Abstract
Whole genome DNA sequencing was used to decrypt the phylogeny of multiple samples from distinct areas of cancer and morphologically normal tissue taken from the prostates of three men. Mutations were present at high levels in morphologically normal tissue distant from the cancer reflecting clonal expansions, and the underlying mutational processes at work in morphologically normal tissue were also at work in cancer. Our observations demonstrate the existence of on-going abnormal mutational processes, consistent with field-effects, underlying carcinogenesis. This mechanism gives rise to extensive branching evolution and cancer clone mixing as exemplified by the coexistence of multiple cancer lineages harboring distinct ERG fusions within a single cancer nodule. Subsets of mutations were shared either by morphologically normal and malignant tissue or between different ERG-lineages, indicating earlier or separate clonal cell expansions. Our observations inform on the origin of multifocal disease and have implications for prostate cancer therapy in individual cases.
Prostate cancer is commonly multifocal1, although the origin of multifocal disease remains controversial. Analyses of patterns of allele loss have suggested the independence of most individual foci2,3. However such studies cannot exclude the presence of common underlying mutations not detected by the methods employed. Recent attempts to unravel the origins of multifocal disease using high-resolution genome technologies have also led to conflicting data with different authors concluding either that all foci in a single prostate are related4 or that all foci are unrelated5. To gain further insights into the mechanism of prostate cancer development particularly the origin of multifocal disease we selected three representative prostate cancers (Fig.1, Supplementary Fig.1) that had been ERG-status mapped using the FISH break-apart method6,7. Twelve cancer samples and three samples designated as morphologically normal prostate based on central pathology review, were analyzed using paired-end massively-parallel DNA sequencing of complete genomes to generate comprehensive catalogues of genetic alterations (for coverage statistics see Supplementary Table 1). For 3D representations of each prostate and clinical characteristics see respectively Supplementary Fig. 2 and Supplementary Table 2. Prostates were named according to their Cancer Research UK project designation: Cases 6, 7 and 8.
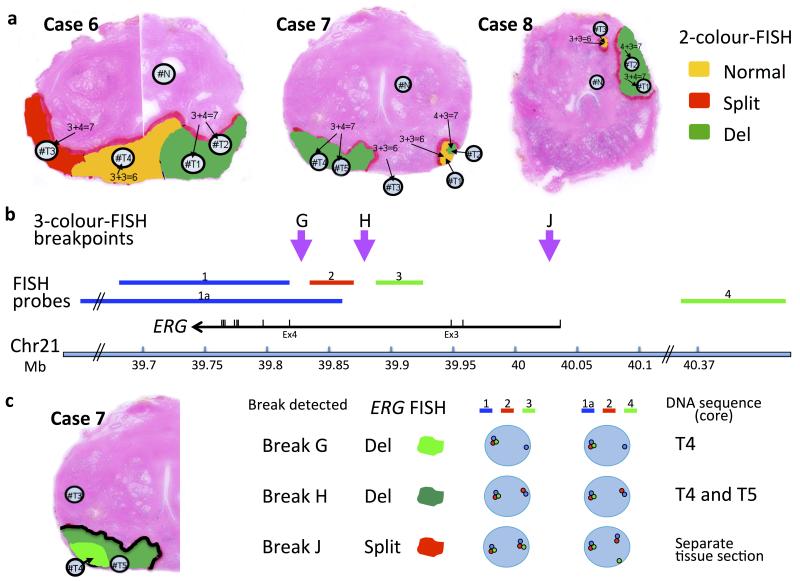
Figure 1.
Prostate samples chosen for whole-genome sequencing. a, ERG rearrangements determined by fluorescence in situ hybridization (FISH). Case 7 is a multifocal cancer containing two separate foci (T1/T2/T4/T5 and T3). Case 8 is also designated as a multifocal cancer,(nodules T1/T2, and T3). Yellow: un-rearranged normal ERG gene; Red, ERG gene split but both 3′ and 5′ ends retained; Green, ERG gene rearranged but only its 3′ end retained. Panels b and c: 3-colour FISH used to distinguish different ERG-locus translocation breakpoints in Case 7. b, Position of the three FISH probes: probe 1 (blue, BAC RP11-164E1, and probe 1a, BACs RP11-95G19, RP11-720N21, CTD-2511E13) was labeled in Aqua (Kreatech 415 Platinum Bright): probe 2 (red, fosmid G248P80319F5 37Kb) labeled with Cy3; and Probe 3 (green, fosmid G248P86592E2 38.5k, and probe 4, BACs RP11-372O17, RP11-115E14, RP11-729O4) labeled with FITC. The purple arrows represent the positions of ERG breakpoints detected in these experiments. For the precise position of the ERG breakpoints G and H see Table 2. c, Left: Tumor areas with ERG locus breaks G and H are indicated as light and dark green respectively. Break J was found in an adjacent prostate section not show in this figure. Right: representations of the ERG FISH patterns. Original FISH images are show in Supplementary Fig. 1. “Split” denotes that 5′ and 3′ ERG signals were separated but retained in the cell. “Del” indicates that 5′ ERG signals were lost from the cell, while 3′ ERG signals were retained.
Somatic mutations, absent from cancer and blood samples, were observed at significant levels in morphologically normal prostate tissue distant from cancer in Case 6 (518 substitutions) and in Case 7 (454 substitutions) (Supplementary Fig. 3), some of which may have potential functional significance (Table 1). The presence of substitution mutations in morphologically normal prostate tissue was confirmed in validation DNA-sequencing experiments to an average read depth of 10,000. Substitutions were present in an estimated ~48%, and ~42% of cells in morphologically normal samples from Case 6 and Case 7 respectively (Supplementary Fig. 3b)), demonstrating clonal expansions of cells within morphologically normal prostate tissue, in agreement with studies using mitochondrially-encoded enzyme cytochrome c oxidase as a marker8.
