Abstract
Though most follicular lymphoma biomarkers rely on tumor features, the host genetic background may also be relevant for outcome. Here we aimed at verifying the contribution of candidate polymorphisms of FCγ receptor, DNA repair and detoxification genes to prognostic stratification of follicular lymphoma treated with immunochemotherapy. The study was based on 428 patients enrolled in the FOLL05 prospective trial that compared three standard-of-care regimens (rituximab-cyclophosphamide-vincristine-prednisone versus rituximab-cyclophosphamide-doxorubicin-vincristine-prednisone versus rituximab-fludarabine-mitoxantrone) for the first line therapy of advanced follicular lymphoma. Polymorphisms were genotyped on peripheral blood DNA samples. The primary endpoint was time to treatment failure. Polymorphisms of FCGR2A and FCGR3A, which have been suggested to influence the activity of rituximab as a single agent, did not affect time to treatment failure in the pooled analysis of the three FOLL05 treatment arms that combined rituximab with chemotherapy (P=0.742, P=0.252, respectively). These results were consistent even when the analysis was conducted by intention to treat, indicating that different chemotherapy regimens and loads did not interact differentially with the FCGR2A and FCGR3A genotypes. The genotype of MLH1, which regulates the genotoxic effect of doxorubicin, significantly affected time to treatment failure in patients in the rituximab-cyclophosphamide-doxorubicin-vincristine-prednisone arm (P=0.001; q<0.1), but not in arms in which patients did not receive doxorubicin (i.e., the rituximab-cyclophosphamide-vincristine-prednisone and rituximab-fludarabine-mitoxantrone arms). The impact of MLH1 on time to treatment failure was independent after adjusting for the Follicular Lymphoma International Prognostic Index and other potential confounding variables by multivariate analysis. These data indicate that MLH1 genotype is a predictor of failure to benefit from rituximab-cyclophosphamide-doxorubicin-vincristine-prednisone treatment in advanced follicular lymphoma and confirm that FCGR2A and FCGR3A polymorphisms have no impact when follicular lymphoma is treated with rituximab plus chemotherapy (clinicaltrials.gov identifier: NCT00774826).
Introduction
The current standard of treatment for advanced follicular lymphoma (FL) is immunochemotherapy, which combines the anti-CD20 monoclonal antibody rituximab with a variety of multiagent chemotherapy regimens incorporating anthracyclines (e.g. doxorubicin), anthracenediones (e.g. mitoxantrone), alkylating agents (e.g. cyclophosphamide), or purine analogues (e.g. fludarabine).1,2 A number of clinical markers have been proposed as tools for refining survival prognostication in FL, most of which rely on the features of the tumor clone.1,2 In contrast, a limited set of biomarkers is available to predict treatment outcome in patients with this lymphoma.3,4
The activity of drugs employed for the treatment of FL may be affected by the patient’s genetic background. The anti-tumor effect of monoclonal antibodies may be modulated by polymorphisms of the FCγ receptors, which are expressed on cells responsible for antibody-dependent cell-mediated cytotoxicity and are devoted to attracting and activating the immune response against antibody-coated tumor cells.5 While FCγ receptor polymorphisms may influence the outcome of rituximab monotherapy in FL, their role in the context of immunochemotherapy is questionable.6–15 The therapeutic activity of doxorubicin may be modulated by a polymorphism of MLH1,16 a molecule that is involved in the induction of cell cycle arrest and apoptosis in response to the DNA damage produced by doxorubicin.17,18 The outcome of doxorubicin-based chemotherapy is also affected by functional polymorphisms of CYBA, a subunit of the NADPH oxidase complex that produces reactive oxygen species in response to chemotherapy.19,20 The therapeutic activity of cyclophosphamide is dependent on polymorphisms of genes deputed to its detoxification, such as GSTA1.19
The FOLL05 study compared three standard-of-care regimens for the first-line therapy of advanced FL.21 Patients were randomized to receive rituximab plus cyclophosphamide, vincristine, and prednisone (R-CVP), or rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP), or rituximab plus fludarabine and mitoxantrone (R-FM). The FOLL05 study showed that R-CHOP and R-FM are superior to R-CVP in terms of time to treatment failure (TTF).21
We took advantage of the FOLL05 study to clarify the role of FCγ receptor polymorphism in advanced FL patients treated with rituximab-based immunochemotherapy and to assess whether MLH1, CYBA and GSTA1 polymorphisms selectively predict the outcome of a specific immunochemotherapy regimen.
