Abstract
Sepsid flies (Diptera: Sepsidae) are important model insects for sexual selection research. In order to develop mitochondrial (mt) genome data for this significant group, we sequenced the first complete mt genome of the sepsid fly Nemopoda mamaevi Ozerov, 1997. The circular 15,878 bp mt genome is typical of Diptera, containing all 37 genes usually present in bilaterian animals. We discovered inaccurate annotations of fly mt genomes previously deposited on GenBank and thus re-annotated all published mt genomes of Cyclorrhapha. These re-annotations were based on comparative analysis of homologous genes, and provide a statistical analysis of start and stop codon positions. We further detected two 18 bp of conserved intergenic sequences from tRNAGlu-tRNAPhe and ND1-tRNASer(UCN) across Cyclorrhapha, which are the mtTERM binding site motifs. Additionally, we compared automated annotation software MITOS with hand annotation method. Phylogenetic trees based on the mt genome data from Cyclorrhapha were inferred by Maximum-likelihood and Bayesian methods, strongly supported a close relationship between Sepsidae and the Tephritoidea.
Introduction
The mitochondrion (mt), descended from an alpha-proteobacterium, is one of the fundamental eukaryotic organelles [1–3] and retains a remnant, bacterial-like genome. The mt genome has been an extensively used marker in phylogenetic studies across broad taxonomic scales [4–8] and in a wide range of taxa since mt genome sequences are often more phylogenetically informative than shorter sequences from individual genes commonly used for shallow or species-level studies [9–12]. Since the first insect mt genome was published in 1985, there has been a rapid accumulation of sequenced insect genomes. Insects have been comprehensively sampled at higher taxonomic levels and mt genomes are available from every insect order [2]. Diptera is one of the most extensively sequenced orders amongst the Insecta, with 93 complete or near-complete Diptera mt genome sequences available on GenBank (as of July 2014), including 57 cyclorrhaphan species (46 complete genomes, 10 near-complete genomes without full control regions, and one partial genomes) representing 13 families (Table 1).
Table 1. Summary of mitogenome sequences from Brachycera.
| Family | Species | Published information | GenBank Accession No. | Length (bp) |
|---|---|---|---|---|
| Tabanidae | Trichophthalma punctata#* | [13] | NC_008755 | 16396 |
| Nemestrinidae | Cydistomyia duplonotata#* | [13] | NC_008756 | 16247 |
| Phoridae | Megaselia scalaris | - | NC_023794 | 15599 |
| Syrphidae | Simosyrphus grandicornis | [13] | NC_008754 | 16141 |
| Fergusoninidae | Fergusonina taylori | [14] | NC_016865 | 16000 |
| Agromyzidae | Liriomyza bryoniae | [15] | NC_016713 | 16183 |
| Liriomyza huidobrensis | [15] | NC_016716 | 16236 | |
| Liriomyza sativae | [16] | NC_015926 | 15551 | |
| Liriomyza trifolii | [17] | NC_014283 | 16141 | |
| Tephritidae | Bactrocera carambolae | [18] | NC_009772 | 15915 |
| Bactrocera correcta | - | NC_018787 | 15936 | |
| Bactrocera cucurbitae | - | NC_016056 | 15825 | |
| Bactrocera dorsalis | [19] | NC_008748 | 15915 | |
| Bactrocera minax | [20] | NC_014402 | 16043 | |
| Bactrocera oleae | [21] | NC_005333 | 15815 | |
| Bactrocera papayae | [18] | NC_009770 | 15915 | |
| Bactrocera philippinensis | [18] | NC_009771 | 15915 | |
| Bactrocera tryoni | [22] | NC_014611 | 15925 | |
| Ceratitis capitata | [23] | NC_000857 | 15980 | |
| Procecidochares utilis | - | NC_020463 | 15922 | |
| Drosophilidae | Drosophila ananassae | [24] | BK006336 (Without CR) | - |
| Drosophila erecta | [24] | BK006335 (Without CR) | - | |
| Drosophila grimshawi | [24] | BK006341 (Without CR) | - | |
| Drosophila littoralis | [25] | NC_011596 | 16017 | |
| Drosophila melanogaster | [26] | NC_001709 | 19517 | |
| Drosophila mojavensis | [24] | BK006339 (Without CR) | - | |
| Drosophila persimilis | [24] | BK006337 (Without CR) | - | |
| Drosophila pseudoobscura | [27] | NC_018348 (Without CR) | - | |
| Drosophila santomea | [28] | NC_023825 | 16022 | |
| Drosophila sechellia | [29] | NC_005780 (Without CR) | - | |
| Drosophila simulans | [29] | NC_005781 (Without CR) | - | |
| Drosophila virilis | [24] | BK006340 (Without CR) | - | |
| Drosophila willistoni | [24] | BK006338 (Without CR) | - | |
| Drosophila yakuba | [30] | NC_001322 | 16019 | |
| Sepsidae | Nemopoda mamaevi | Present study | KM605250 | 15878 |
| Haematobia irritans | - | NC_007102 | 16078 | |
| Muscidae | Musca domestica* | [31] | NC_024855 | 16108 |
| Stomoxys calcitrans | [32] | DQ533708 | 15790 | |
| Scathophagidae | Scathophaga stercoraria* | [31] | NC_024856 | 16223 |
| Calliphoridae | Calliphora vicina | [5] | NC_019639 | 16112 |
| Chrysomya albiceps | [5] | NC_019631 | 15491 | |
| Chrysomya bezziana | [5] | NC_019632 | 15236 | |
| Chrysomya megacephala | [5] | NC_019633 | 15273 | |
| Chrysomya putoria | [33] | NC_002697 | 15837 | |
| Chrysomya rufifacies | [5] | NC_019634 | 15412 | |
| Chrysomya saffranea | [5] | NC_019635 | 15839 | |
| Protophormia terraenovae | [5] | NC_019636 | 15170 | |
| Cochliomyia hominivorax | [34] | NC_002660 | 16022 | |
| Lucilia cuprina | [5] | NC_019573 | 15952 | |
| Lucilia porphyrina | [5] | NC_019637 | 15877 | |
| Lucilia sericata | [5] | NC_009733 | 15945 | |
| Hemipyrellia ligurriens | [5] | NC_019638 | 15938 | |
| Polleniidae | Pollenia rudis | [5] | JX913761 (Partial Genome) | - |
| Oestridae | Dermatobia hominis | - | NC_006378 | 16460 |
| Hypoderma lineatum | [35] | NC_013932 | 16354 | |
| Sarcophagidae | Sarcophaga impatiens | [14] | NC_017605 | 15169 |
| Sarcophaga peregrina | - | NC_023532 | 14922 | |
| Tachinidae | Elodia flavipalpis | [6] | NC_018118 | 14932 |
| Exorista sorbillans | [36] | NC_014704 | 14960 | |
| Rutilia goerlingiana | [5] | NC_019640 | 15331 |
Note: “-” not available (unknown or incomplete data);
“*” species used in phylogenetic analysis;
“#” outgroup.
Sepsidae is a global distributed fly family with more than 320 described species [37]. Sepsid flies are important insect models for sexual selection research for three main reasons: 1. pronounced sexual dimorphisms (strongly modified male forelegs and movable abdominal appendages) [38–40]; 2. complex courtship behaviors (male display, female choice, and sexual conflict) [38, 41–43]; and 3. easily bred under lab conditions (they utilize rotting plant material or animal feces as breeding substrates) [44]. Recently, the transcriptome of a sepsid species Themira biloba has been assembled and analyzed [45] expanding the range of genetic resources for this family, however, no mt genomes are available from this family.
The procedures and software used for insect mt genomes annotation have recently been reviewed by Cameron (2014b) [3] noting that accurate annotations of mt genomes are necessary for all downstream analysis. Since the online implementation of the tRNA prediction software tRNAScan-SE [46] plus alignment with homologous genes is relatively efficient, there are few problems in identifying gene boundaries for tRNAs. However, despite protein-coding genes (PCGs) being used in virtually every phylogenetic and evolutionary biology study of mt genomes, gene boundaries of some PCGs are often difficult to identify. For example, the start codons of CO1 are wildly inconsistent and there are some inaccurate annotations in the GenBank (e.g. 132 incorrect annotations across 36 species of lepidopteran mt genomes [3]). Studies of expression profiles of mt genes should be the most effective way to identify gene boundaries [12, 47–48], however there are few RNAseq datasets for insect species whose mt genomes have also sequenced. In the absence of RNAseq data, comparative alignments of homologous mt genes from all the mt genomes available for a particular taxonomic group is also reliable [3].
Here, we sequenced the complete mt genome of the sepsid fly Nemopoda mamaevi Ozerov, 1996. We annotated with this genome using procedures and quality control methods proposed by Cameron (2014b) [3] and compared these annotation results with the automated annotation software MITOS [49]. We also re-annotated the mt genomes of all Cyclorrhapha species deposited on GenBank, based on comparative analysis of homologous genes, and undertook a statistical analysis of start and stop codons positions in their PCGs. We aligned and analysed two intergenic sequences across Cyclorrapha, which were highly conserved 18-bp motifs for the binding site of mtTERM. The mt genome contributes to reconstruction of the taxonomic positions and evolutionary relationships of the Sepsidae, and will help selecting optimized primer for atypical regions in further molecular research of related taxa. Phylogenetic trees based on the mt genome data from Cyclorrhapha were inferred by both Maximum-likelihood and Bayesian methods, which strongly supported a close relationship between Sepsidae and the Tephritoidea.
Material and Methods
Ethics statement
No specific permits were required for the insects collected for this study. The specimen was collected by using light trap. The field studies did not involve endangered or protected species. The species herein studied are not included in the “List of Protected Animals in China”.
Sampling and DNA extraction
The specimen used for DNA extraction was collected by Yuting Dai from Xiaolongmen (N39°57′55.21″ E115°27′59.58″), Mentougou, Beijing, China, in June 2013. After collection, it was initially preserved in 95% ethanol in the field, and then transferred to -20°C for the long-term storage upon the arrival at China Agricultural University. The specimen was examined and identified by the first author Xuankun Li with ZEISS Stemi 2000–c microscope. It is distinct from other Nemopoda by the absence of setae near the anterior margin of the katepisternum, presence of a large blackish apical spot on the male wing, and absence of setae on male hind femur [50]. Nemopoda mamaevi Ozerov is newly recorded from China. Whole genomic DNA was extracted from the thoracic muscle tissues using TIANamp Genomic DNA Kit (TIANGEN). The quality of PCR products was assessed through electrophoresis in a 1% agarose gel and stained with Gold View (ACME).
