Abstract
While in nucleotide sequencing, the analysis of DNA from complex mixtures of organisms is common, this is not yet true for mass spectrometric data analysis of complex mixtures. The comparative analyses of mass spectrometry data of microbial communities at the molecular level is difficult to perform, especially in the context of a host. The challenge does not lie in generating the mass spectrometry data, rather much of the difficulty falls in the realm of how to derive relevant information from this data. The informatics based techniques to visualize and organize datasets are well established for metagenome sequencing; however, due to the scarcity of informatics strategies in mass spectrometry, it is currently difficult to cross correlate two very different mass spectrometry data sets from microbial communities and their hosts. We highlight that molecular networking can be used as an organizational tool of tandem mass spectrometry data, automated database search for rapid identification of metabolites, and as a workflow to manage and compare mass spectrometry data from complex mixtures of organisms. To demonstrate this platform, we show data analysis from hard corals and a human lung associated with cystic fibrosis.
Keywords: Molecular networking, mass spectrometry, complex mixtures, spectral matching, Cytoscape, database search
1. Introduction
Study of complex mixtures at the molecular level using global untargeted metabolomics contributes a better understanding of the influence of various domains of life on each other, and their environment. Such mixtures may represent environmental samples taken from the soil or sewers; marine communities such as algae, corals, lichens, and diseased human organs such as the gut, oral cavity, lungs, pancreas, and kidneys, to name a few. Due to the enormous diversity of molecules present in such communities, the analysis of mass spectrometry data obtained from such samples often surpasses the capacity of modern informatics analysis. Modern mass spectrometry (MS) represents a powerful tool for studying metabolomics due to its speed, reproducibility, unsurpassed resolution, broad dynamic range, and ability to analyze samples of tremendous complexity (1–3). Rapid development and advancement of mass spectrometry based techniques such as ultra high performance liquid chromatography (UPLC)-MS (4–6), nanoLC-MS (7–8), nano-electrospray ionization-MS (9–10), nanospray desorption ionization-MS (11–12), LTQ and LTQ-Orbitrap hybrid fourier transform ion cyclotron-MS (13–14), paper spray ionization-MS (15), direct analysis in real time-MS (16–17), and imaging-MS (18–20) have made it possible to generate tandem MS data on complex mixtures without a significant investment of time in regards to sample preparation. Although development of multiple high-throughput approaches such as metagenomics, metatrancritptomics, and metaproteomics have made it possible to begin understanding the physiology of complex mixtures, interest in metabolomics has seen a recent surge. Thus, tools for rapid acquisition of MS data are now available, but MS based analysis of complex mixtures is still plagued by the lack of robust tools for data visualization and metabolite identification which can then be used to derive correlations of this data with metagenomics, metatrancritptomics, metaproteomics and KEGG pathway mapping analysis. The unmanageable amount of data, also referred to as “Big Data” generated using these approaches necessitate the development of methodologies to analyze and interpret the large volume of data as well as robust databases to classify and identify molecular features. This has been performed in the past by first prioritizing data using univariate and multivariate statistical analysis such as principal component analysis, partial least square discriminant analysis, t-tests, and hierarchical clustering which entails organizing data into matrices that are compatible with such analysis (21–25). The peak picking and evaluation for this analysis is usually performed by using instrument specific softwares such as Bruker® Data Analysis and Bruker® Profile Analysis for Bruker, MassHunter for Agilent, MarkerLynx for Waters or publically available softwares such as XCMS (26), MZmine, (27) (28–29) and MET-IDEA (30) which can handle data from different instruments. Such analysis helps in organization and classification of data, highlighting important metabolites, intensities of which differ between sample types but does not aid in overall identification of individual molecules or their biological origin which is indeed the major bottleneck in metabolomics of complex mixtures. XCMS Online was developed to overcome this challenge. Herein, following the bioinformatic analysis of datasets, tandem MS data of peaks that differ significantly in intensities between two sample sets are matched with tandem MS data available in the database METLIN (31–32) to aid compound identification. A similar tandem MS mass spectral search approach has also been utilized in the past, for example, to identify metabolites in algal secretions (33) and for identification of lipids from LipidBlast database (34) using NIST MS Search GUI program. Tandem MS containing databases such as NIST (35), METLIN (31), HMDB (36), and MassBank (37) can be utilized for rapid annotation of Big Data generated from complex mixtures. Thus, retention time alignment and similarity in tandem MS spectra are now widely accepted characteristic metabolite features used to generate a high probable structural match. However databases are not comprehensive in the diversity of molecules nor can they be used to begin annotating molecules that are simply related but are not present in the database. Furthermore most molecules that can be found in complex mixtures of organisms have simply never been characterized before.
Thus, analysis of complex mixtures that yields millions of tandem MS spectra requires development of algorithms that organize Big Data in a fashion wherein one can exploit already existing data in the form of public databases or in-house generated databases to identify known molecules as well as new molecules. One such algorithm termed “Molecular networking” exploits the chemical and structural similarity of molecules that is reflected in the similarity of the MS/MS spectra to organize large data sets (38–41). Molecular networks is an effective approach that can be used to tease apart the origin and ID complex samples. Herein, similar MS/MS spectra are grouped into clusters where different clusters represent different classes of molecules. A network of such clusters can be easily visualized and analyzed using Cytoscape or other networking program. In Cytoscape, nodes corresponding to the database hits can be color coded and various relevant attributes such as retention time, parent mass, signal intensity etc. can be visualized in the Cytoscape or equivalent networking display software. Further, use of molecular networking allows rapid identification of molecules/metabolites that are similar to database hits since these molecules cluster together.
Herein, we show that molecular network analysis and automated matching of tandem MS data available in public/in-house database is one representative analysis workflow for molecular analysis of complex mixtures and to compare and contrast data sets of very different origin. In this publication, we demonstrate how one can tease apart complex mixtures at the molecular level by the use of molecular networking and database matching on a cystic fibrosis (CF)-afflicted human lung and hard corals both of which represent complex mixtures of microbial communities.