Methods
Patients
Peripheral blood samples were prospectively obtained from 428/504 (84.9%) untreated advanced FL patients enrolled in the multicenter, randomized FOLL05 study (Table 1; Figure 1).21 The study was designed to assess differences in TTF, which was the primary endpoint of the FOLL05 study (see Online Supplementary Appendix).21,22 The REMARK and STREGA guidelines were followed throughout this study.23,24 FOLL05 (clinicaltrials.gov identifier: NCT00774826) was conducted in compliance with the Declaration of Helsinki, was approved by the appropriate research ethics committee, required each patient to provide written informed consent and also included centralization of DNA from patients’ samples for ancillary studies.
Table 1.
Clinical features by availability of biological samples for genotypinga.
Figure 1.
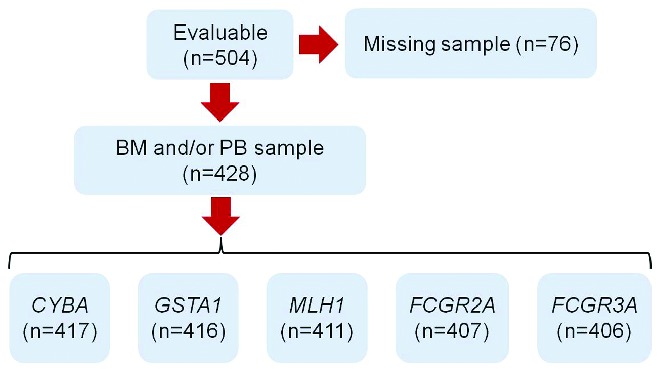
CONSORT diagram representing the number of patients included in the analysis. BM: bone marrow; PB: peripheral blood.
Single nucleotide polymorphism genotyping
Genomic DNA was extracted from peripheral blood samples. Genotyping of the FCGR2A rs1801274, FCGR3A rs396991, CYBA rs4673, and GSTA1 rs3957357 single nucleotide polymorphisms (SNP) was performed on high molecular weight genomic DNA by SNP minisequencing (ABI Prism SNaPshot Multiplex kit, Applied Biosystems, Foster City, CA), after validation of this approach by DNA direct sequencing of each SNP in a pilot panel of cases (n=40). Genotyping of the MLH1 rs1799977 SNP was performed on high molecular weight genomic DNA by Sanger sequencing. Details are provided in the Online Supplementary Appendix. Quality control of genotyping was performed by replicate sample analysis (100% concordance in replicates for all the candidate SNP). Deviation of SNP genotype distribution from the Hardy-Weinberg equilibrium was tested by the χ2 test or Fisher exact test if appropriate. SNP genotyping was performed blind to the study endpoint.