PCR amplification and sequencing
The Nemopoda mamaevi mt genome was amplified in 20 overlapping PCR fragments using NEB Long Taq DNA polymerase (New England BioLabs, Ipswich, MA). The PCR primers used follow Zhao et al (2013) [6]. Several species-specific primers were designed from initial sequencing with the amplification primers and used for internal PCRs (S1 Table).
PCR cycling consisted of an initial denaturation step at 95°C for 30s, followed by 40 cycles of denaturation at 95°C for 10s, annealing at 42–55°C (depending on the primer pair used) for 50s, elongation at 65°C for 1 kb/min (depending on the size of target amplicon) (S1 Table), and a final elongation step at 65°C for 10 min. PCR products were evaluated by agarose gel electrophoreses.
All amplicons were sequenced in both directions using the BigDye Terminator Sequencing Kit (Applied Bio Systems) and the ABI 3730XL Genetic Analyzer (PE Applied Biosystems, San Francisco, CA, USA) using amplification primers and internal primers developed by primer walking.
Bioinformatic analysis
Sequences were proof-read and aligned into contigs in BioEdit version 7.0.5.3 [51]. After fully sequencing the mt genome, it was annotated using both automated annotation methods and by hand. For the automated annotation, we used MITOS [49]. Hand annotation method followed the procedures proposed by Cameron (2014b) [3]. Quality control of the hand alignments was performed by comparing with homologous sequences from previously sequenced Cyclorrhapha mt genomes to identify several tRNAs apparently absent from the N. mamaevi mt genome, and to verify PCGs and rRNAs annotations.
Nucleotide substitution rates, base composition and codon usage were analyzed with MEGA 5.0 [52]. Nucleotide compositional skew was measured using the following formula: AT-skew = (A-T)/(A+T) [53].
Phylogenetic analysis
A total of 20 species of brachyceran insects were used in phylogenetic analysis, including 18 cyclorrhaphans and two outgroup species from Tabanidae and Nemestrinidae. Details of the species used in this study were listed in Table 1.
Sequences of 13 PCGs, two rRNAs and 22 tRNAs were used in phylogenetic analysis. Each PCG was aligned individually based on Hand annotation method followed the procedures proposed by Cameron (2014b) [3]. The sequences of tRNAs and rRNAs were aligned respectively using MEGA 5.0 [52], ambiguous positions in the alignment of RNAs were filtered by hand. Individual genes were concatenated using SequenceMatrix v1.7.8 [54]. We assembled four datasets for phylogenetic analysis: 1) nucleotides of 13 PCGs (PCG123) with 11,058 residues, 2) nucleotides of 13 PCGs, two rRNAs and 22 tRNAs (PCG123RNA) with 14,226 residues, 3) nucleotides of 13 PCGs exclude the third codon sites (PCG12) with 7,372 residues and 4) nucleotides of 13 PCGs exclude the third codon sites, two rRNAs and 22 tRNAs (PCG12RNA) with 10,540 residues. PartitionFinder v1.1.1 [55] was used to select the optimal partition strategy and substitution models for each partition. We created an input configuration file with 63/29 (with 3rd codon positions/ without) pre-defined partitions of the dataset, and used the ‘‘greedy” algorithm with branch lengths estimated as ‘‘unlinked” and Bayesian information criterion (BIC) to search for the best-fit scheme (S2 Table).
We performed maximum likelihood (ML) and Bayesian inference (BI) using the best-fit partitioning schemes recommended by PartitionFinder (S2 Table). For ML analysis, we used RAxML 8.0.0 [56] with 1,000 bootstrap replicates and using the rapid bootstrap feature (random seed value 12345) [57]. The Bayesian analysis was conducted with MrBayes 3.2.2 [58]. Two simultaneous runs of 2 million generations were conducted for the dataset, each with one cold and three heated chains. Samples were drawn every 1,000 Markov chain Monte Carlo (MCMC) steps, with the first 25% discarded as burn-in. When the average standard deviation of split frequencies was below 0.01, we considered the stationarity was reached and stopped run.
Results and Discussion
Genome Organization and Structure
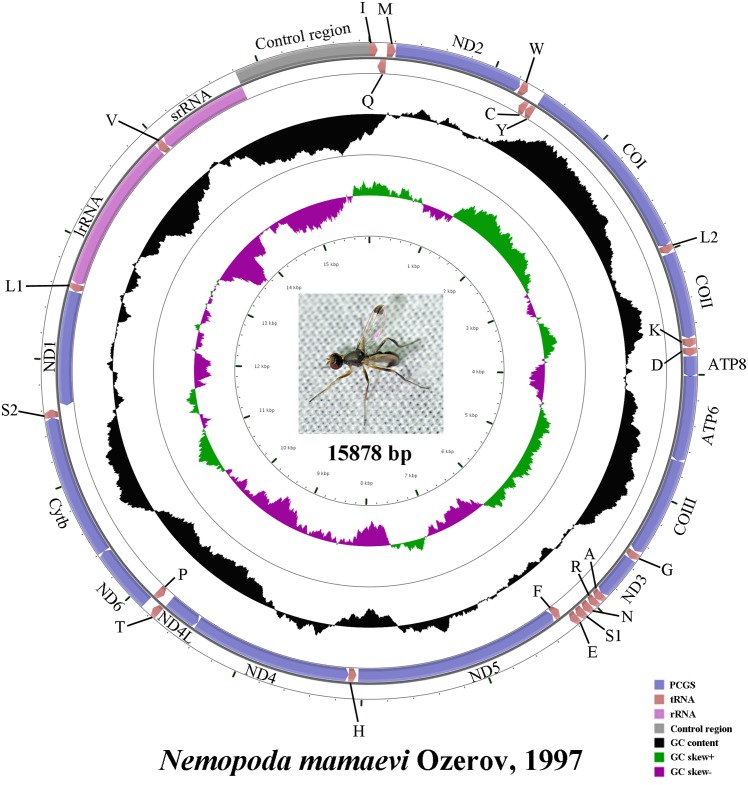
The complete mt genome of N. mamaevi is a typical circular, double-stranded molecule (GenBank accession number: KM605250; Fig 1) 15,878 bp in length. Mt genome length is medium-sized when compared to the mt genomes of other Diptera, that range from 14,922 bp (Sarcophaga peregrine, Sarcophagidae, NC_023794) to 19,517 bp (Drosophila melanogaster, Drosophilidae [26]). The mt genome of N. mamaevi contains the 37 genes, including 13 PCGs, 22 tRNA genes, two rRNA genes and a large control region, that are usually present in bilaterian animals [3]. The gene order is the same as the inferred ancestral insect mt genome pattern, which is conserved amongst all cyclorrhaphan flies sequenced to date. Twenty three genes are encoded on the majority strand (J-strand), while the minority strand (N-strand) encodes the remaining 14 genes.
Fig 1. Mitochondrial map of Nemopoda mamaevi Ozerov.
Circular maps were drawn with CGView [59]. Arrows indicated the orientation of gene transcription. The tRNAs are denoted by the color blocks and are labelled according to the IUPACIUB single-letter amino acid codes (L1: CUN; L2: UUR; S1: AGN; S2: UCN). The GC content was plotted using a black sliding window, as the deviation from the average GC content of the entire sequence. GC-skew was plotted as the deviation from the average GC-skew of the entire sequence. The inner cycle indicated the location of genes in the mt genome.
There are 9 gene boundaries where sequence overlapped between neighboring genes, ranging from 1 to 8 bp in length, and 29 bp in total. The length of intergenic sequences (excluding the control region) was 1–18 bp found at 14 gene boundaries, and totalling 82 bp (Table 2). The longest overlapping sequence belonged to both tRNA Trp and tRNA Cys. The longest intergenic sequence located between tRNA Glu and tRNA Phe.
Table 2. Organization of the mitogenome of Nemopoda mamaevi Ozerov.
| Gene | Direction | Location | Size(bp) | IGN* | Anticodon | Codon | AT% | |
|---|---|---|---|---|---|---|---|---|
| Start | Stop | |||||||
| tRNAIle | F | 1–66 | 66 | GAT | 74.3 | |||
| tRNAGln | R | 67–135 | 69 | 0 | TTG | 81.2 | ||
| tRNAMet | F | 140–208 | 69 | 4 | CAT | 68.1 | ||
| ND2 | F | 209–1231 | 1023 | 0 | ATT | TAA | 74.8 | |
| tRNATrp | F | 1237–1303 | 67 | 5 | TCA | 73.1 | ||
| tRNACys | R | 1296–1359 | 64 | -8 | GCA | 70.4 | ||
| tRNATyr | R | 1360–1426 | 67 | 0 | GTA | 74.6 | ||
| CO1 | F | 1425–2963 | 1539 | -2 | TCG | TAA | 65.8 | |
| tRNALeu(UUR) | F | 2959–3024 | 66 | -5 | TAA | 74.2 | ||
| CO2 | F | 3030–3713 | 684 | 5 | ATG | TAA | 70.6 | |
| tRNALys | F | 3715–3785 | 71 | 1 | CTT | 64.8 | ||
| tRNAAsp | F | 3789–3855 | 67 | 3 | GTC | 86.6 | ||
| ATP8 | F | 3856–4017 | 162 | 0 | ATC | TAA | 74.1 | |
| ATP6 | F | 4011–4688 | 678 | -7 | ATG | TAA | 71.4 | |
| CO3 | F | 4688–5476 | 789 | -1 | ATG | TAA | 65.7 | |
| tRNAGly | F | 5483–5547 | 65 | 6 | TCC | 78.5 | ||
| ND3 | F | 5548–5901 | 354 | 0 | ATC | TAG | 75.7 | |
| tRNAAla | F | 5900–5964 | 65 | -2 | TGC | 72.3 | ||
| tRNAArg | F | 5964–6026 | 63 | -1 | TCG | 71.4 | ||
| tRNAAsn | F | 6029–6094 | 66 | 2 | GTT | 74.2 | ||
| tRNASer(AGN) | F | 6095–6162 | 68 | 0 | GCT | 71.5 | ||
| tRNAGlu | F | 6163–6227 | 65 | 0 | TTC | 86.2 | ||
| tRNAPhe | R | 6246–6311 | 66 | 18 | AAA | 72.8 | ||
| ND5 | R | 6312–8031 | 1720 | 0 | ATT | T | 75.1 | |
| tRNAHis | R | 8047–8112 | 66 | 15 | GTG | 80.3 | ||
| ND4 | R | 8113–9451 | 1339 | 0 | ATG | T | 76.6 | |
| ND4L | R | 9453–9743 | 291 | 1 | ATG | TAA | 80.7 | |
| tRNAThr | F | 9746–9810 | 65 | 2 | TGT | 81.5 | ||
| tRNAPro | R | 9811–9876 | 66 | 0 | TGG | 81.8 | ||
| ND6 | F | 9879–10403 | 525 | 2 | ATT | TAA | 82.5 | |
| CYTB | F | 10403–11539 | 1137 | -1 | ATG | TAG | 69.1 | |
| tRNASer(UCN) | F | 11538–11604 | 67 | -2 | TGA | 77.6 | ||
| ND1 | R | 11622–12569 | 948 | 17 | TTG | TAA | 75.5 | |
| tRNALeu(CUN) | R | 12571–12635 | 65 | 1 | TAG | 81.5 | ||
| lrRNA | R | 12636–13956 | 1321 | 0 | 80.6 | |||
| tRNAVal | R | 13957–14028 | 72 | 0 | TAC | 77.8 | ||
| srRNA | R | 14029–14811 | 783 | 0 | 77.6 | |||
| Control region | – | 14812–15878 | 1067 | 0 | 86.4 | |||
Note:
* IGN: Intergenic nucleotide, minus indicates overlapping between genes. tRNAX: where X is the abbreviation of the corresponding amino acid.
