Abstract
Paris-Trousseau syndrome (PTS; also known as Jacobsen syndrome) is characterized by several congenital anomalies including a dysmegakaryopoiesis with two morphologically distinct populations of megakaryocytes (MKs). PTS patients harbor deletions on the long arm of chromosome 11, including the FLI1 gene, which encodes a transcription factor essential for megakaryopoiesis. We show here that lentivirus-mediated overexpression of FLI1 in patient CD34+ cells restores the megakaryopoiesis in vitro, indicating that FLI1 hemizygous deletion contributes to the PTS hematopoietic defects. FISH analysis on pre-mRNA and single-cell RT-PCR revealed that FLI1 expression is mainly monoallelic in CD41+CD42– progenitors, while it is predominantly biallelic in the other stages of megakaryopoiesis. In PTS cells, the hemizygous deletion of FLI1 generates a subpopulation of CD41+CD42– cells completely lacking FLI1 transcription. We propose that the absence of FLI1 expression in these CD41+CD42– cells might prevent their differentiation, which could explain the segregation of the PTS MKs into two subpopulations: one normal and one composed of small immature MKs undergoing a massive lysis, presumably originating from either FLI1+ or FLI1– CD41+CD42– cells, respectively. Thus, we point to the role of transient monoallelic expression of a gene essential for differentiation in the genesis of human haploinsufficiency-associated disease and suggest that such a mechanism may be involved in the pathogenesis of other congenital or acquired genetic diseases.
Introduction
An autosomal dominant inherited thrombocytopenia associated with giant α granules in a low percentage of platelets and a bone marrow dysmegakaryopoiesis (1, 2) is always present in Paris-Trousseau syndrome (PTS), a variant of Jacobsen syndrome (3). The syndrome is associated with a deletion of 11q23.3-24 region affecting either the paternal or maternal chromosome (4). This deletion includes two ETS (E-twenty-six) transcription factors encoding genes ETS1 and FLI1 (5, 6). Both in vitro and in vivo evidence has demonstrated the critical role played by FLI1 in megakaryocyte (MK) differentiation. FLI1 transactivates several MK-specific genes such as GPIIb (7), GPIX (8), GPIbα (9), and c-mpl (10), associated with terminal MK differentiation. It stimulates GATA-1/FOG-1–dependent transcription of the αIIb gene (11), and its interaction with GATA-1 also mediates synergistic expression of GPIX and GPIbα (12). Moreover, Fli1–/– KO embryos exhibit a dramatic reduction in MK numbers, with the presence of small MKs, similar to those seen in PTS patients (6, 13). ETS1 also binds to the proximal Ets-binding site present in different promoters of MK-specific genes such as those of GPIIb (7), mpl (10), and PF4 (14). In contrast to FLI1, however, ETS1 is dispensable for megakaryopoiesis in vivo because Ets-1–/– mice exhibit normal MK differentiation while displaying other hematopoietic defects (15). Moreover, in vitro overexpression of ETS1 in human pluripotent hematopoietic cell lines favors the development of an erythroid phenotype (16), while that of FLI1 induces MK markers (17). Taken as a whole, these data suggest that FLI1 hemizygous deletion might be at least partly responsible for the platelet/MK defects observed in PTS patients. Here we provide experimental data that directly corroborate this hypothesis. In addition, we explain how the deletion of one FLI1 allele could lead to two distinct MK populations in PTS patients, one population composed of normal MKs and the other of small, immature MKs undergoing massive lysis (2).
Results
Expression of FLI1 and ETS1 in primary human hematopoietic cells.
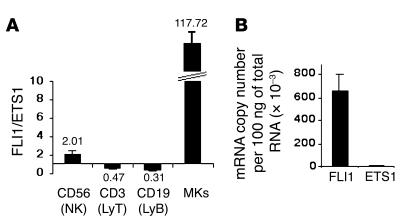
ETS1 and FLI1, which encode two related ETS transcription factors, are on the deleted 11q23 chromosomal region in PTS patients. We have examined their expression level in different human hematopoietic lineages, NK cells, B and T lymphocytes, and MKs using quantitative RT-PCR. We found that FLI1 is expressed two to three times more than ETS1 in NK cells, while the reverse is true in B or T lymphocytes. Importantly, the same study showed that FLI1 mRNA is about 100-fold more abundant than ETS1 mRNA in MKs (Figure 1A): 6.6 × 105 copies of FLI1 mRNA and 5.5 × 103 copies of ETS1 mRNA were found in 100 ng of total MK RNA (Figure 1B). In agreement with previously reported data (6, 13, 15), our results strengthen the conclusion that FLI1 plays a much more important role than ETS1 in megakaryopoiesis. Consequently, we thereafter focused on the possible involvement of FLI1 hemizygous deletion in the PTS-associated dysmegakaryopoiesis.
Figure 1.
Comparison of FLI1 and ETS1 expression in different human hematopoietic lineages: NK cells, lymphocytes T and B (LyT and LyB), and MKs. The absolute expression level of FLI1 and ETS1 in 100 ng of total cell RNA was measured relative to different dilutions (corresponding to 102–106 copies) of plasmids harboring either FLI1 or ETS1 cDNA. (A) The graph represents the relative expression between FLI1 and ETS1 in NK (CD56), T (CD3), and B (CD19) cells and in MKs. The value indicated above or under the columns is the ratio between the FLI1 and the ETS1 copy numbers in cDNA prepared from 100 ng of total cell RNA. (B) The absolute expression level of FLI1 and ETS1 in MKs measured as the number of mRNA copies per 100 ng of total RNA. The error bars represent the SD of the mean of three experiments, each performed in triplicate wells.
Construction of lentiviral vector encoding FLI1 cDNA.
