Abstract
Several reports have shown that statin treatment benefits patients with asthma, however inconsistent effects have been observed. The mir-152 family (148a, 148b and 152) has been implicated in asthma. These microRNAs suppress HLA-G expression, and rs1063320, a common SNP in the HLA-G 3’UTR which is associated with asthma risk, modulates miRNA binding. We report that statins up-regulate mir-148b and 152, and affect HLA-G expression in an rs1063320 dependent fashion. In addition, we found that individuals who carried the G minor allele of rs1063320 had reduced asthma related exacerbations (emergency department visits, hospitalizations or oral steroid use) compared to non-carriers (p=0.03) in statin users ascertained in the Personalized Medicine Research Project at the Marshfield Clinic (n=421). These findings support the hypothesis that rs1063320 modifies the effect of statin benefit in asthma, and thus may contribute to variation in statin efficacy for the management of this disease.
Keywords: pharmacogenetics, HLA-G, statin, asthma, mir-148, mir-152
INTRODUCTION
Statins, or HMG-CoA reductase inhibitors, which are primarily prescribed for the reduction of cardiovascular disease, have well-documented anti-inflammatory effects attributed to the inhibition of both isoprenoid production and prenylation of inflammatory signaling molecules [1]. Thus, statins may be of benefit in the prevention and treatment of asthma, a disease in which inflammation is of central importance [2–5]. Statin exposure has been associated with a reduction in asthma-related hospitalizations, attenuation in the decline of lung function in the elderly and in smokers, and a reduction in the number of Chronic Obstructive Pulmonary Disease (COPD) exacerbations [6–13]. For example, a retrospective cohort study of over 12 million members within the Medco Health Solutions National Integrated database found that adult with asthma and concomitant statin and inhaled corticosteroid therapy had a 33% lower risk of emergency department (ED) visits for asthma compared to non-statin users after adjustment for potential confounding variables [14]. However, two small trials failed to demonstrate improvement in indices of asthma status with statin treatment [15, 16], leading some to question the potential benefit of statins for asthma management [17]. This inconsistency may have a genetic basis as the considerable variability in both the lipid lowering and anti-inflammatory effects of statins is partly due to genetic factors [18, 19].
HLA-G, a non-classic class I HLA molecule with important immunomodulatory properties, has been associated with asthma in multiple independent populations [20–23]. Although the precise mechanism underlying this relationship is unknown, both HLA-G transcript and protein levels have been reported to be increased in the lung and plasma of subjects with asthma [24–26]. In addition, a common SNP (rs1063320 aka HLA-G +3142, 0.46 minor allele frequency,) within the 3’ UTR of the HLA-G gene has been associated with reduced risk of asthma in the children of mothers with asthma [27]. Notably, rs1063320 disrupts the targeting of the mir-152 family (mir-148a, mir-148b and mir-152) to the HLA-G gene such that these microRNAs can only down-regulate HLA-G transcripts carrying the variant G allele, with no effect on transcripts with the C allele. Furthermore, adult asthmatic patients with an asthmatic mother were also shown to have elevated mir-148b levels in airway epithelial cells, and greater soluble HLA-G (sHLA-G) in brochoalveolar lavage fluid in GG homozygotes compared to non-carriers [28]. Thus these results support the hypothesis that allele-specific targeting of mir-148a, mir-148b, and/or mir-152 may contribute to the association of the HLA-G gene with the risk of asthma.
In this study, we report that statin treatment up-regulates expression of mir-148a, mir-148b and mir-152 in vitro, leading to the hypothesis that statin treatment may benefit asthmatic patients that carry the HLA-G G allele by causing a reduction of HLA-G expression in these subjects. Thus, we sought to test if rs1063320 modifies statin effects on asthma-related exacerbations in a population-based cohort of statin users identified from the Marshfield Clinic Personalized Medicine Research Program (PMRP).
MATERIALS, SUBJECTS AND METHODS
Cell culture
Immortalized lymphoblastoid cell lines (LCLs) established from participants of the Cholesterol and Pharmacogenetics (CAP) clinical trial were grown under standard conditions in RPMI supplemented with 10% FBS and either 2.0µM activated simvastatin or sham buffer for 24 hours as previously described[29]. 5×106 HepG2 cells were exposed in replicate to DMEM supplemented with 10% FBS and either 2.0µM activated simvastatin or sham buffer for 24 hours. Simvastatin was obtained as a gift from Merck, and activated as previously described [29].
Gene expression measurements
Total RNA was extracted using the mirVana RNA isolation kit (Ambion, Austin, TX), quantified by NanoDrop (Thermo Scientific, Waltham, MA), and RNA integrity was assessed on the Agilent Technologies 2100 Bioanalyzer. HLA-G was quantified by expression array (GEO accession number: GSE36868) as previously described [30]. mir-148a, mir-148b and mir-152 were quantified by real-time PCR using TaqMan MicroRNA Assays (assay IDs 000470, 000471 and 000475 respectively) according to the manufacturer’s protocol (Applied Biosystems, Foster City, CA). All reactions were performed in triplicate and run on an Applied Biosystems 7900HT Fast Real-Time PCR System. microRNA expression levels were normalized against mir-25, whose expression is unchanged with statin exposure (Figure S1). Statin-induced change in mir-148a, mir-148b and mir-152 was calculated using the delta delta Ct method. Paired two-side t-tests were used to identify differences in HLA-G and miRNA expression levels with statin treatment. HLA-G and microRNA expression levels quantified in LCLs were quantile normalized, split by rs1063320 genotype group, and tested for correlation using linear regression.