Among all sequenced cyclorrhaphan flies, the longest overlapping sequence was 17 bp, between ND4 and ND4L, found in Bactrocera minax (Tephritidae) [20]. The longest intergenic sequence was located between ND1 and tRNA Ser(UCN), (102 bp) in Hypoderma lineatum (Oestridae) [35]. There were four PCG–PCG gene boundaries in every cyclorrhaphan mt genome as in the ancestral insect mt genome: ATP8-ATP6, ATP6-CO3, ND4L-ND4 and ND6-CYTB. ATP8-ATP6 consistently overlapped by 7 bp with a-1 frame shift (A TGA TAA → ATG ATA A). Similarly, ND4L-ND4 usually had a 7 bp overlap, except in the Opomyzoidea, Drosophilidae, and Musca domestica (Muscidae) [31] where these genes overlap by a single base A, in Bactrocera minax (Tephritidae) [20] there is a 17 bp overlap, and in Megaselia scalaris (Phoridae) (NC_023794) an 10 bp overlap. N. mamaevi is unique in that there is actually a single intergenic base between ND4 and ND4L. ATP6-CO3 often overlap by a single base, but is variable across cyclorrhaphan flies ranging from abutting genes without a gap or overlap (Liriomyza sativae, Agromyzidae [16] and Liriomyza trifolii, Agromyzidae [17]) to an intergenic space of up to 52 bp (Drosophila virilis, Drosophilidae [24]). Similarly, ND6-CYTB often overlap by a single base, but is variable across cyclorrhaphan flies ranging from an overlap of up to 8 bp (Fergusonina taylori, Fergusoninidae [14]) to intergenic sequences of up to 4 bp (Liriomyza sativae, Agromyzidae [16]; Liriomyza trifolii, Agromyzidae [17]; Drosophila ananassae, Drosophilidae [24]; Stomoxys calcitrans, Muscidae [32]).
Base composition and codon usage
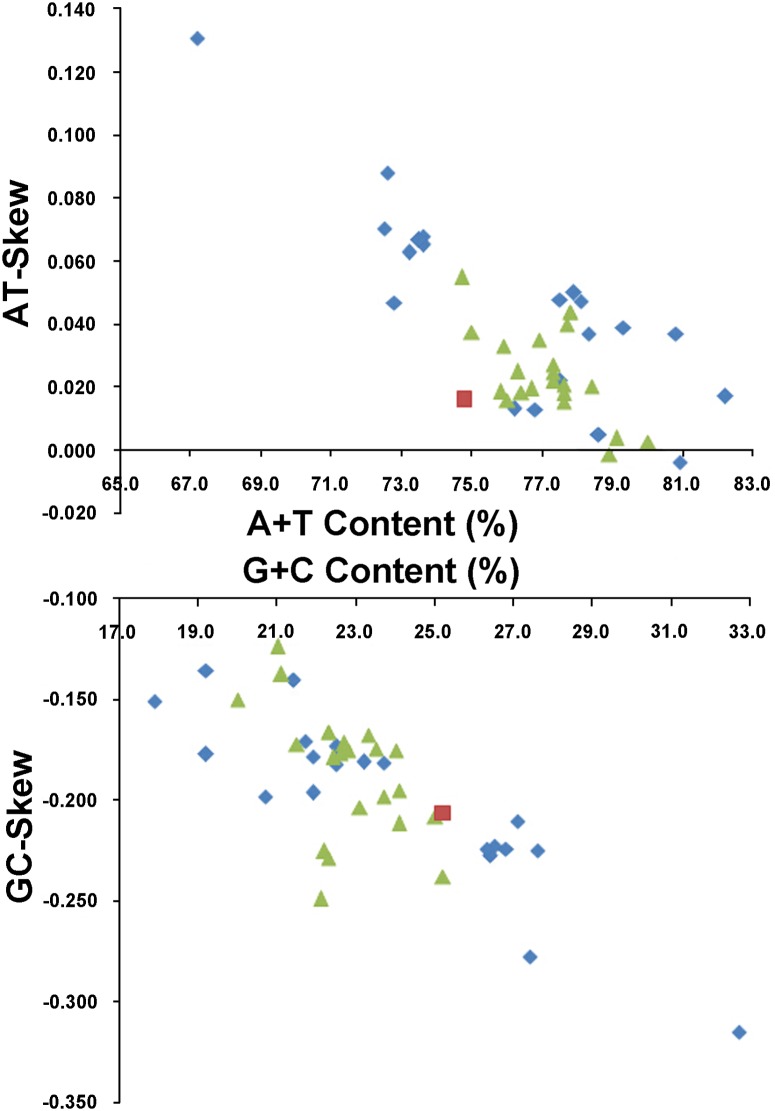
Similar to mt genome of other sequenced cyclorrhaphan flies, and indeed most insects, the nucleotide composition of the N. mamaevi was biased toward A and T (A = 38.0%, T = 36.8%, G = 10.0%, C = 15.2%; Table 3). The overall A+T content of the mt genome was 74.8%, average amongst all reported cyclorrhaphan flies, which range from 67.2% (in Bactrocera minax, Tephritidae [20]) to 82.2% (in Drosophila melanogaster, Drosophilidae [26]). A comparative analysis of nucleotide composition (A+T%, G+C%) versus skew (AT- and GC-skew) across the Cyclorrhapha is shown in Fig 2. The average AT-skew of the cyclorrhaphan mt genomes was 0.033, ranging from -0.004 (Simosyrphus grandicornis, Syrphidae [13]) to 0.131 (Bactrocera minax, Tephritidae [20]), whereas the N. mamaevi mt genome exhibits a weak AT-skew (0.016) (S3 Table). The average GC-skew of cyclorrhaphan mt genomes was -0.190, ranging from -0.315 (in Bactrocera minax, Tephritidae [20]) to -0.124 (in Haematobia irritans, Muscidae, NC_007102), while the N. mamaevi mt genome exhibited a marked GC-skew (-0.206) (S3 Table). AT- and GC- skews of Cyclorrhapha mt genomes are consistent with the strand skew biases found in most metazoan mt genomes (weakly positive AT-skew and strongly negative GC-skew for the J-strand). Within insects the only exceptions are found in three families: Philopteridae (Phthiraptera), Aleyrodidae (Hemiptera) and Braconidae (Hymenoptera), which have positive GC-skew and negative AT-skew on the J-strand [60] and in termites (Isoptera) which have strongly positive AT-skew on the J-strand [61]. In insects, the degree of AT-skew is related to gene direction, replication and codon positions, whereas the degree of GC-skew is affected by reversals in replication orientation [60].
Table 3. Nucleotide composition of the Nemopoda mamaevi mt genome.
| Feature | A% | T% | C% | G% | A+T% | C+G% | AT-skew | GC-skew |
|---|---|---|---|---|---|---|---|---|
| Whole mitgenome | 38.0 | 36.8 | 15.2 | 10.0 | 74.8 | 25.2 | 0.16 | -0.21 |
| PCGs | 30.4 | 42.3 | 13.5 | 13.8 | 72.7 | 27.3 | -0.16 | 0.01 |
| 1st codon position | 30.8 | 36.0 | 12.7 | 20.5 | 66.8 | 33.2 | -0.08 | 0.23 |
| 2nd codon position | 20.4 | 46.0 | 19.5 | 14.2 | 66.4 | 33.7 | -0.39 | -0.16 |
| 3rd codon position | 40.1 | 45.0 | 8.3 | 6.7 | 85.1 | 15.0 | -0.06 | -0.11 |
| tRNA genes | 38.2 | 37.9 | 10.7 | 13.2 | 76.1 | 23.9 | 0.00 | 0.10 |
| lrRNA | 39.1 | 41.5 | 6.4 | 12.9 | 80.6 | 19.3 | -0.03 | 0.34 |
| srRNA | 37.9 | 39.7 | 7.8 | 14.6 | 77.6 | 22.4 | -0.02 | 0.30 |
| Control region | 44.5 | 41.9 | 9.1 | 4.5 | 86.4 | 13.6 | 0.03 | -0.05 |
Note: The A+T and G+C biases of protein-coding genes were calculated by AT-skew = [A-T]/[A+T] and GC-skew = [G-C]/[G+C], respectively.
Fig 2. AT% vs AT-Skew and GC% vs GC-Skew in Cyclorrhapha mt genomes.
Measured in bp percentage (X-axis) and level of nucleotide skew (Y-axis). Values are calculated on full length mt genomes. Blue rhombus, Acalyptratae; red square, Calyptratae; green triangle, Nemopoda mamaevi.
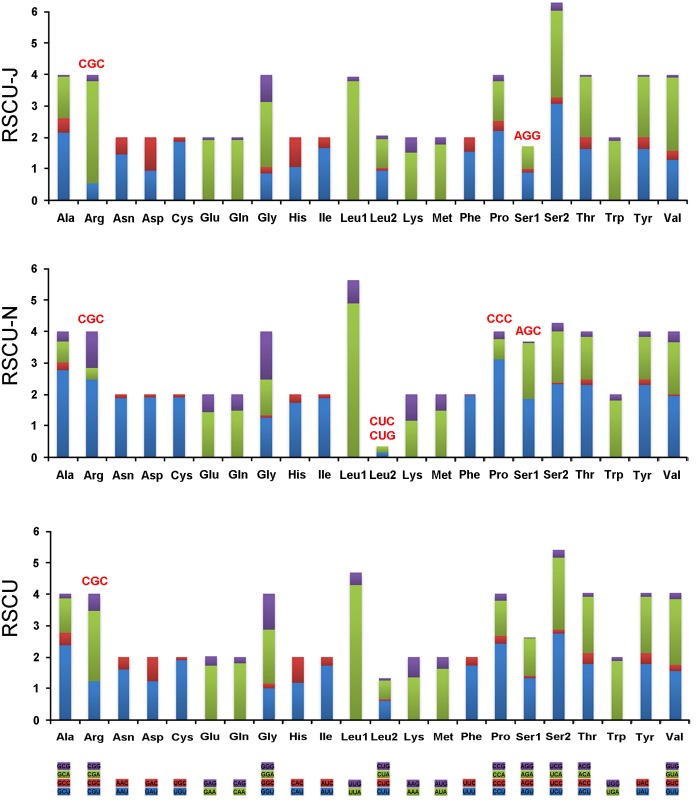
A and T bias is also reflected in relative codon usage by the PCGs. In the mt genome of N. mamaevi, A+T rich codons, such as ATT (Asn), TAA (Leu), AAA (Lys), ATA (Met), TTT (Phe), TAT (Tyr), are more frequently used than G+C rich codons. Among both strands, NNA-form codons were the most frequently used and NNC-type the most infrequently used codons. On J-strand NNA were preponderant and NNG were the least frequent with the reverse found in N-strand encoded PCGs (NNU commonest, NNC least common) (Fig 3, S4 Table).
