Abstract
High-throughput RNA sequencing (RNA-Seq) provides single-nucleotide information that makes it a powerful tool for prediction of RNA editome. A new method, GIREMI, predicts RNA editomes (mainly A-to-I editing) accurately and sensitively using a single RNA-Seq data set, which does not require sample-specific genome sequence data or high sequencing depth. Using GIREMI, we observed prevailing tissue-specificity of RNA editing and interesting evolutionary patterns of editing sites in human population.
Accurate identification of RNA editome is central to better understand the diversity of gene expression and related functional implications1-3. Recently, there has been an extraordinary growth of application of RNA-Seq to identify RNA editing sites (summarized in4). However, many challenges still exist, one of which being the requirement of genome sequence data in order to discriminate RNA editing sites from genomic SNPs. Even with whole-genome sequencing data, some SNPs may still escape identification possibly due to non-uniformity in sequencing coverage or other issues. Thus, it is highly desirable to develop novel methods to predict RNA editomes independent of genome sequencing. Here, we report what is, to our knowledge, the first method to identify RNA editome accurately independent of genome sequence using a single RNA-Seq data set of modest sequencing depth.
The method, GIREMI (Genome-independent Identification of RNA Editing by Mutual Information), builds upon analysis of allelic linkage between single nucleotide variants (SNVs) and further extends the predictive power with generalized linear models. In a typical RNA-Seq data set, there often exists reads (or read pairs in paired-end mode) containing multiple SNVs that may correspond to genomic SNPs, RNA editing sites or experimental errors. A pair of SNPs harbored in the same read maintains the same haplotype in the RNA as in the genomic DNA (Fig. 1a). In contrast, a SNP and an RNA editing site exhibit variable allelic linkage since RNA editing occurs post-transcriptionally to either copy of the gene randomly (unless allele-specific editing exists, which is presumably rare and out of the scope of this study). Similarly, the allelic linkage in RNA-Seq reads for a pair of RNA editing sites may also appear random, although processive editing does exist5 that may lead to allelic bias of multiple editing sites. To examine whether allelic linkage may enable a discrimination of RNA editing sites from SNPs, we calculated the mutual information (MI) associated with SNPs or RNA editing sites in RNA-Seq reads (Online Methods). Indeed, MI values associated with the two types of variants demonstrated a striking difference (Fig. 1b, Supplementary Fig. 1a, Supplementary Note 1), reflecting the discriminative power of this approach. Based on this rationale, the GIREMI method calculates MI of publicly available SNPs (dbSNP) and uncharacterized RNA variants expressed in a given RNA-Seq data set, which is then utilized to predict RNA editing sites and further parameterize a generalized linear model (GLM) for enhanced performance (Supplementary Fig. 1, Online Methods).
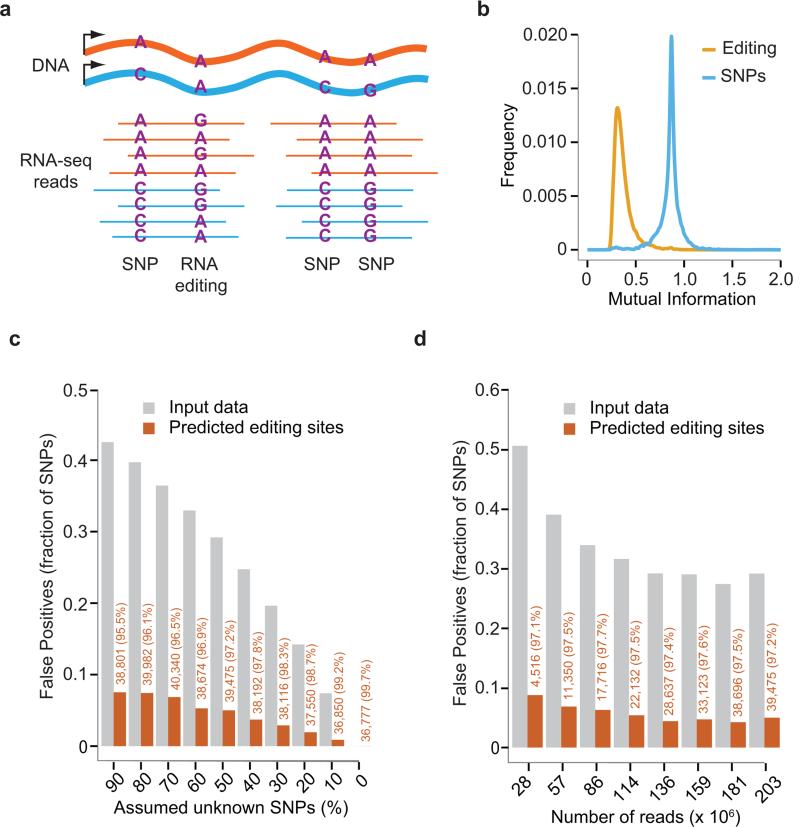
Fig. 1. The GIREMI method.