Statistical analysis
TTF, the primary endpoint of the study, was evaluated according to the intention-to-treat principle and was defined as the time from study entry to last follow-up or to the first of the following events: less than partial remission, a shift to a different therapy for any reason after at least one cycle of treatment, progressive disease, relapse, or death.21 Molecular studies were blinded to the study endpoints. Analysis of TTF was performed by the Kaplan-Meier method using the log-rank test to assess differences between genotype groups.25 The false discovery rate was used to control for multiple statistical testing.26 Cox regression analysis was used to estimate genotype-specific hazard ratios and 95% confidence intervals, adjusting for potentially confounding covariates.27 For each SNP genotype, the hazard ratios were generated using common allele homozygotes as the reference group. For SNP with ten or fewer minor allele homozygotes, only the combination of minor allele homozygotes with heterozygotes was analyzed. If this combined frequency was still less than ten, the SNP was removed from the analysis. Proportional hazard regression assumptions were assessed appropriately. Bias corrected c-index, calibration slope and heuristic shrinkage estimator of the Cox model were calculated.28 Cox model stability was internally validated using bootstrapping procedures.29–31 These approaches provided an estimate of the prediction accuracy of the Cox model to protect against overfitting. Categorical variables were compared by the χ2 test and exact test, when appropriate. All statistical tests were two-sided. Statistical significance was defined as a P value <0.05 and a q value <0.1. The analysis was performed with SPSS v.21.0 and with the R statistical package 3.0.1 (http://www.r-project.org).
Results
FCγ receptor polymorphisms have no prognostic impact when advanced follicular lymphoma is treated with chemoimmunotherapy
The clinical features of the 428 patients with advanced FL available for SNP genotyping (84.9% of the whole FOLL05 study cohort; Figure 1) did not differ from those of the 76 patients not available for genotyping (Table 1). These data indicate that the lack of biological material for genotyping was not due to an unintended selection bias. Out of the 428 genotyped cases, the FCGR2A and FCGR3A polymorphisms were assessable in 407 and 406 patients, respectively (Figure 1). In the remaining cases, the quality and/or quantity of genomic DNA prevented its amplification and sequencing. The distributions of the FCGR2A and FCGR3A polymorphisms were in Hardy- Weinberg equilibrium, thus excluding poor genotyping or population biases (Online Supplementary Table S1). Patients’ characteristics at diagnosis as well as treatment allocation distributed without significant differences across the three genotypes of the FCGR2A and FCGR3A polymorphisms (Online Supplementary Table S2 and S3).
It was planned that all FL patients enrolled in the FOLL05 study, independently of the treatment arm, were to receive eight doses of rituximab combined with chemotherapy.21 The impact of FCGR2A and FCGR3A genotypes on the primary clinical endpoint of the study (i.e. TTF) was, therefore, initially assessed in the whole study cohort. By pooled analysis of the three treatment arms, TTF was not influenced by the FCGR2A (P=0.742) and FCGR3A (P=0.252) genotypes (Table 2; Figure 2). FCGR2A and FCGR3A genotypes did not influence TTF either in clinical subgroups defined by disease bulk or patients’ gender, which might affect disease sensitivity to rituximab, or in groups with different prognoses according to the Follicular Lymphoma International Prognostic Index (FLIPI) (Online Supplementary Figures S1, S2 and S3). The overall response rate also distributed without significant differences across the three genotypes of the FCGR2A (P=0.994) and FCGR3A (P=0.606) polymorphisms. By multivariate analysis, FLIPI and treatment allocation, but not FCGR2A and FCGR3A genotypes, were independent predictors of TTF (Table 2), thus confirming that the FOLL05 study population included in this genotype-phenotype association analysis is representative of patients with advanced FL.
Table 2.
Univariate and multivariate analyses for TTF in the whole study cohort.
Figure 2.