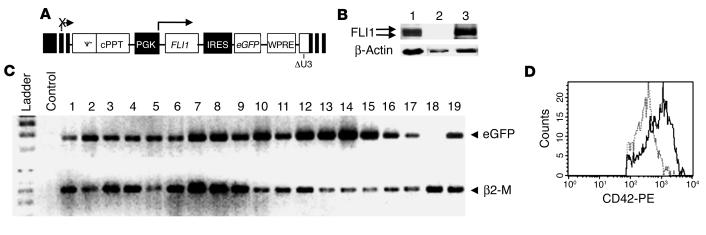
To directly evaluate the role of FLI1 in the pathogenesis of PTS, we constructed a bicistronic lentiviral vector encoding both FLI1 and enhanced GFP (eGFP) cDNAs (Figure 2A) to transduce peripheral blood CD34+ cells of three PTS patients harboring only one FLI1 allele, as demonstrated by FISH analysis (data not shown). To test the construct, 293T cells were transiently transfected with either the empty lentiviral vector or that containing FLI1 cDNA. The synthesis of two forms of FLI1 protein (49 and 51 kDa) was detected (Figure 2B) only in the latter cells, demonstrating that the constructed vector was functional. After transduction of CD34+ cells using this vector, we obtained a low proportion of eGFP-positive cells (10% as measured by flow cytometry), whereas 90% of the cells expressed the bicistronic transcript (as detected by single-cell RT-PCR; Figure 2C). This could be explained by the greater sensitivity of RT-PCR compared with flow cytometry or by the fact that eGFP was poorly translated from the internal ribosome entry site. The same inconsistent internal ribosomal entry site–dependent (IRES-dependent) GFP expression was also observed by others (18). Thus, in following experiments we did not use the eGFP as a reporter for the lentiviral transduction. Importantly, gene transfer resulted in the increased expression of GpIbα (CD42b), a known target gene of FLI1 in MK CD41+ cells (Figure 2D).
Figure 2.
Construction and validation of a lentiviral vector encoding FLI1 cDNA. (A) Construction of a lentiviral expression vector coding for FLI1. FLI1 expression was driven by the PGK promoter. (B) Immunoblot analysis of FLI1 protein in HEL cells expressing endogenous Fli1 (lane 1), in 293T cell line transiently transfected with either the control lentiviral vector (lane 2) or with the lentiviral vector encoding FLI1 cDNA (lane 3). Five hundred × 103 cells were loaded in lane 1 and 50 × 103 in lanes 2 and 3. (C) Single-cell RT-PCR detection of eGFP in transduced CD34+ cells. Peripheral blood CD34+ cells obtained from healthy individuals were investigated for the presence of eGFP 6 days after infection with PGK-Fli1-IRES-eGFP virus. β2-M was used as an internal control of mRNA integrity and cell sorting. Safety control for the experiment was performed in the absence of sorted cells (lane 2: Control). (D) Immunolabeling and flow-cytometry analysis of CD34+ cells transduced with the lentiviral vector. CD34+ cells were stained with anti–CD42-PE and anti–CD41-APC Ab’s 9 days after transduction with the control lentivirus vector (dotted line) or with the FLI1 encoding lentivirus vector (thin line). Analysis of CD42 expression was performed in the cell population expressing high level of CD41 (MKs).
Lentivirus-mediated FLI1 overexpression improves PTS patient megakaryopoieisis in vitro.
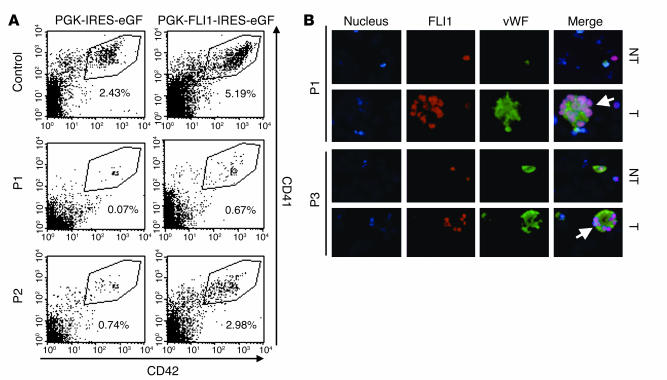
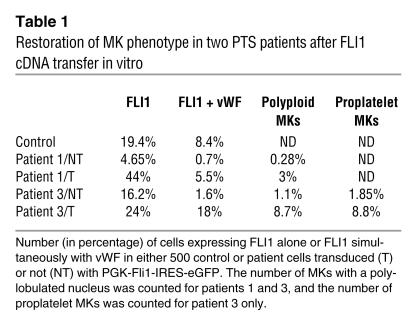
Peripheral blood CD34+ cells from three PTS patients were transduced with the lentiviral vector encoding FLI1 cDNA, and the presence of MKs expressing high levels of CD41 (α2b/β3 integrin) and CD42a (GpIX) was analyzed 9 days later. In patients 1 and 2, the transfer of FLI1 cDNA resulted in a marked increase in the total number of cells obtained in culture, as well as in the percentage of MKs (from 0.07% to 0.67% and from 0.74% to 2.98%, respectively) (Figure 3A). These results were reproduced in another experiment in which the percentage of mature MKs increased from 0.24% to 1.52% in patient 1 and from 0.85% to 3.64% in patient 2 (data not shown). In patient 3, FLI1 overexpression did not increase the percentage of MKs (11% and 12.2% after transduction with control and FLI1 encoding lentivirus, respectively; data not shown). FLI1 overexpression in this patient had three prominent effects, however: (a) it decreased the initial cell death observed with the control vector or in the absence of gene transfer (30% mortality versus 70%) as observed by FACS analysis (data not shown) and thus increased the total (absolute) MK number obtained in culture; (b) it improved MK maturation as demonstrated by the elevation in the number of vWF-expressing cells (this was also observed in patient 1) and restoration of large MKs with a polylobulated nucleus (thus presumably polyploidy) (Figure 3B; Table 1); (c) it enhanced the number of proplatelet-forming MKs (Table 1). Thus, FLI1 overexpression increases MK cell survival, as it does in other cell types, and enhances the MK differentiation of PTS patient cells.
Figure 3.
Restoration of megakaryocytic phenotype in two PTS patients after FLI1 cDNA transfer in vitro. (A) Flow-cytometry analysis of CD41 and CD42 expression in MKs from a control and two patients (P1 and P2) 11 days after transduction of their peripheral blood CD34+ cells with PGK-IRES-eGFP or with PGK-Fli1-IRES-eGFP. Gate R3 and numbers indicated below the gates represent the percentage of mature MKs with high levels of CD41 and CD42 expression. (B) Immunolabeling of FLI1 (red staining) and vWF (green staining) in patient 1 and 3 (P1 and P3) MKs grown from CD34+ cells transduced (T), or not (NT), with PGK-Fli1-IRES-eGFP. Images were captured using epifluorescence microscope (Nikon Eclipse 600) with a ×40 objective. Large, presumably polyploid MKs characterized by a typical polylobulated nucleus (DAPI staining) and by a unique, continuous cytoplasmic membrane (vWF staining) in patient 1 and 3 after FLI1 transduction are indicated by a white arrowhead.
