PKA-C and peptide substrates or inhibitors. (A) Primary sequence of the WT and R14del peptides corresponding to the cytoplasmic region of PLN and the high affinity peptide inhibitor, PKI5–24. (B) X-ray structure of PKA-C (PDB:1ATP) with the architecture of the peptide binding site, with the electrostatic network of interactions between enzyme and substrate.
Results
R14del Causes a Reduction of Binding Affinity and Kinetics.
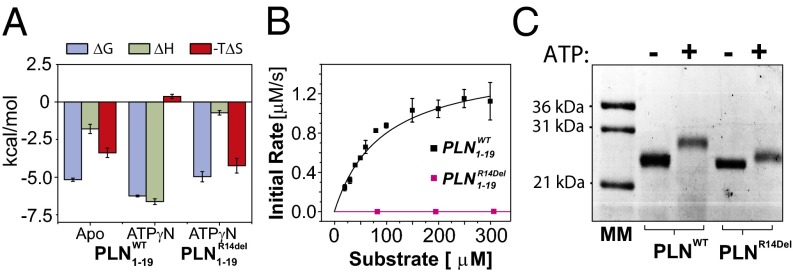
The binding thermodynamics of and to the apo and nucleotide-bound protein kinase (PKA-C/ATPγN complex) were measured using ITC with the synthetic peptides, and , corresponding to the first 19 residues of the cytoplasmic domain of and , respectively, which include the recognition sequence for the kinase. From the ITC measurements carried out in the presence of the nucleotide analog (ATPγN), we derived the binding enthalpy, entropy, dissociation constants (Kd), and substrate binding cooperativity for (SI Appendix, Tables S1 and S3) (14, 16, 17). We found that PKA-C exhibits high substrate affinity for in the presence of ATPγN (Kd = 28 μM) with a slightly unfavorable entropic contribution to binding (TΔS = −0.37 kcal/mol; Fig. 2A). In contrast, binding titrations of under identical experimental conditions resulted in a Kd ∼ 200 μM, with the entropic change TΔS ∼ 4 kcal/mol as the dominating contribution to binding (Fig. 2A). Interestingly, for binding in the absence of ATPγN, we were unable to measure the binding cooperativity observed for and typical of other substrates (SI Appendix, Fig. S1A) (14, 16). These thermodynamic measurements indicate that the affinity and the nature of the interactions are affected by the R14 deletion. To evaluate the efficiency of phosphorylation by PKA-C, we carried out steady-state phosphorylation kinetics using a coupled enzyme assay with and (Fig. 2B). Kinetically, behaves similarly to the standard substrate Kemptide (kcat = 23 ± 1 s−1) (18). In contrast, we did not detect any significant phosphorylation for . On a fourfold increase in PKA-C concentration (from 64 to 256 nM total enzyme) and an increase in substrate from 100 to 900 μM, we observed only marginal phosphorylation (SI Appendix, Fig. S1 B and C). Under these experimental conditions, the kcat is ∼6 times smaller and the catalytic efficiency is ∼300 times lower for than that of (SI Appendix, Table S1). Moreover, we repeated phosphorylation assays using full-length and . On incubation with PKA-C, we detected complete phosphorylation of (five phosphates per pentamer; Fig. 2C). In contrast, was only partially phosphorylated, as shown by the slight band shift in the SDS/PAGE (Fig. 2C). These phosphorylation assays exclude any effect of the transmembrane domains and confirm the sluggish phosphorylation kinetics measured for . These data are in agreement with phosphorylation kinetics carried out with model peptides missing the P-3 site (19) and PLN (20, 21).
Fig. 2.
Thermodynamics and kinetics for and (A) Thermodynamics of and to PKA-C without (apo) or with nucleotide (ATPγN) present. (B) Steady-state phosphorylation kinetics of and . (C) Gel shift assays with the extent of phosphorylation for pentameric and . Note that and oligomerize into stable homo-pentamers and run on SDS/PAGE gels with an apparent molecular weight of ∼26 kDa (55).
NMR Mapping of Substrate Binding to PKA-C.