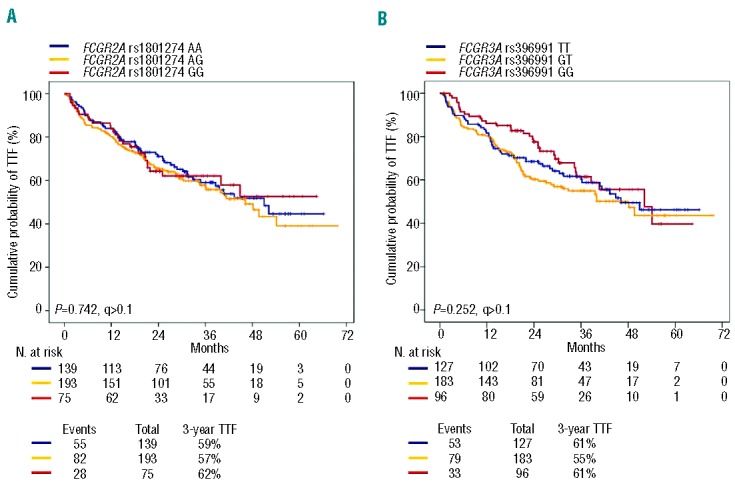
Kaplan-Meier estimates of time to treatment failure in the pooled treatment arms according to FCGR2A and FCGR3A genotypes. (A) Comparison of time to treatment failure (TTF) between patients homozygous for the common FCGR2A rs1801274 allele (blue line), patients heterozygous for the FCGR2A rs1801274 genotype (yellow line), and patients homozygous for the variant FCGR2A rs1801274 allele (red line). (B) Comparison of TTF between patients homozygous for the common FCGR3A rs396991 allele (blue line), patients heterozygous for the FCGR3A rs396991 genotype (yellow line), and patients homozygous for the variant FCGR3A rs396991 allele (red line). P: P values by log-rank test.
Patients enrolled in the FOLL05 study were randomized to receive different loads of chemotherapy combined with rituximab, with the lowest load being in the R-CVP arm.21 In order to verify whether different chemotherapy regimens and loads might interact differentially with FCGR2A and FCGR3A genotypes, the impact of these SNP on TTF was also assessed by treatment arm. However, even when the analysis was conducted by intention-to-treatment arm, TTF did not differ according to FCGR2A and FCGR3A genotypes (Online Supplementary Figure S1 and S2).
LIke FCγ SNP, polymorphisms of GSTA1 and CYBA also had no role in FL outcome prediction (Online Supplementary Figure S4).
The genotype of MLH1 is a predictor of R-CHOP treatment failure in advanced follicular lymphoma
The MLH1 polymorphism was assessable in 411 FL patients (Figure 1), and its distribution was in Hardy-Weinberg equilibrium (Online Supplementary Table S1). Among the drugs utilized in the FOLL05 study, MLH1 is known to regulate the genotoxic effects of doxorubicin.17,18 According to this biological rationale, the clinical impact of the MLH1 polymorphism was initially assessed in FL patients randomized to the R-CHOP arm. Among FL patients allocated to R-CHOP, characteristics at diagnosis distributed without significant differences across the three genotypes of the MLH1 polymorphism, with the sole exception of a trend towards a more frequent involvement of more than one extranodal site in patients homozygous for the variant allele (Online Supplementary Table S4).
Univariate analysis for TTF, controlled for multiple comparisons by false discovery rate testing, identified the MLH1 polymorphism as a predictor of R-CHOP treatment failure in advanced FL (P=0.011; q<0.1) (Table 3; Figure 3A). After R-CHOP treatment, FL patients who carried the homozygous GG variant genotype of MLH1 showed a significantly lower 3-year TTF (30.3%) compared to patients who carried the MLH1 AG (3-year TTF: 66.2%) or AA (3-year TTF: 68.8%) genotypes (P=0.010 and P=0.003, respectively, in the pairwise comparisons) (Figure 3A). Consistent with the selective involvement of MLH1 in doxorubicin pharmacodynamics, the MLH1 polymorphism did not affect the outcome of FL patients treated with regimens lacking this drug (i.e. R-CVP and R-FM) (Online Supplementary Figure S3). By multivariate analysis, FL patients who carried the homozygous GG variant genotype of MLH1 displayed a 2.8-fold increase in risk of failing to benefit from R-CHOP (hazard ratio: 2.81; 95% confidence interval: 1.18–6.73; P=0.020), after adjusting for clinically relevant covariates including FLIPI, number of extranodal sites, bone marrow involvement and raised level of beta-2-microglobulin (Table 3). The increased risk of failing to benefit from R-CHOP translated into a significantly shorter overall survival in FL patients harboring the homozygous GG variant genotype of MLH1 (Figure 3B).
Table 3.
Univariate and multivariate analyses for TTF in patients treated with R-CHOP.