2. Results and Discussion
2.1 Metabolomics workflow of complex mixtures
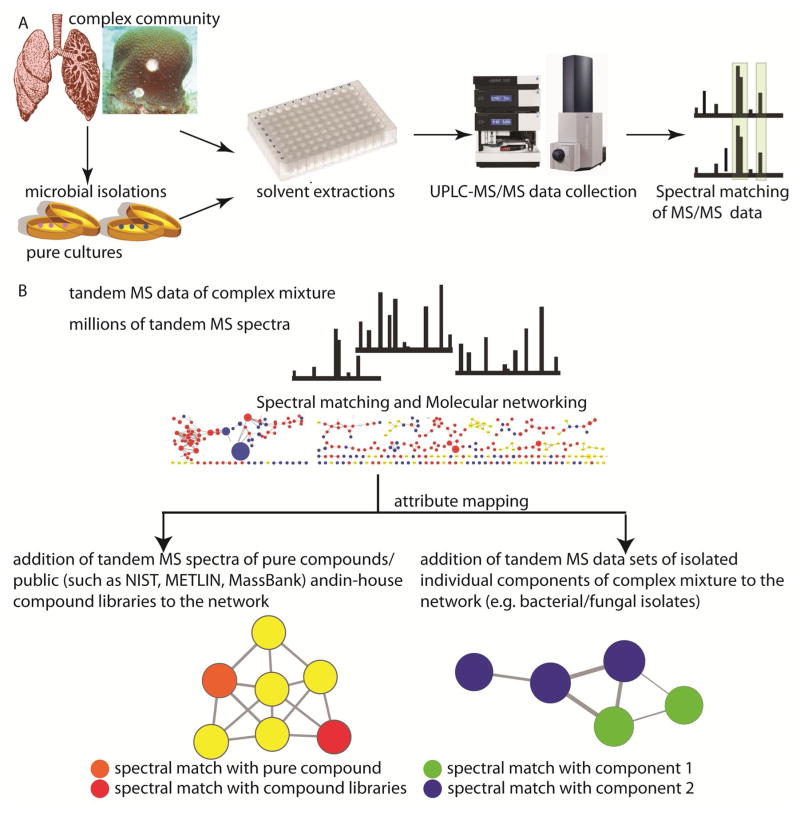
A typical workflow is shown in Figure 1. The first step entails solvent extractions of the complex sample and its individual members. After extraction in appropriate solvents, UPLC-MS and tandem MS data is generated in second step. The Big Data generated in the second step is then organized using molecular networking via spectral matching of tandem MS spectra (Figure 1a). The molecular networks generated via spectral matching create a network of similarity of spectra and are visualized in Cytoscape. The similarity in fragmentation patterns due to similarity in structures under collision-induced dissociation dictates the clustering of metabolites in molecular networking. Each node consists of n number of identical MS/MS spectra present in the samples being analyzed (where n can be any number) and the neighboring nodes consists of MS/MS spectra that are related to each other. The clustering of MS/MS spectra within a node and between nodes is performed using an established algorithm namely “MS-cluster” (42). The nodes are connected by edges where edge thickness is given by cosine score and depicts the relatedness between MS/MS spectra of connecting nodes. The mass spectrometry data analysis of complex mixtures generates thousands to millions of spectra that can generate very complex networks. Visualization in Cytoscape allows attribute mapping where the database hits, the hits from in-house generated pure compound libraries, as well entire datasets of the individual components of the complex mixture are color coded to aid rapid identification (Figure 1b). The neighboring nodes then represent molecules similar to the one obtained from database hit that were not present in the database, and can be dereplicated by comparing mass shifts in MS2 fragments (41). Further, one can add entire data sets of different components of a complex mixture to tease apart the origin of these molecules. In order to demonstrate the potential of molecular networking in utilizing tandem MS data deposited in large databases and acquired on various different biological sample sets, we performed this analysis on anatomically distinct regions of CF-afflicted human lung and hard corals. Molecules with similar structures and hence similar fragmentation patterns clustered together and spectral matching with tandem MS library from NIST11 and METLIN aided identification of metabolites. Further, a combined molecular network of lung tissue and bacterial isolates obtained from the lung tissue was created to tease apart human and bacterial metabolites. The lung tissue and coral LC-MS/MS data was networked together to represent how metabolome of different complex communities can be compared and the commonalities identified using this approach.
Figure 1. Analysis of complex mixture.
A) The first part of workflow is focused at generating tandem MS data. The complex mixture and its isolated members are extracted with appropriate solvents. The extracts then enter the mass spectrometry pipeline where MS1 and MS2 data is simultaneously generated using high-scan-speed mass spectrometer coupled to a UPLC system. The second part of the workflow is focused at data organization which starts with spectral matching of tandem MS data. B) The high density of tandem MS data generated is then organized using molecular networking algorithms. These networks are visualized using Cytoscape, a tool designed to visualize correlations of large data sets. Each node represents one consensus MS/MS spectrum and an edge represents similarity, where thickness of the edge indicates cosine similarity. Molecules with similar structures and hence similar MS/MS fragmentation patterns cluster together. Networking of tandem MS data allows for identification of metabolites by addition of tandem MS data from publically available databases and in-house generated databases to the network. The origin of molecules can be obtained by addition of entire datasets of individual components that are isolated from the complex mixture.
