Abstract
The acetylation and deacetylation of histones play an important role in the regulation of gene transcriptions. Histone acetylation is mediated by histone acetyltransferase (HAT). The resulting modification in the structure of chromatin leads to nucleosomal relaxation and altered transcriptional activation. The reverse reaction is mediated by histone deacetylase (HDAC), which induces deacetylation, chromatin condensation, and transcriptional repression. HDACs are divided into three distinct classes, I, II, and III, on the basis of size, sequence homology, as well as formation of distinct complexes. Among class II HDACs, HDAC4 is implicated in controlling gene expression important for diverse cellular functions. Basic and clinical experimental evidence have well established that HDAC4 performs a wide variety of functions. Understanding the biological significance of HDAC4 will not only provide new insight into the mechanisms of HDAC4 involved in mediating biological response, but also form a platform to develop a therapeutic strategy to achieve clinical implications.
Keywords: HDAC4, HAT, Modification, Function, Regulation
1. Introduction
Lysine acetylation of histones, as well as non-histone proteins, is emerging as a widespread reversible posttranslational modification in regulating gene expression and various cellular processes. Dynamic control of protein acetylation levels in vivo occurs through the opposing actions of HATs and HDACs. Histone acetylation by HATs relaxes the structure of nucleosomes and leads to gene activation, whereas histone deacetylation by HDACs promotes chromatin condensation, favoring the transcriptional repression. Association of HATs and HDACs with sequence-specific DNA-binding proteins allows for gene-specific activation and repression, respectively. Since the identification of HDAC 1 (named HD 1), at least 18 HDACs have been identified in mammals [1–3]. These HDACs can be categorized into three distinct classes based on the specific of their catalytic mechanisms. Class I HDACs consist of HDAC 1, 2, 3, and 8, which are ubiquitously expressed and predominantly located in nuclei. Class II HDACs (4, 5, 6, 7, 9 and 10) are defined based on their homology with Hda1. In contrast to class I, class II HDACs exhibit a tissue specific pattern of expression. Class III HDACs consist of a large family of sirtuins (silent information regulators or SIR) that are evolutionarily distinct, with unique enzymatic mechanisms dependent on NAD+ [4].
Based on their domain organization, class II HDACs are further divided into two subgroups: class IIa (HDAC4, HDA5, HDAC7 and HDAC9) and class IIb (HDAC6 and HDAC10). Genetic evidence has proven that class IIa HDACs play an important role in tissue-specific growth and development. For example, HDAC 4 and HDAC 5 are highly expressed in the heart, brain, and skeletal muscle and shuttle between the nucleus and cytoplasm [5]. For more general information on HDACs and class II HDACs, we respectfully refer the reader to previous excellent reviews [2, 6–8]. HDAC4, a key member of class IIa HDACs, is expressed in multiple tissues [9]. Mice lacking HDAC4 die early during the perinatal period, display abnormal chondrocyte hypertrophy due to ectopic and premature ossification of endochondrial bones [10]. Genome-wide data in humans have documented alterations and mutations of HDAC4 in melanoma and breast cancer [11]. In this review, we place emphasis both on addressing the HDAC4 regulation in posttranscriptional by microRNAs and posttranslational modifications, and also on biological functions in normal development and pathological conditions.
2. Molecular basis of transcriptional and post transcriptional regulation
The human HDAC4 gene, which spans approximately 353.49 kb, is located on chromosome 2q37.3 and produced 8980bp mRNA (NM_006037.3) transcripts. The murine HDAC4 gene, which spans approximately 215.7kb, is located on chromosome 1 and produced 3960 mRNA (NM_207225.1) transcripts. HDAC4 expresses in different tissues and the expression magnitudes have been well established in response to various stimuli. Despite the diversity of pathways modulated by HDAC4 and unique mechanisms regulating the activity of HDAC4, little is known about the mechanisms regulating its expression. Transcription factors Sp1 and Sp3 directly bind to specific consensus GC-rich sequences in the HDAC4 promoter to drive HDAC4 transcription [12]. HDAC4 is not expressed in the nuclei of mouse embryonic stem cells, but is dramatically up-regulated upon differentiation [13].
MicroRNAs (miRNAs) are a class of regulatory RNAs of ~22 nucleotides that post-transcriptionally regulate gene expression. miRNAs are involved in multiple biological responses as well as disease disorders including cancer. microRNAs are implicated in many biological regulations such as proliferation, apoptosis, invasion/metastasis, and angiogenesis. Several miRNAs were characterized to modulate HDAC4, which includes miR-1, miR-29 and miR-140, mir-155, miR-200a, miR-206, miR-365 in different cell types. miR-200a directly targets the 3′-untranslated region of the HDAC4 messenger RNA and represses expression of HDAC4 (Figure 1). Interestingly, HDAC4 also conversely inhibited the expression of miR-200a and its promoter activity and reduced the histone H3 acetylation level at the miR-200a promoter through a Sp1-dependent pathway [14]. Muscle-specific miR-1 promotes myogenesis by targeting HDAC4 3′UTR and represses HDAC4 levels during muscle differentiation [15]. mTOR controls MyoD-dependent transcription of miR-1 through its upstream enhancer, miR-1 suppression of HDAC4 results in production of follistatin and subsequent myocyte fusion [16]. Transient transfection of miR-1 and -499 in cardiomyocyte progenitor cells reduced proliferation rate, and enhanced differentiation into cardiomyocytes in human cardiomyocyte progenitor cells and embryonic stem cells via the repression of HDAC4 [17]. It is also noted that miR-22, down-regulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumorigenicity through targeting HDAC4 [18].
Figure 1. microRNAs target sites localized in the 3′UTR of HDAC4.
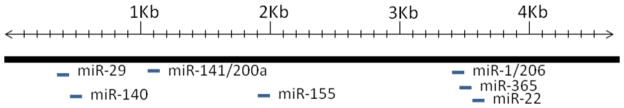
Schematic representation of microRNAs target sites localized in the 4.5kb 3′UTR of HDAC4.
In addition, the overexpression of miR-206 and miR-29 resulted in the translational repression of HDAC4 in the presence or absence of TGF-β via interaction with the HDAC4 3′-UTR. miR-206 and miR-29 implicated in muscle differentiation are down-regulated by TGF-β. TGF-β treatment of myogenic cells is associated with the increased expression of HDAC4 [19]. miR-29b is a key regulator of the development of osteoblast phenotype by targeting HDAC4, TGF-β3, activin receptor type-2A (ACVR2A), beta interacting protein 1 (CTNNBIP1), and dual-specificity phosphatase–2 (DUSP2) proteins. Cartilage specific miR-140 directly targets HDAC4 3′UTR [20]. Mice lacking miR-140 demonstrated the phenotype of dwarf as a consequence of impaired chondrocytes [21]. Likewise, miR-365, a mechanically responsive microRNA, was associated with modulation of chondrocyte differentiation via directly targeting HDAC4 [22]. In miR-155 transgenic mice model, miR-155 targets HDAC4 and subsequently impairs transcriptional activity of B-cell lymphoma 6 gene of B cells. Forced overexpression of HDAC4 in human B-cell lymphoma cells, leads to the reduced miR- 155-induced proliferation, clonogenic potential, and increased apoptosis [23]. Taken together, these evidence suggest that miRNAs specifically target HDAC4 to modulate the cellular responses and biological functions in different cell types.