Table 1.
| Sample | Description | Gene | Protein Description | Type | % reads | Total num reads | MA predicted functional impact | ANNOVAR significant algorithms |
|---|---|---|---|---|---|---|---|---|
| 0006#N | chr9:g.131115799G>A | SLC27A4 | p.V435I | misssense | 13.79 | 58 | low | 1 |
| 0006#N | chr14:g.20389481C>T | OR4K5 | p.T239M | misssense | 13.25 | 83 | high | 4 |
| 0006#N | chr15:g.33873844G>T | RYR3 | p.A525S | misssense | 33.33 | 48 | medium | |
| 0006#N | chr4:g.88766379C>G | MEPE | p.S120* | nonsense | 20.83 | 24 | 2 | |
| 0007#N | chr5:g.150885254A>T | FAT2 | p.S4308T | misssense | 23.4 | 47 | low | 5 |
| 0007#N | chr7:g.150934857G>T | CHPF2 | p.R470L | misssense | 17.24 | 58 | medium | 5 |
| 0007#N | chr8:g.24192995G>A | ADAM28 | p.D470N | misssense | 17.78 | 45 | neutral | 2 |
| 0007#N | chr12:g.24989522G>T | BCAT1 | p.L276M | misssense | 26.47 | 34 | medium |
Mutations and clonal expansions in morphologically normal tissue: point mutations present in exons with indication of functional significance. Missense and nonsense mutations detected and visually confirmed in the adjacent morphologically normal tissue were tested for functional impact using the MutationAssessor.org27 and wANNOVAR28 services. The OR4K5 gene was excluded as a candidate because of the potential to overcall mutations in genes encoding very large proteins29. Since none of the mutations had a high “MA” we considered that epigenetic changes may provide a more likely driver of clonal expansion.
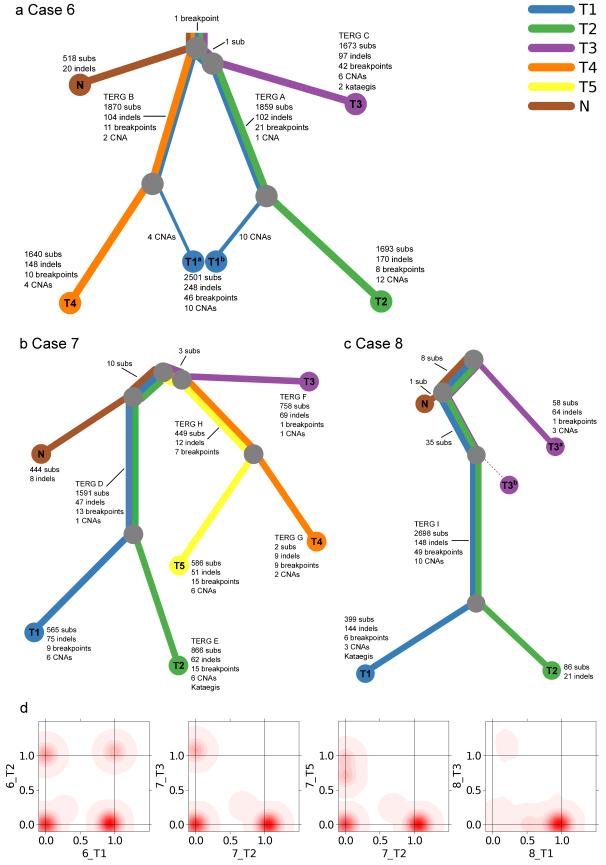
Aiming to understand the tumor subclonal architecture and their phylogeny, we initially constructed phylogenetic trees based on copy number (Supplementary Fig. 4 & 5, Supplementary Data Set 1) and substitution data. We adapted our previously developed Bayesian Dirichlet process to identify clusters of substitutions in n dimensions9, where n is the number of samples from the case, such that shared and unique subclones could be identified between related samples (Fig. 2d and Supplementary Fig. 6). To further explore the fine details and verify the main features of the phylogeny tree and clonal structure, a selection of substitutions from each potential relationship between samples were sequenced to an average read depth of 10,000 in independent DNA sequencing analyses, verifying 279 mutations across all samples. This provided us with our final integrated phylogenetic trees (Fig. 2a-c) and final list of somatic point mutations (Supplementary Data Set 2). The structure of these trees was also supported by verified insertions, deletions and breakpoints (Supplementary Data Set 3 & 4). The single cancer mass from Patient 6 contained three independent cancer clones represented by samples 6_T2, 6_T3 and 6_T4 (Fig. 2a), with a single verified substitution linking 6_T1/6_T2 and 6_T3. Patient 7 contained at least three independent cancer lineages: one (7_T3) representing the smaller cancer nodule and two (7_T1/7_T2 and 7_T4/7_T5) present in the larger cancer mass (Fig. 2b). Ten mutations were common to the morphologically normal prostate sample and to cancer samples 7_T1 and 7_T2, and three mutations joined 7_T4/7_T5 to the separate multifocal lesion 7_T3. These observations show that Prostate 7 contains at least two clones of cells that existed prior to the formation of the distinct cancers lineages. Prostate 8 contained two cancer lineages represented by 8_T1/8_T2 and 8_T3 (Fig. 2c), with 43 substitutions shared between all three tumor samples, 8_T1, 8_T2 and 8_T3, 8 of which were also present in distant morphologically normal sample 8_N.
Figure 2.