2.2 Tandem MS guided molecular networking analysis
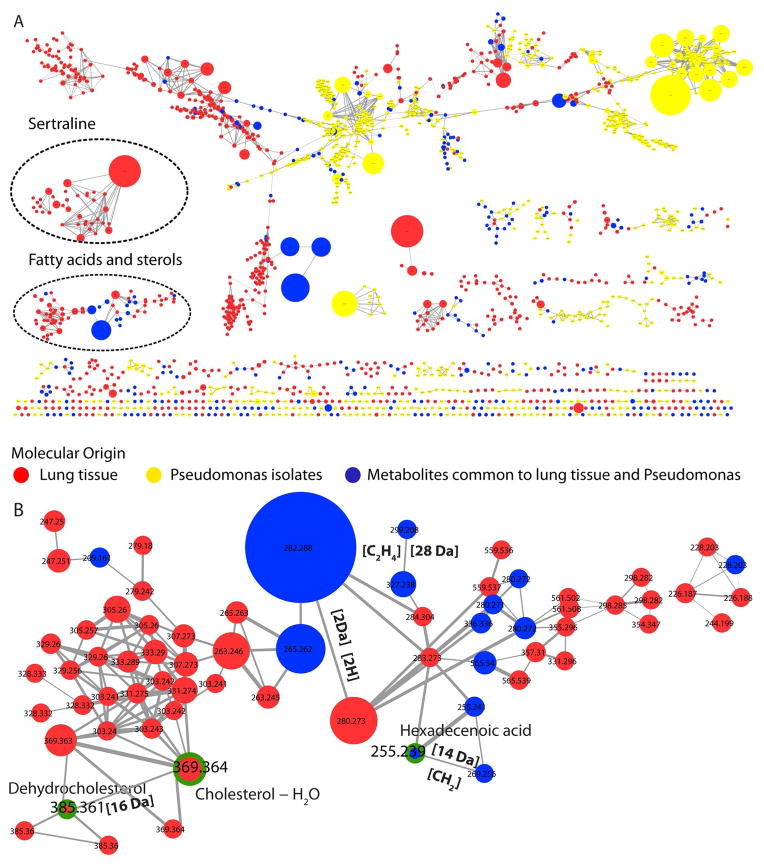
The sample set consisted of organic solvent extractions of ten anatomically distinct locations of ex-plant lung tissue and twenty Pseudomonas spp. isolates from a CF patient. In the current study, UPLC-ESI tandem MS data was collected on methanol and ethyl acetate extracts in the positive mode and organized using molecular networking (Figure 2). The size of nodes represents intensity of the parent ion. As an example, one of the clusters is highlighted in Figure 2b showing mass shifts of 2 Da, 14 Da and 28 Da between nodes suggesting a molecular family of fatty acids or lipids. The molecular networks form the lungs revealed matches to lipids, fatty acids, sterols, as well as drugs that were administered to the patient using spectral matching of tandem MS data generated in this study with tandem MS data available from the database NIST11, METLIN and in-house database generated from commercially available FDA-approved drug library (Selleckchem). The hits generated by spectral matching of tandem MS data are visualized in Cytoscape and color coded in red for ease of identification (Figure 3). For example, hexadecenoic acid, cholesterol, sertraline (antidepressant that was administered to the patient), were identified as a hit to the spectra of hexadecenoic acid, cholesterol, and sertraline available in NIST11 and METLIN database (Nist id 11552, 16480, and 642 respectively) (Figure 2b, 4 and S1).
Figure 2. Molecular network analysis.
The tandem MS data generated from extracted explants lung tissue (red nodes), and agar extractions of Pseudomonas spp. isolates (yellow nodes) obtained from the same lung is organized in molecular networks. The nodes in blue represent the metabolites that were common to lung tissue and Pseudomonas spp. isolates. The size of the node represents the intensity of parent mass in the MS spectrum. This representation is semi-quantitative and should be used as a guide to visualize the abundant metabolites present in the sample set for further investigation such as isolation. The metabolites identified using spectral matching with public databases such as NIST11, METLIN and in-house generated database for Pseudomonas spp. metabolites are labeled (vide infra). The green lines around the nodes highlight few representative database hits.
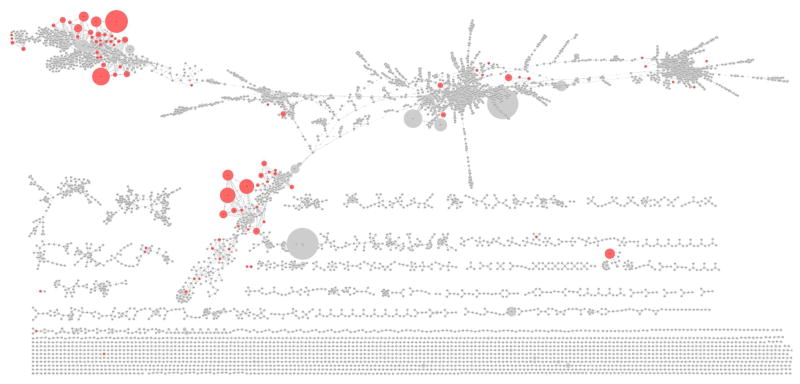
Figure 3. Molecular networking of lung data set with entire tandem MS depository of NIST11 database.
The database hits are imported as an attribute in the network and can be easily visualized. The hits obtained from NIST11 database by matching tandem MS data of lung tissue sections are colored in red.
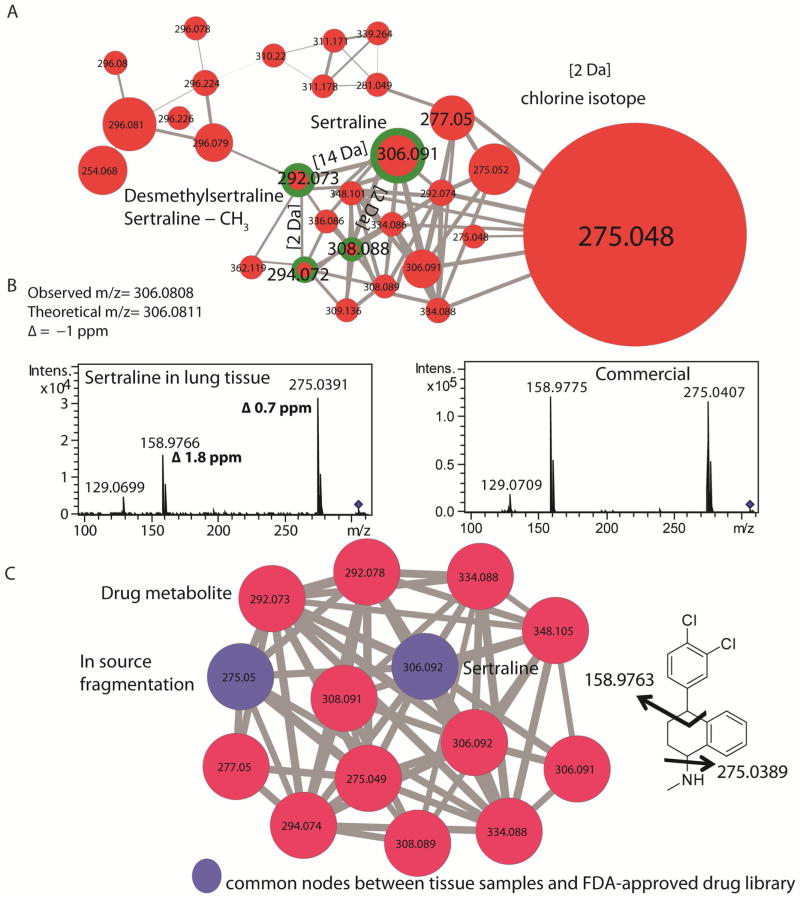
Figure 4. Analysis of cluster corresponding to sertraline.