(a) RNA-Seq reads harboring multiple SNPs and/or RNA editing sites. The allelic combinations of two SNPs in the same reads are the same as their DNA haplotypes. In contrast, a SNP and an RNA editing site (or a pair of RNA editing sites) exhibit variable allelic linkage. (b) Distributions of MI associated with SNPs and RNA editing sites, respectively, estimated using GM12878 RNA-Seq data (ENCODE, cytosolic, polyA+) and its associated genome sequencing data. Our previous genome-dependent method was applied to identify RNA editing sites8. (c) Predicted RNA editing sites by GIREMI in the GM12878 data. Different fractions of genomic SNPs of GM12878 were assumed as unknown by excluding them from dbSNP. For each fraction, the SNPs were selected randomly and the procedure was repeated 9 times. Results shown here are averages of the 9 randomized trials. Gray bars: percentage of GM12878 SNPs among all single-nucleotide mismatches in the mapped RNA-Seq reads after filtering for artifacts (Online Methods). Orange bars: percentage of false positives (GM12878 SNPs) among all predicted editing sites (i.e., FDR). The number of predicted editing sites and % A-to-G editing are shown in orange. (d) Performance of GIREMI at different sequencing depth (down-sampled GM12878 data). Number of mapped reads (singletons) is shown along the x-axis. Fifty percent of the GM12878 SNPs were assumed to be unknown. Labels are similar as in (c).
As a proof of concept, we first applied GIREMI to a deeply sequenced ENCODE RNASeq data set derived from the GM12878 cell line that has associated genome sequencing data6. Overall, 31,660 RNA editing sites (99.6% being the A-to-G type) were predicted by the MI step and 5,117 additional putative A-to-G editing sites were identified by GLM. Since the genome of GM12878 has been well-studied, most of the SNPs in this cell line are already included in dbSNP, which afforded an advantage in predicting RNA editing sites. Thus, to evaluate the performance of GIREMI, we assumed a fraction (10-90%) of the GM12878 SNPs were unknown (Fig. 1c). Strikingly, the false discovery rate (FDR, % GM12878 SNPs in predicted editing sites) was only 3% when 30% of GM12878 SNPs were assumed to be unknown (Fig. 1c). The FDR only increased to 7.6% if assuming 90% of SNPs were unknown, which is an extreme overestimate of the % unknown SNPs in a common human sample given the recent expansion of dbSNP. This performance did not change substantially when a different read mapping method was used (Supplementary Fig. 2, Supplementary Note 2). It should be noted that the FDR defined here assumes that SNPs are the only source of error, without including other possible artifacts, e.g., due to alignment mistakes. Applied to other data sets, GIREMI also outperformed previous methods7 in sensitivity and accuracy (Supplementary Fig. 3, Supplementary Table 1).
It is known that identification of RNA editing sites depends closely on sequencing depth4, 7 and the prediction accuracy may deteriorate with reduced depth. To examine this relationship, we repeated the analysis with down-sampled GM12878 data and with different levels of assumed unknown SNPs (Fig. 1d, Supplementary Fig. 3c, d). As expected, the number of RNA editing sites was lower as the sequencing depth decreased (Fig. 1d). Remarkably, the accuracy of GIREMI was not affected much by sequencing depth, with the FDR remained low (8.8%) even when the sequencing depth was very low (< 30 million singleton reads or 15 million pairs) (Fig. 1d). Robustness to sequencing depth is a highly desirable feature that has not been demonstrated for previous methods. Similar performance was observed for single-end data (Supplementary Fig. 4).
To further evaluate its performance, we compared GIREMI-predicted editing sites to those resulted from the “genome-aware” method that utilizes SNPs identified in whole-genome sequencing data8 (Table 1, Supplementary Table 2, Supplementary Note 3). In addition, we included results of another genome-independent method, the “multiple data sets” method, that calls RNA editing using RNA-Seq data from multiple samples9. For two levels of assumed unknown SNPs (30% and 50%), GIREMI consistently showed higher number of predicted editing sites, higher accuracy (measured as 1-%SNPs among predicted editing sites), higher degree of overlap with the genome-aware method, and higher percentage of A-to-G sites (%AG) than the “multiple data sets” method (Supplementary Note 3). Thus, the performance of GIREMI is highly superior while requiring only minimal data (one RNA-Seq data set, no genome sequence).
Table 1.
Performance of GIREMI compared with other methods applied to the GM12878 data (cytosolic, polyA+ RNA-Seq)
| Genome-aware8 | GIREMI | Multiple data sets method9,c | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Region | Number of sites | %AG | Number of sites | %AG | Accuracya | Overlapb | Number of sites | %AG | Accuracya | Overlapb |
| All | 41,027 | 98.8% | 37,591 | 98.6% | 98.1% | 90.0% | 8,307 | 90.2% | 85.0% | 18.5% |
| Alu | 39,757 | 99.7% | 36,131 | 99.0% | 99.4% | 90.4% | 7,797 | 98.5% | 87.1% | 24.9% |
| Repetitive non-Alu | 260 | 88.6% | 267 | 83.7% | 84.3% | 86.4% | 26 | 65.6% | 65.4% | 14.8% |
| Non-repetitive | 1,010 | 73.5% | 1,193 | 82.8% | 73.8% | 87.6% | 484 | 41.0% | 55.6% | 29.2% |
Accuracy was defined as (1-% SNPs among predicted editing sites in each category); 30% of GM12878 SNPs were assumed to be unknown in applying the GIREMI and multiple data sets methods.