Phylogenies of multi-focal prostate cancers. a-c, Phylogenies revealing the relationships between sample clones for each case. Each line is associated with a clone from a particular sample. The length of each line is proportional to the weighted quantity of variations on a logarithmic scale. The thickness of a line indicates the proportion the clone makes up of that sample i.e. 48%/52% for 6_T1 and 12%/88% for 8_T3. The minor clone of 8_T3b has no detected unique variants. 8_T3 contained 43 mutations present as a 12% subclone (T3a) shared with 8_T1/8_T2. In validation experiments 8_T3 did not contain any of the five ERG and TMPRSS2 rearrangements present in 8_T1/8_T2 (Table 2)) or mutations that were unique to 8_T1/8_T2 (10,000 depth) indicating that it represents an earlier clone of 8_T1/8_T2 seeded into tissue sample 8_T3. The various TMPRSS2-ERG translocations are indicated by their TERG ID (Table 2). d, Example 2D density plots showing the posterior distribution of the fraction of cells bearing a mutation in two samples. The fraction of cells is modeled using a Bayesian Dirichlet processes. These plots illustrate samples that have shared clonal mutations (6_T1/6_T2), and branched (unrelated) mutations (7_T2/T_T3). There are two examples of samples with a subclone. 7_T2/7_T5 has a peak at (0,0.72), which represents subclonal mutations in 72% of cells in 7_T5 that have occurred only in this sample, after divergence from the other samples. Similarly, 8_T1/8_T3 has a peak at (0.54,0), representing subclonal mutations in 54% of cells in T1 only.
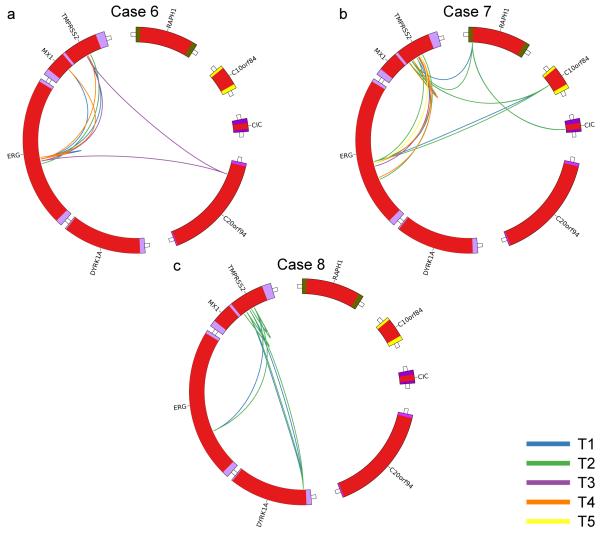
Complex patterns of ERG alteration were observed in samples from Patient 6 and Patient 7 (Fig. 3); each main lineage contained at least one and in some cases two unique TMPRSS2-ERG fusions with distinct breakpoint locations within the TMPRSS2 and ERG genes (Fig. 2, Table2). The presence of multiple distinct TMPRSS2-ERG fusions was demonstrated by direct PCR across the breakpoint and by an ERG FISH break-apart assay (Table 2, Fig. 1b,c, Supplementary Fig. 1). In this respect TMPRSS-ERG fusions could be considered to be similar to the convergent gene alterations observed in kidney cancer where distinct alterations of genes such as SETD2, PTEN, and KDM5C were observed in different parts of the same cancer10. A deletion on Chromosome 8 exhibited a very similar pattern of alterations (Supplementary Fig. 7), but we did not see convergent evolution for other potential driver genes (Supplementary Table 3). Where two TMPRSS2-ERG fusions existed in a single lineage we were unable to determine whether these fusions co-existed at any time in the same cell as reported previously11 and as implied by the phylogenic tree. However the FISH assay (Fig 1b,c) demonstrated that in sample 7_T4 the two TMPRSS2_ERG fusions were present in distinct cell populations at the time that the cancer sample was taken. Moreover, an additional separate ERG breakpoint was detected in a region of the cancer that had not been sampled in the DNA sequencing studies (TERG J). The occurrence of several TMPRSS2-ERG fusions is a single cancer mass is consistent with previous FISH-based studies reporting multiple ETS fusions in a low proportion of individual cancer foci11. ERG alterations are believed to represent a relatively early event in cancer development in agreement with their occurrence in prostatic intraepithelial neoplasia (PIN)6, but our observations suggest that they may not always be present at the very first cellular expansion. Mutations shared either between different ERG-lineages or between cancer and morphologically normal tissue may represent earlier clonal cell expansions on the same lineage (Fig. 2a-c). Alternatively they could represent separate clones of cells within which multiple independent cancer lineages developed.
Figure 3.
Patterns of ERG alterations. a-c, Circos plots highlighting ERG rearrangements present in each prostate. Each color represents a different cancer sample as indicated.
Table 2.