3. Protein domain structure and post translational modifications
3.1 protein domain structure
Human HDAC4 genes encode proteins between 972 to 1084 amino acids and mouse HDAC4 genes encode proteins between 965 to 1076 amino acids. As illustrated in Figure 2, it contains a unique regulatory domain N-terminal through interaction with specific transcription factors and a zinc containing catalytic domain in the C-terminal. Crystal structure analysis has indicated that a properly folded structural zinc-binding domain is required for the formation of the repressor complex. Moreover, the N-terminal of HDAC4 reveals that the glutamine-rich domain of folds into a straight alpha-helix that assembles as a tetramer [24]. C-terminal zinc-binding domain plays a key role in substrate recognition and in the association of HDAC4 with the HDAC3·N-CoR co-repressor complex. The detailed analysis of the crystal packing showed that an intermolecular disulfide bond had formed between cysteine 669, lying within the structural zinc-binding domain, and cysteine 700 from a neighboring molecule [25].
Figure 2. Domain organization and known post-translational modifications of HDAC4.
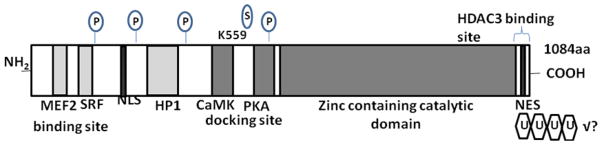
Schematic representation of HDAC4 is shown below. The total number of amino ac id residues (aa) is depicted on the left, and the deacetylase domains are highlighted in gray. Phosphorylated (P), Sumoylated residues (S) and acetylated (A) residues are marked with a circle. Polyubiquitylation (U) is illustrated by multiple hexagons. The positions of modified amino acids are indicated next to each modification. Post-translational modifications that were detected without identification of the exact position are shown on the right and are marked by !?. NLS: nuclear localization
3.2 Post translational modifications
The function, activity, and stability of proteins can be controlled by post-translational modifications. Protein phosphorylation is perhaps the most widely studied and understood modification in which certain amino acid residues are phosphorylated by the action of protein kinases or dephosphorylated by the action of phosphatases. Post translational modifications can influence subcellular localization and partners interaction of HDAC4. It is well known that one of the major functions of HDAC4 is mediating the transcriptional repression through regulating chromatin condensation and structures. Recently, rigorous evidence has revealed that the post-modification of HDAC4 plays a critical role in controlling the cellular response, involved with HDAC4. It is known that the integration of all the post-translational modifications in a coherent and global code is a priority for understanding the regulatory networks operating on these key enzymes in different cellular, developmental, physiologic, and pathologic phenomena. Detailed interpretations of the modulation patterns of HDAC4 at translational levels will advance our knowledge in understating the direct correlation between HDAC4 functions and the fashions of translational modifications. The recent evidence have well demonstrated that HDAC4 can be phosphorylated, sumoylated, carbonylated, ubiquited and proteolytic cleaved in the post-translational modification fashions (Figure 2).
3.2.1 Phosphorylation
Phosphorylation/dephosphorylation efficiently and rapidly couples the repression of class IIa HDACs to environmental signals. Reversible phosphorylation is an essential regulatory mechanism for the function of HDAC4. HDAC4 interacts with the 14-3-3 family of proteins that are known to bind specifically to conserved phosphoserine-containing motifs. Phosphorylation of these specific amino acids creates binding sites for the 14-3-3 chaperone protein, which escorts phospho- HDAC4 from the nucleus to the cytoplasm, with consequent activation of HDAC target genes [26–28]. HDAC4 can be phosphorylated by calcium/calmodulin-dependent protein kinase (CaMK), the extracellular signal-regulated kinases 1 and 2 (ERK1/2), protein kinase A (PKA) and glycogen synthase kinase 3 (GSK3)
Stimulation of CaMK promotes myogenesis by disrupting MEF2-HDAC complexes and causing HDAC nuclear export [29]. CaMKII specifically binds to HDAC4 through a unique docking site. Phosphorylation of HDAC4 at S246, S467, and S632 by CaMKII enhances nuclear export and prevents nuclear import of HDAC4, with the consequent depression of HDAC4 target genes [30]. Signal transduced from endogenous CaMKII is required for agonist-induced cytosolic accumulation of HDAC4 in cardiomyocytes [31, 32]. However, PKA phosphorylates HDAC4 and regulates HDAC4 proteolytic cleavage in tyrosine 207 and antagonizes CaMKII-mediated activation of MEF2 by regulated proteolysis of HDAC4 [33]. The N-terminal HDAC4 cleavage product selectively inhibits activity of MEF2 but not serum response factor (SRF), thereby antagonizing the prohypertrophic actions of CaMKII without affecting cardiomyocyte survival. Activation of the Ras–MAPK pathway by expression of oncogenic Ras or constitutively active MAPK/ERK kinase 1 results in an increased percentage of HDAC4 in the nucleus in C2C12 myoblast cells [34]. Glycogen synthase kinase 3 (GSK3) can phosphorylate HDAC4 at position 298 and 302 to proteasome-mediated degradation of HDAC4 to function as an important element of the signaling pathway governing HDAC4 stability [35].
On the other hand, protein phosphatases also play a critical role in regulating the phosphorylation status of substrates. HDAC4 has been demonstrated to be dephosphorylated in vitro by protein phosphatase 2A (PP2A), in which PP2A interacts with and subsequently dephosphorylates HDAC4. PP2A, via the dephosphorylation of multiple serines including the 14-3-3 binding sites and serine 298, controls HDAC4 nuclear import. Serine 298 also contributes to the nuclear import of HDAC4. PP2A interacts with the N-terminus of HDAC4, and promotes the accumulation of HDAC4 in the nucleus via dephosphorylation of serine 298 [36].
3.2.2 Carbonylation
Carbonylation (or alkylation) is a distinctive post-modification of redox signaling pathways occurring in cells after oxidative stress. Covalent binding of reactive carbonyl species on cysteinyl thiols of substrate proteins is termed carbonylation. Both cysteine-274/cystein-276 in DnaJb5 and cystein-667/cystein-669 in HDAC4 are oxidized and form intramolecular disulfide bonds in response to reactive oxygen species-generating hypertrophic stimuli whereas they are reduced by thioredoxin-1. Where reduction of cystein-274/cystein-276 in DnaJb5 is essential for interaction between DnaJb5 and HDAC4, the reduction of cystein-667/cystein-669 in HDAC4 inhibits its nuclear export independently of its phosphorylation status [37]. Knockdown of Nox4, but not Nox2, attenuated O2− production in the nucleus and prevented phenylephrine-induced oxidation and nuclear exit of HDAC4. HDAC4 oxidation and cardiac hypertrophy were also attenuated in cardiac-specific Nox4 knockout mice [38].
3.2.3 Sumoylation
SUMO (small ubiquitin-like modifier) modification is the covalent attachment of SUMO to lysine residues. Analogous to ubiquination, the conjugation of SUMO proteins (SUMO1, SUMO2, and SUMO3) to lysines of targets substrates plays a critical role in modulating the activity and degradation of targeted proteins. HDAC4 was found to be recognized by SUMO-1 at a single lysine residue (lysine559) which is modified by SUMO-2 chains in vivo [39]. Sumoylated lysine-559 promoted by the E3 SUMO-protein ligase RANBP2 did not affect its subcellular distribution and the binding to some of its known interaction partners [40]. However, HDAC4 at lysine 559 mutation displayed significantly impaired transcriptional repression and enzymatic activities relative to the wild-type HDAC4 protein. Sumoylation of HDAC4 was prevented by phosphorylation by CaMK4.
3.2.4 Ubiquitination
Ubiquitin (Ub) is a small protein conjugated to lysine residues of target substrates through an isopeptide bond, as a single monomer or as a polyubiquitin chain. The reaction involves a well-defined three-step enzymatic cascade (E1 activating enzymes, E2 conjugating enzymes and E3 ubiquitin ligases). Polyubiquitination mainly targets proteins to achieve the degradation via the proteasome machinery, while monoubiquitination acts as a signal for different biological outputs. HDAC4 ubiquitination and proteasomal degradation are regulated by phosphorylation of GSK3β, however, the mechanism and the biological significance of HDAC4 ubiquitination still remains to be clarified [35].