Table 1.
Restoration of MK phenotype in two PTS patients after FLI1 cDNA transfer in vitro
FLI1 expression is transiently monoallelic during normal megakaryopoiesis.
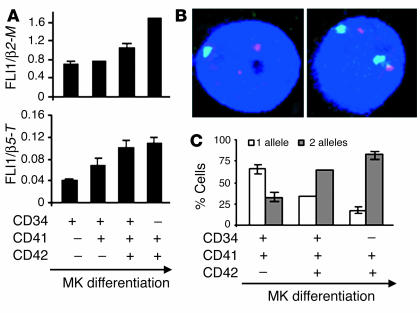
To understand how the loss of one copy of FLI1 could result in the MK defects observed in PTS patients, we first examined the FLI1 expression levels throughout normal MK differentiation in vitro using quantitative RT-PCR. We demonstrated that FLI1 expression continuously increased with the progression of MK differentiation, that is, from CD34+CD41–CD42– to CD34–CD41+CD42+ cell populations (Figure 4A). Because the remaining FLI1 allele in PTS patients encodes a WT protein (data not shown), the hematopoietic defects linked to the FLI1 hemizygous loss could result from a twofold reduction in its expression level in all PTS MKs. Alternatively, as demonstrated for some other genes (19–24), FLI1 could be (at least transiently) monoallelically expressed, leading to a fraction of FLI1–null cells in PTS patients. The latter hypothesis would be in agreement with the generation of the two distinct MK populations in PTS patients. To investigate FLI1 allelic expression throughout normal MK differentiation, we performed the pre–mRNA-FISH analysis on different populations described above (from CD34+CD41–CD42– to CD34–CD41+CD42+). We studied the allelic expression of FLI1 at a given time in cytapheresis samples from three healthy individuals (harboring two FLI1 alleles), and we counted the number of spots representing the FLI1 pre-mRNA (Figure 4B) in 100 nuclei of each cell fraction (Figure 4C). In the most immature cell fraction (CD34+CD41–CD42–), the amount of FLI1 pre-mRNA was at the detection threshold of the technique. In contrast, FLI1 pre-mRNA spots were readily detected in the nucleus of all MKs from the more mature fractions. Importantly, FLI1 expression is monoallelic in the majority (65%) of the intermediately mature CD34+CD41+CD42– cells, but is preferentially biallelic in the more mature CD34+CD41+CD42+ fraction (Figure 4C).
Figure 4.
Fli1 expression throughout normal MK differentiation. CD34+ cells were cultured in the presence of TPO and analyzed at day 6. (A) Fli1 expression level determined by real-time RT-PCR. Diploid MKs were sorted according to their differentiation stages. Fli1 and two housekeeping β2-M and β5-tubulin (β5-T) genes (constant throughout MK differentiation; our unpublished data) were amplified in separate wells in five independent experiments (four using β2-M and one using β5-T). The relative expression level of Fli1 in each fraction was normalized to the corresponding values for β2-M or β5-T. A representative experiment is shown with error bars representing the SD of the mean of triplicate wells. (B) Representative picture of TPO-cultured cells showing the distribution of Fli1 nuclear RNA (green) and chromosome 12 (red) visualized by FISH. Cell with a monoallelic expression of Fli1 is on the left, and cell with its biallelic expression is on the right.The number of green spots indicates the number of transcribed Fli1 alleles. The number of Fli1 alleles in the same MK is determined by the number of red spots. Chromatin is counterstained with DAPI (blue). (C) Statistical analysis of monoallelic/biallelic expression of Fli1 in diploid MKs during their differentiation. Cells were sorted as described in A, and RNA-FISH was performed as described in B. One hundred cells of each population were analyzed on an epifluorescence microscope (Nikon Eclipse 600) using a ×60 objective for the presence of Fli1 pre-mRNA. The error bars represent the SD of the mean of three independent experiments.
Monoallelic expression of FLI1 generates a fraction of FLI1–null cells in PTS patients.
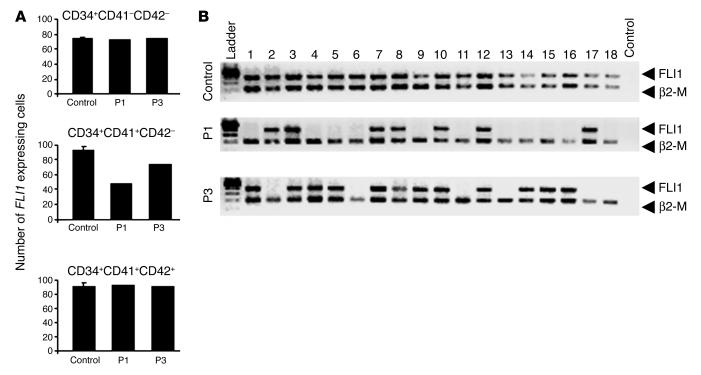
We next investigated whether the existence of such a transient monoallelic stage of FLI1 expression impinges on the MK differentiation in PTS patients who harbor only one FLI1 allele. In PTS cells, the in situ visualization of FLI1 transcription through RNA-FISH is not required since any FLI1 transcript necessarily arises from the remaining allele. Thus, we used the more sensitive single-cell RT-PCR method (25) on sorted cells from different populations as above. The above RNA-FISH analysis on normal individual cells showed that at least one FLI1 allele is always expressed throughout MK differentiation. Accordingly, FLI1 transcription was detected by single cell RT-PCR in nearly all MKs (95%) from the two more mature cell fractions (CD41+CD42– and CD41+CD42+) of the three control individuals (Figure 5A). Importantly, at the stage at which FISH data indicate that only one FLI1 allele is expressed (CD34+CD41+CD42–), single cell RT-PCR show that only 48 from 100 (in patient 1) and 74 from 100 (in patient 3) of the CD34+CD41+CD42– MKs were positive for FLI1 transcripts (Figure 5, A and B) (RT-PCR has not been performed in patient 2 because of an insufficient number of CD34+ cells obtained from peripheral blood). Nearly all patient CD34+CD41+CD42+ cells corresponding to the following stage of MK differentiation expressed FLI1 (Figure 5, A and B), however. In addition, in the more primitive cell fraction (CD34+CD41–CD42–) no difference was noted between patient and control cells, with 75% of the cells expressing FLI1 in both cases (Figure 5A). This suggests that in the most immature MK progenitors FLI1 expression is predominantly biallelic, although weak as indicated by quantitative RT-PCR (Figure 4A). Overall, these data indicate that the transient monoallelic FLI1 transcription, combined with its hemizygous deletion, completely eliminates its expression in a fraction of PTS patient CD34+CD41+C42– cells.