Overlap was calculated relative to the results of the genome-aware method.
Results were derived using two data sets (GM12878 and YH RNA-Seq, Supplementary Note 3). Editing sites were identified in the two data sets separately, and final GM12878 editing sites were called by requiring their presence in YH results. Results of another mode of the multiple data sets method (pooled samples) are included in Supplementary Table 2.
Recent studies identified a large number of editing sites in Alu regions with high confidence10, 11. In contrast, accuracy of predicted non-Alu editing sites was relatively low, especially for those in coding regions9. GIREMI also demonstrated variable accuracy for different types of regions (Supplementary Table 2, Supplementary Notes 3 and 4). Overall, the sensitivity and accuracy of GIREMI are both high compared with existing genome-independent method in pinpointing Alu and non-coding editing sites of non-Alu regions. This observation also applies to the results for a set of human brain RNA-Seq data (Supplementary Table 3), an application mimicking typical individual lab-based projects where a small number of samples were collected, with or without biological replicates.
Compared to non-coding sites, editing sites in coding regions are much less prevalent. Existing RNA-Seq-based methods suffer from low sensitivity and low accuracy in pinpointing non-Alu coding editing events9. On an initial examination, the accuracy of GIREMI is also low (~28% on average) for these sites in non-repetitive regions, although still higher than that of the “multiple data sets” method (5.3% on average, Supplementary Table 2). To gain a detailed evaluation, we examined whether GIREMI could identify previously reported recoding sites12. Since most of recoding sites are highly tissue-specific, we used RNA-Seq data sets derived from a panel of primary human tissues (Supplementary Note 5). Among the 47 recoding sites with adequate read coverage (≥ 5) in at least one sample, 43 were correctly identified by GIREMI, yielding an overall sensitivity of 91.5% and an average per-sample sensitivity of 71.4% (Supplementary Table 4). Given the high sensitivity, the expected small number of non-Alu coding sites, and the likely saturation of such sites in public databases, we can leverage the rapidly expanding sets of known coding sites to improve accuracy. For the GM12878 data, the accuracy in predicting non-repetitive coding sites was 67-80% if only known sites were considered (Supplementary Note 5).
Owing to the genome-independent nature of GIREMI, it can be applied to any RNA-Seq data set without restrictions. We first examined variation of editomes across human tissues, a fundamental question not yet addressed on the genome-wide scale. We used a panel of 38 GTEx RNA-Seq data sets obtained from 5 human subjects and 8 primary tissue types (4 brain regions, heart, skeletal muscle, thyroid, lung)13. The samples were chosen such that each individual had data from nearly all 8 tissues types (Supplementary Table 5). When clustered based on how RNA editing ratios correlate in pairwise comparisons, the samples segregated largely by tissues instead of individuals (Fig. 2a). Three major tissue groups were observed encompassing lung or thyroid, brain regions and muscle (heart and skeletal), respectively. Different brain regions were barely distinguishable based on their editing profiles. This tissue-dominated clustering pattern is especially striking given that the number of predicted editing sites varied greatly across samples largely due to sequencing depth variation (Supplementary Fig. 5, Supplementary Table 6). This result is unlikely a by-product of the expected tissue-dominated clustering of overall gene expression, as the editing ratios are not correlated with gene expression levels (Supplementary Fig. 6). Thus, our observation supports the existence of tissue-specific regulation of RNA editing. In addition, our result is consistent with a recent report of tissue-dominant clustering of editing sites in rhesus macaque14. Notably, in contrast to the previous study, our study only included shallowly sequenced RNA-Seq data (12.3-41.1 M mapped read pairs) without specific genomic data of the samples. This result again attests to the effectiveness of GIREMI.
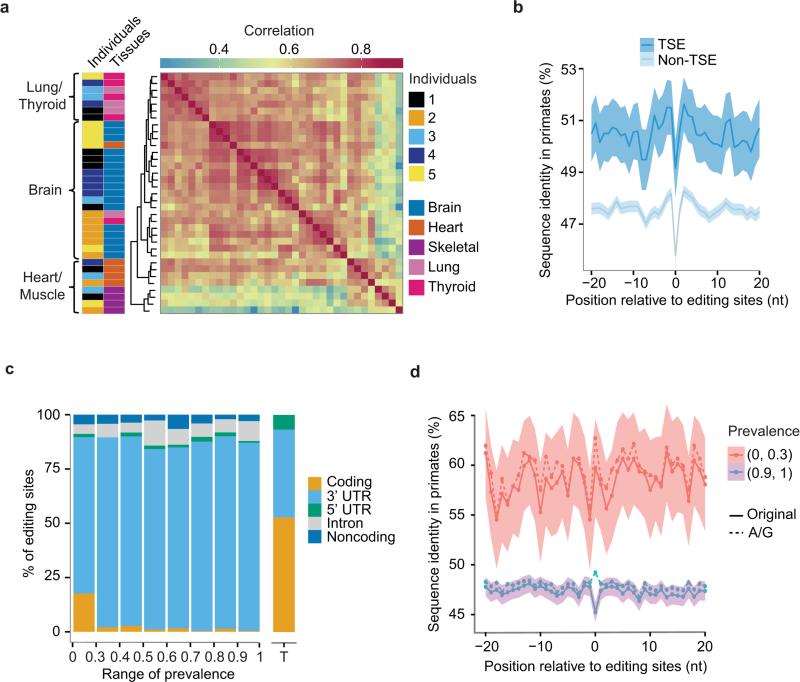
Fig. 2. RNA editomes of human tissues and individuals.
