Abstract
Patched (Ptc) is a twelve-pass transmembrane protein that functions as a receptor for the Hedgehog (Hh) family of morphogens. In addition to Ptc, several accessory proteins including the CDO/Ihog family of co-receptors are necessary for proper Hh signaling. Structures of Hh proteins bound to members of the CDO/Ihog family are known, but the nature of the full Hh receptor complex is not well understood. We have expressed the Drosophila Patched and Mouse Patched-1 proteins in Sf9 cells and find that Sonic Hedgehog will bind to Mouse Patched-1 in isolated Sf9 cell membranes but that purified, detergent-solubilized Ptc proteins do not interact strongly with cognate Hh and CDO/Ihog homologs. These results may reflect a nonnative conformation of detergent-solubilized Ptc or that an additional factor or factors lost during purification are required for high-affinity Ptc binding to Hh.
INTRODUCTION
The Hedgehog (Hh) signaling pathway mediates key tissue patterning events during animal development, and abnormal pathway activity is associated with several cancers [1, 2]. Hh proteins are secreted morphogens that specify cell fates in neighboring tissues in a concentration-dependent manner [3, 4]. The twelve-pass transmembrane protein Patched (Ptc) has been identified as a key Hh receptor in genetic [5, 6, 7, 8] and cell-based binding studies [9, 10, 11]. In the absence of Hh, Ptc constitutively inhibits the activity of Smoothened (Smo) [6, 12], a seven-pass transmembrane protein. The mechanism of this inhibition is unknown but does not appear to involve a direct interaction between Ptc and Smo [12]. In the presence of Hh, this inhibition is relieved and the pathway is activated. For recent reviews, see [13, 14, 15, 16].
Many additional proteins modulate Hh pathway activity, but their presence and activity are not always conserved across phyla. For instance, Gas1 positively regulates Hh signaling in mammals [17], but no Gas1 homolog exists in the fruit fly. Hhip, a cell surface protein that acts as a negative regulator by binding and sequestering Hh proteins in vertebrates [18] also has no apparent ortholog in the fly. The mammalian proteins CDO and BOC are orthologous to the fly proteins Ihog and Boi and each binds its cognate Hh protein, but the manner and co-factor dependence of Hh binding by fly and mammalian orthologs is not conserved [19].
Despite the central importance of the Hh signaling pathway in animal development and the identification of many key pathway components, little is known about the molecular details connecting these components. Ptc is presumed to control Smo activity by transporting a small sterol-like molecule, a hypothesis based on an array of circumstantial evidence: the homology of Ptc to proton antiporters in the RND superfamily; the presence in Ptc of a sterol-sensing domain, which in other eukaryotic homologs is related to cholesterol trafficking; the indirect inhibition of Smo by Ptc, which suggests an intermediate [12]; and recent structural and biochemical studies of Smo showing that sterols bind Smo and modulate its activity [20, 21, 22, 23]. Nevertheless, ligand transport by Ptc has not been conclusively demonstrated, nor has a physiological ligand for Smo been identified. More generally, how Hh proteins modulate the activity of Ptc is not known, and no functional assay for purified Ptc has been established.
A direct interaction between Hh and Ptc is clearly the simplest and most likely interpretation for high-affinity Hh binding to Ptc-expressing cells and Hh modulation of Ptc activity [9, 11]. Assays measuring binding to the cell surface leave open the possibility that other cellular factors could be involved, however. For example, most cell-based binding studies predated knowledge of the importance of accessory proteins Gas1, CDO/Ihog and BOC/Boi for mediating interactions between Hh and the cell surface. These proteins are essential for both fly [24] and mammalian Hh signaling [17, 25]. Members of the CDO family of proteins bind Hh proteins directly with low micromolar affinities [26, 19, 27], whereas the affinity of mammalian Sonic Hedgehog (Shh) for binding to the surface of Ptc-expressing cells is in the low nanomolar range [9, 11]. This difference in affinity, as well as the fact that high-affinity cell-surface binding is dependent on the presence of Ptc, would seem to indicate that Ptc itself directly binds Shh but that the affinity for the binary Shh-Ptc interaction may be weaker than the affinity of Hh for Ptc-expressing cells. The role of the CDO/Ihog family of proteins appears to be to function as co-receptors that enhance binding affinity. In this case, a ternary complex between Ptc, Hh and members of the CDO/Ihog family may represent the initial signaling complex at the cell surface. Evidence for such ternary complexes has been found for Drosophila Ptc and Ihog [24], but the evidence in the mammalian pathway is contradictory. Although CDO, BOC or Gas1 appear to be required along with Ptc for normal signaling in mammals [17, 25], the addition of the soluble Hh-binding domain of CDO (CDOFn3) actually competes for ShhN binding to Ptc on the cell surface [19]. This observation suggests that the binding surfaces for Ptc and CDO on ShhN may overlap. This binding competition has been rationalized with the positive role of both Ptc and CDO in Hh signaling by the observation that physiological Hh is found in multivalent particles [28, 29], allowing simultaneous Ptc and CDO binding. Multivalency is not required for high-affinity binding of Shh to Ptc on cells, however, as monomeric ShhN expressed in E. coli binds Ptc-expressing cells with high affinity [9, 11]. Hh proteins also bind sulfated glycosaminoglycans with affinities that can be in the micromolar range [29, 30], which also likely contributes to high-affinity interactions with the cell surface.
A major barrier to understanding Ptc activity and the nature of its interactions with Hh pathway components has been the difficulty of isolating functional forms of Ptc or Ptc fragments. Most information on the reported molecular mechanisms and binding partners of Ptc has been obtained indirectly, using cell, tissue or whole animal-based studies. We therefore undertook to express and purify mouse and Drosophila Ptc proteins with intact transmembrane and extracellular regions for binding studies with Hh and other Hh pathway components. Sonic Hedgehog will bind to mouse Ptc in isolated Sf9 cell membranes, but we surprisingly find that Ptc proteins extracted and purified in the presence of detergents do not interact with soluble, cognate Hh or CDO/Ihog homologs with high affinity as either binary or ternary complexes.
EXPERIMENTAL PROCEDURES
Materials
All chemicals were purchased from Sigma unless otherwise noted. Detergents were purchased from Affymetrix and included n-dodecyl-β-D-maltopyranoside (DDM), n-dodecylphosphocho-line (fos-choline 12, FC-12), and 2,2-didecylpropane-1,3-bis-β-D-maltopyranoside (lauryl maltose neopentyl glycol, LMNG). Low molecular weight heparin (LMWH, average molecular weight 3000 Da, sodium salt, from porcine sources) was purchased from P212121.com. Heparin Decasaccharide (DH) was purchased from Neoparin Inc. Antibodies for western blotting were mouse α-Myc monoclonal (9E10), which was isolated from hybridoma growth medium; rabbit α-Ihog polyclonal and mouse α-Hh monoclonal antibodies, which were gifts from P. Beachy; and appropriate HRP-conjugated secondary antibodies.
Buffers
Ni2+ Binding Buffer consisted of 35 mM NaH2PO4, 300 mM NaCl, and 15 mM imidazole adjusted to pH 8.0 with NaOH at 25°C. Ni2+ Wash Buffer consisted of 10 mM Tris base, 10 mM Tris HCl, and 300 mM NaCl. Ni2+ Elution Buffer was Ni2+ Wash Buffer with 250 mM imidazole. Strep Wash Buffer was 20 mM MOPS, 10 mM NaOH, and 200 mM NaCl. Strep Elution Buffer was Strep Wash Buffer with 2.5 mM desthiobiotin. Pull-Down Buffer was Strep Wash Buffer containing 0.01% LMNG and 0.2 mM TCEP. CD Buffer was 10 mM NaH2PO4, 150 mM NaF, titrated to pH 7.2 with NaOH. CPM Thermal Stability Buffer was 20 mM HEPES pH 7.5, 200 mM NaCl, 0.02% LMNG.