A) The cluster corresponding to the drug sertraline was pulled out from the network shown in Figure 3. The parent drug sertraline (m/z 306), the metabolite desmethylsertraline (m/z 292), and their chlorine isotopes are highlighted with green circles. The size of the node represents the intensity of parent mass in the MS spectrum. This representation is semi-quantitative and should be used as a guide to visualize the abundant metabolites present in the sample set for further investigation such as isolation. B) The tandem MS spectra for sertraline from the lung section and the in-house FDA library are shown. The ppm errors for the parents mass and the two tandem MS fragments are shown and were below 2 ppm. C) The desmethylamine metabolite was observed only in lung sections (red node) and was not observed in the tandem MS data on commercial sertraline further supporting the annotation of this node as a real drug metabolite.
As expected, sertraline and cholesterol were observed in only human samples (red nodes, Figure 4) where as hexadecenoic acid was observed in both human and Pseudomonas spp. samples (blue nodes, Figure 4). When a node is selected, attributes that are imported into the Cytoscape allow for direct visualization of the parent mass, the sample names that contain tandem MS, intensity of the parent mass as well as the identity of the putative database hit. Visualization of database hits in the form of molecular networking also aids in annotation of neighboring nodes that cluster with the hit due to similarity in fragmentation pattern which in turn is dictated by the similarity in chemical structure. Thus, origin of metabolites can be easily deciphered using color coding in Cytoscape. For sertraline, the neighboring nodes were annotated to be the metabolite N-desmethylsertraline (43) based on loss of 14 Da (Figure 4). The database hits can be further confirmed by comparing the ppm error of the matched fragments, annotation of the observed fragments, and by incorporating tandem MS data of the standard compound if needed (Figure 4b and 4c). The data from lung tissue extracts was co-networked with an in-house database of FDA-approved drug library containing sertraline and similar fragmentation patterns were observed for the drug sertraline from lung tissue sections and commercial sertraline in FDA library (Figure 4b). The tandem MS fragments shown in Figure 3b had ppm error of 1.8 ppm and 0.7 ppm, generating confidence in the observed hit. Further, when the data from lung tissue extracts was co-networked with an in-house database of FDA-approved drug library MS/MS data set, node corresponding to desmethylsertraline was only found in lung tissue sections. The FDA MS/MS database will be made available to the public in the near future and is a part of our global natural product social molecular networking effort to make MS data publicly available. Therefore without the need for clinical information, the use of specific classes of medication could be directly assessed. The node with m/z 275 was identified as common nodes (blue) (Figure 4c) suggesting in-source fragmentation since this peak was also the dominant fragment ion observed in tandem MS data. This suggests that such an analysis may also aid in separation of real metabolites from MS artifacts. Thus, molecular networking combined with spectral matching of tandem MS data and visualization of these results in Cytoscape allows for rapid identification of molecules present in complex mixtures. Molecular networking then also allows for identification of neighboring nodes, the tandem MS data of which may not be present in the databases.
2.3. Identification of common metabolites between coral samples and lung tissue
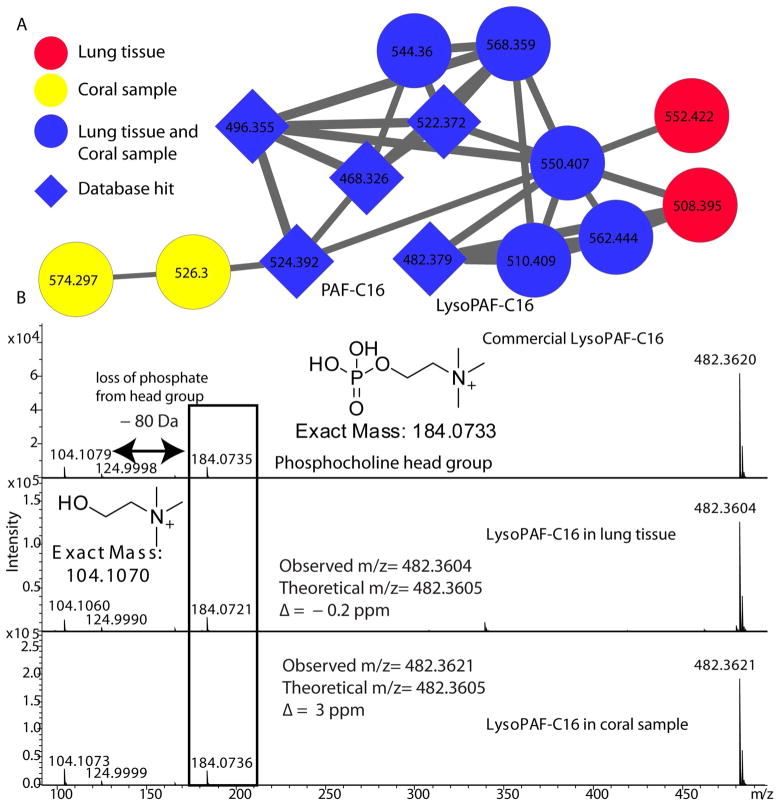
The alignment of DNA sequences from similar or very divergent organisms is commonly used in biology to study associations between different life forms. No such parallels exist in aiding comparisons of large amounts of data generated in mass spectrometry analysis to study associations at the molecular level. Molecular networking can be easily exploited to compare large mass spectrometry based data sets originating from different sources. In order to demonstrate this, UPLC-tandem MS data was collected on eight sections of a hard coral sample. A molecular network of tandem MS data of coral samples and lung tissue samples was created to highlight how molecular features between two different complex mixtures can be compared to identify commonalities and differences between the metabolome (Figure S2). The nodes containing tandem MS data corresponding to lung tissue are colored in red, and nodes containing tandem MS data corresponding to coral samples are colored in yellow. The blue nodes contain tandem MS data from both lung tissue and coral samples. The tandem MS data was matched with the tandem MS data from databases NIST11 and METLIN. The nodes with matches from NIST and METLIN database are shown as diamond shaped nodes. Co-networking of tandem MS data from coral samples with lung data combined with spectral matching with tandem MS data from NIST11 and METLIN enabled identification of phosphocholine head group containing inflammatory lipids (Platelet activating factor (PAF) (44) and lyso-PAF (45)) in coral samples (Figure 5 and Figure S3). In order to confirm the identity of lyso-PAF, this metabolite was isolated by liquid chromatography purification and characterized by NMR. 1H NMR spectrum of the compound from coral samples (in CD3CN) displayed 51 protons. The presence of three singlet methyl protons (δ 3.11) and two methylene protons (δ 4.20 and 3.51) indicated that the compound has a choline group (Figure