Figure 3.
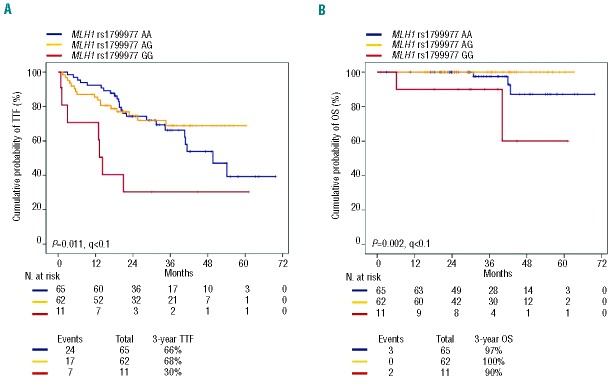
Kaplan-Meier estimates of time to treatment failure and overall survival in patients randomized to the R-CHOP arm according to the MLH1 rs1799977 genotype. (A) Comparison of time to treatment failure (TTF) between patients homozygous for the common MLH1 rs1799977 allele (blue line), patients heterozygous for the MLH1 rs1799977 genotype (yellow line), and patients homozygous for the variant MLH1 rs1799977 allele (red line). (B) Comparison of overall survival (OS) between patients homozygous for the common MLH1 rs1799977 allele (blue line), patients heterozygous for the MLH1 rs1799977 genotype (yellow line), and patients homozygous for the variant MLH1 rs1799977 allele (red line). P: P values by log-rank test; q, q values by false discovery rate.
The genotype of MLH1 predicts reduced benefit from the addition of doxorubicin to treatments for follicular lymphoma
The FOLL05 randomized study demonstrated that R-CHOP significantly improves TTF compared to R-CVP in patients with previously untreated FL, thus documenting a relevant clinical benefit when doxorubicin is added to the R-CVP backbone in this type of lymphoma.21 Consistent with these clinical data, the addition of doxorubicin to the R-CVP backbone resulted in a significant improvement in the 3-year TTF (19% increase; P=0.002) in FL patients harboring the common allele of MLH1 (AA and AG genotypes) (Figure 4A). In contrast, FL patients who carried the homozygous GG variant genotype of MLH1 (~10% of the FOLL05 population) did not gain benefit from doxorubicin (Figure 4B).
Figure 4.
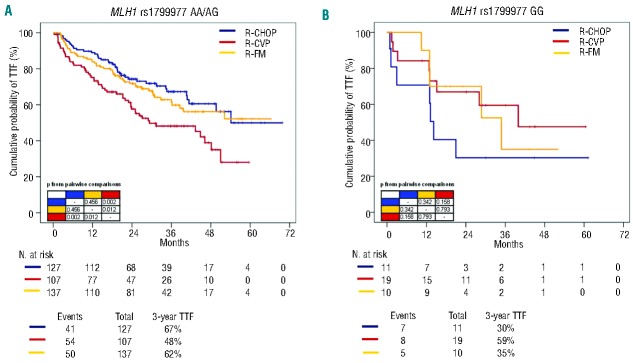
Kaplan-Meier estimates of time to treatment failure stratified according to the MLH1 rs1799977 genotype and treatment randomization. (A) Comparison of time to treatment failure (TTF) between R-CHOP (blue line), R-CVP (red line) and R-FM (yellow line) among patients harboring the MLH1 rs1799977 AA/AG genotype. (B) Comparison of time to treatment failure (TTF) between R-CHOP (blue line), R-CVP (red line) and R-FM (yellow line) among patients harboring the MLH1 rs1799977 GG genotype. P, P values by log-rank test.
Discussion
This large prospective substudy of the FOLL05 trial, shows that: (i) FCγ receptor polymorphisms do not have a prognostic impact when advanced FL is treated with chemoimmunotherapy; and (ii) the MLH1 genotype is a predictor of failure of R-CHOP treatment in FL.