Fig 3. Relative synonymous codon usage (RSCU) in the Nemopoda mamaevi mt genome.
Codon families are provided on the X-axis. Stop codon is not given. Red codon, codon not present in the chain/genome.
Protein-coding Genes
The overall A+T content of the 13 PCGs in N. mamaevi was 72.7%, with individual PCGs ranging from 65.8% (CO1) to 82.5% (ND6) (Table 2). The A+T content of the third codon positions (85.1%) were much higher than either the first (66.8%) or second codon (66.4%). There was moderate negative AT-skew for the PCGs as a whole (-0.16) driven by strongly negative AT-skew at second codon positions (-0.39), with weak skew at first, and third codon positions (-0.08 and -0.06 respectively). The absence of significant GC-skew across the PCGs as a whole (0.01) masks strong skews at each codon position with first codon positions strongly positive (0.23) balanced by strongly negative skews at second and third codon positions (-0.16 and -0.11 respecively) (Table 3).
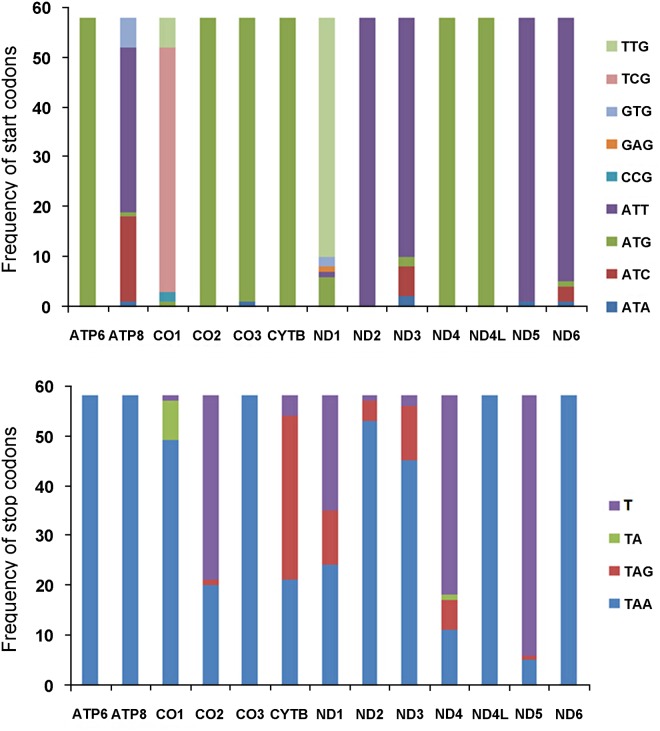
ATN, GTG (Val) and TTG (LeuUUR) are accepted as the canonical start codons for invertebrate PCGs [61], while TCG (SerUCN) has been proposed to be an additional start codon commonly found in flies [13]. All 13 PCGs in the N. mamaevi mt genome used canonical start codons. ND2, ND5 and ND6 started with ATT (Ile); ATC (Ile) were found in ATP8 and ND3; CO1 and ND1 start with TCG (SerUCN) and TTG (LeuUUR), respectively; and ATG (Met) were used in the remaining six PCGs as start codons (Table 2).
Among the cyclorrhaphan flies sequenced to date, ATG (Met) is the most frequently used start codon followed by ATT (Ile). ATG (Met) is almost exclusively used in the ATP6, CO2, CO3, CYTB, ND4 and ND4L genes of almost all cyclorrhaphan species (Procecidochares utilis, Tephritidae, NC_020463) which uses ATA (Met) for CO3). ATT (Ile) is used for ND2 in all species as well as ATP8, ND3, ND5 and ND6 in most species. Other standard start codons such as ATA (Met) and ATC (Ile) are also found in ATP8, CO3, ND3, ND5 and ND6 in a minority of fly species. TTG (LeuUUR) is used in ND1 in most species and TCG (SerUCN) is used in CO1 in most species. (Fig 4, S5 Table). In addition, three non-canonical start codons GTG (Val), CCG (Pro) and GAG (Glu) have been proposed. GTG (Val) is found as the start codon for ATP8 in six Bactrocera species (Tephrididae) and for ND1 in two Liriomyza flies (Agromyzidae). CCG (Pro) is in the start codon of CO1 in Drosophila sechellia and D. simulans (Drosophilidae) [29]. GAG (Glu) was proposed as the start codon for ND1 in Rutilia goerlingiana (Tachinidae) [5]. ATP8, CO1, ND1, ND3 and ND6 therefore collectively utilise three to five different start codons. In contrast, the remaing of the PCGs utilise no more than two and often only one, start codon (Fig 4, S5 Table).
Fig 4. Usage of start and stop codons in complete Cyclorrhapha mt genomes.
(A) Start codons usage of 13 PCGs in Cyclorrhapha. (B) Stop codons usage of 13 PCGs in Cyclorrhapha.
The stop codons most commonly used in N. mamaevi are TAA (ATP6, ATP8, CO1, CO2, CO3, ND1, ND2, ND4L, ND6), or TAG (CYTB, ND3) found in 11 of the 13 PCGs. The remaining two PCGs (ND4, ND5) utilise the partial stop codon T, which has been found in many insect mt genomes and is completed to a full TAA stop codon via post-transcriptional polyadenylation [63]. In all reported cyclorrhaphan flies studied to date, TAA is the most common stop codon used found in every PCG in at least one species, most notably in the ATP6, ATP8, CO3, ND4, ND4L and ND6 genes from all species sequenced. TAG has been found in CO2, CYTB, ND1, ND2, ND3, ND4 and ND5. The incomplete stop codons TA and T are founded in CO2, CYTB, ND1, ND2, ND3, ND4 and ND5 as well as being commonly found in CO1 (Fig 4, S5 Table).
By aligning the homologous genes PCGs among closely related species, we detected an apparent sequencing error in the published mt genomes of Bactrocera minax (Tephritidae) [20]. Base position 896 bp of ND1 in B. minax, a C, is apparently an insertion as it results in a frameshift in the downstream amion-acid sequence that is significantly different from other cyclorrhaphan flies; removal of this C restores the correct reading frame resulting in the same amino acid sequence as in other cyclorrhaphans and the same stop-codon. In addition, CO2 in Procecidochares utilis (Tephritidae) (NC_020463) has an apparent deletion at position 678, resulting in a missing “A” base. As a result, the version of this genome on GenBank is 5 bp too long at the 3’ end.
Intergenic sequences
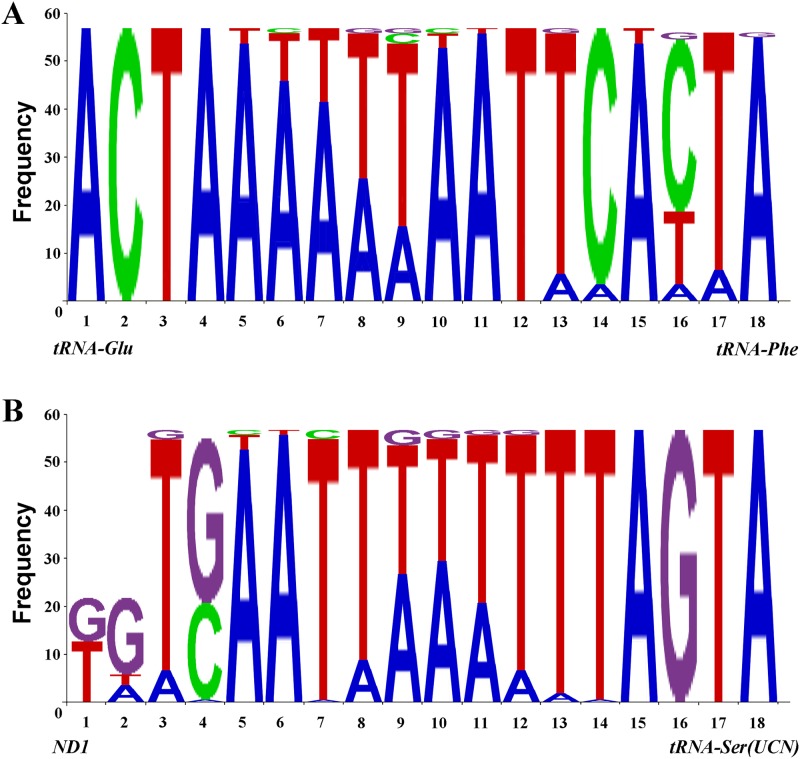
Two conserved intergenic sequences blocks of genes coded on different strands (7 bp between ND1 and tRNA Ser(UCN), 5 bp between tRNA Glu and tRNA Phe) were found in typical insect mt genome [64–65]. Here, we revealed two 18 bp conserved sequences from both spacers across Cyclorrhapha.
The non-coding region between tRNA Glu and tRNA Phe is found in all Cyclorrhapha, although ranging from 65 bp (Fergusonina taylori, Fergusoninidae [14]) to 16 bp (Sarcophaga peregrine, Sarcophagidae, NC_023794), often 18 bp in length (except 17 bp in Hypoderma lineatum, Oestridae [35]; 19 bp in Drosophila grimshawi, Drosophilidae [24]; 20 bp in Drosophila mojavensis, Drosophilidae [25] and 23 bp in Simosyrphus grandicorni, Syrphidae [13], respectively). Sequences from 57 species (Pollenia rudis was excluded because its intergenic sequences were replaced by an 18 bp TATA box) were analysed and presented in Fig 5A.
Fig 5. Two conserved intergenic sequences.
(A) Sequences between tRNA Glu and tRNA Phe, forward sequences. (B) Sequences between ND1 and tRNASer(UCN), reversed sequences.