3.2.5 Proteolytic cleavage
HDAC4 nuclear/cytoplasmic shuttling is also mediated by proteolysis during the occurring apoptosis. HDAC4 is cleaved by caspase-2 and -3 at aspartic acid 289. The caspase-cleaved amino-terminal fragment of HDAC4 containing the nuclear localization signal accumulates in the nucleus, represses transcription and induces cell death and acts as a strong repressor of the MEF2C [41–45]. Relative to other nuclear forms of HDAC4, a caspase-generated nuclear fragment exhibits marked death phenotype coupled with increased repressive effect on Runx2- or SRF-dependent transcription. Caspase-generated N-terminal fragment mainly accumulates in the nuclei and represses Runx2- or SRF-dependent transcription although this fragment does not contain C-terminal Zinc binding domain for substrate recognition and in the association of HDAC4 with the HDAC3·N-CoR co-repressor complex. The caspase-generated fragment is weakly bound to chromatin, whereas an HDAC4 mutant defective in 14-3-3 binding forms a more stable complex with HDAC5 protein [43].
4. Subcelluar HDAC4 and its substrates
The HDAC4 gene expression can be regulated at the levels of transcription, posttranscription, (microRNA and RNA stability) and protein stability (protease degradation). HDAC4 shuttles between the nucleus and cytoplasm, and serves as a nuclear corepressor that regulates bone and muscle development. The activity of HDAC4 is basically regulated through two major mechanisms: subcellular localization and the formation of multi-subunit complexes with other proteins.
4.1 Subcellular distribution
For HDAC4 translocation, HDAC4 alkylation or specific sites (S246, S467, and S632) HDAC4 phosphorylation will interact with 14-3-3 and translocate between cytoplasm and nuclear as discussed above in the HDAC4 protein posttranslational modification. HDAC4 translocation is also regulated through interaction with transport protein CRM1 (also called importin 1) which mediates the nuclear export of cellular proteins (cargos) bearing a leucine-rich nuclear export signal (NES) [41, 46, 47] and nucleoporin 155 (Nup155) that is a major component of the nuclear pore complex (NPC) involved in cellular nucleo-cytoplasmic transport [48]. HDAC4 is thought to act as transcriptional corepressors by deacetylating nucleosomal histones. Since these enzymes do not bind directly to DNA, the current model posits that their deacetylase activity is recruited to specific promoters by sequence-specific DNA-binding proteins. HDAC4 also interacts with different proteins, such as heterochromatin protein 1 (HP1) and histone methytelase distinct transcription factors, thus dictating HDAC4 functions at different tissues. In addition to histones, growing evidence suggests that HAT and HDAC target non-histone proteins, including transcriptional factors, which may represent general regulatory mechanisms in biological signaling. The biological functions of cytoplasmic HDAC4 have been well established, which has fully been discussed in a subsequent section of cancer in this review including Hypoxia-inducible factor 1-alpha (HIF-1α)[49–51], MEKK2, [52] and signal transducer and activator of transcription 1 (STAT1) [53].
4.2. Regulation of histone protein deacetylation
The mechanisms of deacetylation by HDAC4 involve removing acetyl groups from both histones and non-histone proteins by its zinc containing catalytic domain. The reversible acetylation of N-terminal lysine residues at positions 9, 14, 18, and 23 of histone 3 and 5, 8, 12, and 16 of histone 4 mediates decondensation of the nucleosome structure, alters histone and DNA interactions, and facilitates access and binding of transcription factors. Histone acetylation status is balanced by HATs and HDACs. Unlike HDAC6, HDAC4 and HDAC5 interact with HDAC3 and RbAp48 [26]. The catalytic domain of HDAC4 tends to forms a multi-protein complex when it binds with partners such as the SMRT-NCoR ·HDAC3 co-repressor complex [54]. The integrity of the catalytic domain of HDAC4 is necessary for recruiting the HDAC3 N-CoR repressor complex and for subsequent deacetylase activity of the complex. HDAC4 is a deacetylase enzymatically inactive when not associated with HDAC3 [55].
4.3. Regulation of non-histone protein deacetylation
Runx2 serves as a major target of the bone morphogenetic protein (BMP) pathway. BMP-2 signaling stimulates p300- mediated Runx2 acetylation, increasing transactivation activity and inhibiting Smurf1- mediated degradation of Runx2. HDAC4 and HDAC5 deacetylate Runx2, allowing the protein to undergo Smurf-mediated degradation. Inhibition of HDAC increases Runx2 acetylation, potentiates BMP-2-stimulated osteoblast differentiation, and increases bone formation [56]. Recent works accredit HDAC4 deacetylase activity for non histone proteins in cytoplasm: HIF-1α [49–51], MEKK2 [52] and STAT1 [53].
4.4 Demethylation of histone proteins
Histone acetylation and methylation are the most well-characterized epigenetic marks. Trimethylation at H3K4, H3K36, or H3K79 results in an open chromatin configuration and is, therefore characteristic of euchromatin. Euchromatin is also characterized by a high level of histone acetylation, which is mediated by HATs. Conversely, HDACs have the ability to remove this epigenetic mark, which leads to transcriptional repression [57]. Methylated H3K9 provides a binding site for the chromodomain-containing HP1, which induces transcriptional repression and heterochromatinization. HDAC4 is involved in epigenetic gene regulation via interaction with H3K9 metyltransferase SUV39H1 and HP1, providing an efficient mechanism for silencing MEF2 target genes by coupling histone deacetylation and methylation [58, 59]. H3K9 demethylation is closely associated with nucleo-cytoplasmic shuttling of HDAC4 and HP1 dissociation from the promoter regions of Atrial natriuretic peptide (ANP) and B-type Natriuretic Peptide (BNP) and repression of ANP and BNP gene transcription in response to MEF2 [60]. H3K9 trimethylation is enriched significantly after stress at one of the alternative 5′ acetylcholinesterase (AChE) promoters and that the accumulation of this histone mark was associated with HDAC4’s recruiting HP1 and SUV39H1 on the AChE promoter [61]. The Fas promoter in fibroblasts from bleomycin-treated mice exhibited decreased histone acetylation and increased histone 3 lysine 9 trimethylation (H3K9Me3) and this was associated with increased HDAC4 expression [62].
In addition, HDAC4 negatively regulates the transcription factor MEF2 through interaction with the SUMO E2 conjugating enzyme Ubc9. The overexpression of HDAC4 leads to prominent MEF2 sumoylation in vivo, whereas recombinant HDAC4 stimulates MEF2 sumoylation in a reconstituted system in vitro. HDAC4 promotes sumoylation on a lysine residue that is also subject to acetylation by a MEF2 coactivator, the acetyltransferase CBP, suggesting a possible interplay between acetylation and sumoylation in regulating MEF2 activity [63–65]. This proposed model is still debated and further experimental work is necessary to clarify whether HDAC4 directly sumoylates MEF2 or recruits the SUMO E2 conjugating enzyme for MEF2 sumoylation.
5. Physiological functions of HDAC4
HDAC4 plays global roles in the regulation of gene transcription, cell growth, survival and proliferation, and their aberrant expression or activity lead to cancer development.