(a) Comparison of RNA editing sites across human tissues. Hierarchical clustering of Pearson correlation coefficients is shown (calculated for editing ratios of all editing sites that are present in 35 samples). Samples are labeled by the rows with indicated color codes for individuals and tissues, respectively. Different brain regions are represented in the same color given their highly similar editing profiles. (b) Conservation of the immediate neighborhood of tissue specific editing (TSE) sites in 3’ UTRs. Sequence conservation (percentage of sequence identity in primates) of each position flanking editing sites (position 0) is shown. Shaded regions represent 95% confidence interval. A similar plot for non-TSE sites is included for comparison purpose. (c) Distribution of editing sites of 93 human individuals in different types of intragenic regions. Editing sites were grouped according to their prevalence values in this population. “Noncoding” refers to noncoding genes or noncoding transcripts of coding genes. Regional distribution of nucleotides in the entire transcriptome of coding genes (without introns) is shown as a reference (rightmost bar labeled as T). (d) Conservation of 3’ UTR regions flanking two groups of editing sites with different prevalence levels (solid lines), similar as in (b). Dashed lines correspond to the sequence identity if Gs in other genomes were assumed as a conserved base given a reference nucleotide A in human8.
In examining the patterns of tissue-specific editing (TSE), we observed the largest difference in RNA editing between brain and muscle-related tissues, with up to 24% editing sites being specific to brain tissues (Supplementary Fig. 7a). In addition, muscle also demonstrated considerably less editing and lower editing levels compared to thyroid or lung (Supplementary Fig. 7b). The mRNA expression levels of ADAR1 (Supplementary Fig. 7a) approximately explained 77% of the variability in editing levels across tissues (Supplementary Fig. 8a). Similarly notable concordance was not observed for ADAR2 (Supplementary Fig. 8b).
Overall, TSE sites are highly enriched in 3’ UTR regions compared to all editing sites (P < 2.2×10−16, Fisher's Exact test, Supplementary Fig. 9a). Interestingly, higher sequence conservation was observed in 3’ UTR regions harboring TSE compared to those flanking all editing sites (Fig. 2b), supporting existence of selection pressure in TSE regions. We observed a number of distinctive genomic features of 3’ UTR TSEs and their associated genes (Supplementary Fig. 9). In addition, brain-specific editing sites were often in genes related to energy, cellular metabolism and apoptosis, whereas lung or thyroid-specific editing sites were found in genes related to signal peptide processing and response to stimuli (viral or inflammatory) (Supplementary Table 7).
We next examined the level of variability in the editome landscape across human individuals, a fundamental question that has not been addressed on a global scale. To this end, we analyzed RNA-Seq data of lymphoblastoid cells of 93 people in the 1000 Genomes project (GBR population)15. A total of 22,715 editing sites were identified. For each editing site covered by ≥ 10 total reads in ≥ 50% of individuals, we calculated the fraction of these individuals expressing the edited nucleotide. We used this value to represent the prevalence of an editing site in the population and observed that the majority of editing sites (88%) had a prevalence of at least 50% (Supplementary Fig. 10a). Levels of RNA editing varied considerably across the prevalence groups, with an overall trend of enhanced editing as prevalence increased (Supplementary Fig. 10b).
All prevalence groups consisted of editing sites enriched in 3’ UTRs relative to the general composition of the human transcriptome (Fig. 2c). Note that less intronic editing sites were observed here than in the GTEx data set (Supplementary Fig. 9a) possibly due to differences in RNA-Seq protocols. Intriguingly, the group of rare editing sites (the first bin in Fig. 2c) showed a considerably higher enrichment in coding regions than other groups. In addition, rare editing sites occurred in higher conserved 3’ UTR regions than common editing sites (Fig. 2d, Supplementary Fig. 11). Although located in functionally important regions (i.e., coding and highly conserved 3’ UTRs), rare editing sites are unlikely functionally significant, given their low editing levels. Possibly, these editing sites represent random innovations of the transcriptome of few individuals that have not yet undergone long-term selection. Purifying selection may exist to prevent these sites from gaining higher editing levels or higher prevalence in the population.