HLA-G protein quantitation
Soluble HLA-G protein levels were quantified by ELISA (BioVendor, Candler, NC) in cell culture media from in vitro 2.0µM simvastatin versus sham exposed LCLs. All measurements were performed in duplicate and averaged.
Genotyping
Donors of CAP LCLs were subject to genome-wide genotyping on the Illumina HumanHap300 or HumanHap610-Quad genotyping platforms (dbGaP accession number: phs000481), and rs1063320 was imputed as previously described[31]. Genotypes with an imputed probability less than 0.85 were omitted from the analyses. rs1063320 was directly genotyped with a TaqMan SNP Genotyping assay (Applied Biosystems).
Study Subjects
Study subjects were ascertained from the Personalized Medicine Research Project at the Marshfield Clinic in Wisconsin, population-based biobank. PMRP is comprised of 20,000 non-Hispanic whites of Northern European descent (98%) from Central Wisconsin. Enrollment was conducted in accordance with the guidelines established by the Institutional Review Boards of the Marshfield Clinic Research Foundation. Access and evaluation of the PMRP was performed through the Pharmacogenomics Discovery and Replication in Large Populations (PGPop) Consortium.
We identified individuals with asthma using ICD-9 codes (493*), with diagnoses indicated on two separate days. We have previously demonstrated that requiring the presence of such codes on two separate days substantially increases phenotypic accuracy (>95%) and replication for known genetic signals [34, 35]. Exclusion criteria for both cohorts included any individual with a single diagnosis code for chronic obstructive pulmonary disease (496), cystic fibrosis (277*), immunodeficiency (279*), bronchiectasis (494*), hereditary and degenerative diseases of CNS (331), mental retardation (317, 318, 319), congestive heart failure (428*, 429.9), pulmonary hypertension or embolism (415*, 416*, 417*), lupus (710.0), RA (714.0), tuberculosis (511.1), lung cancer (162*), or sarcoidosis (135). These algorithms were validated by manual chart review of 50–100 records per cohort and found to have >90% PPV.
Statin users were then identified as individuals with at least two mentions of statin drugs (simvastatin, fluvastatin, pravastatin, rosuvastatin, atorvastatin, lovastatin or any brand names of these drugs) of the same type and dose within a 6-month time window. We have previously demonstrated the capacity of natural language processing to document statin exposure since this type of data can generate accurate dose response curves despite the lack of information regarding drug compliance [36]. Statin users with asthma were defined as those with asthma drug use or diagnosis indicated after the date of first statin mention. From this analysis we identified 421 subjects (Tables S1 and S2).
Case definition
Asthma related exacerbations were defined as emergency department visits, hospitalizations, and/or use of oral steroids (prednisone, methylprednisone, solumedrol, solu-medrol, orapred, orasone, medrol, depo-medrol, depomedrol, depopred, prednisolone) for which an ICD-9 code for asthma or wheezing (786.07) appeared in the medical record concurrently. Since a high percentage of individuals cease statin use within the first year of prescription, to increase the likelihood that identified individuals were still on statin during the time of the exacerbation, we only included exacerbations that occurred within 6 months (+/− 180 days) of a statin mention in the EMR. This algorithm had a >90% PPV.
Covariate definition
All prescription information, as well as weight, height and smoking status was collected for evaluation as a potential confounding variable. Smoking status was divided into three categories: “current”, “former” or “never smoker”. Confirmed use was validated in a subset of charts by manual review. Subjects were classified as having inhaled corticosteroid (ICS) use if they were prescribed beclomethasone, budesonide, budesonide/formoterol, ciclesonide, flunisolide, fluticasone, fluticasone/salmeterol, mometasone, mometasone/formoterol, triamcinolone.
Statistical Analyses
A general linear model was used to test for a relationship between rs1063320 and statin effects on number of exacerbations (treated as a continuous variable), and included adjustment for gender, smoking status and ICS use. All analyses were performed in JMP 9.1 (SAS Institute, Cary, NC) using an additive model.