Figure 5.
Single-cell RT-PCR analysis of FLI1 expression in peripheral blood CD34+ cells from three controls and two patients, cultured in presence of TPO, IL-3, IL-6, SCF, and FLT3-L. On day 4, the CD34+CD41 –CD42 –, CD34+CD41+CD42 –, and CD34+CD41+CD42+ populations were sorted after immunolabeling at one cell per well in a 96-well plate. Single-cell RT-PCR was performed on 100 individual cells in each population. β2-M was used as an internal control of mRNA integrity, and only cells positive for β2-M were analyzed. (A) The graphs represent the number of FLI1–expressing cells out of 100 analyzed cells from different populations of three healthy individuals (Control) (the error bars represent the SD of the mean of three independent experiments) and of two Paris-Trousseau patients. (B) RT-PCR analysis in a representative 18 (of 100) CD34+CD41+CD42– cells from one healthy individual and two patients. Safety control of experiment was performed in absence of sorted cell (lane 20: Control).
Discussion
PTS is a rare disease; until now, only ten cases have been detected in children (26). PTS patients show a deletion on the long arm of chromosome 11 at 11q23.3, which varies in length and generally includes the FLI1 gene. The possible involvement of the hemizygous loss of the FLI1 gene in the dysmegakaryopoiesis linked to this syndrome was previously proposed (6), but never demonstrated. Here, we investigated (a) whether FLI1 cDNA transfer in PTS CD34+ could rescue the MK differentiation, and (b) how a half reduction in FLI1 gene dosage could lead to haploinsufficiency and MK defects. Transfer of FLI1 cDNA into the progenitor hematopoietic CD34+ cells of three patients via a lentiviral vector restored a normal megakaryocytic phenotype in vitro as demonstrated by the correction of CD41, CD42, and vWF expression, a higher number of mature, proplatelet MKs, and increased ploidization. We cannot completely rule out, however, that the lentivirus-mediated FLI1 overexpression in PTS CD34+ cells bypasses the requirement for other genes localized in the deleted region. Among them, the FLI1 relative ETS1 could be viewed as another possible candidate. Quantitative RT-PCR indicated, however, that FLI1 is expressed about 100-fold more in MKs than is ETS1. Moreover, to date no clear evidence supports a role of ETS1 in megakaryopoiesis, while FLI1 has been shown to be an essential player based on both in vitro and in vivo experiments (6, 11–13). Taken together, these data strongly support the conclusion that FLI1, but not ETS1, hemizygous deletion is involved in the MK defects observed in PTS patients.
Several results obtained in mouse models confirm that MK differentiation is sensitive to a reduction of the Fli1 copy number by half. Kawada et al. showed a reduction in the MK number derived from cultured Fli1+/– aorta-gonad-mesonephros embryonic cells compared with their WT counterparts (27). Moreover, our preliminary data performed on blast cell (BL) colonies derived from ES Fli1+/– cells showed a 57% decrease (compared with WT cells) in CD41+CD42+ cells at day 3 (O. Feraud, personal communication). It should be noted that we have recently reported one PTS patient (out of ten) harboring two FLI1 alleles (26), although whether both of them encode a functional protein is not known.
One feature of the MK defects in PTS is the generation of two distinct populations of MKs, one composed of normal MKs and one of small, immature MKs. To explain how the hemizygous loss of the FLI1 gene could lead to such a heterogeneous haploinsufficiency, we investigated the allelic expression of FLI1 during megakaryopoiesis. RNA-FISH analysis showed that at a given time FLI1 is either transcribed from one or from two alleles, according to the stage of MK differentiation. Indeed, FLI1 expression is biallelic in the majority of CD41+CD42+ MKs, while it appears monoallelic in their CD41+CD42– immediate progenitors. The RNA-FISH method is limited by its relatively weak sensitivity, however. For example, according to RNA-FISH analysis, it has been reported that Pax 5 gene expression switched from a monoallelic to biallelic and again to monoallelic expression during B cell differentiation (28), but this regulation was not confirmed by single cell RT-PCR analysis (25). Single cell RT-PCR analysis was thus also performed on sorted MKs at various stages of differentiation from either three normal individuals (harboring two FLI1 alleles) or two PTS patients (harboring only one FLI1 allele). These experiments showed that FLI1 mRNA is not detected in a significant fraction of CD41+CD42– cells from the two PTS patients, while nearly all the corresponding cells from normal individuals expressed FLI1 mRNA. Consequently, our results suggest that FLI1 haploinsufficiency in PTS is not due to a simple, homogeneous, twofold reduction of FLI1 expression levels in all MKs, but rather arises as a result of the complete lack of FLI1 expression in a fraction of PTS MK progenitors.
How can we explain the monoallelic expression of FLI1 in CD41+42– MK progenitors and its complete absence in a subset of these cells from PTS patients?
An important mechanism accounting for some monoallelic expression is the genomic imprinting, which consists of a differential regulation of the two alleles of a gene according to their parental origin. Some haploinsufficiencies involve a genomic imprinting, such as Prader-Willi syndrome (29), Angelman syndrome (29), and progressive osseous heteroplasia (30). In these patients, the disrupted allele is always of the same parental origin (for a given syndrome), while the other allele is silenced through genomic imprinting. In contrast, in PTS patients, the deletion is of either paternal (in eight patients) or maternal (in six patients) origin (4). Moreover, the concomitance of a deletion of one allele and a shutoff through genomic imprinting on the other should lead to the absence of gene expression in all the cells. Instead, in PTS patients we observed a heterogeneous CD41+42– MK population composed of both FLI1–positive and –negative cells. Thus, the monoallelic expression of FLI1 and its haploinsufficiency in PTS do not rely on a genomic imprinting.