In contrast, common editing sites were associated with relatively high editing levels. This observation argues against the possibility that these sites are randomly occurring transcriptome innovations. Rather, common editing sites should be associated with certain advantage such that evolution has preserved their prevalence. Since these sites are less conserved than TSEs, similarly as non-TSE sites (Fig. 2b vs. 2d), it is unlikely that most of the common editing sites are functionally critical. An alternative hypothesis is that many common RNA editing sites are by-products of the RNA editing machinery carrying out functions to mediate other aspects of gene expression, which is under selection and led to an apparent preservation of the RNA editing sites across population (see Supplementary Note 6 for details).
In summary, we presented a powerful new method, GIREMI, for the identification of RNA editing independent of sample-specific genome sequences. The accuracy of this method is high even given low sequencing depth. Application of the method yielded novel insights about tissue-specific editing and evolutionary implications of RNA editing. We expect that GIREMI will be a powerful method in enabling new discoveries in RNA editing.
Online Methods
Mapping of RNA-seq reads
RNA-seq reads were mapped to the reference human genome (hg19) and transcriptome (Ensembl release 71) using our previously published method8, 16. The method was designed to enable unbiased mapping of alternative RNA alleles corresponding to RNA editing or expressed single nucleotide polymorphisms (SNPs). Briefly, Bowtie17 and Blat18 were used to align all reads to the reference genome and Bowtie was used to align all reads to the transcriptome. Results from the three parallel mapping procedures were merged into a union. Final mapped reads were required to satisfy a dual-filtering scheme such that a read (or a pair of read in paired-end data) maps uniquely with up to n1 mismatches (per read) and does not map to any other regions with up to n2 mismatches (per read) (n2 > n1). For all data sets, n1 and n2 values were set to be about 5% and 12% of the read length, respectively. We previously showed that this mapping method effectively reduced the mapping bias to alternative alleles8, 16 and facilitated relatively accurate quantification of allelic ratios compared to other methods4.
All data sets from ENCODE cell lines, U87MG cells and GTEx human tissues were mapped in the same way as described above. For the 1000 Genomes data sets, we downloaded mapped reads (bam files) directly. However, we implemented an additional filtering step using Blat to remove possible ambiguous mapping (such as those due to existence of pseudogenes or homologs), similarly as in19, 20.
Preprocessing to identify and filter mismatches in RNA-seq reads
The RNA-seq reads were piled up to identify mismatches relative to the reference human genome (hg19). All duplicate reads were removed within each RNA-seq library except the one with the highest-quality score at the mismatch position. Duplicate reads were defined here as (pairs of) reads mapped to exactly the same genomic locations. For each mismatch position, a total read coverage of ≥ 5 was required and the variant allele was required to be present in at least 3 reads. According to previous literature4, 7, 19-23, a number of filters were desirable to remove potential artifacts resulted from sequencing or mapping bias. We thus imposed additional procedures as described in our previous work4 to discard the following types of mismatches: those located in simple repeats regions or homopolymer runs of ≥ 5nt, those associated with reads substantially biased towards one strand, those with extreme variant allele frequencies (> 95% or < 10%), and those located within 4nt of a known spliced junction. To further reduce the impact of sequencing errors, we calculated a log-likelihood ratio (LLR) to examine the likelihood of a mismatch being a sequencing error, as described in8. We only retained mismatches passing an LLR cutoff of 2.
The same procedures as described above (read mapping and mismatch filtering) were applied for all methods included in this study, i.e., the GIREMI, genome-aware, and multiple data sets methods. In addition, known SNPs (dbSNP) were excluded from predicted editing sites by the multiple data sets method.
GIREMI
The method GIREMI combines statistical inference of MI between pairs of single nucleotide variants (SNVs) in RNA-seq reads with machine learning to predict RNA editing sites. The input to GIREMI includes a list of SNVs (mismatches) derived from an RNA-seq data set and known SNPs in public databases such as dbSNP. The output is a collection of predicted RNA editing sites and their editing levels. Except public SNP information, GIREMI carries out all analyses using one RNA-seq data set of interest and does not rely on any other genomic or RNA-seq data sets.
Mutual information (MI) of SNVs and RNA editing prediction
As the first step of GIREMI, we identify known SNPs (dbSNP) in the list of SNVs derived from the RNA-seq reads. We then extract all RNA-seq reads that harbor the known SNPs and the subset of reads (or read pairs in paired-end RNA-seq; required ≥ 5 such reads) that cover more than one SNP. SNP pairs located in the same (pairs of) reads were retained for MI calculation. As an example, in the GM12878 data set, a total of 5,306 SNPs (out of 37,775 SNPs covered by ≥ 5 RNA-seq reads) were involved in this calculation. In another less deeply sequenced RNA-seq data set (GTEx SRR595926, 31M mapped reads), 884 SNPs out of a total of 10,590 RNA-seq-covered SNPs were used for this step. Although the % of SNPs used for the calculation of MI is not high, it is adequate to generate the reference MI distribution (such as that in Fig. 1b) for further prediction of RNA editing sites. The number of RNA editing sites suitable for MI calculation is much larger than that of SNPs. For example, 32,548 editing sites were used to generate the example distribution of MI (Fig. 1b).