RESULTS
Mir-148b and mir-152 are statin responsive and correlated to HLA-G in an rs1063320 dependent fashion
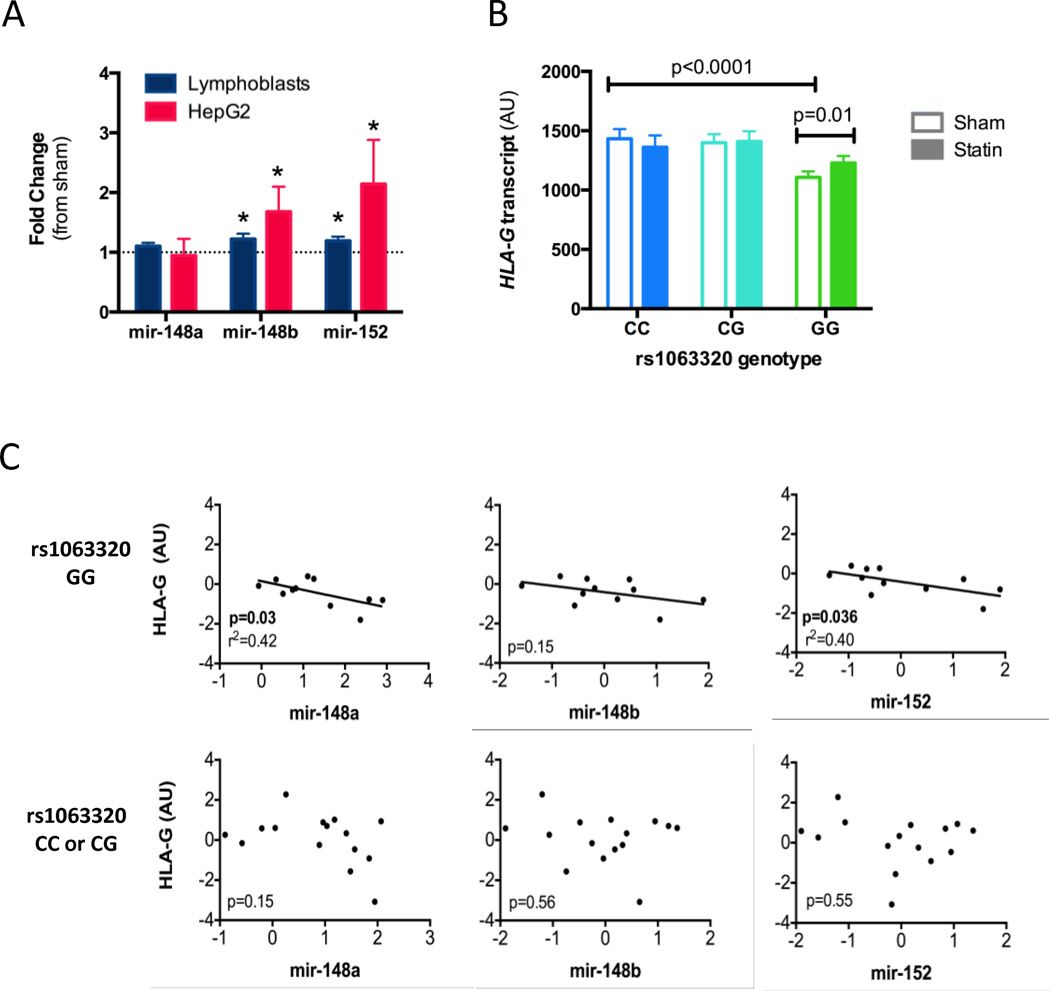
To determine if mir-148a, mir-148b or mir-152 transcript levels are statin responsive, we incubated both LCLs (n=48) and HepG2 cell lines (n=6 replicate experiments) with either 2.0µM simvastatin or sham buffer for 24 hours, and quantified changes in transcript levels using real-time PCR. Mir-148b and mir-152 were upregulated with statin treatment in both cell types (p<0.05, Figure 1A). No change in mir-148a levels was detected.
Figure 1. mir-152 family and HLA-G expression levels are statin responsive.
A) CAP LCLs (n=48) and HepG2 (n=6 replicate experiments) cells were incubated with 2.0 µM simvastatin or sham buffer for 24 hours, and mir-148a, 148b and 152 were quantified by qPCR, with values normalized against mir-25. Values shown are fold change (statin/sham values) ± standard error, *p<0.05. B) Normalized HLA-G transcript level was quantified by expression array in statin or sham treated LCLs (CC n=32, CG n=42, GG n=86) as previously described[30] and tested for association with rs1063320 genotype using an additive model. C) Expression levels of HLA-G and the three microRNAs were measured in sham treated LCLs, quantile normalized, and tested for correlation across the samples split by rs1063320 genotype: GG n=11, CC or CG n=15.
The rs1063320 C allele has been previously reported to disrupt the binding sites of mir-148 and mir-152 in the HLA-G 3’ UTR[27]. Using previously collected genotype data for the LCLs, we confirmed that the C allele carriers had 1.3-fold greater HLA-G transcript (p<0.0001, N=160) than non-carriers (Figure 1B). Furthermore, we found that inter-individual variation in microRNA and HLA-G transcript levels was correlated in the LCLs in an rs1063320 dependent fashion. Specifically, lower expression levels of mir-148a and 152 were correlated with higher HLA-G transcript levels, but only in the LCLs derived from rs1063320 GG homozygotes (p<0.05, n=11). No correlation was seen in LCLs from either CC or CG rs1063220 donors (n=15, Figure 1C). A similar, non-statistically significant, trend was observed between mir-148b and HLA-G.
