Abstract
Developmental biology relies heavily on microscopy to image the finely-controlled cell behaviors that drive embryonic development. Most embryos are large enough that a field of view with the resolution and magnification needed to resolve single cells will not span more than a small region of the embryo. Ascidian embryos, however, are sufficiently small that they can be imaged in toto with fine subcellular detail using conventional microscopes and objectives. Unlike other model organisms with particularly small embryos, ascidians have a chordate embryonic body plan that includes a notochord, hollow dorsal neural tube, heart primordium and numerous other anatomical details conserved with the vertebrates. Here we compare the size and anatomy of ascidian embryos with those of more traditional model organisms, and relate these features to the capabilities of both conventional and exotic imaging methods. We review the emergence of Ciona and related ascidian species as model organisms for a new era of image-based developmental systems biology. We conclude by discussing some important challenges in ascidian imaging and image analysis that remain to be solved.
Keywords: in toto imaging, cell segmentation, ascidian, Ciona, morphogenesis
Introduction
Embryonic patterning and morphogenesis involve the fine spatial and temporal control of gene expression and cell behavior. Developmental biology thus depends heavily on microscopy as a core technique for studying the mechanisms of development in their full spatial and temporal contexts. Advances in developmental biology have frequently been driven by advances in imaging technologies, from the first use of high-quality compound microscopes by the embryologists of the mid to late 19th century, through the development of confocal and two-photon methods in the 1980s and 1990s (Amos and White, 2003; Denk et al., 1990; White et al., 1987), and more recently by the introduction of innovative light sheet microscopies (Huisken et al., 2004; Keller et al., 2008). All of these methods, however, are subject to fundamental constraints between the resolution and field of view of the method, and the size of the embryo being studied. This leads to a disconnect between the 'dissecting scope' view used to visualize gene expression patterns or gross phenotypes at the level of the whole embryo and the '40x' view used to visualize fine details in a subset of embryonic cells. We would argue that much is to be gained by spanning this gap.
The small, simple embryos of the invertebrate chordate Ciona and related species of ascidians present unique opportunities for imaging chordate development with the otherwise challenging combination of subcellular resolution and embryo-wide field of view. Recent molecular phylogenies indicate that it is the tunicates, including the ascidians, and not the cephalochordates that are the true sister clade to the vertebrates (Delsuc et al., 2006). All three chordate subphyla are united by a common embryonic body plan. At its most basic level, this consists of a notochord running down the center of a muscular tail just ventral to a hollow dorsal neural tube. In 1866, Kowalewski noted to some controversy that ascidian embryos had both a notochord and a hollow, dorsal neural tube, which was unexpected for this group of sessile marine filter feeders (Kowalewski, 1866). This chordate embryonic anatomy was confirmed shortly thereafter by Kuppfer and other German embryologists (Dorn, 1871).
Ascidian embryonic size and anatomy
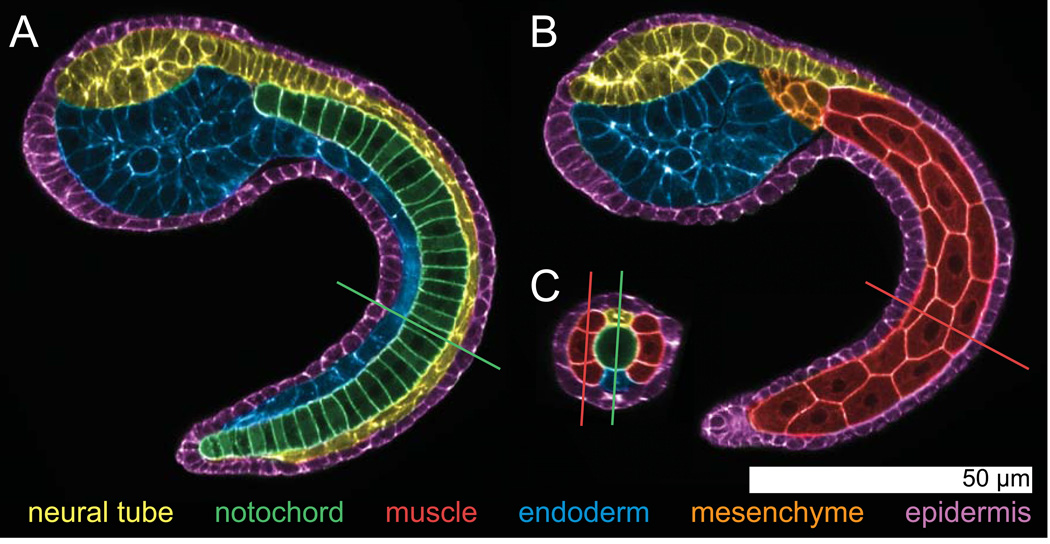
Embryos of Ciona intestinalis, the most widely studied ascidian, can be imaged in their entirety (in toto) with enough resolution to finely resolve the shapes of all embryonic cells and the arrangement of these cells into six simple tissues: epidermis, neural tube, notochord, tail muscle, mesenchyme and endoderm. Figure 1 shows images derived from a confocal stack through a tailbud embryo of Ciona, pseudocolored to highlight these main cell types.
Figure 1. Anatomy of the Ciona intestinalis mid-tailbud embryo.
Pseudocolored images from a confocal volume of a Hotta stage ~22 Ciona intestinalis embryo stained with Bodipy-FL-phallacidin to label the F-actin cytoskeleton, which is largely cortical during embryogenesis. The images were pseudocolored to highlight the 6 main tissues: epidermis, neural tube, notochord, tail muscle, mesenchyme and endoderm. The view in A has been resliced to follow the embryonic midline and shows the notochord in the center of the tail. The view in B is more superficial and shows the tail muscle cells and some mesenchymal cells. The hollow dorsal neural tube, endoderm and epidermis are evident in both A and B. C shows a cross section through the confocal volume at the position indicated by the lines in A and B. Conversely, the lines in C show the positions of the planes in A and B. The central notochord, dorsal neural tube and bilateral muscle cells are all evident in C. The confocal stack for these images was collected with a 40× 1.3NA objective in a single field of view on a standard confocal microscope (Zeiss LSM 700).
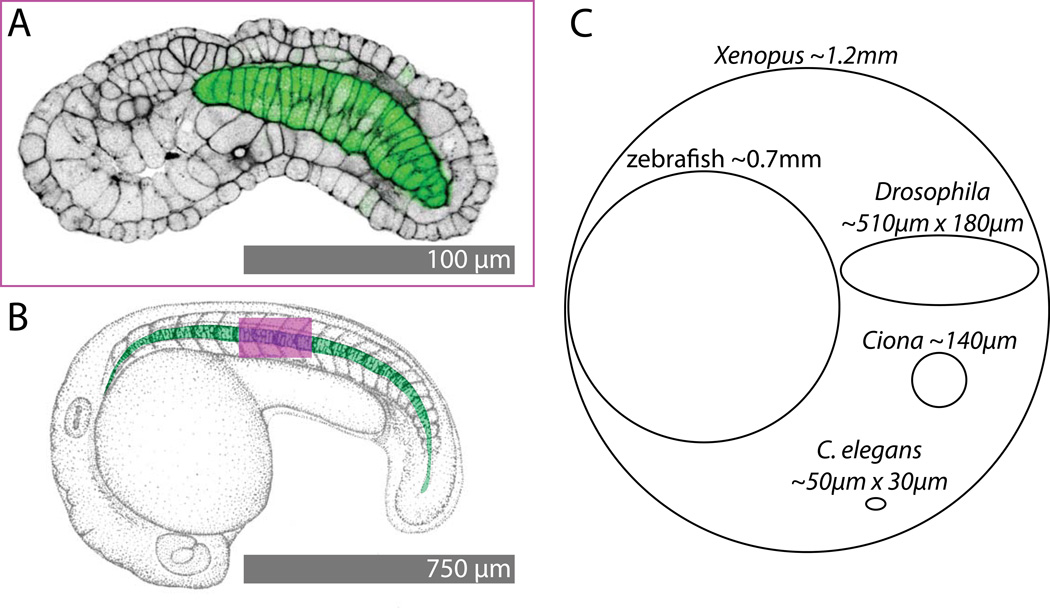
To put the size and scale of the Ciona embryo into perspective, Figure 2A,B compares the size of a Ciona embryo early in tail elongation (Fig. 2A) with a zebrafish embryo at a comparable stage. To match the size of the embryos in image space, the Ciona embryo is shown at 7.5 times the relative size of the zebrafish embryo. The field of view of the Ciona image is shown on the zebrafish as a magenta box. It is apparent that a field of view capable of spanning an entire Ciona embryo will only span the equivalent of a few somites of a zebrafish embryo. Figure 2C shows the relative sizes of Ciona eggs as compared to other model organisms. While larger than the particularly small eggs of C. elegans, Ciona eggs are much smaller than those of other important model organisms such as Xenopus, zebrafish and Drosophila (Begasse and Hyman, 2011; Brown, 2004; Gregory and Veeman, 2013; Kimmel et al., 1995; Markow et al., 2009). For example, the ~140µm Ciona egg is ~120× smaller in volume than the ~700µm zebrafish egg. This has important implications for the use of volumetric imaging methods to capture and quantify embryos in toto.
Figure 2. Size comparisons.
A) Confocal image of phallacidin staining (black, contrast-reversed to match B) and a notochord-specific GFP transgene (green) in a stage ~19 Ciona intestinalis embryo. B) Pen and ink drawing of a zebrafish embryo at a comparable stage of early tail extension (modified from (Kimmel et al., 1995)). The notochord is pseudocolored in green. The magenta box shows the field of view of the Ciona image in A superimposed on the zebrafish embryo. C shows relative egg sizes for Xenopus (Brown, 2004), zebrafish (Kimmel et al., 1995), Drosophila (Markow et al., 2009), Ciona (Gregory and Veeman, 2013) and C. elegans (Begasse and Hyman, 2011).
Despite the remarkably manageable size of the Ciona embryo, its underlying anatomy has much in common with the vertebrates. Here we will briefly review some of the key details:
Notochord
The notochord forms a long, stiff rod of mesodermally-derived cells that has structural roles, especially in species with swimming larval stages, as well as essential signaling roles in many species (Stemple, 2005). The Ciona notochord consists of only 40 cells that converge and extend into a single file column. These convergence and extension movements are driven by mediolateral intercalation and are extremely similar to the movements seen in the axial mesoderm of amphibians and other vertebrates (Jiang et al., 2005; Munro and Odell, 2002b). Unlike in vertebrates, however, the future notochord cells are not thought to act as a dorsal organizing center (Spemann/Mangold organizer) (Passamaneck and Di Gregorio, 2005). The early signaling mechanisms and gene regulatory networks involved in notochord induction have been extensively investigated for both the primary notochord lineage (anterior 32 notochord cells) and the secondary lineage (posterior 8 cells) (Hudson and Yasuo, 2006; Yagi et al., 2004; Yasuo and Satoh, 1998). After intercalation and further elongation, the notochord will later develop a hollow lumen and act as a hydrostatic skeleton in the larval tail (Dong et al., 2011; Dong et al., 2009).