To analyze the structural changes that PKA-C undergoes on binding and , we mapped the amide backbone fingerprint of the enzyme using [1H,15N]-TROSY-HSQC (transverse relaxation optimized spectroscopy–hetereonuclear single quantum correlation spectroscopy) experiments (22). To further validate the assignment of the major conformational equilibria, we performed a full triple resonance assignment on the apo, binary, and closed states (SI Appendix, Figs. S2–S4).The combined 1H and 15N chemical shift perturbations (Δδ) at each amide site of PKA-C report the changes in the local chemical environment (structure, electrostatics, dynamics, etc.) on substrate binding (Fig. 3 and SI Appendix, Fig. S5). Binding of the and peptides to PKA-C results in different chemical shift patterns. Binding of to the binary form of the enzyme (PKA-C/ATPγN complex) retains significant line broadening in many resonances, similar to the nucleotide-bound enzyme (16), primarily along catalytically important motifs. In contrast, binding of to the binary complex renders observable several residues that were previously broadened on nucleotide binding (Fig. 3B and SI Appendix, Fig. S6). Residues belonging to the Gly-rich loop, peptide positioning loop, the hinge region, and the DFG loop, which play critical roles in catalysis, are observed only with the WT substrate, but are absent in the complex formed with the deletion mutant. Because these spectra were compared at saturating conditions, this phenomenon indicates that the conformational dynamics of the ternary complexes are different. To evaluate the structural transition of PKA-C with the addition of the substrates, we used CONCISE (coordinated chemical shifts behavior) (23). This method performs a statistical analysis on linear chemical shift trajectories of each amide resonance to identify the position of each state along the conformational equilibrium. Following ligand binding (i.e., nucleotide and substrate), the PKA-C amide resonances display linear chemical shift, with the resonances corresponding to the apo enzyme populating one extreme and those of the closed state (ternary complex with the inhibitor, PKI5–24) populating the opposite extreme of the linear correlations (Fig. 3). This behavior exemplifies a conformational equilibrium among the three major conformational states of the kinase (open, intermediate, and closed), whose relative populations are modulated by ligand binding (14, 23, 24). The nucleotide shifts the open state population toward the intermediate state, and the substrate shifts the population toward the closed state (14, 17). We found that , as well as Kemptide (SI Appendix, Fig. S7), is able to shift the conformational equilibrium of PKA-C toward a catalytically committed state, whereas is unable to cause a significant conformational change, remaining in an intermediate state (Fig. 3 and SI Appendix, Fig. S5). Dynamic light scattering experiments (SI Appendix, Fig. S8 and Tables S2 and S5) also demonstrate that the enzyme in complex with exhibits a rotational correlation time shorter than the corresponding bound complex, adopting a more open conformation with . Based on this analysis, we conclude that binding shifts the population of the enzyme toward a conformation that is both structurally and dynamically different from the PKA-C/ complex, forming a partially closed state.
Fig. 3.
Shift in conformational equilibria by substrate binding. (A) Colored (purple) residues that undergo concerted chemical shift changes on ligand biding from the CONCISE and CHESCA analysis. (B) Ligand binding gives linear chemical shift changes from Apo (blue), Nucleotide bound (purple), substrate bound (green) to inhibiter bound (red). (C) Probability density plot of the different conformational states obtained using the CONCISE method. Ligand binding shifts the conformation equilibrium from open to closed states.
R14del Mutant Binding Causes Dysfunctional Conformational Dynamics of PKA-C.
The crystal structure of the PKA-C/ complex (14) shows that the arginine residue in P-3 position of PLN forms an electrostatic network with E127, Y330, and the ribose moiety of ATP (Fig. 1B), whereas the P-2 arginine interacts with R133, E230, and Y204. This dense network of interactions positions the substrate in a catalytically committed conformation. We reasoned that the removal of one of the two arginine residues in the recognition sequence may alter the enzyme conformation and motions. To test this hypothesis, we analyzed the conformational dynamics of the complexes using solution NMR spectroscopy. Specifically, we used the [1H,15N] heteronuclear steady-state NOE (nuclear Overhauser effect) experiment (HX-NOE) (25) as a proxy for motions on the picosecond/nanosecond (ns) timescale. The average trimmed HX-NOE values for PKA-C bound to and are 0.84 ± 0.09 and 0.83 ± 0.10, respectively (SI Appendix, Tables S2, S6, and S7), indicating that the overall conformational dynamics of the complexes on the fast timescale are similar. However, significant local variations of HX-NOE values were found between the two complexes. Residues neighboring the Mg2+ positioning, catalytic loop, the αF-αG loop, and the αG-αH loop (D161, R190, V191, D241, Q242, G253, K254, and R256), linked to be involved in substrate binding and regulation (26), are more flexible when PKA-C is bound to , whereas several residues flanking the C-helix, the αA-β1 linker, and the C terminus (S34, Q35, T37, K83, Q96, A97, F347, T348, E349, F350) appear to be more rigid. These changes in the fast dynamic timescale might be related to the changes in the overall binding entropy as measured by ITC (27).