It has been previously suggested that the mir-152 family regulates HLA-G by impacting translation and not mRNA decay [28]. Thus, we quantified both HLA-G transcript and sHLA-G protein levels in LCLs after 24hr in vitro incubation with 2.0µM simvastatin or sham buffer. While we detected no change in HLA-G transcript levels in LCLs that carried the C allele, we found that statin treatment increased HLA-G transcripts levels (p=0.01, Figure 1B) in the GG homozygotes. In contrast, we observed a reduction in HLA-G protein levels in both genetic groups, but there was a trend (p=0.09) of greater reductions of HLA-G protein in the GG versus CC homozygotes (Figure S2). These findings are consistent with our hypothesis that rs1063320 modifies statin effects on HLA-G expression.
rs1063320 genotype is associated with frequency of exacerbations in statin users
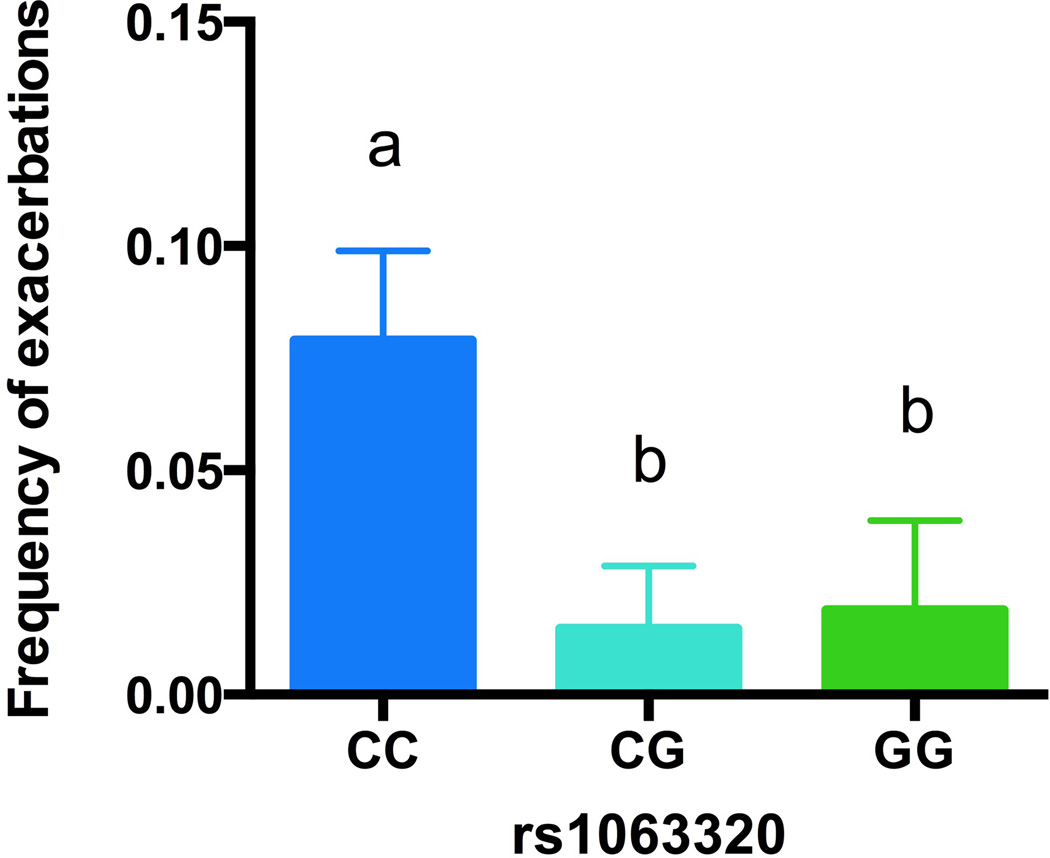
To test our hypothesis that statins may benefit asthma in an rs1063320 dependent fashion, we used logistic regression with adjustment for significantly associated covariates (see Methods). Consistent with our hypothesis, we found that individuals who carried at least one copy of the G minor allele had a reduced frequency of asthma-related exacerbations (emergency department visits, hospitalizations or oral steroid use) compared to non-carriers using an additive genetic model (p=0.03), Figure 2. There was no difference in exacerbation frequency between GG and CG genotypes.
Figure 2. Association between rs1063320 and asthma-related exacerbations.
rs1063320 genotype and frequency of asthma-related exacerbations in statin users was tested using an additive general linear model with adjustment for smoking status, gender and inhaled corticosteroid use (p=0.03). A post-hoc student’s t-test was used to identify differences between genotypic groups as indicated by “a” or “b”. CC n=111, CG n=204, GG n=106. Values shown are least square means ± standard error.