Two other models remain. The first one postulates that, at a single cell level, the same FLI1 allele is transcribed throughout the monoallelic differentiation stage, in a way resembling the allelic exclusion observed in other systems (19, 21, 23–25). Accordingly, FLI1 would be subjected to a random and stage-specific allelic exclusion, leading the CD41+CD42– cells to segregate into two MK subpopulations according to the “choice” of the FLI1 allele. In normal CD41+CD42– cells, FLI1 mRNA (and protein) are produced whatever choice the cells have made between the two FLI1 alleles. In PTS patients, however, the hemizygous FLI1 deletion make these two alternative choices nonequivalent, which could generate two distinct populations of MK progenitors: one FLI1+ (presumably corresponding to normal MKs expressing the remaining FLI1 allele) and one FLI1– (presumably corresponding to small, immature, dying MKs not expressing the remaining FLI1 allele).
Alternatively, it should be recalled that FISH analysis allowed us to visualize the ongoing transcription only at a given time. Thus, it remains possible that the monoallelic expression of FLI1 in CD41+CD42– cells might not be caused by an allelic exclusion but rather reflect an inefficient, intermittent expression of both alleles at this stage, leading a high proportion of these cells to express only one allele at a given time. Indeed, transcription appears often as a “pulsatile,” discontinuous process at a single allele level (31, 32). Thus, the loss of one copy of FLI1 might lead to a stochastic, complete interruption of FLI1 expression in PTS MK progenitors, transiently mimicking a homozygous deletion. This would render the differentiation program more prone to undergo irreversible alterations (33, 34) and hence to generate again an abnormal MK subpopulation, ultimately dying.
Whatever the mechanisms underlying the monoallelic expression of FLI1, this regulation renders the progress of the MK differentiation program from CD42– to CD42+ stages especially sensitive to a reduction in FLI1 gene dosage, in keeping with an essential role of its product in CD42 transactivation (8). Interestingly, while we observed in vitro a reduction in CD42+ cells derived from Fli+/– ES cells (compared with WT cells), their CD42– immediate progenitors occurred at a comparable level in the two populations. A possible explanation is therefore that Fli1 is also mainly monoallelically expressed in mouse CD42– cells, with the subsequent reduction in CD42+ Fli+/– cells corresponding to Fli1–null CD42– cells (i.e., expressing the disrupted Fli1 allele) unable to further differentiate.
In conclusion, our findings agree with the hypothesis that a monoallelic expression of an important gene at a critical stage (through either allelic exclusion or intermittent transcription) might be responsible for a human haploinsufficient disease. It is conceivable that such a monoallelic expression combined with hemizygous loss is operating in other, more frequent genetic diseases, especially in oncogenesis, if it affects tumor suppressor genes (34).
Methods
Patients.
An informed consent for this study was obtained from only three patients for only a peripheral blood venipuncture. Their clinical and biological features have been previously described (1, 2, 26). In summary, all these patients have mental retardation and a congenital thrombocytopenia with giant α granules in a fraction of circulating platelets, bone marrow dysmegakaryopoiesis, and a deletion of the distal part of chromosome 11 (11q23). The deletion of one FLI1 allele in patient samples was confirmed by DNA-FISH (data not shown). The nine exons of the remaining FLI1 allele were sequenced and shown to be to devoid of any mutation, indicating that it encodes a WT FLI1 protein.
Peripheral blood and cytapheresis samples from healthy individuals were obtained with their informed consent.
In vitro growth of MKs from CD34+ cells.
To study FLI1 expression during normal megakaryopoiesis by real-time PCR and RNA-FISH methods, the CD34+ cells were isolated from cytapheresis samples and incubated as previously described by Choi (35) in serum-free medium supplemented with thrombopoietin (TPO) (10 ng/ml).
Patient CD34+ cells were isolated from 80–100 ml of peripheral blood samples and incubated as above in the serum-free medium supplemented with TPO (10 ng/ml), IL-3 (100 U/ml), IL-6 (10 ng/ml), stem cell factor (SCF) (25 ng/ml), and fetal liver tyrosine kinase 3 ligand (FLT3-L) (10 ng/ml).
RNA- and DNA-FISH analysis during normal megakaryopoiesis.
At day 6 of culture, cells obtained from cytapheresis of healthy individuals were first directly analyzed by RNA- and DNA-FISH. For a more rigorous analysis, cells were stained at day 6 with phycoerythrin-conjugated (PE-conjugated) anti-CD34, allophycocyanin-conjugated (APC-conjugated) anti-CD41a, and FITC-conjugated anti-CD42a Ab’s (BD Biosciences, Pont de Claix, France), followed by vital DNA labeling with Hoechst stain (Hoechst 33342; Sigma-Aldrich, Saint Quentin Fallavier, France). Different diploid populations (CD34+CD41–CD42–, CD34+CD41+CD42–, CD34+CD41+CD42+, and CD34–CD41+CD42+) were sorted using a FACSVantage SE cytometer (BD Biosciences), and RNA-FISH analysis was done on these diploid cells.
RNA- and DNA-FISH were performed using two FLI1 genomic probes for pre-mRNA detection and commercial centromeric probe staining chromosome 12 (Oncor Inc., Gaithersburg, Maryland, USA) for DNA detection as previously described (36).
Real-time quantitative RT-PCR during normal megakaryopoiesis.