Although several small studies in FL have shown that FCγ receptor polymorphisms may be useful in predicting response to single agent rituximab, their clinical impact in the setting of immunochemotherapy is still controversial.6–14 Our study is the most complete prospective examination of the effects of FCγ polymorphisms on the outcome of advanced FL patients treated with rituximab combined with chemotherapy. Consistent with the data from the PRIMA study,15 our analysis definitively indicates that FCγ receptor polymorphisms have no prognostic impact when advanced FL is treated with chemoimmunotherapy, independently of the tumor burden and the type and load of drugs that are combined with rituximab. Therefore, FCγ SNP must not be further considered and implemented as biomarkers in the setting of advanced FL treated with immunochemotherapy.
The MLH1 polymorphism is a predictor of R-CHOP treatment failure in advanced FL. Consistent with the selective involvement of MLH1 in doxorubicin pharmacodynamics,16–18 the MLH1 genotype did not affect the outcome of FL patients treated with regimens lacking this drug. The selective association between MLH1 genotype and outcome after R-CHOP has been clinically validated in independent retrospective series of lymphoma patients, including two retrospective cohorts of patients with diffuse large B-cell lymphoma and this prospective FL series.16 Overall, these notions point to MLH1 genotype as a predictor of R-CHOP failure in B-cell lymphoma.
The mechanistic explanation of the phenotype observed in FL patients who carried the homozygous GG variant genotype of MLH1 remains to be clarified. In other disease models, the MLH1 rs1799977 polymorphism associates with reduced MLH1 protein expression in tumor cells.16,32,33 Alternatively, the MLH1 rs1799977 polymorphism might be in linkage disequilibrium with other functionally relevant SNP of the MLH1 gene,34 suggesting that multiple variants within the MLH1 locus may contribute to the risk of treatment failure in FL.
The association between CYBA and GSTA1 SNP and outcome of R-CHOP treatment was not replicated in the current study cohort. It is likely that moderate sample size, inter-subtype and other genetic heterogeneity, as well as small true effect sizes account for the lack of replication. Alternatively, the lack of replication might be the consequence of the false positive report probability that is known to affect candidate gene association studies, and indicates that, at variance with MLH1 rs1799977, neither CYBA nor GSTA1 SNP represent prognosticators in B-cell lymphomas treated with R-CHOP.
Despite the limitations imposed by the sample size, our data provide a signal of reduced benefit from the addition of doxorubicin to the R-CVP backbone in FL patients harboring the homozygous GG variant genotype of MLH1. Conversely, the MLH1 genotype has no clinical relevance in FL patients treated with R-FM, which, in turns, seems equally effective as R-CHOP in the setting of advanced FL. R-FM might, therefore, represent a suitable initial chemotherapy approach for FL patients carrying the homozygous GG variant genotype of MLH1. Replication of these findings in other cohorts of FL patients will be necessary to assess their generalizability.
The FOLL05 study was designed before the establishment of rituximab maintenance as a standard of care for advanced FL.2 Furthermore, the FOLL05 study did not include bendamustine-based regimens among its treatment strategies.2 These facts should prompt investigations aimed at clarifying whether maintenance after initial R-CHOP or bendamustine-based immunochemotherapy might abrogate the prognostic impact of the MLH1 genotype.
Acknowledgments
This study was supported by Special Program Molecular Clinical Oncology 5 × 1000 N. 10007 and My First AIRC Grant N. 13470 Associazione Italiana per la Ricerca sul Cancro Foundation Milan, Italy; Progetto Giovani Ricercatori 2010, Grant N. GR-2010-2317594, Ministero della Salute, Rome, Italy; Compagnia di San Paolo, Grant N. PMN_call_2012_0071, Turin, Italy; Fondazione Cariplo, Grant N. 2012-0689; Futuro in Ricerca 2012 Grant N. RBFR12D1CB, and the Ministero dell’Istruzione, dell’Università e della Ricerca, Rome, Italy.