5.1 Chondrogensis, osteoblast differentiation and chondrocyte hypertrophy
HDAC4, which is expressed in prehypertrophic chondrocytes, regulates chondrocyte hypertrophy and endochondral bone formation by interacting with and inhibiting the activity of Runx2, a transcription factor necessary for chondrocyte hypertrophy. HDAC4− mice display premature ossification of developing bones due to ectopic and early onset chondrocyte hypertrophy, mimicking the phenotype that results from constitutive Runx2 expression in chondrocytes [10]. Runx2 can be acetylated by p300 and acetylated-Runx2 prevents proteins from ubiquitination. HDAC4 and HDAC5 play opposing roles in the bone morphogenetic protein signaling of Runx2 regulation through deacetylated-Runx2, allowing the protein to undergo smurf-mediated degradation [56]. TGF-beta represses osteoblast differentiation through the action of HDAC4 and 5, which are recruited through interaction with Smad3 to the Smad3/Runx2 complex at the Runx2-binding DNA sequence in differentiating osteoblasts [66]. HDAC4 overexpression promotes TGF-beta1- induced synovium-derived stem cell chondrogenesis, but inhibits chondrogenically differentiated stem cell hypertrophy [67].
5.2. Muscle development and cardiovascular diseases
The first stage of myogenesis involves the formation of myoblasts, which express a distinct subset of transcription factors, including the MEF2C. Mice lacking the MEF2C display defective heart morphogenesis, with development arrested at the heart looping stage [68]. HDAC4 directly binds and represses MEF2 function and regulates the specification of mesoderm cells into cardiomyoblasts by inhibiting the expression of GATA4 and Nkx2-5. HDAC inhibitor treatment induces the entry of mesodermal cells into the cardiac muscle lineage, shown by the up-regulation of transcripts Nkx2-5, MEF2C, GATA4 and cardiac alpha-actin. HDACs play a repressive role during the entry of mesoderm cells into the cardiac-muscle lineage [3]. Overexpression of HDAC4 inhibited cardiomyogenesis, shown by the down-regulation of cardiac muscle gene expression [69].
During muscle differentiation, HDAC4-dependent gene regulation showed that HDAC4 mediates gene repression by the recruitment to MEF2 sites in the promoters of repressed genes. The repression of gene transcription by MEF-2/HDAC complexes is relieved due to CaMK-induced translocation of HDAC4 and HDAC5 to the cytoplasm [28, 32, 33, 40, 45, 46, 58, 69–72]. The hearts of transgenic mice overexpressing an activated CaMKIV undergo hypertrophy with the up-regulation of embryonic transcripts such as atrial natriuretic factor and a dramatic enhancement of MEF2C activity [73].
Cardiac hypertrophy is the heart’s response to a variety of extrinsic and intrinsic stimuli that impose increased biomechanical stress. A variety of cardiovascular disorders, including myocardial infarction, arterial hypertension, and altered contractility resulting from mutations of sarcomeric proteins provoke the adult heart to become enlarged because of hypertrophic growth of cardiomyocytes. In cardiomyocytes, CaMKII-phosphorylation of HDAC4 results in hypertrophic growth, which can be blocked by a signal-resistant HDAC4 mutant [29]. In miR-22-null mice, cardiac miR-22 was found to be essential for hypertrophic cardiac growth in response to stress through directly targeting Sirt1 and HDAC4 [74].
HDAC4 plays a role in regulating myofilament contractile activity through the regulation of muscle LIM protein (MLP) deacetylation [75]. HDAC4, and HAT, p300/CBP-associated factor (PCAF), are associated with cardiac myofilaments. Both HDAC4 and PCAF are associated with the Z-disc and I- and A-bands of cardiac sarcomeres. Z-disc-associated protein, MLP, functions as a sensor of cardiac mechanical stretch, as an acetylated target of PCAF and HDAC4.
In muscle cells, all contractions are controlled by the nervous system. HDAC4 is normally concentrated at the neuromuscular junction [76]. Loss of neural input leads to concomitant nuclear accumulation of HDAC4 and transcriptional reduction of MEF2- regulated gene. In the surgical denervation and the neuromuscular disease amyotrophic lateral sclerosis model, the elevated levels of HDAC4 are required for efficient repression of MEF2-dependent structural gene expression. Forced expression of HDAC4 mimics the denervation and activates ectopic acetyl-choline receptor (nAChR) transcription throughout myofibers. Inactivation of HDAC4 prevents denervation-induced synaptic nAChR and muscle-specific receptor tyrosine kinase (MUSK) transcription [77]. HDAC4 is preferentially enriched in myonuclei of fast oxidative skeletal muscle fibers and knockdown of HDAC4 enhances glycolysis in myotubes [78].
5.3. Neuronal survival and mental disease
HDAC4 is present in the perikaryial cytoplasm of most neurons but its nuclear localization is variable. In the dentate gyrus, nuclear expression is not detectable, whereas in other areas some neuronal nuclei contain HDAC4 signal whereas others do not [79]. HDAC4 is normally localized to the cytoplasm in brain tissue and cultured cerebellar granule neurons. HDAC4 rapidly translocates into the nucleus in response to low-potassium or excitotoxic glutamate conditions that induce neuronal cell death. Treatment with the neuronal survival factor (brain-derived neurotrophic factor, BDNF) suppresses HDAC4 nuclear translocation, whereas a proapoptotic CaMK inhibitor stimulates HDAC4 nuclear accumulation. Moreover, ectopic expression of nuclear-localized HDAC4 promotes neuronal apoptosis and represses the transcriptional activities of MEF2 and cAMP response element-binding protein, survival factors [80]. HDACs play an important role in neuronal survival and photoreceptor development. MEF2-HDAC4 transcriptional complex is involved in neuron survival as a target of ataxin-1 [81]. The subcellular localization of HDACs in neurones is specified by neuronal activity. Spontaneous electrical activity was sufficient for nuclear export of HDAC4 but not for HDAC5 [82]. Huntington’s disease (HD) is a neurodegenerative genetic disorder that affects muscle coordination and leads to cognitive decline and psychiatric problems. miR-22 has multipartite anti-neurodegenerative activities including the inhibition of apoptosis and the targeting of genes (including HDAC4, Rcor1 and Rgs2) implicated in the etiology of HD [83].
Reduction in HDAC4 expression during normal retinal development led to apoptosis of rod photoreceptors and bipolar interneurons, whereas overexpression reduced naturally occurring cell death of the BP cells. HDAC4 overexpression in a mouse model of retinal degeneration prolonged photoreceptor survival. The survival effect was due to the activity of HDAC4 in the cytoplasm [84]. HDAC4 works to repress PPARgamma transcription and regulates neuronal death by inhibiting PPARgamma pro-survival activity in cultured cortical neurons under oxidative stress [85].
Defects in HDAC4 are the cause of brachydactyly-mental retardation syndrome [86]. A syndrome resembling the physical anomalies found in Albright hereditary osteodystrophy. Common features are mild facial dysmorphism, congenital heart defects, distinct brachydactyly type E, mental retardation, developmental delays, seizures, autism spectrum disorder, and stocky build. In a study from 278 patients with schizophrenia and 234 normal controls from a Korean population, single nucleotide polymorphisms analysis demonstrates that the HDAC4 gene showed associations with schizophreniain the codominant and dominant models [87]. Ataxia telangiectasia is a neurodegenerative disease caused by mutation of the Atm gene. In the Atm deficiency mice, nuclear accumulation of HDAC4 in neurons promotes neurodegeneration [88].
5.4. Pancreatic beta/delta-cell lineage specification
Class IIa HDAC4, -5, and -9 have unexpected restricted expression in the endocrine beta- and delta-cells of the pancreas. HDAC4, -5, and -9 are key regulators in controlling the pancreatic beta/delta-cell lineage. Analyses of the pancreas of class IIa HDAC mutant mice revealed an increased pool of insulin-producing beta-cells in HDAC5−/− and HDAC9−/− mice and an increased pool of somatostatin-producing delta-cells in HDAC4−/− and HDAC5−/− mice. HDAC4 and HDAC5 overexpression showed a decreased pool of insulin-producing beta-cells and somatostatin-producing delta-cells [89].