At day 6 of culture, cells obtained from cytapheresis of healthy individuals were stained and sorted as described above. For NK, B, and T cells, RNA was extracted directly after cell isolation using magnetic bead–coupled Ab against CD56, CD19, and CD3, respectively (Miltenyi Biotech, Paris, France). Aliquots (1 μg) of total RNA prepared using the Promega kit (Promega France SARL, Charbonnières, France) were treated with RNase-free DNase (Ambion Inc., Huntingdon, United Kingdom), denatured, and reverse transcribed with 200 U of SuperScipt II RNase H– reverse transcriptase (Invitrogen SARL, Cergy Pontoise Cedex, France). Primers and internal probes for amplification of FLI1, ETS1, and β5-tubulin and β2-microglobulin (β2-M) sequences were designed. PCRs were carried out in the ABI Prism GeneAmp 5700 Sequence Detection System using the TaqMan Universal PCR Master Mix (PE Applied Biosystems, Courtaboeuf, France) containing the specific primers (1.2 μM) and the specific probe (0.2 μM). To study the FLI1 expression throughout MK differentiation, FLI1 expression level was measured relative to the expression level of one of the two housekeeping genes. To determine the absolute quantity of FLI1 and ETS1 transcripts in 100 ng of total RNA from different human hematopoietic cell types, their expression level was measured relative to different dilutions of plasmids harboring either FLI1 or ETS1 cDNA, each corresponding to a defined number of plasmid copies (from 102 to 106).
Single-cell RT-PCR analysis.
Control and patient CD34+CD41–CD42–, CD34+CD41+CD42–, and CD34+CD41+CD42+ cells, and CD34+ cells transduced with the lentivirus encoding for FLI1, were sorted using a FACSVantage SE cytometer equipped with an automatic cell deposition unit (BD Biosciences). The single-cell RT-PCR assay was performed as previously described (37). FLI1, β2-M, and GFP cDNAs were amplified by nested PCR. The first PCR round consisted of 30 amplification cycles (45 seconds at 94°C, 1 minute at 57°C, 1 minute at 72°C) with 0.2 μM dNTP, 0.06 U/ml Taq polymerase (ATGC Biotechnologie, Croissy Beaubourg, France), and either 0.235 μM of FLI1 and 0.059 μM of β2-M–specific primers or 0.235 μM of GFP and 0.059 μM of β2-M–specific primers in a volume of 85 μl. Each gene was then amplified separately by a nested PCR for 35 cycles (45 seconds at 94°C, 1 minute at 57°C, 1 minute at 72°C) in a volume of 50 μl containing 0.25 μM dNTP, 0.05 U/μl Taq polymerase (ATGC Biotechnologie), and 0.2 μM specific primers. The 5′ primers for the first PCR were FLI1, 5′-ATCAGCCAGTGAGGGTCAAC-3′; β2-M, 5′-CCTGAAGCTGACAGCATTCGG-3′; eGFP, 5′-ACGTAAACGGCCACAAGTTC-3′. The 3′ primers for the first PCR were FLI1, 5′-GAATTGCCACAGCTGGATCT-3′; β2-M, 5′-CTCCTAGAGCTACCTGTGGAG-3′; eGFP, 5′-TGCTCAGGTAGTGGTTGTCG-3′. The 5′ primers for the nested PCR were FLI1, 5′-GGACTGCAGCGTTAGCAAAT-3′; β2-M, 5′-GTGCTCGCGCTACTCTCTCT-3′; eGFP, 5′-CACATGAAGCAGCACGACTT-3′. The 3′ primers of the nested PCR were FLI1, 5′-GGACTTTTGTTGAGGCCAGA-3′; β2-M, 5′-CAAAGTCACATGGTTCACACG-3′; eGFP, 5′-TTC-ACCTTGATGCCGTTC-3′.
Lentiviral vector cloning.
The 1,359 bp human FLI1 cDNA from the ØEB HA FLI1 plasmid (gift of J. Ghysdael, Centre National de la Recherche Scientifique [CNRS] UMR 146, Orsay, France) was subcloned after having replaced the HA epitope by an ATG codon into a third generation of HIV-derived lentiviral vector (pRRLsin-PGK-IRES2-eGFP-WPRE; Généthon, Evry, France). This vector contains an HIV central polypurine tract, a self-inactivating deletion in the U3 region of the 3′ LTR (ØU3), the eGFP downstream of the encephalomyocarditis virus IRES, and the woodchuck hepatitis virus posttranscriptional regulatory element (WPRE).
Lentiviral production.
Lentiviral stocks were prepared as previously described (38) with slight modifications. Briefly, viral particles were prepared by transient cotransfection in human 293T cells of three plasmids (SIN transfer vector plasmid, with or without FLI1 sequence, the packaging plasmid pCMVØR8.74, and the VSV-G protein envelope plasmid pMD.G). Viral stocks were stored at –80°C, and concentrations of viral particles were normalized according to the p24 (HIV-1 capsid protein) content of supernatants.
Western blot analysis of FLI1 protein.
Cells (293T) were transiently transfected with control lentiviral plasmid or lentivirus encoding for FLI1 cDNA. Two days after transfection, cells were sonicated in Laemmli buffer and proteins were separated on 10% SDS-PAGE, blotted onto a nitrocellulose membrane, and incubated with the anti–FLI1 rabbit polyclonal Ab (sc-356; Santa Cruz Biotechnology Inc., Santa Cruz, California, USA). As a positive control for endogenous FLI1 expression, a total lysate of human erythroleukemia cell line (HEL) was used. Protein levels were quantified using the anti–β-actin mouse mAb (Sigma-Aldrich).
Cell transduction.
CD34+ cells (105/ml) were prestimulated for 24 hours in serum-free medium supplemented with five recombinant human cytokines: TPO (100 ng/ml), IL-3 (1,000 U/ml), IL-6 (100 ng/ml), SCF (250 ng/ml), and FLT3-L (100 ng/ml). Cells (105) were resuspended in 100 μl of serum-free medium containing five cytokines and 2 μg/ml of hexadimethrine bromide (Sigma-Aldrich), and lentiviral particles were added at a concentration corresponding to 125 ng of viral p24/100 μl for 24 hours. Cells were then washed and cultured in vitro in the presence of five cytokines at concentrations indicated above.
Flow-cytometric analysis of MK phenotype.
On day 9 after infection, cells were washed in PBS, stained for 30 minutes at 4°C with an anti–CD41-APC and an anti–CD42-PE (BD Biosciences), and analyzed on a FACSort (BD Biosciences).
Immunofluorescence microscopy of MKs and enumeration of proplatelet and polyploid MKs.