For each SNV si, we consider all possible nucleotides A, C, G, T as the four possible states of the variable si. Thus, for a joint variable representing a pair of mismatches (si, sj), a total of 16 states are possible. Although it is unlikely that all 16 states are present in one RNA-seq data set, we use this scheme because it is general and can accommodate possible existence of sequencing errors or other complexity. The probabilities of observing each state of si, sj or (si, sj) were calculated using the maximum likelihood method. A value of 0.01 was assumed for states that were not observed in the actual data considering existence of sequencing errors of all possible nucleotides and accounting for low sequencing depth in realistic data sets. Incorporation of this pseudo value led to an increase of MI of about 0.2 for both SNPs and editing sites (Fig. 1b), but the final editing predictions with or without this pseudo value are very similar (data not shown). The MI of (si, sj) is thus:
where N = {A, C, G, T} and ni and nj represent the states of si and sj, respectively. We used natural log for the above formula.
Then, the MI of a SNP si is defined as:
where S is the collection of other SNPs paired with si, and T is the total number of pairs in S.
As an example, the distribution of I(si) values for SNP pairs detected in the GM12878 data set is shown (Fig. 1b). In theory, the maximum MI should be log(2) = 0.7 for SNP pairs. However, in practice, larger values were sometimes observed, due to limited read coverage at each site and the numerical difference between joint probability and marginal probability of the states. The marginal probability was estimated using all reads covering the particular SNV, whereas the joint probability was estimated using reads covering both SNVs. Thus, the number of joint reads is often smaller than that of the marginal reads and the joint probability is less accurately estimated than the marginal ones. This discordance sometimes led to MI values larger than the theoretical upper bound.
For each RNA-seq data set, the MI of SNPs is calculated independently. Thus, a data set-specific distribution of I(si, sj) is derived. Subsequently, for a SNV sx that is not a known SNP, an I(sx) value is calculated similarly as described above by examining its relationship with other SNVs (either known SNPs or otherwise). Based on the distribution of I(si) of known SNPs, a P value is calculated for sx to test the null hypothesis that I(sx) is not different from the distribution of I(si) for SNPs. A P value cutoff of 0.05 was used to call RNA editing sites. Correction of P values for multiple testing was not applied due to the discovery nature of this test. As an example, we predicted 31,660 RNA editing sites (99.6% being the A-to-G type) in this step for the GM12878 data set.
Generalized Linear Model (GLM) for the prediction of RNA editing
As the second step of GIREMI, RNA editing sites identified by the MI approach are used to train a GLM to predict additional editing sites. The GLM incorporates two types of features that have discriminative power for SNPs and RNA editing sites. The first feature quantifies the deviation of the allelic ratio of the unknown SNV from an expected allelic ratio reflecting the allelic expression of the respective gene. The second type of features represents the sequence preferences of the neighborhoods of RNA editing sites (mainly A-to-G). It should be noted that the GLM step only analyzes A-to-G mismatches as candidate RNA editing sites, without including other types of SNVs.
To estimate the expected allelic ratio r of a gene g, we extract all expressed heterozygous known SNPs (S) (dbSNP) in gene g (read coverage ≥ 5). The allelic ratio r is calculated by maximizing the log-likelihood function logL(r | D), where D refers to the RNA-seq data for gene g. We assume reads covering a specific SNP sj in gene g follow a binomial distribution. Thus, the estimated allelic ratio r̂ that maximizes logL(r | D) of gene g is:
where m and n refer to the number of reads with alternative and reference alleles, respectively. In practice, the haplotype information for SNPs in S is not known. Thus, we arbitrarily assign msj as the read count for the major allele and nsj as that for the minor allele in the RNA-seq data. This assumption may cause a biased allelic ratio larger than the actual value. Nevertheless, the same directional bias exists in the allelic ratio for a specific SNV that is to be compared to r̂. Thus, the impact of this bias will be largely canceled out. In cases where no SNP is available in gene g, an expected ratio of 0.5 is used assuming the gene has no allelic expression bias.
A heterozygous SNP is expected to have an allelic ratio that is largely consistent as the allelic ratio of the gene. In contrast, RNA editing sites may have allelic ratios that substantially deviate from that of the gene. Thus, we use the absolute difference (d) between the allelic ratio of the unknown SNVs and the estimated r̂ of the gene as one feature in the GLM. This feature has the discriminative power for SNPs and RNA editing sites (Supplementary Fig. 1c), but exceptions do exist. For example, it cannot identify editing sites with editing levels similar to allelic ratios of genetic SNPs in the same gene. In addition, a minor fraction of SNPs may have allelic ratios largely different from that of the entire gene if allele-specific splicing or other local RNA processing events affect the allelic expression of the SNPs16.