5.5. Cancer
Cancer has been considered to be the result of a wide variety of epigenetic dysregulation, chromosome translocations, deletions, and point mutations. In some acute leukaemias, a chromosomal translocation fusing the promyelocyte leukemia zinc finger (PLZF) gene, the promyelocytic leukaemia zinc-finger protein, to that encoding the retinoic acid receptor RARalpha gives rise to a fusion protein, PLZF-RARalpha, thought to be responsible for constitutive repression of differentiation-associated genes. HDAC4 was found to interact with the leukemic PLZF-RARa fusion protein and to mediate repression of differentiation-associated genes in the leukaemic cells [90]. Inhibition of HDAC activities with HDAC inhibitors in clinical and basic experiments indicates potential benefit of HDAC inhibitors in treating cancer. Development of specific HDAC isoform inhibitors by inhibiting its activity to achieve clinical implication in cancer therapy is realized as one of the most promising approach. BCL6 is a member of the POZ/zinc finger family involved in survival and/or differentiation in B cell lymphoma upon chromosomal alteration. HDAC4 associates with BCL6 and PLZF in vivo and in vitro to mediate transcriptional repression [91]. microRNAs are involved in the initiation and progression of cancer. miR-155 is one of the most overexpressed miRNAs in several solid and hematological malignancies. miR-150 can directly target 3′UTR of HDAC4 and inhibit HDAC4 protein translation. Ectopic expression of HDAC4 in humanactivated B-cell-type diffuses large B-cell lymphoma (DLBCL) cells resulting in reduced miR-155-induced proliferation, clonogenic potential, and increased apoptosis [23].
HDAC4 is maximally expressed in the proliferative compartment in normal colonic and small intestinal epithelium, and its expression is down-regulated during differentiation. HDAC4 interacts with Sp1 and reduces histone H3 acetylation at the Sp1/Sp3 binding site-rich p21 (WAF1/Cip1) proximal promoter. Induction of p21 (WAF1/Cip1) mediated by silencing of HDAC4 arrested cancer cell growth and inhibited tumor growth in human glioblastoma model [92, 93]. X-linked tumor suppressor forkhead box P3 (FOXP3) is essential for p21 expression in normal epithelia and the lack of FOXP3 is associated with p21 down-regulation in breast cancer samples. FOXP3 specifically inhibited binding of HDAC4 and increased local histone H3 acetylation[94]. In hepatocellular carcinoma, HDAC4, known to have critical roles in cancer development, was directly targeted and regulated by miR-22. Furthermore, HDAC4 was up-regulated in miR-22-downregulated HCC tissues [18]. In hepatocellular carcinoma cells, miR-200a directly targets HDAC4. However, HDAC4 also inhibited the expression of miR-200a and its promoter activity and reduced the histone H3 acetylation level at the miR-200a promoter through a Sp1-dependent pathway. miR-200a also induced the up-regulation of total acetyl-histone H3 levels and increased the histone H3 acetylation level at the p21WAF/Cip1 promoter miR-22 and miR-200a [14, 18].
Ovarian cancer frequently acquires resistance to platinum chemotherapy, representing a major challenge for improving patient survival. Analysis of the paired tumor biopsies taken before and after development of clinical platinum resistance showed significantly increased HDAC4 expression in resistant tumors [53]. PLU-1/JARID1B, which is expressed in a high proportion of breast cancers, interacts with HDAC4 and is coexpressed in breast cancers [95]. The bladder tumor tissue arrays analysis demonstrated that the frequency of the HDAC4-positive tissue specimens was much higher in the bladder tumors than in the normal bladder tissues. The HDAC4 was also significantly increased in the urinary bladder transitional cell carcinomas in comparison to normal bladder tissues [96]. HIF1α is an essential part of the HIF-1 transcriptional complex that regulates angiogenesis, cellular metabolism, and cancer development [49–51, 97]. HIF1α acetylation can be increased by HDAC4 shRNA but not, HDAC1 or HDAC3 shRNA. Inhibition of HDAC4 decreases both the HIF-1 transcriptional activity and a subset of HIF-1 hypoxia target gene expression, and reduces the resistance to docetaxel chemotherapy.
miR-140 is involved in the chemoresistance by reduced cell proliferation through G(1) and G(2) phase arrest mediated in part through the suppression of HDAC4 in osteosarcoma tumor and colon cancer[98]. Tasquinimod is an orally active antiangiogenic drug that is currently in phase III clinical trials for the treatment of castration-resistant prostate cancer. Tasquinimod directly binds to HDAC4 thereby inhibiting deacetylation of histones and HDAC4 client transcription factors, such as HIF-1α [99].
6. The future perspectives
HDACs appear to be key enzymes in the regulation of gene expression. HDAC function appears to be regulated by the intrinsic features of each HDAC4, including abundance, cellular compartmentalization and association with cofactors. In-depth investigation of the biochemistry and interplay of posttranscriptional and posttranslational modifications of HDAC4 will reveal a novel mechanism(s) by which HDAC4 is involved in many physiological and pathological disorders. Future investigations may provide insight for developing highly selective pharmacological drugs such as inhibitors of the pathology-associated HDAC4 or HDAC modifying enzyme to initiate clinical trials for the treatment of diseases. We need to critically consider the non-histone targets of HDAC4, which contains many important transcription factors. Knowledge from the targets of HDAC4 regulation is expected to improve our comprehension of HDAC4 biological activity and functions. Further studies of both the downstream and the upstream targets of HDAC4 will extend our knowledge of the biological role of HDAC4 and may suggest novel combined therapeutic strategies for human diseases.
Executive Summary
HDAC4, a key member of class IIa HDACs, is expressed in multiple tissues. Recent evidence has demonstrated that HDAC4 plays an important role in modulation of biological responses and pathological disorders. This review primarily focuses on the molecular mechanisms of the HDAC4 regulation and its physiological function.
Table 1. Summary of the PTM code on HDAC4.
Post translational modification
| Enzyme | site | Function | refs | |
|---|---|---|---|---|
| Phosphorylation | CaMKII/IV | Ser-246, Ser-467 and Ser-632 | Stimulation of catalytic activity and binding properties | (42) |
| ERK1/2 | Not determined | Nuclear-cytoplasm translocation | (32) | |
| PKA | Y201 | Proteolytic cleavage | (31) | |
| GSK3 | Ser-298, Ser-302 | Regulating HDAC4 proteolytic cleavage and ubiquitylation | (33) | |
| PP2A | Ser-298 | abrogation of catalytic activity and binding properties | (33,43) | |
| SUMOylation | RanBP2 | K559 | Transcriptional repression and enzymatic activity | (38) |
| Ubiquitination | undetermined | undetermined | Regulayin of protein stability | (33) |
| Carbonylation | Trx1 | Cys-667/Cys-669 | nuclear export | (35,36) |
| Proteolytic cleavage | Caspase-3 | D289 | N-terminal inhibits translocate into Nuclear and bind MEF2 | (39, 40) |
Abbreviations: PTM: Post-transcriptional modulation; CaMK: Calcium/calmodulin-dependent protein kinase; ERK1/2: Extracellular signal-regulated kinases 1 and 2; PKA: Protein kinase A; GSK3: Glycogen synthase kinase 3; PP2A: protein phosphatase 2A; Trx1: Thioredoxin 1.
Executive Summary.
Molecular basis of transcriptional and post transcriptional regulation
HDAC4 is subject to the transcriptional and post transcriptional regulations: transcription factors Sp1 and Sp3 directly bind to specific consensus GC-rich sequence in the HDAC4 promoter to drive HDAC4 transcription. Several miRNAs target HDAC4 to modulate the cellular responses and biological functions in different cell types.
Protein domain structure and post translational modifications
Human HDAC4 genes encode proteins between 972 to 1084 amino acids and mouse HDAC4 genes encode proteins between 965 to 1076 amino acids. HDAC4 itself is post-transcriptionally modified via phophorylation, carbonylation, sumoylation, unbiquitination, and proteolytic cleavage.