The non-coding region located between ND1 and tRNA Ser(UCN) is the binding site of mtTERM, a transcription termination peptide which has a highly conserved 7-bp motif across insects [66]. MtTERM functions to control over-expression of the rRNA genes relative to the protein-coding genes [64, 67].
The annotated sequence length and stop codons of ND1 varies considerably among sequences from cyclorrhaphan flies. For example all Calyptratae sequenced to date have canonical stop codons (TAA or TAG). Most of the sequenced Acalyptratae species lack complete stop codons, resulting in significant length variability. Moreover, some species lack even incomplete stop codons e.g. the bases immediately preceding tRNA Ser(UCN) are neither T nor TA. We aligned the region from ND1 to tRNA Ser(UCN) of all sequenced Cyclorrhapha, and re-annotated ND1 gene based on relatively well conserved sequences in this intergenic spacer and despite large variation in spacer length (e.g. up to 102 bp Hypoderma lineatum, Oestridae [35]). Sequences from 57 species/subspecies populations were aligned and analysed (excluding Bactrocera minax due to the sequencing errors noted above). Although the intergenic sequences between ND1 and tRNA Ser(UCN) ranges from 15–102 bp in size, there is a 16 bp highly conserved sequence, found in all Cyclorrhapha (except Drosophila simulans and Rutilia goerlingiana in which there is a 1 bp deletion). Species that lack complete stop codons in ND1 have two additional, poorly conserved, bases at the 5’ end of this 16 bp conserved region. Nucleotide usage frequency at each position in this conserved region is shown in Fig 5B.
Transfer RNAs
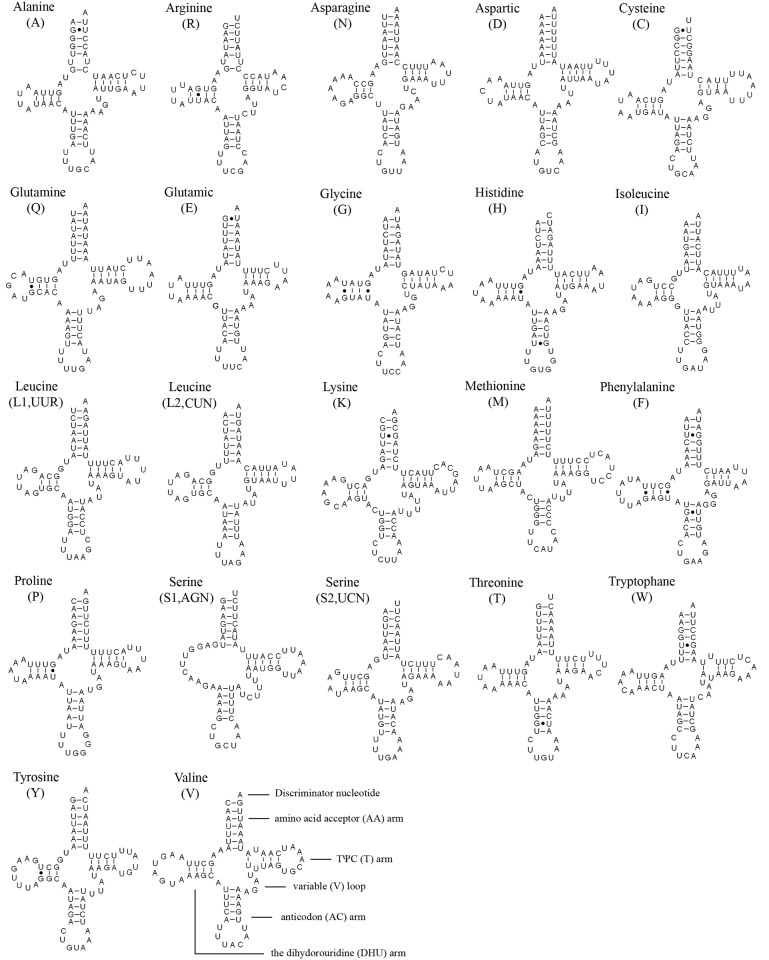
The typical complement of 22 typical tRNAs found in the arthropod mt genomes were found in the N. mamaevi mt genome, ranging in size from 63 bp (tRNA Arg) to 72 bp (tRNA Val) and 1465 bp or 9.23% of the total genome. The overall A+T content the tRNAs was 76.2%, while single genes ranged from 64.8% (tRNA Lys) to 86.6% (tRNA Asp). Fourteen genes are encoded on the J-strand and the remains are encoded on the N-strand. Most tRNAs could be folded into the typical clover-leaf structure (Fig 6), whereas the tRNA Ser(AGN) was an exception lacking a DHU arm as has been observed for this gene in other metazoan mt genomes [62].
Fig 6. Putative secondary structures of tRNAs found in the mitochondrial genome of Nemopoda mamaevi.
All tRNAs can be folded into the usual clover-leaf secondary structure. The tRNAs are labelled with the abbreviations of their corresponding amino acids. Inferred Watson-Crick bonds are illustrated by lines, whereas GU bonds are illustrated by dots.
A total of 17 G–U pairs and two mismatched base U–U pairs were found in N. mamaevi mitochondrial tRNA secondary structures, no A–A pairs or A–C pairs were found. The G–U pairs are located in the AA arm (7 bp), DHU arm (8 bp), AC arm (2 bp), respectively. In contrast, all mismatched base U–U pairs are located in the TψC arm.
Ribosomal RNAs
The rRNAs of N. mamaevi were 1,321 bp for lrRNA and 783 bp for srRNA in length. Their A+T content were 80.6% and 77.6%, respectively. Because rRNA genes lack functional annotation features, analogous to the start and stop codons of PCGs, it is impossible to determine the boundaries from DNA sequence alone [11, 68]. Hence, they were assumed to extend to the boundaries of flanking genes. As in other dipteran species, the lrRNA gene is flanked by tRNA Leu (CUN) and tRNA Val, while the srRNA gene is flanked by tRNA Val and the control region.
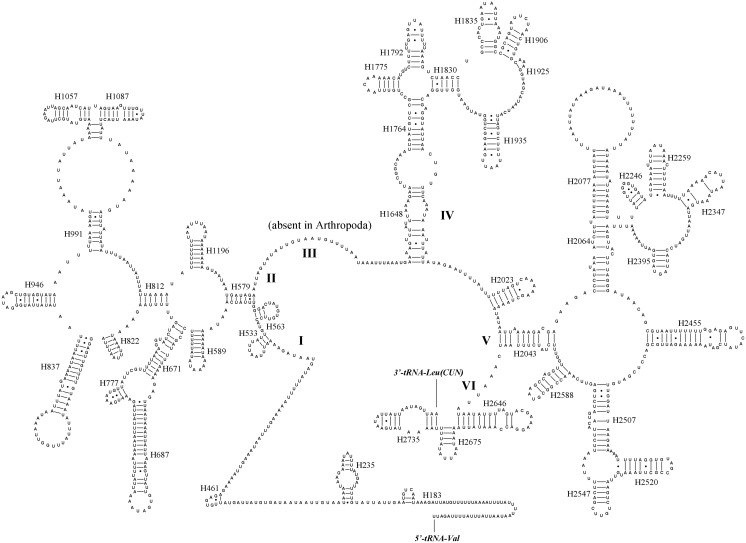
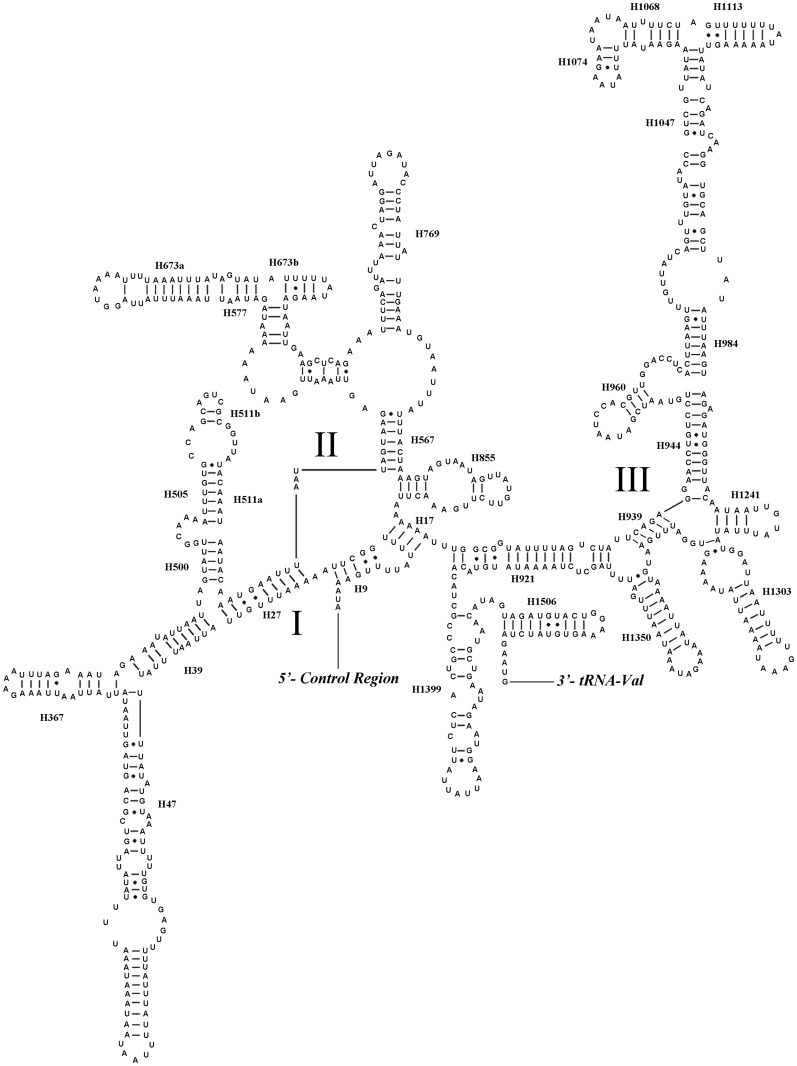
We inferred secondary structures of lrRNA and srRNA of N. mamaevi using the published rRNA secondary structures of a leafminer Liriomyza sativae (Agromyzidae), the only dipteran so analysed to date, as a model [16]. The lrRNA had 49 helices in five structural domains (I-II, IV-VI, domain III is absent as in other insects), similar to L. sativae [16] and typical of arthropods [69]. The secondary structures of srRNA in N. mamaevi included three domains and 33 helices, again similar to other Diptera [16] (Figs 7 and 8).
Fig 7. Predicted secondary structure of the lrRNA gene in Nemopoda mamaevi.
Inferred Watson–Crick bonds are illustrated by lines, GU bonds by dots.
Fig 8. Predicted secondary structure of the srRNA gene in Nemopoda mamaevi.