Proteins and expression vectors
IhogFn1, IhogFn2, IhogFn12 and IhogFn12ΔH are the first, second, or both Type III Fibronectin (FnIII) domains from Ihog, respectively, with ΔH referring to an Ihog surface mutant with reduced heparin binding [26]. BOCFn3, BOCFn23 and BOCFn13 are FnIII domains 3, 2–3, and 1–3 respectively from BOC. These BOC and Ihog fragments were expressed as His-Myc-TEV- (HMT-) fusion proteins using the vector pT7HMT [31]. HhN, ShhN and ShhN-SC (The “Surface C” mutant of ShhN [11], which is deficient in binding to Ptc-expressing cells) were subcloned into a modified version of pMAL-c2x as described [19]. ShhFL and HhFL are the full-length Mouse and Drosophila Hh proteins including native signal sequences and C-terminal self-splicing domains. IhogFn12TM consisted of the native Ihog signal sequence followed by the first and second FnIII domains and the transmembrane region, but with the intracellular region truncated. Ski and Hhat are the entire native Hh acyltransferases from Drosophila and Mouse, respectively. MmHhip consisted of the β-propeller and following two EGF domains of Mouse Hhip. Drosophila Patched and mouse Patched-1 proteins are the tagged Mouse and Drosophila Ptc proteins with their C-termini truncated immediately after the final predicted transmembrane helix (DmPtcT1 and MmPtcT1, respectively). See Supplementary Table 1 for detailed protein sequence specifications.
DmPtcT1 and MmPtcT1 proteins were expressed fused to a concatenated series of N-terminal tags including the Streptavidin Binding Peptide (SBP) [32], an HRV3C protease site, a 6×His tag, a Myc tag, and a TEV protease site. Tags and Ptc proteins were cloned into the transfer vector pFastBac1 (Invitrogen). For co-expression of ShhFL with Hhat, HhFL with Ski, and DmPtcT1 with IhogFn12TM, transfer vectors were constructed from pFastBacDual (Invitrogen). Hhip was cloned into a modified pFastBac1 vector containing the Honeybee Mellitin (HBM) signal sequence to target for secretion, followed by 8×His, SBP, Myc and TEV sequences.
Production of recombinant baculoviruses
Recombinant bacmids and baculoviruses for insect cell expression were constructed using the Bac-to-Bac system (Invitrogen) following manufacturer’s instructions. After transfecting Sf9 cells with bacmid DNA, the secreted virus (designated P1) was amplified two more times (P2 and P3 viruses) following manufacturer’s instructions. P3 virus was used for protein production.
Bacterial expression and purification of HhN, IhogFn1, IhogFn2, IhogFn12, IhogFn12ΔH, ShhN, ShhN-SC, BOCFn3, BOCFn23 and BOCFn13
All expression plasmids were transformed into BL21 and plated on LB with appropriate antibiotics. Single colonies were picked, grown overnight, and used to inoculate TB in baffled flasks at 225 RPM and 37°C. Bacteria were grown to an optical density (600 nm) of around 0.8, at which point the incubator temperature was lowered to 16°C and the cultures allowed to shake for an additional hour. IPTG was then added to a final concentration of 0.5 mM, followed by expression for 24 hr. Bacteria were harvested by centrifugation and cell pellets stored at −20°C or ;80°C.
Individual bacterial pellets were resuspended in ice-cold Ni2+ Binding Buffer containing 0.1 μL/mL Benzonase Nuclease and 1 mM PMSF. Bacteria were then lysed using a French press, and extracts cleared by centrifugation and syringe filtration. All proteins were initially purified using 5 mL HisTrap columns (GE Healthcare) charged with Ni2+. Where appropriate, tags were then removed by addition of His-tagged TEV or HRV3C and overnight incubation at 4°C. HhN, IhogFn12, IhogFn12ΔH, IhogFn1, and ShhN were then further purified by cation exchange chromatography as described [26, 19]. All proteins were desalted if necessary, then separated from tags and proteases by another round of HisTrap purification. All proteins were purified by a final size exclusion chromatography (SEC) step in Pull-Down Buffer, except for BOCFn13 for which the concentration of NaCl was 500 mM.
Insect expression and purification of Hhip, DmPtcT1 and MmPtcT1
Sf9 and High Five cells were adapted and grown in suspension using a modification of the serum-free medium ISFM [33] containing 10 g/L glucose instead of 2.5 g/L. Sf9 or High Five cells were grown or split to a concentration of 2.0×106 cells/mL in air-sparged 1 L spinner flasks before infection with 20 mL of P3 baculovirus (for simultaneous infection with multiple viruses, 20 mL of each virus were added).
Hhip was expressed in High Five cells. After infection, expression was allowed to proceed for 72 hr before removing the cells by centrifugation. The conditioned medium was then adjusted to pH 8.0 with NaOH, resulting in a heavy precipitate of calcium phosphate that was removed by centrifugation and filtration. Up to 6 L of pH-adjusted, clarified medium was flowed by gravity through 15 mL of “6omplete His” resin (Roche) at room temperature overnight, which was then washed and eluted using manufacturer’s recommended conditions. Eluted protein was further purified using a 5 mL StrepTrap column (GE Healthcare), and tags were cleaved by addition of TEV protease overnight. Cut tags, uncleaved fusion protein and TEV protease were then removed using a 5 mL HisTrap column, and the remaining Hhip was further purified by SEC.
MmPtcT1 and DmPtcT1 were expressed in Sf9 cells. After infection, expression was allowed to proceed for 72 hr before harvesting cells by centrifugation. If not used immediately, cell pellets were stored by washing with PBS containing 10% glycerol, followed by snap freezing in liquid nitrogen and storage at −80°C. Cells were resuspended to a final volume of 30 mL per L of culture with Ni2+ Bind Buffer containing 0.5 mM TCEP, 0.1 μL/mL Benzonase and one Complete Protease Inhibitor tablet, EDTA-free (Roche). Cells were lysed by French press, and nuclei and large debris removed by centrifugation at 10,000 rpm for 10 min in an SA-600 rotor. The supernatant was then transferred to a thin-walled ultracentrifuge tube and additional buffer added up to the maximum tube volume, around 40 mL, and centrifuged in an sw-28 rotor at 28,000 rpm for 1 hr to pellet membranes. Isolated membranes were weighed, added to 10 mL of Ni2+ Bind buffer per gram of membrane pellet, and resuspended using a Dounce homogenizer. 1 mL of 10% FC-12 per gram of membrane pellet was then added to the resuspended membranes, which were allowed to solubilize for 30 minutes on ice. Finally, detergent-solubilized protein was clarified with an additional ultracentrifugation spin.
Solubilized Ptc was applied to a 5 mL HisTrap column, washed with 50 mL of Nickel Wash Buffer supplemented with 0.2 mM TCEP and 0.02% LMNG, and eluted with Nickel Elution Buffer supplemented with 0.2 mM TCEP and 0.02% LMNG. A small amount of the reducing agent TCEP was added to prevent unwanted disulfide formation between exposed intracellular cysteines. At this point, purified Ptc could be incubated in batch with a Strep-Tactin bead slurry for use in pulldown assays. Alternatively, to obtain higher concentrations and purity for Circular Dichroism (CD), analytical SEC, or for pulldown by ShhN- or HhN-coupled beads, an additional round of purification was performed using a 5 mL StrepTrap HP column. Purified Ptc from the nickel column was applied to the StrepTrap column, washed with Strep Bind Buffer supplemented with 0.2 mM TCEP and 0.01% LMNG, and eluted with Strep Elution Buffer containing 0.2 mM TCEP and 0.01% LMNG. For Ptc proteins intended for CD, the Strep-Tactin purification buffers were substituted with CD Buffer containing the same amounts of TCEP, LMNG and desthiobiotin. For Ptc proteins intended for 7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin (CPM) thermal melts, Ptc was purified as described above with the following modifications: 2 mg/ml iodoacetamide was added at membrane solublization; all subsequent buffers did not include TCEP; and protein from Nickel column elution was desalted using a 5mL HiTrap desalting column equilibrated in CPM Thermal Stability Buffer. Freshly purified protein was used the same day for CPM thermal melt assays.