Subcelluar HDAC4 and its substrates
The activity of HDAC4 is regulated through subcellular localization and the formation of multi-subunit complexes with other proteins. HDAC4 regulates acetylation and deacetylation of histone and no-histone proteins. It also induces demethylation of histone proteins.
Physiological functions of HDAC4
HDAC4 plays global roles in the regulation of gene transcription, cell growth, survival, and proliferation, and their aberrant expression or activity leads to cancer development. HDAC4 regulates chondrocyte hypertrophy and endochondral bone formation, muscle development, and cardiovascular diseases. Nuclear accumulation of HDAC4 in neurons promotes neurodegeneration, but defects in HDAC4 are the cause of brachydactyly-mental retardation syndrome. HDAC4 overexpression showed a decreased pool of insulin-producing beta-cells and somatostatin-producing delta-cells. HDAC4 is closely associated with the development of tumors in many tissues.
The future perspectives
Investigation of the modifications and functions of HDAC4 will reveal a novel mechanism(s) by which HDAC4 is involved in many physiological and pathological disorders. Further studies of both the downstream and the upstream targets of HDAC4 will extend our knowledge of the biological role of HDAC4 and may suggest novel therapeutic strategies for human diseases.
Acknowledgments
The work is supported by the National Heart, Lung, and Blood Institute Grant (HL089405 and HL115265).
Footnotes
Conflict of interest
None
References
- 1.Thiagalingam S, Cheng KH, Lee HJ, et al. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann N Y Acad Sci. 2003;983:84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- 2*.Grozinger CM, Schreiber SL. Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem Biol. 2002;9:3–16. doi: 10.1016/s1074-5521(02)00092-3. [DOI] [PubMed] [Google Scholar]
- 3.Chen HP, Denicola M, Qin X, et al. HDAC inhibition promotes cardiogenesis and the survival of embryonic stem cells through proteasome-dependent pathway. J Cell Biochem. 2011;112:3246–3255. doi: 10.1002/jcb.23251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007;26:5505–5520. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- 5.Fischle W, Kiermer V, Dequiedt F, et al. The emerging role of class II histone deacetylases. Biochem Cell Biol. 2001;79:337–348. [PubMed] [Google Scholar]
- 6.Martin M, Kettmann R, Dequiedt F. Class IIa histone deacetylases: regulating the regulators. Oncogene. 2007;26:5450–5467. doi: 10.1038/sj.onc.1210613. [DOI] [PubMed] [Google Scholar]
- 7.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barneda-Zahonero B, Parra M. Histone deacetylases and cancer. Mol Oncol. 2012;6:579–589. doi: 10.1016/j.molonc.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang AH, Bertos NR, Vezmar M, et al. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol Cell Biol. 1999;19:7816–7827. doi: 10.1128/mcb.19.11.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vega RB, Matsuda K, Oh J, et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 11.Stark M, Hayward N. Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2007;67:2632–2642. doi: 10.1158/0008-5472.CAN-06-4152. [DOI] [PubMed] [Google Scholar]
- 12.Liu F, Pore N, Kim M, et al. Regulation of histone deacetylase 4 expression by the SP family of transcription factors. Mol Biol Cell. 2006;17:585–597. doi: 10.1091/mbc.E05-08-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Addis RC, Prasad MK, Yochem RL, et al. OCT3/4 regulates transcription of histone deacetylase 4 (Hdac4) in mouse embryonic stem cells. J Cell Biochem. 2010;111:391–401. doi: 10.1002/jcb.22707. [DOI] [PubMed] [Google Scholar]
- 14.Yuan JH, Yang F, Chen BF, et al. The histone deacetylase 4/SP1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology. 2011;54:2025–2035. doi: 10.1002/hep.24606. [DOI] [PubMed] [Google Scholar]
- 15.Chen JF, Mandel EM, Thomson JM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y, Ge Y, Drnevich J, et al. Mammalian target of rapamycin regulates miRNA-1 and follistatin in skeletal myogenesis. J Cell Biol. 2010;189:1157–1169. doi: 10.1083/jcb.200912093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sluijter JP, van Mil A, van Vliet P, et al. MicroRNA-1 and -499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arterioscler Thromb Vasc Biol. 2010;30:859–868. doi: 10.1161/ATVBAHA.109.197434. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Yang Y, Yang T, et al. microRNA-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity. Br J Cancer. 2010;103:1215–1220. doi: 10.1038/sj.bjc.6605895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winbanks CE, Wang B, Beyer C, et al. TGF-beta regulates miR-206 and miR-29 to control myogenic differentiation through regulation of HDAC4. J Biol Chem. 2011;286:13805–13814. doi: 10.1074/jbc.M110.192625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tuddenham L, Wheeler G, Ntounia-Fousara S, et al. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580:4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- 21.Miyaki S, Sato T, Inoue A, et al. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24:1173–1185. doi: 10.1101/gad.1915510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan YJ, Yang X, Wei L, et al. MiR-365: a mechanosensitive microRNA stimulates chondrocyte differentiation through targeting histone deacetylase 4. Faseb J. 2011;25:4457–4466. doi: 10.1096/fj.11-185132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandhu SK, Volinia S, Costinean S, et al. miR-155 targets histone deacetylase 4 (HDAC4) and impairs transcriptional activity of B-cell lymphoma 6 (BCL6) in the EmumiR- 155 transgenic mouse model. Proc Natl Acad Sci U S A. 2013;109:20047–20052. doi: 10.1073/pnas.1213764109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo L, Han A, Bates DL, et al. Crystal structure of a conserved N-terminal domain of histone deacetylase 4 reveals functional insights into glutamine-rich domains. Proc Natl Acad Sci U S A. 2007;104:4297–4302. doi: 10.1073/pnas.0608041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bottomley MJ, Lo Surdo P, Di Giovine P, et al. Structural and functional analysis of the human HDAC4 catalytic domain reveals a regulatory structural zinc-binding domain. J Biol Chem. 2008;283:26694–26704. doi: 10.1074/jbc.M803514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grozinger CM, Schreiber SL. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc Natl Acad Sci U S A. 2000;97:7835–7840. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang AH, Kruhlak MJ, Wu J, et al. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol Cell Biol. 2000;20:6904–6912. doi: 10.1128/mcb.20.18.6904-6912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKinsey TA, Zhang CL, Lu J, et al. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29**.Backs J, Song K, Bezprozvannaya S, et al. CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J Clin Invest. 2006;116:1853–1864. doi: 10.1172/JCI27438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakagawa Y, Kuwahara K, Harada M, et al. Class II HDACs mediate CaMK-dependent signaling to NRSF in ventricular myocytes. J Mol Cell Cardiol. 2006;41:1010–1022. doi: 10.1016/j.yjmcc.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 31.Backs J, Backs T, Neef S, et al. The delta isoform of CaM kinase II is required for pathological cardiac hypertrophy and remodeling after pressure overload. Proc Natl Acad Sci U S A. 2009;106:2342–2347. doi: 10.1073/pnas.0813013106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang T, Kohlhaas M, Backs J, et al. CaMKIIdelta isoforms differentially affect calcium handling but similarly regulate HDAC/MEF2 transcriptional responses. J Biol Chem. 2007;282:35078–35087. doi: 10.1074/jbc.M707083200. [DOI] [PubMed] [Google Scholar]