Roman numerals denote the conserved domain structure. Inferred Watson-Crick bonds are illustrated by lines, GU bonds by dots.
The control region
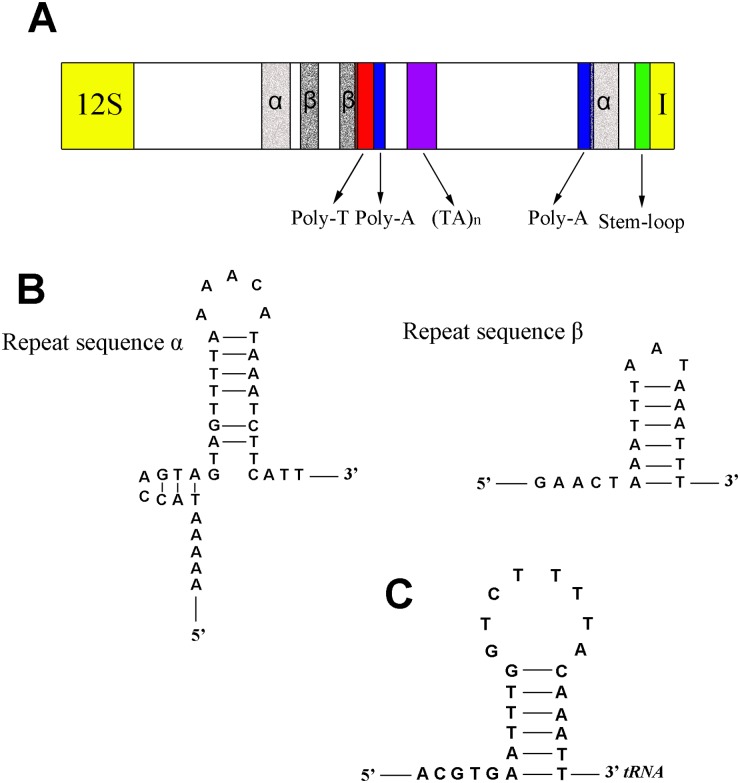
The control region of N. mamaevi is 1067 bp long and located at the conserved insect position between srRNA and tRNA Ile. Control region A+T content was 86.4%, second only to tRNA Asp (86.6%) as the highest A+T content region in the N. mamaevi genome. Of the five conserved structural elements identified by Zhang and Hewitt (1997) [70] across insect mt control regions, we could detect three of them: (1) a 21 bp of poly-T stretch, which may be associated with the control of transcription and replication (Fig 9A); (2) a (TA)n like stretch; (3) a stem-loop structure at 3’-end of control region, however the 5’ ‘TATA’ and a 3’ ‘G(A)nT’ consensus regions were apparently absent (Fig 9C). We identified two additional, structural elements, not conserved in other insects and apparently unique to N. mamaevi: (1) two poly-A stretches (10 bp and 22 bp in length), (Fig 9A); and (2) two non-tandem macro repeats: 5’-AAAAAATACCAGTAGCTGTTTTAAAACATAAATCTTCATT-3’ (from 313 to 352, from 993 to 1,032), and 5’-GAACTAAATTTAATAAAATTT-3’ (from 372 to 392, from 498 to 518). Both macro repeats could be folded into stem-loop structures (Fig 9B) and the second unit of each repeat type overlaps the poly-A and poly-T stretches, respectively (Fig 9A).
Fig 9. Predicted structure elements in the control region of Nemopoda mamaevi.
A. control region structure of Nemopoda mamaevi, B. secondary structures of repeat sequences of Nemopoda mamaevi, C. secondary structures of stem-loop structure at 3’-end.
Automated annotation method with MITOS
MITOS (http://mitos.bioinf.uni-leipzig.de/index.py) [49] is the most advanced automated mt genome annotation pipeline yet produced, and is able to be operated over an external server allowing the batched submission of mt genomes without tying up the user’s computer. However the accuracy of its annotations of protein-coding genes, in particular the location of start and stop codons, has been criticised [3]. We used MITOS to the mt genome of N. mamaevi, and compared the results with our hand annotation results.
MITOS identified all 37 genes in the correct gene order and direction on the mt genome. However not all the PCGs were correctly annotated, only the start codons of ATP8, CO2, CO3, CYTB, ND6 and stop codons of ND1, ND4, ND4L, ND5 were correct (S6 Table). For the tRNA genes, there was only one incorrect annotation: tRNA Ala lacked a base A at 3’-end (S6 Table). In contrast an older, widely used tRNA prediction package tRNAscan-SE Search Server v.1.21 (http://lowelab.ucsc.edu/tRNAscan-SE/) [46] proposed 25 potential tRNAs, and while 20 were correct, tRNA Arg and tRNA Ser(AGN) were not detected, and three spurious tRNA genes were predicted (S7 Table). These results are consistent with previous comparisons in the order Lepidoptera, that found MITOS outperformed tRNAScan for tRNA prediction, but was inferior to hand annotation of PCGs [3].
Phylogeny
Four datasets were used in the phylogenetic analysis, there are 11,058 residues in the PCG123 matrix (containing nucleotides of 13 PCGs), 14,226 residues in the PCG123RNA matrix (containing nucleotides of 13 PCGs, two rRNAs and 22 tRNAs), 7,372 residues in the PCG12 matrix (containing nucleotides of 13 PCGs but excluding the third codon sites) and 10,540 residues in the PCG12RNA matrix (containing nucleotides of 13 PCGs but excluding the third codon sites, two rRNAs and 22 tRNAs).
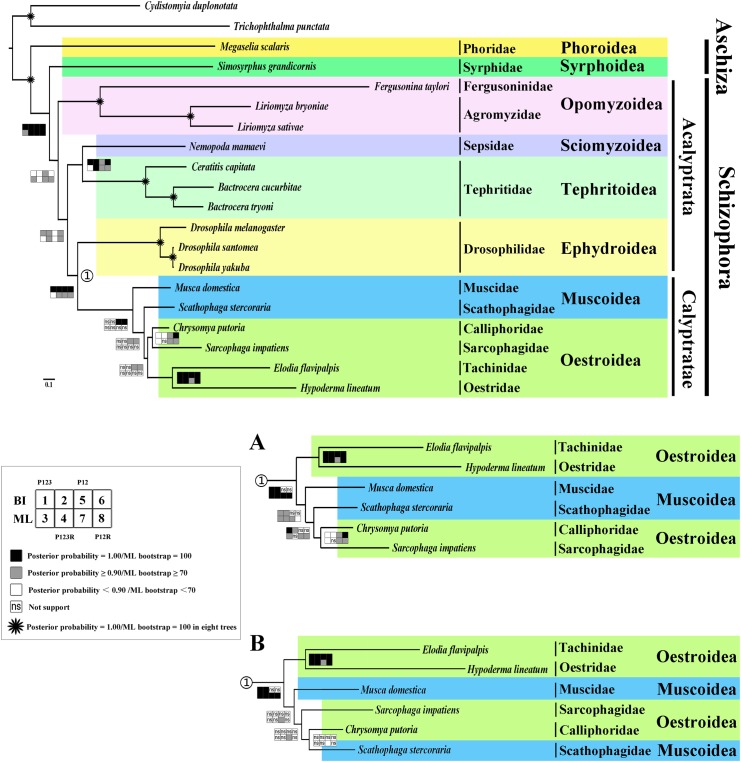
The phylogenetic trees conducted from both Bayesian and ML analyses have very similar topologies across four datasets (Fig 10). The monophyly of Cyclorrhapha, Opomyzoidea, Tephritoidea and Ephydroidea were consistently supported (posterior probability = 1.00, ML bootstrap = 100), as was the monophyly of Schizophora (posterior probability = 1.00, ML bootstrap = 85/100/100/100) and Calyptratae (posterior probability = 1.00, ML bootstrap = 41/73/87/84). Several recent researchers have concluded that ‘Aschiza’ is not monophyletic [71–76], here we add support for this conclusion from mt genome data. Phoridae was sister group of other Cyclorrhapha (posterior probability = 1.00, ML bootstrap = 100), and Syrphoidea was sister group of the Schizophora (posterior probability = 1.00, ML bootstrap = 85/100/100/100). This supports Wiegmann et. al’s findings regarding basal branching events within the Cyclorrhapha [77], however their nodal support for the relationship Phoridae + (remaining Cyclorrhapha) was weaker than that found here. Although only moderately supported (posterior probability = 0.86/0.70/0.98/0.79, ML bootstrap = 78/14/95/82), the position of superfamily Opomyzoidea (Fergusoninidae + Agromyzidae) was sister to the remaining Cyclorrhapha. Wiegmann et al. [77] did not find a monophyletic Opomyzoidea, nor did they find a sister-group relationship between the two representative families included in the present study. Given that there are 14 families recognized within the Opomyzoidea considerable additional data is necessary to firmly resolve their monophyly and relationships to other schizophoran superfamilies. Sepsidae is sister taxon to Tephritoidea (posterior probability = 1.00/1.00/0.92/1.00, ML bootstrap = 33/100/91/99), while Ephydroidea + Calyptratae formed a monophyletic group (posterior probability = 1.00, ML bootstrap = 41/73/87/84). Both relationships have previously been found in Wiegmann et al. [77], however neither had significant nodal support in that study.
Fig 10. Phylogenetic tree of Brachycera families based on mt genome data.
Cladogram of relationships resulting from Bayesian analyses with datasets PCG12 and PCG12RNA, with Cydistomyia duplonotata (Nemestrinidae) and Trichophthalma punctata (Tabanidae) as outgroups. Squares at the nodes are Bayesian posterior probabilities for 1, 2, 5 and 6, ML bootstrap values for 3, 4, 7 and 8. Dataset of PCG123, 1 and 3, PCG123RNA, 2 and 4, PCG12, 5 and 7, PCG12RNA, 6 and 8. Black indicates posterior probabilities = 1.00 or ML bootstrap = 100, gray indicates posterior probabilities ≥ 0.90 or ML bootstrap ≥ 70, white indicates posterior probabilities < 0.90 or ML bootstrap < 70, ‘ns’ indicates not support, * indicates posterior probabilities = 1.00 or ML bootstrap = 100 in eight trees. A. Part of the Bayesian tree of datasets PCG123 and PCG123RNA as well as ML tree of datasets PCG123, PCG12 and PCG12RNA. B. Part of the ML tree of dataset PCG123RNA.