| Donor | Middle | Acceptor | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Samples | Chr | Position | Strand | Type | Seq | Chr | Position | Strand | Breakpoint | Genes | Verification | TERG ID |
| 6_T1, 6_T2 | 21 | 39867180 | + | HOMOLOGY | T | 21 | 42877104 | + | deletion | ERG-TMPRSS2 | CS & P (6_T1); V (6_T1, 6_T2) | A |
| 6_T1, 6_T4 | 21 | 39877208 | + | HOMOLOGY | T | 21 | 42871170 | + | deletion | ERG-TMPRSS2 | P (6_T1); V (6_T1, 6_T4) | B |
| 6_T1, 6_T4 | 21 | 39877355 | − | HOMOLOGY | CC | 21 | 42819405 | − | insertion | ERG-MX1 | CS & P (6_T1); V (6_T1, 6_T4) | |
| 6_T1, 6_T4 | 21 | 39877745 | + | NTS | CAT | 21 | 39880855 | + | deletion | ERG-ERG | CS & P (6_T1); V (6_T1, 6_T4) | |
| 6_T3 | 20 | 10441211 | − | HOMOLOGY | G | 21 | 39872887 | + | translocation | C20orf94-ERG | CS & P & V (6_T3) | |
| 6_T3 | 20 | 10441429 | + | HOMOLOGY | GT | 21 | 42868518 | − | translocation | C20orf94-TMPRSS2 | CS & P & V (6_T3) | |
| 6_T3 | 21 | 39872930 | + | Exact | --- | 21 | 42868510 | + | deletion | ERG-TMPRSS2 | CS & P & V (6_T3) | C |
| 7_T1, 7_T2 | 1 | 205613440 | + | HOMOLOGY | C | 21 | 42857784 | − | translocation | _-TMPRSS2 | V (7_T1, 7_T2) | |
| 7_T1, 7_T2 | 2 | 204298424 | − | HOMOLOGY | A | 21 | 42849002 | + | translocation | RAPH1-TMPRSS2 | V (7_T1, 7_T2) | |
| 7_T1, 7_T2 | 2 | 204298476 | + | Exact | --- | 19 | 42797705 | + | translocation | RAPH1-CIC | P (7_T1); V (7_T1, 7_T2) | |
| 7_T1, 7_T2 | 10 | 120084722 | − | HOMOLOGY | TG | 21 | 42842154 | + | translocation | C10orf84-TMPRSS2 | CS & P (7_T1); V (7_T1, 7_T2) | |
| 7_T1, 7_T2 | 10 | 120084747 | + | HOMOLOGY | AC | 21 | 39872234 | + | translocation | C10orf84-ERG | CS & P (7_T2); V (7_T1, 7_T2) | |
| 7_T1, 7_T2 | 21 | 39872152 | + | HOMOLOGY | A | 21 | 42861527 | + | deletion | ERG-TMPRSS2 | CS & P (7_T1); V (7_T1, 7_T2) | D |
| 7_T1, 7_T2 | 21 | 42842403 | + | Exact | --- | 21 | 42848506 | − | inversion_+ | TMPRSS2-TMPRSS2 | CS & P (7_T1); V (7_T1, 7_T2) | |
| 7_T2 | 21 | 39831266 | + | HOMOLOGY | AAAC | 21 | 42875633 | + | deletion | ERG-TMPRSS2 | CS & P & V (7_T2) | E |
| 7_T3 | 21 | 39861568 | + | NTS | TA | 21 | 42865303 | + | deletion | ERG-TMPRSS2 | CS & P & V (7_T3) | F |
| 7_T4 | 21 | 39835734 | + | HOMOLOGY | G | 21 | 42867100 | + | deletion | ERG-TMPRSS2 | CS & P & V (7_T4) | G |
| 7_T4 | 21 | 42841552 | − | HOMOLOGY | GGCT | 21 | 42851963 | + | inversion_− | TMPRSS2-TMPRSS2 | CS & P & V (7_T4) | |
| 7_T4, 7_T5 | 21 | 39868722 | + | Exact | --- | 21 | 42870051 | + | deletion | ERG-TMPRSS2 | CS & P (7_T4); V (7_T4, 7_T5) | H |
| 8_T1, 8_T2 | 21 | 38745261 | + | HOMOLOGY | T | 21 | 42851601 | − | inversion_+ | DYRK1A-TMPRSS2 | P (8_T1); V (8_T1, 8_T2) | |
| 8_T1, 8_T2 | 21 | 38745286 | − | HOMOLOGY | A | 21 | 42859198 | − | insertion | DYRK1A-TMPRSS2 | CS & P (8_T1); V (8_T1, 8_T2) | |
| 8_T1, 8_T2 | 21 | 39831518 | + | Exact | --- | 21 | 42870497 | − | inversion_+ | ERG-TMPRSS2 | CS (8_T1); P & V (8_T1, 8_T2) | I |
| 8_T1, 8_T2 | 21 | 42844460 | − | HOMOLOGY | T | 21 | 42851648 | + | inversion_− | TMPRSS2-TMPRSS2 | V (8_T1, 8_T2) | |
| 8_T1, 8_T2 | 21 | 42863787 | − | HOMOLOGY | G | 21 | 42870663 | + | inversion_− | TMPRSS2-TMPRSS2 | CS & P (8_T1); V (8_T1, 8_T2) | |
Patterns of ERG alterations. Positions and structure of each ERG breakpoints and related rearrangements. The position and structure of the breakpoint was determined, in the majority of cases, by capillary sequencing using custom-designed PCR across the rearrangement breakpoint as previously described30 (“CS” in column “Verification”), and/or by in-silico reconstruction using local de novo assembly in Brass phase 2. Verification by sizing PCR products across the breakpoint using gel electrophoresis was also performed (“P”). All breakpoints were visually verified (“V”) to ensure the presence of discordant reads and checked that they did not occur in repeat regions.
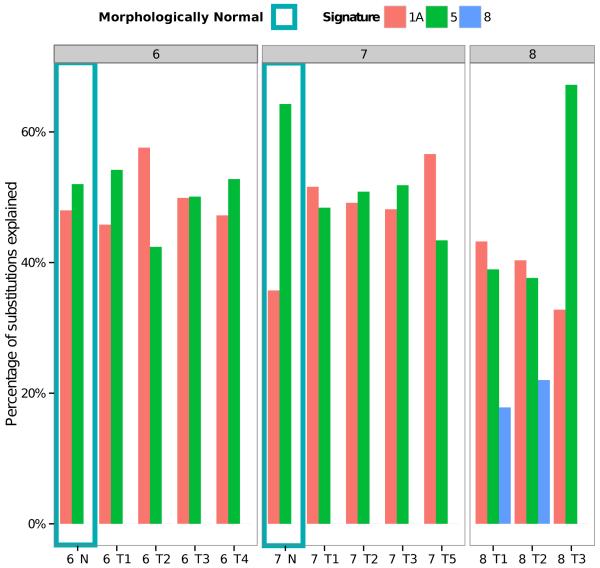
Recently, we identified 21 distinct mutational signatures from 7,042 samples across 30 different cancer types12. The contribution of mutational processes was calculated for prostate cancer as previously described12,13 (Fig. 4). A signature (designated Signature 1A in Ref. 12) associated with spontaneous deamination of 5-methyl-cytosine at CpG sequences explained ~50% of all of our mutations. Two additional signatures with unknown etiology, designated Signature 5 and Signature 8, best explained the remaining somatic mutations. Signature 5, present in all prostate samples may reflect an endogenous mutational process12. Signature 8, present in two cancer samples from a single cancer nodule, is characterized by weak C>A strand bias. Critically these observations show that the same mutational processes, giving rise to Signatures 1a and 5, are detected both in cancer and in matched morphologically normal prostate tissue. We identified clustering of C>T and C>G mutations previously referred to as kataegis14 and complex interdependent translocations and deletions called chromoplexy15 in some cancer lineages (Supplementary Fig. 8 & 9).