Footnotes
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Ghielmini M, Vitolo U, Kimby E, et al. ESMO Guidelines consensus conference on malignant lymphoma 2011 part 1: diffuse large B-cell lymphoma (DLBCL), follicular lymphoma (FL) and chronic lymphocytic leukemia (CLL). Ann Oncol. 2013;24(3):561–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zelenetz AD, Wierda WG, Abramson JS, et al. Non-Hodgkin’s lymphomas, version 1.2013. J Natl Compr Canc Netw. 2013;11(3):257–272. [DOI] [PubMed] [Google Scholar]
- 3.Smith SM. Dissecting follicular lymphoma: high versus low risk. Hematology Am Soc Hematol Educ Program. 2013:561–567. [DOI] [PubMed] [Google Scholar]
- 4.Kridel R, Sehn LH, Gascoyne RD. Pathogenesis of follicular lymphoma. J Clin Invest. 2012;122(10):3424–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ravetch JV, Bolland S. IgG Fc receptors. Annu Rev Immunol. 2001;19:275–290. [DOI] [PubMed] [Google Scholar]
- 6.Cartron G, Dacheux L, Salles G, et al. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood. 2002;99(3):754–758. [DOI] [PubMed] [Google Scholar]
- 7.Weng WK, Levy R. Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J Clin Oncol. 2003;21(21):3940–3947. [DOI] [PubMed] [Google Scholar]
- 8.Ghielmini M, Rufibach K, Salles G, et al. Single agent rituximab in patients with follicular or mantle cell lymphoma: clinical and biological factors that are predictive of response and event-free survival as well as the effect of rituximab on the immune system: a study of the Swiss Group for Clinical Cancer Research (SAKK). Ann Oncol. 2005;16(10):1675–1682. [DOI] [PubMed] [Google Scholar]
- 9.Carlotti E, Palumbo GA, Oldani E, et al. FcgammaRIIIA and FcgammaRIIA polymorphisms do not predict clinical outcome of follicular non-Hodgkin’s lymphoma patients treated with sequential CHOP and rituximab. Haematologica. 2007;92(8):1127–1130. [DOI] [PubMed] [Google Scholar]
- 10.Cartron G, Ohresser M, Salles G, Solal-Céligny P, Colombat P, Watier H. Neutrophil role in in vivo anti-lymphoma activity of rituximab: FCGR3B-NA1/NA2 polymorphism does not influence response and survival after rituximab treatment. Ann Oncol. 2008;19(8):1485–1487. [DOI] [PubMed] [Google Scholar]
- 11.Weng WK, Levy R. Genetic polymorphism of the inhibitory IgGFc receptor FcgammaRIIb is not associated with clinical outcome in patients with follicular lymphoma treated with rituximab. Leuk Lymphoma. 2009;50(5):723–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weng WK, Weng WK, Levy R. Immunoglobulin G Fc receptor polymorphisms do not correlate with response to chemotherapy or clinical course in patients with follicular lymphoma. Leuk Lymphoma. 2009;50(9):1494–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prochazka V, Papajik T, Gazdova J, et al. FcRIIIA receptor genotype does not influence an outcome in patients with follicular lymphoma treated with risk-adapted immunochemotherapy. Neoplasma. 2011;58(3):263–270. [DOI] [PubMed] [Google Scholar]
- 14.Persky DO, Dornan D, Goldman BH, et al. Fc gamma receptor 3a genotype predicts overall survival in follicular lymphoma patients treated on SWOG trials with combined monoclonal antibody plus chemotherapy but not chemotherapy alone. Haematologica. 2012;97(6):937–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghesquières H, Cartron G, Seymour JF, et al. Clinical outcome of patients with follicular lymphoma receiving chemoimmunotherapy in the PRIMA study is not affected by FCGR3A and FCGR2A polymorphisms. Blood. 2012;120(13):2650–2657. [DOI] [PubMed] [Google Scholar]