DISCUSSION
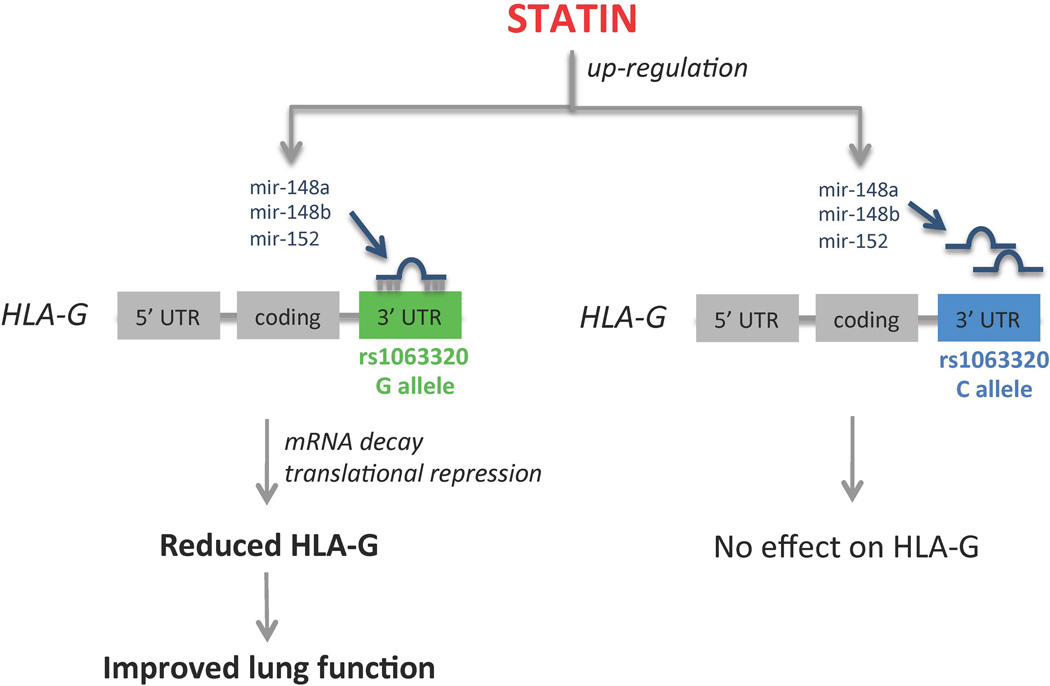
We report evidence that statin treatment increases expression of mir-148b and mir-152, has no impact on HLA-G transcript in rs1063320 CC homozygotes, and appears to cause greater reductions in HLA-G protein levels in GG homozygotes. HLA-G expression is regulated by the mir-152 family of microRNAs (mir-148a, mir-148b and mir-152), and rs1063320 is thought to disrupt this interaction [27, 28, 38]. Since HLA-G levels are increased in asthmatic patients, our identification that statin treatment up-regulates mir-148b and mir-152 and influences HLA-G expression in a rs1063320 dependent manner lead to our hypothesis that statin treatment may show greater benefit in individuals with the rs1063320 G allele (Figure 3). Consistent with this hypothesis, we observed that asthmatic patients with at least one copy of the G allele had a reduced frequency of exacerbations while on statin compared to CC homozygotes. Given the immunomodulatory properties of HLA-G, it has been proposed that understanding gene regulation and the role of polymorphisms (especially those in the 3’ UTR) in HLA-G may help inform the development of personalized medicine [39]. Specifically, rs1063320 is only one of two HLA-G SNPs proposed to have clinical potential, and assays have been developed for quick typing of this polymorphic site [40, 41].
Figure 3. Hypothetical model.
Statin treatment up-regulates mir-148b and mir-152, which target the HLA-G 3’UTR in an rs1063320 dependent fashion, leading to the prediction that statin treatment has greater benefit in individuals with asthma that carry the rs1063320 G allele.
Nicodemus-Johnson et al (2013) recently reported that asthmatic patients with the rs1063320 G allele had reduced sHLA-G in BAL fluid compared to non-carriers [28]. Consistent with this report, we also found that LCLs homozygous for the G allele had reduced sHLA-G protein levels compared to CC homozygotes. Interestingly, they did not identify a statistically significant association between the SNP and HLA-G transcript in the bronchial epithelial cells themselves. In addition, Zhu et al (2010) demonstrated that mir-152 overexpression caused reduced HLA-G protein levels by targeting the 3’ UTR, but had no effect on HLA-G transcript levels [38], leading to the suggestion that the mir-152 family modulates HLA-G at the level of protein translation rather than mRNA decay. Here, we report disparate effects of statin treatment on HLA-G transcript versus protein levels in the rs1063320 GG homozygotes, consistent with the possibility that mir-148 and mir-152 may influence HLA-G translation. It is possible that the increase in HLA-G transcript levels in the GG homozygotes after statin treatment is due to a compensatory upregulation of gene transcription if statin treatment attenuates transcript translation. However, our identification of an association between rs1063320 and HLA-G transcript levels, along with the negative correlation observed between mir-148a, mir-148b and mir-152 expression levels with HLA-G transcripts, strongly suggest that these microRNAs modulate HLA-G transcript levels as well. Thus, further studies are required to ascertain the exact mechanism by which the mir-152 family regulates HLA-G.