The primary difference among the eight phylogenetic trees here is in the relationships of the superfamily Muscoidea (represented by the families Muscidae and Scathophagidae) and Oestroidea (represented by the families Oestridae, Tachinidae, Calliphoridae and Sarcophagidae). In the Bayesian analysis of datasets PCG12RNA and PCG12, the monophyly of Oestroidea was supported (posterior probability = 0.99/0.99), while a paraphyletic Muscoidea formed a grade at base of Calyptratae. Our results provide further evidence that ‘Muscoidea’ is paraphyletic as found in previous studies [77–78]. The Oestroidea was only monophyletic for the datasets that removed third codon positions (PCG12RNA and PCG12) but only when analysed by Bayesian inference. For the datasets that included third codon positions, and all analyses using maximum likelihood as the inference method, Oestroidea was rendered paraphyletic by the muscoid families which formed a clade with Calliphoridae and Sarcophagidae. Two pairs of relationships within Oestroidea were consistently supported by our analyses: Calliphoridae + Sarcophagidae formed a monophyletic group in seven of the eight analyses (posterior probability = 0.70/0.84/0.99/1.00, ML bootstrap = 58/-/98/90), while Oestridae + Tachinidae was strongly supported in all eight analyses (posterior probability = 1.00/1.00/1.00/1.00, ML bootstrap = 100/100/98/100). One of three sets of relationships within Calyptratae were found depending on the combination of dataset and analysis methods: monophyletic Oestroidea (BI-PCG12, BI-PCG12RNA: Fig 10), Muscidae + (Scathophagidae + (Calliphoridae + Sarcophagidae)) (BI-PCG123, BI-PCG123RNA, ML-PCG123, ML-PCG12, ML-PCG12RNA: Fig 10A) or Muscidae + (Sarcophagidae + (Calliphoridae + Scathophagidae)) (ML-PCG123RNA: Fig 10B).
The paraphyly of Oestroidea as found here in the majority of analyses has not been supported in previous studies such as those by Wiegmann et al. [77] and Kutty et al. [78]. Both studies found a monophyletic Oestroidea but with differing relationships between families: Wiegmann et al. [77] inferred ((Oestridae + Sarcophagidae) + (Tachinidae + Calliphoridae)), while Kutty et al. [78] found the relationships (((Tachinidae + Oestridae) + Calliphoridae) + Sarcophagidae) with both Calliphoridae and Tachinidae paraphyletic (Oestridae is nested within Tachinidae, as are some calliphorid subfamilies).
Phylogenetic analyses of mitochondrial genome data are known to be susceptible to compositional bias, particularly of third codon positions in the protein-coding genes [2] particularly in the Diptera [5, 13], and it is likely that the results found here of unstable relationships within the Calyptratae and a paraphyletic Oestroidea are an example of this phenomena. Even if comparisons between the present study and the previous ones by Wiegmann et al. [77] and Kutty et al. [78] are restricted to comparing trees that find a monophyletic Oestroidea, there is considerable variation in inferred relationships with Oestridae + Tachinidae, the only interfamilial relationships found in more than one study (present study and [78]). Considerable additional data is required to reliably resolve relationships within the Oestroidea, while our results suggest the reliability of mt genome data in inferring phylogenetic relationships within the Cyclorrhapha generally, caution is warranted for its use within the Oestroidea given that we have demonstrated susceptibility to third codon biases.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(DOCX)
(DOCX)
Acknowledgments
We express our sincere thanks to Ms. Yuting Dai (Beijing) for collecting the specimens of N. mamaevi, Mr. Yuzhang Jing (Sydney) for helping with early version of this paper and Dr. Hu Li (Lexington) for giving suggestions in phylogenetic analysis.
Data Availability
All data are available from the Genbank database (accession number KM605250).
Funding Statement
DY was funded by the National Natural Science Foundation of China (31320103902) and the National "Twelfth Five-Year" Plan for Science and Technology Support (2012BAD19B00). SLC was funded by an Australian Research Council Future Fellowship (FT120100746). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Koehler CM, Bauer MF. Mitochondrial function and biogenesis. Heidelberg: Springer Verlag; 2004. [Google Scholar]
- 2. Cameron SL. Insect mitochondrial genomics: Implications for Evolution and Phylogeny. Annual Rev Entomol. 2014;59: 95–117. 10.1146/annurev-ento-011613-162007 [DOI] [PubMed] [Google Scholar]
- 3. Cameron SL. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Syst Entomol. 2014;39: 400–411. [Google Scholar]
- 4. Cameron SL, Sullivan J, Song H, Miller KB, Whiting MF. A mitochondrial genome phylogeny of the Neuropterida (lace-wings, alderlies and snakelies) and their relationship to the other holometabolous insect orders. Zool Scr. 2009;38: 575–590. [Google Scholar]
- 5. Nelson LA, Lambkin CL, Batterham P, Wallman JF, Dowton M, Whiting MF, et al. Beyond barcoding: A mitochondrial genomics approach to molecular phylogenetics and diagnostics of blowflies (Diptera: Calliphoridae). Gene. 2012;511: 131–142. 10.1016/j.gene.2012.09.103 [DOI] [PubMed] [Google Scholar]
- 6. Zhao Z, Su TJ, Chesters D, Wang SD, Ho SYW, Zhu CD, et al. The mitochondrial genome of Elodia flavipalpis Aldrich (Diptera: Tachinidae) and the evolutionary timescale of tachinid flies. Plos One. 2013;8: e61814 10.1371/journal.pone.0061814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ma C, Yang PC, Jiang F, Chapuis MP, Shall Y, Sword GA, et al. Mitochondrial genomes reveal the global phylogeography and dispersal routes of the migratory locust. Mol Ecol. 2012;21: 4344–4358. 10.1111/j.1365-294X.2012.05684.x [DOI] [PubMed] [Google Scholar]
- 8. Logue K, Chan ER, Phipps T, Small ST, Reimer L, Henry-Halldin C, et al. Mitochondrial genome sequences reveal deep divergences among Anopheles punctulatus sibling species in Papua New Guinea. Malar J. 2013;12: 64 10.1186/1475-2875-12-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dowton M, Castro LR, Austin AD. Mitochondrial gene rearrangements as phylogenetic characters in the invertebrates: the examination of genome ‘morphology’. Invertebr Syst. 2002;16: 345–356. [Google Scholar]
- 10. Boore JL, Macey JR, Medina M. Sequencing and comparing whole mitochondrial genomes of animals. Meth Enzymology. 2005;395: 311–348. [DOI] [PubMed] [Google Scholar]
- 11. Boore JL. Requirements and standards for organelle genome databases. OMICS. 2006;10: 119–126. [DOI] [PubMed] [Google Scholar]
- 12. Margam VM, Coates BS, Hellmich RL. Agunbiade T, Seufferheld MJ, Sun W, et al. Mitochondrial genome sequences and expression proiling for the Legume Pod Borer Maruca vitrata (Lepidoptera: Crambidae). Plos One. 2011;6: e16444 10.1371/journal.pone.0016444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cameron SL, Lambkin CL, Barker SC, Whiting MF. A mitochondrial genome phylogeny of Diptera: whole genome sequence data accurately resolve relationships over broad timescales with high precision. Syst Entomol. 2007;32: 40–59. [Google Scholar]
- 14. Nelson LA, Cameron SL, Yeates DK. The complete mitochondrial genome of the gall–forming fly, Fergusonina taylori Nelson and Yeates (Diptera: Fergusoninidae). Mitochondrial DNA. 2011;22: 197–199. 10.3109/19401736.2011.632896 [DOI] [PubMed] [Google Scholar]
- 15. Yang F, Du Y, Cao J, Huang F. Analysis of three leafminers’ complete mitochondrial genomes. Gene. 2013;529: 1–6. 10.1016/j.gene.2013.08.010 [DOI] [PubMed] [Google Scholar]
- 16. Yang F, Du Y, Wang L, Cao J, Yu W. The complete mitochondrial genome of the leafminer Liriomyza sativae (Diptera: Agromyzidae): great difference in the A + T-rich region compared to Liriomyza trifolii . Gene. 2011;485: 7–15. 10.1016/j.gene.2011.05.030 [DOI] [PubMed] [Google Scholar]
- 17. Wang S, Lei Z, Wang H, Wang H, Dong B, Ren B. The complete mitochondrial genome of the leafminer Liriomyza trifolii (Diptera: Agromyzidae). Mol Biol Rep. 2011;38: 687–692. 10.1007/s11033-010-0155-6 [DOI] [PubMed] [Google Scholar]
- 18. Ye J, Fang R, Yi JP, Zhou GL, Zheng JZ. The complete sequence determination and analysis of four species of Bactrocera mitochondrial genome. Plant Quarantine. 2010;24: 11–14. [Google Scholar]
- 19. Yu DJ, Xu L, Nardi F, Li JG, Zhang RJ. The complete nucleotide sequence of the mitochondrial genome of the Oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae). Gene. 2007;396: 66–74. [DOI] [PubMed] [Google Scholar]
- 20. Zhang B, Nardi F, Hull-Sanders H, Wan X, Liu Y. The Complete Nucleotide Sequence of the Mitochondrial Genome of Bactrocera minax (Diptera: Tephritidae). Plos One. 2014;9: e100558 10.1371/journal.pone.0100558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nardi F, Carapelli A, Dallai R, Frati F. The mitochondrial genome of the olive fly Bactrocera oleae: two haplotypes from distant geographical locations. Insect Mol Biol. 2003;12: 605–611. [DOI] [PubMed] [Google Scholar]
- 22. Nardi F, Carapelli A, Boore J.L, Roderick GK, Dallai R, Frati F. Domestication of olive fly through a multi–regional host shift to cultivated olives: Comparative dating using complete mitochondrial genomes. Mol Phylogenet Evol. 2010;57: 678–686. 10.1016/j.ympev.2010.08.008 [DOI] [PubMed] [Google Scholar]
- 23. Spanos L, Koutroumbas G, Kotsyfakis M, Louis C. The mitochondrial genome of the Mediterranean fruit fly, Ceratitis capitata . Insect Mol Biol. 2000;9: 139–144. [DOI] [PubMed] [Google Scholar]
- 24. Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450: 203–218. [DOI] [PubMed] [Google Scholar]
- 25. Andrianov B, Goryacheva I, Mugue N, Sorokina S, Gorelova T, Mitrofanov V, et al. Comparative analysis of the mitochondrial genomes in Drosophila virilis species group (Diptera: Drosophilidae). Trend Evol Biol. 2010;2: e4. [Google Scholar]
- 26. Lewis DL, Farr CL, Kaguni LS. Drosophila melanogaster mitochondrial DNA: completion of the nucleotide sequence and evolutionary comparisons. Insect Mol Biol. 1995;4: 263–278. [DOI] [PubMed] [Google Scholar]