S4) (46). Furthermore, 1H-1H COSY analyses revealed the presence of three partial structures (a saturated acyl chain (C16), -O-CH2-(CH-OH)-CH2-O-derived from glycerophosphoric acid and -CH2-CH2- in the choline region). These results strongly suggested that the compound belongs to a phosphatidylcholine family. The previous 1H NMR data is also in good agreement with the molecule isolate from corals (46). Finally, to identify the compound, we compared tandem MS spectra and retention time of the authentic sample (Bachem Biochemicals) using UPLC-MS. As a result, the MS fragment pattern and retention time of authentic sample was identical to that of the compound (Figure 5 and S5). Platelet activating factor (PAF) and lyso-PAF has not been previously identified from corals highlighting the utility of molecular networking for comparing very different datasets and entire databases for rapid identification of unknown molecules in a given complex mixture. Effective comparisons between different data sets would require similar extraction protocols and LC-MS/MS methods. Nonetheless, molecular networking is still capable of quickly identifying individual compounds that are present in one or more data sets, regardless of the source of the samples or how they were prepared/extracted as demonstrated by the identification of lyso-PAF in coral samples on comparison of coral data set with lung data set.
Figure 5. Identification of lyso-PAF in coral dataset.
A) The cluster corresponding to the lyso-PAF was pulled out from the combined network of lung tissue sections, coral sections and database hits (Figure S2). The database hits shown as diamond shaped nodes and the common nodes between lung (red) and coral (yellow) sections are shown in blue. B) The tandem MS spectra of lyso-PAF in lung and coral sections matched with tandem MS spectra of commercial lyso-PAF. Lyso-PAF was isolated from coral sections and the identity was also confirmed by NMR after isolation.
3. Conclusions
Molecular analysis of complex mixtures is a daunting task. The most challenging step in the molecular analysis of complex mixtures involves mining of large data sets to reveal the identity of molecules present and to decipher the sources of these molecules. The availability of tools for bioinformatics and presentation of DNA and RNA sequencing data sets has revolutionized the field of Big Data processing. Using these tools, one can very easily compare various different organisms, query their biosynthetic potential, virulence factors, immune responses, community interactions, and identify healthy vs. diseased environments. Thus, informatics of mass spectrometry underlies development of such tools that can then aid in comparing all of these processes between different organisms at the molecular level. Whereas genomics and proteomics analysis is aided by the availability of well annotated databases and informatics tools, metabolomics analysis suffers from lack of such tools and comprehensive metabolite databases. Furthermore, sequencing can be correlated even when the function of the gene remains unknown. Lack of tools that can organize and present large MS data sets hamper direct comparisons between complex communities at the molecular level. Herein, we have highlighted how this can be achieved by the use of molecular networking algorithms. First, the tandem MS data obtained from complex mixtures is directly compared with tandem MS data of known molecules present in public/in-house generated databases as well tandem MS data obtained on individual components of complex mixtures. We demonstrated this concept by co-networking tandem MS data obtained on a CF-afflicted human lung and tandem MS data obtained on Psuedomonas isolates with tandem MS data available in NIST11 and METLIN database. Second, co-networking of annotated data sets with tandem MS data of unknown data sets can aid in comparisons between different complex mixtures. Comparison of tandem MS data obtained on extracted hard corals with tandem MS data of lung sections enabled identification of a previously unknown inflammatory lipid in coral samples. Using this approach, one can envision the potential of creating libraries of tandem MS data of various classes of molecules such as lipids, peptides, hormones, signaling molecules, drugs, environmental toxins, natural products and entire annotated data sets of complex mixtures aiding automated annotation of molecular features of new complex mixtures. Automation of informatic analysis on these repositories necessitates development of algorithms such as molecular networking that can then be extended to derive molecular relationships and biologically relevant hypothesis between different sample sets as is done by the genome sequencing community.
4. Materials and Methods
4.1. Sample collection and extraction of metabolites from lung sections
The ex-plant CF lung tissue was collected following ex-plant in accordance with University of California Institutional Review Board (HROO 081500) an San Diego State University Institutional Review Board (SDSU IRB#2121). A total of ten sections were cut out from the left lung and each section was further divided into thin slices; two slices were used for mass spectrometry, and one slice was used for isolating bacteria. For isolation of Psuedomonas spp., see section 4.2. For chemical extraction, each section was weighed and incubated in ethyl acetate (10 μL/mg of tissue) for 1 hr. Following centrifugation, ethyl acetate was transferred to fresh vial and evaporated. The remaining tissue was re-extracted in methanol for 1 hr. Following centrifugation, methanol was evaporated. The dried samples were stored at −80 °C until further use.