Recently it has been called into question whether rs1063320 modifies mir-148a, mir-148b and/or mir-152 regulation of HLA-G. It has been independently reported that mir-148 and mir-152 overexpression reduces HLA-G by targeting its 3’ UTR [38, 42], and that this targeting impacts HLA-G function [42]. Although Tan et al (2007) originally reported that rs1063320 modulated luciferase expression from a HLA-G 3’ UTR reporter construct in the context of mir-148a, mir-148b and mir-152 overexpression, Manaster et al (2012) failed to replicate this finding [27, 42]. Although our results are consistent with the possibility that rs1063320 directly impacts HLA-G regulation, we did not test it directly. Thus, it is possible that the associations observed in the LCLs between rs1063320 with HLA-G transcript and protein in both the statin and sham treated states are mediated by an alternate SNP in high linkage disequilbrium with rs1063320.
Statin use has been associated with reduced COPD related exacerbations as well as overall lower mortality [11, 43–45]. Although there is currently no literature supporting a link between COPD and HLA-G, levels of both mir-148a and mir-152 has been reported independently to be differentially expressed in subjects with COPD [46–48], suggesting that HLA-G may also play a role in COPD. Thus, it is possible that rs1063320 may modulate statin benefit in COPD as well.
Despite the strengths of our study, there are a number of caveats to our results. First, all subjects included in the study received statin treatment during the course of routine clinical care, and statin users were identified by natural language processing of the electronic medical records. Thus, we have no direct measures of drug compliance or pharmacy information. However, we did require that the same statin type and dose were mentioned twice in the EMR within a 6-month time window to increase the likelihood that individuals were dose-stabilized and drug compliant. Furthermore, we extensively validated our algorithms using chart review. We have previously used such techniques to identify genetic variation associated with LDL cholesterol response to statin treatment [36]. Second, although we were able to identify a statistically significant association between rs1063320 and asthma-related exacerbations in statin users, replication in independent cohorts is necessary to confirm this association. Notably, using the same algorithms described above, we identified 453 asthmatic statin users in BioVU, a population based biobank at the Vanderbilt University Medical Center in Tennessee. However, only 6 of the 453 subjects identified had a documented exacerbation after the start of statin use, and thus this cohort was severely underpowered to test for replication. Third, we were not able to control for all known covariates. For example, the relationship between rs1063320 and asthma status has been shown to be dependent on maternal asthma status, and we were unable to reliably extract this information from the EMR [27, 28]. In addition, smoking status is particularly challenging to evaluate based on health records alone [49].
Although the rs1063320 C allele is the ancestral allele, its frequency varies between populations: 45.6% in Northern Europeans (CEPH), 58.1% in Han Chinese, 65.5% in Yorubans and 73.3% in Japanese. Thus, if it is successfully validated that the C allele reduces statin benefits for asthma, given the high prevalence of this variant across the global population, these results could help explain discrepancies in studies of statin effects on the severity of these diseases. Importantly, such a relationship could identify a substantial number of asthmatics worldwide for whom statins may be a viable treatment alternative. However, the current results should be considered preliminary observations and are not intended to guide current statin prescription.
In summary, statins up-regulated mir-148b and mir-152, and affect HLA-G expression while modulated by rs1063320. Consistent with this mechanism, we observed that rs1063320 is associated with frequency of asthma-related exacerbations in statin users. These results provide support for genetically-influenced heterogeneity of treatment response to statins for individuals with asthma. Importantly, rs1063320 genotype could inform future statin use, thus introducing another therapeutic option for the management of asthma.
Supplementary Material
Table 1.
PMRP Subject characteristics
| N | 421 |
| Age (median) | 52 |
| % Male | 34.2 |
| % Inhaled corticosteroid use | 66.7 |
| % Non-smoker (never) | 51.4 |
| % With exacerbation (1, 2 or 3)* | 2.8 |
| Statin Type** | |
| Simvastatin (%) | 26.4 |
| Lovastatin (%) | 9.0 |
| Pravastatin (%) | 5.2 |
| Fluvastatin (%) | 3.5 |
| Atorvastatin (%) | 54.5 |
| Rosuvastatin (%) | 1.4 |
Exacerbation defined as asthma-related ER visit, hospitalization or oral steroid use that occurred within 6 months of a statin mention in the EMR.
Either type of statin listed just prior to exacerbation, or first known statin in those without an exacerbation within 6 months of the first statin mention.