Moreover, we investigated conformational dynamics in the microsecond/millisecond timescale by quantifying the contribution of chemical exchange to the transverse relaxation rate (Rex) using the TROSY Hahn-Echo experiment (28). We found striking differences in the Rex values for the two complexes (SI Appendix, Tables S2 and S8–S10). In agreement with the previously reported data, the binary form of the enzyme with ATPγN experiences considerable conformational dynamics (14) (SI Appendix, Fig. S9), especially along catalytic structural elements such as the glycine rich loop, C-helix, peptide positioning loop, and DFG loop (26). When is bound to PKA-C/ATPγN, these structural elements remain considerably dynamic; however, substrate binding decreases the Rex values. Residues along catalytically important motifs such as G55 and M58 in the glycine-rich loop, D166 in the catalytic loop, R190 and V191 near the Mg+2 positioning loop, and E208 belonging to the APE motif all experience a substantial decrease in Rex values. These results indicate that the ternary complex with is still dynamic, although the motions are attenuated with respect to the binary form (SI Appendix, Figs. S9 and S10); in contrast, the PKA-C/ATPγN/ complex displays enhanced conformational dynamics in regions important for catalysis. For instance, residues in the glycine-rich loop (G55, M58, and L59), near the Mg2+ positioning (R190 and V191), and the APE motif (such as E208) all experience Rex values similar to those found in the PKA-C/ATPγN complex (SI Appendix, Figs. S9 and S10). In addition, we observed a substantial increase in conformational dynamics in the microsecond/millisecond timescale in several other regions. Specifically, due to the loss of the P-3 arginine, the acidic patch region positioned along the C-terminal tail (residues 330–334) is no longer electrostatically connected to the active site and experiences an increase in conformational dynamics (Fig. 4). Increased motions occur for residues around D220, which is the conduit for allosteric signaling from the F-helix to the regulatory spine (R-spine; Fig. 4) (16).
Fig. 4.
Allosteric changes in conformational dynamics upon substrate binding. Zoom in of the slow conformational dynamics plotted on the X-ray crystal structure. The PKAC/ATPγN/ complex (Upper) shows attenuated conformational dynamics with respect to the PKA-C/ATPγN/ complex (Lower).
MD Simulation of the Ternary Complexes with and .
To interpret the dynamic effects of the deletion of P-3 arginine in the substrate, we performed atomistic MD simulations in explicit environments on both PKA-C/ and complexes. Starting from the crystal structure of PKA-C (PDB ID code 3O7L), we used the HADDOCK package (29) to dock the substrates ( or ) to the enzyme’s binding groove according to the canonical consensus sequence in the PKA-C/PKI5–24 X-ray structure (26). In general, MD simulations show that the interactions in the proximity of the active site are similar to those observed in the PKA-C/PKI5–24 and PKA-C/ crystal structures (Fig. 1B) (14, 26). In particular, the acidic cluster surrounding Y330 latches on the small lobe, clamping the glycine-rich loop down into the active site. Also, E127 in the linker region connects the small and large lobes. In contrast, the P-2 arginine points toward the large lobe, interacting with E203, Y204, and E230. These electrostatic interactions are part of an allosteric network that ensures structural and dynamic communication within the enzyme (16, 24, 30) (Fig. 5A). During ∼100 ns of MD simulations of the PKA-C/ complex, we found that the P-2 arginine residue (R14) does not change its side chain conformation, thus preserving the interresidue interactions present in the crystal structure (Fig. 5A and SI Appendix, Fig. S12). In contrast, the electrostatic interaction between the R13 guanidino group (P-3 site) and the hydroxyl group of Y330 breaks apart, with an ∼90° rotation of the Arg side chain torsion angles (Fig. 5A). The latter causes the formation of new transient interactions between the guanidino group and the α and β phosphates of ATP (17) (SI Appendix, Fig. S12), with the P-3 Arg (R13) acting as a three-way switch between the small lobe, large lobe, and the ATP molecule. With the PKA-C/ complex, the transient interactions between the guanidine group, Y330, and the phosphates of ATP are lost, pushing the P-3 side chain away from the active site (Fig. 5B). The opening and closing motions of the enzyme on binding the WT and mutated substrate were analyzed using principle component analysis (PCA; Fig. 5C). The first principal component (PC1) reports on the opening and closing motion of the two lobes. When monitored with the Cα distance between S53 and G186 (dS53-G186), PC1 describes the open, intermediate, and closed conformational states of the kinase (17). The PKA-C/ complex samples an ensemble of conformations situated between the open and closed states. In contrast, the ensemble of conformations accessible to the PKA-C/ complex is closer to the open/intermediate basin. The slightly more open state found with the mutated substrate is supported by both the hydrodynamic radii from dynamic light scattering, indicating a less compact conformation of the PKA-C/ complex as well as the chemical shift trajectories (Fig. 3C). Root mean square fluctuation analysis (RMSF) was performed to see the largest change in dynamics upon R14 deletion (SI Appendix, Fig. S12). The analysis demonstrates that the glycine rich, αF-αG, and the αG-αH loops become more dynamic with the mutation, whereas the αA-β1 linker and the C terminus becomes more rigid. This analysis corroborates the HX-NOE data, and, taken together, these results suggest that the missing P-3 guanidino group changes the interactions in the vicinity of the active site, affecting the opening and closing of the kinase.