4.2. Isolation of Pseudomonas spp. and extraction of metabolites
Pseudomonas spp. were isolated on Pseudomonas selective agar (cetrimide agar). Two isolates were randomly selected from each lung section and grown on Luria Broth (LB). Liquid culture was then separated into multiple aliquots to initiate the glycerol stock for storage at −80 °C. Prior to metabolites extraction, the growth of bacteria was initiated from the glycerol stock on cetrimide agar, and colonies were streaked onto ISP2 medium. All isolates were grown overnight at 37 °C. For each isolate, two colonies were excised from the ISP2 agar and transferred to a tube containing either ethyl acetate or methanol and incubated in the respective solvent for 1 hr. Following extraction and centrifugation, the solvent was transferred to a fresh tube, dried, and the dried samples were stored at −80 °C.
4.3. Collection of coral samples and extraction of metabolites
Coral plug samples from Montastraea annularis were flash frozen in liquid nitrogen. Plugs were thawed, weighed, and extracted (10 mL/gram of coral sample) with 70:30 methanol:H2O for 72 hr at 25°C. The extracts were transferred to fresh vials and evaporated. Dried samples were stored at −80°C until further use.
4.4 UPLC-MS/MS analysis
The extracted metabolites were dissolved in acetonitrile and analyzed with UltiMate 3000 UPLC system (Thermo Scientific) using a Kinetex™ 1.7 μm C18 reversed phase UHPLC column (50 × 2.1 mm) and Maxis Q-TOF mass spectrometer (Bruker Daltonics) equipped with ESI source. The gradient employed for chromatographic separation was 10% solvent B (98% acetonitrile, 0.1% formic acid in LC-MS grade water with solvent A as 0.1% formic acid in water) for 1.5 min, a step gradient of 10% B-50% B in 0.5 min, held at 50% B for 2 min, a second step of 50% B-100% B in 6 min, held at 100% B for 0.5 min, 100%-10 % B in 0.5 min and kept at 10% B for 0.5 min at a flow rate of 0.5 mL/min throughout the run. MS spectra were acquired in positive ion mode in the range of 100–2000 m/z. An external calibration with ESI-L Low Concentration Tuning Mix (Agilent technologies) was performed prior to data collection and internal calibrant Hexakis(1H,1H,3H-tertrafluoropropoxy)phosphazene was used throughtout the runs. The capillary voltage of 4500 V, nebulizer gas pressure (nitrogen) of 2 bar, ion source temperature of 200 °C, dry gas flow of 9 L/min source temperature, spectral rate of 3 Hz for MS1 and 10 Hz for MS2 was used. For acquiring MS/MS fragmentation, 10 most intense ions per MS1 were selected and collision induced dissociation energy given in Table 1 was used. Basic stepping function was used to fragment ions at 50% and 125% of the CID calculated for each m/z from Table 1 with timing of 50% for each step. Similarly, basic stepping of collision RF of 550 and 800 Vpp with a timing of 50% for each step and transfer time stepping of 57 and 90 μs with a timing of 50% for each step was employed. MS/MS active exclusion parameter was set to 3 and released after 30 seconds. The mass of internal calibrant was excluded from the MS2 list. The data for coral samples and FDA-approved drug library (Selleckchem) was collected using the same methodology with two modifications. The initial conditions for UHPLC were 5% solventB rather than 10% solvent B and the range for MS acquisition was 50–2000 m/z.
Table 1.
Collision induced energies used for tandem MS data collection.
| Type | Mass | Width | Collision | Charge State |
|---|---|---|---|---|
| Base | 100 | 4 | 22 | 1 |
| Base | 100 | 4 | 18 | 2 |
| Base | 300 | 5 | 27 | 1 |
| Base | 300 | 5 | 22 | 2 |
| Base | 500 | 6 | 35 | 1 |
| Base | 500 | 6 | 30 | 2 |
| Base | 1000 | 8 | 45 | 1 |
| Base | 1000 | 8 | 35 | 2 |
| Base | 2000 | 10 | 50 | 1 |
| Base | 2000 | 10 | 50 | 2 |
4.5 Isolation and NMR characterization of lyso-PAF from hard coral M. annularis
For isolation of lyso-PAF from coral samples, the extracted material was injected onto a reverse phase-HPLC column (Kinetex C18 5 μm, 100 Å, 250×4.6mm). Following injection, the column was kept at 2% solvent A (solvent A = 0.086% Formic Acid in 2% ACN/98% water) for 5 min followed by a gradient of 2–100% of solvent B (0.086% Formic Acid in 100% ACN/0% water) over 60 min. The fractions containing lyso-PAF (identified by LTQ-MS) were lyophilized and analyzed by NMR. 1H-NMR and COSY data was acquired in pyridine-d5 on a 600 MHz Varian instrument.
4.6. Molecular networking and data visualization
The LC-MS/MS raw data files were converted into the mzXML format using Bruker® Data Analysis software version 4.1. Molecular networks were generated using MS-Cluster (42). A mass tolerance of 0.05 Da for parent mass, 0.01 Da for fragment ions was utilized and a cosine score of 0.7 was used to create networks. Visualization of the output from molecular networking was performed in Cytoscape 2.8.1 (47). The edge label width was set to cosine score and nodes were labeled with parent mass. The mass labeled on the node is not lock mass corrected and hence should not be used to calculate ppm error. The discrepancy arises due to the error in the script utilized in the data analysis software to convert raw data to mzXML format. While, the peaks in the spectrum are lock mass corrected but the parent mass in the header of the mzXML file contains the mass observed prior to application of lock mass calibration. The correct calibration is applied to all the spectra including MS1 and MS2 and therefore does not impact the molecular networking itself. Such discrepancy results in multiple nodes of the same m/z when the difference in parent mass is higher than 0.05 Da. Multiple nodes for the same m/z also arise due to the difference in quality of the MS/MS spectra acquired at vastly different intensities of parent MS. The nodes corresponding to the solvent controls and blank extractions were deleted from the network. The network was then organized using FM3 layout. MS/MS data from NIST11 and Metlin in .mgf format were compared to the tandem MS spectra obtained on lung and coral sections. The database hits were displayed in Cytoscape with color code defined in the respective figures.