ACKNOWLEDGEMENTS
The work was supported by NIH grants U19 HL069757, R01 HL104133, UL1TR000427, U01 HG006389, U19 HL065962, U01 HL065899 and K08 HL088046. The PGPop Consortium was initiated and funded as a PGRN Network Resource through the grant NIH U19HL065962 awarded to Vanderbilt University. Additional consortium members include Marshfield Clinic, Kaiser Permanente GA, Kaiser Permanente Hawaii, Harvard Pilgrim, Brigham & Women’s Hospital, and RIKEN Center for Integrative Medical Sciences.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Ridker PM, et al. Rosuvastatin to prevent vascular events in men and women with elevated C-reactive protein. N Engl J Med. 2008;359(21):2195–2207. doi: 10.1056/NEJMoa0807646. [DOI] [PubMed] [Google Scholar]
- 2.Camoretti-Mercado B. Targeting the airway smooth muscle for asthma treatment. Transl Res. 2009;154(4):165–174. doi: 10.1016/j.trsl.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paraskevas KI, et al. Emerging indications for statins: a pluripotent family of agents with several potential applications. Curr Pharm Des. 2007;13(35):3622–3636. doi: 10.2174/138161207782794194. [DOI] [PubMed] [Google Scholar]
- 4.Takizawa H. Novel strategies for the treatment of asthma. Recent Pat Inflamm Allergy Drug Discov. 2007;1(1):13–19. doi: 10.2174/187221307779815101. [DOI] [PubMed] [Google Scholar]
- 5.Walsh GM. Defective apoptotic cell clearance in asthma and COPD--a new drug target for statins? Trends Pharmacol Sci. 2008;29(1):6–11. doi: 10.1016/j.tips.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Stanek E, Aubert R, Xia F. Statin exposure reduces the risk of asthma-related hospitalizations and emergency room visits in asthmatic patients on inhaled corticosteroids. J Allergy Clin Immunol. 2009;123:S65. [Google Scholar]
- 7.Huang CC, et al. Statin use in patients with asthma - a nationwide population-based study. Eur J Clin Invest. 2010 doi: 10.1111/j.1365-2362.2010.02434.x. [DOI] [PubMed] [Google Scholar]
- 8.Alexeeff SE, et al. Statin use reduces decline in lung function: VA Normative Aging Study. Am J Respir Crit Care Med. 2007;176(8):742–747. doi: 10.1164/rccm.200705-656OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keddissi JI, et al. The use of statins and lung function in current and former smokers. Chest. 2007;132(6):1764–1771. doi: 10.1378/chest.07-0298. [DOI] [PubMed] [Google Scholar]
- 10.Dobler CC, Wong KK, Marks GB. Associations between statins and COPD: a systematic review. BMC Pulm Med. 2009;9:32. doi: 10.1186/1471-2466-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janda S, et al. Statins in COPD: a systematic review. Chest. 2009;136(3):734–743. doi: 10.1378/chest.09-0194. [DOI] [PubMed] [Google Scholar]
- 12.Tse SM, et al. Statin exposure is associated with decreased asthma-related emergency department visits and oral corticosteroid use. Am J Respir Crit Care Med. 2013;188(9):1076–1082. doi: 10.1164/rccm.201306-1017OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeki AA, et al. Statin use and asthma control in patients with severe asthma. BMJ Open. 2013;3(8) doi: 10.1136/bmjopen-2013-003314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tse SM, et al. Statin use in asthmatics on inhaled corticosteroids is associated with decreased risk of emergency department visits. Curr Med Res Opin. 2014;30(4):685–693. doi: 10.1185/03007995.2013.865599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hothersall EJ, et al. Effects of atorvastatin added to inhaled corticosteroids on lung function and sputum cell counts in atopic asthma. Thorax. 2008;63(12):1070–1075. doi: 10.1136/thx.2008.100198. [DOI] [PubMed] [Google Scholar]
- 16.Menzies D, et al. Simvastatin does not exhibit therapeutic anti-inflammatory effects in asthma. J Allergy Clin Immunol. 2007;119(2):328–335. doi: 10.1016/j.jaci.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Rubin BK. Statins for the treatment of asthma: a discovery well, dry hole or just snake oil. Thorax. 2009;64(1):4–5. doi: 10.1136/thx.2008.106757. [DOI] [PubMed] [Google Scholar]
- 18.Simon JA, et al. Phenotypic predictors of response to simvastatin therapy among African-Americans and Caucasians: the Cholesterol and Pharmacogenetics (CAP) Study. Am J Cardiol. 2006;97(6):843–850. doi: 10.1016/j.amjcard.2005.09.134. [DOI] [PubMed] [Google Scholar]
- 19.Mangravite LM, Krauss RM. Pharmacogenomics of statin response. Curr Opin Lipidol. 2007;18(4):409–414. doi: 10.1097/MOL.0b013e328235a5a2. [DOI] [PubMed] [Google Scholar]
- 20.Nicolae D, et al. Fine mapping and positional candidate studies identify HLA-G as an asthma susceptibility gene on chromosome 6p21. Am J Hum Genet. 2005;76(2):349–357. doi: 10.1086/427763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou H, et al. Analyses of associations between three positionally cloned asthma candidate genes and asthma or asthma-related phenotypes in a Chinese population. BMC Med Genet. 2009;10:123. doi: 10.1186/1471-2350-10-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ober C. HLA-G: an asthma gene on chromosome 6p. Immunol Allergy Clin North Am. 2005;25(4):669–679. doi: 10.1016/j.iac.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Moffatt MF, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363(13):1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rizzo R, et al. Defective production of soluble HLA-G molecules by peripheral blood monocytes in patients with asthma. J Allergy Clin Immunol. 