Fig. 5.
MD simulations of PKA-C in complex with and . (A) Snapshot of MD simulations showing key interactions between R14 (P-2) and R13 (P-3) of PLN1–19 and the enzyme binding site. (B) Corresponding snapshot for the PKA-C/ complex. (C) Plot of the PC1 vs. dS53-G186 for the two complexes.
Discussion
The R14 deletion mutant of PLN provides a unique opportunity to study the effects of dysfunctional substrates on kinase structural dynamics using NMR spectroscopy. In fact, structural studies on complexes between kinases and their substrates are rather sparse, because the low binding affinity prevents crystallization. As a result, most X-ray structures are obtained in the presence of small drugs or pseudosubstrate inhibitors (31). The R14 deletion is located within the kinase recognition site and eliminates the P-3 site. The P-3 site is crucial for efficient kinetics as assessed in model peptide substrates (19). In fact, the missing electrostatic interactions at the P-3 arginine changes the register of the recognition sequence, with Arg-9 shifted to the P-6 position (Fig. 1). These structural changes hinder binding to PKA-C. However, a reduction in affinity alone is insufficient to explain the sluggish kinetics as most kinase/substrate interactions are weak in nature (31). For instance, the standard PKA-C substrate Kemptide has a Kd of ∼200 μM, yet it displays kinetic parameters common to other PKA-C substrates (16, 30, 32). Although the R14 does not actively participate in the phosphorylation reaction (33), the removal of R14 in PLN has a dramatic effect on the enzyme. Not only does it reduce PLN’s binding affinity, it also affects the organization of the active site. In fact, in the complex with the WT, R13 and R14 act as a bridgehead between the small lobe (through R13/Y330) and large lobe (R13/E127 and R14/E203/Y204). The guanidino group of R13 interacts with the γ-phosphate of ATP, likely facilitating the opening and closing of the active site. Although the guanidino group of the R14 arginine residue is firmly anchored to the large lobe (Fig. 5A), the R13 arginine is more dynamic and establishes transient interactions with the acidic cluster of PKA-C (328–334), a key region for determining opening and closing of the active cleft (26). The role of these interactions in catalysis is also supported by steady-state phosphorylation kinetic measurements, which show that the PKA-CY330F mutant decreased kcat by approximately 60% and increased KM for Kemptide approximately fivefold (34). The dramatic loss of catalytic efficiency for compared with Y330F indicates the loss of P-3 site in PLN is more disruptive than simply the loss of an electrostatic bridge. In addition to a perturbation of the structural and electrostatic preorganization at the active site, the deletion of R14 disrupts the anchoring of the substrate with the small lobe, enhancing the conformational dynamics across the enzyme backbone (Fig. 4 and SI Appendix, Figs. S7 and S8). With completion of the backbone resonance assignment on the three major structural forms of PKA-C (apo, binary, and closed; Fig. 4 and SI Appendix, Fig. S3), we were able to identify several resonances that were previously unassigned and were observable with the addition of . This allowed us to identify that PKA-C bound to substrate remains dynamic, but attenuated with respect to the nucleotide bound form, reflecting an opening and closing of the enzyme (committed conformational dynamics) (14, 17). In contrast, the elimination of the interactions at the P-3 site in the PKA-C/ complex prevents the assembly of the small and large lobes, and the complex adopts a partially closed conformation with dysfunctional dynamics (Fig. 6).
Fig. 6.
Recognition model of the kinase for and . On nucleotide binding, the enzyme’s conformational ensemble shifts toward the intermediate conformation and with dynamics committed to catalysis. With, the two arginine residues of the PKA-C recognition sequence clamp together both lobes of the enzyme to produce a catalytically committed complex, whereas the PKA-C/ complex adopts a partially closed conformation with dysfunctional dynamics.
Recently, we showed that the allosteric mutant of PKA-C, Y204A (35), disrupts the hydrophobic core adjacent to the active site and removes electrostatic interactions with the P-2 arginine, desynchronizing the opening and closing motions globally. The deletion of the P-3 arginine increases the conformational dynamics extending throughout the N terminus (αC-β1-linker), the C-terminal tail (acid patch) (26), and the F-helix, which constitute the central signaling conduit of the kinase connecting the R-spine with the F-helix (36) (Fig. 4 and SI Appendix, Figs. S7 and S8). Although the lack of allosteric transmission from the active site to the C-terminal tail could be anticipated by the loss of the P-3 electrostatic interaction in the active site, the behavior of the conformational dynamics displayed by the central signaling conduit was unexpected. Therefore, this lethal mutation, along with a partial closing of the active cleft and dysfunctional motions, alters the allosteric network with concomitant decrease of the catalytic efficiency (14, 17).