- 27. Torres TT, Dolezal M, Schlötterer C, Ottenwälder B. Expression profiling of Drosophila mitochondrial genes via deep mRNA sequencing. Nucl Acid Research. 2009;37: 7509–7518. 10.1093/nar/gkp856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Llopart A, Herrig D, Brud E, Stecklein Z. Sequential adaptive introgression of the mitochondrial genome in Drosophila yakuba and Drosophila santomea . Mol Ecol. 2014;23: 1124–1136. 10.1111/mec.12678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ballard JWO. Comparative genomics of mitochondrial DNA in members of the Drosophila melanogaster subgroup. J Mol Evol. 2000;51: 48–63. [DOI] [PubMed] [Google Scholar]
- 30. Clary DO, Wolstenholme DR. The mitochondrial DNA molecule of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 1985;22: 252–271. [DOI] [PubMed] [Google Scholar]
- 31. Li X, Wang Y, Su S, Yang D. The complete mitochondrial genomes of Musca domestica and Scathophaga stercoraria (Diptera: Muscoidea: Muscidae and Scathophagidae). Mitochondrial DNA. 2014;8 Available: http://informahealthcare.com/doi/abs/10.3109/19401736.2014.953080?journalCode=mdn. 10.1186/1687-4153-2014-8 [DOI] [PubMed] [Google Scholar]
- 32. Oliveira MT, Azeredo-Espin AML, Lessinger AC. The mitochondrial DNA control region of Muscidae flies: evolution and structural conservation in a dipteran context. J Mol Evol. 2007;64: 519–527. [DOI] [PubMed] [Google Scholar]
- 33. Junqueira ACM, Lessinger AC, Torres TT, Rodrigues F da S, Vettore AL, Arruda P, et al. The mitochondrial genome of the blowfly Chrysomya chloropyga (Diptera: Calliphoridae). Gene. 2004;339: 7–15. [DOI] [PubMed] [Google Scholar]
- 34. Lessinger AC, Martins JAC, Lemos TA, Kemper EL, Da Silva FR, Vettore AL, et al. The mitochondrial genome of the primary screwworm fly Cochliomyia hominivorax (Diptera: Calliphoridae). Insect Mol Biol. 2000;9: 521–529. [DOI] [PubMed] [Google Scholar]
- 35. Weigl S, Testini G, Parisi A, Dantas-Torres F, Traversa D, Colwell DD, et al. The mitochondrial genome of the common cattle grub, Hypoderma lineatum . Med Vet Entomol. 2010;24: 329–335. 10.1111/j.1365-2915.2010.00873.x [DOI] [PubMed] [Google Scholar]
- 36. Shao YJ, Hu XQ, Peng GD, Wang RX, Gao RN, Lin C, et al. Structure and evolution of the mitochondrial genome of Exorista sorbillans: the Tachinidae (Diptera: Calyptratae) perspective. Mol Biol Rep. 2012;39: 11023–11030. 10.1007/s11033-012-2005-1 [DOI] [PubMed] [Google Scholar]
- 37. Ozerov AL. World catalogue of the family Sepsidae (Insecta: Diptera). Zool Stud. 2005;8: 1–74. [Google Scholar]
- 38. Ang Y, Puniamoorthy N, Meier R. Secondarily reduced foreleg armature in Perochaeta dikowi sp. n. (Diptera: Cyclorrhapha: Sepsidae) due to a novel mounting technique. Syst Entomol. 2008;33: 552–559. [Google Scholar]
- 39. Ingram KK, Laamanen T, Puniamoorthy N, Meier R. Lack of morphological coevolution between male forelegs and female wings in Themira (Sepsidae: Diptera: Insecta). Biol J Linn Soc. 2008;93: 227–238. [Google Scholar]
- 40. Puniamoorthy N, Su KFY, Meier R. Bending for love: losses and gains of sexual dimorphisms are strictly correlated with changes in the mounting position of sepsid flies (Sepsidae: Diptera). BMC Evol Biol. 2008;8: 155 10.1186/1471-2148-8-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Puniamoorthy N, Ismail MRB, Tan DSH, Meier R. From kissing to belly stridulation: comparative analysis reveals surprising diversity, rapid evolution, and much homoplasy in the mating behaviour of 27 species of sepsid flies (Diptera: Sepsidae). J Evol Biol. 2009;22: 2146–2156. 10.1111/j.1420-9101.2009.01826.x [DOI] [PubMed] [Google Scholar]
- 42. Baena ML, Eberhard WG. Appearances deceive: female “resistance” behaviour in a sepsid fly is not a test of male ability to hold on. Ethol Ecol Evol. 2007;19: 27–50. [Google Scholar]
- 43. Martin OY, Hosken DJ. The evolution of reproductive isolation through sexual conflict. Nature. 2003;423: 979–982. [DOI] [PubMed] [Google Scholar]
- 44. Pont AC, Meier R. The Sepsidae (Diptera) of Europe. Fauna Entomologica Scandinavica. 2002;37: 1–221. [Google Scholar]
- 45. Melicher D, Torson AS, Dworkin I, Bowsher JH. A pipeline for the de novo assembly of the Themira biloba (Sepsidae: Diptera) transcriptome using a multiple k-mer length approach. BMC Genomics. 2014;15: 188 10.1186/1471-2164-15-188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucl Acid Res. 1997;25: 0955–0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stewart JB, Beckenbach AT. Characterization of mature mitochondrial transcripts in Drosophila and the implications for the tRNA punctuation model in arthropods. Gene. 2009;445: 49–57. 10.1016/j.gene.2009.06.006 [DOI] [PubMed] [Google Scholar]
- 48. Wang HL, Yang J, Boykin LM, Zhao QY, Li Q, Wang XW, et al. The characteristics and expression profiles of the mitochondrial genome for the Mediterranean species of the Bemisia tabaci complex. BMC Genomics. 2013;14: 401 10.1186/1471-2164-14-401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, et al. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 2013;69: 313–319. 10.1016/j.ympev.2012.08.023 [DOI] [PubMed] [Google Scholar]
- 50. Ozerov AL. A new species of the genus Nemopoda Robineau-Desvoidy. 1930 (Diptera, Sepsidae) from the Far East of Russia. Dipt Res. 1997;8: 159–162. [Google Scholar]
- 51. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41: 95–98. [Google Scholar]
- 52. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Perna NT, Kocher TD. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol. 1995;41: 353–358. [DOI] [PubMed] [Google Scholar]
- 54. Vaidya G, Lohman DJ, Meier R. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 2010;27: 171–180. [DOI] [PubMed] [Google Scholar]
- 55. Lanfear R, Calcott B, Ho SYW, Guindon S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analysis. Mol Biol Evol. 2012;29: 1695–1701. 10.1093/molbev/mss020 [DOI] [PubMed] [Google Scholar]
- 56. Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analysis with thousands of taxa and mixed models. Bioinformatics. 2006;22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- 57. Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57: 758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 58. Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 59. Grant JR, Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36: W181–W184. 10.1093/nar/gkn179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wei SJ, Shi M, Chen XX, Sharkey M.J, van Achterberg C, Ye GY, et al. New views on strand asymmetry in insect mitochondrial genomes. Plos One. 2010;5: e12708 10.1371/journal.pone.0012708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cameron SL, Lo N, Bourguignon T, Svenson GJ, Evans TA. A mitochondrial genome phylogeny of termites (Blattodea: Termitoidae): Robust support for interfamilial relationships and molecular synapomorphies define major clades. Mol Phylogen Evol. 2012;65: 163–173. 10.1016/j.ympev.2012.05.034 [DOI] [PubMed] [Google Scholar]
- 62. Wolstenholme DR. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 1992;141: 173–216. [DOI] [PubMed] [Google Scholar]
- 63. Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290: 470–474. [DOI] [PubMed] [Google Scholar]
- 64. Roberti M, Polosa PL, Bruni F, Musicco C, Gadaleta MN, et al. DmTTF, a novel mitochondrial transcription termination factor that recognizes two sequences of Drosophila melanogaster mitochondrial DNA. Nucleic Acids Res. 2003;31: 1597–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Beckenbach AT. Mitochondrial genome sequences of Nematocera (lower Diptera): evidence of rearrangement following a complete genome duplication in a winter crane fly. Genome Biol Evol. 2012;4: 89–101. 10.1093/gbe/evr131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cameron SL, Whiting MF. Mitochondrial genomic comparisons of the subterranean termites from the Genus Reticulitermes (Insecta: Isoptera: Rhinotermitidae). Genome. 2007;50: 188–202. [DOI] [PubMed] [Google Scholar]
- 67. Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. BBA-Mol Basis Dis. 1999;1410: 103–123. [DOI] [PubMed] [Google Scholar]
- 68. Boore JL. Complete mitochondrial genome sequence of the polychaete annelid. Mol Biol Evol. 2001;18: 1413–1416. [DOI] [PubMed] [Google Scholar]
- 69. Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, Du Y, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3: 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhang DX, Hewitt GM. Insect mitochondrial control region: a review of its structure, evolution and usefulness in evolutionary studies. Biochem Syst Ecol. 1997;25: 99–120. [Google Scholar]
- 71. Griffiths GCD. The phylogenetic classification of Diptera Cyclorrhapha, with special reference to the structure of the male postabdomen. The Hague: Junk; 1972. [Google Scholar]
- 72. Griffiths GCD. Book review: Manual of Nearctic Diptera Volume 3. Quaest Entomol. 1990;26: 117–130. [Google Scholar]
- 73. Wada S. Morphologische Indizien für das unmittelbare Schwestergruppenverhältnis der Schizophora mit den Syrphoidea ('Aschiza') in der phylogenetischen Systematik der Cyclorrhapha (Diptera: Brachycera). J Natur Hist. 1991;25: 1531–1570. [Google Scholar]
- 74. Cumming JM, Sinclair BJ, Wood DM. Homology and phylogenetic implications of male genitalia in Diptera-Eremoneura. Entomol Scand. 1995;26: 120–151. [Google Scholar]
- 75. Zatwarnicki T. A new reconstruction of the origin of the eremoneuran hypopygium and its implications for classification (Insecta: Diptera). Genus. 1996;3: 103–175. [Google Scholar]
- 76. Yeates DK, Wiegmann BM, Courtney GW, Meier R, Pape T. Phylogeny and systematics of Diptera: two decades of progress and prospects. Zootaxa. 2007;1688: 565–590. [Google Scholar]
- 77. Wiegmann BM, Trautwein MD, Winkler IS, Barr NB, Kim JW, Lambkin C, et al. Episodic radiations in the fly tree of life. PNAS. 2011;108: 5690–5695. 10.1073/pnas.1012675108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kutty SN, Pape T, Wiegmann BM, Meier R. Molecular phylogeny of the Calyptratae (Diptera: Cyclorrhapha) with an emphasis on the superfamily Oestroidea and the position of Mystacinobiidae and McAlpine’s fly. Syst Entom. 2010;35: 614–635. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(DOCX)
(DOCX)
Data Availability Statement
All data are available from the Genbank database (accession number KM605250).