Cells were resuspended in PBS/0.1% EDTA, cytocentrifuged onto slides, fixed in 4% formaldehyde/PBS, and washed twice in PBS. Cells were then permeabilized with 0.2% Tween-20 (Sigma-Aldrich) before incubation for 1 hour at room temperature with a rabbit anti-human FLI1 polyclonal Ab (sc-356; Santa Cruz Biotechnology Inc.) and a mouse anti-human vWF mAb (a generous gift from D. Meyer, Institut National de la Santé et de la Recherche Médicale [INSERM] U 143, Kremlin-Bicêtre, France). After washing, cells were incubated with TRITC donkey anti-rabbit IgG and FITC donkey anti-mouse IgG (Jackson/Beckman Coulter, Villepinte, France). The slides were mounted using Vectashield with DAPI (Vector Laboratories, Burlingame, California, USA). Polyploid MKs were defined as cells with a typical single polylobulated nucleus. The percentage of polyploid MKs was quantified by enumerating 500 cells per slide after DAPI and vWF staining to distinguish polylobulated MK cells from clumps of single, small cells.
Proplatelet-displaying MKs were defined as cells exhibiting one or more cytoplasmic processes with areas of constriction. The percentage of MKs with such processes was quantified by enumerating 500 cells per well (in 96-well plates) with an inverted microscope at a magnification of ×200.
Image acquisition and analysis.
Immunofluorescence images were acquired using a confocal laser scanning microscope (Leica Microsystems Heidelberg GmbH, Mannheim, Germany) equipped with a Planapochromat 63×/NA1.4 immersion objective or using an epifluorescence microscope (Nikon Eclipse 600; Nikon Corp., Tokyo, Japan) equipped with a ×60 objective and processed using the Adobe Photoshop 6.0 software (Adobe Systems Inc., San Jose, California, USA).
Acknowledgments
We thank the patients and their families for participation in this study. We are grateful to J. Ghysdael (CNRS UMR 146) for the gift of the FLI1 cDNA, to I. Dusanter-Fourt (INSERM U 567) for the gift of the ETS1 cDNA, to D. Meyer (INSERM U 143, Hôpital de Bicêtre) for the mouse anti-human vWF mAb, to Kirin Brewery Co. Ltd. (Tokyo, Japan) for the gift of PEG-rhuMGDF, and to F. Wendling and F. Morle for critical reading of the manuscript and helpful suggestions. This work was supported by grants from the Institut des Maladies Rares, INSERM, and La Ligue Nationale contre le Cancer (équipe labellisée 2004). H. Raslova is supported by a postdoctoral fellowship from the Institut Gustave Roussy and from Amgen, France; E. Komura is supported by a predoctoral fellowship from the Association pour la Recherche sur le Cancer. R. Favier is a recipient of a research fellowship from Assistance Publique–Hôpitaux de Paris–CNRS (grant application PRA 2001-2003) and belongs to the 11q research group.
Footnotes
See the related Commentary beginning on page 17.
Nonstandard abbreviations used: allophycocyanin (APC); blast cell (BL); enhanced GFP (eGFP); fetal liver tyrosine kinase 3 ligand (FLT3-L); human erythroleukemia cell line (HEL); internal ribosomal entry site (IRES); megakaryocyte (MK); β2-microglobulin (β2-M); Paris-Trousseau syndrome (PTS); phosphoglycerate kinase (PGK); phycoerythrin (PE); stem cell factor (SCF); thrombopoietin (TPO); woodchuck hepatitis virus posttranscriptional regulatory element (WPRE).
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Favier R, et al. A novel genetic thrombocytopenia (Paris-Trousseau) associated with platelet inclusions, dysmegakaryopoiesis and chromosome deletion at 11q23. C.R. Acad. Sci. III. 1993;316:698–701. [PubMed] [Google Scholar]
- 2.Breton-Gorius J, et al. A new congenital dysmegakaryopoietic thrombocytopenia (Paris-Trousseau) associated with giant platelet alpha-granules and chromosome 11 deletion at 11q23. Blood. 1995;85:1805–1814. [PubMed] [Google Scholar]
- 3.Jacobsen P, et al. An (11;21) translocation in four generations with chromosome 11 abnormalities in the offspring. A clinical, cytogenetical, and gene marker study. Hum. Hered. 1973;23:568–585. doi: 10.1159/000152624. [DOI] [PubMed] [Google Scholar]
- 4.Penny LA, et al. Clinical and molecular characterization of patients with distal 11q deletions. Am. J. Hum. Genet. 1995;56:676–683. [PMC free article] [PubMed] [Google Scholar]
- 5.Shivdasani RA. Molecular and transcriptional regulation of megakaryocyte differentiation. Stem Cells. 2001;19:397–407. doi: 10.1634/stemcells.19-5-397. [DOI] [PubMed] [Google Scholar]
- 6.Hart A, et al. Fli-1 is required for murine vascular and megakaryocytic development and is hemizygously deleted in patients with thrombocytopenia. Immunity. 2000;13:167–177. doi: 10.1016/s1074-7613(00)00017-0. [DOI] [PubMed] [Google Scholar]
- 7.Lemarchandel V, Ghysdael J, Mignotte V, Rahuel C, Roméo PH. Gata and Ets cis-acting sequence mediate megakaryocyte-specific expression. Mol. Cell. Biol. 1993;13:668–676. doi: 10.1128/mcb.13.1.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bastian LS, Kwiatkowski BA, Breininger J, Danner S, Roth G. Regulation of the megakaryocytic glycoprotein IX promoter by the oncogenic Ets transcription factor Fli-1. Blood. 1999;93:2637–2644. [PubMed] [Google Scholar]
- 9.Hashimoto Y, Ware J. Identification of essential GATA and Ets binding motifs within the promoter of the platelet glycoprotein Ibα gene. J. Biol. Chem. 1995;270:24532–24539. doi: 10.1074/jbc.270.41.24532. [DOI] [PubMed] [Google Scholar]
- 10.Deveaux S, et al. Analysis of the thrombopoietin receptor (MPL) promoter implicates GATA and Ets proteins in the coregulation of megakaryocyte-specific genes. Blood. 1996;87:4678–4785. [PubMed] [Google Scholar]