Case studies on dihydrofolate reductase using artificial mutations have predicted that dysfunctional dynamics (37) or a dynamic KO (38, 39) dramatically reduce enzymatic activity and have been exemplified in other systems as well (40–45). These studies have overlooked the contribution that substrates have on promoting productive motions. The natural occurring R14 deletion in the PKA-C recognition sequence of PLN is a unique example that advocates for a central role of the substrate in the organization of the active site and to promote productive conformational fluctuations. Although these motions might not play a direct role in the chemical step (46) (i.e., phosphoryl transfer), they dictate the opening and closing of the active cleft for phosphoryl transfer and affect the overall catalytic efficiency that can lead to DCM.
Absent or reduced PLN phosphorylation levels cause SERCA to be constitutively down-regulated, leading to reduced Ca2+ uptake in the SR lumen and, ultimately, heart disease (6, 7). Recent in vitro studies demonstrated that several hereditary mutants of PLN constitute a poor substrate for PKA-C (20, 47). Specifically, hypophosphorylation of PLN has been detected for the PLNR9C mutant, where a cysteine substitution occurs at the P-7 site (47), as well as for . However, the molecular etiologies for these mutants’ lack of phosphorylation are dissimilar (21, 48). Although reduced phosphorylation of PLNR9C has been attributed to disulfide bond formation between adjacent monomers in the pentamers, rendering the phosphorylation site inaccessible to PKA-C (47), R14 deletion prevents the formation of the catalytically committed conformation. Nonetheless, in both cases, slow phosphorylation kinetics reduce calcium uptake with concomitant decline of cardiac output, leading to heart failure. Importantly, the fight or flight mechanism, which results in increased PKA-C activity and decreased SERCA inhibition, is severely impaired by the slow phosphorylation of , affecting Ca2+ transients and leading to DCM (10, 13). Our findings emphasize the importance of functional dynamics between the enzyme and substrate to achieve physiological phosphorylation levels for normal cardiac function. These results elucidate a pathway by which dysfunctional conformational dynamics of proteins may result in pathological phenotypes, giving new and exciting evidence of a structural basis of DCM linked to this specific pathway.
Materials and Methods
Sample Preparation.
Recombinant catalytic subunit of PKA was expressed in BL21 DE3 cells by the method described by Studier at 24 °C (49), and 2H/13C/15N and 2H/15N PKA-C was expressed as previously described (16). Purification of PKA-C was performed as previously described using the His6-RIIα(R213K) subunit (50), and a second purification step was performed by using the HiTrap SP cation exchange column. Peptides (//PKI5–24) were synthesized using standard Fmoc chemistry on a CEM Liberty microwave synthesizer, cleaved with Reagent K [trifluoroacetic acid/thioanisole/water/phenol/2–2′-(ethylenedioxy)diethanethiol, 82.5:5:5:5:2.5 (vol/vol)] for 3 h and purified using a semipreparative Supelco C18 reverse-phase HPLC column at 3 mL/min. Molecular weight and the quantity of the peptides were verified by amino acid analysis (Texas Tech).
ITC Measurements.
All ITC data were acquired on a VP-ITC microcalorimeter (Malvern). PKA-C, , and were prepared in 20 mM MOPS, 90 mM KCl, 10 mM DTT, 10 mM MgCl2, and 1 mM NaN3 at pH 6.5. For nucleotide saturation, 2 mM of ATPγN was added. All experiments were performed in 300 K. Data were fit to the Wiseman Isotherm (51) assuming one-site binding using the NanoAnalyze (TA instruments) software. Further details are provided in SI Appendix, SI Materials and Methods.
Enzyme Assays.
The steady-state activity assays of PLN1–19 and R14del1–19 under saturating ATP concentrations were performed spectrophotometrically at 298 K as described by Cook et al. (52). The values of Vmax and Km were obtained from a nonlinear fit of the initial velocities to the Michaelis–Menten equation. The extent of phosphorylation for full-length PLN analogs was monitored by gel-shift assays using 10 μM of substrate (assessed by densitometry), 1,000 U/mg PKA-C in 0.1% octyl-glucoside, 20 mM MOPS (pH 7.25), 0.05 mM PMSF, 0.02% NaN3, and 1 mM MgCl2. The reaction was initiated by adding 1 mM ATP and was incubated for 12 h at 30 °C. The phosphorylation reaction was then stopped by addition of 1% SDS. Tris-tricine polyacrylamide gels (12%) were run with 2 µg of PLN (∼5-µL reaction mixture) and stained with Coomassie brilliant blue.
NMR Measurements.