- 33.Backs J, Worst BC, Lehmann LH, et al. Selective repression of MEF2 activity by PKA-dependent proteolysis of HDAC4. J Cell Biol. 2011;195:403–415. doi: 10.1083/jcb.201105063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou X, Richon VM, Wang AH, et al. Histone deacetylase 4 associates with extracellular signal-regulated kinases 1 and 2, and its cellular localization is regulated by oncogenic Ras. Proc Natl Acad Sci U S A. 2000;97:14329–14333. doi: 10.1073/pnas.250494697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35*.Cernotta N, Clocchiatti A, Florean C, et al. Ubiquitin-dependent degradation of HDAC4, a new regulator of random cell motility. Mol Biol Cell. 2011;22:278–289. doi: 10.1091/mbc.E10-07-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paroni G, Cernotta N, Dello Russo C, et al. PP2A regulates HDAC4 nuclear import. Mol Biol Cell. 2008;19:655–667. doi: 10.1091/mbc.E07-06-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37*.Ago T, Liu T, Zhai P, et al. A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell. 2008;133:978–993. doi: 10.1016/j.cell.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 38.Matsushima S, Kuroda J, Ago T, et al. Increased oxidative stress in the nucleus caused by Nox4 mediates oxidation of HDAC4 and cardiac hypertrophy. Circ Res. 2013;112:651–663. doi: 10.1161/CIRCRESAHA.112.279760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39*.Tatham MH, Jaffray E, Vaughan OA, et al. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 40.Kirsh O, Seeler JS, Pichler A, et al. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. Embo J. 2002;21:2682–2691. doi: 10.1093/emboj/21.11.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu F, Dowling M, Yang XJ, et al. Caspase-mediated specific cleavage of human histone deacetylase 4. J Biol Chem. 2004;279:34537–34546. doi: 10.1074/jbc.M402475200. [DOI] [PubMed] [Google Scholar]
- 42.Paroni G, Fontanini A, Cernotta N, et al. Dephosphorylation and caspase processing generate distinct nuclear pools of histone deacetylase 4. Mol Cell Biol. 2007;27:6718–6732. doi: 10.1128/MCB.00853-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paroni G, Mizzau M, Henderson C, et al. Caspase-dependent regulation of histone deacetylase 4 nuclear-cytoplasmic shuttling promotes apoptosis. Mol Biol Cell. 2004;15:2804–2818. doi: 10.1091/mbc.E03-08-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozhemyakina E, Cohen T, Yao TP, et al. Parathyroid hormone-related peptide represses chondrocyte hypertrophy through a protein phosphatase 2A/histone deacetylase 4/MEF2 pathway. Mol Cell Biol. 2009;29:5751–5762. doi: 10.1128/MCB.00415-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKinsey TA, Zhang CL, Olson EN. Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol Cell Biol. 2001;21:6312–6321. doi: 10.1128/MCB.21.18.6312-6321.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKinsey TA, Kuwahara K, Bezprozvannaya S, et al. Class II histone deacetylases confer signal responsiveness to the ankyrin-repeat proteins ANKRA2 and RFXANK. Mol Biol Cell. 2006;17:438–447. doi: 10.1091/mbc.E05-07-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47**.Wang AH, Yang XJ. Histone deacetylase 4 possesses intrinsic nuclear import and export signals. Mol Cell Biol. 2001;21:5992–6005. doi: 10.1128/MCB.21.17.5992-6005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kehat I, Accornero F, Aronow BJ, et al. Modulation of chromatin position and gene expression by HDAC4 interaction with nucleoporins. J Cell Biol. 2011;193:21–29. doi: 10.1083/jcb.201101046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qian DZ, Kachhap SK, Collis SJ, et al. Class II histone deacetylases are associated with VHL-independent regulation of hypoxia-inducible factor 1 alpha. Cancer Res. 2006;66:8814–8821. doi: 10.1158/0008-5472.CAN-05-4598. [DOI] [PubMed] [Google Scholar]
- 50.Seo HW, Kim EJ, Na H, et al. Transcriptional activation of hypoxia-inducible factor-1alpha by HDAC4 and HDAC5 involves differential recruitment of p300 and FIH- 1. FEBS Lett. 2009;583:55–60. doi: 10.1016/j.febslet.2008.11.044. [DOI] [PubMed] [Google Scholar]
- 51.Geng H, Harvey CT, Pittsenbarger J, et al. HDAC4 protein regulates HIF1alpha protein lysine acetylation and cancer cell response to hypoxia. J Biol Chem. 2011;286:38095–38102. doi: 10.1074/jbc.M111.257055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choi MC, Cohen TJ, Barrientos T, et al. A direct HDAC4-MAP kinase crosstalk activates muscle atrophy program. Mol Cell. 2012;47:122–132. doi: 10.1016/j.molcel.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stronach EA, Alfraidi A, Rama N, et al. HDAC4-regulated STAT1 activation mediates platinum resistance in ovarian cancer. Cancer Res. 2011;71:4412–4422. doi: 10.1158/0008-5472.CAN-10-4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fischle W, Dequiedt F, Hendzel MJ, et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 55.Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jeon EJ, Lee KY, Choi NS, et al. Bone morphogenetic protein-2 stimulates Runx2 acetylation. J Biol Chem. 2006;281:16502–16511. doi: 10.1074/jbc.M512494200. [DOI] [PubMed] [Google Scholar]
- 57.Shinkai Y. Regulation and function of H3K9 methylation. Subcell Biochem. 2007;41:337–50. [PubMed] [Google Scholar]
- 58.Zhang CL, McKinsey TA, Olson EN. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol Cell Biol. 2002;22:7302–7312. doi: 10.1128/MCB.22.20.7302-7312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raychaudhuri N, Raychaudhuri S, Thamotharan M, et al. Histone code modifications repress glucose transporter 4 expression in the intrauterine growth-restricted offspring. J Biol Chem. 2008;283:13611–13626. doi: 10.1074/jbc.M800128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hohl M, Wagner M, Reil JC, et al. Tauchnitz M, Zimmer AM, Lehmann LH, Thiel G, Bohm M, Backs J, Maack C. HDAC4 controls histone methylation in response to elevated cardiac load. J Clin Invest. 2013;123:1359–1370. doi: 10.1172/JCI61084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sailaja BS, Cohen-Carmon D, Zimmerman G, et al. Stress-induced epigenetic transcriptional memory of acetylcholinesterase by HDAC4. Proc Natl Acad Sci U S A. 2012;109:E3687–3695. doi: 10.1073/pnas.1209990110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang SK, Scruggs AM, Donaghy J, et al. Histone modifications are responsible for decreased Fas expression and apoptosis resistance in fibrotic lung fibroblasts. Cell Death Dis. 2013;4:e621. doi: 10.1038/cddis.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gregoire S, Tremblay AM, Xiao L, et al. Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation. J Biol Chem. 2006;281:4423–4433. doi: 10.1074/jbc.M509471200. [DOI] [PubMed] [Google Scholar]
- 64.Zhao X, Sternsdorf T, Bolger TA, et al. Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol Cell Biol. 2005;25:8456–8464. doi: 10.1128/MCB.25.19.8456-8464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gregoire S, Yang XJ. Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol Cell Biol. 2005;25:2273–2287. doi: 10.1128/MCB.25.6.2273-2287.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kang JS, Alliston T, Delston R, et al. Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. Embo J. 2005;24:2543–2555. doi: 10.1038/sj.emboj.7600729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pei M, Chen D, Li J, et al. Histone deacetylase 4 promotes TGF-beta1-induced synovium-derived stem cell chondrogenesis but inhibits chondrogenically differentiated stem cell hypertrophy. Differentiation. 2009;78:260–268. doi: 10.1016/j.diff.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 68.Lin Q, Schwarz J, Bucana C, et al. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karamboulas C, Swedani A, Ward C, et al. HDAC activity regulates entry of mesoderm cells into the cardiac muscle lineage. J Cell Sci. 2006;119:4305–4314. doi: 10.1242/jcs.03185. [DOI] [PubMed] [Google Scholar]