Figure 4.
Relative contributions of mutational signatures to the total mutation burden of each sample. The mutational spectra, as defined by the triplets of nucleotides around each substitution, of each sample were deconvoluted into mutational processes using 22 distinct signatures determined from 7,042 cancers as described previously12,13. The signature designations (1a, 5, 8) match those reported previously12. For sample 7_T4 and 8_N there were too few mutations to be able to accurately identify the contributions of the mutational signatures.
Next generation sequence technologies have previously been used to identify critical genetic processes in prostate cancer development15-19. Our results demonstrate the presence of clonal expansions or fields of cells in the morphologically normal prostate that provide a background against which prostate cancer develops. A recent study on a 115 year old woman identified 424 point mutations, thought to result from somatic mosaicism, in the rapidly dividing tissue blood, but failed to detect any mutations in brain tissue20. The presence of mutations in blood was accompanied by telomere attrition that was not observed in other tissues. Prostate is considered to be a relatively quiescent tissue21, and we found that the telomeres in morphologically normal tissue from Cases 6 and 7 had not undergone attrition, being of comparable length to telomeres in adjacent cancer. The processes at work in morphologically normal prostate therefore appear to be distinct from those reported for blood (see Supplementary Notes for full discussion). Whether the clones of cells observed in morphologically normal prostate are generated by a pathological process or are the product of somatic mosaicism involving unexpectedly high mutation rates, the resulting clonal fields of cells may influence cancer development and/or contribute to multifocality and the presence of multiple cancer lineages in a single cancer mass. Evidence for a field effect in prostate cancer is also supported by studies demonstrating tumor-like alterations in cytomorphology, gene expression, epigenetics in adjacent morphologically normal tissue, and the presence of multifocal disease in a high proportion of prostates. Field effects have also been proposed for oral cancer22, head and neck cancer23 and breast cancer24. Our results have implications for the use of cancer focal therapy when targeting a single nodule of cancer within the prostate25,26 and for potential chemotherapeutic approaches. We propose that (i) focal therapy may only be curative if surrounding clonal cell populations within morphologically normal tissue were also ablated, and (ii) cancer heterogeneity may hinder therapeutic targeting and biomarker investigation.
ONLINE METHODS
Sample Selection and Fluorescence in situ Hybridisation
Samples for analysis were collected from prostatectomy patients at the Addenbrooke’s Hospital (see Supplementary Table 2). The study was approved by the Trent Multicentre Research Ethics Committee. Informed consent was obtained for all patients. Prostates were sliced and processed as described previously31. In brief, a single 5 mm slice of the prostate was selected for research purposes. 4 or 6 mm cores were taken from the slice and frozen. Frozen cores were mounted vertically and sectioned transversely giving a single 5 μm frozen section for H&E staining followed by 6×50 μm sections for DNA preparation. The presence of or complete absence of cancer was confirmed independently by three pathologists in central pathology review of the 5 μm H&E stained tissue slice immediately adjacent to tissue slices used for DNA preparation. The ERG fluorescence in situ hybridisation break-apart assay for assessing ERG gene rearrangement was performed as described previously6, both (i) on whole-mount formalin-fixed sections, taken immediately adjacent to the research slice, and (ii) on the frozen slices, immediately adjacent to the samples selected for DNA sequencing that had been initially subject to H&E staining. In all cases, the ERG status determined by these two methods and shown in Figure 1, were consistent.
DNA sequencing
Samples and Massively Parallel Sequencing
DNA was extracted from 18 samples from 3 patients: 12 prostate cancer samples, 3 adjacent morphologically normal prostate samples and 3 matched bloods. Paired-end whole genome sequencing of the samples was performed at Illumina, Inc. Paired-end libraries were manually generated from 1 μg of gDNA using the Illumina Paired End Sample Prep Kit (Catalog # PE-102-1002). Fragmentation was performed with Covaris E220. After end repair, A-tailing, and adapter ligation as per the sample prep kit instructions, libraries were manually size-selected using agarose gel electrophoresis, targeting 300 bp inserts. Adapter-ligated libraries were PCR amplified for 10 cycles and purified through a second agarose gel electrophoresis. Final libraries were QC’ed on a Agilent Bioanalyzer and quantified by qPCR and/or picogreen fluorimetry. Samples were clustered with Illumina v1.5 flowcells using the Illumina cBot with the TruSeq Paired End Cluster Kit v3. Flowcells were sequenced as 100 base paired-end (non-indexed) reads on the Illumina HiSeq2000 using TruSeq SBS chemistry v3 to a target depth of 50× for the tumour samples and 30× for adjacent morphologically normal and blood samples. The Burrows-Wheeler Aligner (BWA) was used to align the sequencing data from each lane to the GRCh37 reference human genome32. Lanes that pass quality control are merged into a single well-annotated sample BAM file with duplicate reads removed. This data has been submitted to the European Genome-Phenome Archive (EGAD00001000689).