Standard TROSY-select triple resonance, steady-state NOE and HSQC experiments were carried out on an 850-MHz Bruker Advance III spectrometer equipped with a TCI cryoprobe. Concentrations of samples for triple resonance experiments were ∼0.4–0.7 mM, and samples for relaxation experiments were 0.25–0.4 mM; 12 mM of ATPγN was added for the nucleotide-bound form and 1.0–2.0 mM of peptide (/) for the ternary complex. Spectra were collected at 300 K, processed using NMRPipe (53), and visualized using Sparky. Rex values were measured using the TROSY Hahn-Echo pulse sequence (28) and analyzed as described previously (17, 54). Qualitative verification of Rex was done by measuring inverse peak heights as previously described (54). Further details are provided in SI Appendix, SI Materials and Methods.
MD Simulations.
MD simulations were set up using CHARMMc36 and run with the software NAMD (nanoscale molecular dynamics) using an initial docked structure as described in SI Appendix, SI Materials and Methods. All structures were solvated in a TIP3 water box with K+ and Cl− added as counter ions to reach an ionic strength of ∼150 mM. Following an initial equilibration, 80 ns of MD simulations of PKA-C/ were performed at constant temperature and pressure. PCA and RMSF analyses were performed as previously described (17). Further details are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Y. Xia for assistance on the NMR data acquisition and Prof. J. Ervasti and Prof. Geraghty for access to DLS and ITC, respectively. NMR experiments were carried out at the Minnesota NMR Center and MD calculations at the Minnesota Supercomputing Institute. This work was supported in part by National Institutes of Health Grants GM72701 (to G.V.), GM46367 (to J.G.), and T32 AR007612 (to J.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502299112/-/DCSupplemental.
References
- 1.Bers DM. Calcium cycling and signaling in cardiac myocytes. Annu Rev Physiol. 2008;70:23–49. doi: 10.1146/annurev.physiol.70.113006.100455. [DOI] [PubMed] [Google Scholar]
- 2.Lompré AM, et al. Ca2+ cycling and new therapeutic approaches for heart failure. Circulation. 2010;121(6):822–830. doi: 10.1161/CIRCULATIONAHA.109.890954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vandecaetsbeek I, Raeymaekers L, Wuytack F, Vangheluwe P. Factors controlling the activity of the SERCA2a pump in the normal and failing heart. Biofactors. 2009;35(6):484–499. doi: 10.1002/biof.63. [DOI] [PubMed] [Google Scholar]
- 4.Frank KF, Bölck B, Brixius K, Kranias EG, Schwinger RH. Modulation of SERCA: Implications for the failing human heart. Basic Res Cardiol. 2002;97(Suppl 1):I72–I78. doi: 10.1007/s003950200033. [DOI] [PubMed] [Google Scholar]
- 5.MacLennan DH, Kranias EG. Phospholamban: A crucial regulator of cardiac contractility. Nat Rev Mol Cell Biol. 2003;4(7):566–577. doi: 10.1038/nrm1151. [DOI] [PubMed] [Google Scholar]
- 6.Brittsan AG, Kranias EG. Phospholamban and cardiac contractile function. J Mol Cell Cardiol. 2000;32(12):2131–2139. doi: 10.1006/jmcc.2000.1270. [DOI] [PubMed] [Google Scholar]
- 7.MacLennan DH. Ca2+ signalling and muscle disease. Eur J Biochem. 2000;267(17):5291–5297. doi: 10.1046/j.1432-1327.2000.01566.x. [DOI] [PubMed] [Google Scholar]
- 8.Dellefave L, McNally EM. The genetics of dilated cardiomyopathy. Curr Opin Cardiol. 2010;25(3):198–204. doi: 10.1097/HCO.0b013e328337ba52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Movsesian MA. Altered cAMP-mediated signalling and its role in the pathogenesis of dilated cardiomyopathy. Cardiovasc Res. 2004;62(3):450–459. doi: 10.1016/j.cardiores.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 10.Haghighi K, et al. A mutation in the human phospholamban gene, deleting arginine 14, results in lethal, hereditary cardiomyopathy. Proc Natl Acad Sci USA. 2006;103(5):1388–1393. doi: 10.1073/pnas.0510519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Posch MG, et al. Genetic deletion of arginine 14 in phospholamban causes dilated cardiomyopathy with attenuated electrocardiographic R amplitudes. Heart Rhythm. 2009;6(4):480–486. doi: 10.1016/j.hrthm.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 12.van der Zwaag PA, et al. Phospholamban R14del mutation in patients diagnosed with dilated cardiomyopathy or arrhythmogenic right ventricular cardiomyopathy: Evidence supporting the concept of arrhythmogenic cardiomyopathy. Eur J Heart Fail. 2012;14(11):1199–1207. doi: 10.1093/eurjhf/hfs119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haghighi K, et al. The human phospholamban Arg14-deletion mutant localizes to plasma membrane and interacts with the Na/K-ATPase. J Mol Cell Cardiol. 