- 70.McKinsey TA, Zhang CL, Olson EN. Activation of the myocyte enhancer factor- 2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14–3–3 to histone deacetylase 5. Proc Natl Acad Sci U S A. 2000;97:14400–14405. doi: 10.1073/pnas.260501497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miska EA, Langley E, Wolf D, et al. Differential localization of HDAC4 orchestrates muscle differentiation. Nucleic Acids Res. 2001;29:3439–3447. doi: 10.1093/nar/29.16.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu Y, Randall WR, Schneider MF. Activity-dependent and -independent nuclear fluxes of HDAC4 mediated by different kinases in adult skeletal muscle. J Cell Biol. 2005;168:887–897. doi: 10.1083/jcb.200408128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Passier R, Zeng H, Frey N, et al. CaM kinase signaling induces cardiac hypertrophy and activates the MEF2 transcription factor in vivo. J Clin Invest. 2000;105:1395–1406. doi: 10.1172/JCI8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang ZP, Chen J, Seok HY, et al. MicroRNA-22 regulates cardiac hypertrophy and remodeling in response to stress. Circ Res. 2013;112:1234–1243. doi: 10.1161/CIRCRESAHA.112.300682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gupta MP, Samant SA, Smith SH, et al. HDAC4 and PCAF bind to cardiac sarcomeres and play a role in regulating myofilament contractile activity. J Biol Chem. 2008;283:10135–10146. doi: 10.1074/jbc.M710277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cohen TJ, Barrientos T, Hartman ZC, et al. The deacetylase HDAC4 controls myocyte enhancing factor-2-dependent structural gene expression in response to neural activity. Faseb J. 2009;23:99–106. doi: 10.1096/fj.08-115931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cohen TJ, Waddell DS, Barrientos T, et al. The histone deacetylase HDAC4 connects neural activity to muscle transcriptional reprogramming. J Biol Chem. 2007;282:33752–33759. doi: 10.1074/jbc.M706268200. [DOI] [PubMed] [Google Scholar]
- 78.Tang H, Macpherson P, Marvin M, et al. A histone deacetylase 4/myogenin positive feedback loop coordinates denervation-dependent gene induction and suppression. Mol Biol Cell. 2009;20:1120–1131. doi: 10.1091/mbc.E08-07-0759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Darcy MJ, Calvin K, Cavnar K, et al. Regional and subcellular distribution of HDAC4 in mouse brain. J Comp Neurol. 2010;518:722–740. doi: 10.1002/cne.22241. [DOI] [PubMed] [Google Scholar]
- 80.Bolger TA, Yao TP. Intracellular trafficking of histone deacetylase 4 regulates neuronal cell death. J Neurosci. 2005;25:9544–9553. doi: 10.1523/JNEUROSCI.1826-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bolger TA, Zhao X, Cohen TJ, et al. The neurodegenerative disease protein ataxin-1 antagonizes the neuronal survival function of myocyte enhancer factor-2. J Biol Chem. 2007;282:29186–29192. doi: 10.1074/jbc.M704182200. [DOI] [PubMed] [Google Scholar]
- 82.Chawla S, Vanhoutte P, Arnold FJ, et al. Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J Neurochem. 2003;85:151–159. doi: 10.1046/j.1471-4159.2003.01648.x. [DOI] [PubMed] [Google Scholar]
- 83.Jovicic A, Zaldivar Jolissaint JF, et al. MicroRNA-22 (miR-22) overexpression is neuroprotective via general anti-apoptotic effects and may also target specific Huntington’s disease-related mechanisms. PLoS One. 2013;8:e54222. doi: 10.1371/journal.pone.0054222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen B, Cepko CL. HDAC4 regulates neuronal survival in normal and diseased retinas. Science. 2009;323:256–259. doi: 10.1126/science.1166226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang Y, Qin X, Liu S, et al. Peroxisome proliferator-activated receptor gamma is inhibited by histone deacetylase 4 in cortical neurons under oxidative stress. J Neurochem. 2011;118:429–439. doi: 10.1111/j.1471-4159.2011.07316.x. [DOI] [PubMed] [Google Scholar]
- 86.Williams SR, Aldred MA, Der Kaloustian VM, et al. Haploinsufficiency of HDAC4 causes brachydactyly mental retardation syndrome, with brachydactyly type E, developmental delays, and behavioral problems. Am J Hum Genet. 2010;87:219–228. doi: 10.1016/j.ajhg.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim T, Park JK, Kim HJ, et al. Association of histone deacetylase genes with schizophrenia in Korean population. Psychiatry Res. 2012;178:266–269. doi: 10.1016/j.psychres.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 88*.Li J, Chen J, Ricupero CL, et al. Nuclear accumulation of HDAC4 in ATM deficiency promotes neurodegeneration in ataxia telangiectasia. Nat Med. 2013;18:783–790. doi: 10.1038/nm.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lenoir O, Flosseau K, Ma FX, et al. Specific control of pancreatic endocrine beta-and delta-cell mass by class IIa histone deacetylases HDAC4, HDAC5, and HDAC9. Diabetes. 2011;60:2861–2871. doi: 10.2337/db11-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90*.Chauchereau A, Mathieu M, de Saintignon J, et al. HDAC4 mediates transcriptional repression by the acute promyelocytic leukaemia-associated protein PLZF. Oncogene. 2004;23:8777–8784. doi: 10.1038/sj.onc.1208128. [DOI] [PubMed] [Google Scholar]
- 91.Lemercier C, Brocard MP, Puvion-Dutilleul F, et al. Class II histone deacetylases are directly recruited by BCL6 transcriptional repressor. J Biol Chem. 2002;277:22045–22052. doi: 10.1074/jbc.M201736200. [DOI] [PubMed] [Google Scholar]
- 92.Wilson AJ, Byun DS, Nasser S, et al. HDAC4 promotes growth of colon cancer cells via repression of p21. Mol Biol Cell. 2008;19:4062–4075. doi: 10.1091/mbc.E08-02-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mottet D, Pirotte S, Lamour V, et al. HDAC4 represses p21(WAF1/Cip1) expression in human cancer cells through a Sp1-dependent, p53-independent mechanism. Oncogene. 2009;28:243–256. doi: 10.1038/onc.2008.371. [DOI] [PubMed] [Google Scholar]
- 94.Liu R, Wang L, Chen G, et al. FOXP3 up-regulates p21 expression by site-specific inhibition of histone deacetylase 2/histone deacetylase 4 association to the locus. Cancer Res. 2009;69:2252–2259. doi: 10.1158/0008-5472.CAN-08-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barrett A, Santangelo S, Tan K, et al. Breast cancer associated transcriptional repressor PLU-1/JARID1B interacts directly with histone deacetylases. Int J Cancer. 2007;121:265–275. doi: 10.1002/ijc.22673. [DOI] [PubMed] [Google Scholar]
- 96.Xu XS, Wang L, Abrams J, et al. Histone deacetylases (HDACs) in XPC gene silencing and bladder cancer. J Hematol Oncol. 2011;4:17. doi: 10.1186/1756-8722-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Geng H, Harvey CT, Pittsenbarger J, et al. HDAC4 protein regulates HIF1alpha protein lysine acetylation and cancer cell response to hypoxia. J Biol Chem. 2011;286:38095–38102. doi: 10.1074/jbc.M111.257055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Song B, Wang Y, Xi Y, et al. Mechanism of chemoresistance mediated by miR- 140 in human osteosarcoma and colon cancer cells. Oncogene. 2009;28:4065–4074. doi: 10.1038/onc.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Isaacs JT, Antony L, Dalrymple SL, et al. Tasquinimod Is an Allosteric Modulator of HDAC4 survival signaling within the compromised cancer microenvironment. Cancer Res. 2013;73:1386–1399. doi: 10.1158/0008-5472.CAN-12-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]