Cell-free binding assay
Cell-free binding assay was performed as in Zheng et al. [24]. In brief, isolated membrane pellets from uninfected Sf9 cells or cells infected with MmPtcT1 baculovirus were resuspended in ShhN-Renilla conditioned medium [34]. After 1 hr incubation, membranes were reisolated by centrifugation, washed three times with ice cold PBS, and solubilized in 250μl passive lysis buffer (Promega) and 5μL of lysate used to measure Renilla luciferase activity.
Immunostaining and confocal microscopy of Patched-expressing Sf9 cells
Sf9 cells were plated on gelatinized glass coverslips in 6-well plates and simultaneously infected with the indicated viruses (Fig. 1A). Expression was allowed to proceed for 24–36 hours. All steps were performed at room temperature unless otherwise noted. Cells were fixed by adding to the growth medium an equal volume of PFA (4% paraformaldehyde in PBS) for 5 min, followed by aspiration and addition of undiluted PFA for another 5 min. Cells were then permeablized with 0.5% Tween-20 in PBS for 15 min, and blocked with PBST (PBS with 0.1% Tween-20) containing 1% BSA for 30 min. Staining was performed with α-Myc (1:200 in blocking solution) at 4°C overnight. Staining solution was removed and cells washed 3 times with PBST, incubating for 5 minutes each wash. Finally, cells were incubated in the dark for 1 hour with fluorescent secondary antibody solution (Cy3 Goat α-Mouse, 1:1000 in blocking solution), washed 3 times with PBST, and coverslips mounted on glass slides in aqueous mounting medium for confocal microscopy.
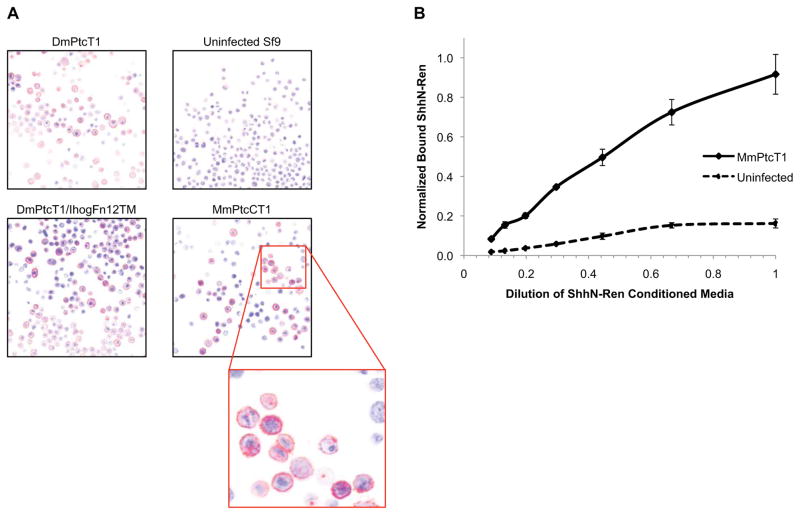
Figure 1. Functional Ptc expresses on the surface of Sf9 cells.
(A) Sf9 cells infected with the indicated viruses were immunostained using anti-Myc (9E10) primary and Cy3 secondary antibodies (red), and imaged by confocal microscopy. Nuclei were stained with DAPI (blue). (B) Membranes from unifected Sf9 cells or cells infected with MmPtcT1 virus were isolated, incubated with varying concentrations of SHhN-Renilla luciferase conditioned medium, washed, and residual luciferase activities determined.
Ptc characterization
The behavior of Ptc proteins was analyzed by CD, CPM fluorescence thermal stability assays, and by analytical SEC using a Superose6 column. For CD analysis, spectra were collected at 4°C between 185 nm and 280 nm, using Ptc proteins in a cuvette with 1 mm pathlength. Thermal unfolding analysis was also performed by following the CD signal at 208 nm while increasing the temperature from 4°C to 95°C. CPM fluorescence thermal stability assays were performed as in [35]. In brief, a 1:40 dilution of a 4 mg/ml solution of CPM dye (Invitrogen) in DMSO was incubated in CPM Thermal Melt Buffer for 5 min at room temperature, protected from light. Ptc protein (10 μg) was diluted in CPM Thermal Melt Buffer to a final volume of 290 μL. After 5 min at room temperature, 10 μL of dilute dye was added and the sample was mixed and transferred to a quartz fluorometer cuvette. The cuvette was transferred to a Fluorolog-3 spectrofluorometer (Horiba Jobin Yvon) equipped with a Peltier sample cooler (F-3004) and heated at a rate of 2°C/min. Emission at 463 nm was monitored with an excitation wavelength of 387 nm from 20°C to 80°C. For SEC analysis, Ptc was injected onto a Superose6 column equilibrated in Pulldown Buffer. Its elution profile was followed by UV absorption at 280 nm, and apparent molecular weights were estimated by comparison to Gel Filtration Standards (Bio-Rad).
Pulldown Assays
Bead-based pulldown assays were used to screen for interactions between purified, detergent-solubilized Ptc and other proteins. All steps were carried out on ice or at 4°C unless otherwise noted. Immediately after performing the nickel purification, Ptc resin was prepared by coupling the SBP-tagged protein to Strep-Tactin resin (IBA). An excess of Ptc protein solution (typically the elution peak from an entire 1 L prep) was added to enough Strep-Tactin resin to give 10 μL of Ptc resin for each desired pulldown condition; typically, this resulted in 2–5 μg of coupled Ptc protein per condition. Ptc was allowed to couple to completion for 2 hr. Resin was then pelleted and resuspended in Pulldown Buffer, divided into separate tubes, and pelleted, such that 10 μL of packed Ptc resin were obtained in each tube. Pulldowns were set up in a total volume of 50 μL using the combinations of components indicated in corresponding Figures. Generally, pulldown conditions contained 10 μM of each soluble protein, and around 0.75 μM of Ptc. These binding reactions were incubated for 1 hr. Ptc beads were then washed 4 times with 1 mL of Pulldown Buffer, and Ptc was eluted by adding 20 μL of Pulldown Buffer supplemented with 2.5 mM biotin. The presence of proteins that bound and eluted with Ptc was then analyzed using Coomassie-stained SDS-PAGE gels and Western blots.
Co-purification of Ptc with other proteins
Ptc proteins were co-expressed with potential partners by infecting Sf9 cells with the following viruses or combinations of viruses: MmPtcT1 virus with ShhFL/Hhat dual expression virus; DmPtcT1/IhogFn12TM dual expression virus; DmPtcT1 virus with HhFL/Ski dual expression virus; and DmPtcT1/IhogFn12TM dual expression virus with HhFL/Ski dual expression virus. In each case, the Ptc protein was fused with N-terminal SbpPHMT tags, and its potential binding partners were untagged. Sequential nickel, Strep-Tactin, and Superose6 chromatography steps were performed as outlined above, and the presence of co-purifying complexes evaluated by Coomassie-stained SDS-PAGE and Western blot.