2012;52(3):773–782. doi: 10.1016/j.yjmcc.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masterson LR, et al. Dynamics connect substrate recognition to catalysis in protein kinase A. Nat Chem Biol. 2010;6(11):821–828. doi: 10.1038/nchembio.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masterson LR, et al. cAMP-dependent protein kinase A selects the excited state of the membrane substrate phospholamban. J Mol Biol. 2011;412(2):155–164. doi: 10.1016/j.jmb.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masterson LR, Mascioni A, Traaseth NJ, Taylor SS, Veglia G. Allosteric cooperativity in protein kinase A. Proc Natl Acad Sci USA. 2008;105(2):506–511. doi: 10.1073/pnas.0709214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masterson LR, et al. Dynamically committed, uncommitted, and quenched states encoded in protein kinase A revealed by NMR spectroscopy. Proc Natl Acad Sci USA. 2011;108(17):6969–6974. doi: 10.1073/pnas.1102701108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adams JA, Taylor SS. Energetic limits of phosphotransfer in the catalytic subunit of cAMP-dependent protein kinase as measured by viscosity experiments. Biochemistry. 1992;31(36):8516–8522. doi: 10.1021/bi00151a019. [DOI] [PubMed] [Google Scholar]
- 19.Adams JA, Taylor SS. Phosphorylation of peptide substrates for the catalytic subunit of cAMP-dependent protein kinase. J Biol Chem. 1993;268(11):7747–7752. [PubMed] [Google Scholar]
- 20.Ceholski DK, Trieber CA, Holmes CF, Young HS. Lethal, hereditary mutants of phospholamban elude phosphorylation by protein kinase A. J Biol Chem. 2012;287(32):26596–26605. doi: 10.1074/jbc.M112.382713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes E, Middleton DA. Comparison of the structure and function of phospholamban and the arginine-14 deficient mutant associated with dilated cardiomyopathy. PLoS ONE. 2014;9(9):e106746. doi: 10.1371/journal.pone.0106746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pervushin K, Riek R, Wider G, Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA. 1997;94(23):12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cembran A, Kim J, Gao J, Veglia G. NMR mapping of protein conformational landscapes using coordinated behavior of chemical shifts upon ligand binding. Phys Chem Chem Phys. 2014;16(14):6508–6518. doi: 10.1039/c4cp00110a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor SS, et al. PKA: A portrait of protein kinase dynamics. Biochim Biophys Acta. 2004;1697(1-2):259–269. doi: 10.1016/j.bbapap.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 25.Palmer AG., 3rd Nmr probes of molecular dynamics: Overview and comparison with other techniques. Annu Rev Biophys Biomol Struct. 2001;30:129–155. doi: 10.1146/annurev.biophys.30.1.129. [DOI] [PubMed] [Google Scholar]
- 26.Johnson DA, Akamine P, Radzio-Andzelm E, Madhusudan M, Taylor SS. Dynamics of cAMP-dependent protein kinase. Chem Rev. 2001;101(8):2243–2270. doi: 10.1021/cr000226k. [DOI] [PubMed] [Google Scholar]
- 27.Frederick KK, Marlow MS, Valentine KG, Wand AJ. Conformational entropy in molecular recognition by proteins. Nature. 2007;448(7151):325–329. doi: 10.1038/nature05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang C, Rance M, Palmer AG., 3rd Mapping chemical exchange in proteins with MW > 50 kD. J Am Chem Soc. 2003;125(30):8968–8969. doi: 10.1021/ja035139z. [DOI] [PubMed] [Google Scholar]
- 29.Dominguez C, Boelens R, Bonvin AM. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125(7):1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 30.Moore MJ, Adams JA, Taylor SS. Structural basis for peptide binding in protein kinase A. Role of glutamic acid 203 and tyrosine 204 in the peptide-positioning loop. J Biol Chem. 2003;278(12):10613–10618. doi: 10.1074/jbc.M210807200. [DOI] [PubMed] [Google Scholar]
- 31.Endicott JA, Noble ME, Johnson LN. The structural basis for control of eukaryotic protein kinases. Annu Rev Biochem. 2012;81(1):587–613. doi: 10.1146/annurev-biochem-052410-090317. [DOI] [PubMed] [Google Scholar]
- 32.Adams JA. Kinetic and catalytic mechanisms of protein kinases. Chem Rev. 2001;101(8):2271–2290. doi: 10.1021/cr000230w. [DOI] [PubMed] [Google Scholar]
- 33.Valiev M, Yang J, Adams JA, Taylor SS, Weare JH. Phosphorylation reaction in cAPK protein kinase-free energy quantum mechanical/molecular mechanics simulations. J Phys Chem B. 2007;111(47):13455–13464. doi: 10.1021/jp074853q. [DOI] [PubMed] [Google Scholar]