- 16.Rossi D, Rasi S, Di Rocco A, et al. The host genetic background of DNA repair mechanisms is an independent predictor of survival in diffuse large B-cell lymphoma. Blood. 2011;117(8):2405–2413. [DOI] [PubMed] [Google Scholar]
- 17.Brown R, Hirst GL, Gallagher WM, et al. hMLH1 expression and cellular responses of ovarian tumour cells to treatment with cytotoxic anticancer agents. Oncogene. 1997;15(1):45–52. [DOI] [PubMed] [Google Scholar]
- 18.Fedier A, Schwarz VA, Walt H, Carpini RD, Haller U, Fink D. Resistance to topoisomerase poisons due to loss of DNA mismatch repair. Int J Cancer. 2001;93(4):571–576. [DOI] [PubMed] [Google Scholar]
- 19.Rossi D, Rasi S, Franceschetti S, et al. Analysis of the host pharmacogenetic background for prediction of outcome and toxicity in diffuse large B-cell lymphoma treated with R-CHOP21. Leukemia. 2009;23(6):1118–1126. [DOI] [PubMed] [Google Scholar]
- 20.Hoffmann M, Schirmer MA, Tzvetkov MV, et al. A functional polymorphism in the NAD(P)H oxidase subunit CYBA is related to gene expression, enzyme activity, and outcome in non-Hodgkin lymphoma. Cancer Res. 2010;70(6):2328–2338. [DOI] [PubMed] [Google Scholar]
- 21.Federico M, Luminari S, Dondi A, et al. R-CVP versus R-CHOP versus R-FM for the initial treatment of patients with advanced-stage follicular lymphoma: results of the FOLL05 trial conducted by the Fondazione Italiana Linfomi. J Clin Oncol. 2013;31(12):1506–1513. [DOI] [PubMed] [Google Scholar]
- 22.Cheson BD, Pfistner B, Juweid ME, et al. Revised response criteria for malignant lymphoma. J Clin Oncol. 2007;25(5):579–586. [DOI] [PubMed] [Google Scholar]
- 23.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol. 2006;100(2):229–235. [DOI] [PubMed] [Google Scholar]
- 24.Little J, Higgins JP, Ioannidis JP, et al. STrengthening the REporting of Genetic Association studies (STREGA): an extension of the STROBE statement. Ann Intern Med. 2009;150(3):206–215. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. Am Stat Assoc. 1958;53(282):457–481. [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling false discovery rate: a practicable and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 27.Cox DR. Regression models and life tables. J R Stat Assoc. 1972;34:187–220. [Google Scholar]
- 28.Harrell FE, Jr, Lee K, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15(4):361–387. [DOI] [PubMed] [Google Scholar]
- 29.Efron B, Tibshirani R. Improvements on cross-validation: the .632_bootstrap method. JASA. 1997;92:548–560. [Google Scholar]
- 30.Van Howelingen JC, le Cessie S. Predictive value of statistical models. Stat Med. 1990;9(11):1303–1325. [DOI] [PubMed] [Google Scholar]
- 31.Chen CH, George SL. The bootstrap and identification of prognostic factors via Cox’s proportional hazards regression model. Stat Med. 1985;4(1):39–46. [DOI] [PubMed] [Google Scholar]
- 32.Curia MC, Palmirotta R, Aceto G, et al. Unbalanced germ-line expression of hMLH1 and hMSH2 alleles in hereditary nonpolyposis colorectal cancer. Cancer Res. 1999;59(15):3570–3575. [PubMed] [Google Scholar]
- 33.Kim JC, Roh SA, Koo KH, et al. Genotyping possible polymorphic variants of human mismatch repair genes in healthy Korean individuals and sporadic colorectal cancer patients. Fam Cancer. 2004;3(2):129–137. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi M, Shimodaira H, Andreutti-Zaugg C, Iggo R, Kolodner RD, Ishioka C. Functional analysis of human MLH1 variants using yeast and in vitro mismatch repair assays. Cancer Res. 2007;67(10):4595–4604. [DOI] [PubMed] [Google Scholar]