To increase the discriminative power, we incorporated sequence-based features into the GLM. Importantly, these features are not based on sequence motifs built from a priori knowledge regarding RNA editing. Instead, they were derived using editing sites predicted by the MI step of GIREMI. Thus, the features are specific to the data set of interest without any a priori assumptions. To this end, we generate a positional weight matrix (PWM) for the sequence neighborhood of the predicted editing sites (Supplementary Fig. 1d). For an unknown SNV, a composite sequence score was calculated using its -1 and +1 nucleotides according to the PWM. It should be noted that putative editing sites predicted by the MI-based approach are mostly (> 97%) of the A-to-G type. Thus, the sequence features derived here largely reflect those of A-to-I editing that is known to demonstrate nucleotide preferences at the -1 and +1 positions8.
Together, for each unknown SNV of the A-to-G type, the GLM estimation is:
where d represents the difference between the allelic ratio of the SNV and the estimated r̂ of the gene and c denotes the composite score for the sequence features. β0, βd and βc are the respective coefficients of the GLM, which are solved using a binomial link function.
The GLM of each RNA-seq data set was trained using the putative editing sites predicted by the MI approach. In addition, a leave-one-out scheme was applied where the genetic allelic ratio was estimated using all expressed heterozygous SNPs except one per gene. These randomly excluded SNPs were used as training data together with the putative editing sites. The training data was then separated into two random subsets of the same size. The first subset was used to parameterize the GLM model. The recall and precision of the predictive model were evaluated using the second subset. To reach a tradeoff between the recall and precision, an F measure was calculated as follows:
In the above F measure, we set β to be 0.5, which puts more emphasis on precision than recall. Finally, a cutoff for the predicted probability of a site being an RNA editing site was chosen to achieve an F measure of 0.75.
It should be noted that, although GLM was designed to predict A-to-G sites only, the MI method was not restricted to identification of A-to-G sites alone. Thus, other types of RNA-DNA mismatches do exist in the final results, but with the vast majority being A-to-G. The biological credibility of the other types of RNA-DNA mismatches is still under debate, which is not a focus of this work.
The two steps in GIREMI demonstrate different efficacies for different types of editing sites. The MI step is most effective for editing sites in close proximity with other editing sites or SNPs (such as A-to-I editing in Alu regions that are known to cluster together). Its sensitivity is lower in predicting editing sites in isolation. In contrast, the GLM step, although contributing a relatively small number of additional sites overall as a second step of GIREMI, is an important procedure to ensure high sensitivity in identifying recoding sites. Thus, both steps are essential for our method.
Code availability
GIREMI was implemented using a combination of R, Perl and C codes. The package is available at https://www.ibp.ucla.edu/research/xiao/GIREMI.html.
RNA-seq and SNP data sets
ENCODE RNA-seq data sets were downloaded from the UCSC Genome Browser (http://genome.ucsc.edu/ENCODE/). In this study, we used the data sets derived from cytosolic polyadenylated RNA. U87MG RNA-seq data (wildtype and ADAR1 knockdown) were obtained from our previous study8. GTEx RNA-seq data sets were downloaded from dbGAP with permission. The 1000 Genomes RNA-seq data were downloaded from the Geuvadis project (http://www.geuvadis.org). SNPs derived from genome sequencing data were obtained from the 1000 Genomes project for GM12878 and a genome sequencing project for U87MG cells24. Public SNP data were obtained from dbSNP (version 137).
Tissue-specific editing (TSE)
In the analysis of the GTEx data, we compared the editomes of any pair of tissues included in this study. Each editing site was required to have a read coverage of at least 10 reads in ≥ 75% of samples (i.e., individuals) in either tissue under comparison. A moderated t-test25 was applied to determine whether the editing levels were significantly different across the two tissues (using samples that meet with the read coverage cutoff; FDR < 5%). Editing sites that passed this test were defined as TSEs.
The heatmap of editomes of different tissues (Fig. 2a) was generated based on Pearson correlation of pairs of samples. For each sample pair, only RNA editing sites with adequate read coverage (≥ 10 reads) in both samples were included. Hierarchical clustering was used to generate the clusters.
Conservation analysis of regions flanking editing sites
The same method as in our previous work8 was used to evaluate the conservation level of each editing site and their flanking regions. Briefly, with the 46-way multiz alignments from the UCSC browser26, we focused on the 10 primates, including Human, Chimp, Gorilla, Orangutan, Rhesus, Baboon, Marmoset, Tarsier, Mouse lemur, and Bushbaby. Based on the multiple sequence alignments, the percent identity at each nucleotide position of interest was calculated, together with a 95% confidence interval.
Gene Ontology (GO) analysis
GO analysis was conducted similarly as in27. Briefly, the GO terms of each gene were obtained from Ensembl. To identify GO categories that are enriched in a specific set of genes, the number of genes in the set with a particular GO term was compared to that in a control gene set. The control gene set was constructed so that the randomly picked controls and the test genes have one-to-one matched transcript length and GC content. Based on 10,000 randomly selected control sets, a P value for enrichment of each GO category in the test gene set was calculated as the fraction of times that Ftest was lower than or equal to Fcontrol, where Ftest and Fcontrol denote, respectively, the fraction of genes in the test set or a random control set associated with the current GO category. A P value cutoff (the smaller of 1/10,000 or 1/total number of GO terms considered) was applied to choose significantly enriched GO terms.