Muscle
Two blocks of tail muscle cells flank the notochord on the left and right sides of the embryo. There are 18 muscle cells on each side of the embryo in Ciona (21 in Halocynthia), which form a stereotyped pattern of hexagonal and pentagonal cells but are not segmented into somites. As cephalochordates have somites, segmentation of the paraxial mesoderm is thought to have been lost in ascidian evolution. The early specification of the muscle lineages is well understood, and there is a key role for a maternal cytoplasmic determinant that has been identified as the transcription factor Macho (Nishida and Sawada, 2001). The cell movements during late gastrula/early neurula stages whereby the muscle cells become lateral to the notochord are not well understood. Later shape changes in the muscle cells are important in tail elongation and have been shown by live imaging to involve anteriorly-polarized protrusive behaviors (Passamaneck et al., 2007).
Central Nervous System
The ascidian CNS forms by a stereotypically chordate process of neurulation whereby a dorsal neural plate rolls up and sinks into the embryo. There are estimated to be fewer than 300 cells in the larval CNS, with only ~100 being neurons (Cole and Meinertzhagen, 2004; Nicol and Meinertzhagen, 1988b). There are several morphologically distinct regions in the CNS. The largest and most anterior derivative of the neural tube is the sensory vesicle, which expresses Otx (Hudson and Lemaire, 2001) and is widely accepted as being homologous to the vertebrate forebrain (Sasakura et al., 2012). Posterior to the sensory vesicle is a narrowed ‘neck’ region that has been previously homologized to the midbrain (Wada et al., 1998) but is currently thought to be homologous to the vertebrate midbrain/hindbrain boundary and hindbrain (Dufour et al., 2006). Posterior to the neck region is a region historically called the visceral ganglion that is becoming more recently and aptly known as the motor ganglion. This region has been homologized to the hindbrain (Wada et al., 1998), but there is now good evidence that it may be homologous to the vertebrate spinal cord (Dufour et al., 2006). The most posterior derivative of the neural tube is the caudal nerve cord running the length of the tail. This region is anatomically reminiscent of a spinal cord, but consists entirely of ependymal cells. The axons innervating the tail muscle cells instead come from the motor ganglion.
Heart
The ascidian heart develops after metamorphosis from progenitor cells that underwent active migration during embryonic stages from their origin in the anterior tail to the ventral trunk region (Christiaen et al., 2008; Davidson et al., 2005). The juvenile/adult heart is a simple valveless tube that periodically reverses its direction of pumping. The early induction and migration of the heart precursor cells appears to involve similar gene regulatory network modules as in vertebrates, and the study of these events in Ciona has led to new hypotheses about the evolutionary origins of the vertebrate primary and secondary heart fields (Stolfi et al., 2010; Tolkin and Christiaen, 2012).
Placodes and Neural Crest
Both cranial ectodermal placodes and neural crest have typically been thought of as vertebrate innovations, but there is now considerable evidence that ascidians definitively have placodes and some evidence that they may have crest-like cells. The best-established placode-like regions are the bilaterally paired epidermal thickenings that give rise to the atrial (excurrent) siphon (Kourakis et al., 2010; Kourakis and Smith, 2007; Manni et al., 2004; Mazet et al., 2005). These form near the motor ganglia and express Pax2/5/8, suggesting that they are homologous to the vertebrate otic placodes/lateral line placodes. Several structures derived from putative ascidian placodes develop sensory neurons, albeit often with different functions than the placode-derived sensory neurons in vertebrates (Manni et al., 2004). The progenitors of certain pigmented cells in the adult body wall have been proposed to have crest-like properties and express many neural crest markers, but these cells have been shown to be mesodermal in origin (Jeffery et al., 2008; Jeffery et al., 2004). More recently the progenitors of the pigmented cells in two sensory organs inside the lumen of the sensory vesicle have been proposed to be crest-like (Abitua et al., 2012). These cells express many neural crest marker genes and are derived from the lateral edges of the neural plate. Whether these sensory organ pigment cells are truly crest-like or simply share ancestry with vertebrate crest remains, however, quite controversial.
Historical Context
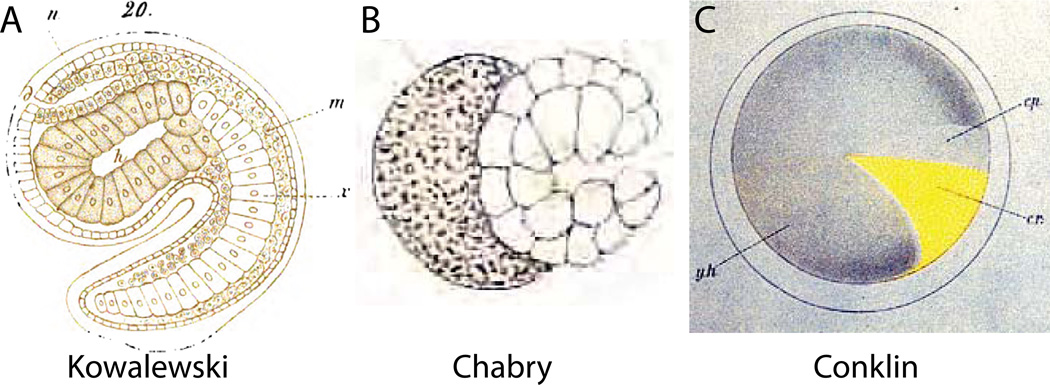
While modern developmental biologists are only beginning to exploit the ascidian embryo for explicitly quantitative, in toto approaches to morphogenesis, classical embryologists took advantage of the ability to visualize cellular detail across entire ascidian embryos. This can be seen in Kowalewski’s first drawings of the ascidian notochord and neural tube (Fig. 3A) from 1866 (Kowalewski, 1866). Some of the details are incorrect, such as the total number of notochord cells, but the overall image is remarkably accurate.
Figure 3. Historical approaches.
A) Hand-drawn image of a Ciona mid-tailbud embryo by Kowaleski (Kowalewski, 1866). The image shows 22 notochord cells instead of 40, and presents an overly regularized view of cell shape in the sensory vesicle and trunk endoderm, but the overall impression of the embryo’s chordate body plan is remarkably true to life. B) Hand-drawn image by Chabry of relatively normal gastrulation in an Ascidiella aspersa embryo where one of the two blastomeres had been killed with a fine needle at the two-cell stage (Chabry, 1887). C) Hand-drawn image by Conklin of the distribution of the yellow crescent material in the Styela egg that plays a key role in determining muscle cell fate (Conklin, 1905a).
Shortly thereafter Laurent Chabry used the ascidian species currently known as Ascidiella aspersa to perform some of the first ever blastomere isolation and ablation experiments (Chabry, 1887; Fischer, 1991). While overshadowed by the better-known sea urchin experiments of Roux and Driesch in 1888 and 1891, Chabry’s 1887 publication was arguably the first truly experimental embryology. Like Kowalewsi, Chabry detailed his findings with both cellular resolution and an embryo-wide field of view (Fig 3B).
Around the turn of the 19th century, Conklin traced the movement and inheritance of a yellow pigment in the eggs and embryos of the ascidian Styela clava (Conklin, 1905a, 1905b). He showed that the yellow pigment correlated perfectly with the segregation of muscle fate, and was instrumental in developing the theory of cytoplasmic determinants of morphogenesis. Conklin’s work again depended on the ability to easily visualize entire ascidian embryos with fine cellular detail (Fig. 3C).
Modern Era
Ascidian developmental biology languished for much of the twentieth century but experienced a tremendous resurgence beginning in the 1980s. Key technical breakthroughs include the full lineage analysis and fatemapping of the early embryo (Nishida, 1987; Nishida and Satoh, 1983, 1985), the sequencing of multiple ascidian genomes (Dehal et al., 2002; Small et al., 2007), the development of robust and straightforward transgenesis methods (Corbo et al., 1997b; Deschet et al., 2003; Sasakura, 2007; Zeller et al., 2006a), and the elucidation of highly detailed early gene regulatory networks (Imai et al., 2006). The early lineages in Ciona and other ascidian species are stereotyped and well understood. The Ciona genome is small (~160Mb) and appears to have diverged before the vertebrate genome duplications. Transient transgenesis by electroporation of fertilized eggs is straightforward, efficient and routine. The gene regulatory network circuitry underlying early patterning and fate specification is arguably better understood than in any other chordate.
In terms of modern, image-based approaches to ascidian developmental biology, there are two pioneering sets of papers that deserve special mention. The first are the early efforts from Ian Meinertzhagen’s group using scanning EM, histological sections and confocal microscopy to decipher cell lineages in the neural plate and early neural tube (Cole and Meinertzhagen, 2004; Nicol and Meinertzhagen, 1988a, 1988b), (Fig 4A,B). These were among the first papers to exploit the small size of the Ciona embryo to begin to characterize an entire differentiating organ, the brain no less, with cellular detail.
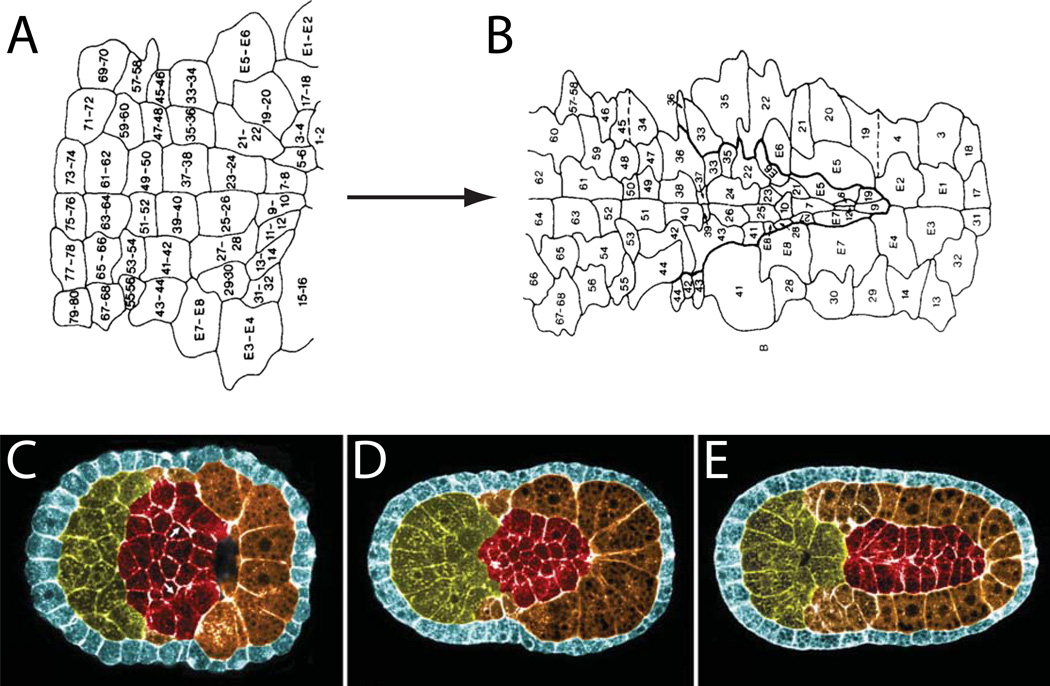
Figure 4. Early in toto approaches to neural plate and notochord development.