Supplementary Material
Highlights.
MS/MS molecular networking was used to analyze complex mixtures.
Data from in-house drug libraries, NIST, METLIN was used for rapid identification.
Lyso-PAF was identified in coral samples using lung data set and NIST database.
Acknowledgments
We would like to acknowledge Mark Vermeiji for help with coral sample collection, Tal Luzzatto Knaan for help with development of UPLC method, and Robert Quinn for his help with tissue homogenization and insights into this work. This work was supported by the NIH as a metabolomics supplement to parent grant GM095384 and from the UCSD Clinical and Translational Research Institute Pilot award. Clifford Kapono is supported by the graduate student fellowship, Tribal Membership Initiative, by UCSD. The visit by Nobuhiro Koyama is supported by School of Pharmacy, Kitasato University, Tokya, Japan.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhang A, Sun H, Wang P, Han Y, Wang X. Modern analytical techniques in metabolomics analysis. Analyst. 2012;137:293–300. doi: 10.1039/c1an15605e. [DOI] [PubMed] [Google Scholar]
- 2.Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13:263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dudley E, Yousef M, Wang Y, Griffiths WJ. Targeted metabolomics and mass spectrometry. Adv Protein Chem Struct Biol. 2010;80:45–83. doi: 10.1016/B978-0-12-381264-3.00002-3. [DOI] [PubMed] [Google Scholar]
- 4.Wilson ID, Nicholson JK, Castro-Perez J, Granger JH, Johnson KA, Smith BW, Plumb RS. High resolution “ultra performance” liquid chromatography coupled to oa-TOF mass spectrometry as a tool for differential metabolic pathway profiling in functional genomic studies. J Proteome Res. 2005;4:591–598. doi: 10.1021/pr049769r. [DOI] [PubMed] [Google Scholar]
- 5.Zhang AH, Wang P, Sun H, Yan GL, Han Y, Wang XJ. High-throughput ultra-performance liquid chromatography-mass spectrometry characterization of metabolites guided by a bioinformatics program. Mol Biosyst. 2013;9:2259–2265. doi: 10.1039/c3mb70171a. [DOI] [PubMed] [Google Scholar]
- 6.Kay RG, Gregory B, Grace PB, Pleasance S. The application of ultra-performance liquid chromatography/tandem mass spectrometry to the detection and quantitation of apolipoproteins in human serum. Rapid Commun Mass Spectrom. 2007;21:2585–2593. doi: 10.1002/rcm.3130. [DOI] [PubMed] [Google Scholar]
- 7.Nagele E, Vollmer M, Horth P. Two-dimensional nano-liquid chromatography-mass spectrometry system for applications in proteomics. J Chromatogr A. 2003;1009:197–205. doi: 10.1016/s0021-9673(03)01034-3. [DOI] [PubMed] [Google Scholar]
- 8.Gaspari M, Cuda G. Nano LC-MS/MS: a robust setup for proteomic analysis. Methods Mol Biol. 2011;790:115–126. doi: 10.1007/978-1-61779-319-6_9. [DOI] [PubMed] [Google Scholar]
- 9.Brugger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc Natl Acad Sci U S A. 1997;94:2339–2344. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korner R, Wilm M, Morand K, Schubert M, Mann M. Nano electrospray combined with a quadrupole ion trap for the analysis of peptides and protein digests. J Am Soc Mass Spectrom. 1996;7:150–156. doi: 10.1016/1044-0305(95)00626-5. [DOI] [PubMed] [Google Scholar]
- 11.Takáts Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306:471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 12.Chen H, Pan Z, Talaty N, Raftery D, Cooks RG. Combining desorption electrospray ionization mass spectrometry and nuclear magnetic resonance for differential metabolomics without sample preparation. Rapid Commun Mass Spectrom. 2006;20:1577–1584. doi: 10.1002/rcm.2474. [DOI] [PubMed] [Google Scholar]
- 13.Nunn BL, Shaffer SA, Scherl A, Gallis B, Wu M, Miller SI, Goodlett DR. Comparison of a Salmonella typhimurium proteome defined by shotgun proteomics directly on an LTQ-FT and by proteome pre-fractionation on an LCQ-DUO. Brief Funct Genomic Proteomic. 2006;5:154–168. doi: 10.1093/bfgp/ell024. [DOI] [PubMed] [Google Scholar]
- 14.Makarov A, Denisov E, Lange O, Horning S. Dynamic range of mass accuracy in LTQ Orbitrap hybrid mass spectrometer. J Am Soc Mass Spectrom. 2006;17:977–982. doi: 10.1016/j.jasms.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Liu J, Cooks RG, Ouyang Z. Paper spray for direct analysis of complex mixtures using mass spectrometry. Angew Chem Int Ed Engl. 2010;49:877–880. doi: 10.1002/anie.200906314. [DOI] [PubMed] [Google Scholar]
- 16.Gross JH. Direct analysis in real time--a critical review on DART-MS. Anal Bioanal Chem. 2014;406:63–80. doi: 10.1007/s00216-013-7316-0. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez FM, Cody RB, Green MD, Hampton CY, McGready R, Sengaloundeth S, White NJ, Newton PN. Characterization of solid counterfeit drug samples by desorption electrospray ionization and direct-analysis-in-real-time coupled to time-of-flight mass spectrometry. ChemMedChem. 2006;1:702–705. doi: 10.1002/cmdc.200600041. [DOI] [PubMed] [Google Scholar]