RESULTS
Ptc Purification and Characterization
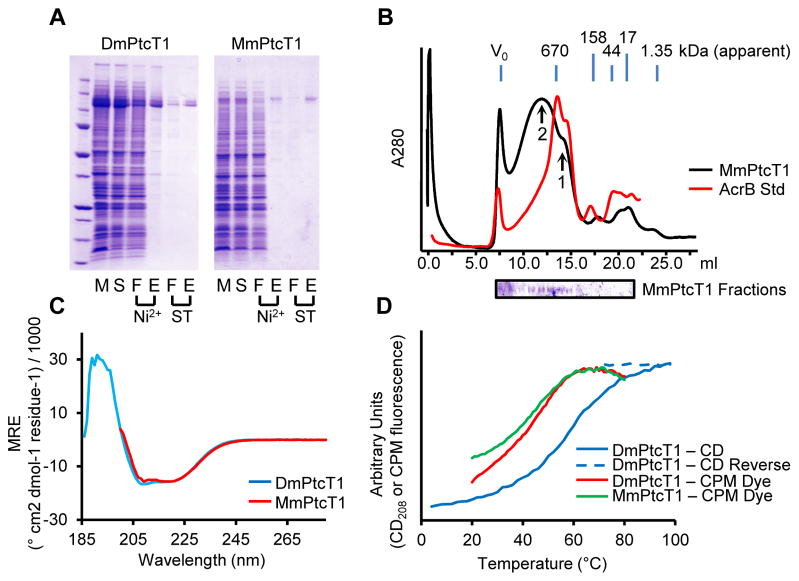
Both Drosophila and Mouse Ptc expressed at levels of 0.5 – 3 mg of protein per liter of Sf9 cell culture. Confocal imaging of infected Sf9 cells expressing Ptc proteins indicated that Ptc was present at the cell surface, and membranes isolated from MmPtcT1-expressing cells bound Shh (Fig. 1). Detergent solubilization trials indicated, however, that Ptc proteins were only solubilized well by the fos-choline class of detergents (Fig. S1), and FC-12 was selected for routine extraction. Solubilized Ptc proteins were purified by a combination of Nickel and Strep-Tactin affinity chromatography (Fig. 2). Yields of purified Ptc ranged from 20–50% of total expressed protein as judged by Coomassie-stained gels and comparison to BSA standards. Losses resulted from incomplete solubilization, incomplete capture, or incomplete elution from Nickel and were dependent on flow rates and the amount of protein present initially. Although most detergents do not solubilize Ptc proteins efficiently, we found that once Ptc had been solubilized, FC-12 could be exchanged for a variety of other detergents during the initial HisTrap purification. We therefore evaluated the behavior of Ptc proteins by SEC after exchange into different detergents (Fig. S2). SEC behavior was qualitatively similar for most detergents tested, with a species eluting as a small oligomer, potentially consistent with a trimer [36] (Fig. 2B, arrow 1); a higher-order oligomer (arrow 2); and a small proportion in the void volume. Although the SEC behavior of Ptc after purification is similar in many detergents, Ptc in LMNG was less prone to precipitation over several days of storage. All binding studies were therefore carried out with Ptc exchanged into LMNG.
Figure 2. Purification and characterization of Ptc proteins.
(A) DmPtcT1 and MmPtcT1 were purified by successive Ni2+ and Strep-Tactin (ST) chromatography steps. Gel Lanes: (M) resuspended membranes; (S) detergent-solubilized membranes; (F) flow-through; (E) elution (B) SEC of purified, concentrated MmPtcT1 (140 kDa calculated) compared to tagged AcrB (123 kDa), a protein standard with the same predicted topology. MmPtcT1 elutes in two peaks: a small oligomer consistent with a potential physiological trimer (arrow 1) and a larger oligomer (arrow 2) (C) CD spectra of DmPtcT1 and MmPtcT1 show substantial alpha-helical character. (D) Thermal unfolding of Ptc proteins as measured by CD at 208 nm (correlated with alpha helical content) and by CPM dye binding to exposed cysteines.
Characterization of purified Ptc proteins by CD spectroscopy revealed spectra typical of proteins with high alpha-helical content (Fig. 2C). The secondary structure content of DmPtcT1 calculated from its CD spectrum was 47% helix and 13% sheet, similar to the values of 44% and 6% predicted from the primary sequence by PSIPRED [37]. CD at 208 nm was used to follow unfolding of DmPtcT1, giving an unfolding midpoint of 58°C. We also followed Ptc unfolding using CPM dye, a measure that is more closely related to tertiary rather than secondary structure. By this method, unfolding midpoints were 36°C and 37°C for MmPtcT1 and DmPtcT1, respectively (Fig. 2D). Deglycosylation of purified MmPtcT1 with PNGaseF also resulted in a small but visible band shift on Coomassie-stained SDS-PAGE (Fig. S3), consistent with the molecular weight of several glycans; MmPtcT1 contains 6 consensus N-linked sites, with total expected glycan mass of 6–7 kDa, assuming pauci-mannose glycosylation profiles typical of Sf9 cells [38]. These results are consistent with proper processing and membrane localization of expressed Ptc proteins.
Pulldown of potential binding partners by MmPtcT1
MmPtcT1-coupled resin was tested for its ability to pull down soluble ShhN, ShhN-SC, HMT-BOCFn3, HMT-BOCFn23 and HMT-BOCFn13 individually as well as in ternary combinations (Fig. 3). 1 mM Ca2+ was included in most conditions as Ca2+ is known to be required for ShhN binding to BOCFn3. 200 μM LMWH was included in one condition with BOCFn13 and ShhN, since both are known to be heparin-binding proteins. We also included conditions with 5 mM EGTA, a Ca2+ chelator, as a control to eliminate ShhN:BOCFn13 binding. Finally, we included Hhip in certain conditions to test its ability to block potential binding of ShhN to MmPtcT1. Strep-Tactin resin without conjugated MmPtcT1 (Fig. 3) or with the tagged, irrelevant membrane protein AcrB conjugated (Fig. S4) was used as a control for nonspecific binding. We used Coomassie-stained SDS-PAGE to detect bound proteins, with Western blots also performed in some cases to detect weakly-bound proteins and confirm the identities of Coomassie-stained protein bands.
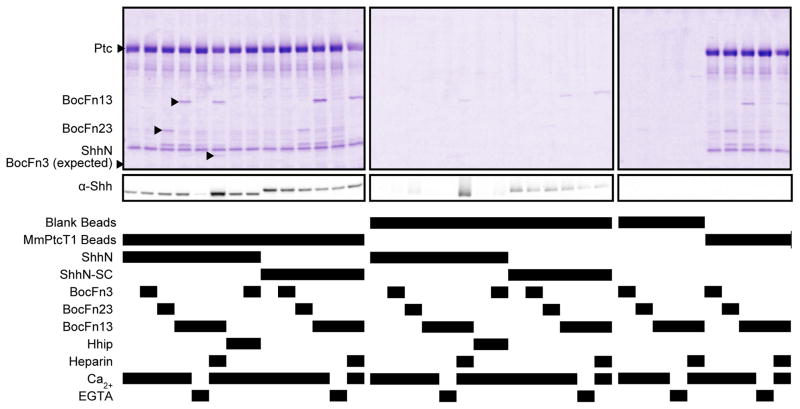
Figure 3. Pulldown of potential binding partners by MmPtcT1.
The indicated proteins and cofactors were incubated with MmPtcT1-coupled Strep-Tactin resin and blank (uncoupled) Strep-Tactin resin. Bound proteins were eluted with biotin and analyzed by Coomassie-stained SDS-PAGE. Bound proteins were eluted with biotin and analyzed by Coomassie-stained SDS-PAGE and anti-Shh Western blot. Shh and BOC fragments, marked with arrows, that appear to co-precipitate with Ptc proved to be due to heparin-dependent precipitation (Shh) or His-tag and Ca2+ dependent association (BOC). See also Fig. S4 and S5.
ShhN did not bind to detectably to MmPtcT1 beads as judged by Coomassie-stained SDS-PAGE gels of the eluted samples. Trace levels of ShhN binding to MmPtcT1 beads were detectable only by Western blotting; this binding was eliminated by EGTA. ShhN and BOCFn13 visibly precipitate in the presence of LMWH and Ca2+, resulting in high nonspecific binding to beads; we could therefore not evaluate specific binding of this complex to MmPtcT1. Since BOCFn13 and ShhN bind each other with micromolar affinity and each independently binds heparin, this precipitation was probably due to heparin-mediated protein crosslinking. Interactions between ShhN and MmPtcT1 beads were not enhanced by the presence of any BOC fragment, whether in the presence or absence of LMWH or Ca2+. Binding of ShhN to MmPtcT1 beads was also unaffected by the presence of Hhip.
By Western blot analysis, ShhN did not adhere to MmPtcT1 resin to a greater degree than the negative control surface mutant ShhN-SC. However, ShhN-SC showed no dependence on EGTA and no precipitation in the presence of LMWH and HMT-BOCFn13, consistent with its inability to bind BOC (Fig. 3).