A,B) An early fate map of the Ciona neural plate through to early neural tube closure derived from histological sectioning and scanning EM of closely spaced timepoints of fixed embryos (Nicol and Meinertzhagen, 1988b). The blastomere numbering system used in these images is now obsolete, but the fundamental insight that the Ciona neural tube could be fate mapped with single cell resolution remains powerful. C,D,E) Pseudocolored confocal images of successive stages of notochord morphogenesis in Boltenia villosa (Munro and Odell, 2002b). These were among the first images to demonstrate how the entire ascidian embryo could be imaged by confocal microscopy to visualize fine details of cell and tissue morphology.
Particular consideration also has to be given to two back-to-back papers by Ed Munro working in Gary Odell’s group at the Friday Harbor Labs. The first paper showed unequivocally that the ascidian notochord converges and extends by mediolaterally-biased intercalation, as had previously been demonstrated only in amphibian embryos (Munro and Odell, 2002b). The second paper took a bold and technically challenging microsurgical approach to show that notochord intercalation requires contact with adjacent muscle cells or neural cells, but not both (Munro and Odell, 2002a). These papers were particularly striking because they were the first to apply confocal imaging to ascidian embryos to produce crisp, insightful images of cell phenotypes and behaviors that had previously been obscured in the murky depths of the embryo (Fig. 4C–E). The labeling protocols developed in these papers, in particular the use of a brisk isopropanol dehydration series to make phalloidin staining of cell cortices compatible with clearing in Murray’s Clear (Benzyl Alcohol/Benzyl Benzoate), are now in widespread use throughout the ascidian community.
Current approaches
3D Virtual Embryos of early cleavage stages
The first effort to systematically and quantitatively analyze cell shapes and sizes across entire ascidian embryos came from the Lemaire lab in 2006 (Tassy et al., 2006). They developed a software framework for manually segmenting cells in confocal or two-photon image volumes and applied it to 19 Ciona and Halocynthia embryos during early cleavage stages (2–44 cells). In this context, segmentation is used not in its normal biological sense but as a term from the computer vision community that refers to identifying which pixels in an image belong to which objects. Manual segmentation of embryonic cells typically involves drawing lines around every cell in every frame of the confocal stack. Often orthogonal views are used to improve the segmentation of the tops and bottoms of cells. Manual segmentation is highly effective but also extremely labor intensive.
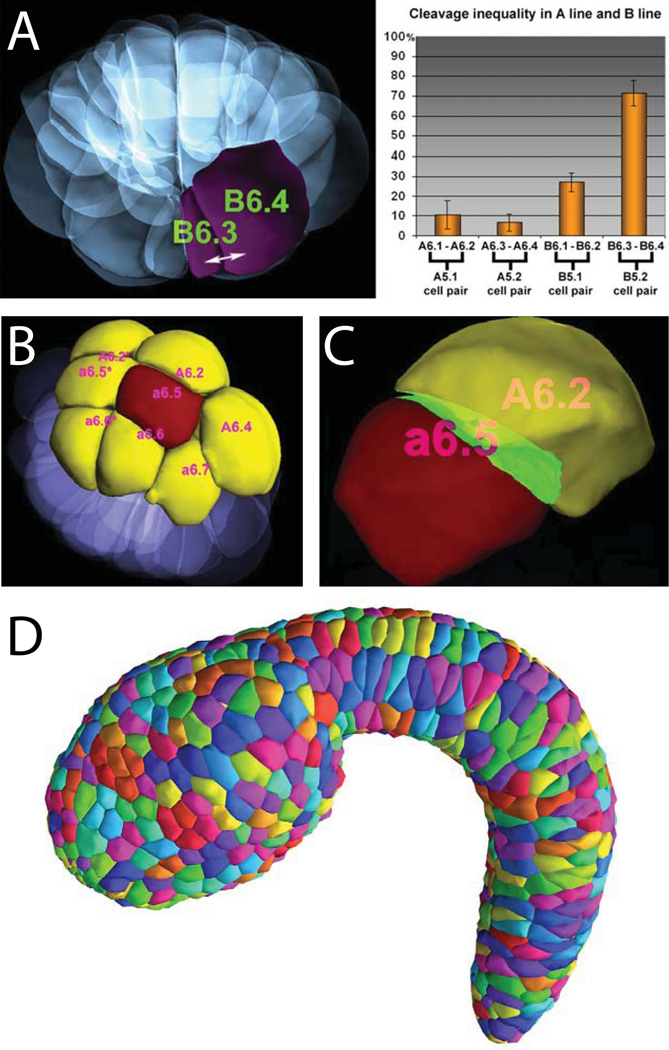
Once a cell a cell has been segmented, then many biologically-meaningful shape parameters can be calculated. These include cell volume, surface area, and various measurements of cell elongation and asymmetry. When entire tissues or embryos have been segmented, then cell-cell contact areas can be calculated as well as measurements of the alignment of cells with each other and with the axes of the embryo. Tassy et al developed an interactive 3D viewer (3D Virtual Embryo) for segmented embryos that incorporates the ability to browse several cell shape and contact parameters and is deeply integrated with cell lineage information. This excellent resource has been incorporated into the ANISEED ascidian community database project (Tassy et al., 2010). The 3D Virtual Embryo project provided important software infrastructure and a vision for the use of Ciona in quantitative, in toto studies of morphogenesis, but also led to important biological insights. In particular, they identified several previously unknown asymmetric cell divisions (Fig. 5A) that correlated with specification of cell fates, and also identified several dynamic areas of cell-cell contact (Fig. 5B,C) that correlated with inductive events (Tassy et al., 2006).
Figure 5. Quantitative approaches to cell shape and tissue architecture in toto.
A–C are from (Tassy et al., 2006)). A) Computationally reconstructed cell shapes for an entire 32-cell stage Ciona embryo. One of the large B6.4 blastomeres and its much smaller B6.3 sibling are highlighted. The chart shows sibling cell volume asymmetries for several sib pairs. B) Rendered view of a 32-cell stage embryo highlighting the a6.5 blastomere in red and all of its immediate neighbors in yellow. C) a6.5 (red) shown next to its neighbor A6.2 (yellow) with touching surface areas highlighted in green. D) Epidermal cells segmented in toto in a mid-tailbud stage Ciona embryo (Nakamura et al., 2012).
3D Virtual Mid-tailbud Embryos
Kohji Hotta and his research group and collaborators have contributed two valuable image-based resources to the Ciona community. The first is the FABA (Four-dimensional Ascidian Body Atlas) staging series (Hotta et al., 2007). The FABA consists of confocal stacks through entire phalloidin stained Ciona embryos at 26 timepoints from the fertilized egg through the hatching larval stage. This has been a tremendous resource for the ascidian community in terms of defining a common reference for staging embryos. The staging series can be appreciated in manuscript form but also as an interactive webpage where the original confocal stacks can be browsed. The second resource is the 3D Mid-Tailbud Virtual Embryo (3DMTVE) (Nakamura et al., 2012). Nakamura and colleagues used the manual segmentation tools developed by Tassy et al to segment two entire mid-tailbud stage embryos (Fig. 5D). Embryos of this stage have approximately 1600 cells, many of which are small or irregularly shaped, making this much more challenging than the far larger cells of the early cleavage embryos segmented by Tassy et al. Manual segmentation on this scale was a yeoman task that required many person-months of effort for each embryo analyzed. Key findings included the identification of several previously undescribed cell types in the tail and the most accurate estimations to date of cell numbers in the embryo’s more complex tissues.
Endoderm invagination
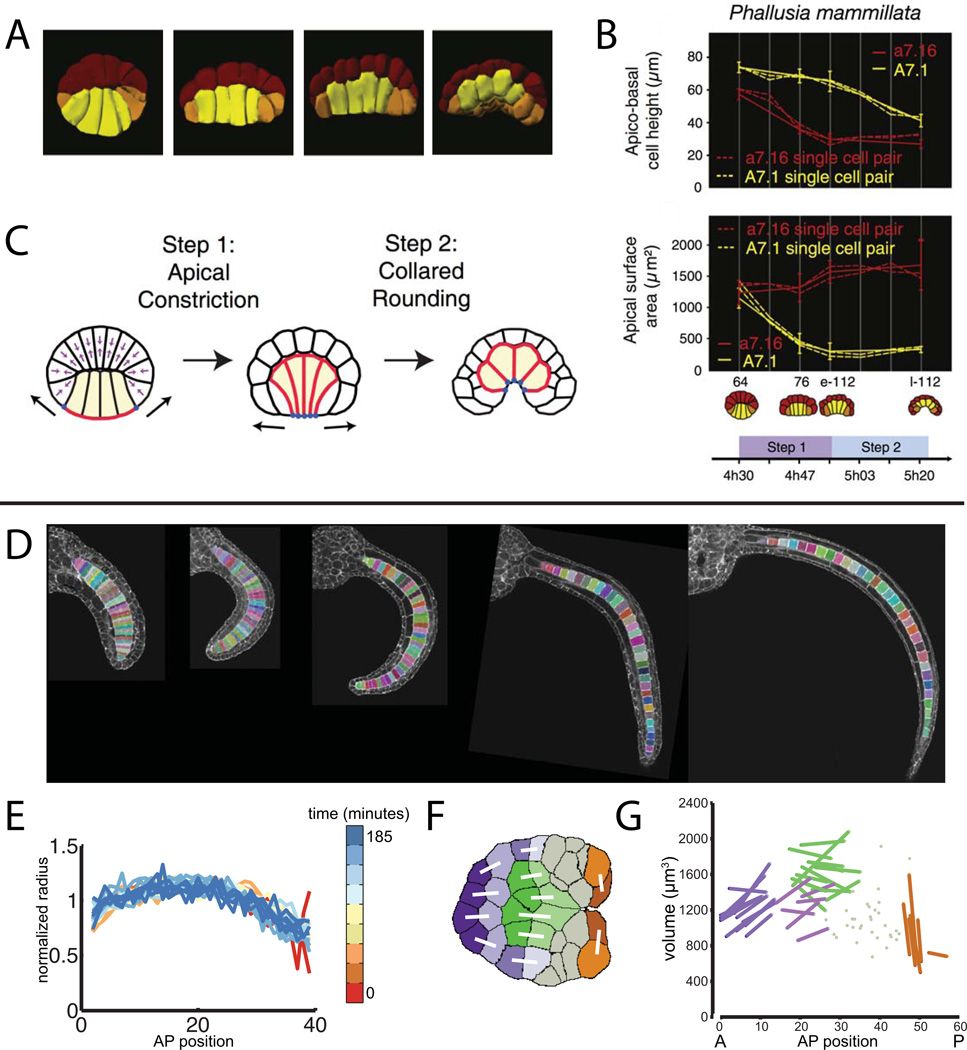
While systematic approaches to analyze cell shapes and tissue relationships in toto remain sufficiently new that descriptive studies are of great importance, these approaches become particularly powerful when integrated into hypothesis-driven, experimental studies. Sherrard et al used in toto imaging and quantitative analysis to great effect in their recent study of the cell shape changes in the endoderm that drive early Ciona gastrulation (Sherrard et al., 2011). They used the 3D Virtual Embryo framework to segment endoderm cells at progressive stages of gastrulation and were able to quantitatively define two distinct phases of localized contractile behavior. The first phase is dominated by apical constriction and drives only the most initial flattening of the embryo, whereas the majority of endoderm invagination is driven by a subsequent phase of ‘collared rounding” where contraction along the apical-basal axis predominates (Fig. 6A–C). Their quantitative in toto imaging approach was integrated with pharmacology, dominant-negative constructs, immunocytochemistry, laser ablation and computational modeling of cell dynamics in what can truly be called developmental systems biology.