Comparison of TSEs with binding sites of RNA binding proteins (RBPs)
Publicly available CLIP-Seq data were collected for hnRNP A1, A2/B1, F, M, U28, hnRNP H29, hnRNP C30, AGO2, IGF2BP1, QKI, PUM231, DGCR832, ELAVL133, EWSR1, FUS, TAF1534, LIN2835, MOV1036, PTB37, SFRS138, TDP4339, TIA1 and TIAL140. CLIP tag clusters were directly downloaded from the above publications or generated using our in-house pipeline. TSEs in 3’ UTRs were then examined for their overlap with CLIP clusters of the above proteins collectively, similarly for non-TSEs.
Supplementary Material
Acknowledgments
We thank members of the Xiao laboratory for helpful comments on this work and for helping with RNA-Seq read mapping. We thank the ENCODE, GTEx and 1000 Genomes projects for generating the data and making their data available to the public. This work was supported in part by US National Institute of Health grants R01HG006264, U01HG007013, and US National Science Foundation grant 1262134.
Footnotes
Author contributions: Q.Z. implemented and developed the GIREMI method, conducted bioinformatic analyses; X.X. initiated the idea, designed and conducted bioinformatic analyses, wrote the paper with input from Q.Z.
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Bass BL. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishikura K. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farajollahi S, Maas S. Trends Genet. 2010;26:221–230. doi: 10.1016/j.tig.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JH, Ang JK, Xiao X. RNA. 2013;19:725–732. doi: 10.1261/rna.037903.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enstero M, Daniel C, Wahlstedt H, Major F, Ohman M. Nucleic Acids Res. 2009;37:6916–6926. doi: 10.1093/nar/gkp731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djebali S, et al. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L. Proc Natl Acad Sci U S A. 2013;110:E2741–2747. doi: 10.1073/pnas.1218884110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bahn JH, et al. Genome Res. 2012;22:142–150. doi: 10.1101/gr.124107.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramaswami G, et al. Nat Methods. 2013;10:128–132. doi: 10.1038/nmeth.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bazak L, et al. Genome Res. 2014;24:365–376. doi: 10.1101/gr.164749.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bazak L, Levanon EY, Eisenberg E. Nucleic Acids Res. 2014;42:6876–6884. doi: 10.1093/nar/gku414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinto Y, Cohen HY, Levanon EY. Genome Biol. 2014;15:R5. doi: 10.1186/gb-2014-15-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Consortium G. Nat Genet. 2013;45:580–585. [Google Scholar]
- 14.Chen JY, et al. PLoS Genet. 2014;10:e1004274. doi: 10.1371/journal.pgen.1004274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abecasis GR, et al. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li G, et al. Nucleic Acids Res. 2012;40:e104. doi: 10.1093/nar/gks280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langmead B, Trapnell C, Pop M, Salzberg SL. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kent WJ. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng Z, et al. Nat Biotechnol. 2012;30:253–260. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- 20.Ramaswami G, et al. Nat Methods. 2012;9:579–581. doi: 10.1038/nmeth.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleinman CL, Majewski J. Science. 2012;335:1302. doi: 10.1126/science.1209658. author reply 1302. [DOI] [PubMed] [Google Scholar]
- 22.Lin W, Piskol R, Tan MH, Li JB. Science. 2012;335:1302. doi: 10.1126/science.1210419. author reply 1302. [DOI] [PubMed] [Google Scholar]
- 23.Pickrell JK, Gilad Y, Pritchard JK. Science. 2012;335:1302. doi: 10.1126/science.1210484. author reply 1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark MJ, et al. PLoS Genet. 2010;6:e1000832. doi: 10.1371/journal.pgen.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smyth GK. In: Bioinformatics and Computational Biology Solutions using R and Bioconductor. Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 26.Dreszer TR, et al. Nucleic Acids Res. 2012;40:D918–923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JH, et al. Circ Res. 2011;109:1332–1341. doi: 10.1161/CIRCRESAHA.111.249433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huelga SC, et al. Cell Rep. 2012;1:167–178. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katz Y, Wang ET, Airoldi EM, Burge CB. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konig J, et al. Nature structural & molecular biology. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hafner M, et al. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macias S, et al. Nature structural & molecular biology. 2012;19:760–766. doi: 10.1038/nsmb.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukherjee N, et al. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoell JI, et al. Nature structural & molecular biology. 2011;18:1428–1431. doi: 10.1038/nsmb.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilbert ML, et al. Mol Cell. 2012;48:195–206. doi: 10.1016/j.molcel.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sievers C, Schlumpf T, Sawarkar R, Comoglio F, Paro R. Nucleic Acids Res. 2012;40:e160. doi: 10.1093/nar/gks697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xue Y, et al. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanford JR, et al. Genome Res. 2009;19:381–394. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tollervey JR, et al. Nat Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z, et al. PLoS Biol. 2010;8:e1000530. doi: 10.1371/journal.pbio.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.