- 11.Wang X, et al. Control of megakaryocyte-specific gene expression by GATA-1 and FOG-1: role of Ets transcription factors. EMBO J. 2002;21:5225–5234. doi: 10.1093/emboj/cdf527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisbacher M, et al. Protein-protein interaction between Fli-1 and GATA-1 mediates synergistic expression of megakaryocyte-specific genes through cooperative DNA binding. Mol. Cell. Biol. 2003;23:3427–3441. doi: 10.1128/MCB.23.10.3427-3441.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spyropoulos DD, et al. Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol. Cell. Biol. 2000;20:5643–5652. doi: 10.1128/mcb.20.15.5643-5652.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu J, Pazin MJ, Ravid K. Properties of Ets-1 binding to chromatin and its effect on platelet factor 4 gene expression. Mol. Cell. Biol. 2004;24:428–441. doi: 10.1128/MCB.24.1.428-441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel FO, Higuchi T, Spyropoulos DD. Mouse models in the study of the Ets family of transcription factors. Oncogene. 2000;19:6443–6454. doi: 10.1038/sj.onc.1204038. [DOI] [PubMed] [Google Scholar]
- 16.Clausen PA, et al. Ets-1 induces increased expression of erythroid markers in the pluripotent erythroleukemic cell lines K562 and HEL. Leukemia. 1997;11:1224–1233. doi: 10.1038/sj.leu.2400735. [DOI] [PubMed] [Google Scholar]
- 17.Athanasiou M, Mavrothalassitis G, Sun-Hoffman L, Blair DG. Fli-1 is a suppressor of erythroid differentiation in human hematopoietic cells. Leukemia. 2000;14:439–445. doi: 10.1038/sj.leu.2401689. [DOI] [PubMed] [Google Scholar]
- 18.Yu X, et al. Lentiviral vectors with two independent internal promoters transfer high-level expression of multiple transgenes to human hematopoietic stem-progenitor cells. Mol. Ther. 2003;7:827–838. doi: 10.1016/s1525-0016(03)00104-7. [DOI] [PubMed] [Google Scholar]
- 19.Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 20.Held W, Roland J, Raulet DH. Allelic exclusion of Ly49-family genes encoding class I MHC-specific receptors on NK cells. Nature. 1995;376:355–358. doi: 10.1038/376355a0. [DOI] [PubMed] [Google Scholar]
- 21.Bix M, Locksley RM. Independent and epigenetic regulation of the interleukin-4 alleles in CD4+ T cells. Science. 1998;281:1352–1354. doi: 10.1126/science.281.5381.1352. [DOI] [PubMed] [Google Scholar]
- 22.Holländer GA, et al. Monoallelic expression of the interleukin-2 locus. Science. 1998;279:2118–2121. doi: 10.1126/science.279.5359.2118. [DOI] [PubMed] [Google Scholar]
- 23.Riviere I, Sunshine MJ, Littman DR. Regulation of IL-4 expression by activation of individual alleles. Immunity. 1998;9:217–228. doi: 10.1016/s1074-7613(00)80604-4. [DOI] [PubMed] [Google Scholar]
- 24.Pereira JP, Girard R, Chaby R, Cumano A, Vieira P. Monoallelic expression of the murine gene encoding TOLL-like receptor 4. Nat. Immunol. 2003;4:464–470. doi: 10.1038/ni917. [DOI] [PubMed] [Google Scholar]
- 25.Rhoades KL, et al. Allele-specific expression patterns of interleukin-2 and Pax-5 revealed by a sensitive single-cell RT-PCR analysis. Curr. Biol. 2000;10:789–792. doi: 10.1016/s0960-9822(00)00565-0. [DOI] [PubMed] [Google Scholar]
- 26.Favier R, et al. Paris-Trousseau syndrome: clinical, hematological, molecular data of ten new cases. Thromb. Haemost. 2003;90:893–897. doi: 10.1160/TH03-02-0120. [DOI] [PubMed] [Google Scholar]
- 27.Kawada H, et al. Defective megakaryopoiesis and abnormal erythroid development in Fli-1 gene-targeted mice. Int. J. Hematol. 2001;73:463–468. doi: 10.1007/BF02994008. [DOI] [PubMed] [Google Scholar]
- 28.Nutt SL, et al. Independent regulation of the two Pax5 alleles during B-cell development. Nat. Genet. 1999;21:390–395. doi: 10.1038/7720. [DOI] [PubMed] [Google Scholar]
- 29.Hanel ML, Wevrick R. The role of genomic imprinting in human developmental disorders: lessons from Prader-Willi syndrome. Clin. Genet. 2001;59:156–164. doi: 10.1034/j.1399-0004.2001.590303.x. [DOI] [PubMed] [Google Scholar]
- 30.Shore EM, et al. Paternally inherited inactivating mutation of the GNAS1 gene in progressive osseous heteroplasia. N. Engl. J. Med. 2002;346:99–106. doi: 10.1056/NEJMoa011262. [DOI] [PubMed] [Google Scholar]
- 31.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 32.Newlands S, et al. Transcription occurs in pulses in muscle fibers. Genes Dev. 1998;12:2748–2758. doi: 10.1101/gad.12.17.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cook DL, Gerber AN, Tapscott SJ. Modeling stochastic gene expression: implications for haploinsufficiency. Proc. Natl. Acad. Sci. U.S.A. 1998;95:15641–15646. doi: 10.1073/pnas.95.26.15641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kemkemer R, Schrank S, Vogel W, Gruler H, Kaufmann D. Increased noise as an effect of haploinsufficiency of the tumor-suppressor gene neurofibromatosis type 1 in vitro. Proc. Natl. Acad. Sci. U. S. A. 2002;99:13783–13788. doi: 10.1073/pnas.212386999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi ES, Nichol JL, Hokom MM, Hornkohl AC, Hunt P. Platelets generated in vitro from proplatelet-displaying human megakaryocytes are functional. Blood. 1995;85:402–413. [PubMed] [Google Scholar]
- 36.Raslova H, et al. Megakaryocyte polyploidization is associated with a functional gene amplification. Blood. 2003;101:541–544. doi: 10.1182/blood-2002-05-1553. [DOI] [PubMed] [Google Scholar]
- 37.Veiga-Fernandes H, Walter U, Bourgeois C, McLean A, Rocha B. Response of naive and memory CD8+T cells to antigen stimulation in vivo. Nat. Immunol. 2000;1:47–53. doi: 10.1038/76907. [DOI] [PubMed] [Google Scholar]
- 38.Naldini L, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]