Figure 6. Integrative uses of quantitative in toto imaging to study endoderm invagination and notochord tapering.
A–C are from (Sherrard et al., 2011). A) Renderings of segmented embryos at successive time points during early gastrulation showing cell shape changes in the endoderm cells (yellow). B) Differential timing and extent of apical constriction and apical-basal shortening in endoderm cells (yellow) versus epidermal cells (red). C) Cartoon model of how early gastrulation is driven by a two step process involving first apical constriction and then subsequent apical-basal shortening. D-G are from (Veeman and Smith, 2012). D) Midvolume planes from confocal volumes of successive stages of notochord elongation. Segmented notochord cells are indicated with random pseudocolors. E) Normalized radius as a function of AP position for several stages of notochord elongation. Different timepoints (minutes after the end of notochord intercalation) are indicated with pseudocolors. F) Midvolume slice through a representative segmented Ciona notochord at the onset of mediolateral intercalation. Identified sibling pairs are connected with white lines. G) Notochord cell volumes as a function of AP position for three embryos segmented at the stage shown in F. Known sibling cells are connected by lines and colored according to the map in F.
Notochord Taper
Veeman and Smith (author MV of this review) took a similarly integrative approach in examining the cellular mechanisms shaping the Ciona notochord (Veeman and Smith, 2012). We imaged a large number of embryos at closely spaced timepoints after notochord intercalation but before lumen formation. We developed semi-automated computational tools to ease the task of segmenting all of the notochord cells in these images (Fig. 6D). It became apparent early in our analysis that cell shape in the Ciona notochord depends strongly on anterior-posterior position (Fig. 6E). Cells in the middle are widest, and the notochord tapers in a very predictable way towards narrower anterior and posterior ends. This notochord taper is seen in many chordates and is obvious in retrospect but was not remarkable until quantified. We used further quantitative imaging experiments in wildtype and mutant Ciona together with microsurgical experiments and simple mathematical modeling to identify two major drivers of the notochord’s tapered shape. The first is the ends-to-middle progression by which Ciona notochord cells intercalate, which gives cells at the ends a ‘head start’ on a subsequent cell shape change that makes them mediolaterally narrower. The second factor is cell volume, which is highest in the middle of the notochord and lowest at the ends. We identified several divisions in the notochord lineage as being asymmetric in ways that would contribute to the observed patterns of cell volume (Fig. 6F,G). Whether asymmetric division is sufficient to account for the overall patterns of cell volume variation is currently being investigated in MV’s lab.
Integrative approaches
While our emphasis here has been on studies that are simultaneously quantitative and in toto in their approach to morphogenesis, there are many other groups who take integrative approaches towards understanding patterning and morphogenesis with single-cell resolution. One important example is the ongoing elucidation of the gene regulatory networks and effector suites controlling distinct migratory and morphogenetic behaviors in the trunk ventral cell-derived heart muscle precursors and atrial siphon muscle precursors (Christiaen et al., 2008; Cooley et al., 2011; Davidson et al., 2005; Norton et al., 2013; Stolfi et al., 2010). A recent study in this context from the Christiaen lab provides an excellent example of how gain and loss of function studies can be integrated with high-resolution imaging (Razy-Krajka et al., 2014), in this case to demonstrate an important but complex role for the transcription factor COE in controlling cell fate in bipotential cardiopharyngeal precursors.
Another great example of the integration of imaging and functional approaches comes from a recent study of notochord cell elongation. Sehring and colleagues combined quantitative imaging, modeling methods and perturbative experiments to show that the formation of a circumferential actomyosin belt important for cell elongation involves equatorial cortical flows of actomyosin reminiscent of the contractile ring in cytokinesis (Sehring et al., 2014). Their data also suggests a role for dynamic blebbiform deformations of the perinotochordal surfaces in this elongation process. These blebbiform movements had been described before in Ascidiella aspersa, but their role had previously only been speculated upon (Veeman et al., 2008).
Also worthy of note in this context is the ongoing work from multiple labs to understand the induction and patterning of the neural plate/neural tube with single-cell detail (Cole and Meinertzhagen, 2004; Corbo et al., 1997a; Dufour et al., 2006; Hudson and Lemaire, 2001; Hudson et al., 2007; Hudson and Yasuo, 2005; Mita and Fujiwara, 2007; Ogura et al., 2011; Ohta and Satou, 2013; Rothbacher et al., 2007; Stolfi et al., 2011; Wada et al., 1998; Wagner and Levine, 2012). There is potentially much to be gained from integrating transcriptomics, GRN analysis and functional genomics with quantitative in toto imaging methods and modeling.
Current challenges
Embryonic cell segmentation
While it is relatively straightforward to acquire high quality confocal stacks through entire fixed, stained ascidian embryos using standard confocal microscopes, there remain major technical challenges restricting the widespread use of quantitative in toto methods. One is the difficulty in achieving high quality segmentation for large datasets. Manual segmentation is surprisingly laborious for more than a modest number of cells, and does not scale well to older stages with larger numbers of cells or to experiments with many replicates, conditions and stages. Fully automated computational tools for embryonic cell segmentation are being actively developed by image analysis experts, but are not yet ready for widespread use by non-specialists.
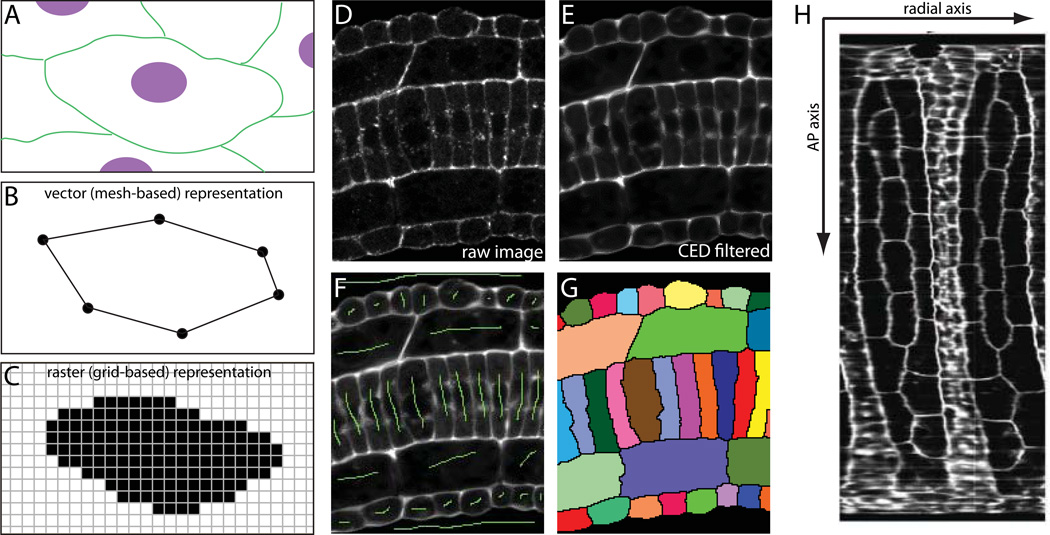
There are two different mathematical representations that can be used for segmented cells derived from volumetric imaging methods such as 3D confocal microscopy (Fig 7A–C). One of these is to represent each cell as a polygonal mesh defined by the location of specific nodes and the edges connecting those nodes (Fig. 7B). The other is to represent each cell as a grid of voxels (a voxel is the 3D equivalent of a pixel) belonging to that particular cell (Fig. 7C). Both the vector (polygonal mesh) and raster (pixel/voxel) representations have their own pros and cons, and cell segmentation data can be transformed from one format to the other with relative ease. The 3D Virtual Embryo framework uses polygonal meshes, which are well suited to surface rendering of cell shapes in a web-based context. Many image analysis pipelines are simpler, however, using grid-based data. Both vector and raster representations can be stored in very compact files and both are subject to certain artifacts depending on the resolution of the image and the segmentation.
Figure 7. Embryonic cell segmentation.
A) Cartoon view of hypothetical cell membranes (green) and nuclei (magenta). B) Cell shapes can be represented in a vector format by lists of points connected by defined edges. C) Cell shapes can also be represented in a raster format by masks of pixels/voxels belonging to each cell. D) Raw confocal image of a section of Ciona tail. E) Preprocessing by membrane-enhancing CED filter. F) Hand-drawn seeds (green) for marker-assisted watershed segmentation. G) Watershed output with segmented cells labeled with random pseudocolors. H) Digitally ‘skinned’ embryo with the muscle/neural/endodermal strand cell layer flattened out to show the two blocks of muscle cells (large hexagonal/pentagonal cells) with neural cells between them (Abdollahian et al., 2011). One axis of this projection follows a curvilinear path along the AP axis. The other represents a radial axis around the embryonic mediolateral/dorsoventral midline.
A common strategy for automated cell segmentation involves imaging cell nuclei in one channel and cell membranes or cortices in another. The nuclear channel can be used to identify a single seed point inside every cell that can be used to initialize some form of propagating front that expands outwards in a way that is modulated by the membrane channel. The cell boundaries are identified as being the places where the propagating fronts from different seeds meet. The simplest method for this form of seeded segmentation is the watershed transform (Vincent and Soille, 1991), which is straightforward to implement but lacks any regularization parameters and is consequently prone to leak through weak points or gaps in the membrane labeling. There is also considerable interest in membrane segmentation methods that do not depend on having a seed point in every cell (Delibaltov et al., 2011; Mosaliganti et al., 2012). Level-set formulations, region merging, graph cuts, parametric active contours, tensor voting and other methods all provide more sophisticated frameworks for cell segmentation, but there is not currently a dominant method emerging (Delibaltov et al., 2011; Dufour et al., 2005; Luengo-Oroz et al., 2012b; Mosaliganti et al., 2012; Obara et al., 2011; Olivier et al., 2010; Zanella et al., 2009).