- 18.Watrous JD, Dorrestein PC. Imaging mass spectrometry in microbiology. Nat Rev Microbiol. 2011;9:683–694. doi: 10.1038/nrmicro2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDonnell LA, Heeren RM. Imaging mass spectrometry. Mass Spectrom Rev. 2007;26:606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- 20.Seeley EH, Caprioli RM. Molecular imaging of proteins in tissues by mass spectrometry. Proc Natl Acad Sci U S A. 2008;105:18126–18131. doi: 10.1073/pnas.0801374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Bekker C, Smith PB, Patterson AD, Hughes DP. Metabolomics reveals the heterogeneous secretome of two entomopathogenic fungi to ex vivo cultured insect tissues. PLoS One. 2013;8:e70609. doi: 10.1371/journal.pone.0070609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ponnusamy K, Lee S, Lee CH. Time-dependent correlation of the microbial community and the metabolomics of traditional barley nuruk starter fermentation. Biosci Biotechnol Biochem. 2013;77:683–690. doi: 10.1271/bbb.120665. [DOI] [PubMed] [Google Scholar]
- 24.Steinfath M, Groth D, Lisec J, Selbig J. Metabolite profile analysis: from raw data to regression and classification. Physiol Plant. 2008;132:150–161. doi: 10.1111/j.1399-3054.2007.01006.x. [DOI] [PubMed] [Google Scholar]
- 25.Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37:W652–660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 27.Katajamaa M, Oresic M. Processing methods for differential analysis of LC/MS profile data. BMC Bioinformatics. 2005;6:179. doi: 10.1186/1471-2105-6-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katajamaa M, Miettinen J, Oresic M. MZmine: toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics. 2006;22:634–636. doi: 10.1093/bioinformatics/btk039. [DOI] [PubMed] [Google Scholar]
- 29.Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Broeckling CD, Reddy IR, Duran AL, Zhao X, Sumner LW. MET-IDEA: data extraction tool for mass spectrometry-based metabolomics. Anal Chem. 2006;78:4334–4341. doi: 10.1021/ac0521596. [DOI] [PubMed] [Google Scholar]
- 31.Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 32.Tautenhahn R, Cho K, Uritboonthai W, Zhu Z, Patti GJ, Siuzdak G. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat Biotechnol. 2012;30:826–828. doi: 10.1038/nbt.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kind T, Meissen JK, Yang D, Nocito F, Vaniya A, Cheng YS, Vandergheynst JS, Fiehn O. Qualitative analysis of algal secretions with multiple mass spectrometric platforms. J Chromatogr A. 2012;1244:139–147. doi: 10.1016/j.chroma.2012.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kind T, Liu KH, Lee do Y, DeFelice B, Meissen JK, Fiehn O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat Methods. 2013;10:755–758. doi: 10.1038/nmeth.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stein SE. Chemical substructure identification by mass spectral library searching. J Am Soc Mass Spectrom. 1995;6:644–655. doi: 10.1016/1044-0305(95)00291-K. [DOI] [PubMed] [Google Scholar]
- 36.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, Scalbert A. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, Oda Y, Kakazu Y, Kusano M, Tohge T, Matsuda F, Sawada Y, Hirai MY, Nakanishi H, Ikeda K, Akimoto N, Maoka T, Takahashi H, Ara T, Sakurai N, Suzuki H, Shibata D, Neumann S, Iida T, Funatsu K, Matsuura F, Soga T, Taguchi R, Saito K, Nishioka T. MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom. 2010;45:703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 38.Phelan VV, Liu WT, Pogliano K, Dorrestein PC. Microbial metabolic exchange--the chemotype-to-phenotype link. Nat Chem Biol. 2012;8:26–35. doi: 10.1038/nchembio.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, van der Voort M, Pogliano K, Gross H, Raaijmakers JM, Moore BS, Laskin J, Bandeira N, Dorrestein PC. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci U S A. 2012;109:E1743–1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guthals A, Watrous JD, Dorrestein PC, Bandeira N. The spectral networks paradigm in high throughput mass spectrometry. Mol Biosyst. 2012;8:2535–2544. doi: 10.1039/c2mb25085c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang JY, Sanchez LM, Rath CM, Liu X, Boudreau PD, Bruns N, Glukhov E, Wodtke A, de Felicio R, Fenner A, Wong WR, Linington RG, Zhang L, Debonsi HM, Gerwick WH, Dorrestein PC. Molecular networking as a dereplication strategy. J Nat Prod. 2013;76:1686–1699. doi: 10.1021/np400413s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frank AM, Bandeira N, Shen Z, Tanner S, Briggs SP, Smith RD, Pevzner PA. Clustering millions of tandem mass spectra. J Proteome Res. 2008;7:113–122. doi: 10.1021/pr070361e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niemi LM, Stencel KA, Murphy MJ, Schultz MM. Quantitative determination of antidepressants and their select degradates by liquid chromatography/electrospray ionization tandem mass spectrometry in biosolids destined for land application. Anal Chem. 2013;85:7279–7286. doi: 10.1021/ac401170s. [DOI] [PubMed] [Google Scholar]
- 44.Archer CB, Cunningham FM, Greaves MW. Actions of platelet activating factor (PAF) homologues and their combinations on neutrophil chemokinesis and cutaneous inflammatory responses in man. J Invest Dermatol. 1988;91:82–85. doi: 10.1111/1523-1747.ep12463296. [DOI] [PubMed] [Google Scholar]
- 45.Kramer RM, Patton GM, Pritzker CR, Deykin D. Metabolism of platelet-activating factor in human platelets. Transacylase-mediated synthesis of 1-O-alkyl-2-arachidonoyl-sn-glycero-3-phosphocholine. J Biol Chem. 1984;259:13316–13320. [PubMed] [Google Scholar]
- 46.Ohno M, Fujita K, Nakai H, Kobayashi S, Inoue K, Nojima S. An enantioselective synthesis of platelet-activating factors, their enantiomers, and their analogues from D-and L-tartaric acids. Chem Pharm Bull (Tokyo) 1985;33:572–582. doi: 10.1248/cpb.33.572. [DOI] [PubMed] [Google Scholar]
- 47.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.