2005;115(3):508–513. doi: 10.1016/j.jaci.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 25.White SR, et al. Levels of soluble human leukocyte antigen-G are increased in asthmatic airways. Eur Respir J. 2010;35(4):925–927. doi: 10.1183/09031936.00164809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng XQ, et al. Analysis of the plasma soluble human leukocyte antigen-G and interleukin-10 levels in childhood atopic asthma. Hum Immunol. 2010;71(10):982–987. doi: 10.1016/j.humimm.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 27.Tan Z, et al. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet. 2007;81(4):829–834. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicodemus-Johnson J, et al. Maternal asthma and microRNA regulation of soluble HLA-G in the airway. J Allergy Clin Immunol. 2013;131(6):1496–1503. doi: 10.1016/j.jaci.2013.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medina MW, et al. Alternative splicing of 3-hydroxy-3-methylglutaryl coenzyme A reductase is associated with plasma low-density lipoprotein cholesterol response to simvastatin. Circulation. 2008;118(4):355–362. doi: 10.1161/CIRCULATIONAHA.108.773267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mangravite LM, et al. A statin-dependent QTL for GATM expression is associated with statin-induced myopathy. Nature. 2013;502(7471):377–380. doi: 10.1038/nature12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barber MJ, et al. Genome-wide association of lipid-lowering response to statins in combined study populations. PLoS One. 2010;5(3):e9763. doi: 10.1371/journal.pone.0009763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie MD, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010;86(4):560–572. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roden DM, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84(3):362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilke RA, et al. Use of an electronic medical record for the identification of research subjects with diabetes mellitus. Clin Med Res. 2007;5(1):1–7. doi: 10.3121/cmr.2007.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denny JC, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31(12):1102–1110. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei WQ, et al. Characterization of statin dose response in electronic medical records. Clin Pharmacol Ther. 2014;95(3):331–338. doi: 10.1038/clpt.2013.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu M, et al. A study of transportability of an existing smoking status detection module across institutions. AMIA Annu Symp Proc. 2012;2012:577–586. [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu XM, et al. Overexpression of miR-152 leads to reduced expression of human leukocyte antigen-G and increased natural killer cell mediated cytolysis in JEG-3 cells. Am J Obstet Gynecol. 2010;202(6):592, e1–e7. doi: 10.1016/j.ajog.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 39.Donadi EA, et al. Implications of the polymorphism of HLA-G on its function, regulation, evolution and disease association. Cell Mol Life Sci. 2011;68(3):369–395. doi: 10.1007/s00018-010-0580-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bortolotti D, et al. An accurate and reliable real time SNP genotyping assay for the HLA-G +3142 bp C>G polymorphism. Tissue Antigens. 2012;80(3):259–262. doi: 10.1111/j.1399-0039.2012.01926.x. [DOI] [PubMed] [Google Scholar]
- 41.Zambra FM, et al. Response to Bortolotti et al. 2012--a re-evaluation of our polymerase chain reaction-restriction fragment length polymorphism genotyping method. Tissue Antigens. 2013;82(4):286–287. doi: 10.1111/tan.12184. [DOI] [PubMed] [Google Scholar]
- 42.Manaster I, et al. MiRNA-mediated control of HLA-G expression and function. PLoS One. 2012;7(3):e33395. doi: 10.1371/journal.pone.0033395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang CC, et al. Statin use and hospitalization in patients with chronic obstructive pulmonary disease: a nationwide population-based cohort study in Taiwan. Clin Ther. 2011;33(10):1365–1370. doi: 10.1016/j.clinthera.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 44.Lahousse L, et al. Statins, systemic inflammation and risk of death in COPD: The Rotterdam study. Pulm Pharmacol Ther. 2012 doi: 10.1016/j.pupt.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 45.Lawes CM, et al. Statin use in COPD patients is associated with a reduction in mortality: a national cohort study. Prim Care Respir J. 2012;21(1):35–40. doi: 10.4104/pcrj.2011.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanfiorenzo C, et al. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PLoS One. 2013;8(1):e54596. doi: 10.1371/journal.pone.0054596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ezzie ME, et al. Gene expression networks in COPD: microRNA and mRNA regulation. Thorax. 2012;67(2):122–131. doi: 10.1136/thoraxjnl-2011-200089. [DOI] [PubMed] [Google Scholar]
- 48.Soeda S, et al. Clinical relevance of plasma miR-106b levels in patients with chronic obstructive pulmonary disease. Int J Mol Med. 2013;31(3):533–539. doi: 10.3892/ijmm.2013.1251. [DOI] [PubMed] [Google Scholar]
- 49.Garies S, et al. Sentinel eye: improving usability of smoking data in EMR systems. Can Fam Physician. 2013;59(1):108, e60–e61. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.