While the development of high-quality fully automated segmentation methods for Ciona embryonic cells is foreseeable in the moderately near future, the question remains of how best to analyze large numbers of embryonic cells today. One strategy is to crowdsource the problem by involving large numbers of undergraduate researchers or citizen scientists. This approach has been used to good effect in certain large-scale biological image analysis problems (Luengo-Oroz et al., 2012a; Mitry et al., 2013), but high quality manual segmentation of embryonic cells is quite skilled work and requires considerable training in embryonic anatomy. Another strategy is to use semi-automated methods that combine interactive human input with computational assistance. In our previous study of notochord taper, for example, we combined interactively generated masks with marker-assisted watershed segmentation to achieve high quality segmentation of more than 50 notochords (~2000 cells) in a reasonable amount of time (Veeman and Smith, 2012).
There are several steps to this interactive segmentation process. We typically preprocess the phalloidin channel with a Coherence Enhancing Diffusion (CED) filter (Weickert, 1999) tuned to enhance membrane features (Fig. 7D,E). We use a Matlab implementation of a generic CED filter with minor changes to enhance planar as opposed to filamentous features (Kroon and Slump, 2009). This step is optional, but usually gives a cleaner segmentation. We then use ImageJ and/or Matlab to draw a 3D mask image (Fig. 7F). This involves making a mark in each cell and building a crude mask around the cells being segmented. The mark in each cell can be a single point, but a line spanning the nucleus helps to avoid artifacts related to perinuclear actin staining. The crude outer mask needs to intersect all of the cells flanking the cells of interest, but does not need to be particularly precise. For segmenting intercalated notochords, we have written Matlab scripts to interactively generate outer masks by using spline interpolation between manually selected points to approximate curvilinear paths of varying radius. These outer masks can also be drawn by hand by using ImageJ to draw a line in a new channel through the middle of the cells flanking the cells of interest in multiple Z planes. This can be repeated along the XZ and/or YZ axes taking care to connect the mask lines in different Z planes. This builds up a gridwork of lines in 3D that are all connected and pass through all of the cells flanking the cells of interest. The lines do not need to be drawn in every plane and do not need to be particularly close to the true outer boundary of the cells of interest, so this is far less laborious than manual segmentation. The next step is to combine the inner markers and outer mask into a single image and use it to seed the marker-assisted watershed transform. We implement this using the Impose Regional Minima (imimposemin) and Watershed (watershed) functions in the Matlab Image Analysis Toolbox (Fig. 7G). The final step is to inspect the resulting segmentation and then iteratively refine the mask/marker image to correct any defects in segmentation. The advantage of this method is that a high quality segmentation can almost always be generated in a fraction of the time needed for a comparable manual segmentation.
While computational methods are valuable for cell segmentation, they also enable powerful new visualization methods. Commercial software such as Imaris (Bitplane AG) can be used for interactive volume rendering where image volumes can be rotated and resliced along arbitrary axes. Open source software such as ImageJ/FIJI also has increasingly powerful tools for exploring multidimensional image data (Schindelin et al., 2012; Schneider et al., 2012). There are also many interesting possibilities for using computational methods to reconstruct views of developing embryos that better represent the axes of the embryo instead of the X, Y and Z axes of the microscope. Fig. 7H, for example shows computationally ‘skinned’ embryos where the epidermal and muscle cell layers have been digitally removed and flattened out (Abdollahian et al., 2011).
Live Imaging
Another major challenge for the ascidian community lies in live imaging. Subject to certain limitations, extensive live imaging is feasible in Ciona (Cooley et al., 2011; Miyamoto and Crowther, 1985; Ogura et al., 2011; Passamaneck et al., 2007; Rhee et al., 2005; Veeman et al., 2008; Zeller et al., 2006b). Timelapse live imaging methods have been used to visualize polarized and dynamic cell behaviors in Ciona muscle cells (Passamaneck et al., 2007), neural cells (Hackley et al.; Ogura et al., 2011; Veeman et al., 2010), heart precursor cells (Cooley et al., 2011; Norton et al., 2013), notochord cells (Miyamoto and Crowther, 1985; Sehring et al., 2014; Veeman et al., 2008) and others. The best live imaging in Ciona has been of relatively superficial cells, or of deeper cells such as notochord at later stages when the embryo is less thick. These studies have been aided by the relative ease of introducing fluorescent protein constructs under the control of tissue specific enhancers (Rhee et al., 2005; Zeller et al., 2006b). Live imaging approaches in Ciona are distinctly limited, however, by the moderate opacity and light-scattering properties of the embryo. Other ascidian species such as Phallusia mammillata and Ascidiella aspersa have crystal-clear embryos that are much better suited to live imaging. Phallusia in particular is becoming increasingly widely used for live imaging approaches to ascidian cell and developmental biology (McDougall et al., 2014; Negishi et al., 2013; Robin et al., 2011). Modern use of Phallusia has largely been pioneered by scientists studying meiosis, fertilization, cortical polarity and asymmetric cell division at the Observatoire Oceanologique, Villefranche-sur-mer, France (McDougall and Sardet, 1995; Paix et al., 2009; Prodon et al., 2010; Prodon et al., 2005; Sardet et al., 1989; Speksnijder et al., 1989a; Speksnijder et al., 1989b). A recent publication of particular relevance from Alex McDougall’s group used extensive timelapse imaging to quantify cell cycle lengths throughout the entire early embryo (Dumollard et al., 2013). They found a key role for beta-catenin signaling in controlling developmentally important changes in cell cycle timing during endodermal cell fate specification.
Another review in this special issue discusses the current state of the art for in toto live imaging of Phallusia by SPIM and two-photon microscopy. There have been impressive recent advances in developing SPIM and related types of fluorescent microscopes with orthogonal illumination and detection axes for live imaging of developing embryos (Hockendorf et al., 2012; Huisken et al., 2004; Keller et al., 2008; Khairy and Keller, 2010). These are being used, for example, to track cell lineages in toto in developing zebrafish embryos (Keller et al., 2008; Khairy and Keller, 2010). It should be noted, however, that the resolution needed to identify nuclei and track cell divisions is much less than the resolution needed to finely resolve the subtleties of cell shape and morphogenetic cell behaviors. Light sheet methods provide advantages in imaging speed, in long-term imaging with minimal bleaching and phototoxicity, and in multiview reconstructions for deep imaging with excellent axial resolution, but the ultimate constraints imposed by the size of the embryo and the field of view of the objective remain. It is likely that an ascidian will be the first chordate to allow a detailed quantitative analysis of embryo-wide cell shape, behavior and tissue architecture based on in toto live imaging.
Integrative challenges for the future
Gene expression data
In toto imaging and quantitative analysis is perhaps best thought of as a powerful phenotyping tool. What is the range of normal cell shapes and behaviors in wild-type embryos? How do they vary in different tissues and across the axes of the embryo? How do they vary in experimentally perturbed embryos? As such, these methods are at their most powerful when deeply integrated with all of the other tools of modern developmental biology. One area of potentially valuable integration is with large-scale gene expression studies. Several high throughput in situ hybridization projects have been carried out in Ciona, which have been tremendously valuable for the community (Hotta et al., 1999; Kusakabe et al., 2002; Ogasawara et al., 2002; Satou et al., 2001), but the typical output is large numbers of low-resolution widefield images. Single cell resolution is sometimes achieved, but only for the earliest stages. Similar screens could be carried out using regular or single molecule fluorescent in situ hybridization imaged by confocal to allow quantitative analysis of in toto patterns of gene expression with single cell resolution. This would be a major undertaking, but is far more feasible in ascidians than in any other chordate.
From the single-cell level to the subcellular
We have emphasized in this review the use of quantitative in toto methods with respect to issues of cell size, shape and tissue architecture, but these methods can also be extended to address the distributions of labeled proteins and other molecules within individual cells. Many ascidian research groups are exploring issues of subcellular localization in contexts as diverse as PAR proteins in early asymmetric divisions (Patalano et al., 2006), PCP proteins in notochord morphogenesis (Jiang et al., 2005) and Talin foci in polarized heart precursor cells (Norton et al., 2013). While these approaches are becoming increasingly quantitative, especially this latter study that made use of extensive cell segmentation methods, they are still often quite narrow in scope. It would be very exciting, for example, to see in toto quantitative analyses of the subcellular localization of small GTPases and other cytoskeletal regulators on the scale of the entire embryo. Subcellular localization can be analyzed in terms of the axes of the embryo, the axes of individual tissues and organs, and the axes of individual anisotropic cells. Subcellular localization can also be analyzed in terms of distinct cell-cell contacts between different cells and tissues. In toto cell segmentation of entire tissues, organs or ideally whole embryos provides an important framework for analyzing gene expression, organelles, tagged protein distributions, or any other feature that can be mapped onto an embryo.
High-throughput phenotyping
Another promising application of these methods lies in applying quantitative in toto imaging to large-scale functional screens. Forward genetic screens (Veeman et al., 2011) and targeted morpholino screens (Hamada et al., 2007; Yamada et al., 2003) have both been performed in Ciona, and emerging genome editing tools (Treen et al., 2014) are likely to make large-scale loss of function projects increasingly feasible. Automated or semiautomated methods for the quantitative phenotyping of perturbed embryonic anatomy would be tremendously valuable in these contexts (Truong and Supatto, 2011). Reliable segmentation methods now exist for the automated segmentation and analysis of massive image sets of cultured cell monolayers grown in multiwall plates and imaged by robotic microscopes (Jones et al., 2008). These methods have been revolutionary in developing new quantitative, image-based approaches to cell biology (Thomas, 2009), and comparable tools will prove similarly powerful when adapted to the more challenging analysis of developing embryos.
Computational modeling
Lastly, the quantitative analysis of in toto images of embryonic development is also fundamental to developing computational models of morphogenesis. Complex processes such as gastrulation, neurulation and mediolateral intercalation ultimately require modeling approaches that integrate knowledge about gene regulatory network circuitry, cell signaling, adhesion, cytoskeletal dynamics and embryonic biophysics (Rizzi and Peyrieras, 2013; Roeder et al., 2011; Trier and Davidson, 2011). Digital 3D or 4D representations of developing embryos provide the natural place to integrate these other layers of information. Quantitative analysis of embryonic details provides real-world data for initializing models of subsequent development. It also provides ‘ground truth’ data for comparing against model predictions. All of the advantages of small size and simple embryonic architecture that make ascidian embryos well suited to in toto imaging also make them well suited to computational modeling approaches.
Conclusion
Developmental biology is intimately concerned with the spatial and temporal control of the molecular and cellular events driving patterning, morphogenesis and differentiation. This puts an emphasis on microscopy and imaging for resolving these spatial and temporal details. Developmental biology is also inherently multi-scale, with functional relationships propagating between many levels of biological organization. Ascidians have conserved chordate embryonic anatomy but their embryos are particularly small and simple, such that development can be imaged in toto at near the limits of conventional optical microscopy. This allows spatial details to be simultaneously resolved from the organismal level of organization down to below the cellular level. The integration of quantitative analysis methods with in toto imaging has already proven valuable in addressing diverse questions in ascidian morphogenesis. While technical challenges remain, there is a bright future for combining the uniquely tractable features of the ascidian embryo with modern imaging and image analysis methods to work towards an integrative understanding of chordate embryogenesis.