HMT-BOCFn13 and HMT-BOCFn23 were not specifically pulled down by MmPtcT1-coupled beads. Although MmPtcT1-coupled beads did pull down BOC proteins as compared to blank beads (Fig. 3), we observed that this binding was dependent on the presence of 1 mM Ca2+ and was eliminated by EGTA, despite the lack of known calcium binding to BOC proteins. We therefore performed additional control experiments comparing the binding and solution behaviors of tagged and untagged BOC proteins in the presence of calcium (Fig. S5). These experiments showed that (i) His-tagged proteins oligomerize in the presence of 1 mM Ca2+ and that (ii) the binding of HMT-BOCFn13 to MmPtcT1 was dependent on the presence of His tags on both proteins. An irrelevant His-tagged protein was pulled down equally well by MmPtcT1 beads, and BOC proteins could also be pulled down equally by beads coupled with the irrelevant His-tagged membrane protein AcrB (Fig. S4). The presence of ShhN or ShhN-SC did not enhance the binding of BOC proteins to MmPtcT1 beads, nor did the addition of LMWH (Fig. 3).
We also performed the pulldown experiments in reverse (Fig. S6) by coupling ShhN and ShhN-SC to beads and attempting to pull down detergent-solubilized MmPtcT1 alone or in the presence of HMT-BOCFn3 and HMT-BOCFn13. Hhip was added to test for its ability to block potential Ptc binding, and soluble ShhN and ShhN-SC were added as specific competitors for Ptc binding to ShhN- and ShhN-SC-coupled resins. Consistent with our previous results (Fig. 3), we observed no binding above background of MmPtcT1 to ShhN resin. Aside from MmPtcT1, the resins pulled down the expected binding partners: ShhN resin pulled down BOCFn3 and HMT-BOCFn13 in a Ca2+ dependent manner, and pulldown of HMT-BOCFn3 by ShhN was blocked by Hhip.
Pulldown of potential binding partners by DmPtcT1
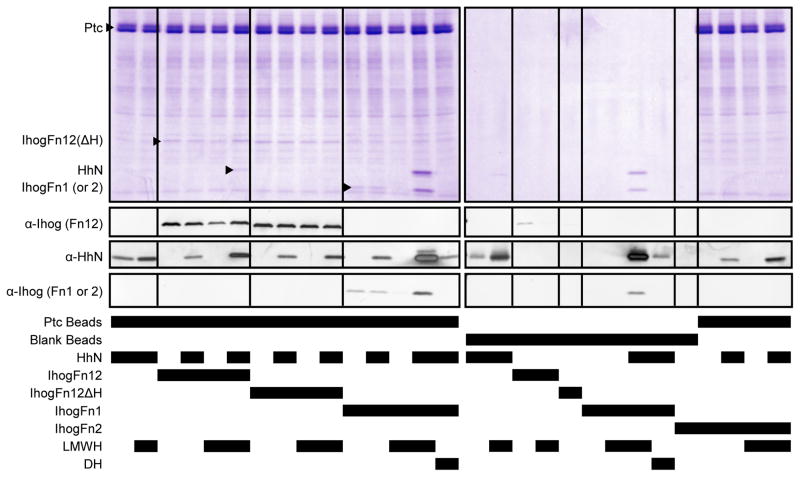
We next used similar pulldown assays to screen for DmPtcT1 binding to potential partners. DmPtcT1-coupled beads were used to pull down HhN, IhogFn12, IhogFn12ΔH, IhogFn1 and IhogFn2. These proteins and their combinations were not found to interact strongly or specifically with DmPtcT1-coupled beads. Although IhogFn1, IhogFn12 and IhogFn12ΔH were pulled down by DmPtcT1 beads in the absence of LMWH and could be detected in Coomassie-stained gels (Fig. 4), this binding was only slightly greater than binding to the irrelevant membrane protein AcrB (Fig. S7). In the presence of 200 μM LMWH, mixing IhogFn1 or IhogFn12 with HhN induced visible precipitation, resulting in high non-specific binding to Strep-Tactin resin. We therefore could not evaluate specific binding to DmPtcT1 resin in these conditions. However, substituting 200 μM decaheparin (DH) for LMWH prevented the precipitation of IhogFn1 and HhN and eliminated binding of IhogFn1 to DmPtcT1. HhN binding to DmPtcT1 beads was not detectable on Coomassie-stained gels except in the cases where precipitation was present. By Western blotting HhN did not bind DmPtcT1 beads specifically or to a greater degree than uncoupled Strep-Tactin beads. The presence of IhogFn1 and DH with HhN did not enhance the binding of HhN to DmPtcT1 beads.
Figure 4. Pulldown of potential binding partners by DmPtcT1.
Proteins and other cofactors were incubated with DmPtcT1-coupled Strep-Tactin resin and blank (uncoupled) Strep-Tactin resin in the indicated combinations.
Finally, we coupled HhN to amylose resin, and attempted to pull down soluble DmPtcT1 (Fig. S8). We also tested for binding to HhN of combinations of DmPtcT1 with IhogFn1, IhogFn2, IhogFn12, and IhogFn12ΔH, and with soluble non-conjugated HhN (as a specific competitor for DmPtcT1 binding to HhN beads). No binding of DmPtcT1 was detectable in any condition, even when assessed by Western blotting. Aside from DmPtcT1, the HhN resin pulled down its known binding partners as expected: IhogFn1 and IhogFn12 in a heparin-dependent manner, and IhogFn12ΔH much more weakly than IhogFn12. As expected, the HhN resin did not pull down IhogFn2.
Co-expression and purification of Ptc proteins with potential binding partners
Since Drosophila Ptc is believed to exist as a co-receptor complex with Ihog [24], we investigated whether the co-expression and purification of IhogFn12TM with Ptc resulted in improved binding or better monodispersity of Ptc as judged by SEC. In addition, since the Hh proteins used in our pulldown assays were expressed in E. coli, they lacked lipid modifications. We therefore also investigated the co-expression and purification of Ptc proteins with dually lipidated Hh proteins, referred to as HhNP and ShhNP. These were expressed using dual-expression HhFL/Ski and ShhFL/Hhat baculoviruses, which simultaneously drive expression of the full-length Hh proteins together with their respective palmitoyltransferases, although we did not determine whether HhNP or ShhNP were palmitoylated.
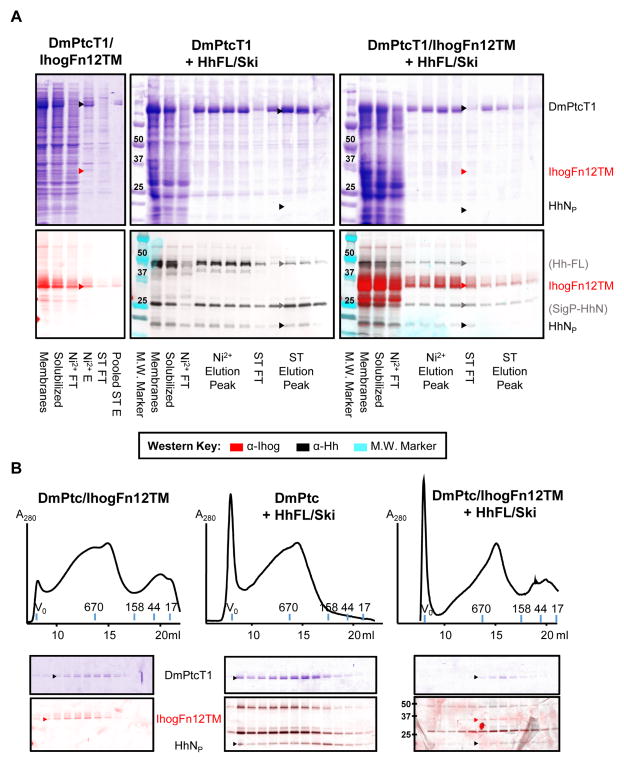
Co-expression of proteins did not result in co-purified stoichiometric complexes that interact strongly. When Sf9 cells were infected with a dual DmPtcT1/IhogFn12TM virus, both proteins were well-expressed. DmPtcT1 was purified using the standard Ni2+/Strep procedure (Fig. 5A), and the resulting purified protein was separated by analytical SEC (Fig. 5B). As judged by Western, some IhogFn12TM bound and eluted with DmPtcT1 during each purification step, but with significant amounts dissociating during each wash step. Comparison of the Ihog Western band intensities to standards (data not shown) indicated that the amount of IhogFn12TM remaining after DmPtcT1 purification was sub-stoichiometric with respect to DmPtcT1. The SEC behavior of DmPtcT1 co-expressed with IhogFn12TM was similar to that of DmPtcT1 expressed alone. Coomassie-stained SDS-PAGE and Western blotting of the SEC fractions indicated that DmPtcT1 and IhogFn12TM elution peaks overlapped, but with IhogFn12TM eluting at slightly higher apparent molecular weight.