Mutation-Calling: Substitutions
CaVEMan (Cancer Variants Through Expectation Maximization), an in-house bespoke algorithm developed at the Sanger Institute, was used for calling somatic substitutions. CaVEMan utilises a Bayesian expectation maximization (EM) algorithm: Given the reference base, copy number status and fraction of aberrant tumor cells present in each cancer sample, CaVEMan generates a probability score for potential genotypes at each genomic position. A ‘somatic’ probability of 95% and above was applied as a cut off. Further post-processing filters were applied to eliminate false positive calls arising from genomic features that generate mapping errors and systematic sequencing artifacts. In addition to the standard filters applied in the Sanger pipeline we designed project-specific filters to improve the positive predictive value of our callers based on results from visually inspecting and calling many hundreds of variants. Visually inspecting involves checking that the variant was in at least three reads, not in any reads of control, no strand bias, no correlation of the reads containing the variant and read quality, not in a location where indels are also detected, not in a poorly mapped region, and not in a repeat region. Substitutions that are found in the WGS data of more than 2.5% of a batch of 465 normal non-malignant samples from a range of tissue types were also removed. Additional visual verification across all samples for a patient was performed for all non-intronic gene substitutions, all substitutions in adjacent morphologically normal samples, potential “field effect” substitutions, substitutions shared between adjacent morphologically normal and neoplastic samples, and the rare predicted substitutions apparently violating the inferred phylogeny.
Mutation-Calling: Insertions/Deletions
Insertions and deletions in the tumor, morphologically normal and matched blood control genomes were called using a modified Pindel version 0.2.0 on the NCBI37 genome build33. As with the substitutions, all standard Sanger pipeline filters were applied, as well as a custom filter built based on results from visually calling identified variants. Indels that were detected by Pindel in more than two samples from a series of hundreds of malignant non-prostate tissue were also removed. If an indel detected by Pindel that does not pass the filters is found in another sample for that patient and does pass all filters, it is also included. From those indels that passed all filters, for each sample, up to one hundred variants were validated by capillary sequencing. In addition, visual verification across all samples for a patient was performed for all indels occurring within genes, all indels in adjacent morphologically normal samples, potential “field effect” indels, those indels that were not supported by the phylogeny and a sampling of variants from each phylogeny relationship.
Mutation-Calling: Structural Variants
Brass (Breakpoints via assembly), an in-house bespoke algorithm developed at the Sanger Institute, was used for detecting structural variants. In Brass phase 1, discordant read pairs are detected and integrated to find regions of interest. These regions of interest are removed if they have been found in the matched blood normal sample, have been detected as germline in PCR validation of any other sample, have a low numbers of reads supporting them or appear to be in a “difficult” region of the genome. For a subset of regions, validation was performed by gel electrophoresis PCR using custom-designed PCR primers across the rearrangement breakpoint as previously described34 and for those products that give a band the precise location and nature of the breakpoint was determined by standard Sanger capillary sequencing methods. In the cases where the PCR experiments failed, Brass phase 2 was applied to the remaining predicted somatic structural variants. This gathers reads around the region, including half-unmapped reads and performs a local de novo assembly using Velvet35. Identifiable breakpoints have a distinctive De Bruijn graph pattern and allowed the breakpoint to be regenerated down to base pair resolution. Any breakpoints where an exact location could not be determined were removed. To ensure that breakpoints shared between samples in a patient were picked up, in-silico and PCR cross-sample experiments were performed. All breakpoints reported have been visually verified to ensure the presence of discordant reads and checked to ensure they were not in repeat regions.
To detect rearrangements involved in chromoplexy, a recently described process generating chained rearrangements we applied ChainFinder15. We used default parameters, selecting the rearrangements from 57 prostate genomes as background. As input copy number data, we used data derived from Affymetrix SNP 6.0 arrays, and processed using ASCAT36. As input structural variants, for each patient, we combined all high confidence breakpoints detected in all samples of that patient. One chained event was manually filtered, as it combined somatic rearrangements present in separate subpopulations in different samples, and hence could not have occurred as one chromoplexy event.
Mutation-Calling: Copy Number
The Battenberg algorithm was used to detect clonal and sub-clonal somatic copy number alterations (CNA) and estimate ploidy and tumour content from the NGS data as previously described9. Briefly, germline heterozygous SNPs are phased using Impute2 and a- and b- alleles assigned. Data is segmented using piecewise constant fitting37 and subclonal copy number segments are identified as those with deviations in the b-allele frequencies from the values expected when all cells have a common copy number in that segment, using a t-test. Ploidy and tumour content are estimated using the same method used by ASCAT36.
Construction of phylogenetic trees
For each patient, phylogenetic trees were constructed separately using (i) copy number aberrations (CNAs) and (ii) point mutations. Clonal and subclonal CNAs were identified using the previously described Battenberg algorithm9. This method achieves high sensitivity for the detection of CNAs found in small proportions of cells by phasing heterozygous SNPs into parent specific haplotype blocks. Joint analysis of SNPs within these blocks, rather than single SNPs, allows the resolution of CNAs found in ~5% of cells, with 30× sequencing depth. Matching of copy number and rearrangement breakpoints, supported by visual inspection of allele frequency and logR plots, was used to identify CNAs common to multiple samples. Point mutations were analysed using an adaptation of a previously described Bayesian Dirichlet process. Mutations within each sample are modelled as deriving from an unknown number of subclones, each of which is present at an unknown fraction of tumour cells and contributes an unknown proportion of all somatic mutations, with all the unknown parameters jointly estimated. In order to identify clusters of mutations that are common to 2 or more samples, the Dirichlet process was extended into 2 dimensions, with the fraction of tumour cells bearing a mutation in each of a pair of samples jointly estimated from the number of reads observed in each sample. The presence of clusters of unique or shared mutations can be inferred from the position of the peaks in the resulting 2-dimensional probability density.