Acknowledgements
We thank the KSU CVM Confocal Microscopy facility and also acknowledge support from K-INBRE (NIH P20GM103418).
Literature cited
- Abdollahian G, Veeman M, Smith W, Manjunath BS. A Curvicylindrical Coordinate System for the Visualization and Segmentation of the Ascidian Tail. Proc IEEE Int Symp Biomed Imaging. 2011:182–186. doi: 10.1109/isbi.2011.5872383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abitua PB, Wagner E, Navarrete IA, Levine M. Identification of a rudimentary neural crest in a non-vertebrate chordate. Nature. 2012;492:104–107. doi: 10.1038/nature11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos WB, White JG. How the confocal laser scanning microscope entered biological research. Biol Cell. 2003;95:335–342. doi: 10.1016/s0248-4900(03)00078-9. [DOI] [PubMed] [Google Scholar]
- Begasse ML, Hyman AA. The first cell cycle of the Caenorhabditis elegans embryo: spatial and temporal control of an asymmetric cell division. Results Probl Cell Differ. 2011;53:109–133. doi: 10.1007/978-3-642-19065-0_6. [DOI] [PubMed] [Google Scholar]
- Brown DD. A tribute to the Xenopus laevis oocyte and egg. J Biol Chem. 2004;279:45291–45299. doi: 10.1074/jbc.X400008200. [DOI] [PubMed] [Google Scholar]
- Chabry L. Embryologie Normale et Teratologique des Ascidies. Faculte des Sciences de Paris. 1887 [Google Scholar]
- Christiaen L, Davidson B, Kawashima T, Powell W, Nolla H, Vranizan K, Levine M. The transcription/migration interface in heart precursors of Ciona intestinalis. Science. 2008;320:1349–1352. doi: 10.1126/science.1158170. [DOI] [PubMed] [Google Scholar]
- Cole AG, Meinertzhagen IA. The central nervous system of the ascidian larva: mitotic history of cells forming the neural tube in late embryonic Ciona intestinalis. Dev Biol. 2004;271:239–262. doi: 10.1016/j.ydbio.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Conklin EG. Mosaic development in ascidian eggs. J. Exp. Zool. 1905a:2. doi: 10.1002/jez.a.87. [DOI] [PubMed] [Google Scholar]
- Conklin EG. The organization and cell lineage of the ascidian egg. J. Acad. Nat. Sci. Phila. 1905b:13. [Google Scholar]
- Cooley J, Whitaker S, Sweeney S, Fraser S, Davidson B. Cytoskeletal polarity mediates localized induction of the heart progenitor lineage. Nat Cell Biol. 2011;13:952–957. doi: 10.1038/ncb2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo JC, Erives A, Di Gregorio A, Chang A, Levine M. Dorsoventral patterning of the vertebrate neural tube is conserved in a protochordate. Development. 1997a;124:2335–2344. doi: 10.1242/dev.124.12.2335. [DOI] [PubMed] [Google Scholar]
- Corbo JC, Levine M, Zeller RW. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development. 1997b;124:589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- Davidson B, Shi W, Levine M. Uncoupling heart cell specification and migration in the simple chordate Ciona intestinalis. Development. 2005;132:4811–4818. doi: 10.1242/dev.02051. [DOI] [PubMed] [Google Scholar]
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, Harafuji N, Hastings KE, Ho I, Hotta K, Huang W, Kawashima T, Lemaire P, Martinez D, Meinertzhagen IA, Necula S, Nonaka M, Putnam N, Rash S, Saiga H, Satake M, Terry A, Yamada L, Wang HG, Awazu S, Azumi K, Boore J, Branno M, Chin-Bow S, DeSantis R, Doyle S, Francino P, Keys DN, Haga S, Hayashi H, Hino K, Imai KS, Inaba K, Kano S, Kobayashi K, Kobayashi M, Lee BI, Makabe KW, Manohar C, Matassi G, Medina M, Mochizuki Y, Mount S, Morishita T, Miura S, Nakayama A, Nishizaka S, Nomoto H, Ohta F, Oishi K, Rigoutsos I, Sano M, Sasaki A, Sasakura Y, Shoguchi E, Shin-i T, Spagnuolo A, Stainier D, Suzuki MM, Tassy O, Takatori N, Tokuoka M, Yagi K, Yoshizaki F, Wada S, Zhang C, Hyatt PD, Larimer F, Detter C, Doggett N, Glavina T, Hawkins T, Richardson P, Lucas S, Kohara Y, Levine M, Satoh N, Rokhsar DS. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Delibaltov D, Ghosh P, Veeman M, Smith W, Manjunath BS. An automatic feature based model for cell segmentation from confocal microscopy volumes; Biomedical Imaging: From Nano to Macro, 2011 IEEE International Symposium on; 2011. pp. 199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- Deschet K, Nakatani Y, Smith WC. Generation of Ci-Brachyury-GFP stable transgenic lines in the ascidian Ciona savignyi. Genesis. 2003;35:248–259. doi: 10.1002/gene.10195. [DOI] [PubMed] [Google Scholar]
- Dong B, Deng W, Jiang D. Distinct cytoskeleton populations and extensive crosstalk control Ciona notochord tubulogenesis. Development. 2011;138:1631–1641. doi: 10.1242/dev.057208. [DOI] [PubMed] [Google Scholar]
- Dong B, Horie T, Denker E, Kusakabe T, Tsuda M, Smith WC, Jiang D. Tube formation by complex cellular processes in Ciona intestinalis notochord. Dev Biol. 2009;330:237–249. doi: 10.1016/j.ydbio.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn A. Review of Kowalewski's 'Embryology of worms and arthropods'. The Academy. 1871;2:496–498. [Google Scholar]
- Dufour A, Shinin V, Tajbakhsh S, Guillen-Aghion N, Olivo-Marin JC, Zimmer C. Segmenting and tracking fluorescent cells in dynamic 3-D microscopy with coupled active surfaces. IEEE Trans Image Process. 2005;14:1396–1410. doi: 10.1109/tip.2005.852790. [DOI] [PubMed] [Google Scholar]
- Dufour HD, Chettouh Z, Deyts C, de Rosa R, Goridis C, Joly JS, Brunet JF. Precraniate origin of cranial motoneurons. Proc Natl Acad Sci U S A. 2006;103:8727–8732. doi: 10.1073/pnas.0600805103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumollard R, Hebras C, Besnardeau L, McDougall A. Beta-catenin patterns the cell cycle during maternal-to-zygotic transition in urochordate embryos. Dev Biol. 2013;384:331–342. doi: 10.1016/j.ydbio.2013.10.007. [DOI] [PubMed] [Google Scholar]
- Fischer JL. Laurent Chabry and the beginnings of experimental embryology in France. Dev Biol (N Y 1985) 1991;7:31–41. doi: 10.1007/978-1-4615-6823-0_2. [DOI] [PubMed] [Google Scholar]
- Gregory C, Veeman M. 3D-printed microwell arrays for Ciona microinjection and timelapse imaging. PLoS One. 2013;8:e82307. doi: 10.1371/journal.pone.0082307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackley C, Mulholland E, Kim GJ, Newman-Smith E, Smith WC. A transiently expressed connexin is essential for anterior neural plate development in Ciona intestinalis. Development. 140:147–155. doi: 10.1242/dev.084681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada M, Wada S, Kobayashi K, Satoh N. Novel genes involved in Ciona intestinalis embryogenesis: characterization of gene knockdown embryos. Dev Dyn. 2007;236:1820–1831. doi: 10.1002/dvdy.21181. [DOI] [PubMed] [Google Scholar]
- Hockendorf B, Thumberger T, Wittbrodt J. Quantitative analysis of embryogenesis: a perspective for light sheet microscopy. Dev Cell. 2012;23:1111–1120. doi: 10.1016/j.devcel.2012.10.008. [DOI] [PubMed] [Google Scholar]
- Hotta K, Mitsuhara K, Takahashi H, Inaba K, Oka K, Gojobori T, Ikeo K. A web-based interactive developmental table for the ascidian Ciona intestinalis, including 3D real-image embryo reconstructions: I. From fertilized egg to hatching larva. Dev Dyn. 2007;236:1790–1805. doi: 10.1002/dvdy.21188. [DOI] [PubMed] [Google Scholar]
- Hotta K, Takahashi H, Erives A, Levine M, Satoh N. Temporal expression patterns of 39 Brachyury-downstream genes associated with notochord formation in the Ciona intestinalis embryo. Dev Growth Differ. 1999;41:657–664. doi: 10.1046/j.1440-169x.1999.00467.x. [DOI] [PubMed] [Google Scholar]
- Hudson C, Lemaire P. Induction of anterior neural fates in the ascidian Ciona intestinalis. Mech Dev. 2001;100:189–203. doi: 10.1016/s0925-4773(00)00528-1. [DOI] [PubMed] [Google Scholar]
- Hudson C, Lotito S, Yasuo H. Sequential and combinatorial inputs from Nodal, Delta2/Notch and FGF/MEK/ERK signalling pathways establish a grid-like organisation of distinct cell identities in the ascidian neural plate. Development. 2007;134:3527–3537. doi: 10.1242/dev.002352. [DOI] [PubMed] [Google Scholar]
- Hudson C, Yasuo H. Patterning across the ascidian neural plate by lateral Nodal signalling sources. Development. 2005;132:1199–1210. doi: 10.1242/dev.01688. [DOI] [PubMed] [Google Scholar]
- Hudson C, Yasuo H. A signalling relay involving Nodal and Delta ligands acts during secondary notochord induction in Ciona embryos. Development. 2006;133:2855–2864. doi: 10.1242/dev.02466. [DOI] [PubMed] [Google Scholar]
- Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- Imai KS, Levine M, Satoh N, Satou Y. Regulatory blueprint for a chordate embryo. Science. 2006;312:1183–1187. doi: 10.1126/science.1123404. [DOI] [PubMed] [Google Scholar]
- Jeffery WR, Chiba T, Krajka FR, Deyts C, Satoh N, Joly JS. Trunk lateral cells are neural crest-like cells in the ascidian Ciona intestinalis: insights into the ancestry and evolution of the neural crest. Dev Biol. 2008;324:152–160. doi: 10.1016/j.ydbio.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Jeffery WR, Strickler AG, Yamamoto Y. Migratory neural crest-like cells form body pigmentation in a urochordate embryo. Nature. 2004;431:696–699. doi: 10.1038/nature02975. [DOI] [PubMed] [Google Scholar]
- Jiang D, Munro EM, Smith WC. Ascidian prickle regulates both mediolateral and anterior-posterior cell polarity of notochord cells. Curr Biol. 2005;15:79–85. doi: 10.1016/j.cub.2004.12.041. [DOI] [PubMed] [Google Scholar]