Figure 5. Co-expression and purification of DmPtcT1 with potential binding partners.
Sf9 cells were co-infected with baculoviruses driving expression of DmPtcT1 along with IhogFn12TM, HhFL/Ski, or both IhogFn12TM and HhFL/Ski. Expected positions of indicated protein bands are marked. (A) DmPtcT1 was purified and detected by Coomassie-stained SDS-PAGE. The presence of co-purifying Hh or Ihog was followed by Western blot. The same Western blot membrane was sequentially probed with Ihog and Hh antibodies. The images were then overlaid and color-coded (see key). Three major Hh bands are observed: one corresponding to the molecular weight of unprocessed Hh (Hh-FL), one corresponding to C-terminally processed Hh retaining its signal sequence (SigP-HhN), and one corresponding to HhNp. (B) The resulting co-purified proteins were subjected to Superose6 SEC, and the fractions analyzed by Coomassie-stained SDS-PAGE (to visualize DmPtcT1) and by Western blot, probing sequentially for Hh and Ihog as in (A).
When DmPtcT1 and HhNP were co-expressed, HhNP was observed to co-purify with DmPtcT1 (Fig. 5A), with some losses during each affinity step. SEC followed by SDS-PAGE and Western blotting showed that the elution profile of HhNP was very broad, and its peak did not track the elution of DmPtcT1. Co-expression of all three proteins (DmPtcT1/IhogFn12TM with HhNP) did not appreciably alter the co-purification or SEC behavior of any protein.
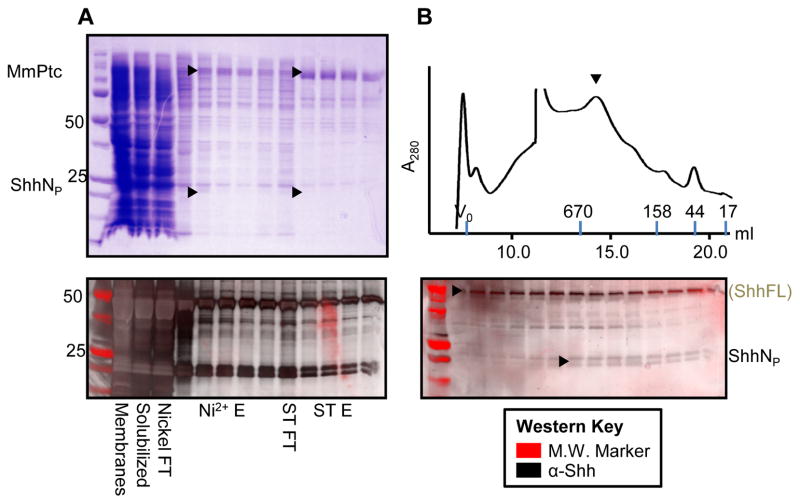
Similarly, MmPtcT1 was co-expressed with ShhNP and purified (Fig. 6A). Significant amounts of ShhNP remained in the flow-through during each affinity step, and although some residual protein co-purified with MmPtcT1 and was detectable by Western, it was not visible on Coomassie-stained gels. By Western, the concentration of ShhNP clearly decreased during each affinity step, while the concentration of MmPtcT1 increased. The behavior of MmPtcT1 on SEC was not significantly altered by co-expression with ShhNP, and the elution peak of ShhNP did not match the peak of MmPtcT1 (Fig. 6B).
Figure 6. Co-expression and purification of MmPtcT1 with Shh.
Sf9 cells were co-infected with baculoviruses driving expression of MmPtcT1 along with ShhFL/Hhat. (A) MmPtcT1 was purified and visualized with Coomassie-stained SDS-PAGE. The presence of co-purifying Shh followed by Western blot. Two major Shh bands are observed: one corresponding to the molecular weight of the unprocessed Shh (ShhFL), and one corresponding to the expected molecular weight of ShhNp. (B) The resulting co-purified proteins were subjected to Superose6 SEC. The fractions were analyzed by Coomassie-stained SDS-PAGE for MmPtcT1 detection, and by Western blot for Shh.
DISCUSSION
In this study, mouse Patched-1 and Drosophila Patched proteins were expressed in Sf9 cells, detergent solubilized, purified, and characterized by SEC and CD. Deglycosylation and confocal microscopy studies suggested that Ptc proteins were glycosylated and transported to the cell surface, and Sonic Hedgehog from conditioned medium bound to MmPtcT1 in membranes isolated from MmPtcT1-expressing Sf9 cells. Detergent-extracted and purified Ptc proteins contained the expected secondary structure content as judged by CD but eluted as multiple species SEC with one peak consistent with a trimer or small oligomer and another of higher apparent molecular weight.
Despite our observations that Ptc proteins behaved as expected by CD, cell surface expression, and deglycosylation analysis, and that some purified protein ran at sizes potentially consistent with physiological oligomers by SEC, high-affinity binding to cognate Hh proteins or Ihog/BOC co-receptors could not be detected. Recent studies indicate that Ptc proteins have an absolute requirement for co-receptors to signal (BOC/CDO/Gas1 in mammals, Ihog/BOI in Drosophila), and that Ptc forms physical complexes with these co-receptors [24, 25, 17]. In addition, high-affinity binding of Shh to cells is Ptc-dependent [11, 9]. We were therefore surprised not to detect high-affinity binding between purified Ptc proteins and any of these putative binding partners, either pairwise or in ternary combinations.
While some bound ShhN was detected in Western blots after saturating amounts were incubated with MmPtcT1 beads, the amount of ShhN detected was not convincingly above levels of nonspecific binding to blank beads (Fig. 3). Based on the clearly observed binding of BOC/Ihog proteins to their cognate Hh proteins in our pulldown assays (Fig. S6 and S8) and their known Kd’s of ~ 2.5–5 μM, we expect interactions between Ptc proteins and any partner would have resulted in the clear observation of Hh on Coomassie-stained gels if their Kd were below ~5 μM. Furthermore, the surface mutant ShhN-SC, which has impaired binding to Ptc-expressing cells [11], showed roughly the same trace levels of binding to MmPtcT1 beads as native ShhN. The addition of Hhip also did not compete with Ptc beads for ShhN binding, further suggesting a lack of specific binding of ShhN to MmPtcT1 beads. Thus, we were not able to reproduce a previous observation of high-affinity binding between ShhN and purified mouse Ptc [39]. In addition, HMT-BOCFn13 and HMT-BOCFn23 did not bind specifically to MmPtcT1. Binding in initial studies in the presence of Ca2+ proved to be mediated by His tags (all BOC and Ptc proteins, but not Ihog proteins, were His-tag fusions) in the presence of Ca2+ ions (Fig. S4-S5). The presence of Shh together with BOC fragments did not alter the apparent binding of any of these proteins to MmPtcT1, providing no evidence for the presence of a ternary complex between soluble forms of BOC and Shh and detergent-solubilized Ptc.