Dirichlet process clustering
We used a previously developed Bayesian Dirichlet process to model clusters of clonal and subclonal point mutations, allowing inference of the number of subclones, the fraction of cells within each subclone and the number of mutations within each clone36. Within this model, the number of reads bearing the ith mutation, yi, is drawn from a binomial distribution
where Ni is the total number of reads at the mutated base and ζi is the expected fraction of reads that would report a mutation present in 100% of tumour cells at that locus. πi ∈ (0, 1), the fraction of tumour cells carrying the ith mutation, is modelled as coming from a Dirichlet process. We use the stick-breaking representation of the Dirichlet process:
where ωh is the weight of the hth mutation cluster, i.e. the proportion of all somatic mutations specific to that cluster. This model was extended into n dimensions, where n is the number of related samples, with the number of mutant reads obtained from each sample modelled as an independent binomial distribution, each with an independent π drawn with a Dirichlet process from a base distribution U(0,1). Gibbs sampling was used to estimate the posterior distribution of the parameters of interest, implemented in R, version 2.11.1. The Markov chain was run for 500 iterations, of which the first 100 were discarded. In order to plot the mutation density, each possible pair of related samples was treated separately. The median of the density was estimated from πh, each weighted by the associated value of ωh, using a bivariate Gaussian kernel, implemented in the R library KernSmooth. Median values were then plotted using the R function ‘levelplot’, using a colour palette graduated from white (low probability of a mutation) to red (high probability of a mutation).
Targeted PCR and MiSeq sequencing of selected mutations and structural variants
PCR primers for somatic substitutions and indels were designed using Primer-Z38, with known SNPs and human repeats masked. All amplicons were designed to be a maximum of 500 bp and all variants of interest were checked to be within a read generated on a 2×250bp MiSeq run. DNA was amplified using Phusion HotStart II DNA polymerase kit (Thermo Fisher Scientific) and thermo cycler. DNA was denatured at 98 °C for 30 seconds followed by 30 cycles of denaturing at 98 °C for 10 seconds, annealing at 65 °C for 20 seconds and extension at 72 °C for 20 seconds. Products were incubated at 72 °C for 5 minutes before cooling to 4 °C. All PCR products were analysed using 96 well 2% agarose E-gel with ethidium bromide (Life Technologies). If no detectable band was present these reactions were repeated using an annealing temperature of 60 °C. 2 μl of PCR mixture for each sample of DNA were pooled. Pooled DNA was diluted 1:10, and tagged with an individual barcode (Fluidigm) using Expand High Fidelity PCR System (Roche), following manufacturers protocol (Access Array System for Illumina Systems User Guide). DNA was denatured at 98 °C for 1 minute followed by 15 cycles of denaturing at 98 °C for 15 seconds, annealing at 60 °C for 30 seconds and extension at 72 °C for 1 minute. Products were incubated at 72 °C for 3 minutes before cooling to 4 °C. Barcoded PCR samples were pooled for each patient and analysed using 2100 Bioanalyzer (Agilent) to determine the average size of the PCR library and by KAPA SYBR FAST qPCR (Anachem) to determine the library concentration. 2 nM of each sample was analysed using MiSeq (Illumina).
The average sequencing depth across all mutations assessed within each patient varied between 4900 (in 8_T1) and 16600 (in 7_T4). However, for around a fifth of the targeted mutations within each patient, the average coverage across all samples from that patient was very much lower, 200 or lower. Many of these low coverage mutations had mutant allele frequencies very different from the values obtained from whole genome sequencing (WGS). These PCRs were considered to have failed and were not included in subsequent analysis.
Due to the very high coverage, a low rate of sequencing errors was observed for most mutations. This manifested as a small percentage of aberrant reads, peaked close to zero and rapidly decaying exponentially with allele fraction. The rate of these errors was evaluated by considering those samples that reported no mutant reads in WGS. For this purpose, only mutations that were identified in samples that were previously identified as being phylogenetically related were included, in order to filter out low quality or questionable calls. Allele frequencies, fs, were converted to mutation copy numbers, nmut, as previously described39.
where ρ, and are, respectively, the tumor purity, the locus-specific copy number in the blood normal cells, inferred from the Battenberg algorithm. Mutation copy numbers correspond to the fraction of cells bearing a mutation multiplied by the number of chromosomal copies bearing the mutation and are more informative than raw allele frequencies as they are adjusted for tumour ploidy and normal cell contamination. The distribution of misreads was then found to have similar distributions for the different patients, with average reported mutation copy numbers of 0.0059 ± 0.0072, 0.0032 ± 0.0070 and 0.0037 ± 0.0035 in patients 6, 7 and 8, respectively. The highest reported mutation copy number for these mutations was 0.041. This value was therefore used as a threshold for distinguishing between mutations present in a small proportion of cells and misreads arising from sequencing errors. It should be noted that a mutation copy number of 0.041 corresponds to an allele frequency of ~1% for most mutations, since most mutations occur in diploid regions of the genome and the average tumour content across the samples is below 50%.
For samples 6_T2, 6_T3 and 6_T4, it was apparent that nearly all mutations that were present in 6_T1 were identified at allele fractions slightly above the threshold used to exclude artefacts (corresponding to a mutation copy number ~0.05). Since these mutations were exclusively those present in 6_T1, it appears that ‘contamination’ of these 3 samples by 6_T1 occurred at some point during the PCR experiment, although whether this contamination is physical or the result of bleed-through of tags used in multiplexing is unknown. Assessment of WGS data, by checking the allele frequency of mutations identified uniquely in 6_T1 in samples 6_T2, 6_T3 and 6_T4, indicated that there may have been some intermixing of the cells 6_T1 with 6_T2, corresponding to a much lower percentage of cells (1.8%) and possibly arising from growth of cells in 6_T1 into the region sampled in 6_T2. Further, no evidence for intermixing of 6_T1 with 6_T3 or 6_T4 was found in WGS data. For this reason, mutations apparently present in the PCR experiment in 6_T2, 6_T3 and 6_T4 and identified in 6_T1 in both WGS and PCR were only considered to be validated if they fell above a higher threshold, set to a mutation copy number of 0.2, that excluded mutant reads arising from the contamination of these samples.
Mutational Signatures
The mutational spectra, as defined by the triplets of nucleotides around each mutation, of each sample was deconvoluted into mutational processes as described12,13.
Clustering of Mutations
We investigated regional clustering of substitution mutations by constructing plots (“rainfall plots”) in which the distance between each somatic substitution, and the substitution immediately before it has been plotted for each mutation. This was achieved exactly as described previously9.