- Jones TR, Kang IH, Wheeler DB, Lindquist RA, Papallo A, Sabatini DM, Golland P, Carpenter AE. CellProfiler Analyst: data exploration and analysis software for complex image-based screens. BMC Bioinformatics. 2008;9:482. doi: 10.1186/1471-2105-9-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EH. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. 2008;322:1065–1069. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- Khairy K, Keller PJ. Reconstructing embryonic development. Genesis. 2010;49:488–513. doi: 10.1002/dvg.20698. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kourakis MJ, Newman-Smith E, Smith WC. Key steps in the morphogenesis of a cranial placode in an invertebrate chordate, the tunicate Ciona savignyi. Dev Biol. 2010;340:134–144. doi: 10.1016/j.ydbio.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourakis MJ, Smith WC. A conserved role for FGF signaling in chordate otic/atrial placode formation. Dev Biol. 2007;312:245–257. doi: 10.1016/j.ydbio.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalewski A. Entwicklungsgeschichte der einfachen Ascidien. Memoires d’Academie des Sciences de St Petersbourgh. 1866;7:1–119. [Google Scholar]
- Kroon DJ, Slump CH. Coherence Filtering to Enhance the Mandibula Canal in Cone-Beam CT Data; Proceedings of the 4th Annual Symposium of the IEEE-EMBS Benelux Chapter; 2009. pp. 41–44. [Google Scholar]
- Kusakabe T, Yoshida R, Kawakami I, Kusakabe R, Mochizuki Y, Yamada L, Shin-i T, Kohara Y, Satoh N, Tsuda M, Satou Y. Gene expression profiles in tadpole larvae of Ciona intestinalis. Dev Biol. 2002;242:188–203. doi: 10.1006/dbio.2002.0538. [DOI] [PubMed] [Google Scholar]
- Luengo-Oroz MA, Arranz A, Frean J. Crowdsourcing malaria parasite quantification: an online game for analyzing images of infected thick blood smears. J Med Internet Res. 2012a;14:e167. doi: 10.2196/jmir.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luengo-Oroz MA, Pastor-Escuredo D, Castro-Gonzalez C, Faure E, Savy T, Lombardot B, Rubio-Guivernau JL, Duloquin L, Ledesma-Carbayo MJ, Bourgine P, Peyrieras N, Santos A. 3D+t morphological processing: applications to embryogenesis image analysis. IEEE Trans Image Process. 2012b;21:3518–3530. doi: 10.1109/TIP.2012.2197007. [DOI] [PubMed] [Google Scholar]
- Manni L, Lane NJ, Joly JS, Gasparini F, Tiozzo S, Caicci F, Zaniolo G, Burighel P. Neurogenic and non-neurogenic placodes in ascidians. J Exp Zool B Mol Dev Evol. 2004;302:483–504. doi: 10.1002/jez.b.21013. [DOI] [PubMed] [Google Scholar]
- Markow TA, Beall S, Matzkin LM. Egg size, embryonic development time and ovoviviparity in Drosophila species. J Evol Biol. 2009;22:430–434. doi: 10.1111/j.1420-9101.2008.01649.x. [DOI] [PubMed] [Google Scholar]
- Mazet F, Hutt JA, Milloz J, Millard J, Graham A, Shimeld SM. Molecular evidence from Ciona intestinalis for the evolutionary origin of vertebrate sensory placodes. Dev Biol. 2005;282:494–508. doi: 10.1016/j.ydbio.2005.02.021. [DOI] [PubMed] [Google Scholar]
- McDougall A, Lee KW, Dumollard R. Microinjection and 4D fluorescence imaging in the eggs and embryos of the ascidian Phallusia mammillata. Methods Mol Biol. 2014;1128:175–185. doi: 10.1007/978-1-62703-974-1_11. [DOI] [PubMed] [Google Scholar]
- McDougall A, Sardet C. Function and characteristics of repetitive calcium waves associated with meiosis. Curr Biol. 1995;5:318–328. doi: 10.1016/s0960-9822(95)00062-5. [DOI] [PubMed] [Google Scholar]
- Mita K, Fujiwara S. Nodal regulates neural tube formation in the Ciona intestinalis embryo. Dev Genes Evol. 2007;217:593–601. doi: 10.1007/s00427-007-0168-x. [DOI] [PubMed] [Google Scholar]
- Mitry D, Peto T, Hayat S, Morgan JE, Khaw KT, Foster PJ. Crowdsourcing as a novel technique for retinal fundus photography classification: analysis of images in the EPIC Norfolk cohort on behalf of the UK Biobank Eye and Vision Consortium. PLoS One. 2013;8:e71154. doi: 10.1371/journal.pone.0071154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto DM, Crowther RJ. Formation of the notochord in living ascidian embryos. J Embryol Exp Morphol. 1985;86:1–17. [PubMed] [Google Scholar]
- Mosaliganti KR, Noche RR, Xiong F, Swinburne IA, Megason SG. ACME: automated cell morphology extractor for comprehensive reconstruction of cell membranes. PLoS Comput Biol. 2012;8:e1002780. doi: 10.1371/journal.pcbi.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro EM, Odell G. Morphogenetic pattern formation during ascidian notochord formation is regulative and highly robust. Development. 2002a;129:1–12. doi: 10.1242/dev.129.1.1. [DOI] [PubMed] [Google Scholar]
- Munro EM, Odell GM. Polarized basolateral cell motility underlies invagination and convergent extension of the ascidian notochord. Development. 2002b;129:13–24. doi: 10.1242/dev.129.1.13. [DOI] [PubMed] [Google Scholar]
- Nakamura MJ, Terai J, Okubo R, Hotta K, Oka K. Three-dimensional anatomy of the Ciona intestinalis tailbud embryo at single-cell resolution. Dev Biol. 2012;372:274–284. doi: 10.1016/j.ydbio.2012.09.007. [DOI] [PubMed] [Google Scholar]
- Negishi T, McDougall A, Yasuo H. Practical tips for imaging ascidian embryos. Dev Growth Differ. 2013;55:446–453. doi: 10.1111/dgd.12059. [DOI] [PubMed] [Google Scholar]
- Nicol D, Meinertzhagen IA. Development of the central nervous system of the larva of the ascidian, Ciona intestinalis L. I. The early lineages of the neural plate. Dev Biol. 1988a;130:721–736. doi: 10.1016/0012-1606(88)90363-6. [DOI] [PubMed] [Google Scholar]
- Nicol D, Meinertzhagen IA. Development of the central nervous system of the larva of the ascidian, Ciona intestinalis L. II. Neural plate morphogenesis and cell lineages during neurulation. Dev Biol. 1988b;130:737–766. doi: 10.1016/0012-1606(88)90364-8. [DOI] [PubMed] [Google Scholar]
- Nishida H. Cell lineage analysis in ascidian embryos by intracellular injection of a tracer enzyme. III. Up to the tissue restricted stage. Dev Biol. 1987;121:526–541. doi: 10.1016/0012-1606(87)90188-6. [DOI] [PubMed] [Google Scholar]
- Nishida H, Satoh N. Cell lineage analysis in ascidian embryos by intracellular injection of a tracer enzymeI. I. Up to the eight-cell stage. Dev Biol. 1983;99:382–394. doi: 10.1016/0012-1606(83)90288-9. [DOI] [PubMed] [Google Scholar]
- Nishida H, Satoh N. Cell lineage analysis in ascidian embryos by intracellular injection of a tracer enzyme. II. The 16- and 32-cell stages. Dev Biol. 1985;110:440–454. doi: 10.1016/0012-1606(85)90102-2. [DOI] [PubMed] [Google Scholar]
- Nishida H, Sawada K. macho-1 encodes a localized mRNA in ascidian eggs that specifies muscle fate during embryogenesis. Nature. 2001;409:724–729. doi: 10.1038/35055568. [DOI] [PubMed] [Google Scholar]
- Norton J, Cooley J, Islam AF, Cota CD, Davidson B. Matrix adhesion polarizes heart progenitor induction in the invertebrate chordate Ciona intestinalis. Development. 2013;140:1301–1311. doi: 10.1242/dev.085548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara B, Veeman M, Choi JH, Smith W, Manjunath BS. Segmentation of ascidian notochord cells in DIC timelapse images. Microsc Res Tech. 2011;74:727–734. doi: 10.1002/jemt.20950. [DOI] [PubMed] [Google Scholar]
- Ogasawara M, Sasaki A, Metoki H, Shin-i T, Kohara Y, Satoh N, Satou Y. Gene expression profiles in young adult Ciona intestinalis. Dev Genes Evol. 2002;212:173–185. doi: 10.1007/s00427-002-0230-7. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Sakaue-Sawano A, Nakagawa M, Satoh N, Miyawaki A, Sasakura Y. Coordination of mitosis and morphogenesis: role of a prolonged G2 phase during chordate neurulation. Development. 2011;138:577–587. doi: 10.1242/dev.053132. [DOI] [PubMed] [Google Scholar]
- Ohta N, Satou Y. Multiple signaling pathways coordinate to induce a threshold response in a chordate embryo. PLoS Genet. 2013;9:e1003818. doi: 10.1371/journal.pgen.1003818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier N, Luengo-Oroz MA, Duloquin L, Faure E, Savy T, Veilleux I, Solinas X, Debarre D, Bourgine P, Santos A, Peyrieras N, Beaurepaire E. Cell lineage reconstruction of early zebrafish embryos using label-free nonlinear microscopy. Science. 2010;329:967–971. doi: 10.1126/science.1189428. [DOI] [PubMed] [Google Scholar]
- Paix A, Yamada L, Dru P, Lecordier H, Pruliere G, Chenevert J, Satoh N, Sardet C. Cortical anchorages and cell type segregations of maternal postplasmic/PEM RNAs in ascidians. Dev Biol. 2009;336:96–111. doi: 10.1016/j.ydbio.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Passamaneck YJ, Di Gregorio A. Ciona intestinalis: chordate development made simple. Dev Dyn. 2005;233:1–19. doi: 10.1002/dvdy.20300. [DOI] [PubMed] [Google Scholar]
- Passamaneck YJ, Hadjantonakis AK, Di Gregorio A. Dynamic and polarized muscle cell behaviors accompany tail morphogenesis in the ascidian Ciona intestinalis. PLoS One. 2007;2:e714. doi: 10.1371/journal.pone.0000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patalano S, Pruliere G, Prodon F, Paix A, Dru P, Sardet C, Chenevert J. The aPKC-PAR-6-PAR-3 cell polarity complex localizes to the centrosome attracting body, a macroscopic cortical structure responsible for asymmetric divisions in the early ascidian embryo. J Cell Sci. 2006;119:1592–1603. doi: 10.1242/jcs.02873. [DOI] [PubMed] [Google Scholar]
- Prodon F, Chenevert J, Hebras C, Dumollard R, Faure E, Gonzalez-Garcia J, Nishida H, Sardet C, McDougall A. Dual mechanism controls asymmetric spindle position in ascidian germ cell precursors. Development. 2010;137:2011–2021. doi: 10.1242/dev.047845. [DOI] [PubMed] [Google Scholar]
- Prodon F, Dru P, Roegiers F, Sardet C. Polarity of the ascidian egg cortex and relocalization of cER and mRNAs in the early embryo. J Cell Sci. 2005;118:2393–2404. doi: 10.1242/jcs.02366. [DOI] [PubMed] [Google Scholar]