Drosophila HhN also did not bind DmPtcT1 beads at levels appreciably above background, as judged by Western blotting. No HhN was detectable on Coomassie-stained gels. IhogFn1, IhogFn12, and IhogFn12ΔH were bound by DmPtcT1 beads in the absence of both heparin and Hh, as detected by Coomassie-stained gels; however, for IhogFn12, roughly comparable binding was also seen to beads coupled with the irrelevant membrane protein AcrB. The addition of HhN and heparin decasachharide eliminated binding of IhogFn1 to DmPtcT1 beads. In contrast with previous observations [24], no binding of IhogFn2 to DmPtcT1 beads was observed. In all cases where some interaction was observed, the relative stoichiometry of the bound protein to Ptc was low, as judged by comparing Coomassie band intensities. Given the low stoichiometry of all observed complexes and the nonspecific binding seen between IhogFn12 and AcrB, we conclude that specific binding of Ihog fragments to Ptc was not observed.
As a final effort at purifying high-affinity, stoichiometric complexes of Ptc proteins with putative ligands, we attempted several co-expression experiments: MmPtcT1 with ShhNP, DmPtcT1 with HhNP, DmPtcT1 with IhogFn12TM, and DmPtcT1 with HhNP and IhogFn12TM. In all cases, as Ptc proteins were purified using His and SBP tags, their expression partners increasingly dissociated with each chromatographic step. A final Superose6 chromatography step showed some proportion of binding partner co-migrating with Ptc in each case, but with most having been lost in preceding purification steps. Qualitatively, this suggests at most a weak interaction between purified Ptc and these binding partners.
The absence of high-affinity interactions between purified Ptc and several potential binding partners observed here could be explained in several ways. Firstly, Ptc may not adopt a native or binding-competent conformation in this expression system, or it may lose these properties during detergent solubilization. The ability of both Ptc proteins to reach the cell surface and of MmPtcT1 to become glycosylated and bind ShhN in Sf9 cell membranes suggests initial folding and biogenesis occurs properly (Fig. 1 and S3), however, and that any non-native structure is more likely to stem from detergent solubilization. Regrettably, no functional assay exists to validate the integrity of purified Ptc—indeed, the central objective of our studies was to find an in vitro activity for Ptc that could be studied—and we can only rely our CD and SEC results to conclude that purified Ptc retains at least some native-like structure.
Although fos-choline detergents are generally considered harsher detergents that may destabilize membrane proteins during solubilization—a particular concern for proteins like Ptc that are only efficiently extracted by fos-choline detergents—many membrane proteins retain function in fos-choline [40, 41, 42]. Proteins have been crystallized directly from fos-choline detergents, for example, including alpha helical membrane proteins [43]. The transmembrane protein SCAP was also only extracted well by a fos-choline detergent and was shown to retain function in detergent-extracted form [44]. SCAP is similar to Ptc in being a multi-pass transmembrane protein (a tetramer of 8-pass subunits) with a sterol-sensing domain and was also expressed in Sf9 cells with N-terminal tags. If Hh proteins interact with a region of the large, presumably independently-folded extracellular loops of Ptc as has been suggested [45], these regions of Ptc may also be less susceptible to destabilization by detergents than the Ptc transmembrane regions.
Another possibility for the absence of strong binding between Ptc and cognate Hh and CDO/Ihog proteins is that additional cell-surface co-factors not present in our assays are required for high-affinity interactions or to maintain Ptc stability. The low concentration of luciferase-tagged Shh in the conditioned medium used to show binding between Shh and mPtc in isolated Sf9 membranes precluded saturation of binding and estimation of a binding constant, but the likely absence of mammalian cofactors in these cells suggests that mPtc itself can bind Shh with measurable affinity in the absence of known co-factors. A requirement for a bilayer environment, particular lipids, or glycans remains a possibility, however. Our inability to observe high-affinity binding between Ptc and cognate Hh or Ihog/CDO homologs thus seems likely to stem from loss of native-like Ptc function during detergent solubilization or detergent disruption of ShhN binding ability. As biochemical characterization of the nature and components of the functional Hh receptor remains a key objective of studies aiming to understand Hh signaling in normal and disease states, it will be important aspect of future efforts will be to find purification conditions that preserve or restore Ptc function. Nonetheless, the ability to produce near milligram amounts of purified Ptc described here establishes a cornerstone for future efforts to characterize the Hh receptor complex and elucidate the role of Ptc in Hh signaling.
Supplementary Material
Acknowledgments
We thank Xiaoyan Zheng and Phil Beachy for providing rabbit polyclonal antiserum against Ihog as well as hybridoma supernatants containing mouse monoclonal antibodies against Drosophila Hh. We thank Hongjin Zheng and Tamir Gonen for advice and assistance with detergent screening by SEC, and for the use of reagents and facilities at the Gonen lab at HHMI Janelia Farms (Ashburn, VA). Supported by NIH GM055545 (DJL) and Graduate Research Fellowship Program NSF DGE-1232825 (JMM).
References
- 1.Johnson RL, Rothman AL, Xie J, Goodrich LV, Bare JW, Bonifas JM, Quinn AG, Myers RM, Cox DR, Epstein EH, Scott MP. Human Homolog of patched, a Candidate Gene for the Basal Cell Nevus Syndrome. Science. 1996;272(5268):1668–71. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 2.Goodrich LV. Altered Neural Cell Fates and Medulloblastoma in Mouse patched Mutants. Science. 1997;277(5329):1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 3.Riddle RL, Johnson RL, Laufer E, Tabin C. Sonic-hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75(7):1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- 4.Heemskerk J, Dinardo S. Drosophila Hedgehog acts as a morphogen in cellular patterning. Cell. 1994;76(3):449–460. doi: 10.1016/0092-8674(94)90110-4. [DOI] [PubMed] [Google Scholar]
- 5.Hidalgo A, Ingham P. Cell patterning in the Drosophila segment: spatial regulation of the segment polarity gene patched. Development. 1990;110:291–301. doi: 10.1242/dev.110.1.291. [DOI] [PubMed] [Google Scholar]
- 6.Ingham PW, Taylor AM, Nakano Y. Role of the Drosophila patched gene in positional signalling. Nature. 1991;353:184–187. doi: 10.1038/353184a0. [DOI] [PubMed] [Google Scholar]
- 7.Forbes AJ, Nakano Y, Taylor AM, Ingham PW. Genetic analysis of hedgehog signalling in the Drosophila embryo. Development. 1993 Jan;(Supplement):115–24. [PubMed] [Google Scholar]
- 8.Marigo V, Scott MP, Johnson RL, Goodrich LV, Tabin CJ. Conservation in hedgehog signaling: induction of a chicken patched homolog by Sonic hedgehog in the developing limb. Development. 1996 Apr;122(4):1225–33. doi: 10.1242/dev.122.4.1225. [DOI] [PubMed] [Google Scholar]