Supplementary Material
ACKNOWLEDGEMENTS
This work is funded by Cancer Research UK Grant C5047/A14835, by the Dallaglio Foundation, and by The Wellcome Trust. We also acknowledge support from the Bob Champion Cancer Trust, The Orchid Cancer appeal, The RoseTrees Trust, The North West Cancer Research Fund, Big C, The King family, The Grand Charity of Freemasons, and the Research Foundation Flanders (FWO). We thank Dave Holland from the Infrastructure Management Team and Peter Clapham from the Informatics Systems Group at the Wellcome Trust Sanger Institute. We acknowledge the Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust supported by the National Institute for Health Research. We acknowledge support of the National Cancer Research Prostate Cancer: Mechanisms of Progression and Treatment (PROMPT) collaborative (Grant G0500966/75466). We thank the National Institute for Health Research, Hutchison Whampoa Limited and the Human Research Tissue Bank (Addenbrookes Hospital), the Cancer Research UK Cambridge Research Institute Histopathology, the In-situ Hybridisation Core Facility, the Genomics Core Facility Cambridge, and the Cambridge University Hospitals Media Studio.
Footnotes
COMPETING FINANCIAL INTEREST
Ros Eeles has received educational grants from Illumina and GenProbe (formerly Tepnel), Vista Diagnostics and Janssen Pharmaceuticals. She has received honoraria from Succint Communications for talks on prostate cancer genetics.
ACCESSION CODES
The sequencing data have been submitted to the European Genome-Phenome Archive (EGAD00001000689).
REFERENCE LIST
- 1.Andreoiu M, Cheng L. Multifocal prostate cancer: biologic, prognostic, and therapeutic implications. Hum. Pathol. 2010;41:781–793. doi: 10.1016/j.humpath.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 2.Cheng L, et al. Evidence of independent origin of multiple tumors from patients with prostate cancer. J. Natl. Cancer Inst. 1998;90:233–237. doi: 10.1093/jnci/90.3.233. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi M, et al. Molecular analysis of multifocal prostate cancer by comparative genomic hybridization. Prostate. 2008;68:1715–1724. doi: 10.1002/pros.20832. [DOI] [PubMed] [Google Scholar]
- 4.Boyd LK, et al. High-resolution genome-wide copy-number analysis suggests a monoclonal origin of multifocal prostate cancer. Genes Chromosomes Cancer. 2012;51:579–589. doi: 10.1002/gcc.21944. [DOI] [PubMed] [Google Scholar]
- 5.Lindberg J, et al. Exome sequencing of prostate cancer supports the hypothesis of independent tumour origins. Eur. Urol. 2013;63:347–353. doi: 10.1016/j.eururo.2012.03.050. [DOI] [PubMed] [Google Scholar]
- 6.Clark J, et al. Complex patterns of ETS gene alteration arise during cancer development in the human prostate. Oncogene. 2008;27:1993–2003. doi: 10.1038/sj.onc.1210843. [DOI] [PubMed] [Google Scholar]
- 7.Attard G, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27:253–263. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaisa NT, et al. Clonal architecture of human prostatic epithelium in benign and malignant conditions. J. Pathol. 2011;225:172–180. doi: 10.1002/path.2959. [DOI] [PubMed] [Google Scholar]
- 9.Nik-Zainal S, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Svensson MA, et al. Testing mutual exclusivity of ETS rearranged prostate cancer. Lab. Invest. 2011;91:404–412. doi: 10.1038/labinvest.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baca SC, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weischenfeldt J, et al. Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell. 2013;23:159–170. doi: 10.1016/j.ccr.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Holstege H, et al. Somatic mutations found in the healthy blood compartment of a 115-yr-old woman demonstrate oligoclonal hematopoiesis. Genome Res. 2014;24:733–742. doi: 10.1101/gr.162131.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mucci NR, et al. Expression of nuclear antigen Ki-67 in prostate cancer needle biopsy and radical prostatectomy specimens. J. Natl. Cancer Inst. 2000;92:1941–1942. doi: 10.1093/jnci/92.23.1941. [DOI] [PubMed] [Google Scholar]
- 22.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 23.Leemans CR, Braakhuis BJM, Brakenhoff RH. The molecular biology of head and neck cancer. Nat. Rev. Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 24.Dworkin AM, Huang TH-M, Toland AE. Epigenetic alterations in the breast: Implications for breast cancer detection, prognosis and treatment. Semin. Cancer Biol. 2009;19:165–171. doi: 10.1016/j.semcancer.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karavitakis M, Ahmed HU, Abel PD, Hazell S, Winkler MH. Tumor focality in prostate cancer: implications for focal therapy. Nat Rev Clin Oncol. 2011;8:48–55. doi: 10.1038/nrclinonc.2010.190. [DOI] [PubMed] [Google Scholar]
- 26.Tareen B, Godoy G, Taneja SS. Focal therapy: a new paradigm for the treatment of prostate cancer. Rev Urol. 2009;11:203–212. [PMC free article] [PubMed] [Google Scholar]
- 27.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang X, Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web. J. Med. Genet. 2012;49:433–436. doi: 10.1136/jmedgenet-2012-100918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 31.Warren AY, et al. Method for sampling tissue for research which preserves pathological data in radical prostatectomy. Prostate. 2013;73:194–202. doi: 10.1002/pros.22556. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Loo P, et al. Allele-specific copy number analysis of tumors. Proc. Natl. Acad. Sci. U.S.A. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nilsen G, et al. Copynumber: Efficient algorithms for single- and multi-track copy number segmentation. BMC Genomics. 2012;13:591. doi: 10.1186/1471-2164-13-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai M-F, et al. PrimerZ: streamlined primer design for promoters, exons and human SNPs. Nucleic Acids Res. 2007;35:W63–5. doi: 10.1093/nar/gkm383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stephens PJ, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.