- Razy-Krajka F, Lam K, Wang W, Stolfi A, Joly M, Bonneau R, Christiaen L. Collier/OLF/EBF-dependent transcriptional dynamics control pharyngeal muscle specification from primed cardiopharyngeal progenitors. Dev Cell. 2014;29:263–276. doi: 10.1016/j.devcel.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee JM, Oda-Ishii I, Passamaneck YJ, Hadjantonakis AK, Di Gregorio A. Live imaging and morphometric analysis of embryonic development in the ascidian Ciona intestinalis. Genesis. 2005;43:136–147. doi: 10.1002/gene.20164. [DOI] [PubMed] [Google Scholar]
- Rizzi B, Peyrieras N. Towards 3D in silico modeling of the sea urchin embryonic development. J Chem Biol. 2013;7:17–28. doi: 10.1007/s12154-013-0101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin FB, Dauga D, Tassy O, Sobral D, Daian F, Lemaire P. Time-lapse imaging of live Phallusia embryos for creating 3D digital replicas. Cold Spring Harb Protoc. 2011;2011:1244–1246. doi: 10.1101/pdb.prot065847. [DOI] [PubMed] [Google Scholar]
- Roeder AH, Tarr PT, Tobin C, Zhang X, Chickarmane V, Cunha A, Meyerowitz EM. Computational morphodynamics of plants: integrating development over space and time. Nat Rev Mol Cell Biol. 2011;12:265–273. doi: 10.1038/nrm3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbacher U, Bertrand V, Lamy C, Lemaire P. A combinatorial code of maternal GATA, Ets and beta-catenin-TCF transcription factors specifies and patterns the early ascidian ectoderm. Development. 2007;134:4023–4032. doi: 10.1242/dev.010850. [DOI] [PubMed] [Google Scholar]
- Sardet C, Speksnijder J, Inoue S, Jaffe L. Fertilization and ooplasmic movements in the ascidian egg. Development. 1989;105:237–249. doi: 10.1242/dev.105.2.237. [DOI] [PubMed] [Google Scholar]
- Sasakura Y. Germline transgenesis and insertional mutagenesis in the ascidian Ciona intestinalis. Dev Dyn. 2007;236:1758–1767. doi: 10.1002/dvdy.21111. [DOI] [PubMed] [Google Scholar]
- Sasakura Y, Mita K, Ogura Y, Horie T. Ascidians as excellent chordate models for studying the development of the nervous system during embryogenesis and metamorphosis. Dev Growth Differ. 2012;54:420–437. doi: 10.1111/j.1440-169X.2012.01343.x. [DOI] [PubMed] [Google Scholar]
- Satou Y, Takatori N, Yamada L, Mochizuki Y, Hamaguchi M, Ishikawa H, Chiba S, Imai K, Kano S, Murakami SD, Nakayama A, Nishino A, Sasakura Y, Satoh G, Shimotori T, Shin IT, Shoguchi E, Suzuki MM, Takada N, Utsumi N, Yoshida N, Saiga H, Kohara Y, Satoh N. Gene expression profiles in Ciona intestinalis tailbud embryos. Development. 2001;128:2893–2904. doi: 10.1242/dev.128.15.2893. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehring IM, Dong B, Denker E, Bhattachan P, Deng W, Mathiesen BT, Jiang D. An equatorial contractile mechanism drives cell elongation but not cell division. PLoS Biol. 2014;12:e1001781. doi: 10.1371/journal.pbio.1001781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrard K, Robin F, Lemaire P, Munro E. Sequential activation of apical and basolateral contractility drives ascidian endoderm invagination. Curr Biol. 2011;20:1499–1510. doi: 10.1016/j.cub.2010.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small KS, Brudno M, Hill MM, Sidow A. A haplome alignment and reference sequence of the highly polymorphic Ciona savignyi genome. Genome Biol. 2007;8:R41. doi: 10.1186/gb-2007-8-3-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speksnijder JE, Corson DW, Sardet C, Jaffe LF. Free calcium pulses following fertilization in the ascidian egg. Dev Biol. 1989a;135:182–190. doi: 10.1016/0012-1606(89)90168-1. [DOI] [PubMed] [Google Scholar]
- Speksnijder JE, Jaffe LF, Sardet C. Polarity of sperm entry in the ascidian egg. Dev Biol. 1989b;133:180–184. doi: 10.1016/0012-1606(89)90309-6. [DOI] [PubMed] [Google Scholar]
- Stemple DL. Structure and function of the notochord: an essential organ for chordate development. Development. 2005;132:2503–2512. doi: 10.1242/dev.01812. [DOI] [PubMed] [Google Scholar]
- Stolfi A, Gainous TB, Young JJ, Mori A, Levine M, Christiaen L. Early chordate origins of the vertebrate second heart field. Science. 2010;329:565–568. doi: 10.1126/science.1190181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolfi A, Wagner E, Taliaferro JM, Chou S, Levine M. Neural tube patterning by Ephrin, FGF and Notch signaling relays. Development. 2011;138:5429–5439. doi: 10.1242/dev.072108. [DOI] [PubMed] [Google Scholar]
- Tassy O, Daian F, Hudson C, Bertrand V, Lemaire P. A quantitative approach to the study of cell shapes and interactions during early chordate embryogenesis. Curr Biol. 2006;16:345–358. doi: 10.1016/j.cub.2005.12.044. [DOI] [PubMed] [Google Scholar]
- Tassy O, Dauga D, Daian F, Sobral D, Robin F, Khoueiry P, Salgado D, Fox V, Caillol D, Schiappa R, Laporte B, Rios A, Luxardi G, Kusakabe T, Joly JS, Darras S, Christiaen L, Contensin M, Auger H, Lamy C, Hudson C, Rothbacher U, Gilchrist MJ, Makabe KW, Hotta K, Fujiwara S, Satoh N, Satou Y, Lemaire P. The ANISEED database: digital representation, formalization, and elucidation of a chordate developmental program. Genome Res. 2010;20:1459–1468. doi: 10.1101/gr.108175.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas N. High-content screening: a decade of evolution. J Biomol Screen. 2009;15:1–9. doi: 10.1177/1087057109353790. [DOI] [PubMed] [Google Scholar]
- Tolkin T, Christiaen L. Development and evolution of the ascidian cardiogenic mesoderm. Curr Top Dev Biol. 2012;100:107–142. doi: 10.1016/B978-0-12-387786-4.00011-7. [DOI] [PubMed] [Google Scholar]
- Treen N, Yoshida K, Sakuma T, Sasaki H, Kawai N, Yamamoto T, Sasakura Y. Tissue-specific and ubiquitous gene knockouts by TALEN electroporation provide new approaches to investigating gene function in Ciona. Development. 2014;141:481–487. doi: 10.1242/dev.099572. [DOI] [PubMed] [Google Scholar]
- Trier SM, Davidson LA. Quantitative microscopy and imaging tools for the mechanical analysis of morphogenesis. Curr Opin Genet Dev. 2011;21:664–670. doi: 10.1016/j.gde.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong TV, Supatto W. Toward high-content/high-throughput imaging and analysis of embryonic morphogenesis. Genesis. 2011;49:555–569. doi: 10.1002/dvg.20760. [DOI] [PubMed] [Google Scholar]
- Veeman MT, Chiba S, Smith WC. Ciona genetics. Methods Mol Biol. 2011;770:401–422. doi: 10.1007/978-1-61779-210-6_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman MT, Nakatani Y, Hendrickson C, Ericson V, Lin C, Smith WC. Chongmague reveals an essential role for laminin-mediated boundary formation in chordate convergence and extension movements. Development. 2008;135:33–41. doi: 10.1242/dev.010892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman MT, Newman-Smith E, El-Nachef D, Smith WC. The ascidian mouth opening is derived from the anterior neuropore: reassessing the mouth/neural tube relationship in chordate evolution. Dev Biol. 2010;344:138–149. doi: 10.1016/j.ydbio.2010.04.028. [DOI] [PubMed] [Google Scholar]
- Veeman MT, Smith WC. Whole-organ cell shape analysis reveals the developmental basis of ascidian notochord taper. Dev Biol. 2012 doi: 10.1016/j.ydbio.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent L, Soille P. Watersheds in digital spaces: An efficient algorithm based on immersion simulations. IEEE PAMI. 1991;13:583–598. [Google Scholar]
- Wada H, Saiga H, Satoh N, Holland PW. Tripartite organization of the ancestral chordate brain and the antiquity of placodes: insights from ascidian Pax-2/5/8, Hox and Otx genes. Development. 1998;125:1113–1122. doi: 10.1242/dev.125.6.1113. [DOI] [PubMed] [Google Scholar]
- Wagner E, Levine M. FGF signaling establishes the anterior border of the Ciona neural tube. Development. 2012;139:2351–2359. doi: 10.1242/dev.078485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert J. Coherence-Enhancing Diffusion Filtering. International Journal of Computer Vision. 1999;31:111–127. [Google Scholar]
- White JG, Amos WB, Fordham M. An evaluation of confocal versus conventional imaging of biological structures by fluorescence light microscopy. J Cell Biol. 1987;105:41–48. doi: 10.1083/jcb.105.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi K, Satou Y, Satoh N. A zinc finger transcription factor, ZicL, is a direct activator of Brachyury in the notochord specification of Ciona intestinalis. Development. 2004;131:1279–1288. doi: 10.1242/dev.01011. [DOI] [PubMed] [Google Scholar]
- Yamada L, Shoguchi E, Wada S, Kobayashi K, Mochizuki Y, Satou Y, Satoh N. Morpholino-based gene knockdown screen of novel genes with developmental function in Ciona intestinalis. Development. 2003;130:6485–6495. doi: 10.1242/dev.00847. [DOI] [PubMed] [Google Scholar]
- Yasuo H, Satoh N. Conservation of the developmental role of Brachyury in notochord formation in a urochordate, the ascidian Balocynthia roretzi. Dev Biol. 1998;200:158–170. doi: 10.1006/dbio.1998.8958. [DOI] [PubMed] [Google Scholar]
- Zanella C, Campana M, Rizzi B, Melani C, Sanguinetti G, Bourgine P, Mikula K, Peyrieras N, Sarti A. Cells segmentation from 3-D confocal images of early zebrafish embryogenesis. IEEE Trans Image Process. 2009;19:770–781. doi: 10.1109/TIP.2009.2033629. [DOI] [PubMed] [Google Scholar]
- Zeller RW, Virata MJ, Cone AC. Predictable mosaic transgene expression in ascidian embryos produced with a simple electroporation device. Dev Dyn. 2006a;235:1921–1932. doi: 10.1002/dvdy.20815. [DOI] [PubMed] [Google Scholar]
- Zeller RW, Weldon DS, Pellatiro MA, Cone AC. Optimized green fluorescent protein variants provide improved single cell resolution of transgene expression in ascidian embryos. Dev Dyn. 2006b;235:456–467. doi: 10.1002/dvdy.20644. [DOI] [PubMed] [Google Scholar]