- 34.Chestukhin A, et al. Functional malleability of the carboxyl-terminal tail in protein kinase A. J Biol Chem. 1996;271(17):10175–10182. doi: 10.1074/jbc.271.17.10175. [DOI] [PubMed] [Google Scholar]
- 35.Srivastava AK, et al. 2014. Synchronous opening and closing motions are essential for cAMP-dependent protein kinase A signaling. Structure 22(12):1735–1743. [Google Scholar]
- 36.Kornev AP, Taylor SS, Ten Eyck LF. A helix scaffold for the assembly of active protein kinases. Proc Natl Acad Sci USA. 2008;105(38):14377–14382. doi: 10.1073/pnas.0807988105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mauldin RV, Carroll MJ, Lee AL. Dynamic dysfunction in dihydrofolate reductase results from antifolate drug binding: Modulation of dynamics within a structural state. Structure. 2009;17(3):386–394. doi: 10.1016/j.str.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhabha G, et al. A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science. 2011;332(6026):234–238. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan Y, Cembran A, Ma S, Gao J. Connecting protein conformational dynamics with catalytic function as illustrated in dihydrofolate reductase. Biochemistry. 2013;52(12):2036–2049. doi: 10.1021/bi301559q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whittier SK, Hengge AC, Loria JP. Conformational motions regulate phosphoryl transfer in related protein tyrosine phosphatases. Science. 2013;341(6148):899–903. doi: 10.1126/science.1241735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruschak AM, Religa TL, Breuer S, Witt S, Kay LE. The proteasome antechamber maintains substrates in an unfolded state. Nature. 2010;467(7317):868–871. doi: 10.1038/nature09444. [DOI] [PubMed] [Google Scholar]
- 42.Tzeng SR, Kalodimos CG. Allosteric inhibition through suppression of transient conformational states. Nat Chem Biol. 2013;9(7):462–465. doi: 10.1038/nchembio.1250. [DOI] [PubMed] [Google Scholar]
- 43.Das R, et al. Dynamically driven ligand selectivity in cyclic nucleotide binding domains. J Biol Chem. 2009;284(35):23682–23696. doi: 10.1074/jbc.M109.011700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akimoto M, et al. Signaling through dynamic linkers as revealed by PKA. Proc Natl Acad Sci USA. 2013;110(35):14231–14236. doi: 10.1073/pnas.1312644110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eisenmesser EZ, et al. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438(7064):117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 46.Pisliakov AV, Cao J, Kamerlin SC, Warshel A. Enzyme millisecond conformational dynamics do not catalyze the chemical step. Proc Natl Acad Sci USA. 2009;106(41):17359–17364. doi: 10.1073/pnas.0909150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ha KN, et al. Lethal Arg9Cys phospholamban mutation hinders Ca2+-ATPase regulation and phosphorylation by protein kinase A. Proc Natl Acad Sci USA. 2011;108(7):2735–2740. doi: 10.1073/pnas.1013987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vostrikov VV, Soller KJ, Ha KN, Gopinath T, Veglia G. Effects of naturally occurring arginine 14 deletion on phospholamban conformational dynamics and membrane interactions. Biochim Biophys Acta. 2015;1848(1 Pt B):315–322. doi: 10.1016/j.bbamem.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41(1):207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 50.Hemmer W, McGlone M, Taylor SS. Recombinant strategies for rapid purification of catalytic subunits of cAMP-dependent protein kinase. Anal Biochem. 1997;245(2):115–122. doi: 10.1006/abio.1996.9952. [DOI] [PubMed] [Google Scholar]
- 51.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179(1):131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 52.Cook PF, Neville ME, Jr, Vrana KE, Hartl FT, Roskoski R., Jr Adenosine cyclic 3′,5′-monophosphate dependent protein kinase: Kinetic mechanism for the bovine skeletal muscle catalytic subunit. Biochemistry. 1982;21(23):5794–5799. doi: 10.1021/bi00266a011. [DOI] [PubMed] [Google Scholar]
- 53.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 54.Fenwick MK, Oswald RE. NMR spectroscopy of the ligand-binding core of ionotropic glutamate receptor 2 bound to 5-substituted willardiine partial agonists. J Mol Biol. 2008;378(3):673–685. doi: 10.1016/j.jmb.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verardi R, Shi L, Traaseth NJ, Walsh N, Veglia G. Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method. Proc Natl Acad Sci USA. 2011;108(22):9101–9106. doi: 10.1073/pnas.1016535108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.