- 9.Marigo V, Davey RA, Zuo Y, Cunningham JM, Tabin CJ. Biochemical evidence that patched is the Hedgehog receptor. Nature. 1996 Nov;384(6605):176–9. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- 10.Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, Johnson RL, Scott MP, Pennica D, Goddard A, Phillips H, Noll M, Hooper JE, de Sauvage F, Rosenthal A. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996 Nov;384(6605):129–34. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 11.Fuse N, Maiti T, Wang B, Porter JA, Hall TM, Leahy DJ, Beachy PA. Sonic hedgehog protein signals not as a hydrolytic enzyme but as an apparent ligand for patched. Proceedings of the National Academy of Sciences of the United States of America. 1999 Sep;96(20):10992–9. doi: 10.1073/pnas.96.20.10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taipale J, Cooper MK, Maiti T, Beachy PA. Patched acts catalytically to suppress the activity of Smoothened. Nature. 2002 Aug;418(6900):892–7. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- 13.Beachy PA, Hymowitz SG, Lazarus RA, Leahy DJ, Siebold C. Interactions between Hedgehog proteins and their binding partners come into view. Genes & Development. 2010 Sep;24(18):2001–12. doi: 10.1101/gad.1951710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingham PW. Hedgehog signaling. Cold Spring Harbor perspectives in biology. 2012;4(6):a011221. doi: 10.1101/cshperspect.a011221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Briscoe J, Thérond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nature reviews Molecular cell biology. 2013;14(7):416–29. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 16.Ryan KE, Chiang C. Hedgehog secretion and signal transduction in vertebrates. The Journal of biological chemistry. 2012;287(22):17905–13. doi: 10.1074/jbc.R112.356006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allen BL, Song JY, Izzi L, Althaus IW, Kang JS, Charron F, Krauss RS, McMahon AP. Overlapping roles and collective requirement for the coreceptors GAS1, CDO, and BOC in SHH pathway function. Developmental Cell. 2011 Jun;20(6):775–87. doi: 10.1016/j.devcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chuang PT, McMahon AP. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature. 1999;397:617–621. doi: 10.1038/17611. [DOI] [PubMed] [Google Scholar]
- 19.McLellan JS, Zheng X, Hauk G, Ghirlando R, Beachy PA, Leahy DJ. The mode of Hedgehog binding to Ihog homologues is not conserved across different phyla. Nature. 2008 Oct;455(7215):979–83. doi: 10.1038/nature07358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C, Wu H, Katritch V, Han GW, Huang XP, Liu W, Siu FY, Roth BL, Cherezov V, Stevens RC. Structure of the human smoothened receptor bound to an antitumour agent. Nature. 2013 May;497(7449):338–43. doi: 10.1038/nature12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nedelcu D, Liu J, Xu Y, Jao C, Salic A. Oxysterol binding to the extracellular domain of Smoothened in Hedgehog signaling. Nature Chemical Biology. 2013 Jul;9(9):557–64. doi: 10.1038/nchembio.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myers BR, Sever N, Chong YC, Kim J, Belani JD, Rychnovsky S, Bazan JF, Beachy PA. Hedgehog pathway modulation by multiple lipid binding sites on the smoothened effector of signal response. Developmental cell. 2013 Aug;26(4):346–57. doi: 10.1016/j.devcel.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nachtergaele S, Whalen DM, Mydock LK, Zhao Z, Malinauskas T, Ingham PW, Covey DF, Siebold C, Rohatgi R. Structure and function of the Smoothened extracellular domain in vertebrate Hedgehog signaling. eLife. 2013 Oct;2:e01340. doi: 10.7554/eLife.01340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng X, Mann RK, Sever N, Beachy PA. Genetic and biochemical definition of the Hedgehog receptor. Genes & Development. 2010 Jan;24(1):57–71. doi: 10.1101/gad.1870310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Izzi L, Lévesque M, Morin S, Laniel D, Wilkes BC, Mille F, Krauss RS, McMahon AP, Allen BL, Charron F. Boc and Gas1 each form distinct Shh receptor complexes with Ptch1 and are required for Shh-mediated cell proliferation. Developmental Cell. 2011 Jun;20(6):788–801. doi: 10.1016/j.devcel.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLellan JS, Yao S, Zheng X, Geisbrecht BV, Ghirlando R, Beachy PA, Leahy DJ. Structure of a heparin-dependent complex of Hedgehog and Ihog. Proceedings of the National Academy of Sciences of the United States of America. 2006 Nov;103(46):17208–13. doi: 10.1073/pnas.0606738103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kavran JM, Ward MD, Oladosu OO, Mulepati S, Leahy DJ. All mammalian Hedgehog proteins interact with cell adhesion molecule, down-regulated by oncogenes (CDO) and brother of CDO (BOC) in a conserved manner. The Journal of Biological Chemistry. 2010 Aug;285(32):24584–90. doi: 10.1074/jbc.M110.131680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen MH, Li YJ, Kawakami T, Xu SM, Chuang PT. Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes & Development. 2004;18(6):641–59. doi: 10.1101/gad.1185804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whalen DM, Malinauskas T, Gilbert RJ, Siebold C. Structural insights into proteoglycan-shaped Hedgehog signaling. Proceedings of the National Academy of Sciences of the United States of America. 2013 Oct;110(41):16420–5. doi: 10.1073/pnas.1310097110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang F, McLellan JS, Ayala AM, Leahy DJ, Linhardt RJ. Kinetic and structural studies on interactions between heparin or heparan sulfate and proteins of the hedgehog signaling pathway. Biochemistry. 46(13):3933–41. doi: 10.1021/bi6025424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geisbrecht BV, Bouyain S, Pop M. An optimized system for expression and purification of secreted bacterial proteins. Protein Expression and Purification. 2006 Mar;46(1):23–32. doi: 10.1016/j.pep.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Keefe AD, Wilson DS, Seelig B, Szostak JW. One-step purification of recombinant proteins using a nanomolar-affinity streptavidin-binding peptide, the SBP-Tag. Protein Expression and Purification. 2001 Dec;23(3):440–6. doi: 10.1006/prep.2001.1515. [DOI] [PubMed] [Google Scholar]
- 33.Inlow D, Shauger A, Maiorella B. Insect cell culture and baculovirus propagation in protein-free medium. Journal of Tissue Culture Methods. 1989 Mar;12(1):13–16. [Google Scholar]
- 34.Ma Y, Erkner A, Gong R, Yao S, Taipale J, Basler K, Beachy PA. Hedgehog-mediated paterning of the mammalian embryo requires transporter-like function of dispatched. Cell. 2002 Oct;111(1):63–75. doi: 10.1016/s0092-8674(02)00977-7. [DOI] [PubMed] [Google Scholar]
- 35.Alexandrov AI, Mileni M, Chien EYT, Hanson MA, Stevens RC. Microscale fluorescent thermal stability assay for membrane proteins. Structure. 2008 Mar;16(3):351–9. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Lu X, Liu S, Kornberg TB. The C-terminal tail of the Hedgehog receptor Patched regulates both localization and turnover. Genes & development. 2006;20(18):2539–51. doi: 10.1101/gad.1461306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16(4):404–5. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 38.Aeed PA, Elhammer AP. Glycosylation of recombinant prorenin in insect cells: the insect cell line Sf9 does not express the mannose 6-phosphate recognition signal. Biochemistry. 1994;33(29):8793–7. doi: 10.1021/bi00195a022. [DOI] [PubMed] [Google Scholar]
- 39.Joubert O, Nehmé R, Fleury D, De Rivoyre M, Bidet M, Polidori A, Ruat M, Pucci B, Mollat P, Mus-Veteau I. Functional studies of membrane-bound and purified human Hedgehog receptor Patched expressed in yeast. Biochimica et biophysica acta. 2009;1788(9):1813–21. doi: 10.1016/j.bbamem.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Chaudhary S, Pak JE, Gruswitz F, Sharma V, Stroud RM. Overexpressing human membrane proteins in stably transfected and clonal human embryonic kidney 293S cells. Nature protocols. 2012;7(3):453–66. doi: 10.1038/nprot.2011.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewinson O, Lee AT, Rees DC. The funnel approach to the precrystallization production of membrane proteins. Journal of molecular biology. 2008;377(1):62–73. doi: 10.1016/j.jmb.2007.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen TA, Lieu SS, Chang G. An Escherichia coli-based cell-free system for large-scale production of functional mammalian membrane proteins suitable for X–ray crystallography. Journal of molecular microbiology and biotechnology. 2010;18(2):85–91. doi: 10.1159/000283512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang W, Black SS, Edwards MD, Miller S, Morrison EL, Bartlett W, Dong C, Naismith JH, Booth IR. The structure of an open form of an E. coli mechanosensitive channel at 3.45 A resolution. Science. 2008;321(5893):1179–83. doi: 10.1126/science.1159262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radhakrishnan A, Sun LP, Kwon HJ, Brown MS, Goldstein JL. Direct binding of cholesterol to the purified membrane region of SCAP: mechanism for a sterol-sensing domain. Molecular cell. 2004;15(2):259–68. doi: 10.1016/j.molcel.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 45.Bosanac I, Maun HR, Scales SJ, Wen X, Lingel A, Bazan JF, de Sauvage FJ, Hymowitz SG, Lazarus RA. The structure of SHH in complex with HHIP reveals a recognition role for the Shh pseudo active site in signaling. Nature Structural and Molecular Biology. 16(7):691–7. doi: 10.1038/nsmb.1632. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.